Abstract
NAD(P)H:Quinone oxidoreductase (NQO1) functions as an important part of cellular antioxidant defense by detoxifying quinones, thus preventing the formation of reactive oxygen species. The aims of our study were to determine if NQO1 is elevated in pancreatic cancer specimens and pancreatic cancer cell lines and if so, would compounds previously demonstrated to redox cycle with NQO1 be effective in killing pancreatic cancer cells. Immunohistochemistry of resected pancreatic specimens demonstrated an increased immunoreactivity for NQO1 in pancreatic cancer and pancreatic intraepithelial neoplasia (PanIN) specimens versus normal human pancreas. Immunocytochemistry and Western immunoblots demonstrated inceased immunoreactivity in pancreatic cancer cells when compared to a near normal immortalized human pancreatic ductal epithelial cell line and a colonic epithelial cell line. Streptonigrin, a compound known to cause redox cycling in the presence of NQO1, decreased clonogenic survival and decreased anchorage-independent growth in soft agar. Streptonigrin had little effect on cell lines with absent or reduced levels of NQO1. The effects of streptonigrin were reversed in pancreatic cancer cells pretreated with dicumarol, a known inhibitor of NQO1. NQO1 may be a therapeutic target in pancreatic cancer where survival is measured in months.
Keywords: NAD(P)H, quinone oxidoreductase, pancreatic cancer, streptonigrin, antioxidants
INTRODUCTION
NAD(P)H:quinone oxidoreductase (NQO1) is primarily a cytosolic enzyme that uses NADH or NADPH to directly reduce quinones to hydroquinones [1]. This two-electron reductase bypasses the highly reactive semi-quinone radical intermediate and prevents redox cycling [1]. Thus, NQO1 prevents the formation of reactive oxygen species and plays an important role in the cellular antioxidant system [2–5]. NQO1 has been demonstrated to be present in varying amounts in most cell types with the exception of the liver [5–7]. NQO1 may be increased during malignant transformation in some tumor types with elevated concentrations found within solid tumors of the breast, ovary, lung, colon, thyroid, and adrenal gland [5,8].
In pancreatic cancer, NQO1 may also be increased. Logsdon et al demonstrate that NQO1 is overexpressed in pancreatic cancer via microarrays that profile gene expression in pancreatic adenocarcinoma, pancreatitis, pancreatic cancer cell lines, and normal pancreas. Pancreatic cancer cells and tumors exhibited a 12-fold increase of NQO1 expression compared to normal pancreas [8]. Additionally, studies from our laboratory demonstrated increased NQO1 messenger RNA, protein, and activity in pancreatic cancer tumor cell lines when compared to normal pancreas [9].
The aim of our present study was twofold. First we wanted to determine by immunohistochemical and immunocytochemical techniques if NQO1 was increased in pancreatic cancer specimens and pancreatic cancer cell lines when compared to normal human pancreas or a nontumorigenic human pancreatic ductal epithelial cell line derived from normal pancreas. Our studies demonstrate a profound increase in immunoreactivity for NQO1 in pancreatic cancer specimens and PanIN specimens when compared to normal human pancreas, and a significant increase in NQO1 in pancreatic cancer cell lines when compared to a nontumorigenic pancreatic ductal epithelial cell line. Thus our second aim was to determine if streptonigrin, a compound known to be efficacious and cause redox cycling in the presence of NQO1 [10], would be effective against pancreatic cancer in vitro. Streptonigrin decreased cell viability, clonogenic survival, and anchorage-independent growth in soft agar in pancreatic cancer cell lines but had little effect on tumorigenic and nontumorigenic cell lines with absent or reduced levels of NQO1.
MATERIALS AND METHODS
Cell Culture
MIA PaCa-2 (undifferentiated), BxPC-3 (poorly differentiated), and AsPC-1 (poorly to moderately differentiated) cells, human primary pancreatic adenocarcinoma cells, and CaCo-2, human colon cancer cells, were obtained from American Type Culture Collection (Manassas, VA). MIA PaCa-2 was maintained in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum and 2.5% horse serum. BxPC-3 was maintained in RPMI 1640 with 10% fetal bovine serum. AsPC-1 was maintained in RPMI 1640 with 20% heat inactivated bovine serum and 1% sodium pyruvate. All media were obtained from Gibco (Grand Island, NY) and all cell lines were maintained at 37°C. CaCo-2 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum and 2.5% horse serum. H6c7 cells, a clonal human pancreatic ductal epithelial cell line with near normal genotype and phenotype of pancreatic duct epithelial cells, were maintained in keratinocyte serum free media and supplemented with epidermal growth factor and bovine pituitary extract [11,12].
Immunohistochemistry
A total of nine human pancreatic specimens were utilized for the present study. Of the nine specimens, there were four normal pancreas specimens and five specimens from resections for pancreatic adenocarcinoma. The normal pancreas specimens were harvested from incidental pancreatectomy after either a gastric resection or splenectomy. All specimens were resected at the University of Iowa Hospitals and Clinics between 1996 and 2001. The protocol to use the tissue blocks from human pancreatic specimens was approved by the University of Iowa Institutional Review Board for Human Subjects on February 12, 2001. Consecutive 4-μm-thick sections were used for immunohistochemical analysis. Representative blocks of tissue were deparaffinized and rehydrated at room temperature. Slides were placed in 3% H2O2 for 1 h to suppress endogenous peroxide. These were then rinsed in Tris-buffered saline (TBS) for 5 min and then heated to 90° C in Antigen Masking Solution. Slides were placed in TBS at 37°C for 30 s and then placed back in the diluted Antigen Masking Solution for 30 min as the solution cooled to room temperature, and were washed in water and TBS and then incubated in normal goat serum for 1 h. The slides were incubated overnight at 4°C in the NQO1 primary antibody. The NQO1 antibody was obtained from Dr. David Ross at the University of Colorado Health Sciences Center, Denver, CO. The primary antibody was diluted 1:200 in Enzyme Label Diluent (Scytek Laboratories, Logan, UT). Negative control slides were incubated in a 1:1000 solution of preimmune serum in Enzyme Label Diluent in the absence of the primary antibody. The following day, slides were rinsed with TBS and incubated in antimouse IgG. These were washed in TBS, incubated with ABC complex (Vector laboratories, Burlingame, CA), rinsed again in TBS, and then stained with diaminobenzidine (DAB). After that the slides were rinsed in distilled water and then counterstained with Harris hematoxylin, soaked in 3% ammonia water for 30 s, rinsed with water, and dehydrated in ETOH and xylene. Slides were finally viewed at X70 on an Olympus BX-51 light microscope, and samples were photographed using SPOT software program.
Tissue Microarray (TMA)
There have been an increasing number of genetic abnormalities in the preinvasive PanIN lesions being reported [13,14]. Thus, to further characterize NQO1 in the early stages of human pancreatic carcinogenesis we examined the protein expression of NQO1 on pancreatic cancer and PanIN TMA that we previously described [15]. The PanIN lesions were graded according to established criteria [13]. Four-micron sections of the TMA blocks were stained immunohistochemically as described [15].
Immunocytochemistry
The cells were grown to confluence in T-75 flasks and were allowed to fix for 10 min in 2% formaldehyde and washed in phosphate buffered saline (PBS). The fixed cells were permeabilized for 10 min in a 0.3% solution of Triton X-100 in PBS, washed twice in PBS, and blocked for 20 min in 1.0% normal goat serum in PBS to eliminate binding of nonspecific proteins. The cells were then incubated overnight at 4°C in NQO1 antibody diluted 1:1 with PBS containing 1% fetal bovine serum. The negative control contained PBS and fetal bovine serum only. Unbound primary antibody was removed with three washes of PBS. A 1:200 dilution of a secondary goat antimouse antibody conjugated to Alexa Fluor 488 (Molecular Probes; Eugene, OR) was applied and allowed to incubate for 30 min. After washing in PBS, the cells were incubated in propidium iodide, a nuclear stain, (1 μg /mL) for 5 min. Finally, cells were rinsed in PBS and mounted in Vectashield on agar-coated slides. Because the cells were in suspension, centrifugation was necessary at 5000g for 2 min between each step. Slides were viewed with a 60× objective lens on an Olympus BX-51 microscope equipped with a mercury lamp, and filter sets transmitting wavelengths of 568 and 488 nm were used to excite propidium iodide and Alexa Fluor 488, respectively. Digitized images were acquired using a Spot CCD camera (Diagnostic Instruments, Inc.).
Digital Imaging Methodology
To determine the content of NQO1 in the immunohistochemically and immunocytochemically stained sections, a semiquantitative digital imaging method was used [16]. Image analysis allows for semiquantitative numerical comparisons of staining intensity based on the gray-level value of the digitally acquired histologic sections because of the linear relationship that exists between staining intensity and antigen concentration. For the digitized images, gray-level values ranged from 0 (white) to 255 (black). Stained tissue sections were viewed under high power by means of a Zeiss light microscope (Zeiss Corporation, Frankfurt, Germany) that was interfaced with a Kodak Image Megagrabber (Eastman Kodak, Rochester, NY) that digitized the light microscopic image. Both units were controlled by a personal computer. Videk Megaplus (Perceptics Corporation, Knoxville, TN) image acquisition software was used to acquire each microscopic field. The video output was amplified and digitized to a 1024- by 1024 matrix. A total of five high-power fields representative of each tissue section were digitized for analysis. NIH Image (National Institutes of Health, Bethesda, MD) was used to perform the semiquantitative analysis on the digitized images. For each high power field, the cytoplasmic regions of five cells were identified and digitized under mouse control. The cytoplasmic area alone was digitized and analyzed;the nuclei were not included in the analysis because their darkness affects the measured staining intensity. Also, in specimens from pancreatic cancer, there was significant fibrosis and an increased inflammatory cell component to some of the histological sections. Fibrotic areas and inflammatory cells were likewise not included in the analysis because their darkness would falsely increase the measured staining intensity. Mean gray-level pixel values were then obtained for each of these cytoplasmic regions. Thus, a total of 25 cytoplasmic regions were acquired from each tissue section of normal pancreas and pancreatic cancer. Pancreatic cancer is generally considered to arise from the pancreatic ductal cells so we averaged the staining intensities of ductal cells alone in our analysis of normal pancreas.
Cell Homogenization and Protein Determination
Cells were washed three times in PBS (pH 7.0), scraped from the dishes using a rubber policeman and then collected in potassium phosphate buffer (pH 7.8). This was followed by sonic disruption on ice for 30 s in 10-s bursts using a VibraCell sonicator (Sonics and Materials, Inc., Danbury, CT) at 100% power. Protein concentration was determined using the Bio-Rad Bradford dye binding protein assay kit (Hercules, CA) according to the manufacturer’s instructions.
Western Blotting
Total protein extracts were prepared and protein concentrations determined. Protein (20 μg/lane) was separated by 12% SDS-polyacrylamide gel electrophoresis and electrotransferred to nitrocellulose membranes. After transfer, membranes were blocked with Tris-buffered saline containing 5% skim milk powder and 1% heat-inactivated fetal bovine serum for 2 h, and then incubated overnight with the anti-NQO1 monoclonal antibody (1:100) at 4°C. Blots were washed in Tris-buffered saline containing 0.05% Tween-20 and incubated for 90 min with 1:4000 dilution of goat antimouse horseradish peroxidase conjugated antibody in Tris-buffered saline containing 1% skim milk powder and 1% heat-inactivated fetal bovine serum. Bands were visualized using an enhanced chemiluminescence detection kit (Amersham Life Science, Buckinghamshire, UK) and exposed to X-ray film as described previously [17].
Cell Fractionation
Cells were bounced for 100 strokes in ice-cold digitonin buffer (0.4 mg digitonin/mL, 2.5 mM EDTA, 250 mM mannitol, 17 mM MOPS) and then spun in a microfuge (14000g for 15–30 min.). Cytosolic and mitochondrial fractions were analyzed by Western blot analysis.
Anchorage-Independent Growth in Soft Agar
MIA PaCa-2 pancreatic cancer cells (5 × 103) were suspended in 3 mL of complete medium containing streptonigrin (0, 50,100 nM) in a solution of 6% agar in double-distilled H2O so that the final concentration of the agar was 0.3%. This suspension was then plated over 3 mL of complete medium made with a 6% agar solution in double-distilled H2O so that the final concentration of the bottom agar was 0.5%. After 16 d, colonies of >0.1 mm in diameter were scored. The clonogenic fraction was determined using the following equation:
Clonogenic Assay
To determine clonogenic survival, MIA PaCa-2 and CaCo-2 cells (5 × 103) were treated with streptonigrin (0, 25, 50, 100 nM) with and without dicumarol (50 μM), a known inhibitor of NQO1. After 2 h, the streptonigrin was removed and replaced with media in 2% serum. One week later, colonies >50 cells were stained and counted. The percent survival was determined using the following equation:
Statistical Analysis
Statistical analysis of mean gray-level pixel values acquired from normal pancreas and pancreatic carcinoma and for the in vitro studies was performed by means of Systat (Systat, Inc., Evanston IL). A single factor ANOVA, followed by post-hoc Tukey test, was used to determine statistical differences between means. All means were calculated from three experiments, and error bars represent standard error of mean (SEM). All western blots were repeated at least twice. All data are expressed as means ± SEM.
RESULTS
NQO1 Immunohistochemistry
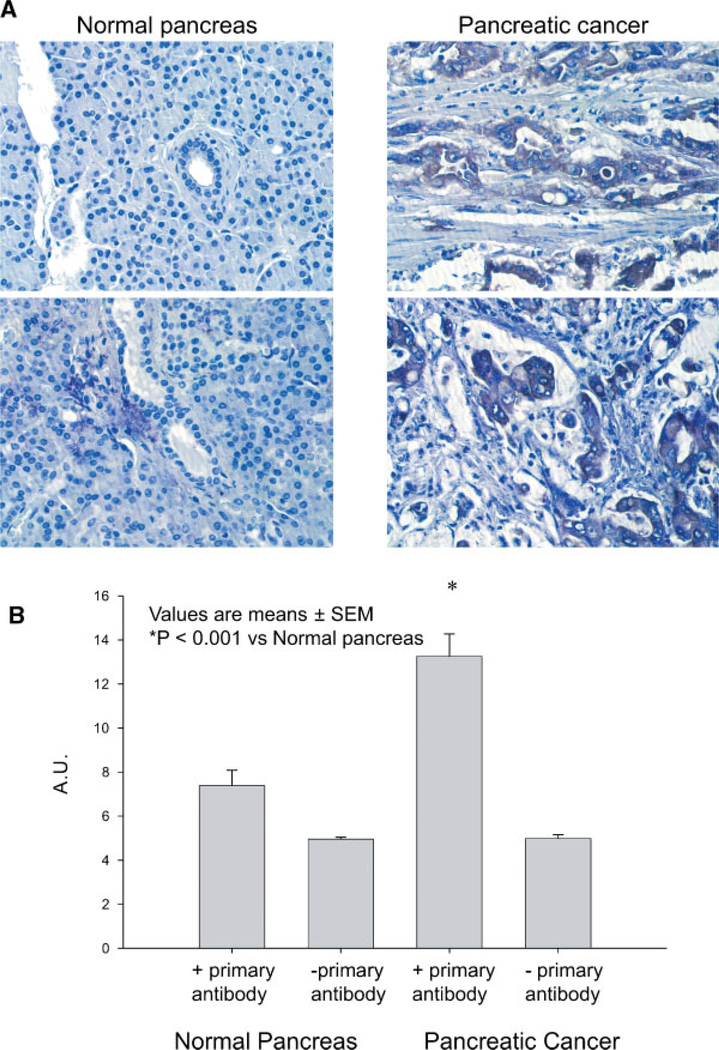
Increased staining intensity for NQO1 was observed in pancreatic cancer cells from pancreatic cancer resections when compared with normal pancreatic ductal epithelial cells. The staining within normal pancreatic ductal epithelial cells was nearly absent, whereas staining throughout the pancreatic cancer cells was more intense (Figure 1A). Image analysis was then performed, which allowed for semiquantitative numerical comparisons of staining intensity based on the gray-level value of the digitally acquired sections because of the linear relationship that exists between staining intensity and antigen concentration. For the NQO1-immunostained sections of pancreatic ductal epithelial cells from normal pancreatic specimens, semiquantitative digital image analysis demonstrated a mean cytoplasmic gray-level value ± SEM of 7.4 ± 0.7 arbitrary units (AU) (Figure 1B). When compared to normal pancreatic ductal epithelial cells, analysis of pancreatic cancer cells demonstrated a significant increase in the mean cytoplasmic gray-level value (13.3 ± 0.001 AU, P < 0.0001).
Figure 1.
(A) NQO1 expression was increased in pancreatic carcinoma cells when compared to normal human pancreas. Light, uniform staining was observed throughout the normal ductal epithelial cells in specimens of normal human pancreas. In contrast, darker, more irregular staining was observed in pancreatic cancer cells throughout the pancreatic cancer specimen. Additionally, specimens from pancreatic cancer demonstrated significant fibrosis and an increased inflammatory cell component to some of the histological sections. (B) NQO1 expression as evaluated by digital image analysis is increased in pancreatic cancer compared with normal pancreas. Image analysis allowed for semi-quantitative numerical comparisons of staining intensity based on the gray-level value of the digitally acquired sections. Analysis of pancreatic cancer cells demonstrated a significant increase in the mean cytoplasmic gray-level value when compared to pancreatic ductal epithelial cells. Values are expressed as AU, means ± SEM, P < 0.0001 versus normal pancreas.
Tissue Microarray
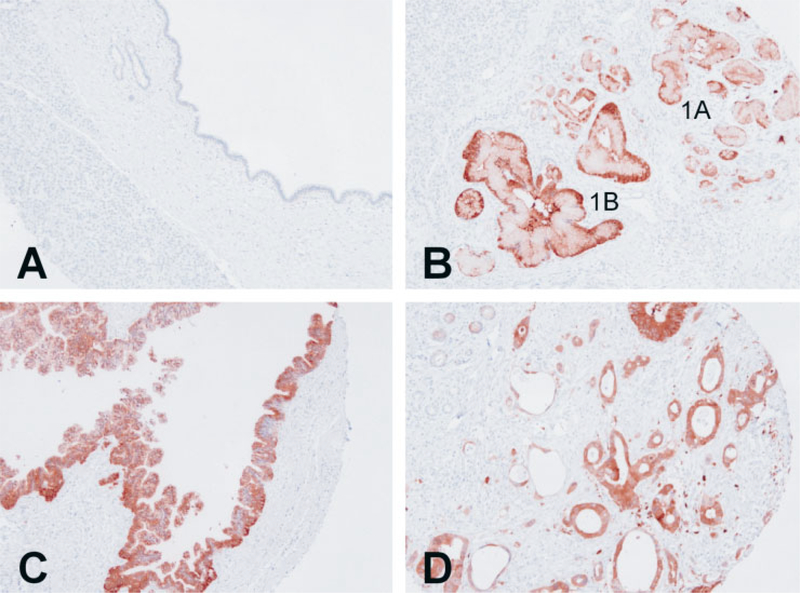
Tissue microarray analysis confirmed our immunohistochemistry results of the normal and pancreatic cancer, and added further information regarding the PanIN lesions. None of the 18 specimens of normal pancreatic duct showed positive staining for NQO1 (Figure 2A). However all of the PanIN and ductal cancer specimens demonstrated moderate to strong staining for NQO1. These included all of 26 PanIN-1A, 9 PanIN-1B, 15 PanIN-2, 14 PanIN-3, and 36 adenocarcinoma specimens.
Figure 2.
Tissue microarray and NQO1 immunohistochemistry of pancreatic ductal tissue. (A) Representative field of tissue microarray that contained normal pancreatic ductal epithelium demonstrating no staining for NQO1. (B) PanIN-1A and PanIN-1B lesions demonstrating strong positive staining for NQO1. (C) Tissue microarray core of PanIN-2 lesion demonstrating strong positive staining for NQO1. (D) Representative field of pancreatic adenocarcinoma also demonstrating strong positive staining for NQO1.
NQO1 Immunocytochemistry
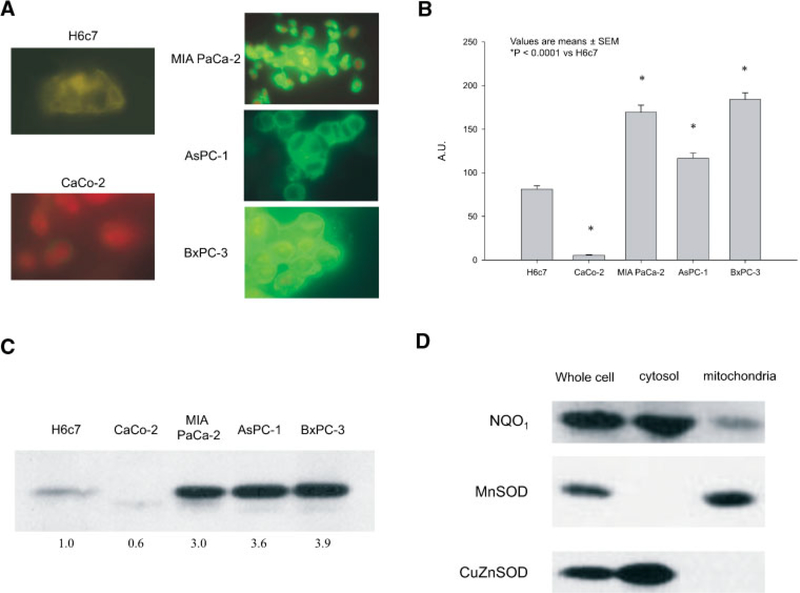
Formaldehyde-fixed pancreatic cancer cell lines, an immortalized normal pancreatic ductal epithelial cell line, and a colon cancer cell line were incubated with a goat antimouse antibody directed against NQO1 and fluorescence detected as described in Materials and Methods. Expression of NQO1 was detected primarily in the cytoplasm of all of the cell lines and was increased in all pancreatic cancer cell lines when compared to either the non-neoplastic pancreatic ductal epithelial cell line or the colon cancer cell line (Figure 3A). Among these, NQO1 staining in the CaCo-2 cell line had the lowest mean cytoplasmic gray-level of (5.5 ± 0.6 AU) (Figure 3B). For the H6c7 cells, semiquantitative digital image analysis demonstrated a mean cytoplasmic gray-level value of 81.2 ± 3.8 AU (Figure 3B). When compared to the H6c7 cell line, there was a significant increase in the mean cytoplasmic gray-level in the pancreatic cancer cell lines AsPC-1 (116.6 ± 5.9 AU), MIA PaCa-2 (169.3 ± 8.1 AU), and BxPC-3 (184.0 ± 7.8 AU) (P< 0.0001).
Figure 3.
(A) Immunocytochemical staining for NQO1 demonstrated increased immunofluorescent protein in the cytoplasm of pancreatic cancer cell lines. MIA PaCa-2, AsPC-1, and BxPC-3 exhibited increased cytoplasmic NQO1 immunofluorescence when compared to the immortalized pancreatic ductal epthielia cell line (H6c7). In addition, the human colon cancer cell line CaCo-2 exhibited little or nearly absent NQO1 immunofluorescence. (B) NQO1 expression as evaluated by digital image analysis was increased in pancreatic cancer cell lines compared with an immortalized pancreatic ductal epithelial cell line and a colon cancer cell line. Cells were fixed, stained, and the digitized images were analyzed. NQO1 staining in the CaCo-2 cell line was nearly absent, increased in the H6c7 cell line, and abundant in the pancreatic cancer cell lines. Values are expressed as AU, means ± SEM, P < 0.0001 versus H6c7. (C) NQO1 immunoreactive protein was increased in pancreatic carcinoma cell lines compared with immortalized pancreatic ductal epithelial cells, and a colon cancer cell line. Total protein extracts were prepared and protein concentrations determined. Protein (20 μg/lane) was separated by 12% SDS-PAGE and electrotransferred to nitrocellulose membranes. After transfer, membranes were blocked and then incubated overnight with anti-NQO1 monoclonal antibody. (D) The majority of NQO1 immunoreactive protein was in the cytoplasm. MIA PaCa-2 cell pellets were suspended in extraction buffer, incubated on ice, homogenized and centrifuged. The supernatants (cytosolic fraction) and the pellets (mitochondrial fraction) were stored at −80°C until ready for Western blotting. Copper zinc superoxide dismutase was used to determine the purity of the cytosolic fraction, while manganese superoxide dismutase was used to determine the purity of the mitochondrial fraction.
Western Analysis for NQO1
Western blot analysis was consistent with the immunocytochemistry in the cell lines. As seen in the immunocytochemistry, CaCo-2 human colon cancer cells had little NQO1 immunoreactive protein (Figure 3C), while the H6c7 cell line had increased immunoreactive protein when compared to the CaCo-2 cells. The pancreatic cancer cell lines MIA PaCa-2, AsPC-1, and BxPC-3 had elevated NQO1 immunoreactive protein compared to the H6c7 normal human pancreatic cells. Immunocytochemistry demonstrated that NQO1 was primarily a cytosolic enzyme. MIA PaCa-2 cells were fractionated to determine the location of NQO1. Western immunoblotting determined that in MIA PaCa-2 pancreatic cancer cells, NQO1 is primarily a cytosolic enzyme with about 20% of the enzyme located in the mitochondria (Figure 3D).
Growth in Soft Agar
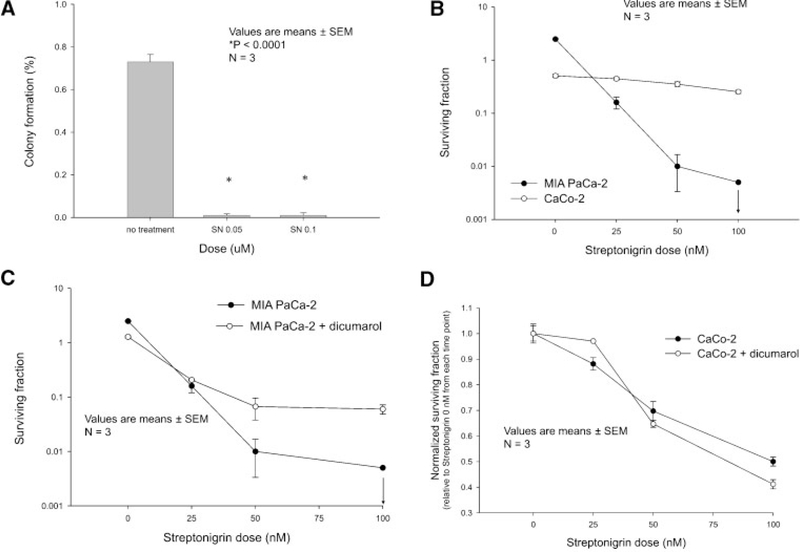
A soft agar assay was performed to determine anchorage-dependent growth. Whereas malignant MIA PaCa-2 cells form colonies in soft agar, normal cells do so in far smaller numbers. Treatment with streptonigrin resulted in significantly decreased MIA PaCa-2 colony formation (Figure 4A). Soft agar plating efficiency was 0.7 ± 0.03% in the nontreated cells and significantly reduced to 0.01 ± 0.01% in cells treated with 100 nM streptonigrin.
Figure 4.
Streptonigrin inhibited the in vitro pancreatic cancer malignant phenotype. (A) Growth in soft agar. MIA PaCa-2 cells treated with streptonigrin (50, 100 nM) demonstrated reductions in soft agar plating efficiency. Mean plating efficiency in soft agar of streptonigrin-treated MIA PaCa-2 cells are shown. Values are means ± SEM, n = 3, P < 0.0001 versus control cells. (B) Streptonigrin decreased pancreatic cancer clonogenic cell survival in a dose-dependent manner but had little effect on colon cancer clonogenic cell survival. A clonogenic assay was performed on MIA-PaCa-2 cells, which have elevated NQO1, and CaCo-2 cells, with little NQO1, treated with streptonigrin (0–100 nM) for 4 h. The arrow in the MIA PaCa-2 cells indicates that no colonies were formed when 5000 cells were plated in the dishes and treated with streptonigrin. Means ± SEM, N=3. (C) Pretreatment with dicumarol, inhibited streptonigrin-induced clonogenic cell death. A clonogenic assay was performed on MIA PaCa-2 cells treated with streptonigrin (0–100 nM) with and without dicumarol (50 μM). Means ± SEM, N = 3. (D) streptonigrin with and without dicumarol had little effect on clonogenic cell survival in CaCo-2 cells. A clonogenic assay was performed on CaCo-2 cells treated with streptonigrin (0–100 nM) with and without dicumarol (50 μM). Means ± SEM, N = 3.
Clonogenic Assay
To determine the clonogenic capacity of streptonigrin-treated cells, a plating efficiency assay was performed in MIA PaCa-2 and CaCo-2 cell lines. Cell lines were treated with streptonigrin (0, 25, 50, and 100 nM), and plating efficiency was reduced in the MIA PaCa-2 pancreatic cancer cell line with elevated NQO1 compared to the CaCo-2 colon cancer cell line with little or no NQO1. Plating efficiency was 0.25 ± 0.02 in the CaCo-2 cells treated with 100 nM of streptonigrin. In the MIA PaCa-2 cells treated with 100 nM of streptonigrin, there were no colonies present when the plates were stained and counted (Figure 4B). To determine if the effect of streptonigrin was due to the presence of NQO1, cells were pretreated with dicumarol (50 μM), a known inhibitor of NQO1. MIA PaCa-2 cells pretreated with dicumarol and 100 nM streptonigrin had a clonogenic survival of 0.06 ± 0.01, which was significantly increased survival compared with MIA PaCa-2 cells treated only with 100 nM streptonigrin where there was no clonogenic survival (Figure 4C). This effect was similar in AsPC-1 human pancreatic cancer cells treated with streptonigrin with and without dicumarol. AsPC-1 cells treated with dicumarol 50 μM and streptonigrin 50 nM had a clonogenic survival of 0.52 ± 0.02, which was significantly reduced to 0.28 ± 0.02 when cells were treated with 50 nM streptonigrin alone (P < 0.001). The same effect was seen at a higher doses of streptonigrin where plating efficiency was 0.63 ± 0.02 in the cells that received dicumarol 50 μM + streptonigrin 100 nM, while streptonigrin 100 nM alone decreased plating efficiency to 0.35 ± 0.01 (P < 0.001). In contrast to the human pancreatic cancer cell lines that have high expression of NQO1, CaCo-2 human colon cancer cells with little NQO1 treated with dicumarol and 100 nM streptonigrin demonstrated no difference in colony formation compared with CaCo-2 cells treated only with 100 nM streptonigrin (Figure 4D).
DISCUSSION
Our present study correlates well with recent studies demonstrating that NQO1 is overexpressed in pancreatic cancer [8,9]. Our immunohistochemical and immunocytochemistry studies demonstrated that pancreatic cancer specimens and cell lines have increased NQO1 expression compared with normal pancreas while Logsdon et al., using microarrays to profile gene expression in pancreatic adenocarcinoma, pancreatic cancer cell lines and normal pancreas, demonstrated a 12-fold increase of NQO1 expression compared to normal pancreas [8]. These data suggest that NQO1 may be useful as a specific target for therapeutic purposes in pancreatic cancer.
Our study also compliments the recent study from Winski et al. that demonstrated the importance of NQO1 in the activation of streptonigrin with BE and BE-NQ7 human colon adenocarcinoma cells [18]. Streptonigrin, an antibiotic derived from Streptomyces flocculus cultures, contains an aminoquinone moiety and has demonstrated antitumor activity in lymphomas, melanomas, and breast, lung, head, and neck cancer [19–23]. Streptonigrin exerts its anti-tumor activity through many cellular mechanisms including inhibition of DNA and RNA synthesis, impairment of cell respiration, and DNA damage [10,24–28]. To produce such effects, streptonigrin must first be activated via a one- or two-electron reductase such as NQO1. Such a reaction produces semiquinones or hydroquinones, which combine with oxygen to form reactive oxygen species [10,18]. Semiquinones are excellent free radical generators, initiating a redox cycle causing formation [29]. dismutates to hydrogen peroxide (H2O2) and molecular oxygen, and H2O2 is further reduced to the hydroxyl radical and OH· via the Fenton reaction [30]. All of these highly reactive free radicals may directly react with DNA or other cellular macromolecules, such as lipids and proteins, causing damage to the cell. Based on these studies, we expected that dicumarol, an inhibitor of NQO1, would reverse the streptonigrin-induced cytotoxicity in NQO1+ cancer cells. In our present study, CaCo-2 human colon cancer cells without NQO1 and MIA PaCa-2 human pancreatic cancer cells containing NQO1 were treated with dicumarol and streptonigrin. The pancreatic cancer cells had decreased clonogenic survival with streptonigrin, which was reversed by coadministration of dicumarol. CaCo-2 human colon cancer cells negative for NQO1 had increased clonogenic cell survival with streptonigrin and minimal effect with coadministration of dicumarol. Although our data suggests that inhibiting NQO1 with dicumarol will reverse the effects of streptonigrin, there may be other pathways activated by streptonigrin other than NQO1.
Our present study compliments previous studies from our laboratory investigating the cytotoxic effect of dicumarol treatment alone. Dicumarol inhibits reductases, including NQO1 by competing with NADH for the binding site of the oxidized NQO1 form [31]. It has been proposed that by preventing the reduction of semiquinones, dicumarol increases the oxidative stress within the cell, which leads to increased cell toxicity. Pancreatic cancer cells treated with dicumarol increased intracellular production of , as measured by hydroethidine staining, and inhibited cell growth [9]. Both of these effects (cell growth inhibition and hydroethidine staining) were blunted with infection of an adenoviral vector containing the cDNA for manganese superoxide dismutase (MnSOD). Dicumarol also inhibited plating efficiency, and growth in soft agar. The difference in the two studies was that in our present study dicumarol was given at a lower dose (50 μM) and for shorter periods of time (4 h), effectively inhibiting NQO1 but reducing the dicumarol-induced cytotoxicity seen with the previous study. We have recently demonstrated that dicumarol has both a time- and dose-dependent cytotoxicity against pancreatic cancer cells [32].
Pancreatic cancer ranks among the most deadly of cancers, with over 31000 deaths per year in the US, and current treatments of radiation therapy, chemotherapy, and resection have been ineffective at improving the 0.4%−4% 5-yr survival rate [33,34]. Furthermore, only 10% of pancreatic cancer patients are eligible for surgical resection [35–37]. A recent study by Neoptolemos et al. randomly assigned 289 pancreatic cancer patients into four treatment groups following surgical resection. Treatments consisted of chemoradiotherapy, chemotherapy, both treatments, and neither treatment; median survival rates ranged from 13.9 mo in the chemoradiotherapy group to 21.6 mo in the chemotherapy group [38]. Although adjuvant treatments have improved survival following surgical resection, the 5-yr survival rates for pancreatic cancer remain low. Therefore, it is necessary to investigate new and more effective therapies such as those designed to target NQO1.
In summary, immunohistochemistry of resected pancreatic specimens demonstrated an increased immunoreactivity for NQO1 in pancreatic cancer and PanIN specimens when compared to normal human pancreas ductal epithelial cells. Pancreatic cancer cell lines also had increased NQO1 immunoreactivity when compared to an immortalized pancreatic ductal epithelial cell line and a colonic epithelial cell line using immunocytochemistry and Western immunoblots. Streptonigrin, a compound known to cause redox cycling in the presence of NQO1 decreased clonogenic survival, and decreased anchorage-independent growth in soft agar. Streptonigrin had little effect on cell lines with absent or reduced levels of NQO1. The effects of streptonigrin were reversed in pancreatic cancer cells pretreated with dicumarol, a known inhibitor of NQO1. Combined, these studies suggest that a novel biochemical characteristic of pancreatic cancer is overexpression of NQO1. Because cancer therapy is based upon the concept that certain cytotoxins kill cancer cells preferentially, relative to normal cells, NQO1 appears to be a novel therapeutic target in pancreatic cancer and a complete understanding of precise mechanisms involved may lead to a therapeutic advantage in pancreatic cancer where survival is measured in months.
ACKNOWLEDGMENTS
Supported by NIH grants DK 60618, CA 66081, the Susan L. Bader Foundation of Hope, the Medical Research Service, Department of Veterans Affairs, and the Canadian Institutes of Health Research Grant MOP-49585 (M.S.T.)
Abbreviations
- NQO1
NAD(P)H:Quinone oxidoreductase
- PanIN
pancreatic intraepithelial neoplasia
- TBS
tris-buffered saline
- TMA
tissue microarray
- PBS
phosphate buffered saline
- SEM
standard error of mean
- Au
arbitrary units
- H2O2
hydrogen peroxide
REFERENCES
- 1.Powis G, Svingen BA, Appel P. Quinone-stimulated superoxide formation by subcellular fractions, isolated hepatocytes, and other cells. Mol Pharmacol 1981;20:387–394. [PubMed] [Google Scholar]
- 2.Chesis PL, Levin DE, Smith MT, Ernster L, Ames BN. Mutagenicity of quinones: Pathways of metabolic activation and detoxification. Proc Natl Acad Sci USA 1984;81:1696–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lind C, Hochstein P, Ernster L. DT-Diaphorase as a quinone reductase: A cellular control device against semiquinone and superoxide radical formation. Arch Biochem Biophys 1982; 216:178–185. [DOI] [PubMed] [Google Scholar]
- 4.Thor H, Smith MT, Hartxell P, Bellomo G, Jewell SA, Orrenius S. The metabolism of menadione (2-methyl-1,4-naphthoquinone) by isolated hepatocytes. A study of the implication of oxidative stress in intact cells. J Biol Chem 1982;257: 12419–12425. [PubMed] [Google Scholar]
- 5.Siegel D, Ross D. Immunodetection of NAD(P)H: Quinone oxidoreductase 1 (NQO1) in human tissues. Free Rad Biol Med 2000;29:246–353. [DOI] [PubMed] [Google Scholar]
- 6.Huggins C, Fukunishi R. Induced protection of adrenal cortex against 7, 12-dimethylbenz(alpha)anthracene: Influence of ethionine: Induction of menadione reductase: Incorporation of thymidine H3. J Exp Med 1964;119:923–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strassburg A, Strassburg CP, Manns MP,Tukey RH. Differential gene expression of NAD(P)H:quinone oxidoreductase and NRH:quinone oxidoreductase in human hepatocellular and biliary tissue. Mol Pharmacol 2002;61:320–325. [DOI] [PubMed] [Google Scholar]
- 8.Logsdon CD, Simeone DM, Binkley C, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Can Res 2003;63:2649–2657. [PubMed] [Google Scholar]
- 9.Cullen JJ, Hinkhouse MM, Grady M, et al. Dicumarol inhibition of NAD(P)H:quinine oxidoreductase (NQO1) induces growth inhibition of pancreatic cancer via a superoxide-mediated mechanism. Can Res 2003;63:5513–5520. [PubMed] [Google Scholar]
- 10.Cone R, Hasan SK, Lown JW, Morgan AR. The mechanism of the degradation of DNA by streptonigrin. Can J Biochem 1976;54:219–223. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao M-S. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts by transfection with the E6E7 gene of human papilloma virus 16. Am J Patho 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang H, Mou L-J, Luk C, Liu N, Squire J, Tsao M-S. Paradiploid human pancreatic duct epithelial cell lines with near normal phenotype. Am J Pathol 2000;157:1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasm: A new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol 2001;25:579–586. [DOI] [PubMed] [Google Scholar]
- 14.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res 2000;6:2969–2972. [PubMed] [Google Scholar]
- 15.Al-Aynati MM, Radulovich N, Riddell RH,Tsao MS. Epithelial-cadherin and β-Catenin expression changes in pancreatic intraepithelial neoplasia. Clin Cancer Res 2004;10:1235–1240. [DOI] [PubMed] [Google Scholar]
- 16.Lam EWN, Hammad HM, Zwack R, et al. Immunolocalization and adenoviral vector-mediated manganese superoxide dismutase gene transfer to experimental oral tumors. J Dent Res 2000;79:1410–1417. [DOI] [PubMed] [Google Scholar]
- 17.Belinsky M, Jaiswal AK. NADPH:quinone oxidoreductase1 (DT-diaphorase) expression in normal and tumor tissues. Cancer Metastasis Rev 1993;12:103–117. [DOI] [PubMed] [Google Scholar]
- 18.Winski SL, Swann E, Hargreaves RHJ, et al. Relationship between NAD(P)H:quinone oxidoreductase 1 (NQO1) levels in a series of stably transfected cell lines and susceptibility to antitumor quinines. Biochem Pharm 2001;61:1509–1516. [DOI] [PubMed] [Google Scholar]
- 19.Beall HD, Murphy AM, Siegel D, Hargreaves RH, Butler F, Ross D Nicotinamide adenine dinucleotide (phosphate):quinone oxidoreductase (DT-diaphorase) as a target for bioreductive antitumor quinones:quinone cytotoxicity and selectivity in human lung and breast cancer cell lines. Mol Pharmacol 1995;48:499–504. [PubMed] [Google Scholar]
- 20.Rao KV, Biemann K, Woodward RB. The structure of streptonigrin. J Am Chem Soc 1963;85:2532–2533. [Google Scholar]
- 21.Oleson JJ, Calderella LA, Mjos KJ, Reith AR, Thie RS, Toplin I. The effects of streptonigrin on experimental tumors. Antibiot Chemother 1961;11:158–164. [PubMed] [Google Scholar]
- 22.Reilly HC, Sigiura K. An antitumor spectrum of streptonigrin. Antibiot Chemother 1961;11:174–177. [PubMed] [Google Scholar]
- 23.Beall HD, Mulcahy RT, Siegel D, Traver RD, Gibson NW, Ross D. Metabolism of bioreductive antitumor compounds. Cancer Res 1994;54:3196–3201. [PubMed] [Google Scholar]
- 24.Mizuno NS. Effects of streptonigrin on nucleic acid metabolism of tissue culture cells. Biochem Biophys Acta 1965;108: 394–403. [DOI] [PubMed] [Google Scholar]
- 25.Levine M, Borthwick M. The action of streptonigrin on bacterial DNA metabolism and on induction of phage production in lysogenic bacteria. Virology 1963;21:568–574. [DOI] [PubMed] [Google Scholar]
- 26.White HL, White JR. Lethal action and metabolic effects of streptonigrin on Escherichia coli. Mol Pharmacol 1968;4: 549–565. [PubMed] [Google Scholar]
- 27.Lown JW, Sim SK. Studies related to anti-tumor antibiotics. Part VIII. Cleavage of DNA by streptonigrin analogues and the relationship to antineoplastic activity. Can J Biochem 1976; 54:446–452. [DOI] [PubMed] [Google Scholar]
- 28.Beall HD, Liu Y, Siegel D, Bolton EM, Gibson NW, Ross D. Role of NAD(P)H:Quinone oxidoreductase (DT-diaphorase) in cytotoxicity and induction of DNA damage by streptonigrin. Biochem Pharm 1996;51:645–652. [DOI] [PubMed] [Google Scholar]
- 29.Llopis J, Ernster L, Cadenas E. Effect of glutathione on the redox transitions of naphthohydroquinone derivatives formed during DT-diaphorase catalysis. Free Radical Res Commun 1990;8:271–285. [DOI] [PubMed] [Google Scholar]
- 30.Buettner GR. The pecking order of free radicals and antioxidants: Lipid peroxidation, alpha-tocopherol, and ascorbate. Arch Biochem Biophys 1993;300:535–543. [DOI] [PubMed] [Google Scholar]
- 31.Dinkova-Kostova AT, Talalay P. Persuasive evidence that quinone reductase type 1 (DT-Diaphorase) protects cells against the toxicity of electrophiles and reactive forms of oxygen. Free Rad Biol Med 2000;29:231–240. [DOI] [PubMed] [Google Scholar]
- 32.Lewis A, Ough M, Li L, et al. Treatment of pancreatic cancer cells with dicumarol induces cytotoxicity and oxidative stress. Clin Can Res 2004;10:4550–4558. [DOI] [PubMed] [Google Scholar]
- 33.Bramhall SR, Allum WH, Jones AG, Allwood A, Cummins C, Neoptolemos JP. Treatment and survival in 13 560 patients with pancreatic cancer, and incidence of the disease, in the West Midlands: An epidemiological study. Br J Surg 1995; 82:111–115. [DOI] [PubMed] [Google Scholar]
- 34.Jemal A, Tiwari RC, Murray T, et al. Cancer statistics, 2004. CA Cancer J Clin 2004;54:8–29. [DOI] [PubMed] [Google Scholar]
- 35.Geer RJ, Brennan MF. Prognostic indicators for survival after resection of pancreatic adenocarcinoma. Am J Surg 1993; 165:68–72. pancreaticoduodenectomy. Surgery 1999;126: 178–183. [DOI] [PubMed] [Google Scholar]
- 36.Sener SF, Fremgen A, Menck HR, Winchester DP. Pancreatic cancer: A report of treatment and survival trends for 100 313 patients diagnosed from 1985–1995, using the National Cancer Database. J Am Coll Surg 1999;189:1–7. [DOI] [PubMed] [Google Scholar]
- 37.Sohn TA, Yeo CJ, Cameron JL, et al. Resected adenocarcinoma of the pancreas- 616 patients: Results, outcomes, and prognostic indicators. J Gastrointest Surg 2000;4:567–579. [DOI] [PubMed] [Google Scholar]
- 38.Neoptolemos JP, Stocken DD, Friess H, et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med 2004;350:1200–1210. [DOI] [PubMed] [Google Scholar]