Abstract
In cancer, both histopathological images and genomic signatures are used for diagnosis, prognosis, and subtyping. However, combining histopathological images with genomic data for predicting prognosis, as well as the relationships between them, has rarely been explored. In this study, we present an integrative genomics framework for constructing a prognostic model for clear cell renal cell carcinoma. We used patient data from The Cancer Genome Atlas (n = 410), extracting hundreds of cellular morphological features from digitized whole-slide images and eigengenes from functional genomics data to predict patient outcome. The risk index generated by our model correlated strongly with survival outperforming predictions based on considering morphological features or eigengenes separately. The predicted risk index also effectively stratified patients in early-stage (stage I and stage II) tumors, whereas no significant survival difference was observed using staging alone. The prognostic value of our model was independent of other known clinical and molecular prognostic factors for patients with clear cell renal cell carcinoma. Overall, this workflow and the shared software code provide building blocks for applying similar approaches in other cancers.
Keywords: prognostic marker, clear cell renal cell carcinoma, histopathological image, gene expression signature, gene co-expression analysis, integrative genomics
Introduction
Histopathological images confer important information for diagnosis, staging, and prognosis for cancers and are being used extensively by pathologists in clinical practice. With the recent availability of digital whole-slide images (1), automated computational histopathological image analysis systems have shown great promise in diagnosis and the discovery of new biomarkers for cancers such as breast (2–4), lung (5,6), brain (7), and colon cancers (8). In comparison with human inspection, computerized image analysis has great potential to improve efficiency, accuracy, and consistency. Besides histopathological images, molecular characteristics, such as genetic alterations and gene expression signatures, are also widely adopted for predicting clinical outcomes for cancers (9,10). Therefore, an interesting scientific question is the relationship between morphological and genomic features while an important translational question is if the integration of these two types of features can lead to more accurate prediction of patient outcome. This has been previously explored in various cancers including breast, ovarian, and glioblastoma, and led to new insights into the relationship between cancer tissue morphology and genetic changes such as PTEN mutations (3,11–13).
To study these issues, matched histopathological images and genomic datasets for cancers are needed. Fortunately, The Cancer Genome Atlas (TCGA) project not only provides an extensive collection of genomics and clinical outcome data for large cohorts of patients of more than 30 types of cancers, but also hosts a large collection of matched histopathological images for solid tumor samples. Currently, more than 24,000 histopathological images are available at the TCGA data portal and can be visualized at the Cancer Digital Slide Archive (CDSA, http://cancer.digitalslidearchive.net/) (14).
Quantitative analysis of these images and integration with genomics data require innovation in integrative genomics and call for techniques from bioimage informatics, genomics, and bioinformatics. We previously developed a computational framework for quantifying morphological features from large histopathological images (4,15) as well as genomics visualization tools for integrating imaging, clinical, and genomic features to predict patient outcomes (4,16,17). Therefore, to further promote this emerging integrative genomics field straddling bioimage informatics and genomics and ensure wide utilization of valuable large datasets, we demonstrate an integrative genomics workflow on the less well-studied renal cancers. The analysis tools are publicly available and can be adopted as building blocks for other integrative genomics workflows (please see Methods section).
Renal cell carcinoma (RCC) is the most common type of malignant neoplasm arising from kidney in adults, responsible for approximately 90–95% of all cases (18). It can be categorized into the following histologic subtypes: clear cell, papillary, chromophobe, collecting duct, and unclassified RCC based on the Heidelberg classification system (19). In this study, we focus on clear cell renal cell carcinoma (ccRCC), which is the most prevalent subtype, accounting for 80–90% of all RCCs (20). In clinical practice, tissue sections are examined under a microscope by pathologists to make a diagnosis and predict prognosis. The clinical behavior of ccRCC is quite diverse, ranging from slow-growing localized tumors to aggressive metastatic disease (9). Therefore, prognostic markers play a crucial role in stratification of patients for personalized cancer management, which could avoid either over-treatment or under-treatment (21). For instance, patients classified into high-risk group may benefit from closer follow-up, more aggressive therapies, and advance care planning (5,22). Currently, prognostic markers for ccRCC in routine clinical use consist mainly of tumor stage, nuclear grade, and presence of necrosis (23–25). However, cancer is a highly heterogeneous disease. The prediction accuracy of traditional clinical factors remains limited for individual patients, especially for early-stage patients, and also relies on the experience of pathologists. Therefore, there is a need for more effective markers for predicting prognosis of ccRCC.
Using the large cohort of ccRCC patients from TCGA, hundreds of cellular morphological features can be extracted from hematoxylin and eosin (H&E) stained whole-slide images, characterizing nucleus size, shape, texture, and the spatial relationship between nuclei. In this paper, we demonstrate how image features correlate with co-expressed gene signatures and developed an automated prognostic model that could predict patient’s survival risk for patient stratification, using a combination of quantitative image features and eigengenes. To the best of our knowledge, this is the first study to couple histopathological images and genomic data to predict ccRCC clinical outcome and our results indicate that the integration of imaging and genomic features can lead to improved prognosis prediction for early-stage (stages I and II) ccRCC patients than existing clinical markers.
Materials and methods
Data and codes availability
Processed data (extracted quantitative imaging features, combined gene expression data, etc.) and code with annotations, comments and instructions are available at https://github.com/chengjun583/image-mRNA-prognostic-model.
Data source and selection
ccRCC patient samples used in our study included matched H&E stained whole-slide images, transcriptome, somatic mutation, and clinical information, which were acquired from TCGA data portal at NCI Genomic Data Commons (26). Patients with missing or too short (i.e., less than 30 days) follow-up were excluded. Microscopic images (20X and 40X magnification) were obtained from TCGA. The demographic and clinical characteristics for the selected 410 patients are summarized in Table 1.
Table 1.
Demographic and clinical characteristics.
| Characteristics | Summary |
|---|---|
| Patient No. | 410 |
| Age (years) | |
| Range | 26–90 |
| Median | 60 |
| Gender | |
| Female | 140 (34.2%) |
| Male | 270 (65.8%) |
| Follow-up (months) | |
| Range | 1.3–112.6 |
| Median | 37.8 |
| Death | 135 (32.9%) |
| Grade | |
| G1 | 7 (1.7%) |
| G2 | 171 (41.7%) |
| G3 | 169 (41.2%) |
| G4 | 63 (15.4%) |
| Stage | |
| Stage I | 202 (49.3%) |
| Stage II | 41 (10%) |
| Stage III | 98 (23.9%) |
| Stage IV | 69 (16.8%) |
One challenge for this study was the lack of other large cohorts of ccRCC with matched histological image and genomic data. Thus, instead of using a second dataset for validation, we applied cross-validation in every step of downstream of the machine learning analysis as described below.
Data analysis and integration workflow
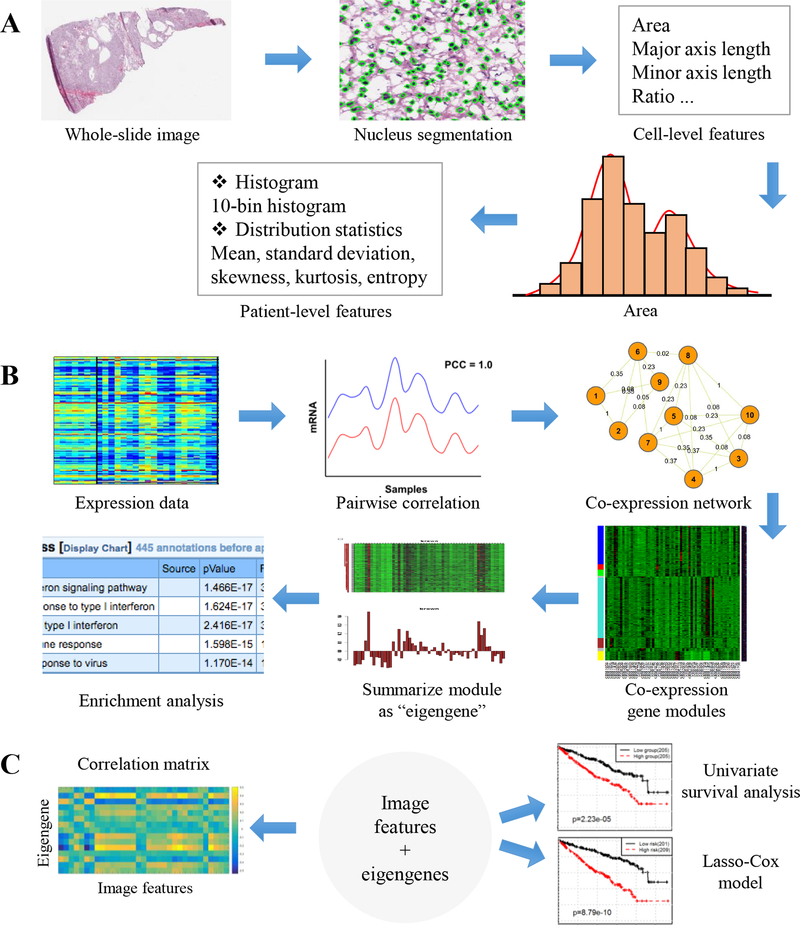
Fig. 1 outlines our data analysis workflow for both imaging and genomic data for both univariate and multivariate analyses with details of each major step being described in the following sections.
Figure 1.
Data analysis and integration workflow. (A) Cellular morphological feature extraction pipeline. (B) Schematic diagram for gene co-expression analysis and summarization. (C) Integrative analysis of image features with eigengenes. Univariate survival analysis is used for an initial selection of survival-associated variables, and then these variables are used to train a lasso-Cox prognostic model. Correlation between image features and eigengenes is also explored.
Histopathological image features
Our image feature extraction pipeline consists of three steps: nucleus segmentation, cell-level feature extraction, and aggregation of cell-level features into patient-level features (Fig. 1A). Rich pathological information is present in stained cell nuclei that requires segmentation to facilitate subsequent analyses. For this task, a recently proposed approach by Phoulady et al (27) was employed, which is an unsupervised segmentation method requiring no parameter learning or training data because the parameters are set adaptively. Next, ten types of cell-level features were extracted for each segmented nucleus, characterizing nucleus size, shape, texture, and distance to neighbors. These cell-level features are nuclear area (denoted as area), lengths of the major and minor axes of cell nucleus and the ratio of major axis length to minor axis length (major, minor, and ratio), mean pixel values of nucleus in RGB three channels respectively (rMean, gMean, and bMean), and mean, maximum, and minimum distances (distMean, distMax, and distMin) to neighboring nuclei in the Delaunay triangulation graph (28). The Delaunay triangulation graph was constructed based on the locations of segmented nuclei. In this graph, each nucleus was a node and connected to neighboring nuclei. Finally, for each type of cell-level features, a ten-bin histogram and five distribution statistics (i.e. mean, standard deviation, skewness, kurtosis, and entropy) were adopted to aggregate the numerous cell-level features extracted from a patient into patient-level features; 150 patient-level features were generated in total. Taking the cell-level feature, area, as an example, corresponding 15 patient-level features were denoted as area_bin1 to area_bin10 for the 10 histogram features, and area_mean, area_std, area_skewness, area_kurtosis, and area_entropy for the 5 distribution statistics. For other cell-level features, corresponding patient-level features were named in the same way. Area_bin1 represents the percentage of very small nuclei over the entire slide for a patient while area_bin10 indicates the percentage of very large nuclei in the patient sample. Skewness is a measure of the asymmetry of the data distribution around the sample mean, kurtosis is a measure of how outlier-prone a distribution is, and entropy is a statistical measure of randomness.
Additional description about aggregation of cell-level features into patient-level features is provided in the Supplemental Material. A qualitative example of nucleus segmentation results is shown in Fig. S1.
Gene co-expression analysis and summarization
mRNA expression profiles for the ccRCC tumors in TCGA were transformed from Illumina HiSeq 2000 RNA-seq readcounts to normalized RPKM (reads per kilobase per million). While our first goal was to establish the relationships between gene expression data and the imaging features, the large number of genes posed a challenge to obtaining sufficient statistical power. Therefore instead of focusing on individual genes, we first carried out gene co-expression network analysis (GCNA) to cluster genes into co-expressed modules and summarized each module as an “eigengene” using the protocol described in (29) (Fig. 1B). Modules are clusters of highly interconnected/correlated genes. The eigengene of a module is defined as the first principle component, which can be considered a representative of the gene expression profiles in a module. This approach not only substantially improves statistical power (30), but also allows us to focus on important biological processes or genetic variations associated with the co-expressed gene modules, making the results more interpretable than individual genes as the co-expressed modules are often strongly associated with a specific gene group participating in the same biological process or located on the same chromosomal band.
While there are many algorithms for performing GCNA including the well-known WGCNA package (31), we applied our recently developed weighted network mining algorithm called local maximum quasi-clique merging (lmQCM) (32). Unlike WGCNA, which uses hierarchical clustering and does not allow overlap between modules, our algorithm is a greedy approach allowing genes to be shared among multiple modules, consistent with the fact the genes often participate in multiple biological processes. In addition, we have shown that lmQCM can find smaller co-expressed gene modules that are often associated with structural mutations such as copy number variation in cancers (32). The lmQCM algorithm has four parameters γ, α, t, and β. Among these parameters, γ is the most influential, as it determines if a new module can be initiated by setting the weight threshold for the first edge of the module as a subnetwork. In the lmQCM algorithm, we transformed the absolute values of the Spearman correlation coefficients between expression profiles of genes into weights using a normalization procedure adopted from spectral clustering for which we have shown to be effective in previous studies (33). In practice, we found with γ = 0.30, t = 1, α = 1, and β = 0.4 the algorithm yielded 15 co-expressed gene modules (Table S1) with balanced sizes and clear biological interpretation based on enrichment analysis (Table S2).
Machine-learning methods for prognosis prediction
We built a lasso-regularized Cox proportional hazards (lasso-Cox) model (R package “glmnet”) to calculate the risk index of each patient (34), based on the cellular morphological features and eigengenes (Fig. 1C). Lasso penalty (i.e. L1 penalty) can induce sparsity and thus select an informative subset of features. To validate our method, we used a two-level cross validation (CV) strategy. After each patient was used as a test sample and classified into a low-risk or high-risk group, we used Kaplan-Meier estimator and log-rank test to test if these two groups had distinct survival.
Additional description of the training and prediction process is provided in Supplemental Material.
Statistical methods and enrichment analysis
To screen survival-associated features, for each patient-level feature we divided patients into two groups (low and high groups) where the median of each feature was used as a cut-off point. Kaplan-Meier estimator was used for patient stratification, and p value was calculated with the log-rank test, where p < 0.05 was considered significant. For the initial survival analysis, since our initial goal was screening, we did not apply multiple test compensation such as FDR control in order to obtain more candidate features. The lasso-Cox model was learned on the selected survival-associated features. Cox proportional hazards regression model was fitted, and 95% confidence intervals were computed to determine the prognostic values of our lasso-Cox risk indices and other known prognostic factors. Correlation was computed using Spearman rank correlation coefficients. Enrichment analysis of co-expressed gene modules was carried out using Toppgene (35). All the survival analyses were performed using R package “survival.”
Results
Both image and gene expression data identify poor-prognosis subtype with high percentage of tumor stroma
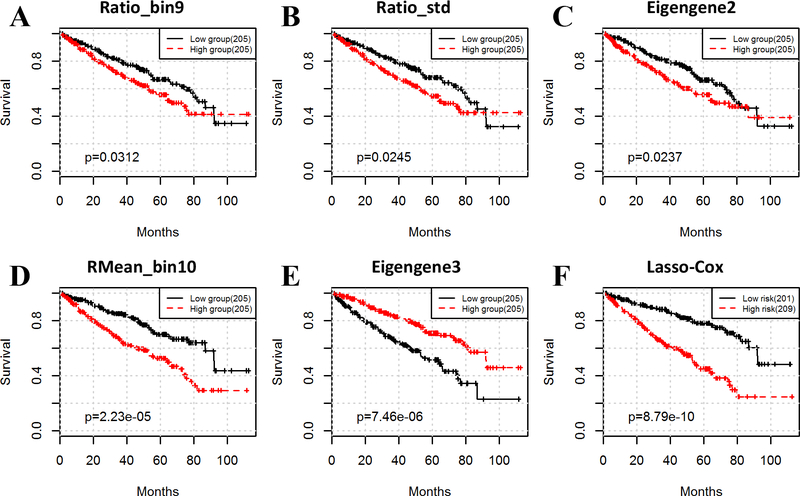
To investigate which specific image features and eigengenes are associated with patient survival, we tested for each feature the statistical significance of difference in overall survival between low and high risk groups that were stratified by the median of feature values. Log-rank test results revealed that 33 image features and 6 eigengenes were significantly related to prognosis (p < 0.05). The log-rank test results of all survival-related variables are listed in Table 2 and the Kaplan-Meier survival curves for some variables are shown in Fig. 2A–E.
Table 2.
Survival-associated image features and eigengenes, identified by Kaplan-Meier estimator and log-rank test (p < 0.05). For each variable, patients were stratified into low and high groups using the median as cut-off point. For P/N, P means positive relation to survival (i.e., patients with high feature values have good prognosis), whereas N means negative relation to survival.
| Feature | P value | P/N | Feature | P value | P/N |
|---|---|---|---|---|---|
| rMean_bin10 | 2.23e-5 | N | gMean_entropy | 0.0194 | N |
| rMean_bin6 | 8.55e-5 | P | ratio_std | 0.0245 | N |
| rMean_std | 1.18e-4 | N | rMean_kurtosis | 0.0269 | P |
| rMean_entropy | 2.45e-4 | N | ratio_bin8 | 0.0297 | N |
| gMean_std | 7.70e-4 | N | ratio_bin9 | 0.0312 | N |
| rMean_bin5 | 0.0010 | P | area_std | 0.0319 | N |
| major_bin9 | 0.0022 | N | ratio_bin5 | 0.0322 | N |
| major_entropy | 0.0028 | N | ratio_mean | 0.0324 | N |
| area_bin5 | 0.0056 | P | major_bin1 | 0.0333 | N |
| major_bin4 | 0.0058 | P | major_bin2 | 0.0337 | N |
| ratio_bin6 | 0.0059 | N | bMean_bin10 | 0.0338 | N |
| major_bin8 | 0.0060 | N | major_bin10 | 0.0366 | N |
| major_std | 0.0072 | N | bMean_std | 0.0407 | N |
| area_bin7 | 0.0089 | P | eigengene3 | 7.46e-6 | P |
| rMean_bin9 | 0.0097 | N | eigengene9 | 1.19e-4 | N |
| major_bin5 | 0.0113 | P | eigengene13 | 9.39e-4 | P |
| gMean_bin10 | 0.0113 | N | eigengene11 | 0.0013 | N |
| area_bin6 | 0.0124 | P | eigengene1 | 0.0217 | N |
| bMean_entropy | 0.0164 | N | eigengene2 | 0.0237 | N |
| ratio_bin7 | 0.0176 | N |
Figure 2.
Image features and eigengenes predict the survival outcomes of ccRCC patients. Both image features (A and B) and eigengenes (C) identify poor-prognosis subtypes with high percentage of stroma. Gene module 2 is enriched with extracellular matrix genes. RMean_bin10 (D) and eigengene3 (E) are the most significant variables for image features and eigengenes, respectively. Integrative analysis of histopathological images and genomic data using lasso-Cox can significantly improve the prognosis prediction power (F).
After examining these survival-associated variables, we found many of them were connected to stroma tissue. Stromal cells such as fibroblasts are typically spindle-shaped with elongated nuclei and therefore characterized by long major axes and/or large ratio between major and minor axes. As shown in Table 2, Fig. 2A and B, image features such as major_bin8, major_bin9, ratio_bin8, ratio_bin9, ratio_std, and major_std, were negatively related to prognosis, that is, patients with large values of these variables had worse prognosis than other patients. Large values of these variables imply a high percentage of stromal cell nuclei in whole-slide images (in terms of major_std, and ratio_std, large values of these variables mean that the major axis length and the ratio of major axis length to minor axis length are spread out in a wide range, indicating a high percentage of stromal cell nuclei). In other words, patients with high percentage of stromal tissue are related to poor prognosis for ccRCC in our study.
In addition to histopathological images, gene expression data also corroborated that stroma played an important role in tumor prognosis. Enrichment analysis showed that gene module 2 was enriched with extracellular matrix genes (Table S2), which is consistent with our knowledge that the tumor microenvironment plays critical roles in tumor development (2,3). Kaplan-Meier survival curves demonstrated distinctly different outcomes for low- and high-expression groups (log-rank test p value = 0.024), where high expression of eigengene 2 was associated with poor prognosis.
Integrative analysis enhances prognostic prediction power
In the previous sections, we showed that many individual features derived from histopathological images and genomic data stratified patients with distinct prognosis. We next investigated if the integration of all identified survival-associated features would provide better prognostic prediction. We built a lasso-regularized Cox proportional hazards model to select the most informative features and calculate a risk index for each patient. Based on the risk indices, patients were divided into a low- or high-risk group by the median. The lasso-Cox model provided significantly better patient stratification than that using individual features (Fig. 2D–F, log-rank test p values = 2.23e-5, 7.46e-6, and 8.79e-10 for the most significant image feature, rMean_bin10, the most significant eigengene expression, eigengene3, and lasso-Cox model, respectively). Among the 33 survival-associated image features and six survival-associated eigengenes, eight image features and five eigengenes were selected: rMean_bin6, major_bin9, area_bin5, gMean_bin10, ratio_bin7, ratio_bin8, ratio_bin9, major_bin1, eigengene1, eigengene3, eigengene9, eigengene11, and eigengene13 (Enrichment analyses of survival-related gene modules are listed in Table S2). Both image features and eigengenes appeared in the final selected feature set, and most of the pairwise mutual information values between them are smaller than the ones between significantly correlated image features and eigengenes (Fig. S2), suggesting that histopathological images and genomic data complement each other in predicting survival outcome.
Survival-associated image features correlate with eigengenes
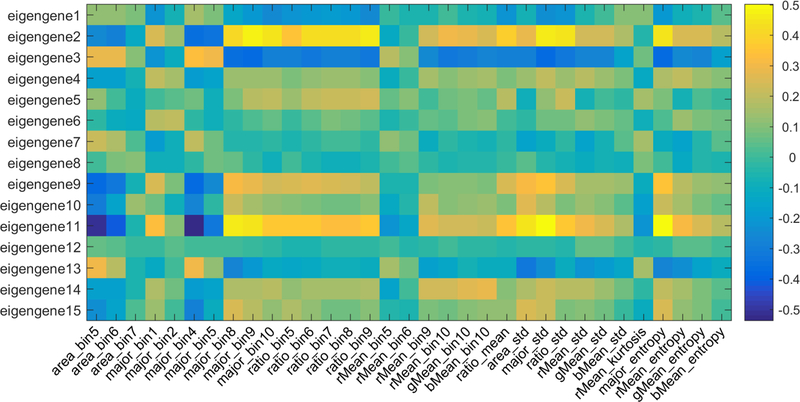
Genotype is one of the three factors that determine phenotype, the other two being inherited epigenetic factors and non-inherited environmental factors. Therefore, tumor characteristics or morphology is very likely to have some relationships with gene expression data. To find out these relationships, we calculated Spearman rank correlation coefficients between each pair of 33 survival-associated image features and all eigengenes for the 15 modules. The heat map of the correlation matrix is shown in Fig. 3.
Figure 3.
Pairwise correlation heat map between 33 survival-associated image features and all 15 eigengenes, using Spearman rank correlation.
As can be seen from the heat map, eigengenes 2, 3, 9, and 11 significantly correlated with many image features (statistically significant after Bonferroni correction). The gene module 2 was enriched with extracellular matrix genes, which explained why it positively correlated with image features such as ratio_bin8, ratio_bin9, ratio_std, major_bin9, and major_std that describe the percentage of stromal cells. Gene module 3 was enriched with acid metabolic process and transmembrane transporter activity. Genes in this module play a central role in renal functions such as organic anion transport (36). Patients with low expression of this eigengene were related to poor prognostic outcome (log-rank test p value = 7.46e-6, Fig. 2E), implying impaired renal function. This eigengene also negatively correlated with images features representing the amount of stromal cells such as ratio_bin9, major_bin9, ratio_std, and major_std. Gene module 9 was highly enriched with cell cycle and mitosis genes. In fact, genes in this module are frequently observed to co-express in multiple types of cancers (37). High expression of this eigengene indicates that the tumor is more aggressive, and it was negatively related to patient prognosis (log-rank test p value = 1.19e-4). Cells become bigger when they come into mitotic phase, which was in line with our observation that the gene module 9 was significantly and positively correlated with image features such as area_bin5, area_bin6, and area_std. The top molecular functions of gene module 11 by were frizzled binding and G-protein coupled receptor binding. G-protein-coupled receptors (GPCRs) represent the largest family of cell-surface molecules involved in signal transduction. Experimental and clinical data indicate that GPCRs have a crucial role in cancer progression and metastasis (38). Patients with high expression of gene module 11 had significantly worse outcome than other patients (log-rank p value = 1.33e-3). Similar to gene module 2, module 11 also significantly correlated with many image features that describe stroma cells, such as ratio_bin8, ratio_bin9, and ratio_std. Survival analysis results and enrichment analysis results for all survival-associated eigengenes are summarized in Table 2 and Table S2, respectively.
Lasso-Cox risk index is independent of known prognostic factors
Using univariate and multivariate Cox proportional hazards analysis, we performed a comprehensive comparison between the lasso-Cox risk index and other known prognostic biomarkers, including two clinical variables, grade (G1+G2 vs. G3+G4), stage (I+ II vs. III+ IV), six gene expression signatures (39,40), CSNK2A1, SPP1, DEFB1, CD31, EDNRB, TSPAN7, and five somatic mutation genes (26,41–46), VHL, PBRM1, BAP1, SETD2, TP53. Patient subtyping for gene expression signatures was carried out by using the median as cut-off point. In terms of genes with somatic mutation, patients were classified as mutant or wild-type. Of these factors, only grade, stage, lasso-Cox risk index, DEFB1, EDNRB, and TSPAN7 were associated with survival by univariate Cox proportional hazards analysis (Table 3). DEFB1 encodes beta-defensin, which belongs to a family of antimicrobial peptides produced by white blood cells and epithelial cells. Rabjerg (40) suggested that DEFB1 might be a tumor suppressor gene, but our results revealed that high expression of this gene predicted a worse prognosis with very weak significance (p = 4.99e-2, hazard ratio = 1.41, and 95% confidence interval = [1.00, 1.98]). EDNRB is a member of the endothelin axis, and TSPAN7 is a member of the transmembrane 4 superfamily. Wuttig (39) showed that EDNRB and TSPAN7 might be suppressors of tumor progression and metastatic tumor growth, which is in agreement with our results that high expression of these two genes predicted a better prognosis. Subsequently, multivariate Cox proportional hazards analysis demonstrated that lasso-Cox risk index was an independent prognostic factor (p = 2.31e-4, hazard ratio = 2.26, 95% confidence interval 1.46–3.49), as well as stage and TP53 (Table 3).
Table 3.
Univariate and multivariate Cox proportional hazards analysis of the prognostic values of lasso-Cox risk index and other prognostic factors. HR, hazard ratio. CI, confidence interval.
| Univariate Cox regression | Multivariate Cox regression | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P value | HR (95% CI) | P value |
| Lasso-Cox | 3.06 (2.10–4.45) | 5.02e-9 | 2.26 (1.46–3.49) | 2.31e-4 |
| Clinical | ||||
| Grade | 2.38 (1.63–3.5) | 8.45e-6 | 1.46 (0.95–2.23) | 8.22e-2 |
| Stage | 3.68 (2.57–5.27) | 1.12e-12 | 3.00 (2.00–4.49) | 9.23e-8 |
| Gene expression | ||||
| CSNK2A1 | 0.90 (0.64–1.26) | 5.34e-1 | 1.07 (0.74–1.56) | 7.11e-1 |
| SPP1 | 1.15 (0.82–1.61) | 4.14e-1 | 1.10 (0.75–1.63) | 6.20e-1 |
| DEFB1 | 1.41 (1.00–1.98) | 4.99e-2 | 1.36 (0.95–1.95) | 9.71e-2 |
| PECAM1 | 0.77 (0.55–1.09) | 1.40e-1 | 1.04 (0.69–1.58) | 8.45e-1 |
| EDNRB | 0.50 (0.35–0.71) | 9.10e-5 | 0.96 (0.59–1.57) | 8.77e-1 |
| TSPAN7 | 0.54 (0.38–0.76) | 5.12e-4 | 1.03 (0.64–1.67) | 9.07e-1 |
| Somatic mutation | ||||
| VHL | 0.99 (0.70–1.38) | 9.33e-1 | 1.23 (0.86–1.75) | 2.57e-1 |
| PBRM1 | 0.85 (0.58–1.24) | 3.94e-1 | 1.03 (0.69–1.54) | 8.85e-1 |
| BAP1 | 1.49 (0.78–2.85) | 2.22e-1 | 1.49 (0.74–3.00) | 2.60e-1 |
| SETD2 | 1.29 (0.77–2.14) | 3.29e-1 | 1.03 (0.62–1.74) | 9.00e-1 |
| TP53 | 2.26 (1.00–5.15) | 5.13e-2 | 2.86 (1.19–6.86) | 1.85e-2 |
Predicting survival in early-stage ccRCC
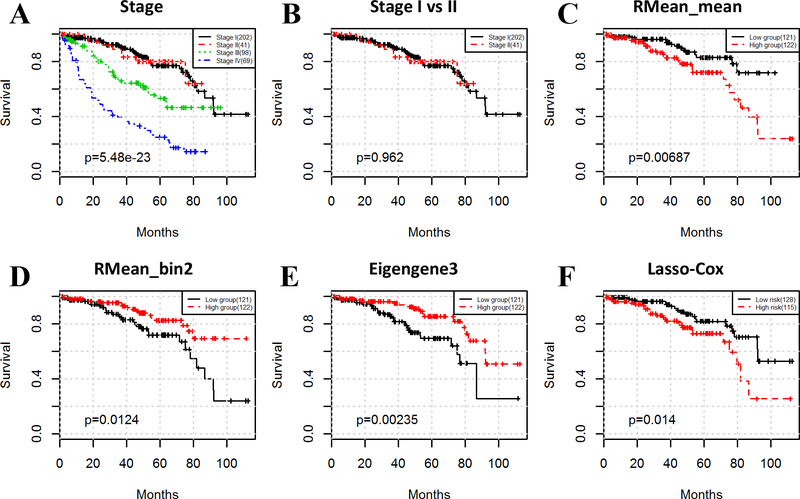
As shown in Table 3, tumor stage is the most effective prognostic factor, but its capability of stratifying early-stage (i.e. stage I and II) ccRCC patients is very limited (Fig. 4A and B). The Kaplan-Mejer curves of stages I and II are intertwined (log-rank test p value = 0.962), which may be attributed to the less significant morphological differences between stages I and II tumors and/or large subtyping variations among pathologists.
Figure 4.
Image features and eigengenes predict the survival outcomes in early-stage (stage I and II) ccRCC patients. Stage is strongly associated with survival (A) but cannot stratify early-stage patients (B). However, image features (C, D), eigengenes (E), and lasso-Cox model (F) are significantly related to survival in early-stage patients.
However, the image features and eigengenes can successfully stratify early-stage patients with distinct survival outcomes. Log-rank testing of each of the 165 variables (150 image features and 15 eigengenes) revealed that 13 image features and 2 eigengenes were associated with survival (Table S3). Survival curves of 3 variables are shown in Fig. 4C–E. In addition, we also trained a lasso-Cox prognostic model using the above 15 variables related to survival. Fig. 4F shows the survival curves stratified by the lasso-Cox risk index (log-rank test p value = 0.014). Compared to individual variables, integrating image features and eigengenes did not improve the accuracy of prognostic prediction for early-stage patients while there indeed was a very significant improvement when using all patients. This is because the death rate in early-stage patients is much lower than that in all patients (18.5% vs 32.9%), and high death rate is key to ensuring prediction accuracy of lasso-Cox model. If all patients were used in the lasso-Cox model to predict early-stage patient prognosis, the performance was improved (log-rank test p value = 8.65e-3). The two eigengenes associated with the prognosis of the early-stage patients corresponded to co-expressed gene modules 3 and 13. The gene module 3 was highly enriched with genes related to kidney functions such as organic acid metabolic process (p = 5.702 × 10−18), ammonium ion metabolic process (p = 6.612 × 10−9), and anion transport (p = 5.994 × 10−8). This observation suggests that the physiological functions for kidney can be potential prognostic markers for early-stage patients. Besides gene module 3, gene module 13 contains 10 genes. Interestingly, all the 10 genes locate on the same chromosome, straddling chromosome 14q11 to 14q32, implying potential copy number variation on 14q may be related to the prognosis of kidney patients.
Sensitivity analysis
Since our analysis relies on parameters for the machine learning algorithms and choices of cross validation (CV) methods, we also examined the choice of various parameters, especially the choice of number of clusters K for the cellular features. Fig. S3 shows the log-rank test p value as a function of the number of clusters in K-means algorithm. Fig. S3 suggests that lasso-Cox model can achieve very low p values when K ranges from 8 to 14. We also compared leave-one-out CV with k-fold CV. Fig. S4 shows that as k increases, the p value tends to continuously decline. This is because in k-fold CV a large k means we have more training samples, and thus the learned model is likely to perform better especially when the whole data set is not very large. As a result, we chose K=10 in the K-means algorithm, and we used leave-one-out CV in our experiments.
Discussion
To our knowledge, this is the first study to predict the survival outcomes of ccRCC patients using a combination of quantitative morphological features extracted from whole-slide tissue images and gene expression signatures. In this study, we developed an automatic image analysis pipeline to extract hundreds of cellular morphological features, and found cellular morphology was highly linked to co-expressed gene signatures. For example, image features characterizing the amount of stromal cells positively correlated with extracellular matrix genes. Standard deviation of nuclear area correlated with genes that regulate cell cycle and mitosis. In addition, a powerful prognostic model was built to predict the survival outcomes of ccRCC patients using these two types of data. The performance of the integrated prognostic model significantly outperformed that of individual image or genomic features, which indicates that image data are complementary to genomic data for predicting patient prognosis. Using multivariate Cox regression, we verified that the risk index generated by our model was a prognostic factor independent of tumor grade, stage, and other known molecular markers. For early-stage patients, besides the imaging data, the genomic data suggests that the kidney functions and status of 14q may be predictors of the survival time for these patients.
Recent studies have underscored the important contribution of stromal gene expression and morphologic phenotypes to cancer growth and progression for breast cancer (2,3,47). The implication of tumor stroma to prognosis could be different for different cancer types. For instance, high percentage of tumor stroma is associated with poor prognosis in triple-negative disease but good prognosis in estrogen receptor-positive disease (48,49). Here, we found in this study that for ccRCC both image features and gene expression signatures revealed that a large percentage of tumor stroma predicted poor prognosis.
The high resolution of whole-slide tissue images poses a great computational challenge to researchers. For this reason, many previous studies only focused on selected views in tissue microarrays or a few representative image tiles in whole-slide images (2,5). Since tumor is a highly heterogeneous disease, image features extracted from a much larger area of the tumor would be more likely to ensure the robustness of the derived prognostic model. Our prognostic model was established on the fully automated quantitative image features that were extracted from whole-slide histopathological images, which could avoid biases or discrepancies arising from only using a small portion of the tumor.
Our study is limited to only one large ccRCC patient cohort as it is difficult to find other cohorts that have matched histopathological images, gene expression profiles, and survival information. However, the performance of our prognostic model was strictly assessed by cross validation. The model selection was performed by 10-fold cross validation on the training set, and then the selected model was applied to the held-out test samples to predict risk indices. Another technical contribution of this work lies in the fact that we used only the cryohistological images from TCGA. Usually for each TCGA solid tumor sample, two histopathological images are generated – the H&E stained diagnostic image and the cryohistological image from a slice of tissue immediately adjacent to the tissue used for generating the omics data. Thus, due to spatial proximity, the cryohistological image is a more accurate reflection of the molecular profiles of the tissue for the omics data. However, due to processing artifact, many of these images appear damaged and cannot be processed for tissue features using previous methods, preventing accurate characterization on the tumor morphology. Here we showed that the cell nucleic features suggestive of stromal cells indeed correlated well with the gene expression profiles of extracellular matrix and stromal genes, suggesting in such images, albeit for the artifacts affecting texture analysis, the cell nucleic features can still be used.
Finally, although our study focused on predicting survival for ccRCC patients, we believe that the workflow of integrative analysis of histopathological images with genomic data could be easily applied to other cancer types or to predict response of specific treatments, which would allow for better patient management and cancer care.
Supplementary Material
Acknowledgments
Financial support: This work was partially supported by an NCI ITCR grant 5 U01CA188547 (K. Huang), Leidos grant 15×014 (K. Huang), and the Science and Technology Project of Guangdong Province, China (No. 2015B010131011) (Q. Feng).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Hipp J, Flotte T, Monaco J, Cheng J, Madabhushi A, Yagi Y, et al. Computer aided diagnostic tools aim to empower rather than replace pathologists: Lessons learned from computational chess. J Pathol Inform. 2011;2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, et al. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Sci Transl Med. 2011;3:108ra113–108ra113. [DOI] [PubMed] [Google Scholar]
- 3.Yuan Y, Failmezger H, Rueda O, Ali H, Graf S, Chin S, et al. Quantitative Image Analysis of Cellular Heterogeneity in Breast Tumors Complements Genomic Profiling. Sci Transl Med. 2012;4:157ra143–157ra143. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Pecot T, Zynger DL, Machiraju R, Shapiro CL, Huang K. Identifying survival associated morphological features of triple negative breast cancer using multiple datasets. J Am Med Inform Assoc. 2013;20:680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu K-H, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Xing F, Su H, Yang L, Zhang S. High-throughput histopathological image analysis via robust cell segmentation and hashing Med Image Anal. Elsevier B.V; 2015;26:306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong J, Cooper LAD, Wang F, Gao J, Teodoro G, Scarpace L, et al. Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PLoS One. 2013;8:e81049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gultekin T, Koyuncu CF, Sokmensuer C, Gunduz-Demir C. Two-tier tissue decomposition for histopathological image representation and classification. IEEE Trans Med Imaging. 2015;34:275–83. [DOI] [PubMed] [Google Scholar]
- 9.Gulati S, Martinez P, Joshi T, Birkbak NJ, Santos CR, Rowan AJ, et al. Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers. Eur Urol. European Association of Urology; 2014;66:936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maroto P, Rini B. Molecular biomarkers in advanced renal cell carcinoma. Clin Cancer Res. 2014;20:2060–71. [DOI] [PubMed] [Google Scholar]
- 11.Martins FC, Santiago I De, Trinh A, Xian J, Guo A, Sayal K, et al. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol. 2014;15:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colen R, Foster I, Gatenby R, Giger ME, Gillies R, Gutman D, et al. NCI workshop report: Clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Transl Oncol. Elsevier B.V; 2014;7:556–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper LAD, Kong J, Gutman DA, Dunn WD, Nalisnik M, Brat DJ. Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab Invest. 2015;95:366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutman DA, Cobb J, Somanna D, Park Y, Wang F, Kurc T, et al. Cancer Digital Slide Archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J Am Med Inform Assoc. 2013;20:1091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosaliganti K, Pan T, Ridgway R, Sharp R, Cooper L, Gulacy A, et al. An imaging workflow for characterizing phenotypical change in large histological mouse model datasets. J Biomed Inform. 2008;41:863–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding H, Wang C, Huang K, Machiraju R. iGPSe: a visual analytic system for integrative genomic based cancer patient stratification. BMC Bioinformatics. 2014;15:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding H, Wang C, Huang K, Machiraju R. GRAPHIE: graph based histology image explorer. BMC Bioinformatics. 2015;16 Suppl 1:S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.American Cancer Society. Cancer Facts & Figures 2016. Cancer Facts Fig 2016. 2016;1–9. [Google Scholar]
- 19.Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, Delahunt B, et al. The Heidelberg classification of renal cell tumours. J Pathol. 1997;183:131–3. [DOI] [PubMed] [Google Scholar]
- 20.Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F, Hora M, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur. Urol. 2015. page 913–24. [DOI] [PubMed] [Google Scholar]
- 21.Chen J-M, Qu A-P, Wang L-W, Yuan J-P, Yang F, Xiang Q-M, et al. New breast cancer prognostic factors identified by computer-aided image analysis of HE stained histopathology images. Sci Rep. Nature Publishing Group; 2015;5:10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HL, Seligson D, Liu X, Janzen N, Bui MHT, Yu H, et al. Using protein expressions to predict survival in clear cell renal carcinoma. Clin Cancer Res. 2004;10:5464–71. [DOI] [PubMed] [Google Scholar]
- 23.Zisman a, Pantuck a J, Dorey F, Said JW, Shvarts O, Quintana D, et al. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol. 2001;19:1649–57. [DOI] [PubMed] [Google Scholar]
- 24.Tang PA, Vickers MM, Heng DYC. Clinical and molecular prognostic factors in renal cell carcinoma: What we know so far. Hematol. Oncol. Clin. North Am. 2011. page 871–91. [DOI] [PubMed] [Google Scholar]
- 25.Motzer RJ, Mazumdar M, Bacik J, Berg W, Amsterdam A, Ferrara J. Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. J Clin Oncol. 1999;17:2530–40. [DOI] [PubMed] [Google Scholar]
- 26.Creighton C, Morgan M, Gunaratne P, Wheeler D, Gibbs R, Gordon Robertson A, et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmady Phoulady H, Goldgof DB, Hall LO, Mouton PR. Nucleus segmentation in histology images with hierarchical multilevel thresholding. Proc SPIE 9791, Med Imaging 2016 Digit Pathol. 2016. page 979111. [Google Scholar]
- 28.Yang Y, Li F, Gao L, Wang Z, Thrall MJ, Shen SS, et al. Differential diagnosis of breast cancer using quantitative, label-free and molecular vibrational imaging. Biomed Opt Express. 2011;2:2160–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. BioMed Central; 2007;1:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. [DOI] [PubMed] [Google Scholar]
- 31.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Huang K. Normalized lmQCM : an Algorithm for Detecting Weak Quasi-clique Modules in Weighted Graph with Application in Functional Gene Cluster Discovery in Cancer. Cancer Inform. 2016;1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng A, Jordan M, Weiss Y. On spectral clustering: Analysis and an algorithm. Adv Neural Inf Process Syst. 2002; [Google Scholar]
- 34.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekine T, Miyazaki H, Endou H. Molecular physiology of renal organic anion transporters. Am J Physiol - Ren Physiol. 2006;290:F251–61. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Lu K, Xiang Y, Islam M, Kotian S, Kais Z, et al. Weighted Frequent Gene Co-expression Network Mining to Identify Genes Involved in Genome Stability. PLoS Comput Biol. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutkind JS, Dorsam RT. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. [DOI] [PubMed] [Google Scholar]
- 39.Wuttig D, Zastrow S, Füssel S, Toma MI, Meinhardt M, Kalman K, et al. CD31, EDNRB and TSPAN7 are promising prognostic markers in clear-cell renal cell carcinoma revealed by genome-wide expression analyses of primary tumors and metastases. Int J Cancer. 2012;131. [DOI] [PubMed] [Google Scholar]
- 40.Rabjerg M, Bjerregaard H, Halekoh U, Jensen BL, Walter S, Marcussen N. Molecular characterization of clear cell renal cell carcinoma identifies CSNK2A1, SPP1 and DEFB1 as promising novel prognostic markers. Apmis. 2016;124:372–83. [DOI] [PubMed] [Google Scholar]
- 41.Kim JH, Jung CW, Cho YH, Lee J, Lee SEH, Kim HOY, et al. Somatic VHL alteration and its impact on prognosis in patients with clear cell renal cell carcinoma. Oncol Rep. 2005;13:859–64. [PubMed] [Google Scholar]
- 42.Yao M, Yoshida M, Kishida T, Nakaigawa N, Baba M, Kobayashi K, et al. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. J Natl Cancer Inst. 2002;94:1569–75. [DOI] [PubMed] [Google Scholar]
- 43.Kapur P, Peña-Llopis S, Christie A, Zhrebker L, Pavía-Jiménez A, Rathmell WK, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: A retrospective analysis with independent validation. Lancet Oncol. 2013;14:159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen YB, Gonen M, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19:3259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–7. [DOI] [PubMed] [Google Scholar]
- 46.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C. Mutational landscape and significance across 12 major cancer types. Nature. 2013;503:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh E-Y, Christensen SM, Ghanta S, Jeong JC, Bucur O, Glass B, et al. Extensive rewiring of epithelial-stromal co-expression networks in breast cancer. Genome Biol. Genome Biology; 2015;16:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dekker TJA, Van De Velde CJH, Van Pelt GW, Kroep JR, Julien JP, Smit VTHBM, et al. Prognostic significance of the tumor-stroma ratio: Validation study in node-negative premenopausal breast cancer patients from the EORTC perioperative chemotherapy (POP) trial (10854). Breast Cancer Res Treat. 2013;139:371–9. [DOI] [PubMed] [Google Scholar]
- 49.Downey CL, Simpkins SA, White J, Holliday DL, Jones JL, Jordan LB, et al. The prognostic significance of tumour-stroma ratio in oestrogen receptor-positive breast cancer Br J Cancer. Nature Publishing Group; 2014;110:1744–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.