Abstract
The prevalence of IgE-mediated food allergy is increasing at a rapid pace in many countries. The association of high food allergy rates with Westernized lifestyles suggests the role of gene-environment interactions, potentially underpinned by epigenetic variation, in mediating this process. Recent studies have implicated innate immune system dysfunction in the development and persistence of food allergy. These responses are characterized by increased circulating frequency of innate immune cells and heightened inflammatory responses to bacterial stimulation in food allergic patients. These signatures mirror those described in trained immunity, whereby innate immune cells retain a “memory” of earlier microbial encounters, thus influencing subsequent immune responses. Here, we propose that a robust multi-omics approach that integrates immunological, transcriptomic, and epigenomic datasets, combined with well-phenotyped and longitudinal food allergy cohorts, can inform the potential role of trained immunity in food allergy.
Subject Areas: Immunology, Systems Biology, Omics
Graphical Abstract
Immunology; Systems Biology; Omics
Introduction
A relatively new concept, termed Trained Immunity (TRIM), describes the plasticity of innate immune cells and their capacity to develop a “memory” in response to specific exogenous exposures via specific metabolic and epigenetic reprogramming (Netea et al., 2016). TRIM was originally identified in human monocytes in response to infection and microbe exposures (Kleinnijenhuis et al., 2012, Quintin et al., 2012) but has subsequently been associated with diet and certain metabolites in the TCA cycle (Arts et al., 2016a, Christ et al., 2018). Recently, the potential role of TRIM in a wide range of complex non-communicable diseases is becoming appreciated (Arts et al., 2018b, Thiem et al., 2019). Here, we make the case for a similar role of TRIM in the development of food allergy, a disease that has been increasing in prevalence at an alarming rate in association with gene-environment interactions, potentially mediated by epigenetic reprogramming (Martino and Prescott, 2010).
Long thought to be a disease of the adaptive immune system, several recent studies have shown dysfunction of innate immunity in food allergy (Neeland et al., 2018, Zhang et al., 2016). Similar phenotypic changes in monocytes to those observed in atherosclerosis, chronic inflammation, and autoimmune diseases have also been reported in food allergy. In specific conditions, the hyperinflammatory phenotype of monocytes remain after treatment and amelioration of disease, showing that memory of disease in the monocyte may influence future responses to specific environmental exposures or infection (Bekkering et al., 2019). In this review, we summarize the latest research in the field of TRIM, with a focus on epigenetic and chromatin remodeling required for the establishment of inflammatory memory. We discuss a body of work that has explored TRIM in a range of diseases and make the argument that the same strategies be employed to understand food allergy.
Trained Immunity
The immune system is classically divided into the innate and adaptive compartments (Janeway et al., 2001). The long-accepted characterization of innate immunity is of a rapid, non-specific response to pathogens, unaffected by previous exposures. In contrast, adaptive immunity is a slower-acting immune response that retains a memory of previous microbial encounters to mount a highly specific response when re-challenged. Although the unchanging nature of the innate immune response has become one of its defining characteristics, this concept has been challenged in recent years by data demonstrating a capacity for adaptation of the innate immune response following previous pathogen exposure (Natoli and Ostuni, 2019, Netea et al., 2016). It is now clear that certain pathogenic and other stimuli “prime” the innate immune system to alter its response to subsequent microbial exposures. This adaptability of the innate immune system has been termed TRained IMmunity (TRIM) or innate immune memory (Netea et al., 2016). The initial priming of innate cells may result in two opposing states: training, whereby the cell elicits an enhanced response upon further exposure, or tolerance, characterized by an attenuated immune response. In contrast to genetically mediated adaptive immune memory, innate immune memory is underpinned by epigenetic reprogramming of cells, in association with altered cellular and metabolic pathways (Cheng et al., 2014, Saeed et al., 2014). As an epigenetic phenomenon, TRIM is potentially reversible, making these pathways appealing drug targets (Mulder et al., 2019).
Although TRIM provides enhanced protection against subsequent microbial infection (Arts et al., 2016b, Garcia-Valtanen et al., 2017, Leonhardt et al., 2018) the hyper-responsive, hypermetabolic, and proinflammatory phenotype of trained cells may be deleterious in diseases characterized by inappropriate inflammation, such as atherosclerosis, autoimmune diseases, and autoinflammatory diseases (Arts et al., 2018b) (Bekkering et al., 2019). Such diseases draw parallels with food allergy, in terms of etiology and immune signatures, implicating both the innate and adaptive immune systems (outlined in Table 1). A variety of inflammatory disorders have been associated with an altered epigenetic pattern in innate immune cells, and several studies have identified similarities between patient-derived cells and trained cells generated in vitro (Figure 1).
Table 1.
Common Features of Food Allergy, Chronic Inflammation/Autoinflammatory, and Autoimmune Diseases
| Food Allergy | Chronic inflammation/Autoinflammatory Diseases | Autoimmune Diseases | |
|---|---|---|---|
| Adaptive immune dysfunction | + | – | + |
| Innate immune dysfunction | + | + | + |
| Hyperinflammatory innate immune response upon activation | + | + | ? |
| Epigenetic mechanisms | + | + | + |
| Environmental influences | + | + | + |
| Trained immunity | ? | + | + |
Figure 1.
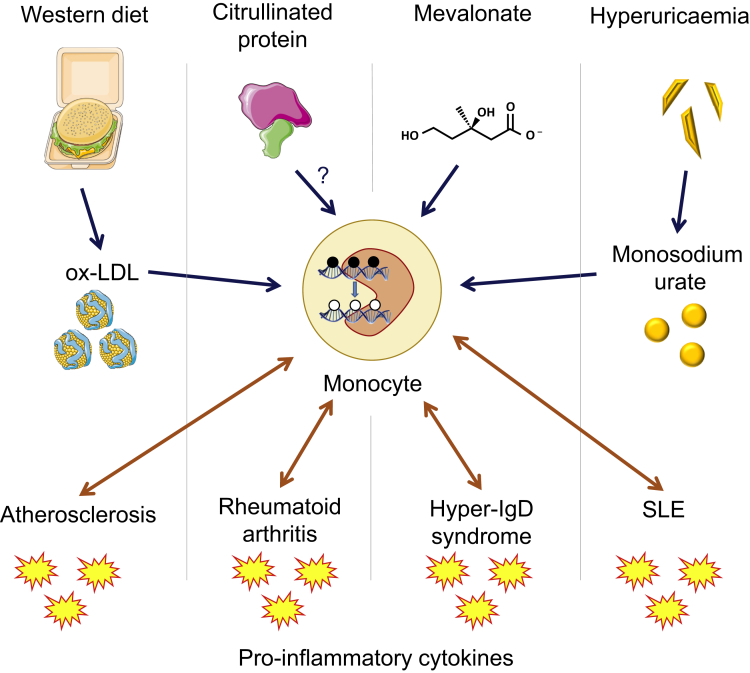
Proposed Role of TRIM in Chronic Inflammatory Diseases
Sterile inflammation is a hallmark of autoimmune diseases, autoinflammatory diseases, and inflammatory diseases such as atherosclerosis. Several danger-associated molecular patterns (DAMPs) have been shown to induce inflammation in innate immune cells. Metabolic intermediates including oxidized low-density lipoprotein (ox-LDL) and mevalonate induce epigenetic modifications and metabolic rewiring in monocytes and macrophages, resulting in a hyperinflammatory, hypermetabolic state. These are produced in high levels in atherosclerosis and hyper-IgD syndrome, respectively, and therefore TRIM may play an integral role in the chronic inflammation seen in these diseases. Other DAMPs have been proposed as inducers of TRIM in autoimmunity, including citrullinated histones in RA, and monosodium urate in SLE patients with cardiovascular comorbidities. The epigenetic reprogramming in TRIM results in an increased production of pro-inflammatory cytokines, which is also consistently reflected in the serum levels of patients with autoimmune and autoinflammatory diseases.
Monocytes and Macrophages: The Prototypical Cells in Trained Immunity
The capacity for memory of previous inflammatory exposures has been observed in a range of innate immune cells, including human monocytes, mouse NK cells (Cooper et al., 2009, Romee et al., 2016, Sun et al., 2009, Sun et al., 2012), and mouse neutrophils (Chen et al., 2014), and has been shown at the level of mouse hematopoietic stem cells (Kaufmann et al., 2018, Mitroulis et al., 2018). Non-immune cells, such as epithelial stem cells and endothelial cells, may also remember inflammatory insults (Naik et al., 2017, Wolff et al., 1998), suggesting that this is a fundamental property of a range of cells following exposure to external stimuli. This review will focus on monocytes and macrophages, in which TRIM has been most well characterized.
Constituting approximately 5%–10% of the white blood cell population in humans, monocytes are significant in both health and disease. Monocytes exhibit diverse roles ranging from effector functions (directly acting on pathogens) to regulatory roles (secretion of cytokines to modulate immune mechanisms) (Karlmark et al., 2012). The differentiation of monocytes into dendritic cells and macrophages further diversifies their functionality and potential role in various immune disorders (Karlmark et al., 2012). Monocytes serve both protective and causative roles in disease, having been implicated in a range of chronic inflammatory and autoimmune disorders. Monocyte abundance, cytokine profiles, and recruitment have all been linked to specific disease states. For example, monocytosis and altered monocyte cytokine landscapes are features of multiple sclerosis, systematic lupus erythematosus, cardiovascular disease, acute myocardial infarction, and atherosclerosis (Karlmark et al., 2012, Ma et al., 2019, Shahid et al., 2018, Woollard and Geissmann, 2010).
Exposures that Induce Trained Immunity
TRIM has been implicated in the heterologous (non-specific) effects of vaccines, which are those exerted beyond the induction of antigen-specific memory in B and T cells against their target disease (Flanagan et al., 2013). Epidemiological and clinical trial data have described heterologous effects for the bacille Calmette-Guerin (BCG) (Aaby et al., 2011) and DTaP vaccines (Aaby et al., 2012), with the former associated with significantly lower all-cause mortality rates in Africa, an effect that cannot be entirely attributed to tuberculosis protection alone (Biering-Sorensen et al., 2017, Kristensen et al., 2000). Indeed, in recent years, researchers have identified BCG as an inducer of trained immunity with epigenetic alterations as the underlying mechanism (Arts et al., 2018c, Bannister et al., 2020, Kaufmann et al., 2018, Kleinnijenhuis et al., 2012). Results from BCG-vaccinated healthy volunteers showed a significant increase of monocyte-derived interferon (IFN) gamma and pro-inflammatory cytokines (TNF, IL-1β, and IL-6) at 2 weeks (Kleinnijenhuis et al., 2012), 1 month (Arts et al., 2018c), and 3 months (Kleinnijenhuis et al., 2012) following vaccination. The hyperinflammatory response observed in monocytes following BCG exposure was attributed to the activation of the intracellular pattern recognition receptor, NOD2, by mycobacterial peptidoglycans, causing NF-κβ translocation and downstream epigenetic changes (Kleinnijenhuis et al., 2012).
Infection by yeast and bacterial pathogens can also remodel monocytes to induce memory. Bacterial sepsis and lipopolysaccharide (LPS)-induced endotoxemia are both associated with monocyte reprogramming to induce a tolerized (immune-compromised) phenotype (Biswas and Lopez-Collazo, 2009, Kox et al., 2014, Shalova et al., 2015). A landmark study showed that after in vitro exposure to LPS, TLR-dependent genes can be divided into two subsets, toleriseable (transcriptionally non-responsive) genes and non-toleriseable genes that evade repression and remain transcriptionally active (Foster et al., 2007). On the other side of the priming spectrum, Candida albicans exposure remodels monocytes for a stronger response to a range of pathogens (Quintin et al., 2012). β-glucan, an integral cell wall component of various fungi, was found to be the key stimulus leading to TRIM, mediated through the dectin-1 receptor and activation of the mTOR pathway (Arts et al., 2016a, Cheng et al., 2014).
A wide range of stimuli are capable of inducing TRIM in innate immune cells, thereby conferring a long-term “mark” on these cells. Although the most widely studied are microbe-associated molecular patterns or MAMPs (Netea et al., 2016), specific dietary components (Christ et al., 2018, Gianfrancesco et al., 2019) and danger-associated molecular pattern (DAMPs) molecules (Arts et al., 2018a, Di Gioia et al., 2020, Jentho et al., 2019) are also capable of inducing the hallmarks of TRIM by initiating functional reprogramming of monocytes. Although each stimulus triggers a unique signaling pathway, they all influence the production of metabolites affecting core metabolic pathways such as glycolysis, glutaminolysis, and cholesterol synthesis (Arts et al., 2016a, Bekkering et al., 2018, Dominguez-Andres et al., 2019b). Overall, TRIM induction through epigenetic and metabolic reprogramming has been shown in monocytes in vitro and in vivo (Arts et al., 2016b, Arts et al., 2018c, Kleinnijenhuis et al., 2012), tissue-specific macrophages (Haley et al., 2019), and bone-marrow myeloid progenitors (Kaufmann et al., 2018, Mitroulis et al., 2018), suggesting that this process is fundamental to a wide-range of organ functions and diseases.
Epigenetic Mechanisms Underpinning Trained Immunity
Epigenetic Mechanisms in Inflammatory Cells
The underlying mechanisms that confer trained immunity and inflammatory memory occur at the level of epigenetic marks (Natoli and Ostuni, 2019). The term “epigenetics” is defined as “above DNA” and refers to the study of molecular interactions that influence chromosome structure and gene activity. The epigenetic “state” of a cell is determined by a combination of DNA and histone modifications and open chromatin patterns. The functional unit of chromatin is the nucleosome, which is comprised of core histones, the tails of which can be post-translationally modified by specific enzymes (Bannister and Kouzarides, 2011). This epigenetic state dictates both the cell's transcriptional profile and its response to stimuli (Bonasio et al., 2010). Although most gene promoters remain open during differentiation, distal regulatory elements (for example, enhancers) are much more dynamic and show cell-type-specific patterns (Heinz et al., 2015). Lineage determining transcription factors (LDTFs) remodel chromatin during differentiation, establishing a regulatory landscape within which stimulus determining TFs (SDTFs) can act to influence gene expression in response to exogenous exposures (Figure 2) (Glass and Natoli, 2016). Around ∼35% of all enhancers present in lymphoid and myeloid lineages are established during hematopoiesis and predict the transcriptional programs of differentiated cells (Lara-Astiaso et al., 2014). In monocytes/macrophages, PU.1, C/EBPs, AP-1 factors, RUNX1, and IRFs are LDTFs that establish the regulatory landscape, and the vast majority of genomic regions that are activated in response to inflammatory stimuli are marked by their motifs (DNA-binding sequences) and priming histone marks, such as histone 3 lysine 4 mono-methylation (H3K4me1) (Ghisletti et al., 2010).
Figure 2.
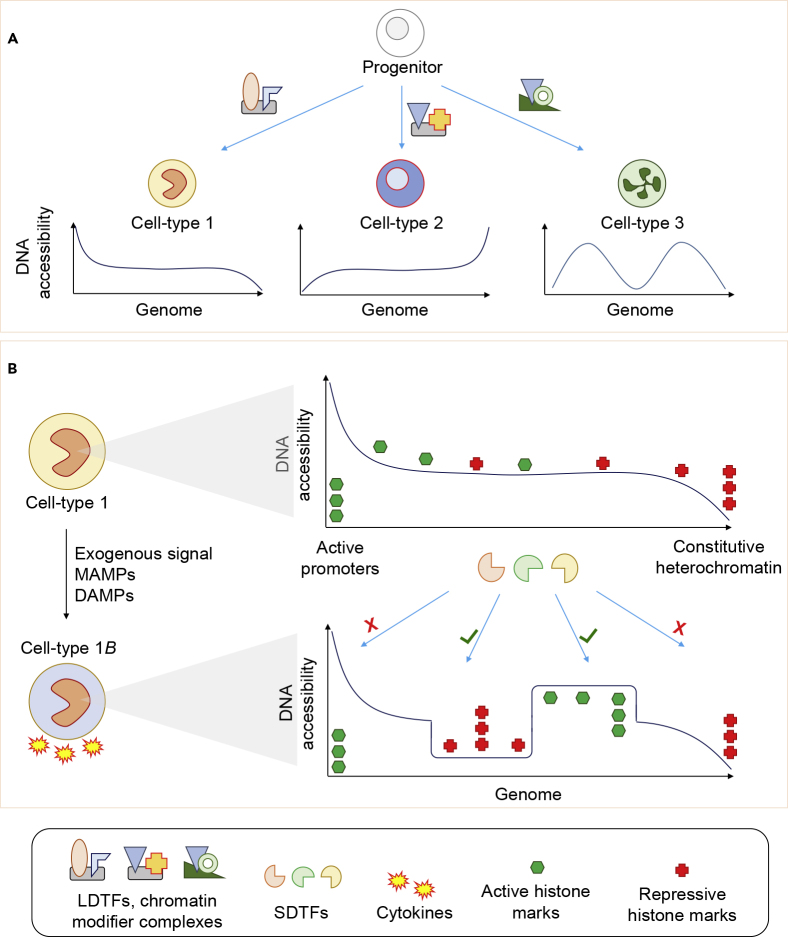
Transcription Factors Remodel DNA Accessibility and Epigenetic Marks to Establish Cell Identity and Response to Exogenous Signals
(A) LDTFs establish cell-type-specific DNA accessibility landscapes.
(B) Exposure of cells to environmental signals such as MAMPs and DAMPs induces remodeling of chromatin by SDTF and chromatin remodeling complexes. This remodeling generally occurs at intermediately accessible regions, with constitutively active promoters and heterochromatin not affected. The newly established active or inactive regions influence the cell's response to future environmental signals.
Transcription Factors and the Establishment of Accessible Chromatin in Macrophages
Only around 2% of all human DNA (genome) is ever accessible to interactions with regulatory proteins (such as TFs) in any given cell type, with a high degree of cell-type specificity (Klemm et al., 2019, Thurman et al., 2012). The genome exists in an accessibility continuum that ranges from tightly closed (compacted) chromatin to open and highly dynamic chromatin, dictated by a process known as “epigenetic remodeling” that involves the interplay between transcription factors and epigenetic marks (Figure 2) (Klemm et al., 2019). Epigenetic mechanisms such as the post-translational modifications of histones, DNA methylation, 3D chromatin structure, and non-coding RNAs have all, to some extent, been profiled in the context of innate immune differentiation, response to stimulus, and TRIM (Fanucchi and Mhlanga, 2019, Phan et al., 2017, van der Heijden et al., 2018).
During macrophage polarization by stimuli such as IL-4, IFN-γ, TNF, or LPS, an extensive network of SDTFs bind primed enhancers and orchestrate their activation or repression (Czimmerer et al., 2018a, Kang et al., 2017, Kang et al., 2019, Kusnadi et al., 2019, Park et al., 2017), by recruiting enzymes, known as chromatin remodelers, which add or remove chemical groups from histone tail residues, and further modulate the chromatin landscape to induce enhanced or tolerized transcriptional responses to subsequent stimuli (Czimmerer et al., 2018a). There is an extensive functional overlap between TF binding (Czimmerer et al., 2018b, Daniel et al., 2018, Wang et al., 2019) and a core TF complex likely involved in opposing inflammatory and alternative activation programs (Piccolo et al., 2017).
Role of Histone Modifications in Innate Immune Memory
The “histone code” posits that histone modifications influence gene expression by attracting TFs and other proteins (Jenuwein and Allis, 2001). Although the actual histone code is extremely complex (with many potential combinations of modifications), six specific marks have been used to establish a reference epigenome, including histone H3 lysine-4 trimethylation (H3K4me3) that specifies active promoters, H3K4me1 (primed enhancers), H3K27ac (active promoter or enhancer), H3K9me3 (repressive), H3K27me3 (repressive), and H3K36me3 (active gene bodies) (Stunnenberg et al., 2016).
The role of histone modifications in regulating innate immune memory was demonstrated by showing that genes downstream of TLR4 can be both primed or tolerized by LPS, indicating that the local chromatin landscape, and not the signaling pathway per se, determines the priming state of specific genes (Foster et al., 2007). Bromodomain and extra terminal domain (BET) family proteins are capable of regulating gene expression upon recognition of acetylated histone domains. Studies have demonstrated that the synthetic histone mimic, iBET, interferes with the recruitment of BET family proteins to H3K27ac-marked genomic regions and blocks inflammation (Nicodeme et al., 2010). Accordingly, blocking the initial inflammatory transcriptional response with iBET also blocks the establishment of TRIM and tolerance by fungal antigens and LPS, respectively (Dominguez-Andres et al., 2019a, Novakovic et al., 2016).
Across different studies, it appears that ∼15% of active enhancers (marked by H3K4me1 and H3K27ac) in human monocytes can be remodeled by exogenous signals, such as LPS or different TRIM stimuli (Novakovic et al., 2016, Ostuni et al., 2013), suggesting that this is the approximate limit of the number of remodeling events. Further, it was recently shown that most of these stimulus-induced remodeling events occur at poorly positioned nucleosomes (Comoglio et al., 2019), which suggests that strong promoters and enhancers are essential for macrophage identity and cannot be remodeled by signals from the environment. Other TRIM stimuli have subsequently been shown to induce remodeling of H3K27ac in vitro, such as the metabolites fumarate and mevalonate (Arts et al., 2016a, Bekkering et al., 2018), danger-signals such heme (Jentho et al., 2019), and in vivo exposure to BCG (Arts et al., 2018c). The extent of remodeling is dependent on the stimulus, with receptor-mediated TRIM induction leading to a higher number of epigenetic changes, whereas metabolite exposure induces a modest change in the H3K27ac signal.
The Emerging Role of Non-coding RNA and 3D Chromatin Structure in Innate Immunity
Estimates suggest that more than 85% of the human genome is transcribed (Consortium, 2012, Hangauer et al., 2013), with the majority (∼70%) of RNA polymerase II genomic enrichment found at enhancer regions (De Santa et al., 2010). Genome-wide transcription of enhancers is induced during macrophage polarization by IL-4 or IFN-γ (Denisenko et al., 2017), as well as during exposure to mycobacterium tuberculosis infection (Denisenko et al., 2019). In addition, long non-coding RNAs (lncRNAs) have been shown to mediate cholesterol homeostasis by altering the 3D chromatin structure at specific gene loci (Sallam et al., 2018). Finally, a recent study defined a novel type of immune gene–priming lncRNAs (IPLs), which facilitate deposition of H3K4me3 at primed genes by recruiting chromatin remodels, such as MLL1, to gene promoters in humans (Fanucchi et al., 2019). In a series of elegant experiments, it was shown that the absence of a specific IPL in mice is associated with loss of priming, which is restored by insertion of the human IPL (Fanucchi et al., 2019). Collectively, these studies show that lncRNAs are fundamental to establishing epigenetic memory in TRIM (Fanucchi and Mhlanga, 2019, Fok et al., 2018).
Genetic Influence on Epigenetics and Immune Responses
Innate immune responses, such as cytokine release, are under strong genetic influence (Li et al., 2016) and are therefore considered expression quantitative trait loci (eQTLs) (Barreiro et al., 2012, Fairfax et al., 2014). Similarly, epigenetic marks are also influenced by genetic variation (e.g. methylation QTLs and histone QTLs) (Chen et al., 2016, McVicker et al., 2013), and these in turn influence gene expression. A recent study explored the relationship between inflammatory response eQTLs and chromatin accessibility QTLs (caQTLs), revealing a 37% overlap, with genetic variation influencing both priming of macrophage-specific enhancers as well as establishment of new enhancers (Alasoo et al., 2018). These studies highlight the need for next-generation studies aimed at fully understanding TRIM by incorporating genetic, epigenetic, transcriptional, and phenotypic data.
IgE-Mediated Food Allergy
IgE-mediated food allergy is a rising global health concern. Approximately 5% of the population in developed countries suffers from the most common eight food allergies: peanuts, tree nuts, cow's milk, egg, soy, wheat, shellfish, and fish (Loh and Tang, 2018). It is clear that food allergy begins in early life, with up to 10% of infants affected by 12 months of age (Osborne et al., 2011). Some food allergies, such as egg and milk, are often outgrown in childhood, whereas others, including peanut and tree nut allergies, can persist into later life, often in association with severe and life-threatening reactions (Berin, 2019, Sasaki et al., 2018).
The rising incidence of food allergy is occurring more rapidly than evolutionary changes to the genome allow, suggesting the causes are multifactorial, and likely involve multiple gene-environmental interactions that alter the immune response. These contributing factors have recently been summarized by three separate hypotheses: the dual allergen exposure hypothesis, the hygiene hypothesis, and insufficient vitamin D (Peters et al., 2017). However, despite the high burden of disease, the standard of care for patients with food allergy is sub-optimal, consisting primarily of food allergen avoidance, which can be difficult to achieve and may result in severe reactions upon accidental ingestion, requiring emergency treatment. As such, substantial research efforts are now being employed to understand the causes of food allergy at a mechanistic level, identify the immune factors governing the development of tolerance, and inform primary prevention and therapeutic strategies.
Immunology of Food Allergy
The induction of an allergic immune response to foods begins with a process known as sensitization, reviewed extensively in Sampson et al. (2018). Briefly, protein food antigens are processed by dendritic cells and presented to antigen-specific naive CD4 T cells in local lymph nodes. A combination of co-stimulation and the production of IL-4 leads to the differentiation and proliferation of antigen-specific Th2 cells. These antigen-specific Th2 cells and associated cytokines provide “help” to B cells, leading to the production of antigen-specific IgE antibodies. Following re-exposure to the antigen in a food-sensitized individual, it is this specific IgE, bound to high-affinity receptors on mast cells and basophils, that triggers degranulation and release of histamine and other mediators of an allergic response (Satitsuksanoa et al., 2018). Food allergic reactions have several clinical manifestations, including gastrointestinal (nausea, abdominal pain, vomiting), respiratory (wheezing), skin (urticaria, angioedema), and systemic responses. Anaphylaxis, the most severe reaction, is a multi-system response and can be life-threatening (Yu et al., 2016).
The immune mechanisms responsible for the persistence and resolution of food allergy throughout the life course remain unclear. Due to their crucial role in initiating the allergen-specific response, CD4 T cells and their functional subtypes are by far the most studied cell population, and the focus of many expert reviews (Chinthrajah et al., 2016, Sampson et al., 2018, Satitsuksanoa et al., 2018, Yu et al., 2016). Accordingly, changes in the transcriptional pattern and genome-wide DNA methylation landscape has been reported in T cells from food allergic children (Do et al., 2020, Kosoy et al., 2016, Martino et al., 2014, Martino et al., 2018). The induction of immune tolerance is thought to involve regulatory CD4 T cells (Tregs) and B cells (Bregs), as well as the generation of allergen-specific IgG4. Increased frequency and tolerogenic function of Tregs has been observed in children who naturally outgrow their food allergy (Qamar et al., 2015, Shreffler et al., 2009). Likewise, greater frequency and proliferation of IL-10+ B regulatory cells has been observed in non-allergic patients when compared with those with food allergy (Neeland et al., 2019, Noh et al., 2010).
Endotoxin Exposure and Allergic Outcomes
In support of the association between the hygiene hypothesis and allergic disease, a seminal study comparing children from two genetically similar US farming populations, the Amish and the Hutterites, showed that Amish children, who practice traditional farming and are exposed to a microbe-rich environment, show lower rates of asthma. Further mechanistic work revealed that innate immune responses to endotoxin were significantly different between the two populations, with monocytes from Amish children responding in a more regulated and anti-inflammatory manner compared with monocytes from the Hutterite children (Stein et al., 2016). A recent study investigated the association between levels of endotoxin present in house dust and sensitization to milk, egg, and peanut (Tsuang et al., 2020). Household endotoxin level was positively associated with sensitization (not clinical allergy) to milk and egg, but not peanut. Interestingly, endotoxin-stimulated PBMCs from children allergic to milk or egg, but not peanut, produced less of the Th1 cytokine IFNγ than healthy controls, highlighting potential differential responses to endotoxin depending on the type of food allergy.
A Potential Role for Trained Immunity in Food Allergy
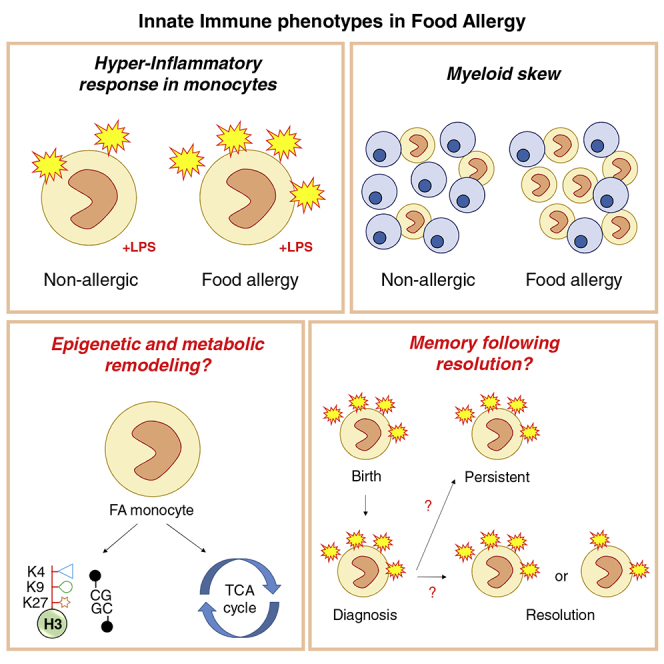
In addition to T- and B-cell-mediated responses, several studies have investigated innate immune function in food allergy. One of the first studies to investigate this reported an exaggerated innate immune response at birth in cord blood mononuclear cells from allergic children (inclusive of patients with food allergy, eczema, and/or asthma) relative to non-allergic controls (Tulic et al., 2011). This included elevated production of IL-6, IL-1β, and TNFα following in vitro TLR stimulation. Unlike their non-allergic counterparts, allergic children showed an age-related decline in production of inflammatory cytokines, such that TLR responses were attenuated by the age of five. An important finding of this study was that hyper-inflammatory responses in the perinatal period significantly correlated with the propensity for allergen-specific Th2 responses at birth and during the first year of life, particularly for IL-13 production (Tulic et al., 2011).
Findings from this study were replicated in 2016, where cord-blood derived monocytes from children who go on to develop food allergy showed elevated production of IL-6, IL-1β, and TNFα following stimulation with endotoxin (TLR-4 stimulation) (Zhang et al., 2016). This study also showed that stimulating cord-blood derived naive CD4 T cells with commercially available IL-1β or TNFα (both in combination with the regulatory cytokine TGF-β) could skew these naive CD4 T cells to produce Th2-associated cytokines. The authors thus hypothesized that the increase in monocyte-derived inflammatory cytokines in the cord blood of food allergic children directly contribute to the Th2 skew associated with allergic disease. However, it was not clear from this study if naive CD4 T cell responses to stimulation with these inflammatory cytokines differed between food allergic and non-allergic children or if the monocytes themselves were capable of skewing naive CD4 T cells toward an allergic pathway.
More recently, a study comparing single-sensitized egg-allergic one-year-old infants with non-sensitized healthy controls showed increased frequency of circulating monocytes and dendritic cells in infants with egg allergy (Neeland et al., 2018). Circulating monocytes from egg allergic infants also expressed higher levels of the activation marker HLA-DR relative to non-allergic controls. Within the egg-allergic infants, those who remained egg-allergic at follow-up (age 2–4 years had higher circulating levels of monocytes at age one than infants who naturally resolved their egg allergy by 4 years old. When assessing the non-T cell fraction for functional responses, this study showed that non-T cells from egg-allergic infants were hyper-responsive to stimulation with endotoxin, producing more TNFα, IL-6, IL-1β, and IL-8 relative to non-allergic controls. This effect was more pronounced in infants with persistent egg allergy outcomes, particularly for TNF-α and IL-1β, when compared with infants with transient egg allergy. Interestingly, constitutive production of IL-6, IL-1β, and IL-8 remained elevated into childhood in children with persistent egg allergy relative to children whose allergy had naturally resolved by this age. As this study assessed a mixed population of non-T cells, it remained unclear if this heightened inflammatory response was due to the monocyte fraction. However, further studies have revealed that purified monocytes from egg-allergic infants are hyper-responsive to stimulation with LPS relative to non-allergic infants, confirming that the global inflammatory response observed in this study was a monocyte-specific signature (manuscript in preparation).
Another study from this group recently showed that infants with peanut allergy show greater production of TNFα following non-specific stimulation of total PBMCs relative to non-allergic infants (Neeland et al., 2020). This increased cytokine production was observed within the monocyte and DC fractions, suggesting that increased innate inflammation is present in both egg and peanut allergy during the first year of life. Interestingly, infants with peanut-sensitized tolerance showed greater numbers of circulating plasmacytoid dendritic cells, known for their tolerogenic immune properties against innocuous antigens, relative to non-allergic controls. Combined, these findings confirm a central role for innate immune cells in the development of allergic immune responses in early life, as well as highlight the potential role for a subset of dendritic cells in averting clinical allergy in the presence of allergen-specific IgE.
Emerging evidence therefore reveals a remarkable similarity of food allergic immune cell phenotypes to those that define trained immunity, primarily the skew toward higher myeloid cells in the circulation and a hyper-inflammatory phenotype (Netea et al., 2020). Combined with the observation that several features of allergic disease overlap with TRIM-associated diseases, including the increasing trends in prevalence of TRIM disease in westernized countries (Table 1), it is reasonable to hypothesize a similar role for TRIM in atopic diseases. However, this field is in its infancy and further work is required. At the forefront of this will be investigation of epigenetic reprogramming of innate immune cells from food-allergic patients following in vitro or environmental exposure to MAMPs. It will also be important to determine the role of specific innate immune cells—monocytes, dendritic cells, natural killer cells—in food allergy over the lifecourse, as these still remain relatively unexplored. It will also be interesting to study resolved allergy cases to see if innate immune memory persists in these individuals, as has been shown in mice upon cessation of western diet (Christ et al., 2018) and after in hypercholesterolemia patients following several months of lipid lowering with statins (Bekkering et al., 2019). This analysis would indicate if allergy induced longer term TRIM, which may have implications for future microbial responses.
Potential Applications of Trained Immunity in Food Allergy
The potential role of TRIM-related epigenetic remodeling in food allergy provides an opportunity to improve the diagnosis of food allergy and may even help in the development of primary prevention and treatment strategies. Current allergy diagnostic methods rely on measuring allergen-specific IgE, either through a skin prick test or in the serum, followed by a formal oral food challenge. Tests of IgE sensitization can lead to over-diagnosis, as they are not necessarily indicative of clinical reactivity to allergens (Heinzerling et al., 2013) and should be interpreted carefully in light of the patient's history. On the other hand, the gold standard test, the oral food challenge, is associated with a risk of adverse outcomes, including anaphylaxis, and is usually reserved for testing for the development of tolerance. In addition, there are no clinically approved treatments for food allergy, and current management strategies can result in severe reactions upon accidental exposure to allergenic foods. Emerging therapies of food allergy include oral immunotherapy (OIT), which involves giving gradually increasing amounts of food allergen under medical supervision and continued daily ingestion of the allergen. However, a recent meta-analysis has shown that this form of treatment results in more frequent allergic reactions, including anaphylaxis (Chu et al., 2019). If future studies show that innate cell frequency or activation status in early life could accurately predict the development or persistence of food allergy, this could be used in combination with clinical testing to improve currently suboptimal diagnostic and management strategies.
TRIM-based vaccine strategies, which would specifically exploit the heterologous effects of certain widely used vaccines, are of increasing interest in the prevention of disease (Sánchez-Ramón et al., 2018). For example, the non-specific benefits of the BCG vaccine range from the prevention of cancer recurrence in superficial bladder cancer (Sylvester et al., 2002), to protection against all-cause mortality in infants in high-mortality settings (Higgins et al., 2016). There is also growing evidence from both human and animal studies that BCG protects against allergic diseases. BCG is proposed to act as an early life microbial exposure, thus promoting the switch to a T helper (Th1) immune response and thereby preventing the persistent Th2 responses associated with development of allergic disease (Figure 3) (Gouveia et al., 2013, Herz et al., 1998, Hopfenspirger and Agrawal, 2002, Libraty et al., 2014, Ozer et al., 2003, Yang et al., 1999). Nevertheless, a meta-analysis of 22 studies found that BCG vaccination had no significant effect on specific IgE or allergic sensitization as determined by skin prick test (SPT) (Linehan et al., 2014). More recent RCTs have come to the same conclusion, showing that neonatal BCG vaccination had no effect on allergic sensitization or asthma/wheeze (Thostesen et al., 2017a, Thostesen et al., 2017b). Another existing vaccine that may influence allergy is the whole-cell killed pertussis (wP) vaccine (Estcourt et al., 2019). In a recently published Australian case-control study, infants who received the first dose of wP were 23% less likely to be diagnosed with food allergy (Estcourt et al., 2019). Many factors are likely to contribute to the variability seen in the potential for a vaccine to prevent allergy including age at vaccination, vaccine strain used (Shann, 2016), maternal vaccination status (Berendsen et al., 2020), as well as environmental and population (e.g. genetic) factors. Thus, although there remains potential for BCG and other vaccines to be developed as a preventative strategy for allergy, further determination of its potential efficacy and optimal usage are needed.
Figure 3.
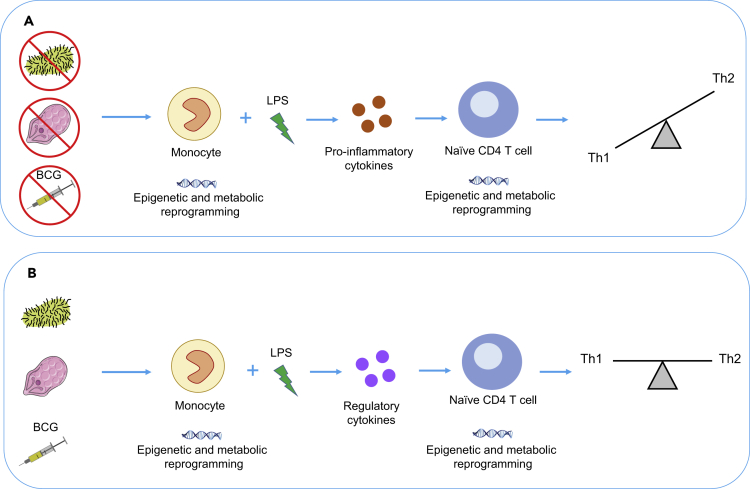
The Potential Role of Microbial Exposures and the Trained Immune Phenotype in Skewing the Th2 Phenotype
(A) As shown by the proinflammatory response to LPS in allergic cells that have not had sufficient microbial exposures, causing epigenetic and metabolic rewiring of naive CD4 T cells, leading to improper development of the Th1 pathway and bias toward the Th2 phenotype. (B) The possible preventative role of BCG and microbial exposures in allergy, by inducing production of anti-inflammatory cytokines to ensure appropriate development of Th1/Th2 balance.
Synthetic drug development that targets TRIM pathways at multiple levels—the immunological/cellular landscape, the metabolome, and the epigenome (Mourits et al., 2018)—is a possibility for potential food allergy treatment. TRIM is mediated through epigenetic remodeling, and the phenotype can be offset at the epigenetic level by histone mimics (such as iBETs) (Dominguez-Andres et al., 2019a, Nicodeme et al., 2010). At the metabolic level, a number of key pathways may be targeted, including glutaminolysis and glycolysis, to alter the innate immune phenotype (Cheng et al., 2016). A wide-range of drug targets for trained immunity pathways in various diseases have already been proposed, discussed in detail in a recent review (Mulder et al., 2019).
Future Studies
As outlined in this review, there is compelling evidence linking certain aspects of the TRIM phenotype to food allergy. However, the epigenetic and metabolic basis for this potential link remains poorly characterized, requiring further research into the immune cell profiles of allergic individuals. Likewise, further work is required to determine if innate hyper-responsiveness continues into later life in persistently food-allergic children or whether this signature returns to baseline if food allergy naturally resolves in childhood. Differences between innate immune responses in the different types of food allergies and their associated pathologies will also be important to investigate. This work demands a thorough multi-omics approach, integrating immunological data along with genetic, transcriptomic, epigenomic, and phenotypic profiling of each cell type in food-allergic, allergic/sensitized but clinically tolerant and non-allergic individuals over the lifecourse. With recent advances in high-throughput technologies and computational techniques, it is now possible to simultaneously quantify a large number of analytes or cellular components within a single sample (Dhondalay et al., 2018, Suprun et al., 2019, Wen and Rothenberg, 2019). Applying these approaches, in combination with well-defined sample collection, clinical phenotyping, and environmental data, will rapidly accelerate our understanding of TRIM in food allergy, as well as aid in discovery of new diagnostic tools and potential treatment strategies. TRIM-based therapies may represent novel, safe, and effective measures for the prevention and potential treatment of allergic disorders.
Acknowledgments
RS is supported by an NHMRC Project Grant (1165073); BN is supported by an NHMRC Project Grant (1157556) and an NHMRC Investigator Grant (1173314). MN is supported by an MCRI Lifecourse Fellowship. KPP is supported by a Melbourne Children's Clinician Scientist Fellowship. MGN is supported by an ERC Advanced Grant and a Spinoza grant from the Netherlands Organisation for Scientific Research (NWO). Figures were adapted from Servier Medical Art.
References
- Aaby P., Benn C., Nielsen J., Lisse I.M., Rodrigues A., Ravn H. Testing the hypothesis that diphtheria-tetanus-pertussis vaccine has negative non-specific and sex-differential effects on child survival in high-mortality countries. BMJ Open. 2012;22:e000707. doi: 10.1136/bmjopen-2011-000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaby P., Roth A., Ravn H., Napirna B.M., Rodrigues A., Lisse I.M., Stensballe L., Diness B.R., Lausch K.R., Lund N. Randomized trial of BCG vaccination at birth to low-birth-weight children: beneficial nonspecific effects in the neonatal period? J. Infect. Dis. 2011;204:245–252. doi: 10.1093/infdis/jir240. [DOI] [PubMed] [Google Scholar]
- Alasoo K., Rodrigues J., Mukhopadhyay S., Knights A.J., Mann A.L., Kundu K., Consortium H., Hale C., Dougan G., Gaffney D.J. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J., Novakovic B., Ter Horst R., Carvalho A., Bekkering S., Lachmandas E., Rodrigues F., Silvestre R., Cheng S.C., Wang S.Y. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 2016;24:807–819. doi: 10.1016/j.cmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Carvalho A., La Rocca C., Palma C., Rodrigues F., Silvestre R., Kleinnijenhuis J., Lachmandas E., Goncalves L.G., Belinha A. Immunometabolic pathways in BCG-induced trained immunity. Cell Rep. 2016;17:2562–2571. doi: 10.1016/j.celrep.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Huang P.K., Yang, Joosten L.A.B., van der Meer J.W.M., Oppenheim J.J., Netea M.G., Cheng S.C. High-mobility group nucleosome-binding protein 1 as endogenous ligand induces innate immune tolerance in a TLR4-sirtuin-1 dependent manner in human blood peripheral mononuclear cells. Front. Immunol. 2018;9:526. doi: 10.3389/fimmu.2018.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Joosten L.A.B., Netea M.G. The potential role of trained immunity in autoimmune and autoinflammatory disorders. Front. Immunol. 2018;9:298. doi: 10.3389/fimmu.2018.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Moorlag S., Novakovic B., Li Y., Wang S.Y., Oosting M., Kumar V., Xavier R.J., Wijmenga C., Joosten L.A.B. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89–100.e5. doi: 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister S., Messina N.L., Novakovic B., Curtis N. The emerging role of epigenetics in the immune response to vaccination and infection: a systematic review. Epigenetics. 2020;21:381–395. doi: 10.1080/15592294.2020.1712814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L.B., Tailleux L., Pai A.A., Gicquel B., Marioni J.C., Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. U S A. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S., Arts R.J.W., Novakovic B., Kourtzelis I., van der Heijden C., Li Y., Popa C.D., Ter Horst R., van Tuijl J., Netea-Maier R.T. Metabolic induction of trained immunity through the mevalonate pathway. Cell. 2018;172:135–146 e139. doi: 10.1016/j.cell.2017.11.025. [DOI] [PubMed] [Google Scholar]
- Bekkering S., Stiekema L.C.A., Bernelot Moens S., Verweij S.L., Novakovic B., Prange K., Versloot M., Roeters van Lennep J.E., Stunnenberg H., de Winther M. Treatment with statins does not revert trained immunity in patients with familial hypercholesterolemia. Cell Metab. 2019;30:1–2. doi: 10.1016/j.cmet.2019.05.014. [DOI] [PubMed] [Google Scholar]
- Berendsen M.L.T., Oland C.B., Bles P., Jensen A.K.G., Kofoed P.E., Whittle H., De Bree C., Netea M.G., Martins C., Benn C.S. Maternal priming: Bacillus calmette-guerin (BCG) vaccine scarring in mothers enhances the survival of their child with a BCG vaccine scar. J. Pediatr. Infect Dis Soc. 2020;9:166–172. doi: 10.1093/jpids/piy142. [DOI] [PubMed] [Google Scholar]
- Berin M.C. Mechanisms that define transient versus persistent food allergy. J. Allergy Clin. Immunol. 2019;143:453–457. doi: 10.1016/j.jaci.2018.12.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biering-Sorensen S., Aaby P., Lund N., Monteiro I., Jensen K.J., Eriksen H.B., Schaltz-Buchholzer F., Jorgensen A.S.P., Rodrigues A., Fisker A.B. Early BCG-Denmark and neonatal mortality among infants weighing <2500 g: A randomized controlled trial. Clin. Infect. Dis. 2017;65:1183–1190. doi: 10.1093/cid/cix525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.K., Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Bonasio R., Tu S., Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Wu W., Millman A., Craft J.F., Chen E., Patel N., Boucher J.L., Urban J.F., Jr., Kim C.C., Gause W.C. Neutrophils prime a long-lived effector macrophage phenotype that mediates accelerated helminth expulsion. Nat. Immunol. 2014;15:938–946. doi: 10.1038/ni.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martin D., Watt S., Yan Y., Kundu K., Ecker S. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.-C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H.A., Rao N.A., Aghajanirefah A. mTOR- and HIF-1α–mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.C., Scicluna B.P., Arts R.J., Gresnigt M.S., Lachmandas E., Giamarellos-Bourboulis E.J., Kox M., Manjeri G.R., Wagenaars J.A., Cremer O.L. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat. Immunol. 2016;17:406–413. doi: 10.1038/ni.3398. [DOI] [PubMed] [Google Scholar]
- Chinthrajah R.S., Hernandez J.D., Boyd S.D., Galli S.J., Nadeau K.C. Molecular and cellular mechanisms of food allergy and food tolerance. J. Allergy Clin. Immunol. 2016;137:984–997. doi: 10.1016/j.jaci.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ A., Gunther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K., Scholz C.J., Oosting M., Haendler K., Bassler K. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell. 2018;172:162–175 e114. doi: 10.1016/j.cell.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Wood R.A., French S., Fiocchi A., Jordana M., Waserman S., Brożek J.L., Schünemann H.J. Oral immunotherapy for peanut allergy (PACE): a systematic review and meta-analysis of efficacy and safety. Lancet. 2019;393:2222–2232. doi: 10.1016/S0140-6736(19)30420-9. [DOI] [PubMed] [Google Scholar]
- Comoglio F., Simonatto M., Polletti S., Liu X., Smale S.T., Barozzi I., Natoli G. Dissection of acute stimulus-inducible nucleosome remodeling in mammalian cells. Genes Dev. 2019;33:1159–1174. doi: 10.1101/gad.326348.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.A., Elliott J.M., Keyel P.A., Yang L., Carrero J.A., Yokoyama W.M. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. U S A. 2009;106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czimmerer Z., Daniel B., Horvath A., Ruckerl D., Nagy G., Kiss M., Peloquin M., Budai M.M., Cuaranta-Monroy I., Simandi Z. The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity. 2018;48:75–90.e6. doi: 10.1016/j.immuni.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czimmerer Z., Nagy Z.S., Nagy G., Horvath A., Silye-Cseh T., Kriston A., Jonas D., Sauer S., Steiner L., Daniel B. Extensive and functional overlap of the STAT6 and RXR cistromes in the active enhancer repertoire of human CD14+ monocyte derived differentiating macrophages. Mol. Cell Endocrinol. 2018;471:63–74. doi: 10.1016/j.mce.2017.07.034. [DOI] [PubMed] [Google Scholar]
- Daniel B., Nagy G., Czimmerer Z., Horvath A., Hammers D.W., Cuaranta-Monroy I., Poliska S., Tzerpos P., Kolostyak Z., Hays T.T. The nuclear receptor PPARgamma controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity. 2018;49:615–626.e6. doi: 10.1016/j.immuni.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K., Muller H., Ragoussis J., Wei C.L., Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E., Guler R., Mhlanga M., Suzuki H., Brombacher F., Schmeier S. Transcriptionally induced enhancers in the macrophage immune response to Mycobacterium tuberculosis infection. BMC Genomics. 2019;20:71. doi: 10.1186/s12864-019-5450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E., Guler R., Mhlanga M.M., Suzuki H., Brombacher F., Schmeier S. Genome-wide profiling of transcribed enhancers during macrophage activation. Epigenetics Chromatin. 2017;10:50. doi: 10.1186/s13072-017-0158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondalay G.K., Rael E., Acharya S., Zhang W., Sampath V., Galli S.J., Tibshirani R., Boyd S.D., Maecker H., Nadeau K.C. Food allergy and omics. J. Allergy Clin. Immunol. 2018;141:20–29. doi: 10.1016/j.jaci.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Di Gioia M., Spreafico R., Springstead J.R., Mendelson M.M., Joehanes R., Levy D., Zanoni I. Endogenous oxidized phospholipids reprogram cellular metabolism and boost hyperinflammation. Nat. Immunol. 2020;21:42–53. doi: 10.1038/s41590-019-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do A.N., Watson C.T., Cohain A.T., Griffin R.S., Grishin A., Wood R.A., Burks W.A., Jones S.M., Scurlock A., Leung D.Y.M. Dual transcriptomic and epigenomic study of reaction severity in peanut allergic children. J. Allergy Clin. Immunol. 2020;145:1219–1230. doi: 10.1016/j.jaci.2019.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Andres J., Ferreira A.V., Jansen T., Smithers N., Prinjha R.K., Furze R.C., Netea M.G. Bromodomain inhibitor I-BET151 suppresses immune responses during fungal-immune interaction. Eur. J. Immunol. 2019;49:2044–2050. doi: 10.1002/eji.201848081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Andres J., Novakovic B., Li Y., Scicluna B.P., Gresnigt M.S., Arts R.J.W., Oosting M., Moorlag S., Groh L.A., Zwaag J. The itaconate pathway is a central regulatory node linking innate immune tolerance and trained immunity. Cell Metab. 2019;29:211–220.e5. doi: 10.1016/j.cmet.2018.09.003. [DOI] [PubMed] [Google Scholar]
- Estcourt M.J., Campbell D.E., Gold M.S., Richmond P., Allen K.J., Quinn H.E., Marsh J.A., Peters R.L., Valerio C., Dai D. Whole-cell pertussis vaccination and decreased risk of IgE-mediated food allergy: a nested case-control study. J. Allergy Clin. Immunol. Pract. 2019 doi: 10.1016/j.jaip.2019.12.020. S2213-2198(19)31054-2. [DOI] [PubMed] [Google Scholar]
- Fairfax B.P., Humburg P., Makino S., Naranbhai V., Wong D., Lau E., Jostins L., Plant K., Andrews R., McGee C. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi S., Fok E.T., Dalla E., Shibayama Y., Borner K., Chang E.Y., Stoychev S., Imakaev M., Grimm D., Wang K.C. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat. Genet. 2019;51:138–150. doi: 10.1038/s41588-018-0298-2. [DOI] [PubMed] [Google Scholar]
- Fanucchi S., Mhlanga M.M. Lnc-ing trained immunity to chromatin architecture. Front. Cell Dev. Biol. 2019;7:2. doi: 10.3389/fcell.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan K.L., van Crevel R., Curtis N., Shann F., Levy O., Optimmunize N. Heterologous ("nonspecific") and sex-differential effects of vaccines: epidemiology, clinical trials, and emerging immunologic mechanisms. Clin. Infect. Dis. 2013;57:283–289. doi: 10.1093/cid/cit209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok E.T., Davignon L., Fanucchi S., Mhlanga M.M. The lncRNA connection between cellular metabolism and epigenetics in trained immunity. Front. Immunol. 2018;9:3184. doi: 10.3389/fimmu.2018.03184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S.L., Hargreaves D.C., Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Garcia-Valtanen P., Guzman-Genuino R.M., Williams D.L., Hayball J.D., Diener K.R. Evaluation of trained immunity by beta-1, 3 (d)-glucan on murine monocytes in vitro and duration of response in vivo. Immunol. Cell Biol. 2017;95:601–610. doi: 10.1038/icb.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S., Barozzi I., Mietton F., Polletti S., De Santa F., Venturini E., Gregory L., Lonie L., Chew A., Wei C.L. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Gianfrancesco M.A., Dehairs J., L'Homme L., Herinckx G., Esser N., Jansen O., Habraken Y., Lassence C., Swinnen J.V., Rider M.H. Saturated fatty acids induce NLRP3 activation in human macrophages through K(+) efflux resulting from phospholipid saturation and Na, K-ATPase disruption. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:1017–1030. doi: 10.1016/j.bbalip.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Glass C.K., Natoli G. Molecular control of activation and priming in macrophages. Nat. Immunol. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia A.C., Brugiolo A.S., Alves C.C., Silva F.M., Mesquita F.P., Gameiro J., Ferreira A.P. Th2 responses in OVA-sensitized BALB/c mice are down-modulated by Mycobacterium bovis BCG treatment. J. Clin. Immunol. 2013;33:235–245. doi: 10.1007/s10875-012-9746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley M.J., Brough D., Quintin J., Allan S.M. Microglial priming as trained immunity in the brain. Neuroscience. 2019;405:47–54. doi: 10.1016/j.neuroscience.2017.12.039. [DOI] [PubMed] [Google Scholar]
- Hangauer M.J., Vaughn I.W., McManus M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Romanoski C.E., Benner C., Glass C.K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzerling L., Mari A., Bergmann K.-C., Bresciani M., Burbach G., Darsow U., Durham S., Fokkens W., Gjomarkaj M., Haahtela T. The skin prick test – European standards. Clin. Translat. Allergy. 2013;3:3. doi: 10.1186/2045-7022-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz U., Gerhold K., Gruber C., Braun A., Wahn U., Renz H., Paul K. BCG infection suppresses allergic sensitization and development of increased airway reactivity in an animal model. J. Allergy Clin. Immunol. 1998;102:867–874. doi: 10.1016/s0091-6749(98)70030-2. [DOI] [PubMed] [Google Scholar]
- Higgins J.P., Soares-Weiser K., Lopez-Lopez J.A., Kakourou A., Chaplin K., Christensen H., Martin N.K., Sterne J.A., Reingold A.L. Association of BCG, DTP, and measles containing vaccines with childhood mortality: systematic review. BMJ. 2016;355:i5170. doi: 10.1136/bmj.i5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfenspirger M.T., Agrawal D.K. Airway hyperresponsiveness, late allergic response, and eosinophilia are reversed with mycobacterial antigens in ovalbumin-presensitized mice. J. Immunol. 2002;168:2516–2522. doi: 10.4049/jimmunol.168.5.2516. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Jr., Travers P., Walport M., Shlomchik M.J. Immunobiology: The Immune System in Health and Disease. Fifth Edition. Garland Science; 2001. Principles of innate and adaptive immunity. [Google Scholar]
- Jentho E., Novakovic B., Ruiz-Moreno C., Kourtzelis I., Martins R., Chavakis T., Soares M.P., Kalafati L., Guerra J., Roestel F. Heme induces innate immune memory. bioRxiv. 2019 doi: 10.1101/2019.12.12.874578. [DOI] [Google Scholar]
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kang K., Bachu M., Park S.H., Kang K., Bae S., Park-Min K.H., Ivashkiv L.B. IFN-gamma selectively suppresses a subset of TLR4-activated genes and enhancers to potentiate macrophage activation. Nat. Commun. 2019;10:3320. doi: 10.1038/s41467-019-11147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Park S.H., Chen J., Qiao Y., Giannopoulou E., Berg K., Hanidu A., Li J., Nabozny G., Kang K. Interferon-gamma represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity. 2017;47:235–250.e4. doi: 10.1016/j.immuni.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlmark K.R., Tacke F., Dunay I.R. Monocytes in health and disease - Minireview. Eur. J. Microbiol. Immunol. (Bp) 2012;2:97–102. doi: 10.1556/EuJMI.2.2012.2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E., Sanz J., Dunn J.L., Khan N., Mendonca L.E., Pacis A., Tzelepis F., Pernet E., Dumaine A., Grenier J.C. BCG educates hematopoietic stem cells to generate protective innate immunity against. Tuberculosis. Cell. 2018;172:176–190 e119. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis J., Quintin J., Preijers F., Joosten L.A., Ifrim D.C., Saeed S., Jacobs C., van Loenhout J., de Jong D., Stunnenberg H.G. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U S A. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm S.L., Shipony Z., Greenleaf W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- Kosoy R., Agashe C., Grishin A., Leung D.Y., Wood R.A., Sicherer S.H., Jones S.M., Burks A.W., Davidson W.F., Lindblad R.W. Transcriptional profiling of egg allergy and relationship to disease phenotype. PLoS One. 2016;11:e0163831. doi: 10.1371/journal.pone.0163831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kox M., van Eijk L.T., Zwaag J., van den Wildenberg J., Sweep F.C., van der Hoeven J.G., Pickkers P. Voluntary activation of the sympathetic nervous system and attenuation of the innate immune response in humans. Proc. Natl. Acad. Sci. U S A. 2014;111:7379–7384. doi: 10.1073/pnas.1322174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen I., Aaby P., Jensen H. Routine vaccinations and child survival: follow up study in Guinea-Bissau, West Africa. BMJ. 2000;321:1435–1438. doi: 10.1136/bmj.321.7274.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnadi A., Park S.H., Yuan R., Pannellini T., Giannopoulou E., Oliver D., Lu T., Park-Min K.H., Ivashkiv L.B. The cytokine TNF promotes transcription factor SREBP activity and binding to inflammatory genes to activate macrophages and limit tissue repair. Immunity. 2019;51:241–257.e9. doi: 10.1016/j.immuni.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt J., Grosse S., Marx C., Siwczak F., Stengel S., Bruns T., Bauer R., Kiehntopf M., Williams D.L., Wang Z.Q. Candida albicans beta-glucan differentiates human monocytes into a specific subset of macrophages. Front. Immunol. 2018;9:2818. doi: 10.3389/fimmu.2018.02818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Oosting M., Deelen P., Ricano-Ponce I., Smeekens S., Jaeger M., Matzaraki V., Swertz M.A., Xavier R.J., Franke L. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat. Med. 2016;22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libraty D.H., Zhang L., Woda M., Acosta L.P., Obcena A., Brion J.D., Capeding R.Z. Neonatal BCG vaccination is associated with enhanced T-helper 1 immune responses to heterologous infant vaccines. Trials Vaccinol. 2014;3:1–5. doi: 10.1016/j.trivac.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan M.F., Nurmatov U., Frank T.L., Niven R.M., Baxter D.N., Sheikh A. Does BCG vaccination protect against childhood asthma? Final results from the Manchester Community Asthma Study retrospective cohort study and updated systematic review and meta-analysis. J. Allergy Clin. Immunol. 2014;133:688–695.e14. doi: 10.1016/j.jaci.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Loh W., Tang M.L.K. The epidemiology of food allergy in the global context. Int. J. Environ. Res. Public Health. 2018;15:2043–2051. doi: 10.3390/ijerph15092043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.-T., Gao F., Gu K., Chen D.-K. The role of monocytes and macrophages in autoimmune diseases: a comprehensive review. Front. Immunol. 2019;10:1140. doi: 10.3389/fimmu.2019.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D., Joo J.E., Sexton-Oates A., Dang T., Allen K., Saffery R., Prescott S. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics. 2014;9:998–1006. doi: 10.4161/epi.28945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D., Neeland M., Dang T., Cobb J., Ellis J., Barnett A., Tang M., Vuillermin P., Allen K., Saffery R. Epigenetic dysregulation of naive CD4+ T-cell activation genes in childhood food allergy. Nat. Commun. 2018;9:3308. doi: 10.1038/s41467-018-05608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D.J., Prescott S.L. Silent mysteries: epigenetic paradigms could hold the key to conquering the epidemic of allergy and immune disease. Allergy. 2010;65:7–15. doi: 10.1111/j.1398-9995.2009.02186.x. [DOI] [PubMed] [Google Scholar]
- McVicker G., van de Geijn B., Degner J.F., Cain C.E., Banovich N.E., Raj A., Lewellen N., Myrthil M., Gilad Y., Pritchard J.K. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 2018;172:147–161.e12. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourits V.P., Wijkmans J.C.H.M., Joosten L.A.B., Netea M.G. Trained immunity as a novel therapeutic strategy. Curr. Opin. Pharmacol. 2018;41:52–58. doi: 10.1016/j.coph.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Mulder W.J.M., Ochando J., Joosten L.A.B., Fayad Z.A., Netea M.G. Therapeutic targeting of trained immunity. Nat. Rev. Drug Discov. 2019;18:553–566. doi: 10.1038/s41573-019-0025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Larsen S.B., Gomez N.C., Alaverdyan K., Sendoel A., Yuan S., Polak L., Kulukian A., Chai S., Fuchs E. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature. 2017;550:475–480. doi: 10.1038/nature24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G., Ostuni R. Adaptation and memory in immune responses. Nat. Immunol. 2019;20:783–792. doi: 10.1038/s41590-019-0399-9. [DOI] [PubMed] [Google Scholar]
- Neeland M.R., Andorf S., Manohar M., Dunham D., Lyu S.C., Dang T.D., Peters R.L., Perrett K.P., Tang M.L.K., Saffery R. Mass cytometry reveals cellular fingerprint associated with IgE+ peanut tolerance and allergy in early life. Nat. Commun. 2020;11:1091. doi: 10.1038/s41467-020-14919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeland M.R., Koplin J.J., Dang T.D., Dharmage S.C., Tang M.L., Prescott S.L., Saffery R., Martino D.J., Allen K.J. Early life innate immune signatures of persistent food allergy. J. Allergy Clin. Immunol. 2018;142:857–864 e853. doi: 10.1016/j.jaci.2017.10.024. [DOI] [PubMed] [Google Scholar]
- Neeland M.R., Martino D.J., Dang T.D., Koplin J.J., Peters R.L., Grishin A., Dharmage S.C., Tang M.L., Sampson H.A., Saffery R. B-cell phenotype and function in infants with egg allergy. Allergy. 2019;74:1022–1025. doi: 10.1111/all.13707. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Dominguez-Andres J., Barreiro L.B., Chavakis T., Divangahi M., Fuchs E., Joosten L.A.B., van der Meer J.W.M., Mhlanga M.M., Mulder W.J.M. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020;4:1–14. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A., Latz E., Mills K.H., Natoli G., Stunnenberg H.G., O'Neill L.A., Xavier R.J. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E., Jeffrey K.L., Schaefer U., Beinke S., Dewell S., Chung C.W., Chandwani R., Marazzi I., Wilson P., Coste H. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J., Lee J.H., Noh G., Bang S.Y., Kim H.S., Choi W.S., Cho S., Lee S.S. Characterisation of allergen-specific responses of IL-10-producing regulatory B cells (Br1) in cow milk allergy. Cell Immunol. 2010;264:143–149. doi: 10.1016/j.cellimm.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Novakovic B., Habibi E., Wang S.Y., Arts R.J.W., Davar R., Megchelenbrink W., Kim B., Kuznetsova T., Kox M., Zwaag J. Beta-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell. 2016;167:1354–1368 e1314. doi: 10.1016/j.cell.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne N.J., Koplin J.J., Martin P.E., Gurrin L.C., Lowe A.J., Matheson M.C., Ponsonby A.L., Wake M., Tang M.L., Dharmage S.C. Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J. Allergy Clin. Immunol. 2011;127:668–676.e1-2. doi: 10.1016/j.jaci.2011.01.039. [DOI] [PubMed] [Google Scholar]
- Ostuni R., Piccolo V., Barozzi I., Polletti S., Termanini A., Bonifacio S., Curina A., Prosperini E., Ghisletti S., Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Ozer A., Tukenmez F., Biricik A., Barlan I.B., Cirakoglu B., Basaran M.M. Effect of BCG vaccination on cytokine mRNA expression in atopic children with asthma. Immunol. Lett. 2003;86:29–35. doi: 10.1016/s0165-2478(02)00261-4. [DOI] [PubMed] [Google Scholar]
- Park S.H., Kang K., Giannopoulou E., Qiao Y., Kang K., Kim G., Park-Min K.H., Ivashkiv L.B. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 2017;18:1104–1116. doi: 10.1038/ni.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R.L., Neeland M.R., Allen K.J. Primary prevention of food allergy. Curr. Allergy asthma Rep. 2017;17:52. doi: 10.1007/s11882-017-0718-x. [DOI] [PubMed] [Google Scholar]
- Phan A.T., Goldrath A.W., Glass C.K. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity. 2017;46:714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo V., Curina A., Genua M., Ghisletti S., Simonatto M., Sabo A., Amati B., Ostuni R., Natoli G. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol. 2017;18:530–540. doi: 10.1038/ni.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar N., Fishbein A.B., Erickson K.A., Cai M., Szychlinski C., Bryce P.J., Schleimer R.P., Fuleihan R.L., Singh A.M. Naturally occurring tolerance acquisition to foods in previously allergic children is characterized by antigen specificity and associated with increased subsets of regulatory T cells. Clin. Exp. Allergy. 2015;45:1663–1672. doi: 10.1111/cea.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J., Saeed S., Martens J.H.A., Giamarellos-Bourboulis E.J., Ifrim D.C., Logie C., Jacobs L., Jansen T., Kullberg B.J., Wijmenga C. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romee R., Rosario M., Berrien-Elliott M.M., Wagner J.A., Jewell B.A., Schappe T., Leong J.W., Abdel-Latif S., Schneider S.E., Willey S. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl Med. 2016;8:357ra123. doi: 10.1126/scitranslmed.aaf2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S., Quintin J., Kerstens H.H., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.C., Ratter J., Berentsen K., van der Ent M.A. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam T., Jones M., Thomas B.J., Wu X., Gilliland T., Qian K., Eskin A., Casero D., Zhang Z., Sandhu J. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 2018;24:304–312. doi: 10.1038/nm.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson H.A., O'Mahony L., Burks A.W., Plaut M., Lack G., Akdis C.A. Mechanisms of food allergy. J. Allergy Clin. Immunol. 2018;141:11–19. doi: 10.1016/j.jaci.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Sánchez-Ramón S., Conejero L., Netea M.G., Sancho D., Palomares Ó., Subiza J.L. Trained immunity-based vaccines: a new paradigm for the development of broad-spectrum anti-infectious formulations. Front. Immunol. 2018;9:2936. doi: 10.3389/fimmu.2018.02936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Koplin J.J., Dharmage S.C., Field M.J., Sawyer S.M., McWilliam V., Peters R.L., Gurrin L.C., Vuillermin P.J., Douglass J. Prevalence of clinic-defined food allergy in early adolescence: the SchoolNuts study. J. Allergy Clin. Immunol. 2018;141:391–398.e4. doi: 10.1016/j.jaci.2017.05.041. [DOI] [PubMed] [Google Scholar]
- Satitsuksanoa P., Jansen K., Globinska A., van de Veen W., Akdis M. Regulatory immune mechanisms in tolerance to food allergy. Front. Immunol. 2018;9:2939. doi: 10.3389/fimmu.2018.02939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid F., Lip G.Y.H., Shantsila E. Role of monocytes in heart failure and atrial fibrillation. J. Am. Heart Assoc. 2018;7:e007849. doi: 10.1161/JAHA.117.007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalova I.N., Lim J.Y., Chittezhath M., Zinkernagel A.S., Beasley F., Hernandez-Jimenez E., Toledano V., Cubillos-Zapata C., Rapisarda A., Chen J. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1alpha. Immunity. 2015;42:484–498. doi: 10.1016/j.immuni.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Shann F. Substantial benefits from finding the most effective BCG strain. Lancet Respir. Med. 2016;4:e35. doi: 10.1016/S2213-2600(16)30108-4. [DOI] [PubMed] [Google Scholar]
- Shreffler W.G., Wanich N., Moloney M., Nowak-Wegrzyn A., Sampson H.A. Association of allergen-specific regulatory T cells with the onset of clinical tolerance to milk protein. J. Allergy Clin. Immunol. 2009;123:43–52 e47. doi: 10.1016/j.jaci.2008.09.051. [DOI] [PubMed] [Google Scholar]
- Stein M.M., Hrusch C.L., Gozdz J., Igartua C., Pivniouk V., Murray S.E., Ledford J.G., Marques Dos Santos M., Anderson R.L., Metwali N. Innate immunity and asthma risk in amish and hutterite farm children. N. Engl. J. Med. 2016;375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunnenberg H.G., International Human Epigenome, C. Hirst M. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167:1897. doi: 10.1016/j.cell.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., Lanier L.L. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Madera S., Bezman N.A., Beilke J.N., Kaplan M.H., Lanier L.L. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med. 2012;209:947–954. doi: 10.1084/jem.20111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suprun M., Getts R., Raghunathan R., Grishina G., Witmer M., Gimenez G., Sampson H.A., Suarez-Farinas M. Novel Bead-Based Epitope Assay is a sensitive and reliable tool for profiling epitope-specific antibody repertoire in food allergy. Sci. Rep. 2019;9:18425. doi: 10.1038/s41598-019-54868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester R.J., van der M.A., Lamm D.L. Intravesical bacillus Calmette-Guerin reduces the risk of progression in patients with superficial bladder cancer: a meta-analysis of the published results of randomized clinical trials. J. Urol. 2002;168:1964–1970. doi: 10.1016/S0022-5347(05)64273-5. [DOI] [PubMed] [Google Scholar]
- Thiem K., Stienstra R., Riksen N.P., Keating S.T. Trained immunity and diabetic vascular disease. Clin. Sci. (Lond) 2019;133:195–203. doi: 10.1042/CS20180905. [DOI] [PubMed] [Google Scholar]
- Thostesen L.M., Kjaer H.F., Pihl G.T., Nissen T.N., Birk N.M., Kjaergaard J., Jensen A.K.G., Aaby P., Olesen A.W., Stensballe L.G. Neonatal BCG has no effect on allergic sensitization and suspected food allergy until 13 months. Pediatr. Allergy Immunol. 2017;28:588–596. doi: 10.1111/pai.12748. [DOI] [PubMed] [Google Scholar]
- Thostesen L.M., Stensballe L.G., Pihl G.T., Kjaergaard J., Birk N.M., Nissen T.N., Jensen A.K.G., Aaby P., Olesen A.W., Jeppesen D.L. Neonatal BCG vaccination has no effect on recurrent wheeze in the first year of life: a randomized clinical trial. J. Allergy Clin. Immunol. 2017;140:1616–1621.e3. doi: 10.1016/j.jaci.2016.12.990. [DOI] [PubMed] [Google Scholar]
- Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang A., Grishin A., Grishina G., Do A.N., Sordillo J., Chew G.L., Bunyavanich S. Endotoxin, food allergen sensitization, and food allergy: A complementary epidemiologic and experimental study. Allergy. 2020;75:625–635. doi: 10.1111/all.14054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulic M.K., Hodder M., Forsberg A., McCarthy S., Richman T., D’Vaz N., van den Biggelaar A.H.J., Thornton C.A., Prescott S.L. Differences in innate immune function between allergic and nonallergic children: New insights into immune ontogeny. J. Allergy Clin. Immunol. 2011;127:470–478.e1. doi: 10.1016/j.jaci.2010.09.020. [DOI] [PubMed] [Google Scholar]
- van der Heijden C., Noz M.P., Joosten L.A.B., Netea M.G., Riksen N.P., Keating S.T. Epigenetics and trained immunity. Antioxid. Redox Signal. 2018;29:1023–1040. doi: 10.1089/ars.2017.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Nanni L., Novakovic B., Megchelenbrink W., Kuznetsova T., Stunnenberg H.G., Ceri S., Logie C. Extensive epigenomic integration of the glucocorticoid response in primary human monocytes and in vitro derived macrophages. Sci. Rep. 2019;9:2772. doi: 10.1038/s41598-019-39395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T., Rothenberg M.E. Cell-by-cell deciphering of T cells in allergic inflammation. J. Allergy Clin. Immunol. 2019;144:1143–1148. doi: 10.1016/j.jaci.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff B., Burns A.R., Middleton J., Rot A. Endothelial cell "memory" of inflammatory stimulation: human venular endothelial cells store interleukin 8 in Weibel-Palade bodies. J. Exp. Med. 1998;188:1757–1762. doi: 10.1084/jem.188.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard K.J., Geissmann F. Monocytes in atherosclerosis: subsets and functions. Nat. Rev. Cardiol. 2010;7:77–86. doi: 10.1038/nrcardio.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wang S., Fan Y., Zhu L. Systemic mycobacterial infection inhibits antigen-specific immunoglobulin E production, bronchial mucus production and eosinophilic inflammation induced by allergen. Immunology. 1999;98:329–337. doi: 10.1046/j.1365-2567.1999.00856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Freeland D.M.H., Nadeau K.C. Food allergy: immune mechanisms, diagnosis and immunotherapy. Nat. Rev. Immunol. 2016;16:751–765. doi: 10.1038/nri.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Collier F., Naselli G., Saffery R., Tang M.L., Allen K.J., Ponsonby A.L., Harrison L.C., Vuillermin P., Group B.I.S.I. Cord blood monocyte-derived inflammatory cytokines suppress IL-2 and induce nonclassic "T(H)2-type" immunity associated with development of food allergy. Sci. Transl Med. 2016;8:321ra328. doi: 10.1126/scitranslmed.aad4322. [DOI] [PubMed] [Google Scholar]