Abstract
The transient receptor potential melastatin member 8 (TRPM8) ion channel is the primary detector of environmental cold and an important target for treating pathological cold hypersensitivity. Here, we present cryo-electron microscopy structures of TRPM8 in ligand-free, antagonist-bound, or calcium-bound forms, revealing how robust conformational changes give rise to two non-conducting states, closed and desensitized. We describe a malleable ligand-binding pocket that accommodates drugs of diverse chemical structures, and we delineate the ion permeation pathway, including the contribution of lipids to pore architecture. Furthermore, we show that direct calcium binding mediates stimulus-evoked desensitization, clarifying this important mechanism of sensory adaptation. We observe large rearrangements within the S4-S5 linker that reposition the S1-S4 and pore domains relative to the TRP helix, leading us to propose a distinct model for modulation of TRPM8 and possibly other TRP channels.
Transient receptor potential melastatin member 8 (TRPM8) is a cold- and menthol-activated ion channel that plays an essential role in the detection of environmental temperatures (1–5). It is also targeted by synthetic cooling agents in personal care products and confectionaries (6) and by antagonists that may be useful for reducing cold hypersensitivity resulting from nerve damage (7, 8). TRPM8 blockers may also be beneficial in treating chronic cough, asthma, or other airway hyperactivity syndromes that are exacerbated by cold (9, 10), further highlighting interest in understanding how these compounds interact with the channel.
TRPM8, like many other TRP subtypes, is a polymodal, non-selective cation channel with substantial permeability to calcium ions (PCa2+/PNa+ ~3) (1). Moreover, cold- or cooling agent-evoked responses show calcium-dependent desensitization (a phenomenon that likely contributes to cold adaptation), and the synthetic “super cooling” agent icilin activates the channel in a calcium-dependent manner (11). Indeed, a conserved TRPM channel calcium-binding site has been identified in close proximity to a TRPM8 agonist-binding pocket (12–14), but whether this site contributes to agonist-evoked desensitization has not been determined.
Recently published TRPM8 structures from the collared flycatcher (cfTRPM8), either ligand-free or in complex with cooling agents and the positive regulator, phosphatidylinositol-4,5-bisphosphate (PIP2), describe the overall architecture of the channel and reveal binding sites for these ligands (14, 15). However, the local resolution of the transmembrane domain in the cryo-electron microscopy (cryo-EM) maps was insufficient for unambiguous and complete model building, and regions corresponding to the selectivity filter and outer pore loop were invisible, limiting our understanding of the molecular mechanisms of channel gating at the selectivity filter or lower gate. To learn how ligand binding and channel gating are coupled, we determined cryo-EM structures of TRPM8 alone or in complex with antagonists or calcium.
Visualizing TRPM8 in distinct conformational states
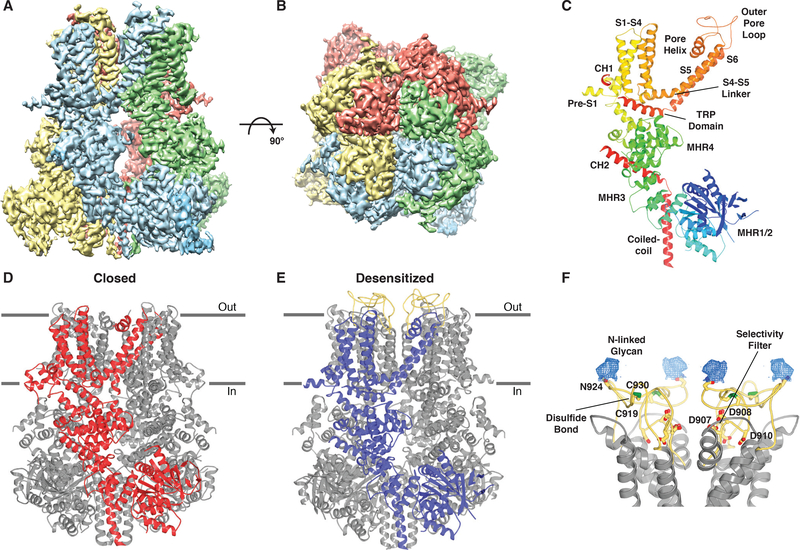
We found that channels from birds exhibited superior conformational homogeneity compared with other vertebrate species, in agreement with previous studies (15). We also found that TRPM8 from Parus major (great tit; pmTRPM8) produced the highest resolution reconstructions. TRPM8 is remarkably well conserved across vertebrate species, with human and avian orthologs sharing >80% and 85% amino acid sequence identity and similarity, respectively (Fig. S1). Recombinant full-length pmTRPM8 protein was purified in detergent supplemented with cholesterol hemisuccinate (CHS) and reconstituted into amphipols. Structures in ligand-free, antagonist-bound (AMTB or TC-I 2014), and calcium-bound states were determined using cryo-EM to average resolutions of 3.6 Å, 3.2 Å, 3.0 Å, and 3.2 Å, respectively (Fig. S2–S9). The cryo-EM density maps are of high quality, allowing for construction of atomic models with good stereochemistry and correlation with the density (Fig. 1A, B; S10 and Table S1).
Fig. 1. Structure of TRPM8 in two distinct states.
(A-B) Side view (A) and top view (B) of the cryo-EM density map of the desensitized state of TRPM8 with subunits differentiated by color. (C) Monomer of the desensitized state with secondary structure elements colored blue-to-red from N-terminus to C-terminus and domains labeled. (D) Ribbon representation of the closed state of TRPM8 (TC-I 2014-bound) with a single subunit colored red. Horizontal lines indicate the approximate boundaries of the cell membrane. (E) Ribbon representation of the desensitized state of TRPM8 with a single subunit colored blue. The outer pore loop is colored yellow. (F) Close-up view of the outer pore loop. Conserved residues important for selectivity (Asp907, Asp908, and Asp910), disulfide bond formation (Cys919 and Cys930), and N-linked glycosylation (Asp924) are drawn as sticks. Nitrogen, blue; oxygen, red; sulfur, green. Density (blue mesh, 4σ contour) for the N-linked glycan is shown.
Overall, our structures conform to the characteristic homotetrameric arrangement described for cfTRPM8 and other TRPM family members, including N-terminal homology regions (MHR1-MHR4), six transmembrane helices (S1-S6) arranged in a domain swap architecture, and a C-terminal coiled-coil (Fig. 1; S11A–C) (13, 15–17). The pore of TRPM8 is formed by S5 and S6, as well as the intervening pore helix and outer pore loop (Fig. 1C). The 44-residue outer pore loop was disordered in the cfTRPM8 structures but is clearly visible in our calcium-bound state (Fig. 1A–C, E), allowing for the ion conduction pathway to be modeled in its entirety. The pore can be divided into two portions: the negatively charged selectivity filter (Fig. S8I) that occupies the outer leaflet of the membrane and the lower gate within the inner leaflet. The outer pore loop, which is essential for channel functionality, is both disulfide bonded and glycosylated (Fig. 1F) (18). Unexpectedly, our structures in distinct conformational states reveal large gating movements, including an unusual formation and dissolution of the S4-S5 linker and a repositioning of the TRP domain. Although we describe the most highly resolved and complete structure obtained for each condition, these likely represent conformational sub-states sampled by this thermosensitive channel (our final reconstructions are derived from ~3% of particles) (Fig. S3; S5; S7; S9). For example, our dataset obtained in the presence of calcium yielded additional high-resolution structures that indicate heterogeneity within the outer pore loop and the region of calcium binding (S2-S3) (Fig. S12).
An adaptable binding pocket for modulators
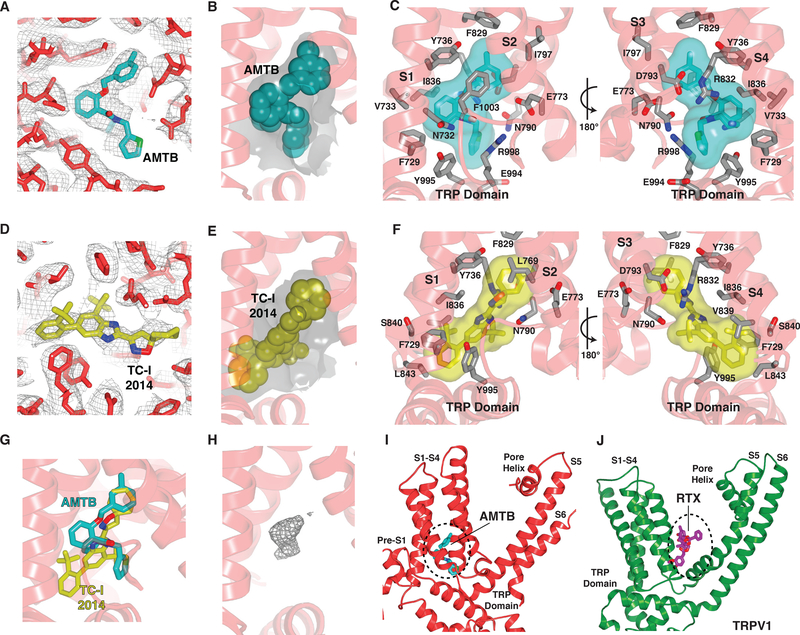
All known TRPM8 ligands (agonists and antagonists, natural or synthetic) have as their chemical core one or more pentameric or hexameric rings, but they bear a range of substituents (19), suggesting that adaptability of the binding pocket is a main determinant of high affinity binding. To explore this concept further, we determined structures in the presence of two chemically distinct antagonists, AMTB and TC-I 2014 (20, 21) (Fig. S4H; S6H), both of which inhibit menthol-evoked responses in oocytes expressing pmTRPM8 (Fig. S4I; S6I). The antagonists are nestled in a membrane-embedded cleft formed by the lower half of the S1-S4 domain, near the membrane-cytosol interface, where they are encapsulated in a pocket that is lined by both hydrophobic and charged residues contributed by the S1-S4 helices and the TRP domain (Fig. 2A–F). Substituents on the rings do not form specific ionic interactions with charged side chains, substantiating shape complementarity as the major determinant of ligand recognition (Fig. 2B–C, E–F). Furthermore, although both drugs bind within this same pocket, they dock in distinct orientations (Fig. 2G), which is facilitated by complementary rearrangement of the side chains that define cavity shape. This ligand-binding pocket also accommodates agonists (although their precise binding orientations were not ascertained) (14) (Fig. S13A, B). The overall structure of TRPM8 remains unaltered in ligand-free or antagonist-bound maps, suggesting that these chemically distinct inhibitors mediate their effects by binding to this promiscuous pocket and locking the channel in its ligand-free configuration.
Fig. 2. Binding site for hydrophobic modulators of TRPM8.
(A) Density within the AMTB-binding site. The map is contoured at 3σ (gray mesh). Stick representation of AMTB-bound TRPM8 with the antagonist AMTB colored teal. (B) Gray surface indicates the shape of the binding pocket, as dictated by residues lining the cavity. AMTB is shown as spheres. (C) Interactions with AMTB (transparent surface and sticks with teal carbon atoms). Nitrogen, blue; oxygen, red. (D) Density within the TC-I 2014-binding site. The map is contoured at 3σ (gray mesh). Stick representation of TC-I 2014-bound TRPM8 with the antagonist TC-I 2014 colored yellow. (E) Gray surface indicates the shape of the binding pocket, as dictated by residues lining the cavity. TC-I 2014 is shown as spheres. (F) Interactions with TC-I 2014 (transparent surface and sticks with yellow carbon atoms). (G) The antagonists AMTB and TC-I 2014 adopt distinct poses in the ligand-binding pocket. (H) Unassigned density (gray mesh, 8σ contour) observed in the ligand-binding pocket of ligand-free TRPM8. (I) Binding site for hydrophobic modulators in TRPM8. The dashed region demarcates the binding pocket. (J) Structure of TRPV1 highlighting the resiniferatoxin (RTX) binding site. The dashed region demarcates the binding site for hydrophobic modulators in TRPV1.
In the absence of an exogenous ligand we observed a discrete density in the ligand-binding pocket (Fig. 2H). Although we cannot identify the molecule corresponding to this density, its presence suggests that ligands do not bind to an empty cavity, but rather displace an endogenous molecule. This observation may have potential physiologic significance and is important to bear in mind when assigning density in this location to an exogenous ligand. This is reminiscent of the vanilloid-binding pocket in TRPV1, which is occupied by a phosphatidylinositol lipid in its apo state (22, 23). This similarity notwithstanding, the location of the TRPM8 ligand-binding pocket is distinct from that described for other TRP channels. For example, vanilloid ligands bind to TRPV1 in an elbow between the S1-S4 and S5-S6 domains and above the S4-S5 linker (Fig. 2I, J) (22, 23), and nucleotide ligands bind within cytoplasmic regions of TRPM2 and TRPM4 (Fig. S11A, B) (13, 16).
Structural mechanism of calcium-dependent modulation
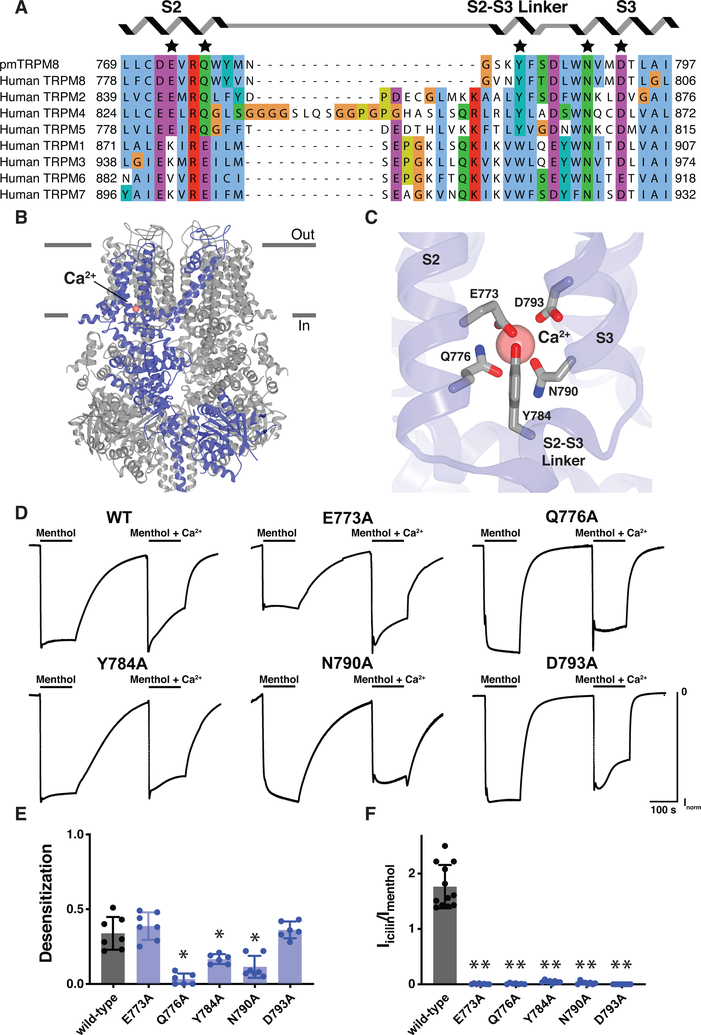
TRPM8 currents show pronounced calcium-dependent desensitization during continuous agonist application (11). We therefore determined the structure of TRPM8 in the presence of calcium to identify the major site(s) of interaction and ascertain whether divalent cation binding is associated with specific conformations, particularly a desensitized state. We found that calcium is bound in a manner analogous to that previously described for a subset of TRPM channels (Fig. S11A, B) (12–14), whereby the ion is coordinated by four negatively charged residues (Glu773, Gln776, Asn790, and Asp793) from the S2 and S3 transmembrane helices (Fig. 3A–C), adjacent to the ligand-binding pocket described above. We also observed involvement of a fifth side chain belonging to Tyr784 within the S2-S3 linker (Fig. 3A, C).
Fig. 3. TRPM8 calcium-binding site.
(A) Sequence alignment of the calcium-binding site within the TRPM subfamily. The conserved binding site is shared by TRPM2, TRPM4, TRPM5, and TRPM8. Residues that coordinate the calcium ion are indicated with a star. Alignment was made with ClustalW. (B) Ribbon representation of calcium-bound TRPM8 with a single subunit colored blue and the calcium ion colored pink (sphere). Horizontal lines indicate the approximate boundaries of the cell membrane. (C) Interactions with calcium (pink sphere). Nitrogen, blue; oxygen, red. (D and E) Structure-function analysis of the role of the calcium-binding site in TRPM8 desensitization. In oocytes expressing pmTRPM8, application of menthol (100 μM) evoked inward currents for which desensitization was observed in the presence of calcium. However, upon mutating several of the residues important for calcium coordination, TRPM8 desensitization was reduced. The A796G mutation was introduced to all constructs to sensitize avian TRPM8 to the agonist icilin. Voltage was held at −60 mV. Desensitization is reported as Ifinal/Iinitial (maximum currents ranged from 0.5–4 mA) for the application of menthol in the presence of calcium. Data represent n=6 or 7 oocytes. Asterisk indicates p < 0.01 comparing wild type and each mutant construct with unpaired two-tailed Student’s t-tests. Representative traces are shown in (D). (F) Structure-function analysis of the role of the calcium-binding site in co-agonism with icilin. In oocytes expressing pmTRPM8, application of menthol (100 μM) or icilin (10 μM), in the presence of calcium, evoked inward currents. However, upon mutating residues proposed to be important for calcium coordination, there was no longer a response to icilin. The A796G mutation was introduced to all constructs to sensitize avian TRPM8 to the agonist icilin. Voltage was held at −60 mV. Data represent n=7 to 12 oocytes. Two asterisks indicate p < 1×10−8 comparing wild type and each mutant construct with unpaired two-tailed Student’s t-tests.
To determine whether this calcium-binding site is responsible for calcium-dependent desensitization, we analyzed menthol-evoked responses for TRPM8 channels bearing alanine substitutions at the identified calcium-binding residues (Fig. 3D, E). Several of these mutations diminished desensitization, with the most pronounced effects associated with Gln776 and Asn790, the acidic residues that form the base of the binding site. By introducing a A796G mutation into the pmTRPM8 channel (a modification that renders avian TRPM8 icilin-sensitive) (11) we could also show that all five calcium-binding residues are required for icilin sensitivity (Fig. 3F). Mutation of Glu773 and Asp793 abrogated icilin-evoked responses without impairing desensitization, suggesting that these actions require high and low calcium-binding affinities, respectively.
Conformational states associated with ligand and lipid binding
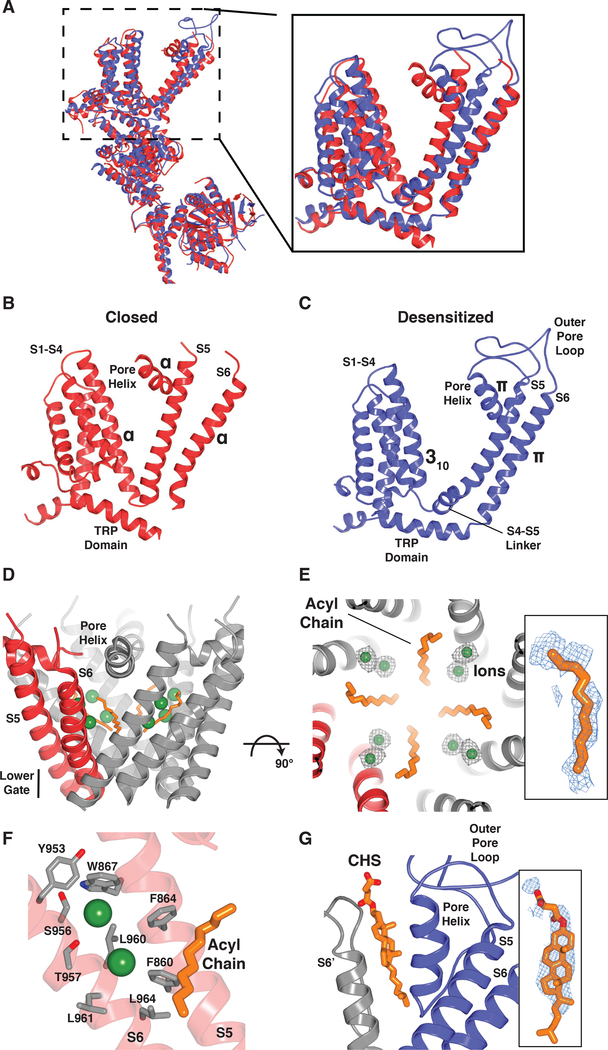
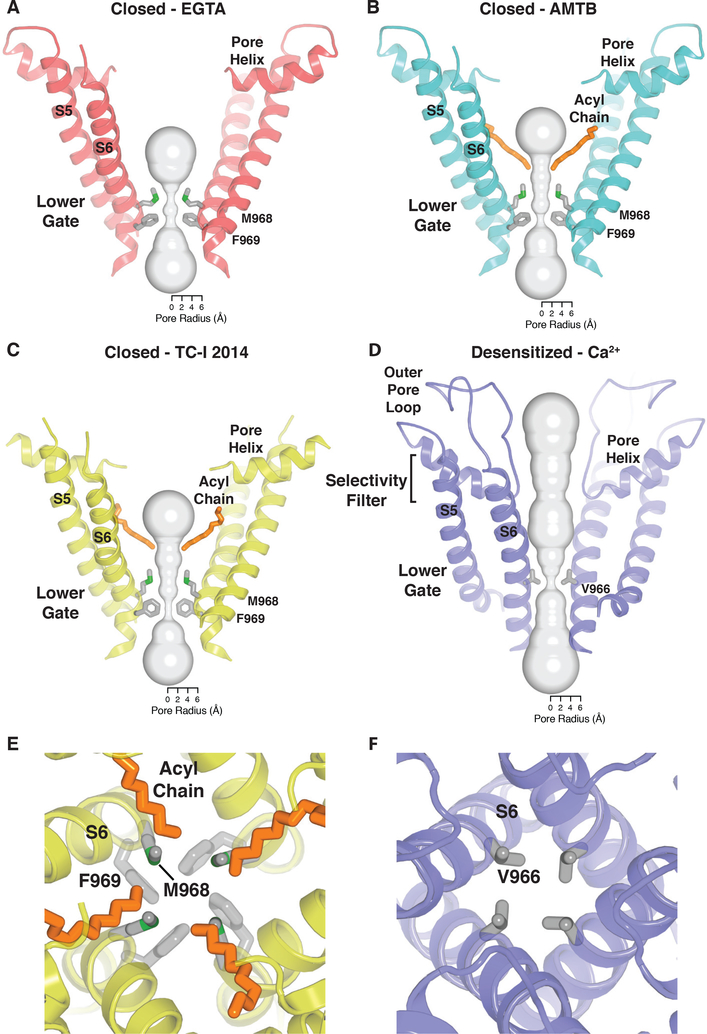
Our high-quality maps (~2.5–3 Å in the transmembrane domain) are sufficient to allow for unambiguous placement of all residues in S1-S6, enabling us to define two distinct conformational states of TRPM8: a closed state in the presence of an antagonist, and a desensitized state in the presence of an agonist (cold) and calcium (Fig. 4). With the improved map quality, we can correct previous assignment of the lower gate in the closed state from Leu973 (Leu964 in pmTRPM8) (14, 15) to an extended restriction comprising two hydrophobic residues, Met968 and Phe969 (Fig. 5A–C, E). In the presence of antagonists, we also observed a lipid tail protruding into the ion conduction pathway, where it forms a hydrophobic barrier that narrows the ion permeation path to ~3 Å in diameter (Fig. 4D–F; 5B–C, E). Indeed, we see ions collecting at this constriction (Fig. 4D–E), which is consistent with the idea that lipids create a vestibule below the selectivity filter, where ions accumulate along a negatively charged face of S6 when the channel is closed (Fig. 4F). This is reminiscent of two-pore domain potassium channels, some voltage-dependent sodium channels, and mitochondrial calcium uniporters, where lipids similarly form a hydrophobic barrier along the ion conduction pathway (24–26). Another unusual feature of the closed state is lack of a canonical S4-S5 linker (Fig. 4B), which plays a critical role in activation of voltage-gated channels (27) but is relatively static in TRP channel structures reported to-date (28).
Fig. 4. Conformational changes associated with ligand binding to a shared pocket.
(A) Superimposition of the closed (TC-I 2014-bound; red) and desensitized (blue) TRPM8 structures. (B and C) Structural changes associated with TRPM8 channel gating. Transition from the closed (B) to desensitized (C) state is accompanied by formation of a S4-S5 linker, a local α-to-π-helical transition in S6, and rearrangement of the TRP domain, which alter the lower gate. (D) In the closed state, an ordered acyl chain (orange) is present along the ion permeation pathway, thereby increasing the hydrophobicity of the pore. Green spheres indicate ions observed in the vicinity of the acyl chains. (E) Close-up view, orthogonal to (D) with ion densities (gray mesh; 3σ contour). Inset shows density for acyl chain (blue mesh, 3σ contour). (F) Cation binding sites (green spheres) are in close proximity to negatively charged residues contributed by S6. Nitrogen, blue; oxygen, red. (G) In the desensitized state, a stabilizing lipid packs between the pore helix and S6 of the neighboring subunit (modeled as CHS). Inset shows lipid density (blue mesh, 4σ contour).
Fig. 5. Ion pore.
(A - D) Ion conduction pathway with front and rear subunits removed for clarity and a representation (gray surface) of the minimal radial distance from the center of the pore to the nearest van der Waals protein or lipid contact. Residues lining the selectivity filter and lower gate are shown as sticks for the EGTA-bound (A), AMTB-bound (B), TC-I 2014-bound (C), and calcium-bound (D) states. (E - F) Close-up view of the lower gate of closed (TC-I 2014-bound) (E) and desensitized (F) TRPM8. The residues and acyl chain (closed confirmation only) that form the hydrophobic seal are shown as sticks.
In contrast to the closed TRPM8 structure, our desensitized structure shows the typical S4-S5 linker architecture seen in other TRP channels (Fig. 4C). In this desensitized state, we see that the side chain from a single hydrophobic residue (Val966) reduces the radius of the lower gate to <1 Å (Fig. 5D, F). In other TRPM channel conformations with similar overall architecture (Fig. S11A–C), the lower gate is formed by two residues, producing a more extended restriction, which is consistent with these structures representing the closed state (Fig. S11D–F) (16, 17, 29). Introduction of a positively-charged residue at this position in TRPM2 (I1045K) or TRPM8 (V966K) is reported to alter ion selectivity, as would be expected if this residue plays a critical role in controlling ion permeation (30). We posit that this minimal, efficient constriction in the presence of calcium defines the desensitized state of TRPM8. We also found that the outer pore loop is structurally resolved in this state, where the presence of calcium induces a large rearrangement leading to the formation of an exposed crevice between the pore helix and S6 of an adjacent subunit. This crevice is occupied by a lipid, modeled as CHS on the basis of the characteristic shape of its density (Fig. 4G), which presumably stabilizes the outer pore domain in this functional state.
Another notable feature (of both states) pertains to the packing of lipids at subunit interfaces, where they are cradled by the pre-S1 elbow and S1 helix of one subunit and the S5 helix of its neighbor (Fig. S14). In this pocket we observed a mixture of lipids, including some that can be modeled as CHS. PIP2, a modulator of TRPM8 function (31, 32), has recently been speculated (33) and shown (14) to bind in this pocket. Whereas a single lipid molecule has been observed in this location for other TRP channels containing a similar pre-S1 elbow (12, 34, 35), here we see clusters of lipids occupying the entire region. In the closed conformation in which the cavity is larger, more lipid molecules were observed (Fig. S14). Cryo-EM image processing suggests that the transmembrane domain of TRPM8 is very conformationally dynamic (Fig. S3; S5; S7; S9), and this tight packing of well-ordered lipids may have enhanced channel stability for structure determination.
Ligand-induced TRPM8 gating movements
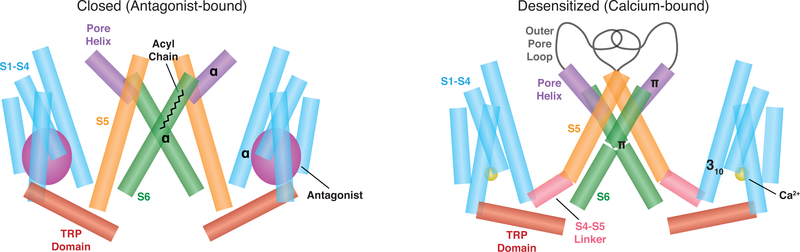
By modeling the transition from the closed to desensitized state, we observed a constellation of conformational changes that provide insight into TRPM8 gating mechanisms (Fig. 4A–C; 6). First, as previously observed upon binding of icilin (14), the S1-S4 domain undergoes a rigid-body tilt away from the central axis that is accompanied by transition of an α-helical turn in S4 (Thr830 - Arg832) to a 310-helix (Fig. 4A–C; 6). We now show that this transition is stabilized by several interactions (Arg832 forms ionic bonds with Tyr736 and Asp793; His835 stacks with Trp789) and leads to formation of a canonical S4-S5 linker (Fig. 4A–C; 6). This is accompanied by large shifts of S5, the pore helix (including introduction of a π-helix), and S6, pivoting these regions into the central pathway (Fig. 4A–C; 6) and adopting an overall conformation distinct from the icilin-PIP2-calcium-bound structure (Fig. S13C) (14), but resembling that observed for other TRP channels. Furthermore, the previously disordered and invisible outer pore loop (14, 15) is now stabilized and well resolved (Fig. 4A–C; 6), perhaps reflecting its conformationally dynamic nature, reminiscent of TRPV1, another temperature-sensitive channel (22). This is consistent with the fact that the loop is not seen in all subclasses of the desensitized state (Fig. S12).
Fig. 6. Gating movements in TRPM8.
In the presence of antagonists (left panel), TRPM8 is closed and density corresponding to an acyl chain is present within the ion conduction pathway. In the presence of calcium (right panel), the channel is desensitized. Structural rearrangements associated with transitioning from closed to desensitized states include a rigid-body tilt of the S1-S4 domain; formation of a canonical S4-S5 linker; shifts of S5, the pore helix, and S6; stabilization of the outer pore loop; and tilting of the TRP domain, such that it is parallel to the membrane bilayer. This transition is accompanied by introduction of a 310-helix in S4 and a π-helix in both the pore helix and S6.
In comparing our two states, we also see interesting rearrangements near the lower gate (Fig. 4A–C; 6). For example, the section spanning residues Ser956-Leu961 in S6 undergoes a transition from an α-helix to π-helix, shifting the register of the lower gate and reducing the constriction at the hydrophobic seal – a structural mechanism underlying channel opening in some TRPV channels (36, 37). Finally, a large tilting movement (~25°) is seen in the conserved and functionally essential TRP domain, rendering it parallel to the membrane bilayer (Fig. 4A–C; 6). The TRP domain is known to play an important role in the allosteric modulation of many TRP channel family members (28). Furthermore, the TRP domain is stacked between the overlying S4-S5 linker and the underlying MHR4 domain within the cytoplasmic N-terminus, resembling the highly integrated allosteric nexus observed in the TRPA1 channel (38). Whether this arrangement reflects a mechanism by which TRPM8 detects and/or integrates cytoplasmic signals remains to be determined.
Close inspection of the S1-S4 ligand-binding pocket provides mechanistic insight into how the binding of modulators promotes key conformational transitions, including movements of the TRP domain and the S4-S5 linker, which control the lower gate. For example, the antagonist TC-I 2014 nestles deep within a pocket present only in the closed state that is formed in part by residues of S4 and the TRP domain (Fig. S15A). Features of the closed state (i.e. absence of the S4-S5 linker and relative position of the TRP domain) allow antagonists to lock the channel in this closed conformation. In the desensitized state, the pocket as defined by these interactions dissolves and cannot accommodate antagonist molecules (Fig. S15A), but can be fit by icilin (14). Thus, we conclude that the ligand-binding pocket associated with closed or open states primarily reflects the position of the TRP helix relative to S1-S4, rather than intrinsic rearrangements to the S1-S4 domain. This is reminiscent of gating movements recently described for TRPM2, where TRP helix movement is also observed but initiated from below by binding of an activator to the soluble domain (13). Furthermore, comparison of our structures in EGTA- and calcium-bound states shows why calcium stabilizes the desensitized state: its binding disrupts an important ionic interaction between Arg998 (TRP domain) and Gln776 (S2 helix), enabling the TRP domain to assume the location described above for the desensitized state (Fig. S15B).
Discussion
Calcium plays a multifaceted role in TRP channel physiology, serving as a co-factor for stimulus-evoked gating and/or desensitization (39). In regard to the latter, it remains unknown for many TRP subtypes whether desensitization is mediated through direct interaction of calcium with the channel, by auxiliary proteins such as calmodulin, or via calcium-sensitive pathways, such as phospholipase C-mediated depletion of PIP2 (32, 40). Our results demonstrate that desensitization of TRPM8 occurs through direct calcium binding at a site that is conserved among a subset of TRPM channels. Calcium binding to this same site is also required for activation by icilin, consistent with the fact that icilin-evoked responses always desensitize. This stands in contrast to some TRPV channel subtypes, where calmodulin is the effector for calcium-mediated desensitization (41). We have shown that closed and desensitized states are distinct and define distinct conformational rearrangements. Although structures corresponding to open states have been difficult to visualize for TRP channels, we propose that the calcium-bound, non-conducting desensitized structure described here closely resembles the TRPM8 open state, except for closure of the gate by a single hydrophobic residue (Val966) as a minimal energetic step defining this transition. However, it is also possible that the open state is unrelated, but sufficiently unstable for structural characterization, perhaps explaining why the closed state is observed even in the presence of cold or cooling compounds (14). Ultimately, visualization of all major functional states will be required to unequivocally assign specific structures to precise physiological states.
Our results support an emerging concept in which TRP channels fall nominally into two main classes, one exemplified by TRPV and TRPML subtypes that show relatively modest conformational changes associated with distinct functional states (22, 36, 37, 41–44), and another represented by TRPM subtypes, in which much larger conformational changes are observed throughout the protein (13, 22). Studies of TRPV and TRPML channels show that the S1-S4 domain remains stationary during channel opening, and ligands bind between this domain and the pore region (S5-S6) to affect gating (22, 36, 37, 41–44). This stands in contrast to voltage-gated channels, where channel opening is promoted by conformational rearrangements within the S1-S4 domain (45). Our results show that TRPM8 channels exhibit an intermediate behavior in which ligands (agonists, antagonists, and calcium) bind within the S1-S4 domain to alter its position relative to the TRP helix without changing its overall structure. At the same time, thermosensitive TRPM8 and TRPV1 channels show dynamic conformational changes within the outer pore region, which has been implicated in sensing chemical and thermal stimuli (46–48).
Both agonists and antagonists of TRPM8 have sensorial and therapeutic applications (49), with antagonists being explored for management of cold hypersensitivity associated with neuropathic pain (50). Our structures suggest that the ligand-binding pocket is malleable and can adopt different contours through subtle side chain movements to accommodate a range of small molecule structures and orientations. This local flexibility might be exploited to introduce moieties that enhance drug stability, solubility, or availability. It is also intriguing to see a density in the ligand-binding pocket in the absence of an exogenous ligand, and it will be interesting to determine the identity of this agent and whether it behaves as a stabilizing co-factor, a natural antagonist, or an inverse agonist. In any case, our findings provide a mechanistic rationale for understanding how ligands affect channel gating, which may facilitate the design of drugs that selectively modulate aberrant channel activity under pathophysiological conditions.
Supplementary Material
Acknowledgements
We thank Y. Guo and J. Osteen for their contributions to initial stages of this project; members of the Cheng and Julius laboratories for discussions; and the staff of the Keck Advanced Microscopy Laboratory of the University of California, San Francisco, D. Bulkley and A. Myasnikov, for help with data collection.
Funding: This work was supported, in part, by an A.P. Giannini Foundation Postdoctoral Fellowship (M.M.D.) and by grants from the National Institutes of Health (R35NS105038 to D.J. and R01GM098672, S10OD020054, and S10OD021741 to Y.C.). Y.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Cryo-EM density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-0631 (ligand-free TRPM8), EMD-0636 (AMTB-bound TRPM8), EMD-0638 (TC-I 2014-bound TRPM8), and EMD-0639 (calcium-bound TRPM8). Atomic coordinates have been deposited in the Protein Data Bank under IDs 6O6A (ligand-free TRPM8), 6O6R (AMTB-bound TRPM8), 6O72 (TC-I 2014-bound TRPM8), and 6O77 (calcium-bound TRPM8). All DNA constructs described in this study are available upon request.
References and Notes
- 1.McKemy DD, Neuhausser WM, Julius D, Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature 416, 52–58 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Peier AM et al. , A TRP Channel that Senses Cold Stimuli and Menthol. Cell 108, 705–715 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Bautista DM et al. , The menthol receptor TRPM8 is the principal detector of environmental cold. Nature 448, 204–208 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Dhaka A et al. , TRPM8 Is Required for Cold Sensation in Mice. Neuron 54, 371–378 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Colburn RW et al. , Attenuated Cold Sensitivity in TRPM8 Null Mice. Neuron 54, 379–386 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Bharate SS, Bharate SB, Modulation of Thermoreceptor TRPM8 by Cooling Compounds. ACS Chem Neurosci 3, 248–267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing H, Chen M, Ling J, Tan W, Gu JG, TRPM8 Mechanism of Cold Allodynia after Chronic Nerve Injury. J Neurosci 27, 13680–13690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Descoeur J et al. , Oxaliplatin-induced cold hypersensitivity is due to remodelling of ion channel expression in nociceptors. EMBO Mol Med 3, 266–278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonvini SJ, Belvisi MG, Cough and airway disease: The role of ion channels. Pulm Pharmacol Ther 47, 21–28 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Liu Q, Hua L, Pan J, Inhibition of transient receptor potential melastatin 8 alleviates airway inflammation and remodeling in a murine model of asthma with cold air stimulus. Acta Biochim Biophys Sin (Shanghai) 50, 499–506 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Chuang H-H, Neuhausser WM, Julius D, The super-cooling agent icilin reveals a mechanism of coincidence detection by a temperature-sensitive TRP channel. Neuron 43, 859–869 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Autzen HE et al. , Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 359, 228–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Winkler PA, Sun W, Lü W, Du J, Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium. Nature 562, 145–149 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Yin Y, Hsu A, Borgnia MJ, Yang H, Lee S-Y, Structural basis of cooling agents and lipid sensing by the cold-activated TRPM8 channel. Science 363, eaav9334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin Y et al. , Structure of the cold- and menthol- sensing ion channel TRPM8. Science 359, 237–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo J et al. , Structures of the calcium-activated, non-selective cation channel TRPM4. Nature 552, 205–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan J et al. , Structure of the mammalian TRPM7, a magnesium channel required during embryonic development. Proc. Natl Acad. Sci. USA 115, E8201–E8210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pertusa M, Madrid R, Morenilla-Palao C, Belmonte C, Viana F, N-Glycosylation of TRPM8 Ion Channels Modulates Temperature Sensitivity of Cold Thermoreceptor Neurons. J Biol Chem 287, 18218–18229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Journigan VB, Zaveri NT, TRPM8 Ion Channel Ligands for New Therapeutic Applications and as Probes to Study Methnol Pharmacology. Life Sci 92, 425–437 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Lashinger ESR et al. , AMTB, a TRPM8 channel blocker: evidence in rats for activity in overactive bladder and painful bladder syndrome. American Journal of Physiology-Renal Physiology 295, F803–F810 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Parks DJ et al. , Design and Optimization of Benzimidazole-Containing Transient Receptor Potential Melastatin 8 (TRPM8) Antagonists. Journal of medicinal chemistry 54, 233–247 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Cao E, Liao M, Cheng Y, Julius D, TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Cao E, Julius D, Cheng Y, TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature, 347–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller AN, Long SB, Crystal structure of the human two-pore domain potassium channel K2P1. Science 335, 432–436 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Payandeh J, Scheuer T, Zheng N, Catterall WA, The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baradaran R, Wang C, Siliciano AF, Long SB, Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature 559, 580–584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long SB, Campbell EB, Mackinnon R, Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science (New York, NY) 309, 897–903 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Madej MG, Ziegler CM, Dawning of a new era in TRP channel structural biology by cryo-electron microscopy. Pflugers Arch 470, 213–225 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Toth B, Structure of a TRPM2 channel in complex with Ca2+ explains unique gating regulation. eLife 7, e36409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kühn FJP, Knop G, Lückhoff A, The Transmembrane Segment S6 Determines Cation versus Anion Selectivity of TRPM2 and TRPM8. J Biol Chem 282, 27598–27609 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Liu B, Qin F, Functional control of cold- and menthol-sensitive TRPM8 ion channels by phosphatidylinositol 4,5-bisphosphate. J Neurosci 25, 1674–1681 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohacs T, Lopes CMB, Michailidis I, Logothetis DE, PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nature Neurosci 8, 626–634 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Zheng W et al. , Direct Binding between Pre-S1 and TRP-like Domains in TRPP Channels Mediates Gating and Functional Regulation by PIP2. Cell Rep 22, 1560–1573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin P et al. , Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan C, Choi W, Sun W, Du J, Lü W, Structure of the human lipid-gated cation channel TRPC3. eLife 7, e36852 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGoldrick LL et al. , Opening of the human epithelial calcium channel TRPV6. Nature 553, 233–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh AK, McGoldrick LL, Sobolevsky AI, Structure and gating mechanism of the transient receptor potential channel TRPV3. Nature Struct Mol Biol 25, 805–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen CE, Armache J-P, Gao Y, Cheng Y, Julius D, Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasan R, Zhang X, Ca2+ Regulation of TRP Ion Channels. Int J Mol Sci 19, 1256–1216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarria I, Ling J, Zhu MX, Gu JG, TRPM8 acute desensitization is mediated by calmodulin and requires PIP2: distinction from tachyphylaxis. J Neurophysiol 106, 3056–3066 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes TET et al. , Structural insights on TRPV5 gating by endogenous modulators. Nature Comm 9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dosey TL et al. , Structures of TRPV2 in distinct conformations provide insight into role of the pore turret. Nat Struct Mol Biol 26, 40–49 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmiege P, Fine M, Blobel G, Li X, Human TRPML1 channel structures in open and closed conformations. Nature Publishing Group, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X et al. , Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states. Nature structural & molecular biology 6, a016865–016813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swartz KJ, Sensing voltage across lipid membranes. Nature 456, 891–897 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matos-Cruz V et al. , Molecular Prerequisites for Diminished Cold Sensitivity in Ground Squirrels and Hamsters. Cell Rep 21, 3329–3337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grandl J et al. , Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat Neurosci 13, 708–714 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertusa M, Rivera B, González A, Ugarte G, Madrid R, Critical role of the pore domain in the cold response of TRPM8 channels identified by ortholog functional comparison. J Biol Chem 293, 12454–12471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knowlton WM, McKemy DD, TRPM8: From Cold to Cancer, Peppermint to Pain. Curr Pharm Bioltechnol 12, 68–77 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Weyer A, Lehto S, Development of TRPM8 Antagonists to Treat Chronic Pain and Migraine. Pharmaceuticals 10, 37–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawate T, Gouaux E, Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Goehring A et al. , Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Prot 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zakharian E, Cao C, Rohacs T, Gating of transient receptor potential melastatin 8 (TRPM8) channels activated by cold and chemical agonists in planar lipid bilayers. J Neurosci 30, 12526–12534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Metods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K, Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandez-Leiro R, Scheres SHW, A pipeline approach to single-particle processing in RELION. Acta Crystallogr D Struct Biol 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF et al. , UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Rosenthal PB, Henderson R, Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. J Mol Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Kucukelbir A, Sigworth FJ, Tagare HD, Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Struct Biol 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Struct Biol 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Struct Biol 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS, HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.