Abstract
Cisplatin resistance is a major barrier in the effective treatment of lung cancer. Cisplatin resistant (CR) lung cancer cells do not primarily use glucose, but rather consume amino acids such as glutamine and tryptophan (TRP) for survival. CR cells activate the kynurenine (KYN) pathway (KP) to cope with excessive reactive oxygen species (ROS) and maintain homeostasis for growth and proliferation. Consequently, indoleamine 2,3-dioxygenase-1 (IDO1) becomes an essential enzyme for CR cells’ survival since it initiates and regulates the first step in the KP. Increased IDO1 activities and ROS levels are found in CR cells vs. cisplatin sensitive lung cancer. Importantly, significantly greater KYN/TRP ratio (p=0.005) is detected in serum of patients who fail cisplatin when compared to naive treatment. Knocking down IDO1 using shRNA or IDO1 inhibitors heightens ROS levels and results in a significant growth inhibitory effect only on CR cells and not cisplatin sensitive cells. Exposing CR cells to antioxidant (TIRON) results in suppression of IDO1 activity and confers resistance to IDO1 inhibition, indicating an interrelationship between ROS and IDO1. Since KYN plays a critical role in reprogramming naïve T-cells to the immune suppressive regulatory T-cell (T-reg) phenotype, we observed higher expression of T-reg (TGFβ, FoxP3 and CD4+CD25+) in mice bearing CR tumors compared to tumors from cisplatin sensitive counterparts.
Keywords: Lung cancer; Reactive oxygen species; Kynurenine; indoleamine 2,3-dioxygenase-1; Cisplatin resistance
INTRODUCTION
Surgery is the best treatment approach for early stage lung cancer; nonetheless, most patients already have locally advanced or metastatic disease at the time of diagnosis. Anti-PD1 (pembrolizumab) in combination with chemotherapy was recently approved as first-line treatment of non-small cell lung cancer (NSCLC) patients with stage 3B who are not candidates for surgical resection. However, patients’ tumors must express PD-L1 and have no EGFR or ALK mutation in order to receive this therapy. While these combination therapies offer a longer duration of response compared to other second line chemotherapy, the overall response rate in NSCLC is low (15–20%) (1). Subsequently, cisplatin-based chemotherapy alone and in combination with radiation remains the primary modality of treatment. Despite early positive responses to cisplatin, all lung cancer patients will develop drug resistance, thus cisplatin resistance remains the major obstacle for the effective treatment of lung cancer. Our research focus has been to elucidate molecular differences in metabolic heterogeneity observed among patients with sensitive and resistant tumors.
We have previously reported that cisplatin resistant (CR) lung cancer cells possessed increased numbers of mitochondria and consumed higher rates of oxygen than non-resistant cancer cells, resulting in significantly higher (2–3 fold) basal levels of reactive oxygen species (ROS) (2, 3). In addition, we showed that CR tumors are no longer addicted to glucose for energy and do not utilize classic aerobic glycolysis (Warburg effect (4, 5)). By switching to oxidative metabolism (OXMET), CR cells increase cellular energy or metabolic demand and outstrip the glutamine supply; making glutamine the conditionally essential amino acid for cell survival. Besides glutamine, CR cells are also auxotrophic for arginine due to the lack of argininosuccinate synthetase (ASS), which is a key enzyme in the synthesis of arginine from citrulline (6–8). Lack of ASS expression therefore makes arginine an essential amino acid for CR tumors as well. Due to these phenomena, CR cells are collectively dependent more on OXMET for survival and are susceptible to amino acid deprivation.
Here we show that these metabolic derangements have led us to the discovery that L-tryptophan (TRP) catabolism is also upregulated in CR cells. While TRP is an essential amino acid, required for protein synthesis and as the precursor of serotonin and melatonin, the catabolism of TRP can also generate kynurenine (KYN) via the kynurenine pathway (KP). This pathway is responsible for the catabolism of approximately 95% of ingested TRP not used for protein synthesis or the serotonin pathway (9, 10), and not limited to its originally derived role in the biogenesis of nicotinamide adenine dinucleotide (NAD+) (11). At the first step, TRP is oxidized through indoleamine 2,3-dioxygenase-1 and 2 (IDO1 and IDO2) or tryptophan 2,3-dioxygenase-2 (TDO2) to form formylkynurenine which is later degraded to KYN (12). However, TDO2 is primarily present in the liver and is not expressed in lung tissue or lung tumor (13). IDO2 is expressed in human tumors (14), and recent genetic data have implicated it functionally in drug-resistant pancreatic ductal adenocarcinoma resistant to DNA damaging radiotherapy (15). But the significance of the relationship between cisplatin resistance and IDO2 activity in lung cancer remains elusive and needs to be explored in future.
IDO1 is the unique enzyme similar to superoxide dismutase-1 (SOD1) that exploits superoxide as a substrate (16, 17). We have previously reported that CR cells which possessed elevate basal levels of ROS also expressed higher superoxide dismutase-1 (SOD1) when compared to parental cell counterparts (3). Increasing SOD1 levels and IDO1 activities are crucial mechanisms for the capture of ROS/superoxide and maintenance of homeostasis in CR cells. Importantly, increased IDO1 activity results in greater KYN production which plays a key role in reprogramming naïve T-cells to the immune suppressive regulatory T-cell (T-reg) phenotype (18, 19). Thus, it appears that adaptations in CR cells confer a survival advantage in an environment with elevated basal levels of ROS which may otherwise be lethal to normal cells (3). These findings are supported by recent reports that IDO levels impact outcomes in patients with lung cancer and gliomas receiving radiotherapy that damages DNA as well as immunoradiotherapy (20, 21). Hence, links have been established between metabolic alterations in oxidative processes and amino acid catabolism and tumor responsiveness to treatment that may be utilized to affect outcomes.
Here, we show that IDO1 activity is dependent on ROS levels in CR cells. Increased basal level of ROS heightens IDO1 activity and in turn hyper-activates the kynurenine pathway (KP) in CR cells in vitro, in vivo, and in patients who failed cisplatin treatment. Inhibiting KP using IDO1 inhibitor further increases ROS in CR cells beyond their tolerance limit which ultimately leads to cell death. By investigating a new paradigm between CR cells and their microenvironment, and by targeting their distinct metabolic features, we believe that our studies will identify a population of lung cancer patients who will be benefit greatly from future chemo-immunotherapy and have improved outcomes.
MATERIALS AND METHODS
Cell Lines and reagents
Two pairs of parental vs. cisplatin resistant human NSCLC cells (A vs. ALC and FA vs. FC) and a pair of mouse Lewis lung cells (LLC vs. LLC-CR) were used. Cell line “A” was established with pleural fluid from a patient with adenocarcinoma. FA was established from a poorly differentiated squamous cell carcinoma. Cellular characteristics have been previously characterized (22–25) and routinely tested for mycoplasma infection (MycoAlert; Lonza cat#LT07–710). These cell lines were not derived from the patients reported here in this study. LLC and LL24 were purchased (ATCC). We established their cisplatin resistant variants using previously published methods (23, 24). Note: ALC and FC exhibit 7 fold resistance to cisplatin. LLC-CR exhibits 3 fold resistance. LL24 is normal lung fibroblast. Supplementary Table1 shows the ID50 values for the sensitive and resistant cell lines. Recombinant human IFNγ was purchased from Biolegend (cat#570204). IDO1 inhibitors (Epacadostat (cat#T3545), PF-06840003 (cat#T4307), NLG-919 (cat#T1806), and Indoximod (cat# T6543)) were purchased from TargetMol. Dimethoxyflavone (DMF; cat#D6571), CH-223191 (cat#C8124), TIRON (4,5-dihydroxy-1,3-benzenedisulfonic; cat#172553), and NAD+ were purchased from Sigma.
Measurement of patients’ plasma
Patients undergoing cisplatin treatment were recruited in this study at the Miami VA Healthcare System. All procedures were approved by the Institution Review Board (IRB; ref.no.1190748–1). Participants prospectively provided informed consent for the use of their specimen in research.
Blood samples were drawn and centrifuged at 1000g for 10 min to extract plasma and then stored at −80°C. Plasma samples (100μl) were placed into a centrifugal tube and the same volume of 5% perchloric acid solution was added. Next, the samples were placed in a vortex mixer for 30sec and then maintained at room temperature for 15min to allow plasma protein precipitation. Finally, the samples were centrifuged at 12,000 × g for 5min and 20μl samples of supernatant were collected for analysis. Samples were analyzed by Agilent 1290 Infinity high performance liquid chromatography system (HPLC) on phenomenex kinetex biphenyl column (2.1mm × 50mm; 2.6μm). HPLC conditions: buffer A: 0.1% Trifluoroacetic acid in water, buffer B: 0.1% Trifluoroacetic acid, 70% Acetonitrile in water, UV detector: 225nm/230nm, linear gradient from 5%B to 50%B in 5min.
Animal studies
Procedures and mice protocol were approved by the Institutional Animal Care and Use Committee (IACUC) of the Miami VA Healthcare System (Animal Welfare Assurance Number: A3739–01). Male or female C57BL6/N mice (age 5 weeks, Jackson laboratory) were used to establish in vivo allograft using LLC vs. LLC-CR. Mice were inoculated subcutaneously with 2.5×106 cells on the dorsal lumbosacral region. Tumor growth was evaluated twice a week by measuring tumor volume according to the following formula: tumor volume = width2 × length × 0.5. Experiment was ended when either W or L reached the final set value of 10 mm.
Growth inhibition and cytotoxicity assay
Cells were seeded in 24-well dishes and treated with various concentrations of IDO1 inhibitors (i.e. Epacadostat). The procedure was described previously (22, 26). Briefly, the culture media as well as the trypsinized cells were collected and this mixture was centrifuged at 400 × g for 5 min. The supernatant was discarded, re-suspended in 1 mL of Hank’s buffer, and assayed for live cells and death cells using trypan blue exclusion method.
Western Blot analysis
Cells were seeded at 1×105/ml onto 60 mm dishes, treated, collected, lysed and immunoblotted with indicated antibody. Detailed procedure was described in our previous publications (22, 26). Briefly, cell lysis was completed by sonication and the total protein was separated on an SDS-PAGE, transferred onto a PVDF membrane (Millipore) and immunoblotted with indicated primary antibody. Antibody to IDO1 (cat#NBP1–87702) was purchased from Nuvous. Antibody to HIF1α (cat#610958) was purchased from BD bioscience. Phospho-AHR (cat#GTX113124) and FoxP3 (GTX107737) were purchased from Genetex. AHR (cat# A1451) was purchased from Abclonal. Antibody to LAT1 (cat#5347) was purchased from Cell Signaling. All antibody dilutions were at 1:1000, except for Actin (Sigma; cat#A5441) which was diluted at 1:10000. Bands were measured using a molecular imager Chemidoc system with Quality One software (Bio-Rad).
Qualitative real-time PCR
qRT-PCR was carried out as previously described (2). Briefly, 1μg of RNA was used for cDNA synthesis. The primers for qRT-PCR were designed with Perlprimer for SYBR Green fluorophore (See Supplement Method). Forty cycle amplification was used and the data were analyzed with CFX manager software from Bio-Rad. To calculate the relative mRNA level, we used the ΔΔCt method. The level of mRNA was corrected with that of GAPDH or actin.
Knockdown experiment
For stable knockdown (shRNA) of IDO1, ALC cells were transfected with 1μg of pGFP-C-shLenti plasmid (Origene) expressing shIDO1 (NM_002164) or scrambled control using lipofectamine 2000 (Thermo Fisher) transfection reagent (see Supplemental Method). After 24h, transfection medium Opti-MEM was exchanged to RPMI1640 containing 5μg/ml of G418. GFP as a reporter was used to evaluate target gene knocked down efficiency. For siRNA, cell lines A were transfected with 1nM of HIF1α SMARTpool® siRNA (NM_005566) or siCONTROL®(Dharmacon) using INTERFERin™ transfection reagent (Polyplus) as previously described (26).
Real time assay of oxygen consumption
Simultaneous multi-parameter metabolic analysis of cell populations in culture was performed in the Seahorse XFe24 extracellular flux analyzer (Seahorse Bioscience) as described by Wu et al. (27). All cell lines were cultured in growth medium for 24h in plate before real-time metabolic analysis. At the start of the assay, growth medium was removed and replaced with assay medium. Basal OCR (oxygen consumption rate) and ECAR (extracellular acidification rate) of the cells were measured. Oligomycin, FCCP, and rotenone were used to inhibit ATP synthase, uncouple OXPHOS, and inhibit mitochondrial complex 1, respectively. After XF assay, cells were harvested by trypsin-EDTA treatment and counted. The number of cells per well were used to normalize OCR and ECAR.
Assay of Intracellular ROS/H2O2
As previously described (3), cells were collected and intracellular H2O2 was measured by incubating with 10 μM of CM-H2DCFDA (Life Technologies; cat#C2938) at 37°C for 30 min in the dark. Then the cells were washed once with PBS and centrifuged to remove impermeable reagents. Cells were resuspended in 500 μL of PBS and analyzed either by fluorimeter (FLUOstar OPTIMA, BMG Labtech (excitation at 485 nm and emission at 520 nm)) or flow cytometer (CytoFlex, Beckman).
LAT1 localization
For cell surface LAT1 staining, fluorochrome-conjugated antibodies were used to detect LAT1/CD98 (Biolegend; cat#315603). Cells were washed in PBS and re-suspended in 250 μl of FACS buffer (PBS, 1% fetal calf serum, 0.1% NaN3) with LAT antibody. Cells were incubated for 30 min on ice, followed by three washes, and resuspended in 500μL of PBS. Live cells were gated according to their forward scatter and side scatter. Data were acquired using CytoFLEX (Beckman) flow cytometer.
Tryptophan uptake
Seeded cells were cultured in complete RPMI media for 24 hours, at which point media was replaced with fresh tryptophan and serum free RPMI media with “hot” (5μCi L-[5-3H(N)] tryptophan) (2). Next, the hot medium was removed, and cells were washed three times with fresh, “cold” RPMI serum-free medium with tryptophan. Then cells were lysed with 500 μL of 1 N NaOH, collected in 1.5 mL Eppendorf tubes and gently vortexed (2). Protein analysis was conducted on 250 μL samples using the Micro BCA Protein Assay Kit (Thermo Scientific; cat#23235). Radioactivity in the remaining sample was measured as counts per minute (CPM), and normalized to protein, using a PerkinElmer Tri-Carb 2810TR liquid scintillation spectrometer with QuantaSmart (2).
IDO1 activity assay
An IDO1 assay kit (Biovision; cat#K972–100) was used to measure IDO1 activity. Briefly, 5×106 cells were seeded onto 100 mm dishes, treated, collected, and homogenized with IDO1 assay buffer. IDO1 activities were calculated by the amount of N-formylkynurenine (NFK) produced / (Reaction time x Amount of protein in well). Samples were measure at Ex/Em = 402/488nm.
NAD+ concentration assay
A NAD/NADH assay kit (Cayman Chemical; cat#600480) was used to measure total cellular NAD+ concentration. Cell were seeded in 96-well at 1×104 and grow overnight. Total NAD+ was detected at 450 nm according to manufacturer instruction.
Amino acid analyzer
Physiological fluids were collected and the Biochrom 30 amino acid analyzer (using ion exchange chromatography) was used to analyze free amino acids (30–40 in the complete panel) in samples (2). Values were reported compared to normal ranges in controls. Samples used in ion exchange chromatography are separated in an analytical fashion with an ion-exchange column and buffers of increasing pH and ionic strength. After post-column derivatization with Ninhydrin, and colorimetric intensity recordings at 570 nm and 440 nm, the amino acids separated by chromatography were detected (2). Amino acid concentration was calculated by comparing the peak area of a particular amino acid to the peak area of an internal standard-AEC of known concentration, and then multiplying by its specific response factor from calibration.
Immunohistochemistry staining
Immunohistochemical staining was performed according to our routine methodology with some modification (2). Target retrieval solution (citric buffer; pH 6.0) was used to enhance the staining. Samples were incubated overnight with CD4 (Genetex; cat#GTX50984), CD25 (Biorbyt; cat#orb389314), and TGFβ (Cell Signaling; cat#3709) primary antibodies at 1:100 in antibody dilution solution (Dako S3022). Anti-IDO-1 Antibody (clone 4B7; Millipore cat# MABF850) which has been validated in IDO−/− mouse was diluted at 1:50. Secondary antibody solution (LSAB™2 Kits Dako, biotinylated antibody solution; cat#K0675) was added for 30 min, washed, and streptavidin conjugated to peroxidase (HRP) solution was added for 30 min. DAB chromogen (Dako, cat#K3466) was used and then counter-stained with hematoxylin.
Immunofluorescent staining
Cells were treated with and without AHR antagonist, washed with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were incubated with PBS containing 0.2%TritonX-100 (antibody diluent) for 15 min at room temperature and subsequently blocked with PBS containing 5% BSA for 1hr. Cells were incubated with anti-AHR antibody (cat#GTX113124 at 1:1000 in antibody diluent,) overnight at 4degC. Secondary antibody (1:250; Alexa Fluor 555) were added for 1hr. DAPI (Vector Laboratories; cat#H1500) were used as nuclei staining. Cells were examined under a fluorescence microscope (Keyence BZ-X710) to determine localization of AHR. Our Keyence microscope also equipped with hybrid cell count (an algorithm software which allows accurate quantification for phase contrast).
Statistical analysis
All statistical analyses were performed from three separate biological replicates which were isolated and analyzed in technical triplicates. Separate measurements using the two-tailed t-test and the results were expressed as mean ± standard deviation. A p-value of less than 0.01 or 0.05 was considered as statistically very significant and significant, respectively. The medium or plasma concentrations of KYN or TRP, as well as KYN/TRP ratios approximated a parametric distribution; therefore, data were presented as means ± SD. Due to the small sample size, the associations between serum biomarker levels and time point variables were evaluated with a Mann-Whitney U test.
RESULTS
The kynurenine pathway (KP) contributes to higher KYN/TRP ratio in patients who fail cisplatin
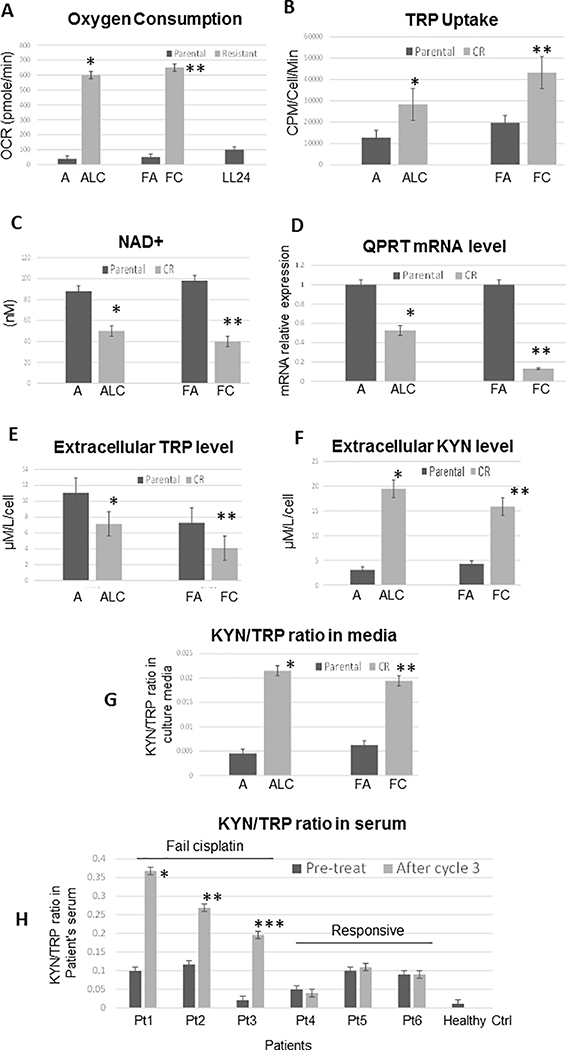
We have reported that CR lung cancer cells were less dependent on the glycolytic pathway, but instead utilized mitochondria for biogenesis to catabolize glutamine as an alternative carbon skeleton source (2, 7). Here, we first compared oxygen consumption among resistant and sensitive cells using the Seahorse flux analyzer. In response to adding oligomycin, FCCP, and rotenone (Supplementary Figure S1A), CR cells consumed significantly higher rates of oxygen (Figure 1A), and thus had higher levels of ATP production when compared to their parental cells counterparts (Supplementary Figure S1B). Adding to these metabolic alterations and the reliance on amino acids, we further discovered that CR cells also uptake significantly higher amounts of tryptophan (TRP) (Figure 1B). It is noteworthy that even though TRP can be used for de novo biosynthesis of NAD+ (essential co-enzyme of all organisms’ redox system), TRP is less efficient and a poor NAD+ precursor in vivo. TRP will only be diverted to NAD+ synthesis when its supply exceeds enzymatic capacity (28). Thus, the main source of NAD+ is from salvage pathways which require the uptake of other NAD+ precursors such as nicotinic acid. Consistent with these findings, we have found that while uptake of TRP is significantly increased in CR cells as showed in Figure 1B, the NAD+ levels and QPRTase (quinolate acid phosphoribosyltransferase, a key enzyme in NAD+ biosynthesis) expressions were significantly decreased when compared to parental cell counterparts (Figure 1C&D). Hence, these findings demonstrate that CR cells actively utilized KP, but did not engage in de novo synthesis of NAD+.
Figure 1.
CR lung cancer cells relied on OXMET and utilized KP which contributed to the production of kynurenine (KYN) in CR tumors. (A) Parent (A and FA) and CR (ALC and FC) cells were assayed for baseline oxygen consumption using Seahorse XFe24 extracellular flux analyzer. The rate of oxygen consumption (OCR) was 6 times higher in CR than parental cells (*p=0.009 and **P=0.003). LL24 was used as control. (B) Tryptophan (TRP) uptake was determined in parental vs CR cells using L-[5-3H(N)] tryptophan. The rate of TRP uptake was higher in CR cells (*p=0.02, **p=0.007). (C) Total cellular NAD+ concentrations were detected in parental vs. CR cell lines. CR possessed lower basal levels of NAD+ (*p=0.02; **p=0.006). (D) Relative mRNA levels of QPRTase. QPRTase mRNA was significantly decreased in CR cells. GAPDH was used as internal control. The results shown in the graph were calculated with the ΔΔCt method by setting the mRNA level of parental cells as 1 (*p=0.04, **p=0.01). (E) Extracellular TRP concentration in culture medium were determined by amino acid analyzer. Lower levels of TRP were found in CR cells’ culture media (*p=0.04, **p=0.02). (F) Extracellular KYN concentration in culture medium were determined by amino acid analyzer. Higher levels of KYN were found in CR cells’ culture media (*p=0.01, **p=0.005). (G) Extracellular KYN/TRP ratio in culture medium were determined by HPLC. CR cells possessed significantly greater KYN/TRP ratio (*p=0.003, **p=0.01) than control. (H) KYN/TRP ratio in patients’ serum (HPLC). Patients 1–3 failed cisplatin after third cycle of cisplatin (*p=0.002, **p=0.004, ***p=0.005). Patients 4–6 continued to respond to cisplatin after the third cycle of treatment. (Note: Pt = patient).
Next, we evaluated TRP levels in cell culture and found that TRP concentrations were significantly reduced in CR cells’ media (Figure 1E). We then assayed for extracellular kynurenine (KYN) levels (Figure 1F), as well as using the KYN/TRP ratio as an indicator of IDO1 activity. A significantly higher mean KYN/TRP ratio in CR cells’ media (as compared to parental cell counterparts), implied that IDO1 activity was elevated in CR cells (Figure 1G). To gain insight into the physiological role of KP, we assayed the KYN/TRP ratio in cancer patients’ serum prior to and after cisplatin/radiation therapy. All cancer patients from our study possessed higher basal levels of KYN/TRP ratio when compared to healthy controls. Patients (patients 1 to 3) who failed cisplatin possessed significantly higher KYN/TRP ratios when compared to the pre-treatment baseline (Figure 1H). Consistently, we did not find an increase in KYN/TRP ratio in patients (patients 4 to 6) who continued to respond to cisplatin treatment.
Together, our data illustrate that CR cells consumed TRP at a higher rate specifically for KYN production, but not for production of NAD+. Importantly patients who failed cisplatin therapy, in our small sampling, possessed significantly higher ratio of KYN/TRP, the indicator of IDO1 activity. These results suggest that increased IDO activity is involved in disease progression in lung cancer, possibly through immunosuppressive effects.
The KYN/AHR/ARNT axis is a crucial modulator of metabolism in CR cells
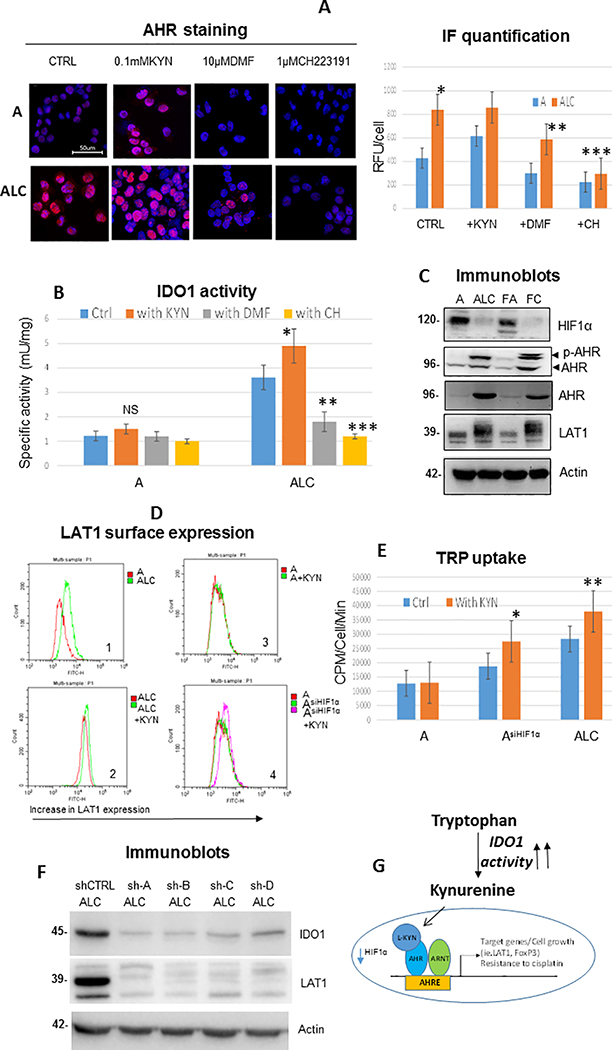
Recent discoveries revealed that KYN can serve as an endogenous ligand for the aryl hydrocarbon receptor (AHR) in cancer cells (29). KYN formed a complex with AHR and translocated into the nucleus to modulate their target genes as a ligand-activated transcription factor (30). To further elucidate the unique role of AHR, immunofluorescence staining showed that AHR localized with high intensity to the nucleus of CR cells, whereas less accumulation was observed in parental counterparts, implicating that KP is specifically active in CR cells (Figure 2A). To determine that AHR can be activated by KYN, we added exogenous KYN (100μM) and assessed AHR localization. As depicted, treatment with KYN increased the nuclear distribution of AHR in both parental and CR cells. We then determined that KYN was specific to AHR activation in CR cells by inhibiting AHR translocation with either 10μM of dimethoxyflavone (DMF; an AHR antagonist) or 1μM CH-223191 (AHR inhibitor). AHR were significantly less accumulated in the nucleus after treatments. Importantly, exposure with AHR inhibitors also resulted in suppression of IDO1 activities, whereas addition of KYN increased IDO1 activity only in CR cells (Figure 2B).
Figure 2.
Modulator role of KYN/AHR/ARNT axis in CR cells metabolism. (A) Immunofluorescence (IF) staining of cells with 1:1000 AHR antibody (red) and DAPI (blue nuclei). CR cells (ALC) possessed significantly higher intensity of AHR expression in the nucleus when compared to parental cells (*p=0.008). DMF or CH223191 significantly reduced AHR accumulation in CR cells (**p=0.007, ***p=0.001; respectively) while exposure to KYN markedly increased AHR accumulation in the nucleus of both parental and CR cells. Bar graph indicated quantification of IF intensity (RFU/cell) using hybrid cell count. (B) Addition of 100μM of KYN increased IDO1 activity in CR cells substantially (*p=0.03), but not significantly (NS) in parental cells. Treatment of 10μM of DMF or 1μM of CH223191 resulted in significant suppression of IDO1 activities (**p=0.006, ***p=0.002, respectively). (C) Immunoblot of lung cancer cell lines showed that resistant variants did not possess HIF1α, but expressed higher levels of AHR and LAT1. Actin was used as a loading control. (D) Flow cytometry analysis of surface LAT1 in lung cancer cell lines. D1:CR cells possessed higher surface LAT1 when compared to parental counterparts. D2:Treatment of KYN at 100μM for 48h further enhanced LAT1 expression in CR cells. D3:KYN treatment did not increase LAT1 expression in parental cells. D4:Knocking down HIF1α in parental cells resulted in increased LAT1 expression after KYN treatment. (E) Knocking down HIF1α in parental cells increased TRP uptake and further increased TRP uptake upon exposure to KYN (100μM). Treatment of KYN further heightened TRP uptake in CR cells (48h; *p=0.02, **p=0.006). (F) Knocking down IDO1 (shIDO1) in ALC suppressed LAT1 expression. Sh-A to D represent 4 unique shRNA sequences. (G) Diagram illustrating the binding partners of ARNT (HIF1β). When HIF1α is down-regulated, ARNT formed a new binding partner with AHR/KYN and initiated the transcription of genes that favored proliferation and increased TRP uptake which can lead to further KYN secretion in CR cells.
Furthermore, we have previously demonstrated that CR cells possess very low levels of hypoxia-inducible factor-1α (HIF1α) due to metabolic re-programming (7) (Figure 2C). ARNT or HIF1β is a known binding partner of both HIF1α and AHR (30, 31). Thus, in the absence of HIF1α, it is possible that ARNT is now available to bind and form a new partner with KYN/AHR and initiate the transcription of genes which favor the survival/proliferation of CR cells. Immunoblot showed that CR cells expressed higher basal level of AHR as well as its phosphorylated form (Figure 2C). This is interesting since others have shown that AHR can be fully activated upon phosphorylation (32). To characterize the role of the KYN/AHR/ARNT axis, we assayed the expression of the AHR-target gene LAT1 (L-type amino acid transporter-1; a known amino acid transporter for large neutral amino acids such as arginine, leucine, and TRP (33, 34)) with and without adding KYN. As shown in Figure 2C, LAT1 protein expressions were higher in CR cells, then we assayed for functional LAT1 that localized to the cell surface using the flow cytometer (Figure 2D1). LAT1 was further augmented upon addition of KYN in CR cells (Figure 2D2), while no increase in LAT1 was observed after exposing parental cells to KYN (Figure 2D3), suggesting that HIF1α still maintained its complex with ARNT in parental cells. To determine the role of HIF1α in parental cells, we knocked down HIF1α using siRNA and exposed AsiHIF1α to KYN. Increased LAT1 expression was observed in AsiHIF1α after treatment (Figure 2D4) and increased TRP uptake was found in CR cell after KYN treatment as anticipated. But importantly, TRP uptake was increased in AsiHIF1α and exposure to KYN further significantly increased TRP uptake (Figure 2E), suggesting that downregulation of HIF1α levels will lead to an increase in AHR/ARNT complex formation (Figure 2G). We used 4 different unique sequences to silence IDO1 expression in order to determine efficacy of silencing and consistency of effect (Figure 2F). All knock downs of IDO1 (shIDO1) suppressed LAT1 (Figure 2F) and CYP1B1 (also a known AHR-target gene; Supplementary Figure S2) expressions in CR cells. Thus, our findings strongly suggest that KYN/AHR/ARNT axis plays the unique modulator role in CR cells’ metabolism, and increased TRP uptake can lead to further KYN secretion in CR cells.
CR tumors possess higher IDO1 activity and immunosuppressive phenotype in vivo.
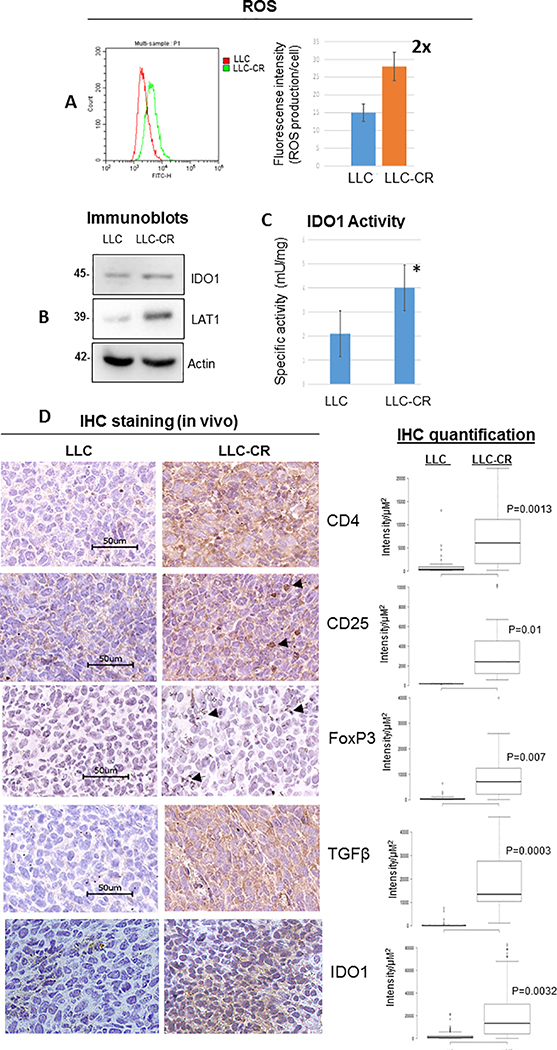
The tumor microenvironment with increase KYN levels can facilitate differentiation of naïve T-cells into T-reg cells (18, 19) creating a highly immunosuppressive environment which favors the growth of CR cells. To determine that our in vitro cell model is indeed representative of in vivo settings, we created mouse CR cells (LLC-CR) from the Lewis lung mouse cell line (LLC). LLC-CR possessed 3–4 fold resistance to cisplatin (Supplementary Figure S3) and yielded 2 fold higher basal levels of ROS (Figure 3A). LAT1 and IDO1 activity were increased in LLC-CR, thus suggesting a strong evidence of KP utilization (Figure 3B&C).
Figure 3.
Increase IDO1 activity and immune suppressive phenotype were found in CR tumor. (A) ROS analysis of mouse (LLC vs LLC-CR) cells. LLC-CR expressed 2 fold higher basal level of ROS. (B) Immunoblot showed that LLC-CR also expressed higher levels of LAT1 protein. (C) LLC-CR possessed higher IDO1 activity (*p=0.04). (D) Immunohistochemistry (IHC) staining of intratumoral CD4+, CD25+, FoxP3+ (arrow), TGFβ, and IDO1. Higher T-reg densities were found in mice bearing CR tumor than control. Box graph indicated quantification of IHC (intensity/μM2) using hybrid cell count.
LLC and LLC-CR were then allografted into C57BL6/N mice. After tumors developed, we then assayed for T-reg phenotypes. CD4+CD25+, and FoxP3 (forkhead family transcriptional regulator) which are the lineage-specific marker of T-reg cells were higher in LLC-CR when compared to parental counterpart which further indicated the involvement of KP in CR tumors (Figure 3D). Furthermore, we also found higher expression of TGFβ in LLC-CR tumors. TGFβ is a well-known immunosuppressive cytokine that is released by T-reg (35) and has been shown to activate IDO1 biosynthesis (36, 37). In fact IDO1 expression was elevated in mouse allografts of LLC-CR cells compared to allografts of LLC as seen through IHC with a mouse validated antibody (Figure 3D; IDO1). Together, our data suggest that increased IDO1 activity found in CR cells can create a highly immunosuppressive environment which favors the growth of tumors, thus making CR tumors good candidates for IDO1 treatment therapy.
Evaluation of IDO1 expression alone may be insufficient to determine clinical benefits
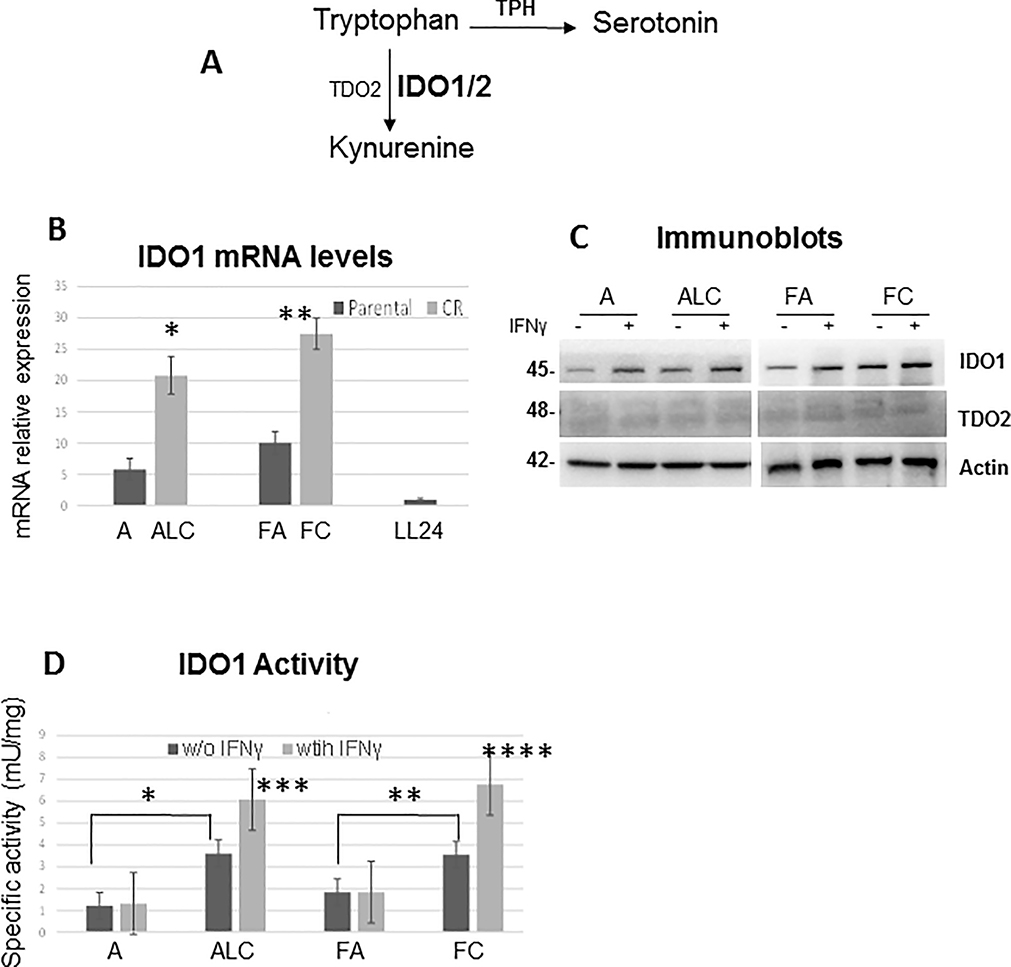
Depletion of TRP induces signaling events in T-cells leading to anergy and since TRP is a precursor for both serotonin and the kynurenine pathway (KP) (Figure 4A), we assessed the expression of tryptophan hydroxylase (TPH), the rate-limiting step in the serotonin pathway. TPH mRNA levels were extremely low and downregulated in our NSCLC cell line models (Supplementary Figure S4A). We then assayed for IDO1, IDO2 and TDO2 (the rate-limiting step in KP) expressions. Even in the absence of IFNγ (cytokine inducer of IDO1), IDO1 mRNA expression was significantly higher in CR cells when compared to parental counterparts (Figure 4B). However to our surprise, a surge in CR cells’ mRNA expression did not significantly increase IDO1 protein levels (Figure 4C). IDO2 mRNA expressions were higher in CR cells but not as significant when compare to IDO1 (Supplementary Figure S4B). As for TDO2, this enzyme was not expressed in NSCLC cells, but only expressed in brain cancer A172 cells utilized as positive controls (Supplementary Figure S4C), thus indicating that TDO2 was not required in the formation of KYN in lung cancer cells. We then assayed for IDO1 activity to determine whether increased TRP uptake in CR cells was being utilized by the KP. As depicted in Figure 4D, all CR cells possessed significantly higher IDO1 activity which was further enhanced with IFNγ treatment, but not in parental cells.
Figure 4.
IDO1 expression alone is not the only factor in determining its activity. (A) Diagram illustrated that tryptophan can be used to generate serotonin and KYN. (B) Relative mRNA levels of IDO1. Total RNAs extracted from these cells were reverse-transcribed and subsequently used as template for real-time quantitative PCR. GAPDH was used as internal control. The results shown in the graph were calculated with the ΔΔCt method by setting the IDO1 mRNA level of LL24 cells as 1 (*p=0.005, **p=0.01). (C) Immunoblot of lung cancer cell lines treated with and without IFNγ (20ng/ml) for 24h. No significant differences in IDO1 protein expression levels were observed between parental and CR cells. (D) Significantly higher IDO1 activity was found in CR cells when compared to parental cell counterparts (*p=0.02, **p=0.03) and was further enhanced with IFNγ (20ng/ml) treatment for 24h (***p=0.03, ****p=0.008).
Our data strongly support the concept that the level of IDO1 expression alone may not be the crucial factor in determining its activity. Consequently, these results may explain why many studies have shown insignificant correlation between IDO1 expressions and clinic-pathological parameters.
CR cells were hypersensitive to IDO inhibitors.
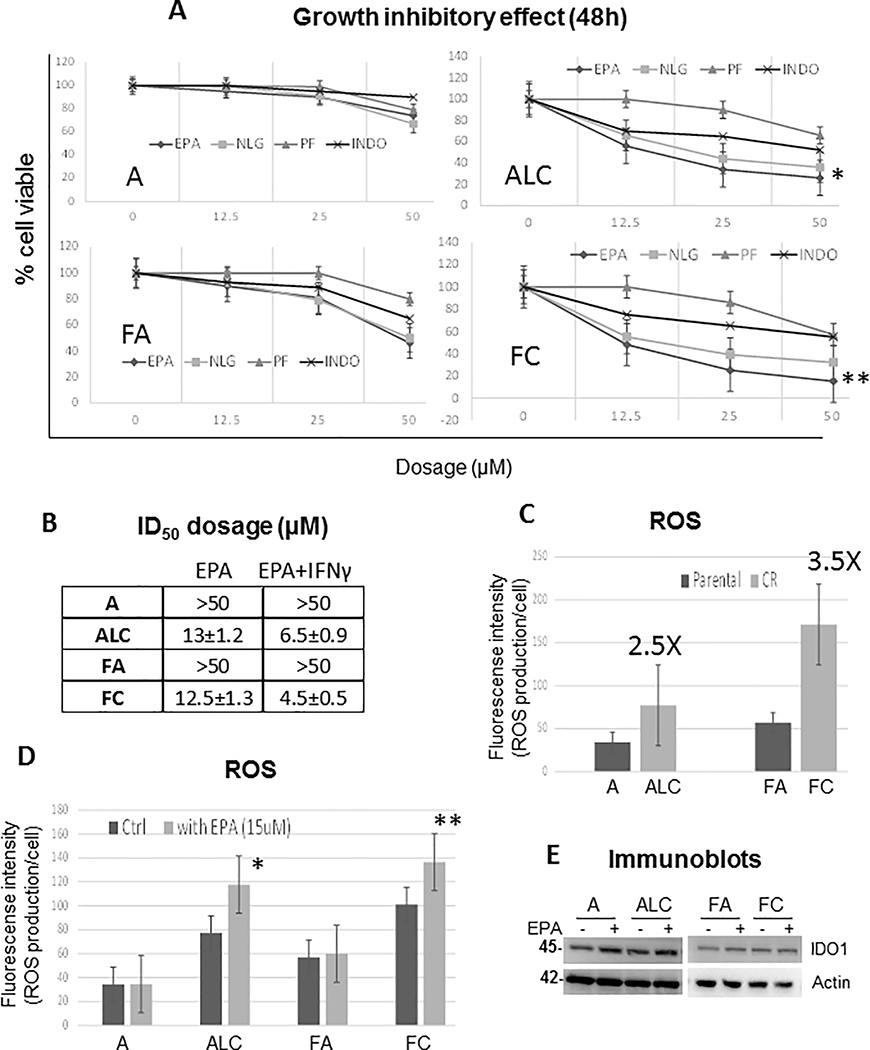
To determine that IDO1 plays an important role in CR cell proliferation and survival, we assayed for growth inhibitory effects using commercially available IDO1 inhibitors (Epacadostat, PF-06840003, and NLG-919), as well as indoximod which targets IDO2 (38). Only CR cells were hypersensitive to IDO1 inhibitors with Epacadostat (EPA) showing to have the best efficacy for growth inhibition at 12μM±0.5 (Figure 5A). In addition, IFNγ treatment further enhanced the cytotoxicity of EPA only in CR cells (Figure 5B) providing additional validation of CR cell’s dependence on IDO. Henceforth, we selected the use of EPA to carry out the subsequent experiments of IDO1 inhibition. We then determined the basal level of ROS in parental vs. CR cells. ALC and FC cells possessed 2.5 and 3.5 fold higher basal level of ROS (Figure 5C, respectively), and inhibiting IDO1 led to significantly higher ROS accumulation only in CR cells (Figure 5D). It is noteworthy that EPA did not affect IDO1 protein level as determined by Western blot analysis (Figure 5E), suggesting that EPA inhibits IDO1 enzymatic activity and not its expression in cells.
Figure 5.
Antitumor activity of IDO inhibitors in CR cells. (A) Growth inhibitory effect of various IDO inhibitors (Epacadostat: EPA, NLG-919: NLG, PF-06840003: PF, or Indoximod: INDO) for 48hrs showed that CR cells were sensitive to IDO1 inhibitors with EPA yielding the best efficacy (*p=0.03, **p=0.02). (B) ID50 dosage of EPA alone and in combination with 20ng/ml of IFNγ. Combination treatment enhance EPA toxicity only in CR cells. (Mean SD of three experiments, 48h). (C) ROS analysis detected by CM-H2DCFDA probe indicated that CR cells expressed higher basal levels of ROS. (D) ROS levels were heightened when treated with 15μM of EPA for 48hrs (*p=0.009, **p=0.005). Bar graph represents the relative fluorescent units/cell via fluorometer plate reader. (E) Immunoblot of lung cancer cell lines treated with and without EPA (15μM/ml) for 48h.
Sensitivity to IDO1 inhibitor is correlated with higher ROS levels.
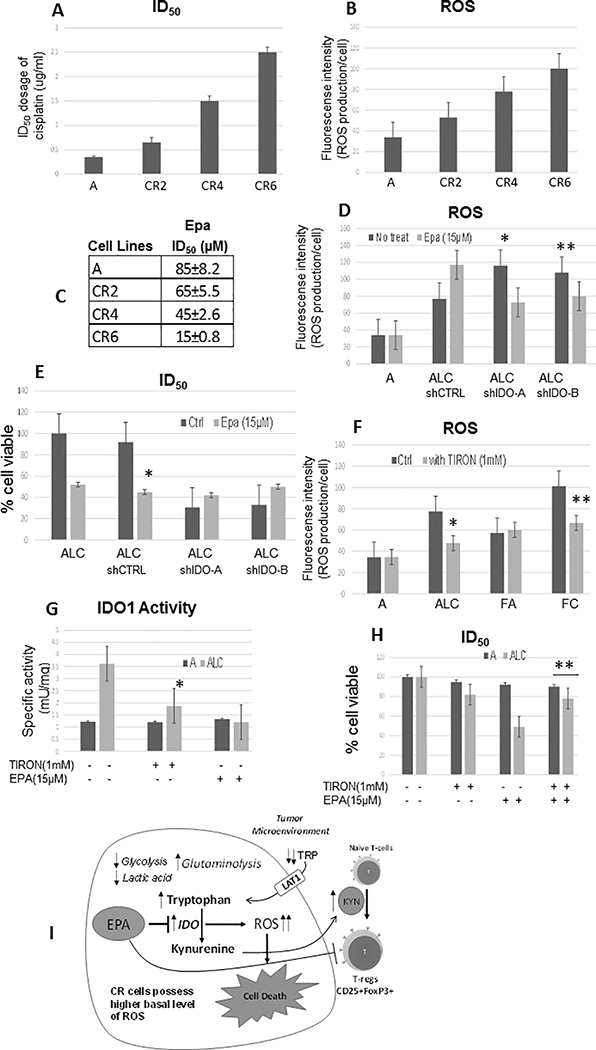
It is also noteworthy that IFNγ raised IDO1 protein expression slightly but not significantly in both parental and CR cell counterparts. We postulate here that the expression levels of IDO1 alone may not be a sole indicator of IDO1 inhibitor sensitivity, and the efficacy of IDO1 inhibition may depend on ROS levels. In further support of this notion, we found that a stepwise increase in cisplatin resistance (Figure 6A) is correlated with the progressive increase in ROS production (Figure 6B), and escalating sensitivity to EPA (Figure 6C). These findings suggested that when cancer cells acquired greater resistance to cisplatin, ROS production also progressively increased. Consequently, activation of KP is amplified leading to increasing sensitivity to IDO1 inhibitor.
Figure 6.
Sensitivity to IDO1 inhibitor is dependent with higher ROS levels. (A) ID50 dosage of cisplatin in parental cell line “A” and its cisplatin resistance variants. (B) Intracellular ROS production measured by fluorescence intensity using CM-H2DCFDA probe. (C) ID50 dosage of EPA and its cisplatin resistance variants. CR2, CR4, and CR6 possessed 2, 4, and 6 fold resistance to cisplatin, respectively (Mean SD of three experiments, 48h). (D) Knocking down of IDO1 (shIDO-A and shIDO-B) further enhanced ROS level in CR cells (*p=0.02,**p=0.03); however, treatment with 15μM of EPA for 48hrs did not produce additional ROS accumulation in ALCshIDO. (E) Growth inhibitory effect of EPA (48hrs) on ALC and ALCshIDO. EPA significantly suppressed ALC cells’ growth (*P=0.002), and no significant growth inhibition were observed in the knocked down clones. (F) Intracellular ROS production. Antioxidant (TIRON) suppressed ROS production in CR cells (*p=0.003, **p=0.006). (G) Treatment of TIRON (48hrs) attenuated IDO1 activity (*p=0.03) in CR cells. (H) TIRON treatment conferred resistance to EPA in CR cells (48h; **p=0.02). (I) CR cells do not primarily utilize glucose, but rather consume amino acids such as glutamine and tryptophan for survival. This metabolic switch is due to increased ROS production hyper-activating kynurenine pathway (KP) to balance oxidative stress and maintain cellular growth and proliferation. Kynurenine (KYN) is oxidized through indoleamine 2,3-dioxygenase (IDO), and plays a key role in reprogramming naïve T-cells to the immune suppressive regulatory T-cell (T-reg) phenotype. Further increase in ROS by interfering with tumor metabolism via IDO1 inhibiting will selectively target these cisplatin resistant lung cancer cells.
To establish the relationship between IDO1 and ROS, in our lung cancer and CR models, we inhibited IDO1 using shRNA as previously shown (Figure 2F). As we expected, knock down (shIDO1) enhanced ROS production in ALCshIDOA&B since IDO metabolizes ROS as a substrate (Figure 6D). The addition of EPA to inhibit IDO only increased ROS in shCTRL and found no further effect in the absence of IDO (ALCshIDOA&B), consistent with the specific modulation of ROS by IDO in these cells. More importantly, silencing IDO suppressed CR cell growth which further implicated the important role of KP in proliferation of CR tumors (Figure 6E). Moreover, the chemical inhibition of IDO with EPA in already knocked down cells did not result in further cell death indicating the specificity of the inhibitor for the IDO1 enzyme. Exposing CR cells to 1mM of TIRON (mitochondria-localized antioxidant; (39)), resulted in significant suppression of ROS production (Figure 6F). Importantly, treatment of TIRON decreased IDO1 activity (Figure 6G) and conferred resistance to EPA (Figure 6H) in CR cells. Taken together, our results indicated that the IDO1 activity, as well as sensitivity to IDO1 inhibitor were dependent on ROS levels.
DISCUSSION
Over the past few years, we have discovered that metabolic reprogramming in CR cells contributes to various mechanisms of resistance to treatment (2, 7). Among these metabolic changes is the use of glutamine and fatty acids by CR cells as a carbon skeleton source in metabolism instead of glucose (2, 7). Another important finding was the detection of significantly higher basal levels of ROS accumulation in proliferating CR cells (3). Here, we have uncovered a unique metabolic difference wherein CR cells possess higher basal levels of ROS which in turn activates the kynurenine pathway (KP) for tryptophan (TRP) catabolism. Importantly, KP generates kynurenine (KYN), which has been shown to reprogram naïve T-cells to the immune suppressive regulatory T-cell (T-reg) phenotype (Figure Diagram 6I).
Since ROS is a substrate for IDO1 in the oxidation reactions yielding KYN, in this study we inhibited IDO1 (i.e. IDO inhibitor (EPA) or shIDO) leading to higher ROS (Figure 6). This induction in ROS accumulation pushed CR cells beyond a threshold of ROS tolerance resulting in greater growth inhibition. Moreover, EPA did not suppress IDO1 protein expression levels, suggesting that EPA inhibits IDO1 enzymatic activity and not its expression in cells. Nevertheless, these results clearly illustrate that survival of CR cells can be influenced by changes in ROS levels.
To further investigate the specific role of TRP in CR cells, we investigated the production of KYN and NAD+. As described above, TRP is oxidized through IDO1 to generate KYN. Here, we reported that CR cells consumed TRP at a higher rate and generated more KYN, but not NAD+. In fact, we have also previously reported that NMNAT2 (a cytosolic enzyme that represents the final step in the biosynthesis of NAD+ in both de novo and salvage pathways) expression was significantly lower in CR cells (2). These findings strongly confirm that TRP is being used as a precursor for KYN production in CR cells. The TRP consumption rate can also be utilized to stratify tumors based on this metabolic attribute.
Increased amino acid transport (via evaluation of LAT1 (CD98)), has also been reported in many types of cancers including lung cancer, providing some insight into molecular modifications in tissue metabolism along tumor development (40, 41). In fact, all our CR cells possess higher LAT1 levels when compared to parental cell counterparts, and treatment with the IDO1 inhibitor suppressed LAT1 expression. Interestingly, we previously reported that CR cells possessed higher expression of the glutamate/cystine/xCT anti-transporter pump (2), and LAT1 is a major component of this plasma membrane antiporter. LAT1 facilitates the cellular uptake of extracellular cystine in exchange for intracellular glutamate and plays a key role in glutathione (GSH) synthesis. The activity of CD98/xCT-mediated cystine uptake in cancer cells is known to be highly associated with tumor growth and chemo-resistance (42, 43). Thus, the important role of LAT1 in altered tumor metabolism is not limited to TRP uptake, but also extends to the homeostatic regulation of antioxidant and ROS.
Interestingly, we did not find an increase in LAT1 levels in parental cells after KYN treatment, even though more AHR was translocated into nucleus. This may due to the fact that parental cells possessed significantly higher level of HIF1α which has a much better binding affinity to HIF1β than AHR (31, 44). Thus, in the presence of HIF1α, ARNT (HIF1β) is not readily available to bind and form a complex with KYN/AHR. In addition, we have previously reported that CR cells secrete less lactate (2, 7) which may very well impact the fate of T-cells. Lactic acid can be taken up by neighbor cells, including immune cells, to perpetuate the glycolytic pathway and upregulate HIF1α (45, 46). In fact, effector T-cells (T-eff) rely on glycolysis to expand and this is growth is greatly dependent on HIF1α (47). A diminished amount of lactate may prevent a robust immune response to the neighboring tumor. Furthermore, mice deficient in HIF1α fail to mount a strong T-cell response and have increased T-reg populations (48, 49). All of these findings support a concept wherein the metabolic modifications in CR tumors provide a growth advantage which in turn impacts immune cells in the tumor microenvironment.
While high IDO expression is a suggested predictor for poor prognosis and not associated with improved survival (50–52), it is often expressed in late stage lung tumors with no study conducted in CR tumors (Supplementary Figure S5). In addition, many studies have reported no significant correlation between IDO1 expression and clinico-pathological parameters (53, 54), although the concept of potential therapeutic efficacy for IDO1 inhibitors has been promoted (47). But the higher basal levels of ROS we have reported in CR cells will lead to a higher efficacy of the IDO1 inhibitor to decrease the level of KYN. Our studies have evaluated the metabolic background of these tumors, adding another novel level of molecular effectors that can influence IDO1 activity and any acquired resistance to the immune response. Thus, we believe that IDO1 expression level alone should not be used as a sole factor in the determination of clinical benefit/tumor response to IDO1 inhibitor.
Activation of IDO1 enzymatic activity requires ROS as a co-factor, hence activity along with expression should be evaluated in the clinic. In previous work, we reported that parental (or naïve treatment) cancer cells can upregulate both GSH and thioredoxin-1 (TRX1) antioxidant activities to keep ROS in check where CR cells cannot (3). Therefore, in the presence of active antioxidants, our findings here may explain why less IDO1 activity was found in parental tumors despite the expression of IDO1 protein. As for IDO2, a recent report showed that indoximod, which acts downstream of IDO1/2 to suppress mTORC1 activity, exerted a variety of benefits as an immuno-metabolic adjuvant (55). This is interesting since we have previously found that certain CR cells possessed higher mTORC1 activities, and inhibiting mTOR with CCI-779 can restore cisplatin sensitivity (23) in these CR cells. In the future, we plan to investigate the role of IDO2 in CR tumors by inhibiting both IDO2 and mTOR in our tumor models.
A recent phase III ECHO-301/KEYNOTE-252 (NCT02752074) clinical trial tested the efficacy of EPA in combination with pembrolizumab in patients with advanced melanoma previously untreated with PD-1 or PD-L1 checkpoint inhibitors (56). The trial evaluated the potential enhancement of anti-PD-1 therapy via IDO1 inhibition by gathering data on progression-free survival, overall survival, and adverse events but did not yield satisfactory results and was stopped early showing no indication that epacadostat provided an increased benefit. Various limitations to the study parameters have been proposed such as the degree of adequate IDO1 inhibition within the tumor based on dosage and through sufficient drug exposure, use of combination therapies with DNA damaging modalities, or the use of IDO and TDO inhibition including dual inhibitors and AHR inhibitors (57). Perhaps future studies of such IDO and other inhibitors should include the evaluation of some metabolic factors such as elevated catabolism of TRP that affect the tumor microenvironment and the suppression of immunity (58). Here in we have gathered specific evidence to describe the effects of IDO, ROS, the KYN/TRP ratio in lung mediated modifications of the tumor microenvironment and the potential suppression of antitumor immunity. We believe that higher ROS levels together with significantly increased serum KYN/TRP ratios can be used to select patients for future treatment using IDO1 inhibitors. Such a stratification or patient grouping may increase efficacy in EPA trials. It is also noteworthy that even though cancer patients from our study possessed higher basal levels of KYN/TRP ratio when compared to healthy controls, these levels were negligible when compared to patients who failed cisplatin and radiation therapy. Thus, the question remains on how high is high enough to use the KYN/TRP ratio as a biomarker for predicting the efficacy of IDO inhibitors. Our work has some limitations with only six patients, but we are currently recruiting more patients to our study who have failed cisplatin and radiation therapy. In the context of a secondary treatment compendium based on IDO inhibition, the KYN/TRP ratio may simultaneously signal a need for a shift away from cisplatin therapy for some patients, in anticipation of the development of resistance. We firmly believe that IDO inhibitors may be more beneficial in the treatment of lung cancer patients who failed cisplatin therapy than naïve treatment patients. The data presented herein, provide a more comprehensive molecular investigation of metabolic pathway components in lung tumor cells that are resistant to cisplatin, characterizing variations that are proving to be valuable in survival mechanisms in resistant lung cancer cells.
Supplementary Material
Implications.
Findings suggest that the enzyme inhibitory activity and anti-tumor efficacy of IDO1 inhibitors rely in part on ROS levels, arguing that IDO1 expression alone may be insufficient to determine the clinical benefits for this class of experimental cancer drugs. Importantly, IDO1 inhibitors maybe more suitable to treat lung cancer patients who failed cisplatin therapy than naïve treatment patients.
ACKNOWLEDGEMENTS
The authors thank Drs. Mustafa Tekin and Jingyu Huang (Division of Clinical and Translational Genetics, University of Miami) for helping us with amino acid levels analysis. This work is supported by BLR&D Merit Review Award (1BX003328) to Savaraj N; by BLR&D Career Development Award-2 (1K2BX001289) and Merit Review Award (1I01BX004371) to Wangpaichitr M.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Lim SH, Sun JM, Lee SH, Ahn JS, Park K, Ahn MJ. Pembrolizumab for the treatment of non-small cell lung cancer. Expert opinion on biological therapy. 2016;16(3):397–406. [DOI] [PubMed] [Google Scholar]
- 2.Wangpaichitr M, Wu C, Li YY, Nguyen DJM, Kandemir H, Shah S, et al. Exploiting ROS and metabolic differences to kill cisplatin resistant lung cancer. Oncotarget. 2017;8(30):49275–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wangpaichitr M, Wu C, You M, Maher JC, Dinh V, Feun LG, et al. N1,N3-Dimethyl-N1,N3-bis(phenylcarbonothioyl) Propanedihydrazide (Elesclomol) Selectively Kills Cisplatin Resistant Lung Cancer Cells through Reactive Oxygen Species (ROS). Cancers. 2009;1(1):23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warburg O On the origin of cancer cells. Science. 1956;123(3191):309–14. [DOI] [PubMed] [Google Scholar]
- 5.Warburg O On respiratory impairment in cancer cells. Science. 1956;124(3215):269–70. [PubMed] [Google Scholar]
- 6.Wangpaichitr M, Wu CJ, Li Y, Nguyen DJM, Shah S, Kandemir H, et al. Exploiting ROS and Metabolic Differences to Kill Cisplatin Resistant Lung Cancer Oncotarget. 2017:Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wangpaichitr M, Sullivan EJ, Theodoropoulos G, Wu C, You M, Feun LG, et al. The relationship of thioredoxin-1 and cisplatin resistance: its impact on ROS and oxidative metabolism in lung cancer cells. Mol Cancer Ther. 2012;11(3):604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long Y, Tsai WB, Chang JT, Estecio M, Wangpaichitr M, Savaraj N, et al. Cisplatin-induced synthetic lethality to arginine-starvation therapy by transcriptional suppression of ASS1 is regulated by DEC1, HIF-1alpha, and c-Myc transcription network and is independent of ASS1 promoter DNA methylation. Oncotarget. 2016;7(50):82658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters JC. Tryptophan nutrition and metabolism: an overview. Adv Exp Med Biol. 1991;294:345–58. [DOI] [PubMed] [Google Scholar]
- 10.Prendergast GC, Malachowski WJ, Mondal A, Scherle P, Muller AJ. Indoleamine 2,3-Dioxygenase and Its Therapeutic Inhibition in Cancer. Int Rev Cell Mol Biol. 2018;336:175–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colabroy KL, Begley TP. Tryptophan catabolism: identification and characterization of a new degradative pathway. Journal of bacteriology. 2005;187(22):7866–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badawy AA. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int J Tryptophan Res. 2017;10:1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Baren N, Van den Eynde BJ. Tryptophan-degrading enzymes in tumoral immune resistance. Front Immunol. 2015;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lob S, Konigsrainer A, Zieker D, Brucher BL, Rammensee HG, Opelz G, et al. IDO1 and IDO2 are expressed in human tumors: levo- but not dextro-1-methyl tryptophan inhibits tryptophan catabolism. Cancer Immunol Immunother. 2009;58(1):153–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nevler A, Muller AJ, Sutanto-Ward E, DuHadaway JB, Nagatomo K, Londin E, et al. Host IDO2 Gene Status Influences Tumor Progression and Radiotherapy Response in KRAS-Driven Sporadic Pancreatic Cancers. Clin Cancer Res. 2019;25(2):724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis I, Liu A. What is the tryptophan kynurenine pathway and why is it important to neurotherapeutics? Expert Rev Neurother. 2015;15(7):719–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosell FI, Kuo HH, Mauk AG. NADH oxidase activity of indoleamine 2,3-dioxygenase. J Biol Chem. 2011;286(33):29273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platten M, von Knebel Doeberitz N, Oezen I, Wick W, Ochs K. Cancer Immunotherapy by Targeting IDO1/TDO and Their Downstream Effectors. Front Immunol. 2014;5:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton R, Priyadharshini B, Turka LA. Immunometabolism of regulatory T cells. Nat Immunol. 2016;17(6):618–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Huang L, Jin JY, Jolly S, Zang Y, Wu H, et al. IDO Immune Status after Chemoradiation May Predict Survival in Lung Cancer Patients. Cancer Res. 2018;78(3):809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladomersky E, Zhai L, Lenzen A, Lauing KL, Qian J, Scholtens DM, et al. IDO1 Inhibition Synergizes with Radiation and PD-1 Blockade to Durably Increase Survival Against Advanced Glioblastoma. Clin Cancer Res. 2018;24(11):2559–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wangpaichitr M, Wu C, You M, Kuo MT, Feun L, Lampidis T, et al. Inhibition of mTOR restores cisplatin sensitivity through down-regulation of growth and anti-apoptotic proteins. Eur J Pharmacol. 2008;591(1–3):124–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C, Wangpaichitr M, Feun L, Kuo MT, Robles C, Lampidis T, et al. Overcoming cisplatin resistance by mTOR inhibitor in lung cancer. Mol Cancer. 2005;4(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savaraj N, Wu C, Wangpaichitr M, Kuo MT, Lampidis T, Robles C, et al. Overexpression of mutated MRP4 in cisplatin resistant small cell lung cancer cell line: collateral sensitivity to azidothymidine. Int J Oncol. 2003;23(1):173–9. [PubMed] [Google Scholar]
- 25.Xu J, Tyan T, Cedrone E, Savaraj N, Wang N. Detection of 11q13 amplification as the origin of a homogeneously staining region in small cell lung cancer by chromosome microdissection. Genes Chromosomes Cancer. 1996;17(3):172–8. [DOI] [PubMed] [Google Scholar]
- 26.Wangpaichitr M, Savaraj N, Maher J, Kurtoglu M, Lampidis TJ. Intrinsically lower AKT, mammalian target of rapamycin, and hypoxia-inducible factor activity correlates with increased sensitivity to 2-deoxy-D-glucose under hypoxia in lung cancer cell lines. Mol Cancer Ther. 2008;7(6):1506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292(1):C125–36. [DOI] [PubMed] [Google Scholar]
- 28.Henderson LM, Gross CJ. Metabolism of niacin and niacinamide in perfused rat intestine. The Journal of nutrition. 1979;109(4):654–62. [DOI] [PubMed] [Google Scholar]
- 29.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511(7508):184–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–34. [DOI] [PubMed] [Google Scholar]
- 31.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12(12):5447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pongratz I, Stromstedt PE, Mason GG, Poellinger L. Inhibition of the specific DNA binding activity of the dioxin receptor by phosphatase treatment. J Biol Chem. 1991;266(25):16813–7. [PubMed] [Google Scholar]
- 33.Fukasawa Y, Segawa H, Kim JY, Chairoungdua A, Kim DK, Matsuo H, et al. Identification and characterization of a Na(+)-independent neutral amino acid transporter that associates with the 4F2 heavy chain and exhibits substrate selectivity for small neutral D- and L-amino acids. J Biol Chem. 2000;275(13):9690–8. [DOI] [PubMed] [Google Scholar]
- 34.Yanagida O, Kanai Y, Chairoungdua A, Kim DK, Segawa H, Nii T, et al. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochim Biophys Acta. 2001;1514(2):291–302. [DOI] [PubMed] [Google Scholar]
- 35.Chen ML, Pittet MJ, Gorelik L, Flavell RA, Weissleder R, von Boehmer H, et al. Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-beta signals in vivo. Proc Natl Acad Sci U S A. 2005;102(2):419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. [DOI] [PubMed] [Google Scholar]
- 37.Mbongue JC, Nicholas DA, Torrez TW, Kim NS, Firek AF, Langridge WH. The Role of Indoleamine 2, 3-Dioxygenase in Immune Suppression and Autoimmunity. Vaccines (Basel). 2015;3(3):703–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metz R, Duhadaway JB, Kamasani U, Laury-Kleintop L, Muller AJ, Prendergast GC. Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound D-1-methyl-tryptophan. Cancer Res. 2007;67(15):7082–7. [DOI] [PubMed] [Google Scholar]
- 39.Oyewole AO, Birch-Machin MA. Mitochondria-targeted antioxidants. FASEB J. 2015;29(12):4766–71. [DOI] [PubMed] [Google Scholar]
- 40.Tomblin JK, Arthur S, Primerano DA, Chaudhry AR, Fan J, Denvir J, et al. Aryl hydrocarbon receptor (AHR) regulation of L-Type Amino Acid Transporter 1 (LAT-1) expression in MCF-7 and MDA-MB-231 breast cancer cells. Biochem Pharmacol. 2016;106:94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai H, Kaira K, Oriuchi N, Yanagitani N, Sunaga N, Ishizuka T, et al. L-type amino acid transporter 1 expression is a prognostic marker in patients with surgically resected stage I non-small cell lung cancer. Histopathology. 2009;54(7):804–13. [DOI] [PubMed] [Google Scholar]
- 42.Lo M, Wang YZ, Gout PW. The x(c)- cystine/glutamate antiporter: a potential target for therapy of cancer and other diseases. J Cell Physiol. 2008;215(3):593–602. [DOI] [PubMed] [Google Scholar]
- 43.Lo M, Ling V, Wang YZ, Gout PW. The xc- cystine/glutamate antiporter: a mediator of pancreatic cancer growth with a role in drug resistance. Br J Cancer. 2008;99(3):464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270(3):1230–7. [DOI] [PubMed] [Google Scholar]
- 45.Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118(12):3930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cascone T, McKenzie JA, Mbofung RM, Punt S, Wang Z, Xu C, et al. Increased Tumor Glycolysis Characterizes Immune Resistance to Adoptive T Cell Therapy. Cell metabolism. 2018;27(5):977–87 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14(11):1173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schalper KA, Carvajal-Hausdorf D, McLaughlin J, Altan M, Velcheti V, Gaule P, et al. Differential Expression and Significance of PD-L1, IDO-1, and B7-H4 in Human Lung Cancer. Clin Cancer Res. 2017;23(2):370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theate I, van Baren N, Pilotte L, Moulin P, Larrieu P, Renauld JC, et al. Extensive profiling of the expression of the indoleamine 2,3-dioxygenase 1 protein in normal and tumoral human tissues. Cancer Immunol Res. 2015;3(2):161–72. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, Shen Z, Wang Z, Wang X, Zhang H, Qin J, et al. Increased expression of IDO associates with poor postoperative clinical outcome of patients with gastric adenocarcinoma. Sci Rep. 2016;6:21319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karanikas V, Zamanakou M, Kerenidi T, Dahabreh J, Hevas A, Nakou M, et al. Indoleamine 2,3-dioxygenase (IDO) expression in lung cancer. Cancer Biol Ther. 2007;6(8):1258–62. [DOI] [PubMed] [Google Scholar]
- 54.Gao YF, Peng RQ, Li J, Ding Y, Zhang X, Wu XJ, et al. The paradoxical patterns of expression of indoleamine 2,3-dioxygenase in colon cancer. J Transl Med. 2009;7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fox E, Oliver T, Rowe M, Thomas S, Zakharia Y, Gilman PB, et al. Indoximod: An Immunometabolic Adjuvant That Empowers T Cell Activity in Cancer. Front Oncol. 2018;8:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long GV, Dummer R, Hamid O, Gajewski TF, Caglevic C, Dalle S, et al. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase 3, randomised, double-blind study. Lancet Oncol. 2019. [DOI] [PubMed] [Google Scholar]
- 57.Muller AJ, Manfredi MG, Zakharia Y, Prendergast GC. Inhibiting IDO pathways to treat cancer: lessons from the ECHO-301 trial and beyond. Semin Immunopathol. 2019;41(1):41–8. [DOI] [PubMed] [Google Scholar]
- 58.Lemos H, Huang L, Prendergast GC, Mellor AL. Immune control by amino acid catabolism during tumorigenesis and therapy. Nature reviews. 2019;19(3):162–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.