Abstract
The emerging pharmacological target soluble epoxide hydrolase (sEH) is a bifunctional enzyme exhibiting two different catalytic activities that are located in two distinct domains. Although the physiological role of the C-terminal hydrolase domain is well-investigated, little is known about its phosphatase activity, located in the N-terminal phosphatase domain of sEH (sEH-P). Herein we report the discovery and optimization of the first inhibitor of human and rat sEH-P that is applicable in vivo. X-ray structure analysis of the sEH phosphatase domain complexed with an inhibitor provides insights in the molecular basis of small-molecule sEH-P inhibition and helps to rationalize the structure−activity relationships. 4-(4-(3,4-Dichlorophenyl)-5-phenyloxazol-2-yl)butanoic acid (22b, SWE101) has an excellent pharmacokinetic and pharmacodynamic profile in rats and enables the investigation of the physiological and pathophysiological role of sEH-P in vivo.
Graphical Abstract

INTRODUCTION
Homodimeric soluble epoxide hydrolase (sEH) consists of two functionally different domains that are connected by a flexible proline-rich linker.1 The C-terminal domain catalyzes the conversion of epoxyeicosatrienoic acids (EETs) and related epoxy fatty acids (EpFAs) to the corresponding α,β-diols. The role of this hydrolase activity of sEH is well-studied, and inhibitors of the C-terminal domain (sEHIs) hold promising therapeutical potential for various diseases, including diabetes, fibrosis, chronic pain, and cardiovascular and neurodegenerative diseases.2 The very broad therapeutical scope of sEHIs is based on the hypothesis that while an accumulation of EpFAs has no or little effect under physiological conditions, they can prevent mitochondrial dysfunction and reduce subsequent reactive oxygen species (ROS) formation and endoplasmic reticulum stress conditions.3 Accordingly, sEHIs targeting the hydrolase domain are in the focus of drug discovery, and two candidates have already reached clinical development.2,4
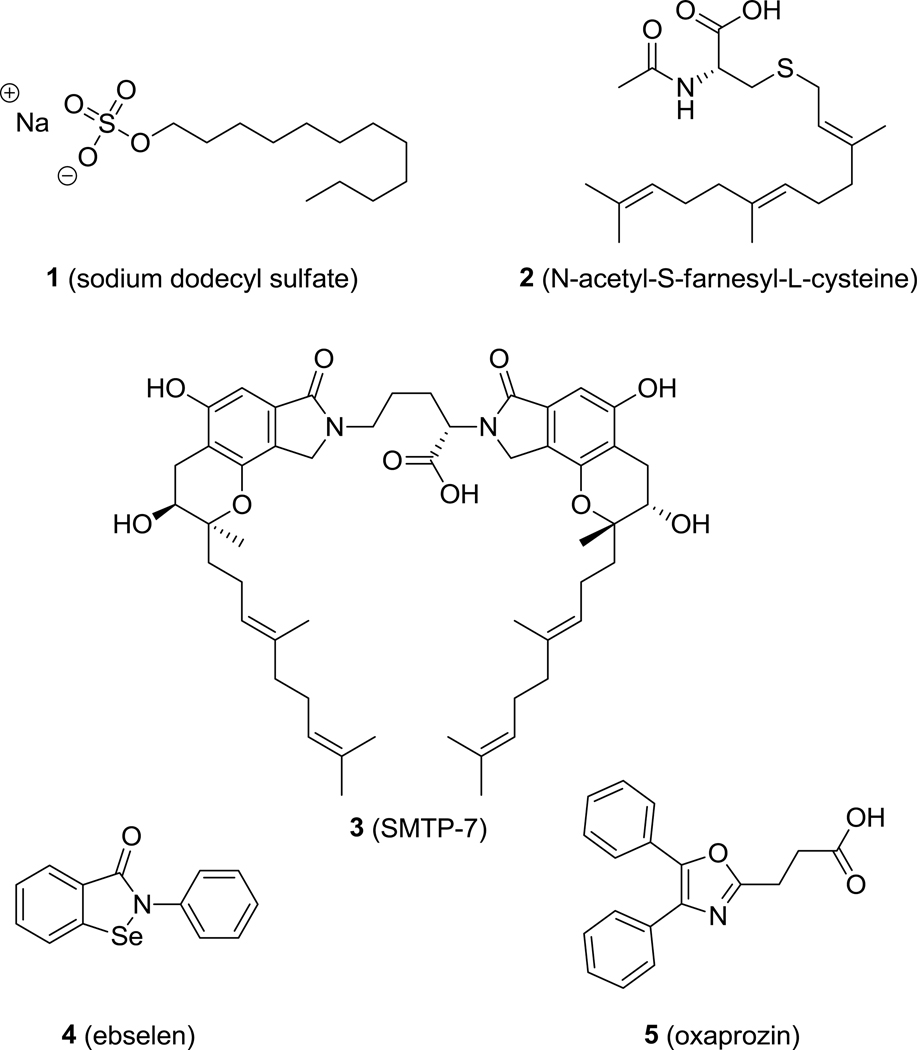
In contrast to the well-understood function of the C-terminal hydrolase domain, the role of the N-terminal phosphatase domain remains unclear. In 2003, Cronin et al.5 and Newman et al.6 independently reported the phosphatase activity of the N-terminal domain of sEH (sEH-P). In vitro, sEH-P is able to hydrolyze diverse lipid phosphates, including farnesyl pyrophosphate,7 sphingosine-1-phosphate, and lysophosphatidic acid.8,9 The mechanism of lipid phosphate hydrolysis is well-investigated.10 However, neither the endogenous substrate nor its physiological and possibly pathophysiological role have yet been identified.11 Several molecules with sEH-P inhibitory activity have been identified to date (Figure 1). Sodium dodecyl sulfate (1, SDS)7 and N-acetyl-S-farnesyl-L-cysteine (2)12,13 are lipid-like inhibitors of sEH-P. SMTP-7 (3) is an investigational thrombolytic drug for treatment of ischemic stroke that simultaneously inhibits both the hydrolase and phosphatase activities of sEH.14 The development of high-throughput screening assays for sEH-P led to the discovery of ebselen (4)15 and oxaprozin (5)16 as novel sEH-P inhibitors. While 4 also affects the sEH hydrolase activity, 5 is a selective but weak inhibitor of sEH-P (IC50 = 5μM). In this study, we investigated the structure–activity relationships (SARs) of oxazole-based sEH-P inhibitors using the nonsteroidal antirheumatic drug 5 as a starting point.
Figure 1.
Structures of previously identify sEH-P inhibitors 1–5.
RESULTS AND DISCUSSION
Chemistry.
The 2,4,5-trisubstituted oxazole scaffold of compound 5 is accessible via Blümlein–Lewy synthesis or starting from α-acyloxy ketones. In both methods, an α-bromodeoxybenzoin or benzoin derivative can be employed as the starting material to generate the heterocycle. However, crossed benzoin derivatives with two different aryl moieties are not easily accessible, whereas asymmetric deoxybenzoins can be obtained from O-protected cyanohydrins, as reported by Stork et al.,17 and are subsequently converted into α-bromoketones. This approach allows very flexible alteration of the oxazole scaffold at all three substituent positions and enables a comprehensive SAR analysis.
To investigate the importance of the carbon in the alkyl linker of 5, thioglycolic acid derivative 9 was synthesized in two steps via the reaction of 4,5-diphenyloxazole-2-thiol (6) and methyl 2-bromoacetate (7) to give methyl ester 8, which was subsequently hydrolyzed (Scheme 1).
Scheme 1a.
aReagents and conditions: (i) Et3N, THF, rt, 24 h; (ii) KOH, MeOH/H2O/THF, 70 °C, 15 min μw.
Amide derivative 12 was obtained from the reaction of commercially available α-bromodeoxybenzoin (10) and succinamic acid (11) under basic conditions to afford an α-acyloxy ketone, which under heating in acetic acid in the presence of excess of ammonium acetate cyclized to give oxazole 12 (Scheme 2).
Scheme 2a.
aReagents and conditions: (i) 1. Et3N, acetonitrile, 45 °C, 6 h; 2. NH4OAc, AcOH, reflux, 3 h.
The synthesis of oxaprozin derivatives 20a–m, with alterations in the 4-phenyl moiety of the oxazole core structure, started from benzaldehydes 13a–m, as shown in Scheme 3. The substitution strategy followed the Topliss optimization scheme.18 Benzaldehydes 13a−m were treated with trimethylsilyl cyanide (TMSCN, 14) in the presence of a catalytic amount of triethylamine under neat conditions to yield silylprotected cyanohydrins 15a–m. As described by Stork et al.,17 O-protected cyanohydrins are easily deprotonated at the α-position by LDA in THF to obtain stable nucleophiles at low temperatures, which can react with an electrophile, in this case benzyl bromide (16). The desired deoxybenzoins 17a–m were subsequently obtained by cleaving the remaining silyl ether group of the intermediate with tetrabutylammonium fluoride (TBAF) solution. Deoxybenzoin derivatives 17a–m were then transformed to the appropriate α-bromodeoxybenzoins 18a–m under heating with bromine in dichloromethane. The respective α-bromodeoxybenzoins 18a–m were subsequently reacted with succinic acid (19) under basic conditions to produce α-acyloxy ketones, which under heating in acetic acid in the presence of excess of ammonium acetate yielded oxaprozin derivatives 20a–m.
Scheme 3a.
aReagents and conditions: (i) Et3N, 24 h; (ii) 1. LDA, THF, −78 °C → rt, 24 h; 2. TBAF, 30 min; (iii) Br2, CHCl3, reflux, 2 h; (iv) 1. Et3N, MeCN, 45 °C, 6 h; 2. NH4OAc, AcOH, reflux, 3 h.
The synthesis of oxaprozin derivatives 22a–e with alterations of the oxazole 2-substituent is depicted in Scheme 4. Starting from intermediate 18l, 22a–e were obtained by the same strategy as described above for oxaprozin derivatives 20a–m using dicarboxylic acids 21a–e with different lengths and rigidities.
Scheme 4a.
aReagents and conditions: (i) 1. 21a–e, Et3N, MeCN, 45 °C, 6 h; 2. NH4OAc, AcOH, reflux, 3 h.
For alteration of the 5-phenyl moiety of the oxazole ring, a different synthetic approach was chosen to reduce the synthetic effort. The adjusted strategy (Scheme 5) employed the reaction of 3,4-dichlorophenylacyl bromide (23) and glutaric acid (21b) under basic conditions to obtain α-acyloxy ketone 24, which in turn reacted under heating in BF3·Et2O in the presence of excess acetamide to form oxazole 25. The oxazole 5-position was then activated for the subsequent Suzuki cross-coupling by the introduction of bromine using NBS in acetonitrile, yielding key intermediate 26 for final modifications. Oxazole 26 was successfully coupled with the appropriate boronic acid derivatives 27a–k in DMF/water with tetrakis(triphenylphosphine)palladium(0) as the catalyst and potassium phosphate as the base to afford oxaprozin derivatives 28a–k.
Scheme 5a.
aReagents and conditions: (i) 21b, Et3N, acetone, TLC control; (ii) acetamide, BF3·Et2O, 140 °C, 30 h; (iii) NH4OAc, NBS, acetonitrile, rt, 1.5 h; (iv) 27a–k, K3PO4, 2:1 DMF/H2O, Pd(PPh3)4, 55–80 °C, 30–48 h.
Structure−Activity Relationships.
The exploration of the SARs of substituted oxazoles was performed using a fluorescence-based enzyme activity assay as reported previously.16 In this kinetic assay, the conversion of the fluorogenic substrate fluorescein diphosphate (FDP) by the recombinant sEH-P domain is monitored to assess the inhibitory potency of the test compounds. Bioisosteric replacement of the β-methylene bridge by sulfur (9) slightly diminished the potency, while replacement of the carboxylate by a primary amide (12) resulted in complete loss of activity (Figure 2). Replacement of the oxazole core by a pyrazole core (29) was also not tolerated.
Figure 2.
Initial diversification of 5.
The next optimization round addressed the 4-position of the oxazole ring (Table 1). Ortho-substituted phenyl moieties (as present in compounds 20a and 20b) were poorly tolerated, while meta-substituted phenyl residues (20c and 20d) retained similar potency as 5. The SAR of para-substituted phenyl residues at the 4-position of the oxazole core was more sophisticated. While fluorine (20e) only slightly diminished the potency, larger electron-withdrawing moieties such as trifluoromethyl (20f) and nitrile (20g) led to a loss of activity. A chlorine substituent at this position (20h) slightly enhanced inhibitory potency, while an electron-releasing methoxy moiety (20i) decreased the activity. Following the Topliss optimization scheme,18 a second chlorine substituent was introduced at the meta position of the phenyl ring (20k), which led to a more than 10-fold increase in sEH-P inhibition. Other variants of the chlorine substitution pattern (20l and 20m) did not lead to potency improvement.
Table 1.
Optimization of the 4-position of the oxazole core.
 |
We then addressed optimization of the oxazole 2-position to adjust the interactions of the important carboxylate moiety (Table 2). While rigidization of the flexible ethyl linker by a double bond resulted in a 2-fold loss of potency (22a), the introduction of an additional methylene (22b) further improved the inhibitory activity. Replacement of the propyl bridge by a rigid phenyl linker (22d) demonstrated that optimal interactions of the carboxylate are achieved at a distance of four bonds from the oxazole core. The conformationally restrained ortho- and para-substituted benzoic acids did not retain this high potency (22c and 22e).
Table 2.
Optimization of the linker in 2-position of the oxazole core.
 |
Finally, the impact of the phenyl substituent at the 5-position of the oxazole was investigated while the 3,4dichlorophenyl substituent at the 4-position and the propionic acid at the 2-position were maintained (Table 3). When the phenyl substituent was omitted, the potency dropped dramatically (25), but it was partially restored by introduction of a bromine substituent (26). Pyridine moieties were not favored (28a and 28b), while almost every substituent at the ortho or meta position was well-tolerated, with a slight preference for fluorine (compound 28e) or linear substituents such as nitrile (28g) or ethynyl (28j). However, no further potency improvement over 22b and 22d was achieved, rendering these oxaprozin derivatives as the most active sEH-P inhibitors for further characterization.
Table 3.
Optimization of the 5-position of the oxazole core.
 |
In Vitro Characterization of 22b and 22d.
To verify the binding of the most potent compound, 22b, to sEH-P, orthogonal biophysical assays were performed. Differential scanning fluorimetry (DSF), also known as the thermal shift assay, was conducted with the N-terminal domain and the C-terminal domain of sEH. The N-terminal domain was stabilized by 22b, while the reference inhibitor of the sEH hydrolase activity, N-cyclohexyl-N′-(4-iodophenyl)urea (CIU),19 had no stabilizing effect on the phosphatase domain (Figure 3A). A full list of the DSF experiments with additional reference inhibitors is included in the Supporting Information. In contrast, 22b did not stabilize the C-terminal domain, whose melting point was, however, significantly increased by CIU (Figure 3B). Isothermal titration calorimetry revealed tight binding of 22b to the N-terminal domain of sEH with a Kd value of 0.3 μM (Figure 3C). Analysis of the thermodynamic properties of binding revealed balanced enthalpic and entropic contributions to the binding free energy (Figure 3C inset).
Figure 3.
Biophysical characterization of 22b. (A) 22b stabilized the N-terminal domain of sEH in the differential scanning fluorimetry (DSF) assay, but the sEH hydrolase inhibitor CIU did not. (B) 22b did not stabilize the C-terminal domain of sEH, in contrast to CIU. The concentration of the inhibitors in the DSF experiments was 50 μM. The statistical significance of the measured melting points compared with the DMSO control was determined by a two-tailed t test using GraphPad Prism (version 7.05; GraphPad Software, Inc.). (C) ITC measurements of the formation of the complex between 22b and the sEH N-terminal domain. Shown are (top) raw binding heats of each injection and (bottom) normalized binding heats fitted to a single binding site model (solid line). The thermodynamic parameters of this titration experiment are shown in the inset.
Compound 5 (oxaprozin) is an approved nonselective inhibitor of cyclooxygenases 1 and 2. Therefore, the inhibitory potency of 22b toward cyclooxygenases was investigated (Table 4). 22b exhibited slightly lower potency toward both cyclooxygenase isoforms compared with 5, which inhibits COX-1 with an IC50 of 0.74 μM (29% CV) and COX-2 with an IC50 of 0.11 μM (38% CV). Furthermore, agonistic potency toward peroxisome proliferator-activated receptor gamma (PPARγ) and retinoid X receptor alpha (RXRα) was observed, which needs to be considered when using 22b in cellular assays.
Table 4.
Selectivity Screen of 22b and Oxaprozin (5) toward Diverse Targets of Fatty Acid Mimeticsa
| effect |
||
|---|---|---|
| target | 22b | 5 |
| COX-1 | IC50 = 1.2 μM (20% CV) | IC50 = 0.74 μM (29% CV) |
| COX-2 | IC50 = 0.42 μM (9.6% CV) | IC50 = 0.11 μM (38% CV) |
| PPARα | inactive at 10 μM | inactive at 10 μM |
| PPARβ/δ | inactive at 10 μM | inactive at 10 μM |
| PPARγ | 29 ± 1% at 10 μM | inactive at 10 μM |
| FXR | inactive at 10 μM | inactive at 10 μM |
| LXRα | inactive at 10 μM | inactive at 10 μM |
| LXRβ | inactive at 10 μM | inactive at 10 μM |
| RXRα | 77 ± 2% at 10 μM | 14.4 ± 0.4% at 10 μM |
All values were measured at least twice as duplicates (n ≥ 2). Values for nuclear receptors are mean ± SD% transactivation of the reference agonist; “inactive” denotes no statistically significant reporter transactivation at the indicated concentration.
In order to understand the structural basis of sEH-P inhibition by oxazole-based compounds, experiments involving cocrystallization of the sEH-P domain with oxazole inhibitors 22b and 22d were performed. While the most affine inhibitor 22b did not yield crystals, 22d cocrystallized with sEH-P. Figure 4A shows the N-terminal domain with the bound compound 22d, while Figure 4B and Figure 4C show details of the inhibitor interaction. The main interaction of 22d with the protein is mediated by its carboxyl group, which forms hydrogen bonds with residues Asn124 and Asn189 as well as three water molecules. Two of these water molecules are tightly coordinated by a magnesium ion that is present in the active site. In addition, the nitrogen of the oxazole ring forms a water-mediated hydrogen bond with the backbone NH of Val19. The aromatic oxazole system exhibits aromatic edge-to-face interactions with the side chain of Phe41. The phenyl substituents fit tightly into deeply buried hydrophobic pockets. Figure 4C shows that the phenyl substituent at the 5-position of the oxazole core of the inhibitor is in close proximity to the amino acids Trp126, Phe92, Leu60, Trp63, Leu53, and Phe41, which constitute the most hydrophobic surface area of the binding site. Furthermore, these residues belong to a region in the structure where the most prominent structural changes upon inhibitor binding were observed. The environment around the 3,5-dichlorophenyl group at the 4-position of the oxazole core comprises three hydrophobic amino acids (Ile96, Val19, and Trp126) in close proximity to this ring system. Here the importance of the chlorine substituents can be explained by contacts toward backbone carbonyl oxygens of Phe92 and Ile 96, comparable to ones observed by Heroven et al.20 and Falke et al.21 (Figure 4D). The phenyl ring at the 2-position of the oxazole core carrying the carboxyl group is positioned in close proximity to the entrance of the active site. The amino acids Phe20, Phe41, Val19, and Thr50 shield one side of the phenyl ring, while the other side is mainly solvent-exposed.
Figure 4.
X-ray structure of 22d in complex with the N-terminal domain of sEH (PDB code 5MWA). (A) 2Fo − Fc electron density map at 1σ of 22d in the binding pocket (blue mesh) and the polder omit map at 3σ (green mesh). (B) Interactions of the carboxylate headgroup with the Mg2+ ion mediated by water molecules. (C) Hydrophobic interaction of the phenyl residues. (D) Interactions of the chlorine substituents with backbone carbonyl atoms.
In order to investigate the applicability of 22b in typical model organisms in vivo, the inhibitory potencies against full-length (fl) human, mouse, and rat sEH were compared (Table 5). 22b inhibited the phosphatase activity of the full-length human enzyme, but its potency was significantly decreased compared with that for the isolated N-terminal domain. Furthermore, the sEH-H activity was also affected, showing an IC50 of about 9 μM. Most strikingly, 22b did not inhibit either the hydrolase or phosphatase activity of mouse sEH, while rat sEH-P was selectively impaired.
Table 5.
Inhibitory Activities of Oxaprozin (5) and 22b against sEH Enzymes from Different Species
| human fl sEH |
mouse fl sEH |
rat fl sEH |
||||
|---|---|---|---|---|---|---|
| sEH-H | sEH-P | sEH-H | sEH-P | sEH-H | sEH-P | |
| 5 IC50(μM) | >300 | 70.0 ± 3.0 | >300 | 35.5 ± 5.8 | >300 | 62.5 ± 1.9 |
| 22b IC50 (μM) | 9.5 ± 0.5 | 4 ± 1 | >30 | >30 | >30 | 2.8 ± 2.2 |
In order to explain the lack of activity against mouse sEH, residues in close proximity to the inhibitor in the enzyme were investigated. Five amino acids that differed in the human and mouse enzymes were identified, namely, Val19Ile, Phe20Ala, Phe41Tyr, Leu60Phe, and Ile96Met. The rat protein features the same sequence changes as mouse protein except for Phe41, which remains unchanged in the rat protein compared to human sEH. To investigate the influence of these five sequence changes on inhibitor binding, in silico mutagenesis of the sEHP complex structure was performed using the MOE software suite. Figure 5 shows the influence of the mutations on the shape of the binding site. We identified Phe41Tyr as the most significant difference between the human and mouse enzymes, while Leu60Phe might also have a slight influence on the inhibitor binding. The reason for the prominent effect of the Phe41Tyr mutation on inhibitor binding is the addition of the hydroxyl group, which collides with the oxazole core of 22d, efficiently blocking the inhibitor binding. Phe41 is not altered in rat sEH, which supports the model shown in Figure 5.
Figure 5.
Analysis of sequence changes in different species and their implications for inhibitor potency. The crystal structure of the human sEH-P was used to calculate the surface of the receptor binding site (with hydrophobic and hydrophilic areas shown in green and pink, respectively) with the bound inhibitor 22d. In addition, the five amino acids in the binding pocket that differ between human and mouse sEH were mutated in silico and are shown in the picture. The two most significant changes, Tyr41 and Phe60, are shown in the space-filling representation.
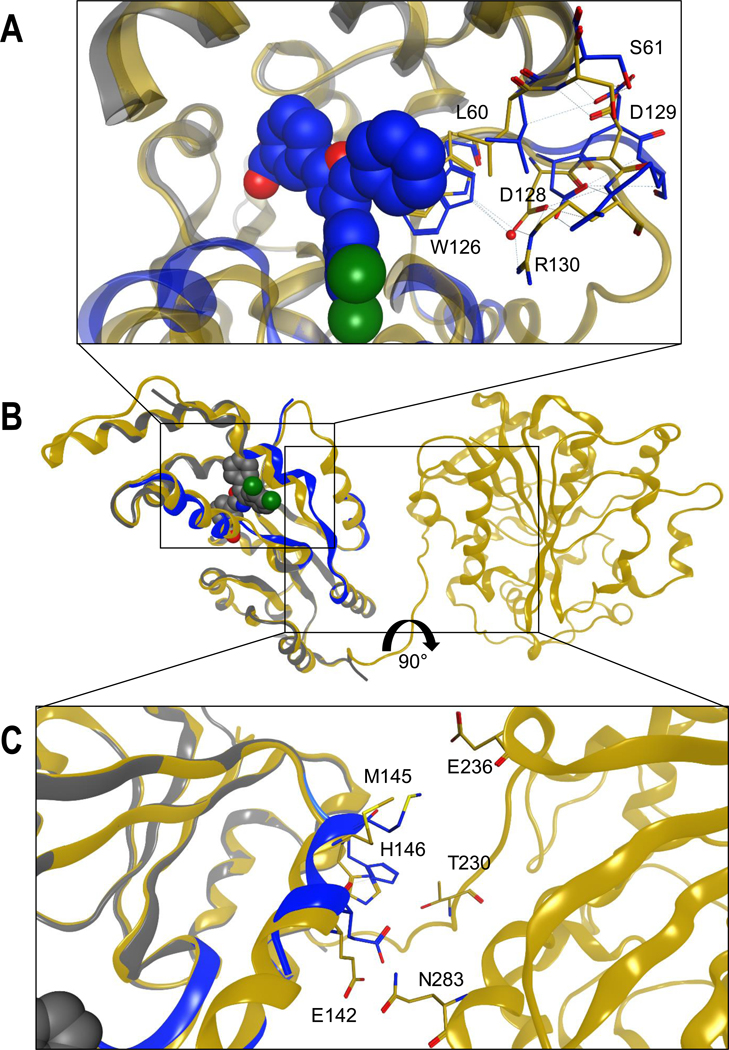
Structural analysis of the binding of 22d to sHE-P suggests a possible explanation of the pronounced different between inhibition of the full-length enzyme and the isolated sEH-P domain. Although the structure was refined at high resolution (1.55 Å), the positions of amino acids 74−84 and 131−138 could not be determined because of disorder, resulting in a lack of interpretable electron density in these regions. A comparison of the crystal structure with the crystallized full-length protein revealed that the areas that were not modeled in our structure have quite high B-factors in full length sEH (PDB code 5ALU).22 Additionally, the protein sequences of the human and mouse enzymes show high sequence variability in the areas 82−88 and 89−100 as well as 130−146. Comparison with one of the published full-length sEH crystal structures (PDB code 5ALU)22 showed that binding of the inhibitor led to a prominent induced fit in the binding site (Figure 6A). The phenyl ring at the 5-position of the oxazole core pushes Leu60 and Trp126 toward the interface between the N-terminal and C-terminal domains. This leads to movement of the interacting residues Asp128 and Arg130. These structural changes lead to shifts in residues Met145, His146, and Glu142, which are in contact with the interface between the N-terminal and C-terminal domains (Figure 6B,C).
Figure 6.
Induced-fit binding of 22d to the N-terminal domain leads to structural distortion of the interface with the C-terminal domain. (A) Binding site of 22d. (B) Global view of the superposed proteins (PDB codes 5MWA and 5ALU). (C) Interface between the N-terminal and C-terminal domains.
The structural data suggest that there is a probability that the C-terminal domain hinders the binding of 22d to the N-terminal domain in the context of the full-length enzyme. Therefore, we performed kinetic experiments to validate the mode of inhibition and determine the Ki of 22b toward the isolated N-terminal domain and full-length sEH (Figure 7). We used DiFMUP, an sEH-P substrate with only one phosphate moiety, as a substrate and determined KM values of 219.9 μM for the isolated N-terminal domain and 628.5 μM for full-length sEH. We were able to demonstrate that 22b is a competitive inhibitor under both conditions tested. We determined Ki values of 0.05 μM for the isolated N-terminal domain and 4.8 μM for full-length sEH, in agreement with the discrepancy we observed for the IC50 values using FDP as the substrate.
Figure 7.
Determination of Ki for 22b on (A) the isolated sEH-P domain and (B) full-length sEH by a mixed-model inhibition fit in GraphPad Prism (version 7.05).
Because of the decreased activity of 22b against full length sEH-P compared with the isolated N-terminal domain as well as its inhibitory activity against sEH-H in the full-length enzyme (Table 5), we also evaluated and compared the inhibitory behaviors of previously published sEH-P inhibitors. Initial DSF experiments showed that only compounds 2 and 22b are able to stabilize the N-terminal domain of sEH (Table 6; also see the Supporting Information). SDS (1) displays inhibitory potency against N-terminal and full-length sEH-P while leaving sEH-H almost unaffected. N-Acetyl-S-farnesyl-Lcysteine (2) potently inhibits the phosphatase activity of the single N-terminal domain and full-length sEH without affecting the hydrolase activity. In contrast, ebselen (4) potently and unselectively inhibits the activities of both separated sEH domains and the full-length enzyme. 22b is the most potent inhibitor of the phosphatase activity of the N-terminal domain. However, as mentioned above, the activity toward full-length sEH-P is markedly reduced, and in the full-length protein, both domains are inhibited with similar IC50 values (Table 6). These results were surprising because the initial screening hit oxaprozin (5) inhibited the N-terminal domain while leaving the sEH-H unaffected.16 Taken together, these results show that the weak activity of current inhibitors limits their applications as chemical probes in cellular assay systems and that interspecies differences in the sEH-P active site need to be considered in murine model systems. Furthermore, reported post-translational modifications of sEH by nitration23 or phosphorylation12 might also influence inhibitor binding.
Table 6.
DSF Experiments with the Isolated sEH N-Terminal Domain and Inhibitory Activities of sEH-P Inhibitors against the Isolated Human sEH Domains and Full-Length Human sEH Enzyme
| compd | sEH-P DSF (°C) | sEH-H IC50 (μM) | sEH-P IC50 (μM) | fl sEH-H IC50 (μM) | fl sEH-P IC50 (μM) |
|---|---|---|---|---|---|
| l | −0.6 ± 1.8 | 101 ± 22 | 2.3 ± 0.2 | >300 | 5.5 ± 0.4 |
| 2 | 3.4 ± 0.7 | >300 | 0.55 ± 0.01 | >300 | 2.04 ± 0.13 |
| 4 | n.d. | 0.11 ± 0.01 | 0.29 ± 0.01 | 0.30 ± 0.01 | 0.42 ± 0.03 |
| 22h | 6.6 ± 0.8 | 12 ± 1 | 0.058 ± 0.005 | 9.5 ± 0.5 | 4±1 |
In order to characterize 22b as a suitable inhibitor to study the role of sEH-P in vivo, a pharmacokinetic/pharmacodynamic study was conducted. After acute administration of 30 mg/kg p.o. to male Sprague-Dawley (SD) rats, the evolution of the plasma concentration of 22b was monitored over 8 h. 22b displayed excellent exposure, reaching a maximum concentration of 48 μg/mL (128 μM) and a calculated plasma half-life of 7 h (Figure 8A). Despite the inhibition of the conversion of the artificial fluorogenic substrate PHOME by sEH-H observed in vitro (Table 6), treatment with 22b did not influence the ratio of the main epoxy fatty acids metabolized by sEH-H to their respective diols, as quantified in rat plasma by LC−MS/MS (Figure 8B). However, the ratios of different monoacylglycerols (MAGs) to the corresponding lysophosphatidic acids (LPAs) were significantly shifted toward the LPA species, as expected (Figure 8C). This is a strong indication that the role of sEH-P is determined by the degradation of different LPAs, as suggested in two independent studies by Morisseau8 and Oguro.9 In order to confirm that the results we obtained in vivo are transferable to a human system, we incubated human peripheral blood mononuclear cells (PMBCs) with 30 μM 22b. In this system we also observed a reduction of the MAG/LPA ratio (Figure S2). Still, properly chosen control substances are required for use of 22b in cellular and in vivo experiments because of its residual activity toward COX enzymes and some nuclear receptors (Table 4).
Figure 8.
In vivo studies of 22b in male SD rats. (A) Evolution of the plasma concentration of 22b after acute administration of 30 mg/kg p.o. in SD rats (n = 3) with a peak concentration (Cmax) of 128 μM. (B) Ratios of the epoxy fatty acids 14,15-epoxyeicosatrienoic acid (14,15-EET), 12,13-epoxyoctadecenoic acid (12,13-EpOME), and 19,20-epoxydocosapentaenoic acid (19,20-EpDPA) to their respective diols 14,15-dihydroxyeicosatrienoic acid (14,15-DHET), 12,13dihydroxyoctadecenoic acid (12,13-DiHOME), and 19,20-dihydroxydocosapentaenoic acid (19,20-EpDPA) as quantified in rat plasma 8 h after acute oral administration of 22b or vehicle (n = 3 per group). (C) Ratios of 1-oleyl lysophosphatidic acid (sn1−18:1 LPA), 1linoleyl LPA (sn1−18:2 LPA), and 1-arachidonoyl LPA (sn1−20:4 LPA) to their corresponding monoacylglycerol (MAG) species for 22b and vehicle 8 h after administration as quantified in rat plasma (n = 3 per group).
CONCLUSIONS
In this study, we have reported the optimization of oxazole-based ligands of the N-terminal domain of sEH. The structure−activity relationships revealed an important role of the 3,4-dichlorophenyl substituent at the 4-position of the oxazole core. Optimization led to the identification of 22b and 22d as potent inhibitors of the sEH-P activity in vitro. The reported X-ray structure of the potent ligand 22d bound to the sEH-P domain provided first insights into inhibitor binding to sEH-P. A prominent induced fit upon inhibitor binding was observed. Significant interspecies differences in the sEH-P active site that need to be considered when using sEH-P inhibitors can be rationalized by the X-ray structure. In vivo studies confirmed that 22b is active in rats with favorable pharmacokinetics and revealed the involvement of sEH-P in LPA hydrolysis. Hence, this study has reported the first in vivo active inhibitor to study the elusive physiological and pathophysiological role of sEH-P.
EXPERIMENTAL SECTION
Chemistry Materials and General Procedures.
All of the solvents and chemicals were purchased from Sigma-Aldrich Chemie GmbH (Munich, Germany), Acros Organics (Geel, Belgium), AlfaAesar GmbH & Co KG (Karlsruhe, Germany), TCI Europe (Zwijndrecht, Belgium), or Apollo Scientific Ltd. (Manchester, England) and used without further purification. Analytical thin-layer chromatography (TLC) was performed with F254 TLC plates (Merck KGaA, Darmstadt, Germany) visualized with ultraviolet light (254 nm). Column chromatography was performed with technical-grade solvent mixtures specified in the corresponding experiments with Fluka silica gel 60 (230−400 mesh ASTM). 1H NMR spectra were recorded on a DPX 250 (250 MHz) or AV300 (300 MHz) spectrometer (Bruker, Karlsruhe, Germany). 1H NMR data are reported in the following order: chemical shift (δ) in parts per million downfield relative to tetramethylsilane (internal reference nondeuterated solvent); multiplicity (br, broad; s, singlet; d, doublet; dd, doublet of doublets; t, triplet; q, quadruplet; m, multiplet); number of protons; approximate coupling constant (J) in hertz. 13C NMR spectra were recorded on an AV300 (75 MHz) or AV500 (125 MHz) spectrometer (Bruker). HPLC and mass analyses were performed on a LCMS 2020 system (Shimadzu, Duisburg, Germany) with a MultoHigh UC column (50 mm × 2 mm) from CS Chromatography-Service GmbH (Langerwehe, Germany) or a Luna 10 μ C18(2) 100A column (250 mm × 4.60 mm) for analytical purposes and a Luna 10 μ C18(2) (250 mm × 21.20 mm) column for preparative purposes from Phenomenex LTD Deutschland (Aschaffenburg, Germany). Conditions were as follows: acetonitrile/0.1% aqueous formic acid as the eluent at a flow rate of 0.5 mL/min (UPLC), 0.1 mL/min (Scout column), or 21 mL/min (semipreparative) at room temperature. UV absorption was monitored at 254 and 280 nm, while the electrospray ionization (ESI) detector produced positive ion (+) as well as negative ion (−) spectra. The purities of all final compounds were 95% or higher as determined by HPLC using the following method (except in the case of compound 9): gradient of 50% to 10% acetonitrile over 10 min and then holding at 10% acetonitrile for another 10 min. Compound 9 was monitored with an isocratic method with 50% acetonitrile. Integrated peaks of the chromatogram at λ = 254 and 280 nm were used to define the purity. MALDI-HRMS was performed on a MALDI LTQ Orbitrap XL instrument (Thermo Scientific, USA).
Methyl 2-((4,5-Diphenyloxazol-2-yl)thio)acetate (8).
To a solution of 4,5-diphenyloxazole-2-thiol (0.75 g, 2.94 mmol, 1.0 equiv) in 10 mL of THF was added 0.83 mL of triethylamine (5.87 mmol, 2.0 equiv) at room temperature. After 10 min, methyl 2-bromoacetate (0.31 mL, 3.23 mmol, 1.1 equiv) was added, and the mixture was stirred overnight followed by filtration over Celite and concentration of the filtrate under reduced pressure. The residue was purified via column chromatography, eluting with 5:1 hexane/ethyl acetate (Hex/ EtOAc), to yield 0.85 g of the product (2.58 mmol, 88% yield). Rf: 0.28 (8:1 Hex/EtOAc). 1H NMR (250 MHz, CD3OD): δ = 7.51 (m, 10H, Ph), 4.24 (s, 2H, −CH2−), 3.73 (s, 3H, −CH3). ESI-MS: m/z = 326.0 [M + H]+.
2-((4,5-Diphenyloxazol-2-yl)thio)acetic Acid (9).
In a microwave vial, 8 (0.83 g, 2.58 mmol, 1 equiv) was dissolved in 4 mL of a 1:2:1 MeOH/water/THF mixture, and potassium hydroxide powder (0.43 g, 7.65 mmol, 3 equiv) was added. The vial was capped and heated to 70 °C for 15 min in the microwave. The solvent was removed, and the residue was taken up in 7 mL of water, acidified with 10% HCl to pH 5, and lyophilized. Recrystallization from EtOAc yielded 0.52 g of the raw product (1.67 mmol, 65% yield). Rf (RP): 0.64 (3:1 MeOH/water). 1H NMR (250 MHz, CDCl3): δ = 7.46 (m, 10H, Ph), 3.94 (s, 2H, −CH2−). 13C NMR (75 MHz, CDCl3): 171.3, 158.1, 146.4, 135.0, 130.5, 127.7, 127.6, 127.4, 127.2, 126.8, 125.3, 35.5. HRMS: calcd, m/z = 312.06889; found, m/z = 312.06931.
General Procedure for the Synthesis of Cyanohydrin Compounds 15a–m.
To the appropriate benzaldehyde derivative 13a–m (1 equiv) was added trimethylsilyl cyanide (14, TMSCN) (1 equiv) followed by triethylamine (0.2 equiv). If the starting material did not dissolve within 10 min, 1 mL of DCM was added. The solution was stirred overnight at room temperature. The mixture was diluted with DCM and washed with concentrated ammonium chloride solution and brine. The organic layer was filtered over silica gel and rinsed with 100 mL of DCM. The solvent was removed under reduced pressure, and the crude product was used without further purification. In most cases, a liquid was isolated.
2-(2-Fluorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15a).
Yield: 1.28 g (colorless liquid; 96%). Rf: 0.76 (5:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.64 (td, 1H, J = 7.6 Hz, 1.6 Hz, Ph-4H), 7.45−7.35 (m, 1H, Ph-6H), 7.23 (td, 1H, J = 7.6 Hz, 1.1 Hz, Ph-2H), 7.61 (td, 1H, J = 8.7 Hz, 1.1 Hz, Ph-5H), 5.76 (s, 1H, −CH−), 0.24 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 195.8 [M − CN]−.
2-(2-Chlorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15b).
Yield: 607 mg (colorless liquid; quant.). Rf: 0.78 (9:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.74−7.70 (m, 1H, Ph-5H), 7.42− 7.32 (m, 3H, Ph-3H,4H,6H), 5.80 (s, 1H, −CH−), 0.26 (s, 9H, 3 × −CH3). ESI-MS: m/z = 211.75 [M − CN]−.
3-(Cyano((trimethylsilyl)oxy)methyl)benzonitrile (15c).
Yield: 1.30 g (pale-yellow liquid; 94%). Rf: 0.09 (9:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.78 (s, 1H, Ph-2H), 7.70−7.68 (m, 2H, Ph-4H,5H), 7.56 (d, 1H, J = 7.7 Hz, Ph-6H), 5.52 (s, 1H, −CH−), 0.28 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 202.9 [M − CN]−.
2-(3-Fluorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15d).
Yield: 1.35 g (pale-yellow liquid; quant.). Rf: 0.71 (5:1 Hex/ EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.43−7.34 (m, 1H, Ph-5H), 7.26−7.18 (m, 2H, Ph-2H,4H), 7.09 (td, 1H, J = 8.5 Hz, 2.5 Hz, Ph-6H), 5.49 (s, 1H, −CH−), 0.25 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 195.8 [M − CN]−.
2-(4-Fluorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15e).
Yield: 1.41 g (colorless liquid; quant.). Rf: 0.56 (9:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.49−7.41 (m, 2H, Ph-2H,6H), 7.10 (pt, 2H, J = 8.6 Hz, Ph-3H,5H), 5.47 (s, 1H, −CH−), 0.24 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 195.8 [M − CN]−.
2-(4-(Trifluoromethyl)phenyl)-2-((trimethylsilyl)oxy)acetonitrile (15f).
Yield: 601 mg (pale-yellow liquid; 88%). Rf: 0.80 (3:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.69 (d, 2H, J = 8.6 Hz, Ph-3H,5H), 7.61 (d, 2H, J = 8.6 Hz, Ph-2H,6H), 5.55 (s, 1H, −CH−), 0.27 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 245.8 [M − CN]−.
4-(Cyano((trimethylsilyl)oxy)methyl)benzonitrile (15g).
Yield: 517 mg (yellow liquid; 90%). Rf: 0.57 (9:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.73 (d, 2H, J = 8.5 Hz, Ph-3H,5H), 7.60 (d, 2H, J = 8.6 Hz, Ph-2H,6H), 5.54 (s, 1H, −CH−), 0.27 (s, 9H, 3 × −CH3). ESI-MS: m/z = 202.0 [M − CN]−.
2-(4-Chlorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15h).
Yield: 2.24 g (yellow liquid; 93%). Rf: 0.88 (9:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.04 (ps, 4H, Ph), 5.46 (s, 1H, −CH−), 0.24 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 211.8 [M − CN]−.
2-(4-Methoxyphenyl)-2-((trimethylsilyl)oxy)acetonitrile (15i).
Yield: 604 mg (colorless liquid; quant.). Rf: 0.57 (5:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.38 (d, 2H, J = 8.8 Hz, Ph-2H,6H), 6.92 (d, 2H, J = 8.8 Hz, Ph-3H,5H), 5.43 (s, 1H, −CH−), 3.82 (s, 3H, O−CH3), 0.21 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 209.3 [M − CN]−.
2-(4-Chloro-3-(trifluoromethyl)phenyl)-2-((trimethylsilyl)oxy)acetonitrile (15j).
Yield: 722 mg (yellow liquid; 89%). Rf: 0.87 (5:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.79 (s, 1H, Ph-2H), 7.59 (ps, 2H, Ph-5H,6H), 5.51 (s, 1H, −CH−), 0.27 (s, 9H, 3 × −CH3). ESI-MS: m/z = 279.7 [M − CN]−.
2-(3,4-Dichlorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15k).
Yield: 673 mg (orange liquid; 98%). Rf: 0.69 (3:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.57 (sd, 1H, J = 2.0 Hz, Ph-2H), 7.50 (d, 1H, J = 8.3 Hz, Ph-5H), 7.30 (dd, 1H, J = 8.3 Hz, 2.1 Hz, Ph-6H), 5.44 (s, 1H, −CH−), 0.26 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 245.7 [M − CN]−.
2-(2,4-Dichlorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15l).
Yield: 680 mg (yellow liquid; 99%). Rf: 0.82 (5:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.65 (d, 1H, J = 8.4 Hz, Ph-6H), 7.42 (sd, 1H, J = 2.0 Hz, Ph-3H), 7.35 (dd, 1H, J = 8.4 Hz, 2.0 Hz, Ph-5H), 5.73 (s, 1H, −CH−), 0.26 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 248.1 [M − CN]−.
2-(3,5-Dichlorophenyl)-2-((trimethylsilyl)oxy)acetonitrile (15m).
Yield: 600 mg (brown liquid; 88%). Rf: 0.88 (5:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.39−7.35 (m, 3H, Ph), 5.43 (s, 1H, −CH−), 0.27 (s, 9H, 3 ×−CH3). ESI-MS: m/z = 245.7 [M − CN]−.
General Procedure for the Synthesis of Deoxybenzoin Compounds 17a–m.
The appropriate cyanohydrin (2 mmol; 1 equiv) was dissolved in 3 mL of absolute THF and cooled to −78 °C. LDA (2.1 mmol, 1.05 equiv) was added, and the mixture was left to stir for 30 min, followed by the addition of benzyl bromide (16) (2.2 mmol, 1.1 equiv) in 1 mL of THF. The mixture was stirred for 2 h, allowed to warm to room temperature (RT), and stirred overnight. To the mixture was added 1 M TBAF solution (2 mL, 1 equiv), and the resulting mixture was stirred for another half an hour. The mixture was diluted with ethyl acetate, washed with saturated ammonium chloride solution and 0.5 M NaOH solution, and dried, and the solvent was removed. The yellow-orange residue (ca. 600 mg) was purified by column chromatography, eluting with 9:1 Hex/EtOAc. The purified product solidified upon standing.
1-(2-Fluorophenyl)-2-phenylethan-1-one (17a).
Yield: 247 mg (yellow oil; 58%). Rf: 0.70 (5:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 7.86 (td, 1H, J = 7.6 Hz, 1.7 Hz, Ph-F-4H), 7.70− 7.61 (m, 1H, Ph-F-6H), 7.38−7.15 (m, 7H, Ph, Ph-F-3H,5H), 4.31 (s, 2H, −CH2−). ESI-MS: m/z = 214.8 [M + H]+.
1-(2-Chlorophenyl)-2-phenylethan-1-one (17b).
Yield: 202 mg (pale-yellow liquid; 44%). Rf: 0.68 (9:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 7.72 (dd, 1H, J = 7.8 Hz, 1.6 Hz, Ph-Cl-6H), 7.53−7.48 (m, 2H, Ph-Cl-3H,4H), 7.72 (td, 1H, J = 6.7 Hz, 1.9 Hz, Ph-Cl-5H), 7.33−7.29 (m, 2H, Ph-3H,5H), 7.25−7.21 (m, 3H, Ph2H,4H,6H), 4.29 (s, 2H, −CH2−). ESI-MS: m/z = 230.8 [M]+.
3-(2-Phenylacetyl)benzonitrile (17c).
Yield: 320 mg (tan solid; 72%). Rf: 0.14 (9:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 8.49 (s, 1H, Ph-CN-2H), 8.30 (d, 1H, J = 8.0 Hz, Ph-CN-4H), 8.11 (d, 1H, J = 7.8 Hz, Ph-CN-6H), 7.52 (t, 1H, J = 7.8 Hz, Ph-CN-5H), 7.37−7.23 (m, 5H, Ph), 4.46 (s, 2H, −CH2−). ESI-MS: m/z = 219.9 [M − H]−.
1-(3-Fluorophenyl)-2-phenylethan-1-one (17d).
Yield: 200 mg (yellow solid; 47%). Rf: 0.60 (5:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 7.90 (d, 1H, J = 7.7 Hz, Ph-F-6H), 7.81 (dd, 1H, J = 7.7 Hz, 1.5 Hz, Ph-F-4H), 7.62−7.56 (m, 1H, Ph-F-2H), 7.49 (td, 1H, J = 9.2 Hz, 2.5 Hz, Ph-F-5H), 7.35−7.29 (m, 3H, Ph-2H,4H,6H), 7.27−7.23 (m, 2H, Ph-3H,5H), 4.41 (s, 2H, −CH2−). ESI-MS: m/z = 214.9 [M + H]+.
1-(4-Fluorophenyl)-2-phenylethan-1-one (17e).
Yield: 399 mg (pale-yellow solid; 93%). Rf: 0.43 (9:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 8.13 (t, 2H, J = 8.6 Hz, Ph-F-3H,5H), 7.39−7.22 (m, 7H, Ph, Ph-F-2H,6H), 4.38 (s, 2H, −CH2−). ESI-MS: m/z = 214.8 [M + H]+.
2-Phenyl-1-(4-(trifluoromethyl)phenyl)ethan-1-one (17f).
Yield: 236 mg (white solid; 45%). Rf: 0.70 (3:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 8.22 (d, 2H, J = 8.0 Hz, Ph-CF3-2H,6H), 7.90 (d, 2H, J = 8.1 Hz, Ph-CF3-3H,5H), 7.35−7.22 (m, 5H, Ph), 4.47 (s, 2H, −CH2−). ESI-MS: m/z = 264.8 [M + H]+.
4-(2-Phenylacetyl)benzonitrile (17g).
Yield: 213 mg (white solid; 45%). Rf: 0.39 (5:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 8.20 (d, 2H, J = 8.6 Hz, Ph-CN-2H,6H), 8.03 (d, 2H, J = 8.6 Hz, Ph-CN-3H,5H), 7.38−7.23 (m, 5H, Ph), 4.47 (s, 2H, −CH2−). ESIMS: m/z = 219.9 [M − H]−.
1-(4-Chlorophenyl)-2-phenylethan-1-one (17h).
Yield: 192 mg (pale-yellow oil; 55%). Rf: 0.75 (9:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 7.93 (d, 2H, J = 8.6 Hz, Ph-Cl-2H,6H), 7.42 (d, 2H, J = 8.6 Hz, Ph-Cl-3H,5H), 7.35−7.23 (m, 5H, Ph), 4.25 (s, 2H, −CH2−). ESI-MS: m/z = 230.8 [M + H]+.
1-(4-Methoxyphenyl)-2-phenylethan-1-one (17i).
Yield: 286 mg (yellow liquid; 63%). Rf: 0.36 (5:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.99 (d, 2H, J = 8.5 Hz, Ph-OCH3-2H,6H), 7.30−7.20 (m, 5H, Ph), 6.92 (d, 2H, J = 8.5 Hz, Ph-OCH3-3H,5H), 4.23 (s, 2H, −CH2−), 3.85 (s, 3H, −CH3). ESI-MS: m/z = 226.9 [M + H]+.
1-(4-Chloro-3-(trifluoromethyl)phenyl)-2-phenylethan-1-one (17j).
Yield: 213 mg (colorless liquid; 45%). Rf: 0.53 (5:1 Hex/ EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 8.32 (pd, 2H, Ph3CF3,4Cl-2H,6H), 7.92 (d, 1H, J = 8.8 Hz, Ph-3CF3,4Cl-5H), 7.36− 7.21 (m, 5H, Ph), 4.49 (s, 2H, −CH2−). ESI-MS: m/z = 299.7 [M + H]+.
1-(3,4-Dichlorophenyl)-2-phenylethan-1-one (17k).
Yield: 258 mg (pale-yellow solid; 49%). Rf: 0.59 (3:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 8.23 (sd, 1H, J = 2.0 Hz, Ph-3,4Cl-2H), 7.99 (dd, 1H, J = 8.3 Hz, 2.1 Hz, Ph-3,4Cl-6H), 7.81 (d, 1H, J = 8.3 Hz, Ph-3,4Cl-5H), 7.34−7.21 (m, 5H, Ph), 4.42 (s, 2H, −CH2−). ESI-MS: m/z = 265.9 [M + H]+.
1-(2,4-Dichlorophenyl)-2-phenylethan-1-one (17l).
Yield: 167 mg (pale-yellow liquid; 31%). Rf: 0.65 (9:1 Hex/EtOAc). 1H NMR (250 MHz, CDCl3): δ = 7.73 (sd, 1H, J = 2.0 Hz, Ph-2,4-diCl-3H), 7.35− 7.19 (m, 7H, Ph, Ph-2,4-diCl-5H,6H), 4.24 (s, 2H, −CH2−). ESI-MS: m/z = 265.8 [M + H]+.
1-(3,5-Dichlorophenyl)-2-phenylethan-1-one (17m).
Yield: 151 mg (pale-yellow liquid; 26%). Rf: 0.60 (9:1 Hex/EtOAc). 1H NMR (250 MHz, DMSO-d6): δ = 8.01 (sd, 2H, J = 1.9 Hz, Ph-3,5-diCl2H,6H), 7.92 (sd, 1H, J = 1.9 Hz, Ph-3,5-diCl-4H), 7.36−7.23 (m, 5H, Ph), 4.44 (s, 2H, −CH2−). ESI-MS: m/z = 265.9 [M + H]+.
General Procedure for the Synthesis of α-Bromoketone Compounds 18a–m.
The deoxybenzoin derivative (1 equiv) was dissolved in chloroform, and bromine (1 equiv), dissolved in chloroform as well, was slowly added. The mixture was heated to reflux for 2 h, diluted with dichloromethane, washed with water and brine, dried, and concentrated under reduced pressure. The resulting liquid was used immediately without further purification.
General Procedure for the Synthesis of Oxaprozin Derivatives 12, 20a–m, and 22a–e.
Succinamic acid (11), succinic acid (19), or the corresponding dicarboxylic acid 21a–e (3 equiv) was dissolved in acetonitrile and treated with triethylamine (3 equiv). After 15 min, the α-bromoketone (1 equiv), dissolved in a minimal amount of acetonitrile, was added, and the mixture was stirred at 45 °C for 6 h. The solvent was removed, and the residue was diluted with ethyl acetate and washed with water and brine. A white resin was isolated. The residue was taken up in acetic acid (10 mL), and ammonium acetate (10 equiv) was added. The mixture was heated to reflux for another 3 h and then thrown on ice and extracted with ethyl acetate, and the residue was purified via column chromatography, eluting with 3:1 Hex/EtOAc + 3% AcOH). Yields were calculated for the two-step reaction starting from the deoxybenzoin. The products were further purified by preparative HPLC, yielding white solids.
3-(4,5-Diphenyloxazol-2-yl)propanamide (12).
11 was used starting from α-bromodeoxybenzoin. Yield: 296 mg (68%). Rf: 0.22 (98:2 DCM/MeOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.57−7.32 (m, 11H, Ph, 1 ×−NH2), 6.87 (br, 1H, 1 ×−NH2), 3.04 (t, 2H, J = 7.3 Hz, CH2−CONH2), 2.62 (t, 2H, J = 7.3 Hz, CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 172.4, 162.73, 144.4, 134.2, 132.0, 128.8, 128.7, 128.5, 128.4, 128.0, 127.3, 126.2, 31.3, 23.1. HRMS: calcd, m/z = 293.12845; found, m/z = 293.12880.
3-(4-(2-Fluorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20a).
Yield: 58 mg (34%). Rf: 0.39 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.31 (s, 1H,−COOH), 7.60−7.46 (m, 2H, Ph-F-4H,6H), 7.59−7.39 (m, 7H, Ph, Ph-F-3H,5H), 3.09 (t, 2H, J = 7.0 Hz, −CH2−COOH), 2.79 (t, 2H, J = 7.0 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 162.4, 131.3, 131.3, 130.8, 130.7, 128.7, 128.4, 127.9, 124.8, 124.8, 124.7, 116.1, 115.8, 30.1, 22.8. HRMS: calcd, m/z = 312.10305; found, m/z = 312.10411.
3-(4-(2-Chlorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20b).
Yield: 82 mg (29%; hygroscopic). Rf: 0.49 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.33 (s, 1H, −COOH), 7.69−7.28 (m, 9H, Ph-Cl, Ph), 3.06 (t, 2H, J = 7.0 Hz, CH2−COOH), 2.78 (t, 2H, J = 7.0 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 163.0, 141.8, 135.7, 133.3, 132.4, 131.8, 131.1, 130.1, 128.5, 128.0, 127.8, 127.8, 125.8, 30.2, 22.9. HRMS: calcd, m/z = 328.07350; found, m/z = 328.07482.
3-(4-(3-Cyanophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20c).
Yield: 39 mg (18%). Rf: 0.76 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.36 (s, 1H, −COOH), 7.93 (s, 1H, Ph-CN-2H), 7.85 (d, 1H, J = 7.9 Hz, Ph-CN-4H), 7.81 (d, 1H, J = 7.9 Hz, Ph-CN-6H), 7.61 (t, 1H, J = 7.8 Hz, Ph-CN-5H), 7.55−7.51 (m, 3H, Ph-2H,4H,6H), 7.49−7.40 (m, 2H, Ph-3H,5H), 3.07 (t, 2H, J = 7.0 Hz, −CH2−COOH), 2.79 (t, 2H, J = 7.0 Hz, −CH2− CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 162.8, 145.7, 133.2, 132.3, 131.7, 130.4, 130.0, 129.3, 129.1, 128.8, 127.8, 127.5, 126.6, 118.4, 111.8, 30.1, 22.8. HRMS: calcd, m/z = 319.10772; found, m/z = 319.10865.
3-(4-(3-Fluorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20d).
Yield: 57 mg (24%). Rf: 0.23 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.33 (s, 1H, −COOH), 7.55−7.53 (m, 2H, Ph-3H,5H), 7.48−7.41 (m, 4H, Ph2H,4H,6H, Ph-F-2H), 7.39 (d, 1H, J = 7.8 Hz, Ph-F-6H), 7.31 (dd, 1H, J = 9.4 Hz, 1.1 Hz, Ph-F-4H), 7.18 (td, 1H, J = 7.8 Hz, 1.1 Hz, Ph-F-5H), 3.06 (t, 2H, J = 6.9 Hz, −CH2−COOH), 2.78 (t, 2H, J = 7.0 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 162.5, 145.2, 134.3, 132.9, 130.7, 128.9, 128.0, 126.6, 123.1, 115.0, 113.8, 30.2, 22.8. HRMS: calcd, m/z = 312.10305; found, m/z = 312.10417.
3-(4-(4-Fluorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20e).
Yield: 53 mg (19%). Rf: 0.30 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.31 (s, 1H, −COOH), 7.60−7.56 (m, 2H, Ph-F-2H,6H), 7.53−7.50 (m, 2H, PhF-3H,5H), 7.47−7.39 (m, 3H, Ph-2H,4H,6H), 7.49 (t, 2H, J = 8.9 Hz, Ph-3H,5H), 3.06 (t, 2H, J = 7.1 Hz, −CH2−COOH), 2.78 (t, 2H, J = 7.1 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 163.3, 162.3, 160.1, 144.4, 133.3, 129.4, 129.3, 128.9, 128.8, 128.4, 128.3, 128.2, 126.2, 115.7, 115.4, 30.1, 22.8. HRMS: calcd, m/z = 312.10305; found, m/z = 312.10410.
3-(5-Phenyl-4-(4-(trifluoromethyl)phenyl)oxazol-2-yl)propanoic Acid (20f).
Yield: 71 mg (23%). Rf: 0.51 (3:1 hexane/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.31 (s, 1H,−COOH), 7.77 (ps, 4H, Ph-CF3), 7.57−7.44 (m, 5H, Ph), 3.08 (t, 2H, J = 7.0 Hz, CH2−COOH), 2.79 (t, 2H, J = 6.9 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 162.8, 145.84, 136.0, 132.7, 129.2, 129.0, 127.9, 127.6, 126.7, 125.6, 125.5, 30.1, 22.8. HRMS: calcd, m/z = 362.09985; found, m/z = 362.10073.
3-(4-(4-Cyanophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20g).
Yield: 24 mg (15%). Rf: 0.32 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.32 (s, 1H, −COOH), 7.86 (d, 2H, J = 8.6 Hz, Ph-CN-3H,5H), 7.73 (d, 2H, J = 8.6 Hz, Ph-CN-2H,6H), 7.57−7.46 (m, 5H, Ph), 3.07 (t, 2H, J = 6.9 Hz, CH2−COOH), 2.78 (t, 2H, J = 6.9 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 163.0, 146.4, 136.5, 132.6, 129.5, 129.1, 127.8, 127.6, 126.9, 118.6, 110.3, 30.1, 22.8. HRMS: calcd, m/z = 319.10772; found, m/z = 319.10820.
3-(4-(4-Chlorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20h).
Yield: 19 mg (34%). Rf: 0.46 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.31 (br, 1H, −COOH), 7.59−7.39 (m, 9H, Ph, Ph-Cl), 3.06 (t, 2H, J = 7.1 Hz, −CH2−COOH), 2.78 (t, 2H, J = 7.0 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 162.5, 144.9, 133.1, 132.5, 130.7, 128.9, 128.9, 128.7, 128.6, 128.4, 128.1, 126.4, 30.1, 22.8. HRMS: calcd, m/z = 328.07350; found, m/z = 328.07469.
3-(4-(4-Methoxyphenyl)-5-phenyloxazol-2-yl)propanoic Acid (20i).
Yield: 58 mg (16%). Rf: 0.12 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.33 (s, 1H, −COOH), 7.55−7.36 (m, 7H, Ph, Ph-OCH3-2H,6H), 6.99 (d, 2H, J = 8.5 Hz, PhOCH3-3H,5H), 3.85 (s, 3H, −CH3), 3.06 (t, 2H, J = 7.0 Hz, CH2− COOH), 2.79 (t, 2H, J = 6.9 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.1, 162.1, 159.0, 143.6, 134.2, 128.9, 128.7, 128.6, 128.5, 126.01, 124.2, 114.0, 55.0, 30.2, 22.9. HRMS: calcd, m/z = 324.12303; found, m/z = 324.12406.
3-(4-(4-Chloro-3-(trifluoromethyl)phenyl)-5-phenyloxazol-2-yl)propanoic Acid (20j).
Yield: 52 mg (11%). Rf: 0.32 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.40 (s, 1H,−COOH), 8.01 (sd, 1H, J = 1.9 Hz, Ph-4Cl-3CF3-2H), 7.88 (dd, 1H, J = 8.4 Hz, 1.9 Hz, Ph-4Cl-3CF3-6H), 7.80 (d, 1H, J = 8.4 Hz, Ph-4Cl3CF3-5H), 7.63−7.51 (m, 5H, Ph), 3.13 (t, 2H, J = 6.8 Hz, CH2− COOH), 2.84 (t, 2H, J = 6.8 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 162.9, 145.9, 132.1, 132.0, 131.7, 131.5, 129.5, 129.1, 127.7, 126.9, 30.1, 22.8. HRMS: calcd, m/z = 396.06088; found, m/z = 396.06074.
3-(4-(3,4-Dichlorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20k).
Yield: 68 mg (21%). Rf: 0.43 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.31 (s, 1H,−COOH), 7.74 (sd, 1H, J = 2.0 Hz, Ph-3,4-diCl-2H), 7.66 (d, 1H, J = 8.3 Hz, Ph-3,4-diCl-5H), 7.56−7.44 (m, 6H, Ph-3,4-diCl-6H, Ph), 3.06 (t, 2H, J = 7.0 Hz, CH2−COOH), 2.78 (t, 2H, J = 6.9 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 172.9, 162.7, 145.6, 132.5, 131.8, 131.3, 130.9, 130.4, 129.3, 129.0, 128.6, 127.8, 127.0, 126.7, 30.1, 22.8. HRMS: calcd, m/z = 362.03453; found, m/z = 362.03564.
3-(4-(2,4-Dichlorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20l).
Yield: 50 mg (25%). Rf: 0.36 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.30 (s, 1H, −COOH), 7.86 (sd, 1H, J = 1.0 Hz, Ph-2,4-diCl-3H), 7.62−7.58 (m, 2H, Ph-2,4-diCl5H,6H), 7.45−7.26 (m, 5H, Ph), 3.04 (t, 2H, J = 7.0 Hz, CH2− COOH), 2.78 (t, 2H, J = 7.0 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 172.9, 163.2, 140.6, 136.2, 135.5, 134.3, 133.5, 130.9, 129.8, 128.53, 128.5, 128.0, 127.9, 127.4, 126.9, 126.0, 30.2, 22.8. HRMS: calcd, m/z = 362.03453; found, m/z = 362.03563.
3-(4-(3,5-Dichlorophenyl)-5-phenyloxazol-2-yl)propanoic Acid (20m).
Yield: 23 mg (11%). Rf: 0.35 (3:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.31 (s, 1H,−COOH), 7.61−7.48 (m, 8H, Ph, Ph-3,5-diCl), 3.07 (t, 2H, J = 6.8 Hz, CH2−COOH), 2.76 (t, 2H, J = 6.8 Hz, −CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.0, 162.9, 146.1, 135.3, 134.3, 131.4, 129.6, 129.1, 127.6, 127.4, 126.9, 125.3, 30.1, 22.8. HRMS: calcd, m/z = 362.03453; found, m/z = 362.03575.
3-(4-(3,4-Dichlorophenyl)-5-phenyloxazol-2-yl)acrylic Acid (22a, Isomeric Mixture).
Instead of succinic acid, maleic acid (21a) was used. Yield: 112 mg (yellow solid; 21%). Rf: 0.56 (2:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.88 (sd, 1H, J = 2.0 Hz, Ph-3,4-diCl-2H), 7.78 (sd, 0.4H, J = 2.0 Hz, Ph-3,4-diCl-2H), 7.69 (d, 1H, J = 8.4 Hz, Ph-3,4-diCl-5H), 7.69 (d, 1H, J = 8.4 Hz, Ph3,4-diCl-5H), 7.67 (d, 0.4H, J = 8.4 Hz, Ph-3,4-diCl-5H), 7.65−7.45 (m, 9H, 2 × Ph, 2 × Ph-3,4-diCl-6H), 7.32 (d, 1.4H, J = 15.9 Hz, −CH=CH−), 6.87 (d, 1H, J = 15.9 Hz, =CH−COOH), 6.77 (d, 0.4H, J = 15.9 Hz, =CH−COOH). 13C NMR (75 MHz, DMSO-d6): δ = 166.2, 166.2, 157.7, 157.5, 147.3, 143.8, 138.3, 134.5, 131.9, 131.9, 131.8, 131.5, 131.2, 131.1, 131.1, 130.8, 130.0, 129.2, 129.1, 129.0, 128.0, 128.0, 127.8, 127.5, 127.2, 127.1, 127.0, 126.7, 126.3. HRMS: calcd, m/z = 360.01888; found, m/z = 360.01937.
4-(4-(3,4-Dichlorophenyl)-5-phenyloxazol-2-yl)butanoic Acid (22b).
Instead of succinic acid, glutaric acid (21b) was used. Yield: 121 mg (27%). Rf: 0.57 (2:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.40 (s, 1H, −COOH), 7.75 (sd, 1H, J = 1.9 Hz, Ph-3,4-diCl-2H), 7.66 (d, 1H, J = 8.4 Hz, Ph-3,4-diCl-5H), 7.56−7.44 (m, 6H, Ph, Ph-3,4-diCl-6H), 2.87 (t, 2H, J = 7.5 Hz, CH2−COOH), 2.39 (t, 2H, J = 7.1 Hz, oxazole−CH2−CH2), 1.98 (q, 2H, J = 7.5 Hz, −CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 174.0, 163.2, 145.7, 132.6, 131.9, 131.4, 131.0, 130.4, 129.4, 129.1, 128.7 127.9, 127.1, 126.8, 32.6, 26.5, 21.69. HRMS: calcd, m/z = 376.05018; found, m/z = 376.05145.
2-(4-(3,4-Dichlorophenyl)-5-phenyloxazol-2-yl)benzoic Acid (22c).
Instead of succinic acid, phthalic acid (21c) was used. Yield: 70 mg (pale-yellow solid; 14%). Rf: 0.56 (2:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 13.40 (s, 1H,−COOH), 8.00 (dd, 1H, J = 6.8 Hz, 1.9 Hz, Ph-3,4-diCl-6H), 7.85 (sd, 1H, J = 1.9 Hz, Ph-3,4-diCl-2H), 7.78−7.57 (m, 7H, Ph-COOH, Ph-3H,5H, Ph-3,4-diCl-6H), 7.54−7.47 (m, 3H, Ph-2H,4H,6H). 13C NMR (75 MHz, DMSO-d6): δ = 169.1, 159.3, 146.8, 133.4, 133.3, 132.4, 131.6, 131.1, 130.9, 130.9, 129.7, 129.3, 129.2, 128.9, 127.6, 127.3, 126.9, 124.8. HRMS: calcd, m/z = 410.03453; found, m/z = 410.03509.
3-(4-(3,4-Dichlorophenyl)-5-phenyloxazol-2-yl)benzoic Acid (22d).
Instead of succinic acid, isophthalic acid (21d) was used. Yield: 53 mg (11%). Rf: 0.48 (2:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): 13.40 (bs, 1H, −COOH), 8.60 (t, 1H, J = 1.4 Hz, Ph-COOH-5H), 8.35−8.32 (m, 2H, Ph-COOH-2H,4H), 8.13−8.09 (m, 1H, Ph-COOH-6H), 7.88 (sd, 1H, 2.0 Hz, Ph-Cl-2H), 7.75−7.69 (m, 4H, Ph-Cl-5H, Ph-2H,4H,6H), 7.62 (dd, 1H, J = 8.4 Hz, 2.0 Hz, Ph-Cl-6H). 13C NMR (75 MHz, DMSO-d6): δ = 166.1, 159.0, 146.7, 133.7, 132.4, 131.8, 131.6, 131.1, 131.0, 130.3, 129.9, 129.8, 129.3, 129.1, 127.5, 127.5, 127.1, 126.7, 126.6. HRMS: calcd, m/z = 410.03453; found, m/z = 410.03460.
4-(4-(3,4-Dichlorophenyl)-5-phenyloxazol-2-yl)benzoic Acid (22e).
Instead of succinic acid, terephthalic acid (21e) was used. Yield: 57 mg (pale-yellow solid; 12%). Rf: 0.53 (2:1 Hex/EtOAc + 3% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 13.29 (bs, 1H, −COOH), 8.26−8.20 (m, 2H, Ph-COOH-3H,5H), 8.12−8.10 (m, 2H, Ph-COOH-2H,6H), 7.91 (dd, 1H, J = 8.7 Hz, 1.8 Hz, Ph-3,4diCl-6H), 7.75−7.65 (m, 3H, Ph-3,4-diCl-2H, Ph-3H,5H), 7.62−7.77 (m, 1H, Ph-3,4-diCl-5H), 7.53−7.46 (m, 3H, Ph-2H,4H,6H). 13C NMR (75 MHz, DMSO-d6): δ = 166.7, 159.2, 143.5, 137.9, 132.7, 132.3, 131.9, 131.6, 131.2, 130.1, 129.7, 129.2, 129.0, 128.4, 127.9, 127.5, 127.0, 126.5, 126.3, 126.3. HRMS: calcd, m/z = 410.03453; found, m/z = 410.03478.
5-(2-(3,4-Dichlorophenyl)-2-oxoethoxy)-5-oxopentanoic Acid (24).
To a solution of 21b (3.3 g, 24.6 mmol, 3 equiv) in 150 mL of acetone was added triethylamine (3.5 mL, 24.6 mmol, 3 equiv), and the mixture was stirred at room temperature. After 30 min, 3,4dichlorophenyl bromomethyl ketone (23) (2.3 g, 8.2 mmol, 1 equiv) was added in small portions, and the reaction was monitored by TLC. The mixture was concentrated under reduced pressure and taken up in 50 mL of 1 N hydrochloric acid, followed by extraction with dichloromethane. The solvent was dried over MgSO4 and removed under reduced pressure, and the was residue purified via column chromatography, eluting with 98:2 DCM/MeOH + 1% AcOH, yielding 2.69 g of the product as a tan solid (97% yield). Rf: 0.20 (7:3 Hex/EtOAc + 0.1% AcOH). 1H NMR (250 MHz, CDCl3): δ = 8.01 (sd, 1H, J = 2.0 Hz, Ph-2H), 7.74 (dd, 1H, J = 8.4 Hz, 2.0 Hz, Ph-6H), 7.59 (d, 1H, J = 8.2 Hz, Ph-5H), 5.29 (s, 2H, C(O)−CH2−O), 2.57 (dt, 4H, J = 18.3 Hz, 7.2 Hz, 2 × C(O)−CH2−CH2−), 2.06 (quin, 2H, J = 7.1 Hz, CH2−CH2−CH2). ESI-MS: m/z = 316.9 [M − 2H]−.
4-(4-(3,4-Dichlorophenyl)oxazol-2-yl)butanoic Acid (25).
24 (1.2 g, 3.7 mmol, 1 equiv), acetamide (1.1 g, 18.5 mmol, 5 equiv), and boron trifluoride diethyl etherate (0.5 mL, 3.7 mmol, 1 equiv) were mixed under neat conditions for 30 h at 140 °C. The mixture was poured into 50 mL of 1 N hydrochloric acid, followed by extraction with ethyl acetate. The combined organic phases were washed with brine and dried over MgSO4, and the solvent was removed under reduced pressure. The residue was purified via column chromatography, eluting with 4:1 Hex/acetone, yielding 0.64 g of the product as a brown solid (58% yield). Rf: 0.20 (7:3 Hex/EtOAc + 0.1% AcOH). 1H NMR (250 MHz, DMSO-d6): δ = 12.13 (bs, 1H,−OH), 8.63 (s,1H, oxazolyl-5H), 7.99 (sd, 1H, J = 1.9 Hz, Ph-2H), 7.77−7.66 (m, 2H, Ph-5H,6H), 2.83 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.35 (t, 2H, J = 7.3 Hz, oxazole−CH2−), 1.94 (quin, 2H, J = 7.4 Hz, CH2−CH2− CH2). 13C NMR (75 MHz, DMSO-d6): δ = 178.3, 164.7, 138.6, 134.0, 133.0, 131.7, 131.0, 130.7, 127.3, 124.6, 32.9, 27.2, 21.8. HRMS: calcd, m/z = 300.01888; found, m/z = 300.01896.
4-(5-Bromo-4-(3,4-dichlorophenyl)oxazol-2-yl)butanoic Acid (26).
To a solution of 25 (0.7 g, 2.3 mmol, 1.0 equiv) and ammonium acetate (0.2 g, 0.2 mmol, 0.1 equiv) in 20 mL of acetonitrile was added NBS (0.4 g, 2.4 mmol, 1.05 equiv) in small portions. The mixture was stirred for 1.5 h at room temperature, after which the solvent removed under reduced pressure and the residue was suspended in 100 mL of water. The solidified product was isolated by filtration and purified via column chromatography, eluting with 7:3 Hex/EtOAc + 1% AcOH, yielding 0.58 g of the title compound (67%). Rf: 0.29 (7:3 Hex/EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.97 (sd, 1H, J = 2.0 Hz, Ph-2H), 7.82 (dd, 1H, J = 8.5 Hz, 2.1 Hz, Ph-6H), 7.71 (d, 1H, J = 8.5 Hz, Ph5H), 2.81 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.34 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 1.91 (quin, 2H, J = 7.4 Hz, CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 174.4, 165.9, 134.0, 132.0, 131.5, 131.2, 130.6, 127.6, 126.1, 118.0, 32.9, 27.0, 21.8. HRMS: calcd, m/z = 379.92734; found, m/z = 379.92693.
General Procedure for the Synthesis of Oxaprozin Derivatives 28a−k.
A vial filled with 127 mg of K3PO4, 2 mL of water, and 1 mL of DMF was capped and degassed with argon using an ultrasonic bath. Afterward, tetrakis(triphenylphosphine)palladium(0) (23 mg, 0.02 mmol, 0.1 equiv), 26 (76 mg, 0.2 mmol, 1.0 equiv), and the appropriate boronic acid derivative 27a−k (0.2 mmol, 1.0 equiv) were added to the solution. The resulting mixture was degassed again and heated to 80 °C for 48 h. The mixture was diluted with 20 mL of EtOAc and filtered over silica gel. After the silica gel was rinsed several times with EtOAc, the solution was washed with 1 N hydrochloric acid and brine, dried over MgSO4, and concentrated under reduced pressure. The residue was purified via preparative HPLC.
4-(4-(3,4-Dichlorophenyl)-5-(pyridin-4-yl)oxazol-2-yl)butanoic Acid (28a).
Pyridin-4-ylboronic acid (27a) was used as the coupling reagent. Yield: 2 mg (3%). Rf: 0.21 (98:2 DCM/MeOH + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 8.64 (d, 2H, J = 6.1 Hz, pyridinyl-3H,5H), 7.82 (sd, 1H, J = 2.0 Hz, Ph-2H), 7.73 (d, 1H, J = 8.4 Hz, Ph-5H), 7.56 (dd, 1H, J = 8.4 Hz, 2.0 Hz, Ph-6H), 7.45−7.53 (m, 2H, pyridinyl-2H,6H), 2.91 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.39 (t, 2H, J = 7.1 Hz, oxazole−CH2−), 1.99 (quin, 2H, J = 7.4 Hz, CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 164.6, 150.5, 143.0, 140.7, 139.2, 137.8, 135.3, 134.9, 132.1, 131.7, 131.4, 131.2, 129.6, 127.9, 119.9, 32.8, 26.6, 21.7. HRMS: calcd, m/z = 377.04542; found, m/z = 377.04579.
4-(4-(3,4-Dichlorophenyl)-5-(pyridin-3-yl)oxazol-2-yl)butanoic Acid (28b).
Pyridin-3-ylboronic acid (27b) was used as the coupling reagent. Yield: 8 mg (11%). Rf: 0.26 (98:2 DCM/MeOH + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 8.74 (sdd, 1H, J = 2.2 Hz, 0.7 Hz, pyridinyl-2H), 8.63 (dd, 1H, J = 4.8 Hz, 1.6 Hz, pyridinyl-4H), 7.91−8.01 (m, 1H, pyridinyl-6H), 7.77 (sd, 1H, J = 2.0 Hz, Ph2H), 7.68 (d, 1H, J = 8.4 Hz, Ph-5H), 7.46−7.55 (m, 2H, Ph-6H, pyridinyl-5H), 2.90 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.40 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 1.92−2.08 (m, 2H, CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 174.0, 164.1, 150.0, 147.4, 143.1, 134.4, 133.4, 132.2, 131.6, 131.2, 130.9, 128.9, 127.2, 124.4, 124.1, 32.7, 26.6, 21.7. HRMS: calcd, m/z = 377.04542; found, m/z = 377.04623.
4-(4-(3,4-Dichlorophenyl)-5-(2-fluorophenyl)oxazol-2-yl)butanoic Acid (28c).
(2-Fluorophenyl)boronic acid (27c) was used as the coupling reagent. Yield: 14 mg (18%). Rf: 0.42 (3:2 Hex/ EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.64 (sd, 1H, J = 1.5 Hz, Ph-3,4-diCl-2H), 7.45−7.07 (m, 6H, Ph-3,4-diCl5H,6H, Ph-2-F), 2.89 (t, 2H, J = 7.4 Hz, −CH2−C(O)), 2.48 (t, 2H, J = 6.9 Hz, oxazole−CH2−), 2.12 (quin, 2H, J = 7.3 Hz, CH2−CH2− CH2). 13C NMR (75 MHz, DMSO-d6): δ = 178.1, 163.9, 161.1, 157.7, 140.7, 134.9, 132.6, 131.8, 131.5, 130.5, 130.3, 128.8, 126.0, 124.6, 116.7, 116.4, 32.9, 27.2, 21.80. HRMS: calcd, m/z = 394.04075; found, m/z = 394.04059.
4-(4-(3,4-Dichlorophenyl)-5-(2,5-difluorophenyl)oxazol-2-yl)butanoic Acid (28d).
(2,5-Difluorophenyl)boronic acid (27d) was used as the coupling reagent. Yield: 22 mg (27%). Rf: 0.53 (3:2 Hex/ EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.70 (sd, 1H, J = 2.0 Hz, Ph-3,4-diCl-2H), 7.64 (d, 1H, J = 8.4 Hz, Ph-3,4diCl-5H), 7.50−7.59 (m, 1H, Ph-3,4-diCl-6H), 7.33−7.49 (m, 3H, Ph-2,5-diF), 2.89 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.40 (t, 2H, J = 7.3 Hz, oxazole−CH2−), 1.99 (quin, 2H, J = 7.4 Hz, CH2−CH2− CH2). 13C NMR (75 MHz, DMSO-d6): δ = 174.0, 164.6, 159.8, 156.6, 153.4, 138.9, 134.6, 132.0, 131.4, 130.9, 130.7, 128.1, 126.5, 119.0, 116.8, 32.7, 26.6, 21.7. HRMS: calcd, m/z = 412.03133; found, m/z = 412.03118.
4-(4-(3,4-Dichlorophenyl)-5-(3-fluorophenyl)oxazol-2-yl)butanoic Acid (28e).
(3-Fluorophenyl)boronic acid (27e) was used as the coupling reagent. Yield: 31 mg (40%). Rf: 0.44 (3:2 Hex/ EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 12.19 (s, 1H, −COOH), 7.78 (sd, 1H, J = 2.0 Hz, Ph-3,4-diCl-2H), 7.69 (d, 1H, J = 8.4 Hz, Ph-3,4-diCl-5H), 7.47−7.58 (m, 2H, Ph-3,4-diCl-6H, Ph-3-F-2H), 7.38 (pd, 2H, J = 7.4 Hz, Ph-3-F-4H,6H) 7.22−7.34 (m, 1H, Ph-3-F-5H), 2.88 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.40 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 1.99 (quin, 2H, J = 7.3 Hz, CH2−CH2− CH2). 13C NMR (75 MHz, DMSO-d6): δ = 178.2, 164.5, 163.4, 161.2, 144.9, 133.5, 132.9, 132.4, 131.9, 130.6, 130.1, 129.8, 127.0, 122.1, 115.9, 113.6, 32.9, 27.1, 21.8. HRMS: calcd, m/z = 394.04075; found, m/z = 394.03928.
4-(4-(3,4-Dichlorophenyl)-5-(3-methoxyphenyl)oxazol-2-yl)butanoic Acid (28f).
(3-Methoxyphenyl)boronic acid (27f) was used as the coupling reagent. Yield: 41 mg (50%). Rf: 0.24 (7:3 Hex/ EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.77 (sd, 1H, J = 2.0 Hz,, Ph-3,4-diCl-2H), 7.65 (d, 1H, J = 8.4 Hz, Ph-3,4diCl-5H), 7.52 (dd, 1H, J = 8.4 Hz, 2.0 Hz, Ph-3,4-diCl-6H), 7.38 (t, 1H, J = 7.7 Hz, Ph-3-OCH3-5H), 6.96−7.14 (m, 3 H, Ph-3-OCH32H,4H,6H), 3.74 (s, 3H, −CH3), 2.86 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.39 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 1.98 (quin, 2H, J = 7.3 Hz, CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 174.0, 163.2, 159.5, 145.5, 132.7, 132.1, 131.4, 130.9, 130.5, 130.4, 129.1, 128.9, 127.3, 119.1, 115.2, 112.0, 55.2, 32.8, 26.6, 21.8. HRMS: calcd, m/z = 406.06074; found, m/z = 406.06096.
4-(5-(3-Cyanophenyl)-4-(3,4-dichlorophenyl)oxazol-2-yl)butanoic Acid (28g).
(3-Cyanophenyl)boronic acid (27g) was used as the coupling reagent. Yield: 32 mg (40%). Rf: 0.17 (7:3 Hex/ EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 8.01 (s, 1H, Ph-3,4-diCl-2H), 7.84 (d, 1H, J = 8.1 Hz, Ph-3,4-diCl-5H), 7.89 (d, 1H, J = 7.7 Hz, Ph-3,4-diCl-6H), 7.76 (st, 1H, J = 1.9 Hz, Ph-3CN-2H), 7.60−7.72 (m, 2H, Ph-3-CN-4H,5H), 7.49 (dd, 1H, J = 8.4 Hz, 2.0 Hz, Ph-3-CN-6H), 2.89 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.40 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 2.00 (quin, 2H, J = 7.3 Hz, CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 174.0, 163.9, 143.6, 133.4, 132.7, 132.1, 131.6, 131.2, 131.1, 131.0, 130.4, 130.1, 129.2, 129.0, 127.3, 118.1, 112.4, 32.7, 26.6, 21.7. HRMS: calcd, m/z = 401.04542; found, m/z = 401.04548.
4-(4-(3,4-Dichlorophenyl)-5-(3-(trifluoromethyl)phenyl)oxazol-2yl)butanoic Acid (28h).
(3-(Trifluoromethyl)phenyl)boronic acid (27h) was used as the coupling reagent. Yield: 11 mg (12%). Rf: 0.51 (3:2 Hex/EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.86−7.77 (m, 4H, Ph-3,4-diCl, Ph-3-CF3-6H), 7.73−7.67 (m, 2H, Ph-3-CF3-2H,5H), 7.52 (dd, 1H, J = 8.4 Hz, 2 Hz, Ph-3-CF3-4H), 2.90 (t, 2H, J = 7.4 Hz, −CH2−C(O)), 2.40 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 1.99 (quin, 2H, J = 7.4 Hz, CH2−CH2−CH2). 13C NMR (125 MHz, DMSO-d6): δ = 173.8, 163.6, 143.8, 133.0, 132.0, 131.3, 130.8, 130.7, 130.3, 130.2, 129.7, 129.5, 128.7, 127.2, 125.5, 124.6, 122.8, 32.5, 26.3, 21.5. HRMS: calcd, m/z = 444.03756; found, m/z = 444.03752.
4-(4-(3,4-Dichlorophenyl)-5-(m-tolyl)oxazol-2-yl)butanoic Acid (28i).
m-Tolylboronic acid (27i) was used as the coupling reagent. Yield: 17 mg (21%). Rf: 0.16 (98:2 DCM/MeOH + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.76 (sd, 1H, J = 2.0 Hz, Ph-3,4diCl-2H), 7.64 (d, 1H, J = 8.4 Hz, Ph-3,4-diCl-5H), 7.51 (dd, 1H, J = 8.4 Hz, 2.0 Hz, Ph-3,4-diCl-6H), 7.22−7.41 (m, 4H, Ph-3-CH3), 2.86 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.39 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 2.32 (s, 3H, −CH3), 1.98 (quin, 2H, J = 7.4 Hz, CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 173.9, 163.2, 145.9, 138.5, 132.7, 131.8, 131.4, 130.9, 130.4, 130.0, 129.0, 128.6, 127.9, 127.3, 127.0, 124.0, 32.7, 26.6, 21.8, 20.9. HRMS: calcd, m/z = 390.06583; found, m/z = 390.06567.
4-(4-(3,4-Dichlorophenyl)-5-(3-ethynylphenyl)oxazol-2-yl)butanoic Acid (28j).
(3-Ethynylphenyl)boronic acid (27j) was used as the coupling reagent. Yield: 8 mg (10%). Rf: 0.30 (7:3 Hex/EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.79 (sd, 1H, J = 2.0 Hz, Ph-3,4-diCl-2H), 7.46−7.73 (m, 6H, Ph-3,4-diCl-5H,6H, Ph-3-C≡CH), 4.30 (s, 1H, C≡CH), 2.90 (t, 2H, J = 7.5 Hz, −CH2−C(O)), 2.41 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 2.01 (quin, 2H, J = 7.3 Hz, CH2−CH2−CH2). 13C NMR (75 MHz, DMSO-d6): δ = 174.0, 163.6, 144.6, 132.6, 132.4, 132.3, 131.8, 131.6, 131.5, 131.0, 128.9, 128.7, 128.5, 127.3, 127.2, 122.6, 82.5, 81.8, 32.7, 26.6, 21.7. HRMS: calcd, m/z = 400.05018; found, m/z = 400.05005.
4-(5-(3-Chlorophenyl)-4-(3,4-dichlorophenyl)oxazol-2-yl)butanoic Acid (28k).
(3-Chlorophenyl)boronic acid (27k) was used as the coupling reagent. Yield: 4 mg (5%). Rf: 0.47 (3:2 Hex/EtOAc + 0.1% AcOH). 1H NMR (300 MHz, DMSO-d6): δ = 7.78 (sd, 1H, J = 2.0 Hz, Ph-3,4-diCl-2H), 7.69 (d, 1H, J = 8.4 Hz, Ph-3,4-diCl-5H), 7.58−7.63 (m, 1H, Ph-3,4-diCl-6H), 7.47−7.54 (m, 4H, Ph-3-Cl), 2.88 (t, 2H, J = 7.4 Hz, −CH2−C(O)), 2.39 (t, 2H, J = 7.2 Hz, oxazole−CH2−), 1.99 (quin, 2H, J = 7.3 Hz, CH2−CH2−CH2). 13C NMR (125 MHz, DMSO-d6): δ = 174.0, 163.7, 144.2, 133.8, 133.0, 132.4, 131.6, 131.1, 130.8, 129.9, 129.1, 129.0, 127.4, 126.3, 125.3, 39.9, 32.7, 26.6, 21.7. HRMS: calcd, m/z = 410.01120; found, m/z = 410.01126.
Cloning of the sEH N-Terminal Domain.
The cloning of the construct encoding amino acids 1−222 of sEH with a hexa-His tag at the N-terminus as well as a thrombin cleavage site followed by a second hexa-His tag at the C-terminus was published previously16 and was used for activity assays.
For the crystallization, DSF, and ITC experiments, a new construct was created. The sEH-P sequence covering the amino acids 2−224 of sEH was amplified by PCR (forward primer, ccccGGTACCCTGCGCGCGGCCGTCTTCG; reverse primer, CCCCCTCGAGTTAGGTATTGAGAAGCTGGATTCCGG) from the full-length sEH construct published by Hahn et al.24 The PCR product was cloned into a pET29b derivative (in which the expressed ORF was changed to the one from pBH4 with a TEV site) using KpnI and XhoI. The target protein was expressed as a fusion protein with an N-terminal hexa-His tag followed by a TEV recognition and cleavage site. The plasmid carried a kanamycin resistance gene.
Protein Expression and Purification.
The sEH-P for activity assays and the sEH-H used in the performed activity assays and DSF experiments were expressed and purified as published previously by Klingler et al.16 and Lukin et al.25 The proteins for the assay were flash-frozen in liquid nitrogen after the addition of 20% v/v glycerol and stored at −80 °C.
The expression of sEH-P for the crystallization, DSF, and ITC experiments was done according to previously published expression of full-length sEH,24 while for the purification a modified protocol was used. In brief, 10 mL of the preculture of the BL21(DE3) cells, cotransformed with the sEH-P plasmid as well as a coplasmid for the expression of rare codon tRNAs, was used to inoculate a 1 L culture of autoinduction medium ZYP505226 supplemented with chloramphenicol (35 μg/mL) and kanamycin (100 μg/mL). The cultures were incubated at 37 °C and 180 rpm for 3−4 h, and then the temperature was reduced to 16 °C. The cultures were harvested after 36 h by centrifugation (5554g, 4 °C, 20 min). The resulting pellets were stored at −20 °C or directly processed. For ITC measurements, expression pellets were resuspended in buffer A (50 mM Bis-Tris pH 7.2 (HCl), 500 mM NaCl, 5 mM β-ME, 5% v/v glycerol, 20 mM imidazole pH 7 (HCl)) with one tablet of Complete EDTA Free Protease Inhibitor Mix (Roche, Basel, Switzerland) and a trace amount of DNase I (Applichem, Darmstadt, Germany) and then purified as follows. After passing 20 times through an Invensys APV1000 homogenizer (APV Systems, Denmark), the resulting solution was subsequently centrifuged at 43992g for 60 min at 4 °C to pellet the debris. The supernatant was filtered through a 0.45 μm syringe filter and loaded onto a 5 mL HisTrap HP column (GE Healthcare, Solingen, Germany) pre-equilibrated with buffer A. The column was washed with 1% v/v buffer B (buffer A with 400 mM instead of 20 mM imidazole pH 7 (HCl)), and then the protein was eluted at 50% v/v buffer B. The protein fraction was concentrated to a final volume of 5 mL using a Centriprep YM-3 filter with a molecular weight cutoff (MWCO) of 3000 Da (Millipore, Germany), filtered through a 0.45 μm syringe filter, and applied to a Superdex 200 HiLoad 16/600 column (GE Healthcare, Germany) equilibrated and run at 1 mL/min in phosphatse assay buffer (50 mM acetate, 10 mM MgCl2, pH 5.75 (HCl)) supplemented with an additional 5% v/v glycerol. The concentration of the protein for ITC measurements was determined by absorption measurement with a Nanodrop 2000C micro volume spectrophotometer spectrophotometer (Thermo Fisher Scientific, USA).
For crystallization expression, pellets were resuspended in buffer C (50 mM Bis-Tris pH 7.2 (HCl), 500 mM NaCl, 1 mM TCEP, 5% v/v glycerol) with recommended volume of Protease Inhibitor Mix. The cells were lysed by sonification before 3 mL of 5% PEI pH 7.5 (HCl) solution was added to 100 mL of cell solution, and the solution was subsequently centrifuged to pellet the debris. The supernatant was loaded onto 5 mL of cobalt beads (GE Healthcare, self-loaded with Co) preequilibrated with buffer C. The beads were subsequently washed with buffer C supplemented with 20, 40, 60, and 250 mM imidazole pH 7 (HCl). The protein mainly eluted with the 60 and 250 mM wash fractions. These fractions were concentrated (Amicon Ultra Centrifugal Filters, MWCO = 3000 Da, Merck Millipore) to a final volume of 5 mL and filtered with a 0.45 μm syringe filter, and the sample was loaded on a Superdex 200 HiLoad 16/600 column (GE Healthcare, Germany) equilibrated and run with crystallization buffer (20 mM Tris, 100 mM NaCl, 0.5 mM TCEP, pH 7.5 (HCl)), The concentration of the protein was determined with a Nanodrop 2000C spectrophotometer. The identity of the protein was verified by intact mass spectrometry (data not shown).
The full-length human, mouse, and rat sEH were expressed in a baculovrius/insect cell system and purified by affinity as described previously.27 The concentrated purified enzymes appeared as single bands upon SDS-PAGE separation and Coomassie Brilliant Blue staining with an estimated purity above 95%. The final preparations had no detectable esterase or glutathione S-transferase activity.28
FDP-Based Phosphatase Assay.
The fluorescein diphosphate assay was performed as published previously by Klingler et al.16 using frozen sEH-P as well as full-length human, mouse, and rat sEH. In brief, the assay was performed in a black 96-well microplate in which 1 μL of compound in DMSO (or DMSO only in the case of the positive control) was mixed with 89 μL of protein solution (final concentration of 0.1 μM for sEH-P, full-length human sEH, and full-length rat sEH and 0.01 μM for full-length mouse sEH) in phosphatase assay buffer (50 mM acetate, 10 mM MgCl2, pH 5.75) supplemented with 0.01% w/v Triton X-100. For the blank, assay buffer without protein was used. The plates were incubated at RT for 30 min to allow binding of the inhibitor. To start the reaction, 10 μL of FDP solution (10 μM final concentration) in assay buffer was added, and the fluorescence changes (λex = 485 nm, λem = 525 nm) were detected at 30 time points over the duration of 30 min using an Infinite F200 PRO plate reader (Tecan, Crailsheim, Germany). All of the samples were measured in triplicate on the plate as well as in three separate experiments. The percent inhibition was calculated relative to the activities of the blank well (without protein) and positive control well (without inhibitor). To calculate IC50 values, data obtained from measurements with at least six different inhibitor concentrations were fitted with a sigmoidal dose−response function using GraphPad Prism software (version 5.03; GraphPad Software, Inc.).
DiFMUP-Based Phosphatase Assay.
The 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) (Life Technologies) assay was done to determine the KM of DiFMUP as well as the Ki value of 22b for the sEH-P and full-length human sEH. In brief, the assay was performed in a black 96-well microplate in which different substrate concentrations (final concentations in 100 μL: 600, 400, 300, 200, 100, 30, 10, 3, 1, 0.3, 0.1, and 0 μM) of substrate dissolved in DMSO (or pure DMSO in the case of 0 μM) and acetate buffer (50 mM acetate, 10 mM MgCl2, pH 5.75) were mixed with a protein inhibitor solution (or pure DMSO in case of the control or Km measurements). The solution contained protein (50 nM final concentration for sEH-P as well as for full-length human sEH) in phosphatase assay buffer (50 mM acetate, 10 mM MgCl2, pH 5.75) supplemented with Triton X100 to a final concentration of 0.01% w/v as well as different inhibitor concentrations (final concentrations of 22b: 10, 3, 1, 0.3, 0.1, 0.03, 0.01 μM as well as pure DMSO). The protein inhibitor solution was incubated at RT for 30 min to allow binding of the inhibitor, and then the reaction was started by adding the inhibitor solution to the substrate mix in the plate. The total concentration of DMSO in the assay volume was 3.5% v/v. The fluorescence changes (λex = 360 nm, λem = 465 nm) were detected at 45 time points over the duration of 45 min using an Infinite F200 PRO plate reader. All conditions were measured in triplicate on the plate. The fluorescence changes in the linear phase of the reaction were determined by a linear fit, and the mean and standard deviation were calculated in Microsoft Excel. Values of the different measurements were exported to GraphPad Prism (version 7.05) and analyzed by a mixed-model inhibition fit. The KM value for the DiFMUP was determined from the data of the DMSO measurement using a Michaelis−Menten fit.
PHOME-Based Hydrolase Assay.
The fluorescence-based sEH hydrolase activity assay was performed as described previously25,29 in a 96-well format with some minor modifications.
In short, sEH (0.1 μM final concentration for all tested proteins) was dissolved in hydrolase assay buffer (Bis-Tris buffer, pH 7) supplemented with 0.1 mg/mL BSA as well as Triton-X 100 to a final concentration of 0.01%. Protein solution (89 μL) was incubated with 1 μL of different concentrations of compounds (final DMSO concentration 1%) for 30 min at room temperature. The reaction was started by addition of 10 μL of a cyano(6-methoxy-2naphthalenyl)methyl 3-phenyloxiran-2-ylacetate (PHOME) solution (to a final concentration of 50 μM). Hydrolysis of the nonfluorescent substrate by sEH to release fluorescent 6-methoxynaphthaldehyde was measured (λex = 330 nm, λem = 465 nm) using a Tecan Infinite F200 PRO plate reader. The reaction was monitored over a duration of 45 min (one point every minute). A blank control (no protein and no compound) and a positive control (no compound) were carried out as well. All of the measurements were performed in triplicate.
DSF Experiments.
Differential scanning fluorimetry30 was performed in a PCR plate at a total volume of 40 μL. sEH-H and sEH-P (Cfinal = 5 μM), Triton X-100 (Cfinal = 0.001% w/v), and SYPRO Orange (Thermo Fisher Scientific GmbH, Braunschweig, Germany) (Cfinal = 2.5×) were mixed with either phosphatase assay buffer (50 mM acetate, 10 mM MgCl2, pH 5.75) or hydrolase assay buffer (Bis-Tris buffer, pH 7) and the corresponding inhibitors. All of the inhibitors were tested at a fixed concentration of 50 μM (or pure DMSO in the case of the control). To determine the background fluorescence, a sample without protein was prepared, as well as a protein control without inhibitor. The plate was measured on an Icycler IQ single-color real-time PCR system (MyiQiCycler, BioRad) with an excitation wavelength of 490 nm and emission wavelength of 570 nm. The emission was monitored while the temperature was increased at 0.2 °C per 24 s (25−79.80 °C). Data from the sEH-P measurements were analyzed by determining the maximum of the first derivative curve given by MyIQ 1.0 software in Microsoft Excel after calculation of the mean of the first derivative curves of the triplicates and data normalization. The temperature at the local maximum was assumed to be the melting point. For sEH-H samples, the raw data were analyzed directly in GraphPad Prism (version 5.03) using a Boltzmann sigmoidal fit. The V50 value of the fit was considered as the melting temperature. All of the samples were measured in triplicate on the plate as well as in three separate experiments. The statistical significance of the measured melting points with respect to the DMSO control was determined by a two-tailed t test using GraphPad Prism (version 7.05).
Isothermal Titration Calorimetry.
ITC experiments were performed on a Nano ITC (TA Instruments, Waters GmbH) in inverse mode, with the protein solution in the 250 μL syringe and the inhibitor sample in the 940 μL sample cell. Protein and inhibitor samples were prepared in assay buffer (50 mM acetate, 10 mM MgCl2, 5% v/v glycerol, pH 5.75) supplemented with DMSO to a final concentration of 2%. The fresh protein solution used in the experiments had a fixed concentration of 185 μM. All of the measurements were performed at 25 °C with a stirring speed of 350 rpm. The number of titration steps (20−25) and the volume (8−10 μL) were adjusted according to the specific measurement. The delay between the injections was 200 s. All of the tested inhibitors were prepared as 50 mM DMSO stock solutions. The inhibitor samples were prepared with assay buffer, adjusting the final DMSO concentration in the sample to 2% v/v, while the concentration of inhibitor was chosen to optimize the shape of the curve. The gained data were analyzed using NanoAnalyze Data Analysis (version 3.5.0, TA Instruments).
Crystallization.
Fresh sEH-P was concentrated to a concentration of 30 mg/mL, and then 22d was added as a 50 mM DMSO stock solution to a final DMSO concentration of 2.08% v/v in the sample. After a 1 h incubation on ice, the solution was centrifuged at maximum speed for 10 min (4 °C). The supernatant was used in 96well screens (JCSG7) via the sitting-drop vapor diffusion method, mixing protein and reservoir buffer in three different ratios (50 nL + 100 nL, 75 nL + 75 nL, and 100 nL + 50 nL). Crystals appeared in the 20 °C screen after a few days in the well containing 25% w/v PEG3350, 0.1 M Bis-Tris pH 5.5 as the reservoir buffer. The crystals were transferred into precipitant solution supplemented with 25% ethylene glycol for 1 min before flash-freezing in liquid nitrogen. Diffraction of the crystals was checked in house at the SGC.
X-ray diffraction data for good diffracting crystals were collected at beamline station I02 at the Diamond Light Source (Oxfordshire, England). All of the diffraction data were obtained from a single crystal, and the data reduction was done with xia231 (0.4.0.291ga780859-dials-1.1) while the scaling was done with SCALA.32,33 The initial protein structure was determined at a resolution of 1.55 Å by molecular replacement with the PHASER program34 within the PHENIX software package35 using a truncated version of the full-length sEH structure (PDB code 5ALU) in which coordinates of heteroatoms (water and ligands) were excluded from the starting model.
After several iterative rounds of model building with COOT36 into the 2Fo − Fc electron density map, the model containing the polypeptide chain and the ligand was refined37 to final Rwork and Rfree values of 0.2034 and 0.2305, respectively, using the phenix.refine program38 in the PHENIX software package (version: 1.10.1_2155). The graphical representations were made using MOE (Chemical Computing Group, Montreal, Canada). Statistics of data collection and structural refinement were generated using the “Phenix table one” tool and are summarized in Table 7. The coordinates and structure factor amplitudes for 22d have been deposited in the Protein Data Bank as entry 5MWA.
Table 7.
Data Collection and Refinement Statisticsa
| wavelength | 0.979490 Å |
| resolution range | 42.86−1.55 (1.605−1.55) |
| space group | P212121 |
| unit cell | 52.09 54.11 75.44 90 90 90 |
| total reflections | 197971 (19739) |
| unique reflections | 31021 (1506) |
| multiplicity | 6.4 (6.6) |
| completeness (%) | 0.89 (0.48) |
| mean I/σ(I) | 15.21 (2.68) |
| Wilson B-factor | 14.31 |
| Rmerge | 0.05117 (0.4581) |
| Rmeas | 0.05583 (0.4973) |
| CC1/2 | 0.999 (0.935) |
| CC* | 1 (0.983) |
| reflections used in refinement | 28201 (1506) |
| reflections used for Rfree | 1420 (90) |
| Rwork | 0.2034 (0.2306) |
| Rfree | 0.2305 (0.2527) |
| CCwork | 0.942 (0.853) |
| CCfree | 0.927 (0.800) |
| no. of non-H atoms | 1766 |
| no. of macromolecules | 1634 |
| no. of ligands | 29 |
| no. of protein residues | 209 |
| RMS (bonds) | 0.014 |
| RMS (angles) | 1.23 |
| Ramachandran favored (%) | 98.52 |
| Ramachandran allowed (%) | 1.48 |
| Ramachandran outliers (%) | 0 |
| rotamer outliers (%) | 0.56 |
| clash score | 2.98 |
| average B-factor | 18.49 |
| macromolecules B-factor | 18.22 |
| ligands B-factor | 9.40 |
| solvent B-factor | 25.35 |
Statistics for the highest-resolution shell are shown in parentheses.
Cyclooxygenase Assay.
COX-1 and COX-2 inhibitory activity and selectivity assays were carried out using human platelets and monocytes, respectively.39,40 Briefly, washed human platelets, treated with increasing concentrations of the tested compounds, were challenged with calcium ionophore A23187 in order to stimulate COX-1-dependent thromboxane A2 (TXA2) production. TXB2 (a stable TXA2 metabolite) was evaluated in the supernatant.
COX-2 activity was measured in monocytes resuspended in HBSS in order to avoid compound binding to plasma protein. COX-2 inhibition was evaluated by quantifying prostaglandin E2 (PGE2) production in LPS-challenged preparations pretreated with increasing concentrations of the tested compounds. The PGE2 and TXB2 concentrations were evaluated by liquid chromatography−tandem mass spectrometry using isotopic dilution of the deuterated internal standards (ISs) PGE2-d4 and TXB2-d4 as previously described.39 Briefly, samples were spiked with IS and an aliquot injected into an Agilent 1100 liquid chromatograph. Chromatography was carried out using a reversed phase eluted with a linear gradient from 25 to 100% solvent B (65:35 methanol/acetonitrile) over 10 min (solvent A: 0.05% acetic acid pH 6 with ammonia). The effluent from the HPLC column was directly infused into an API4000 triple-quadrupole mass spectrometer operated in negative ion mode, monitoring the following specific transitions: m/z 351 → 271 for PGE2, m/z 355 → 275 for PGE2-d4, m/z 369 → 169 TXB2, and m/z 373 → 173 for TXB2-d4. Quantitation was carried out using standard curves obtained with synthetic standards.
Hybrid Reporter Gene Assays for PPARα/γ/δ, LXRα/β, RXRα, and FXR.
Plasmids.
The Gal4 fusion receptor plasmids pFA-CMV-hPPARα-LBD,41 pFA-CMV-hPPARγ-LBD,41 pFA-CMV-hPPARδ-LBD,41 pFA-CMV-hLXRα-LBD,42 pFA-CMV-hLXRβ-LBD,42 pFACMV-hRXRα-LBD,43 and pFA-CMV-hFXR-LBD coding for the hinge region and ligand binding domain (LBD) of the canonical isoform of the respective nuclear receptor were reported previously. pFR-Luc (Stratagene) was used as the reporter plasmid, and pRLSV40 (Promega) was used for normalization of the transfection efficiency and cell growth.
Assay Procedure.
HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) high glucose supplemented with 10% FCS, sodium pyruvate (1 mM), penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37 °C and 5% CO2. The day before transfection, HEK293T cells were seeded in 96-well plates (2.5 × 104 cells/well). Before transfection, the medium was changed to OptiMEM without supplements. Transient transfection was carried out using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer’s protocol with pFR-Luc (Stratagene), pRL-SV40 (Promega), and pFA-CMV-hNR-LBD. Five hours after transfection, the medium was changed to Opti-MEM supplemented with penicillin (100 units/mL) and streptomycin (100 μg/mL) and now additionally containing 0.1% DMSO and the respective test compound or 0.1% DMSO alone as an untreated control. Each concentration was tested in triplicate, and each experiment was repeated independently at least three times. Following overnight (12−14 h) incubation with the test compounds, the cells were assayed for luciferase activity using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol. Luminescence was measured with an Infinite M200 luminometer (Tecan Deutschland GmbH). Normalization of the transfection efficiency and cell growth was done by dividing the firefly luciferase data by the Renilla luciferase data and multiplying the value by 1000, resulting in relative light units (RLUs). Fold activation was obtained by dividing the mean RLU of a test compound at a given concentration by the mean RLU of the untreated control. Relative activation was obtained by dividing the fold activation of a test compound at a given concentration by the fold activation of the respective reference agonist at 1 μM (PPARα, GW7647; PPARγ, pioglitazone; PPARδ, L165,041; LXRα/β, T0901317; RXRα, bexarotene; FXR, GW4064). All of the hybrid assays were validated with the above-mentioned reference agonists, which yielded EC50 values in agreement with the literature.
Supplementary Material
ACKNOWLEDGMENTS
The crystal diffraction experiments were performed on beamline station I02 at the Diamond Light Source (Oxfordshire, England). We thank Dr. F. J. Sorrell and Dr. N. Fox for assistance and support in using the beamline.
Funding
This work was supported by Deutsche Forschungsgemein-schaft (DFG) (Sachbeihilfe PR1405/2-2; SFB 1039 Teilprojekt A07; Heisenberg-Professur PR1405/4-1) and by the research funding program Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) of the State of Hessen, Research Center for Translational Medicine and Pharmacology (TMP). This work was supported in part by NIEHS Grant ES02710 and NIEHS Superfund Grant P42 ES04699. A.C. acknowledges funding by the SFB autophagy. A.C. and S.K. are grateful for support by the German Cancer Network DKTK and the SGC, a registered charity (no. 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through the Ontario Genomics Institute [OGI-055], Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD Grant 115766], Janssen, Merck KGaA, MSD, Novartis Pharma AG, the Ontario Ministry of Research, Innovation and Science (MRIS), Pfizer, Saõ Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. This work was cosupported in part for the in vivo study by the French National Research Agency (ANR-16-CE17-0012), European Union, and Normandie Regional Council. Europe gets involved in Normandie with the European Regional Development Fund (ERDF).
ABBREVIATIONS USED
- BCA assay
bicinchoninic acid assay
- CIU
N-cyclohexyl-N′-(4iodophenyl)urea
- COX-1
cyclooxygenase 1
- COX-2
cyclooxygenase 2
- CV
coefficient of variance
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- DSF
differential scanning fluorimetry
- EET
epoxyeicosatrienoic acid
- EpFA
epoxy fatty acid
- FCS
fetal calf serum
- FDP
fluorescein diphosphate
- FXR
farnesoid X receptor
- HTS
high-throughput screening
- IS
internal standard
- ITC
isothermal titration calorimetry
- LBD
ligand binding domain
- LDA
lithium diisopropylamide
- LXR
liver X receptor
- NBS
N-bromosuccinimide
- ORF
open reading frame
- PGE2
prostaglandin E2
- PPAR
peroxisome proliferator-activated receptor
- RLU
relative light units
- ROS
reactive oxygen species
- RXR
retinoid X receptor
- SAR
structure−activity relationship
- SDS
sodium dodecyl sulfate
- sEH
soluble epoxide hydrolase
- sEH-H
C-terminal/hydrolase domain of sEH
- sEHI
inhibitors of the C-terminal domain of sEH
- sEHP
N-terminal/phosphatase domain of sEH
- TBAF
tetrabutylammonium fluoride
- TCEP
tris(2-carboxyethyl)phosphine
- TEV
tobacco etch virus
- THF
tetrahydrofuran
- TMSCN
trimethylsilyl cyanide
- TXA2
thromboxane A2
- TXB2
thromboxane B2
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publication Web site at DOI: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.9b00445.
DSF experiments; experimental procedures of the performed animal experiments, including details about animals, surgery, treatment overview, sample collection, quantification of 22b, and determination of rat plasma levels of sEH substrates and metabolites; and procedures and results of PBMC experiments (PDF)
Molecular SMILES strings and IC50 values (CSV)
Accession Codes
Coordinates and structure factors for 22d have been deposited in the Protein Data Bank with code 5MWA.
The authors declare no competing financial interest.
REFERENCES
- (1).Arand M; Cronin A; Oesch F; Mowbray SL; Alwyn Jones T. The Telltale Structures of Epoxide Hydrolases. Drug Metab. Rev 2003, 35 (4), 365–383. [DOI] [PubMed] [Google Scholar]
- (2).Wagner KM; McReynolds CB; Schmidt WK; Hammock BD Soluble Epoxide Hydrolase as a Therapeutic Target for Pain, Inflammatory and Neurodegenerative Diseases. Pharmacol. Ther 2017, 180, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Inceoglu B; Bettaieb A; Haj FG; Gomes AV; Hammock BD Modulation of Mitochondrial Dysfunction and Endoplasmic Reticulum Stress Are Key Mechanisms for the Wide-Ranging Actions of Epoxy Fatty Acids and Soluble Epoxide Hydrolase Inhibitors. Prostaglandins Other Lipid Mediators 2017, 133, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lazaar AL; Yang L; Boardley RL; Goyal NS; Robertson J; Baldwin SJ; Newby DE; Wilkinson IB; Tal-Singer R; Mayer RJ; Cheriyan J. Pharmacokinetics, Pharmacodynamics and Adverse Event Profile of GSK2256294, a Novel Soluble Epoxide Hydrolase Inhibitor. Br. J. Clin. Pharmacol 2016, 81 (5), 971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cronin A; Mowbray S; rk H; Homburg S; Fleming I; Fisslthaler B; Oesch F; Arand M. The N-Terminal Domain of Mammalian Soluble Epoxide Hydrolase Is a Phosphatase. Proc. Natl. Acad. Sci. U. S. A 2003, 100 (4), 1552–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Newman JW; Morisseau C; Harris TR; Hammock BD The Soluble Epoxide Hydrolase Encoded by EPXH2 Is a Bifunctional Enzyme with Novel Lipid Phosphate Phosphatase Activity. Proc. Natl. Acad. Sci. U. S. A 2003, 100 (4), 1558–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Tran KL; Aronov PA; Tanaka H; Newman JW; Hammock BD; Morisseau C. Lipid Sulfates and Sulfonates Are Allosteric Competitive Inhibitors of the N-Terminal Phosphatase Activity of the Mammalian Soluble Epoxide Hydrolase. Biochemistry 2005, 44 (36), 12179–12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Morisseau C; Schebb NH; Dong H; Ulu A; Aronov PA; Hammock BD Role of Soluble Epoxide Hydrolase Phosphatase Activity in the Metabolism of Lysophosphatidic Acids. Biochem. Biophys. Res. Commun 2012, 419 (4), 796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Oguro A; Imaoka S. Lysophosphatidic Acids Are New Substrates for the Phosphatase Domain of Soluble Epoxide Hydrolase. J. Lipid Res 2012, 53 (3), 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).De Vivo M; Ensing B; Dal Peraro M; Gomez GA; Christianson DW; Klein ML Proton Shuttles and Phosphatase Activity in Soluble Epoxide Hydrolase. J. Am. Chem. Soc 2007, 129 (2), 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kramer J; Proschak E. Phosphatase Activity of Soluble Epoxide Hydrolase. Prostaglandins Other Lipid Mediators 2017, 133, 88–92. [DOI] [PubMed] [Google Scholar]
- (12).Hou H-H; Liao Y-J; Hsiao S-H; Shyue S-K; Lee T-S Role of Phosphatase Activity of Soluble Epoxide Hydrolase in Regulating Simvastatin-Activated Endothelial Nitric Oxide Synthase. Sci. Rep 2015, 5, 13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Enayetallah AE; Grant DF Effects of Human Soluble Epoxide Hydrolase Polymorphisms on Isoprenoid Phosphate Hydrolysis. Biochem. Biophys. Res. Commun 2006, 341 (1), 254–260. [DOI] [PubMed] [Google Scholar]
- (14).Matsumoto N; Suzuki E; Ishikawa M; Shirafuji T; Hasumi K. Soluble Epoxide Hydrolase as an Anti-Inflammatory Target of the Thrombolytic Stroke Drug SMTP-7. J. Biol. Chem 2014, 289 (52), 35826–35838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Morisseau C; Sahdeo S; Cortopassi G; Hammock BD Development of an HTS Assay for EPHX2 Phosphatase Activity and Screening of Nontargeted Libraries. Anal. Biochem 2013, 434 (1), 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Klingler F-M; Wolf M; Wittmann S; Gribbon P; Proschak E. Bacterial Expression and HTS Assessment of Soluble Epoxide Hydrolase Phosphatase. J. Biomol. Screening 2016, 21 (7), 689–694. [DOI] [PubMed] [Google Scholar]
- (17).Stork G; Maldonado L. Anions of Protected Cyanohydrins as Acyl Carbanion Equivalents and Their Use in a New Synthesis of Ketones. J. Am. Chem. Soc 1971, 93 (20), 5286–5287. [Google Scholar]
- (18).Topliss JG Utilization of Operational Schemes for Analog Synthesis in Drug Design. J. Med. Chem 1972, 15 (10), 1006–1011. [DOI] [PubMed] [Google Scholar]
- (19).Argiriadi MA; Morisseau C; Goodrow MH; Dowdy DL; Hammock BD; Christianson DW Binding of Alkylurea Inhibitors to Epoxide Hydrolase Implicates Active Site Tyrosines in Substrate Activation. J. Biol. Chem 2000, 275 (20), 15265–15270. [DOI] [PubMed] [Google Scholar]
- (20).Heroven C; Georgi V; Ganotra GK; Brennan P; Wolfreys F; Wade RC; Fernandez-Montalván, A. E.; Chaikuad, A.; Knapp, S.´ Halogen−Aromatic π Interactions Modulate Inhibitor Residence Times. Angew. Chem., Int. Ed 2018, 57 (24), 7220–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Falke H; Chaikuad A; Becker A; Loaec N; Lozach O; Abü Jhaisha S; Becker W; Jones PG; Preu L; Baumann K; Knapp S; Meijer L; Kunick C. 10-Iodo-11H-Indolo[3,2-c]Quinoline-6-Carboxylic Acids Are Selective Inhibitors of DYRK1A. J. Med. Chem 2015, 58 (7), 3131–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Öster L; Tapani S; Xue Y; Kack H. Successful Generation̈ of Structural Information for Fragment-Based Drug Discovery. Drug Discovery Today 2015, 20 (9), 1104–1111. [DOI] [PubMed] [Google Scholar]
- (23).Barbosa-Sicard E; Fromel T; Keserü B; Brandes RP; Morisseau C; Hammock BD; Braun T; ger M; Fleming I. Inhibition of the Soluble Epoxide Hydrolase by Tyrosine Nitration. J. Biol. Chem 2009, 284 (41), 28156–28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hahn S; Achenbach J; Buscató E; Klingler F-M; Schroeder M; Meirer K; Hieke M; Heering J; Barbosa-Sicard E; Loehr F; Fleming I; Doetsch V; Schubert-Zsilavecz M; Steinhilber D; Proschak E. Complementary Screening Techniques Yielded Fragments That Inhibit the Phosphatase Activity of Soluble Epoxide Hydrolase. ChemMedChem 2011, 6 (12), 2146–2149. [DOI] [PubMed] [Google Scholar]
- (25).Lukin A; Kramer J; Hartmann M; Weizel L; Hernandez-Olmos V; Falahati K; Burghardt I; Kalinchenkova N; Bagnyukova D; Zhurilo N; Rautio J; Forsberg M; Ihalainen J; Auriola S; Leppanen J; Konstantinov I; Pogoryelov D; Pröschak E; Dar’in D; Krasavin M. Discovery of Polar Spirocyclic Orally Bioavailable Urea Inhibitors of Soluble Epoxide Hydrolase. Bioorg. Chem 2018, 80, 655–667. [DOI] [PubMed] [Google Scholar]
- (26).Studier FW Protein Production by Auto-Induction in High Density Shaking Cultures. Protein Expression Purif. 2005, 41 (1), 207–234. [DOI] [PubMed] [Google Scholar]
- (27).Morisseau C; Beetham JK; Pinot F; Debernard S; Newman JW; Hammock BD Cress and Potato Soluble Epoxide Hydrolases: Purification, Biochemical Characterization, and Comparison to Mammalian Enzymes. Arch. Biochem. Biophys 2000, 378 (2), 321–332. [DOI] [PubMed] [Google Scholar]
- (28).Morisseau C; Merzlikin O; Lin A; He G; Feng W; Padilla I; Denison MS; Pessah IN; Hammock BD Toxicology in the Fast Lane: Application of High-Throughput Bioassays to Detect Modulation of Key Enzymes and Receptors. Environ. Health Perspect 2009, 117 (12), 1867–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wolf NM; Morisseau C; Jones PD; Hock B; Hammock BD Development of a High-Throughput Screen for Soluble Epoxide Hydrolase Inhibition. Anal. Biochem 2006, 355 (1), 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Niesen FH; Berglund H; Vedadi M. The Use of Differential Scanning Fluorimetry to Detect Ligand Interactions That Promote Protein Stability. Nat. Protoc 2007, 2 (9), 2212–2221. [DOI] [PubMed] [Google Scholar]
- (31).Winter G; Lobley CMC; Prince SM Decision Making in Xia2. Acta Crystallogr., Sect. D: Biol. Crystallogr 2013, 69 (7), 1260–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Winn MD; Ballard CC; Cowtan KD; Dodson EJ; Emsley P; Evans PR; Keegan RM; Krissinel EB; Leslie AGW; McCoy A; McNicholas SJ; Murshudov GN; Pannu NS; Potterton EA; Powell HR; Read RJ; Vagin A; Wilson KS Overview of the CCP4 Suite and Current Developments. Acta Crystallogr., Sect. D: Biol. Crystallogr 2011, 67 (4), 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Evans PR An Introduction to Data Reduction: Space-Group Determination, Scaling and Intensity Statistics. Acta Crystallogr., Sect. D: Biol. Crystallogr 2011, 67 (4), 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).McCoy AJ; Grosse-Kunstleve RW; Adams PD; Winn MD; Storoni LC; Read RJ Phaser Crystallographic Software. J. Appl. Crystallogr 2007, 40 (4), 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung L-W; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66 (2), 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Emsley P; Lohkamp B; Scott WG; Cowtan K. Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66 (4), 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Moriarty NW; Grosse-Kunstleve RW; Adams PD Electronic Ligand Builder and Optimization Workbench (ELBOW): A Tool for Ligand Coordinate and Restraint Generation. Acta Crystallogr., Sect. D: Biol. Crystallogr 2009, 65 (10), 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Afonine PV; Grosse-Kunstleve RW; Echols N; Headd JJ; Moriarty NW; Mustyakimov M; Terwilliger TC; Urzhumtsev A; Zwart PH; Adams PD Towards Automated Crystallographic Structure Refinement with phenix.refine. Acta Crystallogr., Sect. D: Biol. Crystallogr 2012, 68 (4), 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hoxha M; Buccellati C; Capra V; Garella D; Cena C; Rolando B; Fruttero R; Carnevali S; Sala A; Rovati GE; Bertinaria M. In Vitro Pharmacological Evaluation of Multitarget Agents for Thromboxane Prostanoid Receptor Antagonism and COX2 Inhibition. Pharmacol. Res 2016, 103, 132–143. [DOI] [PubMed] [Google Scholar]
- (40).Bertinaria M; Shaikh MAAG; Buccellati C; Cena C; Rolando B; Lazzarato L; Fruttero R; Gasco A; Hoxha M; Capra V; Sala A; Rovati GE Designing Multitarget Anti-Inflammatory Agents: Chemical Modulation of the Lumiracoxib Structure toward Dual Thromboxane Antagonists-COX-2 Inhibitors. ChemMedChem 2012, 7 (9), 1647–1660. [DOI] [PubMed] [Google Scholar]
- (41).Rau O; Wurglics M; Paulke A; Zitzkowski J; Meindl N; Bock A; Dingermann T; Abdel-Tawab M; Schubert-Zsilavecz M. Carnosic Acid and Carnosol, Phenolic Diterpene Compounds of the Labiate Herbs Rosemary and Sage, Are Activators of the Human Peroxisome Proliferator-Activated Receptor Gamma. Planta Med. 2006, 72 (10), 881–887. [DOI] [PubMed] [Google Scholar]
- (42).Heitel P; Achenbach J; Moser D; Proschak E; Merk D. DrugBank Screening Revealed Alitretinoin and Bexarotene as Liver X Receptor Modulators. Bioorg. Med. Chem. Lett 2017, 27 (5), 1193–1198. [DOI] [PubMed] [Google Scholar]
- (43).Flesch D; Cheung S-Y; Schmidt J; Gabler M; Heitel P; Kramer J; Kaiser A; Hartmann M; Lindner M; Lüddens-Dämgen K; Heering J; Lamers C; ddens H; Wurglics M; Proschak E; Schubert-Zsilavecz M; Merk D. Nonacidic Farnesoid X Receptor Modulators. J. Med. Chem 2017, 60 (16), 7199–7205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.