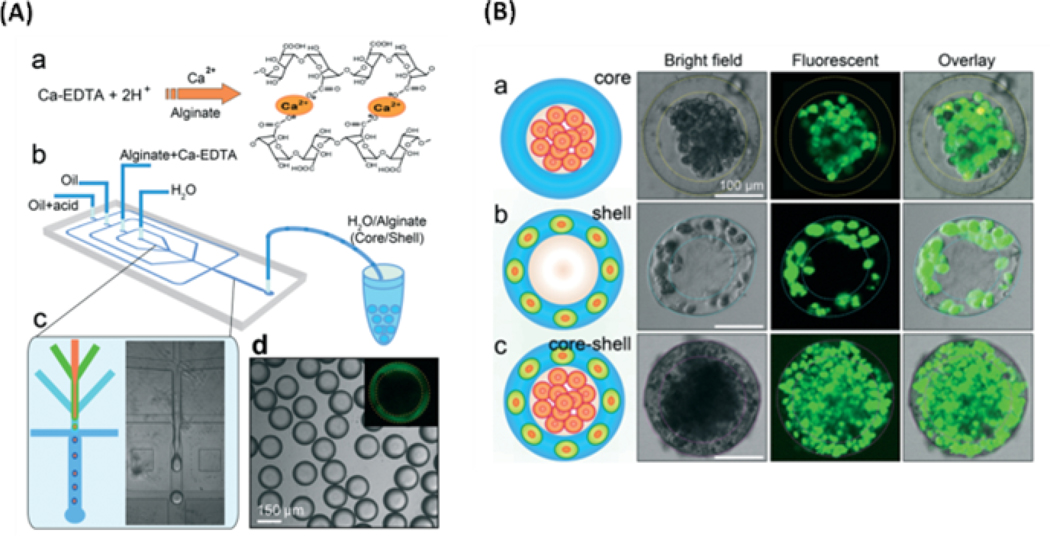
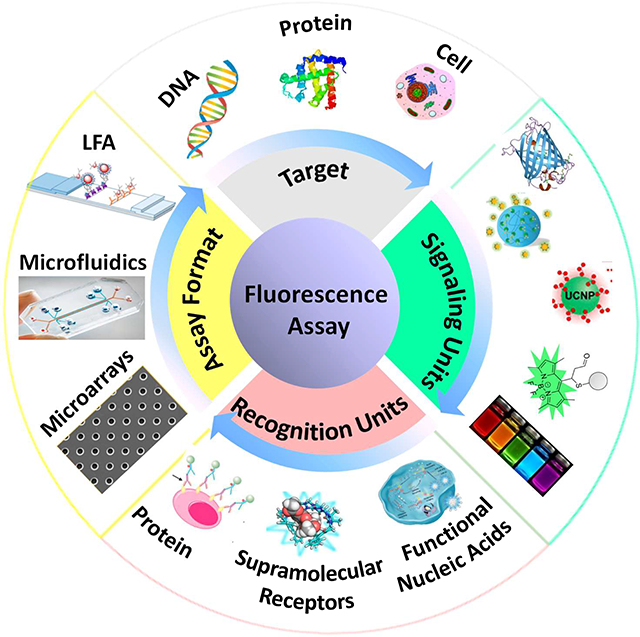
Graphical Abstract
1. INTRODUCTION
Significant improvements in disease management have been achieved in the past few decades. Using cancer as an example, survival rates have increased dramatically in economically developed countries, according to the World Cancer Report published by WHO in 2014.1 One of the highlights is the average 5-year survival rate of children diagnosed before the age of 15 years in Britain increased from less than 30% in 1966–70 to almost 80% in 1996–2000. The improvement can be attributed to the establishment of early screening and awareness programs, identification of a large number of genetic or non-genetic risk factors or markers, and availability of diverse therapeutic strategies including surgery and treatments using radioactive reagents, small molecules, or biological drugs. Still, cancer remains one of the major causes of deaths worldwide, affecting millions of people. Survival rates in developing countries are poor, and the global and regional burden of cancer in terms of both incidence and mortality keep increasing along with the growing population and aging society. Further improvement heavily relies on early diagnosis, wide access to treatment options, and discovery of more effective biomarkers as well as anticancer drugs with high efficacy. Such demands are also applied to diagnosis and cure for all other diseases, calling for rapid advancements in high-throughput screening (HTS) technologies that can efficiently test a large number of targets and samples, monitor diverse enzymatic processes in situ, and effectively identify biologically active substances.2
However, it is challenging to design effective HTS methods that meet the demands of health care and diagnostics. For example, effective disease markers need to be identified and validated to improve early disease diagnosis, which require highly sensitive assays for screening trace levels of biomarkers with minimum consumption of the biospecimen. Diagnosis approaches that are simple, easily accessible, and operable with simple instruments and expertise are needed to reduce the disease-cure gap between economically developed and developing countries. Assays that can monitor the functions of more diverse chemicals in complex biological processes are not yet adequate.
Fluorescence-based assays are one of the dominant measurement methods in HTS, because of their high sensitivity, good tolerance to interference, fast signaling speed, high versatility, simplicity, and non-destructive way of tracking or analyzing targets.3–5 Rapid advancements in preparation of fluorescent materials with unique properties have been achieved to gain ultrahigh assay sensitivity and selectivity, expand the spectral range of the excitation and emission wavelengths, and improve spatial and temporal resolution in fluorescence testing.6–8 Innovative optical instruments have been designed to enhance the levels of automation and miniaturization.9 Advanced fluorescent sensors that allow imaging of multiple targets within living cells, and even organisms, have been constructed.10–13 To circumvent the limitations faced when using traditional fluorescent probes, modern synthetic chemistry and bioengineering have been employing nucleic acids, proteins, supramolecular receptors, and nanomaterials for biochemical analytes.14,15 All these technology advancements in fluorescence-based assays permit better portability, higher flexibility in assay design, easier operation, and enhance the numbers of events and targets to be examined simultaneously.
A variety of reviews have been published on the recent advances of specific classes of fluorescent biosensors, fluorescent techniques, or fluorescence technologies applied to a certain field, as cited above. Different than the cited work, our review summarizes the recent (2016–2018) developments in designing fluorescence-based assays that are highly amendable for HTS applications and covers the three important aspects of fluorescence-based assays: the signaling unit, the target recognition unit, and the assay platform. Diverse types of molecules, including nucleic acids, proteins, and synthetic receptors, have been employed in fluorescence-based assays to enhance the specificity and efficiency in target recognition. Recordable fluorescence signals would be produced upon target recognition; and integration of the target recognition and signaling events could occur in homogeneous solutions or take place on solid surfaces.
Based on the types of fluorescent signals to be collected, fluorescence-based assays encompass diverse techniques that either macroscopically record the total fluorescence intensity, fluorescence polarization, fluorescence resonance energy transfer, time-resolved fluorescence, etc.; or microscopically detect fluorescence from single fluorescent molecules. This review does not endeavor to cover such a tremendous scope of the fluorescence-based applications. Instead, it focuses on the assays using total fluorescence intensity to reveal the presence of target molecules, while the fluorescence could be the native property of the signaling entity or be induced upon energy transfer (ET) between several signaling units.
2. SIGNALING UNITS
Modern developments in synthetic chemistry and nanotechnology have resulted in many different kinds of fluorescent entities that largely increase the selection of the signaling units used in fluorescence-based assays and greatly improve detection flexibility and performance.16–19 Small organic dyes, bulkier fluorescent biomolecules, and fluorescent nanomaterials are all included in our discussion. We also review the progress in applications of fluorogenic dyes or “stimuli-responsive” nanomaterials in fluorescence-based assays.
2.1. Fluorescent dyes
Synthetic organic dyes, such as fluorescein, were the first fluorescent compounds used in biological research.16,20,21 They continue to be a major class of signaling molecules employed in fluorescence-based assays, in particular, as the label conjugated to the target recognition unit.22,23 Bioconjugation is often carried out by chemically crosslinking the fluorophore to biological macromolecules like protein receptors and antibodies; and the relatively small physical size of organic fluorophores produces little interference to the structure and function of the macromolecules, representing a big benefit compared to the bulkier nanomaterial or biological labels.24,25 To simplify the process of dye conjugation to biomolecules, derivatives of the original fluorescent compounds have also been developed, such as fluorescein isothiocyanate (FITC) and tetramethyl rhodamine isothiocyanate (TRITC), as well as the commercial variants that are directly added to nucleic acid or protein products by manufacturers, all exhibiting excellent detection performance.26,27
However, employing fluorescent dyes in homogeneous assays is not possible unless certain kinds of energy transfer process exist, because the free dyes cannot be removed and are not distinguishable from those bound to the targets. Moreover, the fluorescence intensity from organic fluorophores is typically not high. To improve detection performance and reduce background in homogeneous assays, quenchers are often used to initially quench the dye fluorescence, which would be turned back on in the presence of the target molecules.28,29 Such strategies can solve the issue of background noise from the free dye, generating “target-responsive” fluorescence, i.e. the desired fluorescence is only turned on after the signaling molecule becomes stimulated via target recognition or changes in the surrounding environment.30,31 Dimethylaminophenylazobenzoic acid (DABCYL) and Black Hole are commonly used quenchers for many fluorophores.32,33 The most well-known application of the quencher-fluorophore pair is its use the molecular beacons (MB), which bring the quencher and fluorophore in proximity through hairpin formation.34,35 The MB can be opened by target nucleic acids, turning on the fluorescence of the fluorophore by separation of the quencher-fluorophore pair.
To further improve the detection sensitivity, strategies that could add multiple amplification steps to the assay have been widely developed while still using the fluorescent dyes and their quenchers as the signaling probes.36,37 Zhang et al. developed a PCR-free fluorescence-based assay for detection of telomerase activity using a fluorescein tagged MB.37 The fluorescence from fluorescein was initially quenched by a quencher on the MB. Telomerase, if present in the sample, could extend the substrate (TS) primer to form a long product which contained several TTAGGG repeats. Each of these repeats released one trigger DNA (t-DNA) via strand displacement reaction with a DNA probe. Thus, one long product could produce multiple copies of the t-DNA to subsequently open the MB and restore the fluorescence of fluorescein, forming the first amplification cycle. Hybridization between the t-DNA and the MB also produced the cutting sites for the nicking endonuclease to digest the MBs and open more MBs through repeated hybridization with the t-DNA, leading to a second amplification cycle. Owing to the double amplification strategy, the method allowed measurement of telomerase activity in crude cell extracts obtained from as few as 5 HeLa cells and 10 CCRF-CEM cells.
In addition, physical strategies have also been used to allow the incorporation of multiple fluorescent dyes in bioassays.38,39 For example, Linz et al. relied on the difference in photostability of the organic dyes to develop a new method for production of beads with different optical “codes” so they can be employed in multiplexed assays.38 The beads were encoded by two dyes with overlapping spectral properties but different photostabilities. The dyes were mixed at various ratios to permit bead differentiation based on the simple photobleaching rate measurement. All beads initially exhibited similar fluorescence intensities; however, following an appropriate photoexposure procedure, the emission intensity of the less stable dye would drop faster than the more stable one. By comparing the original fluorescence emission intensity to that obtained after photobleaching, multiple bead populations could be reliably identified. Using only a single excitation/emission band, two different initial intensity levels were optimized to produce six uniquely identifiable bead populations whereas only two could have been achieved with the conventional intensity measurement approach.
2.2. Fluorogenic molecules
The background issue of employing fluorescent dyes in homogeneous assays can also be solved by the smart design of fluorogenic molecules that possess no or very low intrinsic fluorescence, but would be turned on by the presence of the target analyte and emit fluorescence. The negligible background from the fluorogenic dyes allows the assay to be performed in the one-pot, mix-and-measure procedure, greatly simplifying the assays and improving the signal-to-noise ratios. Various classes of fluorogenic dyes have been developed to detect metal cations, reactive oxygen, nitrogen and sulfur species, and small metabolomic or drug molecules like glucose, ATP, antibiotics.17,40,41 The fluorogenic dyes response to microenvironment changes have also been extensively studied and applied in fluorescence-based assays. The ground state, excitation state, or nonradiative relaxation process can be affected by the surrounding conditions, thus altering its fluorescence intensity.17,32,42
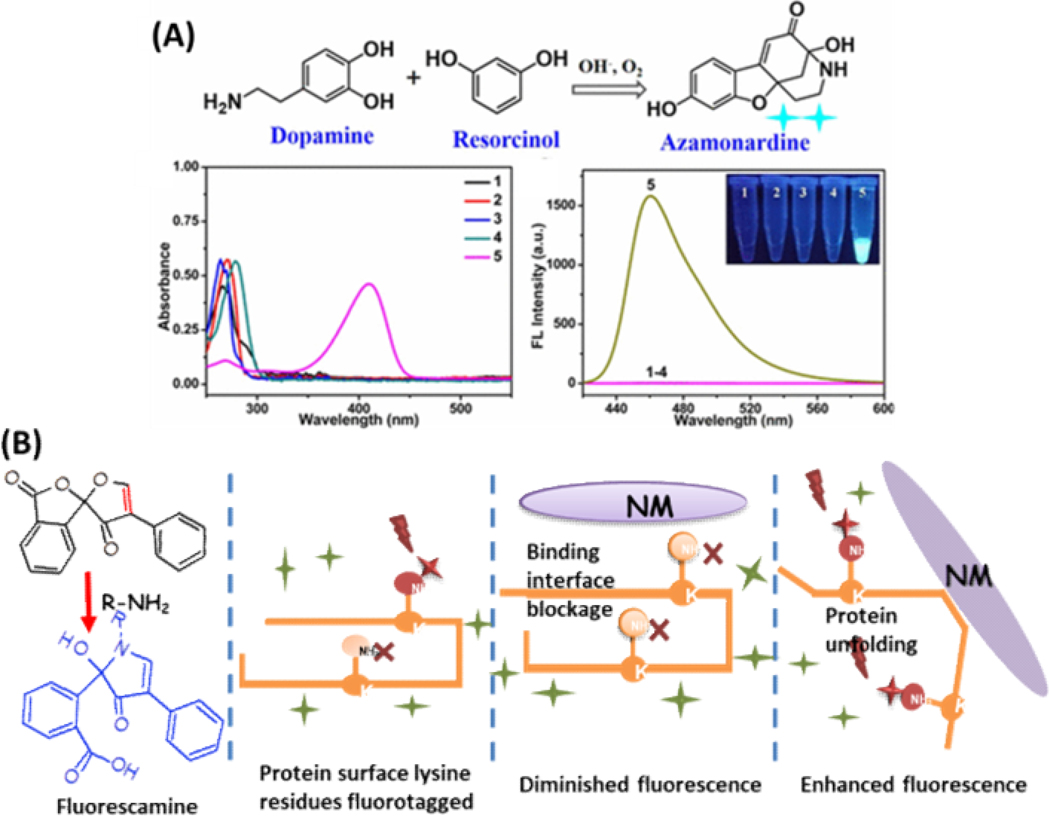
Some fluorogenic dyes can be substrates for enzymes and employed to detect enzymes. One strategy is to build a dye-enzyme hybrid system. A voltage-sensitive dye was designed to be the substrate of porcine liver esterase (PLE), and its fluorescence increased after being cleaved by PLE. PLE can be selectively expressed in cells of interest (e.g. neurons), so only these cells can have PLE on their surface to catalyze the dye and show fluorescence.43 Zhao et al. described an exquisite label-free fluorescent and colorimetric dual-readout assay of tyrosinase activity, showing a good example of employing enzymes to convert the non-fluorescent substrates into fluorescent products. In this assay, tyrosinase catalyzed the oxidation of monophenolamine and converted it to catecholamine, which in turn could react with the fluorogenic dye of resorcinol and turn on its fluorescence.39 As shown in Figure 1A, the tyramine solution incubated with tyrosinase could emit an intense blue fluorescence under ultraviolet light in the presence of resorcinol. At the same time, the resultant fluorescent azocine product of the sensing system exhibits an obvious pale yellow emission under visible light and possesses characteristic absorption peak centered at 420 nm to form the colorimetric signal. Such a convenient, rapid, cost-effective, and highly sensitive assay exhibits high promise in detecting enzyme activity in biospecimens and screening potential tyrosinase inhibitors.
Figure 1.
A) Schematic representation of dopamine reacting with resocinol in synthesis of fluorescent azamonardine; and the absorption and fluorescence spectra of tyramine (1), dopamine (2), resorcinol (3), mixture of tyramine and resorcinol (4) and mixture of dopamine and resorcinol (5) under the same conditions, respectively. Reproduced from Zhao, J.; Bao, X.; Wang, S.; Lu, S.; Sun, J.; Yang, X. Anal. Chem. 2017, 89, 10529–10536 (ref 39). Copyright 2017 American Chemical Society. (B) Molecule structure of fluorescamine and its reaction with primary amine, which can be utilized to probe protein binding to the NPs: protein surface blocked by the binding interface with nanomaterials will result in diminished fluorescence; and protein unfolding induced by nanomaterials will result in enhanced fluorescence.
Chemical reactions with active groups can also be used to turn on fluorogenic dyes. One common way is to design the fluorogenic dye as an electrophile alkene, which can react with electron rich thiol or lysine groups by Michael addition to form a fluorescent product.44 Our group has employed one such dye, fluorescamine, to detect interactions between nanoparticles (NPs) and proteins. NP-protein interaction can form a protein layer, so called the protein corona, on the surface of NPs present in biological matrices, which can influence the interaction of nanomaterials with cells and tissues. This can ultimately alter and determine their uptake, distribution, toxicity, etc.45–47 The study of protein corona formation on nanomaterials with high intendancy of human exposure is important for the improvement of their biocompatibility, efficacy, safety and sustainability.48 To accommodate the large variations of NPs produced nowadays and the high diversity of proteins they could encounter, our group developed a high-throughput platform to profile NP−protein interactions using fluorescamine. Fluorescamine has no fluorescence until it reacts with primary amines (Figure 1B).49,50 Thus, it can specifically target the surface amines of proteins with fast reaction rates. NP-protein interaction could either block the surface reaction sites or induce protein conformational changes to expose more primary amines, changing the resultant fluorescence from fluorescamine labeling (Figure 1B). By simply adding this fluorogenic dye to the protein-nanoparticle mixture, we collected the resultant fluorescence profiles from various types of nanoparticles, including silica, polystyrene, and iron oxide nanoparticles, which showed good differentiation of the nanoparticles by their physicochemical properties like size, surface coating, and core material. Our results revealed the close relationship between protein adsorption and particle characteristics. In the follow-up work, we further proved the capability of the resultant fluorescence indicative of the binding strength and revealed that protein adsorption onto nanoparticles could be strongly affected by structure flexibility.50 Since high degrees of conformation change could potentially influence protein function, our method could be a fast HTS approach to evaluate the biocompatibility of nanoparticles, besides its potential in rapidly screening protein-nanoparticle interaction.
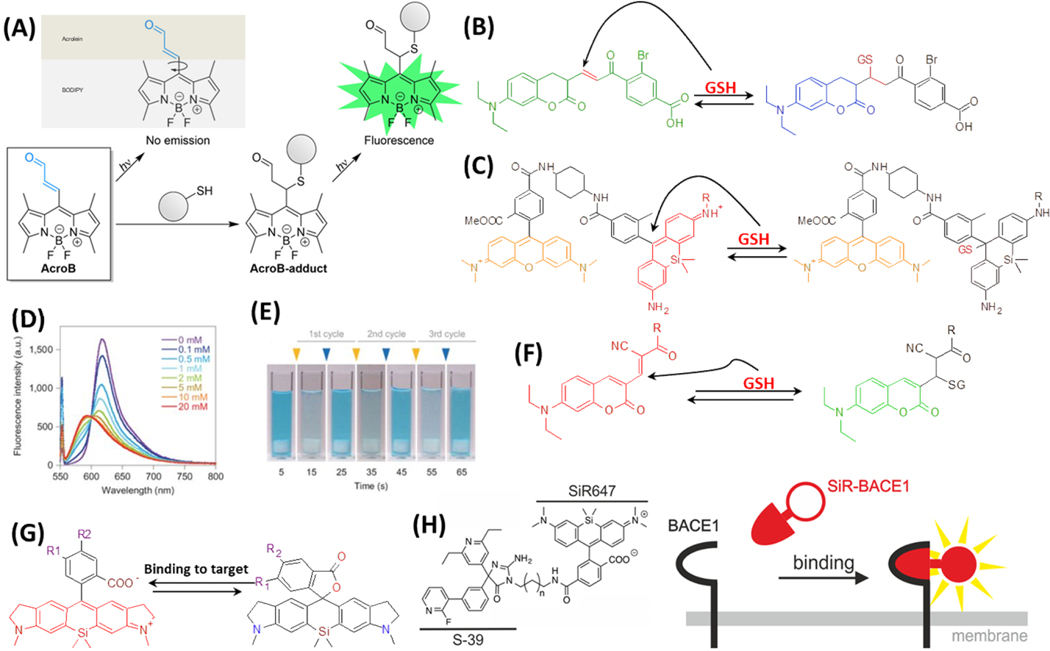
Another feasible way of turning on a fluorogenic dye through chemical reactions is to conjugate a reactive warhead to the dye to quench its fluorescence. After the warhead reacts with the target, the quenching effect would be removed, turning on the fluorescence. One example is AcroB that contains the reactive warhead of acrolein and the fluorophore structure of BODIPY (Figure 2A). The conjugate is not-fluorescent due to the free rotation of acrolein.51 Acrolein is one of the lipid-derived electrophiles (LDE) and can react with the active thiol groups on proteins, increasing the fluorescence of AcroB by more than 350 fold. This probe was applied to enable high resolution spatiotemporal imaging of mitochondria.52 To improve the specificity of this kind of covalently reactive fluorogenic probe, fragment-based HTS in native biological systems has been used to identify the structures that can covalently and selectively label target proteins.53 The vinyl sulfone was used as the warhead for active thiol groups, and the successful hit (DNS-pE2) showed high specificity toward 3-phosphoglycerate dehydrogenase (PHGDH), which is a potential drug target for cancer.
Figure 2.
(A) Molecule structure and mechanism of action of AcroB. Reproduced from Lincoln, R.; Greene, L. E.; Zhang, W.; Louisia, S.; Cosa, G. J. Am. Chem. Soc. 2017, 139, 16273–16281 (ref 51). Copyright 2017 American Chemical Society. Molecule structures and reactions of reversible fluorogenic probes for GSH: (B) TQ green, (C) SiR and TMR FRET system, and (F) QG-1. (D) The fluorescence intensity changes of SiM-TMR FRET system, with adding different concentration of GSH. (E) The visualization of reversible colour-changing of the SiM probe. GSH and NEM (N-ethylmaleimide) are added alternatively at 10-second intervals. The concentration of the dye is 20 μM. The final concentrations of GSH and NEM after each cycle are 5 mM. (D) and (E) are reprinted by permission from Macmillan Publishers Ltd: NATURE, Umezawa, K.; Yoshida, M.; Kamiya, M.; Yamasoba, T.; Urano, Y. Nat. Chem. 2017, 9, 279–286 (ref 55). Copyright 2017. (G) Examples of one SiR dye undergoing ground-state isomerization. R1 and R2 are possible sites to add targeting moieties for various applications. (H) Molecule structure and mechanism of action of SiR647 conjugated to S-39 that targets BACE1 protein. Reproduced from Karch, S.; Broichhagen, J.; Schneider, J.; Böning, D.; Hartmann, S.; Schmid, B.; Tripal, P.; Palmisano, R.; Alzheimer, C.; Johnsson, K.; Huth, T. J. Med. Chem. 2018, 61 (14), 6121–6139 (ref 62). Copyright 2018 American Chemical Society.
To develop reversible labeling for in vivo imaging or sensing, ThiolQuant (TQ) Green, which contained one Michael addition moiety between two fluorophores and emitted green fluorescence, was designed to react with glutathione (GSH).54 The blue fluorescence intensity at 463 nm was enhanced after the reaction (Figure 2B), however, this probe suffered from slow kinetics and dynamics and could not be applied to detect GSH at physiological levels. To improve it, a silicon-rhodamine based fluorophore, i.e., a donor, was linked to a fluorescence donor (tetramethylrhodamine, or TMR) to make a FRET system (Figure 2C). As a result, the reaction with GSH could induce a large enhancement in the fluorescence intensity of TMR (Figure 2D).55 This probe has shown quick and reversible responses to multiple cycles of GSH additions (Figure 2E). On the other hand, QG-1, another reversible probe for GSH, was designed by modifying a cysteine-targeted kinase inhibitor, via adding a cyanide (CN) group to the α-carbon of the unsaturated enal (Figure 2F).56 This molecule emits red light; and with GSH added to the double bond, only one side of the molecule remains as a fluorophore, causing it to emit green light instead. Conjugating a moiety targeting mitochondria could achieve real time and dynamic detection of GSH in the mitochondria.57
Recently, some new mechanisms have been added to the toolkit to tune the fluorogenic properties of the dyes, including ground-state isomerization, aggregation-induced emission (AIE), and motion-induced change in emission (MICE).58,59 The silicon-rhodamine based fluorophore (SiR) is one good example that utilizes the mechanism of ground-state isomerization, which achieves an equilibrium between the fluorescent zwitterionic form and non-fluorescent spirolactone form to control fluorescence intensity (Figure 2G).60 Traditionally, the equilibrium was viewed as a disadvantage, and chemists were trying to modify the structure of SiR to shift the equilibrium and keep the dye in the zwitterionic form, which could fluoresce. This prospect changed after the successful demonstration of how the equilibrium of SiR can be used as a spontaneously blinking property for super-resolution imaging. 61 Later on, SiR was also explored as a fluorogenic dye for live-cell imaging, based on the phenomenon that binding to a target molecule can keep SiR in the zwitterionic state. 60 The SiR molecule itself has little specificity, so different probes can be linked to SiR and achieve specificity towards target analytes. For example, SiR647 conjugated to the inhibitor of the β-site APP-cleaving enzyme 1 (BACE1) could specifically bind to BACE1, allowing the monitoring of the physiological and pathophysiological progress of BACE1 in Alzheimer’s disease (Figure 2H).62
The specificity of fluorogenic probes is very important for their applications. By utilizing rational design, probes with different spectral ranges and high specificity toward lipid droplets have been synthesized and used for multicolor imaging and tracking.63 Another example fused a methyl-CpG-binding domain (MBD) with a DNA-binding fluorogenic dye to achieve selectivity over methylated DNA.64
2.3. Fluorescent nanomaterials
To improve the intensity, photostability and multiplex capacity of fluorescent labels, many fluorescent nanomaterials have been developed, such as quantum dots, upconversion nanoparticles, semiconducting polymer dots.65,66 Their size- or shape-controllable optical characteristics diversify the selection of diverse probes for enhanced assay throughput. In addition, nanomaterials can simplify fluorescence-based assays by acting as the solid support for the target recognition unit.67–70 Nanomaterials are also adopted for functions other than signaling, including enriching target molecules and quenching interferences.71–73 Therefore, fluorescent nanomaterials have been gradually replacing conventional fluorophores as the signaling units in fluorescence-based assays.
Quantum dots (QDs) are dominantly employed in fluorescence-based assays because of their brightness, tunable optical and electronic properties.74–76 However, one obstacle QDs encounter in biological applications is their surface property may not be compatible with the high salt content of physiological buffers, which can cause aggregation or even dissolution. In addition, biological samples are often overwhelmed with interferents that could non-specifically adsorb to the QDs. To overcome these issues, the surface properties of QDs have been improved to possess high aqueous solubility, stability, and reduced nonspecific binding through several surface modifications, such as coating with oxides, organosilica, or polymers.77–79 For example, Bala et al. developed a novel detection platform for malathion using CdTe@CdS QDs.77 The designed nanoprobe was comprised of the QDs, poly(N-(3-guanidinopropyl)methacrylamide) homopolymer (PGPMA) and the malathion-specific aptamer. In the absence of malathion, the cationic polymer interacted with the aptamer via electrostatic interactions, leaving the fluorescence of QDs unaffected. However, when malathion is present and competes with the polymer for aptamer binding, the free polymer interacts with and quenches the QDs. The assay exhibited excellent sensitivity towards malathion with a detection limit of 4 pM. A logarithmic correlation was observed over a wide range of malathion concentrations, from 0.01 nm to 1 μM, facilitating quantitative determination of the analyte.
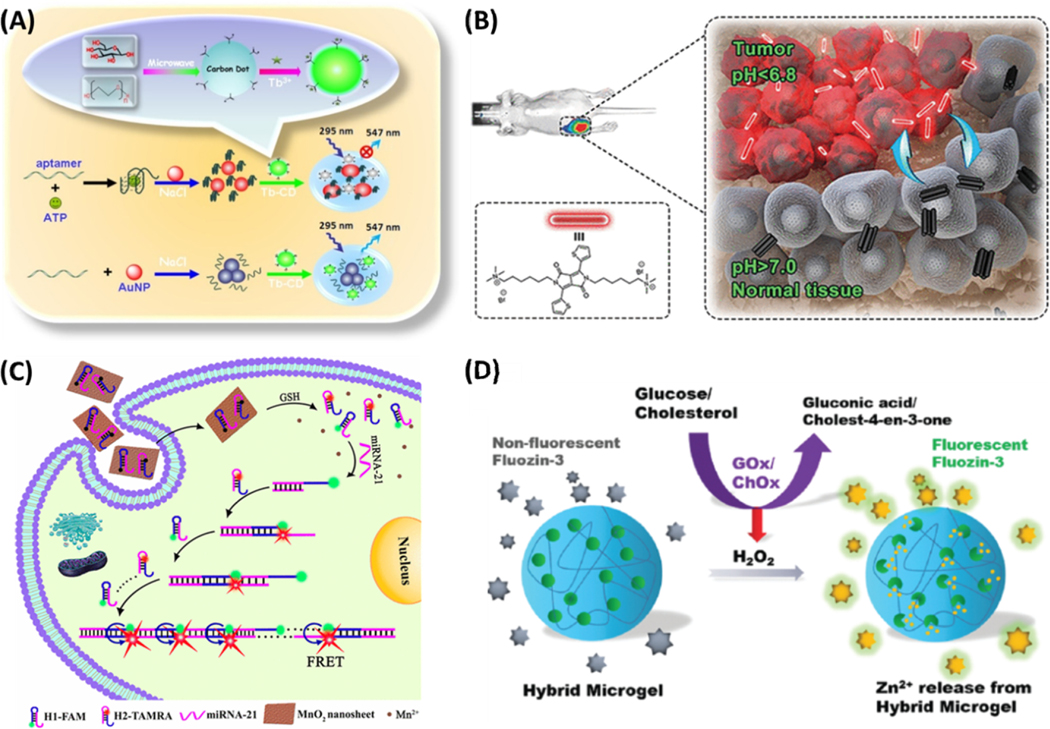
Another issue QDs pose is that they often contain heavy metals and are thus potentially toxic in biological applications. Carbon dots (CDs) are a class of QDs that also exhibit fluorescence due to quantum confinement but are more biocompatible.80,81 CDs can be mixed with fluorescent dyes and metal ions to achieve ratiometric detection, improving the quantitation capability of the fluorescence-based assays.82,83 Their biocompatibility is an attractive advantage. For example, thiazole orange (TO) modified CDs were used for detection of G-Quadruplex and double-stranded DNA.84 In this application, a specific DNA probe modified by TO was covalently linked to the surface of CDs, acting as both the recognition element and the fluorescence response unit. In the absence of the target DNA, the nanoprobe only emitted the blue fluorescence of CDs, while TO was mostly nonfluorescent due to the quenching effect from the CDs. Upon addition of the target DNA, hybridization produced the rigid dsDNA and moved the TO away from the CDs, which turned on the emission of TO at 530 nm. While the TO emission intensity increased along with the increasing concentrations of the target DNA, the fluorescence of the CDs was unchanged, realizing ratiometric detection. The CD–TO nanoprobe showed good selectivity to the parallel G-quadruplex (G4) and double-stranded (ds) DNA over antiparallel G4 and single-stranded DNA. Moreover, the ratiometric fluorescence nanoprobe exhibited high sensitivity for detection of ssab (a dsDNA) and c-myc (a parallel G4), achieving a detection limit as low as 0.90 and 3.31 nM, respectively. Metal ion doping can also produce ratiometric CD-based probes.85 As shown in Figure 3A, terbium ion-coordinated carbon dots (Tb-CDs) can detect ATP through the dispersive/agglomerative interaction of gold nanoparticles (AuNPs) with Tb-CDs. Other novel functional CDs include carbon dots–MnO2 nanocomposites and CDs-encapsulated breakable organosilica nanocapsule for the sensitive detection of malathion, glutathione, bacteria, etc.
Figure 3.
(A) Schematic illustration of fluorescent aptasensing platform for the quantitative monitoring of adenosine 5′-triphosphate (ATP) by interaction of unmodified gold nanoparticles (AuNPs) with terbium ion-functionalized carbon dots (Tb-CDs). Reprinted from Biosens. Bioelectron., vol. 86, Xu, M.; Gao, Z.; Zhou, Q.; Lin, Y.; Lu, M.; Tang, D. Terbium Ion-coordinated Carbon Dots for Fluorescent Aptasensing of Adenosine 5’-Triphosphate with Unmodified Gold Nanoparticles, pp. 978–984 (ref. 85). Copyright 2016, with permission from Elsevier. (B) Schematic illustration of pH switchable nanoassembly for imaging a broad range of malignant tumors. Reproduced from Liu, Y.; Qu, Z.; Cao, H.; Sun, H.; Gao, Y.; Jiang, X. ACS Nano 2017, 11, 12446–12452 (ref 18). Copyright 2017 American Chemical Society. (C) Schematic illustration of MnO2 nanosheet-mediated in cell HCR signal enhancement for sensitively detecting miRNA-21 in living cells. Reproduced from Li, J.; Li, D.; Yuan, R.; Xiang, Y. ACS Appl. Mater. Interfaces 2017, 9, 5717–5724 (ref 100). Copyright 2017 American Chemical Society. (D) Scheme of microgels for the detection of glucose and cholesterol. Reproduced from Liu, Y.; Wang, Y.-M.; Sedano, S.; Jiang, Q.; Duan, Y.; Shen, W.; Jiang, J.-H.; Zhong, W. Chem. Commun. 2018, 54, 4329–4332 (ref 105), with permission of The Royal Society of Chemistry.
Compared to QDs and CDs, fluorescent silica NPs prepared from doping the colloidal silica particles with organic fluorophores or QDs provide several advantages. For instance, wider selection of particle sizes (spanning from a few to hundreds of nanometers), less strict size control during NP synthesis, better water solubility and higher biocompatibility have been reported advantages that reinforce the rising popularity of NPs in fluorescence-based assays in the past few years.86,87 Surface functionalization is also easier for silica NPs than QDs because of the availability of diverse silylation derivatization reagents.88 The reactive amines on APTES-modified mesoporous silica NPs, for example, allow easy attachment of the synthetic fluorescent antioxidant flavone analogs through the sulfonamide linkage.89 The flavone moiety, when attached onto the flavone-modified fluorescent silica (FMFS) NP surface, impaired its characteristic fluorescence and antioxidant activities. Moreover, the NPs are highly biocompatible, as evidenced from their cytotoxicity assay on the normal lung cells (L132). The fluorescence activity of these biocompatible NPs was further utilized to study their interaction with a biomolecule, Bovine Serum Albumin (BSA), which alters the fluorescence behavior of FMFS NPs. On the other hand, the intrinsic fluorescence activity of BSA was also significantly modified due to its interaction with the FMFS NPs. Thus, the sensing and detection of biomolecules, like BSA in presence of FMFS NPs, can be accomplished by monitoring changes in the fluorescence behavior of either FMFS NPs or BSA. Furthermore, these FMFS NPs retained their intrinsic fluorescence behavior in cellular medium, supporting their possible use as biocompatible cell imaging agents in the future.
Besides inorganic nanomaterials, polymeric NPs that are either composed of intrinsically fluorescent polymers or doped with fluorophores have been developed. Because of their exceptional brightness due to incorporation of multiple dyes, potential in HTS applications, remarkable stability in biological environments, and well-controlled surface properties, polymer NPs are particularly promising in fluorescence-based assays.90–93 Liu et al. developed the pH switchable polymeric nanoassembly for imaging a broad range of malignant tumors by using a small molecule (DPP-thiophene-4) composed of a diketopyrrolopyrrole (DPP) core and two alkyl chains terminated with quaternary ammonium.18 As shown in Figure 3B, DPP-thiophene-4 can self-assemble into a nonfluorescent nanoassembly when the pH is >7.0 but reversibly disassembles back to fluorescent monomers at pH <6.8. Meanwhile, its fluorescence emission increases by 10-fold within a 0.2 pH unit change. Such a fluorogenic nanoassembly can precisely differentiate a number of malignant tumors among normal tissues in vivo due to the slight acidity within tumor microenvironments. Furthermore, the nanoassembly shows satisfactory biocompatibility and an effective clearance from the body.
Sonawane and co-workers developed the fluorescent polystyrene microbeads as the invisible security ink to make an optical vapor sensor for 4-Nitrotoluene (4-NT).94 In their design, the color-tunable solid-state emitting polystyrene (PS) microbeads were developed by dispersion polymerization. Pyrene and perylenebisimide were incorporated into the PS backbone as the acrylate monomer and acrylate cross-linker, respectively. Dispersion of the polymer beads in ethanol produced a fluorescent security “invisible” ink, which became visible only under ultraviolet light. The color of the ink could be tuned by the amounts of the pyrene and perylenebisimide. More than 80% of the emission from pyrene was quenched upon exposure of the polymer to the vapors of 4-NT, while the emission of perylenebisimide was unaffected. The limit of detection was estimated to be 2.7 ppm in vapor. The simplicity of material synthesis, along with the invisible ink characteristics and nitro aromatic vapor detection, opens up new opportunities for exploring the application of these PS-based materials as optical sensors and fluorescent ink for security purposes.
One remarkable development of the polymeric NPs is the water-soluble, fluorescent, cytocompatible polymer single-walled carbon nanotube (SWNT) that was prepared by sonication in aqueous solution leading to the final polymer-SWNT complexes.95 This material was developed to overcome the challenges of applying fluorescent nanomaterials in complex biological samples, such issues as water solubility, cell permeability, biodistribution, and toxicity. The polymer backbone solubilizes the SWNTs, decorates them with fluorescent perylene bisimide (PBI) and strongly improves their cytocompatibility by wrapping around the SWNT scaffold. In photophysical measurements and biological in vitro studies, sulfated complexes exhibit superior optical properties, cellular uptake, and intracellular staining over their hydroxylated analogs. A toxicity assay confirms the highly improved cytocompatibility of the polymer-wrapped SWNTs toward surfactant-solubilized SWNTs. In microscopic studies the complexes allow direct imaging of the cellular uptake of SWNT via the emission from both the PBI and SWNT. These findings substantiate the polymer-SWNT complexes as valuable systems for a broad range of fluorescence bioimaging studies because of their nanometer size, dual fluorescence, multiple charges, and high cytocompatibility.
2.4. “Stimuli-responsive” signaling units
With thoughtful design, quenchers can make fluorescent dyes only turn on by target molecules. However, the quenching efficiency is extremely sensitive to the distance between the quencher and the fluorophore, reducing signal-to-noise ratios.96 In addition, covalently tethering organic quenchers to nucleic acids is an expensive procedure that is vulnerable to cleavage by nonspecific enzymes in biospecimen. Thus, a variety of novel fluorescent nanomaterials have been employed to solve these problems.
One approach is to utilize energy transfer between nanomaterials and fluorescent molecules, using nanomaterials as the energy acceptors to quench the fluorescence of diverse dye molecules, i.e. donors. Since the efficiency of ET is highly dependent on the spatial distance between donors and acceptors, quenching could be reversed and used as a measure of target response once the dye molecules are desorbed, which can be stimulated by the target recognition event. The unique properties of nanomaterials, such as large specific surface areas, high extinction coefficients, and broad ranges of the excitation/absorption wavelengths can greatly improve the ET effects and increase the flexibility of assay design. Typical nanomaterials employed in such systems are noble metal nanoparticles such as Au NPs and 2D nanomaterials like graphene oxides (GO).97–99 For example, Sun and co-workers have developed a GO-based ET sensor for rapid and specific detection of unfolded collagen fragments.98 The fluorescein-labeled collagen-mimic, triple helical peptide was initially adsorbed onto the surface of GO via π–π stacking and hydrophobic interaction, bringing the fluorescein to close to the GO and resulting in efficient fluorescence quenching. In the presence of the unfolded single-stranded collagen fragments, which could bind to the triple helical peptide and remove it from the GO surface, the fluorescence can be turned back on. This yields conformation-sensitive and sequence-specific detection of unfolded collagen fragments at the nM level.
The MnO2 nanosheets have also been used as the energy acceptors for detection of microRNAs in live cells. As illustrated in Figure 3C, the MnO2 nanosheets can adsorb onto the dye labeled hairpin probes and exhibit excellent quenching. As they are biodegradable, the MnO2 nanosheets feature highly reduced cytotoxicity to the target cells.100 Upon entering cells, the surface-adsorbed FAM- and Tamra (TMR)-conjugated hairpins can be released due to the displacement reactions by other proteins or nucleic acids and the degradation of the MnO2 nanosheets by cellular GSH. Subsequently, the down-regulated target miRNA-21 triggers cascaded assembly of the two hairpins into long dsDNA polymers, which bring the fluorescence resonance energy transfer (FRET) pair, FAM (donor) and TMR (acceptor), into proximity to generate significantly enhanced FRET signals for detecting trace miRNA-21 in living cells. By carefully tailoring the sequences of the hairpins, the developed method can offer new opportunities for monitoring various trace intracellular miRNA targets with low expression levels in living cells.
ET can also occur between nanomaterials. For instance, fluorescent CDs can be quenched efficiently by MnO2 nanosheeets if they are located on the nanosheet surface.101 However, the presence of reducing analytes, like butyrylcholinesterase (BChE) and acetylthiocholine (ACh), could convert the MnO2 nanosheets to Mn2+ and subsequently release the CDs, recovering the fluorescence. This assay demonstrated high selectivity toward analytes ranging from 0.05 to 5 ng mL−1 with a detection limit of 0.015 ng mL–1.102 Diverse nanomaterial-based donors and acceptors, like QDs, upconversion nanoparticles, graphene, graphene-like two-dimensional (2D) nanomaterials, and silica nanoparticles-based composites have been produced to expand the FRET spectra and applied in bioassays to improve assay sensitivity and selectivity.103,104
Another approach of generating “stimuli-responsive” fluorescence is through release or dissolution of the nanomaterial components upon target recognition. Built upon our previously developed signal amplification strategy based on cation-exchange in nanocrystals, we recently constructed the hydrogel microparticles that encapsulated ionic nanoparticles, like ZnS and CuS.105 As shown in Figure 3D, selective reactive oxygen species (ROS) release the enclosed cations from the microgel and subsequently turn on the fluorogenic dyes to emit intense fluorescence, permitting rapid detection of ROS or ROS-producing molecules.
Tan et al. developed a fluorescent sensing platform based on graphene oxide (GO) hydrogel for antibiotic detection, in which the adenosine and aptamer worked as the co-crosslinkers to connect GO sheets and then form the three-dimensional (3D) macrostructures.106 The as-prepared hydrogel showed high mechanical strength and thermal stability. The optimal hydrogel had a linear response of 25–1000 μg/L for oxytetracycline (OTC) and a limit of quantitation (LOQ) of 25 μg/L. Together with the high affinity of the aptamer for its target, this assay exhibited excellent sensitivity and selectivity.
Ma et al. constructed the target-responsive DNA hydrogel for enzyme- and label-free detection of glucose.107 In this work, the glucose-responsive hydrogel was prepared using the target aptamer and its two short complementary DNA strands grafted onto a linear polyacrylamide chain as cross-linkers. AuNPs modified with thiol-PEG were encapsulated in the gel and used as the output signal for visible detection. The complex of glucose and its boronic acid derivative ligands (Shinkai’s receptor) can bind with the aptamer to disrupt the hydrogel, releasing the AuNPs to the supernatant to generate a distinct red color. By this method, glucose can be detected with the naked eyes, and the sensor has a detection limit of 0.44 mM in buffer with the help of UV-Vis spectrophotometry. The aforementioned stimuli-responsive platforms offer high sensitivity because of low or no background, and high specificity owing to the specific reactions or interactions that trigger fluorescence emission, providing great promises for HTS in biomedical applications.
2.5. Fluorescent proteins
Genetically encoded fluorescent proteins (FPs) have become important tools that enable visualization of proteins in living cells.108–110 As the products of genetic coding, biological labels can be tailored for tracking signaling dynamics in living systems over multiple length and time scales, and are also noninvasive and nondestructive in nature.111–113 The field has matured greatly in recent years, insofar that diverse strategies have been developed to allow expression of fluorescent proteins in specific locations within live cells, produce fusion with different targets for study of specific cellular activities, cover the full visible spectrum, and accommodate diverse detection schemes, as highlighted by the recently published reviews.107,114,115
While the main applications of fluorescent proteins are for imaging cellular activities, they have also been employed in cell-free bioassays for target detection. For example, Liao et.al constructed a specific and biocompatible fluorescent sensor based on the hybrid of GFP chromophore and peptide for HSA detection.116 A coelenteramide-containing fluorescent protein (CLM-CFP) was also employed in a simple bioassay for damage assessment of gamma radiation exposure, which would destroy the microenvironment of the coelenteramide, impact the photochemical proton transfer in the coelenteramide-apoprotein complex, and thus change the fluorescent output.117 Another interesting work employed a cell-free protein synthesis system to generate green fluorescence proteins (GFP) in response to the amino acid targets in samples.118 The fluorescence intensity of the produced GFP was linearly proportional to the AA target concentration, achieving a limit of detection of 100 nM. Successful quantification of disease-related amino acids was also demonstrated in biological samples.119 This development reduces the cost and eliminates the necessity to produce and purify the fluorescence proteins, which are two of the obstacles preventing their wide applications in bioassays.
3. TARGET RECOGNITION UNITS
In fluorescence-based assays, target recognition is commonly carried out by antibodies and protein receptors, which can bind specifically and strongly to diverse analytes spanning from macromolecules like proteins to small metabolites and ligands.112,120 However, obtaining specific antibodies is not an easy task; batch-to-batch reproducibility in antibody or protein expression is a big concern, and the cost of production remains to be high. They also suffer from selectivity, stability and cross reactivity issues. To overcome part of these limitations in using antibodies in HTS, the animal-free approach of phage display has been ad opted to engineer antibodies through an in vitro and high-throughput selection process. Phage display allows for selective enrichment of library pools and high-throughput screening of resulting clones.121,122 The selected proteins can have the desired properties including increased affinity, specificity, stability, and new enzymatic activity; furthermore, the conditions of the binding selection can be tightly controlled during the in vitro selection process.
Protein-based affinity probes demand stringent storage conditions and could interfere with techniques that show high response to proteins like mass spectrometry. Thus, aptamers, the non-protein based affinity probes, have emerged as attractive alternatives to antibodies in diagnostic, therapeutic, imaging and targeting applications. Aptamers can be discovered via systematic evolution of ligands by exponential enrichment (SELEX).123 They are considered as chemical, or synthetic, “antibodies” because of their ability to mimick antibodies in target binding with high affinity and specificity. However, they offer several unique characteristics and advantages compared to antibodies.124,125 Firstly, aptamers can be selected in vitro for any given targets, without the use of cell lines or animals. Therefore, aptamers can be selected against toxic or non-immunogenic targets since the process is independent of the introduction of an animal immune system, which is necessary for antibody production.126 More importantly, SELEX can be conducted in regular chemistry or biomedical research labs to produce the affinity probes for the specific targets of interest. Secondly, aptamers, once selected, can be chemically synthesized with high reproducibility and purity in a large quantity. With the aptamer sequence known, any lab can easily attain and employ them for development of diverse bioassays. Lastly, aptamers are very stable and can recover their active conformation after thermal denaturation, which is not the case for antibodies. All these features greatly expand the accessibility and applicability of these affinity probes. To date, many high-affinity aptamers have been selected for a broad range of targets including metal ions, peptides, drugs, proteins, and even whole cells or pathogenic particles. Since their discovery, aptamers have garnered tremendous attention for the design of biosensors, target imaging agents and drug delivery.
On the other hand, synthetic chemistry has made tremendous progress in preparation of the biomimetic “receptors”, i.e., the “synthetic receptors”, through chemical reactions as well as diverse molecularly imprinted polymers using the target biomolecules as the templates.127,128 Synthetic receptors may not offer comparable affinity to their targets as antibodies and aptamers do; but their production is much easier and cheaper. They are also much more stable during long-term storage and in complex or harsh assay conditions compared to the biomolecule-based probes, which greatly simplifies assay design. In addition, the target recognition capability and specificity of synthetic receptors are highly tunable by modifying chemical structure and employing accessory chemicals in the binding environment, which produce diverse sensing elements to be adopted to statistical analysis models for target differentiation. All these features make them highly adaptable to HTS to enable rapid screening of large numbers of samples for identification of potential hits to be analyzed in follow-up tests using the more expensive and complicated approaches.
Although there are large varieties of affinity probes employed in fluorescence-based assays, this review, due to space limitations, only concentrates on the protein- and nucleic acid-based target recognition units, as well as the synthetic receptors developed and applied in bioassays in recent years.
3.1. Functional nucleic acids
Nucleic acid-based target recognition probes used in fluorescence-based assays can usually be categorized into three groups: the complimentary probes used to detect gene sequences via DNA or RNA hybridization; the aptamers that can bind and recognize non-nucleic acid targets; and the DNAzymes, the activity of which is turned on in the presence of the targets. The common feature of the nucleic acid-based target recognition probes is their high structural flexibility. This enables the design of innovative sensors by utilizing conformational change in the nucleic acids upon target binding to trigger a series of downstream events for signal amplification.
3.1.1. Hybridization probes
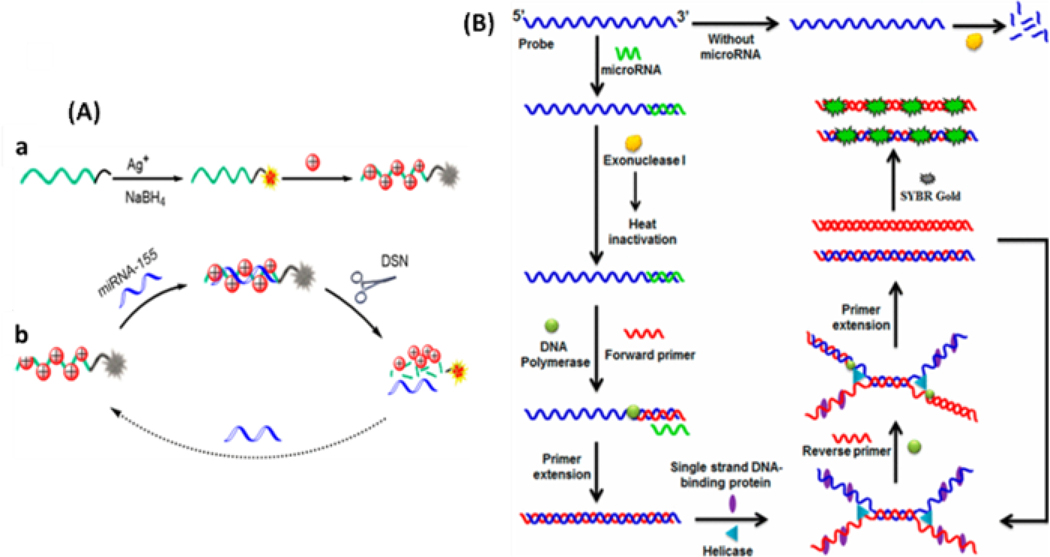
Conventionally, recognition of nucleic acid targets like genes, mRNAs, and non-coding RNAs can be done by fully complementary binding between the targets and the recognition probes, such as MBs, which unwinds the hybridized region on the MB and releases the fluorophore from the quencher, turning on strong fluorescence.129–131 As discussed in Section 2.4, nanomaterials are excellent quenchers; thus, they are often employed in MB construction to improve the detection limits of fluorescence-based assays. A label-free platform for microRNA detection was constructed based on fluorescence quenching of silver nanoclusters by the positively charged AuNPs.132 As shown in Figure 4A, DNA-stabilized Ag nanoclusters (DNA/AgNCs) were introduced as fluorescent probes, and DNA−RNA heteroduplexes were formed upon the addition of target miRNA-155. Meanwhile, the positively charged AuNPs could be electrostatically adsorbed on the negatively charged single-stranded DNA (ssDNA) or DNA−RNA heteroduplexes to quench the fluorescence signal. In the presence of duplex-specific nuclease (DSN), DNA−RNA heteroduplexes are substrates for the enzymatic hydrolysis of the DNA strand to yield a fluorescence signal due to the diffusion of AgNCs away from the AuNPs.
Figure 4.
(A) a - Formation of DNA/AgNCs and the adsorption of AuNPs on DNA/AgNCs for fluorescence quenching; b - Schematic representation for fluorescent detection of miRNA-155 based on DSN-Assisted Target Recycling Signal Amplification. Reproduced from Miao, X.; Cheng, Z.; Ma, H.; Li, Z.; Xue, N.; Wang, P. Anal. Chem. 2018, 90, 1098–1103 (ref 132). Copyright 2018 American Chemical Society. (B) Scheme of isothermal helicase-dependent amplification for miRNA. Reproduced from Ma, F.; Liu, M.; Tang, B.; Zhang, C.-y. Anal. Chem. 2017, 89, 6182–6187 (ref 138). Copyright 2017 American Chemical Society.
To amplify the signal from targets at trace levels, enzymatic reactions like polymerase chain reaction (PCR) can be employed. Even with the recent advancement of the next-generation sequencing (NGS) technologies, PCR remains the most dominant method in detection of specific nucleic acid strains in biomedical labs during marker discovery and validation owing to its robustness, simplicity in operation, and high amplification efficiency. For example, by using quantitative reverse transcription polymerase chain reactions (qRT-PCR), Peng et al. discovered the expression level of the circulating long non-coding RNA (lncRNA) MALAT1 in serum was different between samples collected from non-small cell lung cancer (NSCLC) patients and those from healthy controls.133 Another potential lncRNA biomarker for NSCLC, AFAP1-AS1, was also found with the same method by Li et al.,134 proving the effectiveness of RT-PCR in such applications.
To avoid the temperature cycles required by PCR and to enable highly sensitive detection of nucleic acids in ill-equipped labs, diverse isothermal amplification approaches have also been developed and adopted in fluorescence-based assays.135 Among the diverse isothermal amplification techniques, loop-mediated isothermal amplification (LAMP) stands out to be a reliable approach and is employed by several groups in their works. Recently, Kong et. al. developed a LAMP strategy on lambda DNA and achieved a 69-fold increase in signal above background, which is 20-fold higher than the gold standard and yields an overall limit of detection of 25 copies/μL within an hour via a mobile phone-based platform.136 Reverse-transcription-loop-mediated isothermal amplification (RT-LAMP) was also proposed as an enabling technology for simplified diagnostic tests for RNA viruses.137
Besides polymerase, other enzymes have also been employed in isothermal amplification of nucleic acids. Ma and co-workers developed a helicase-dependent amplification strategy for label-free detection of low-abundance microRNAs.138 The target microRNA specifically hybridizes with the 3’-terminus of the linear probe to form a DNA-microRNA heteroduplex, protecting the probes from digestion by Exonuclease I (Fig. 4B). The remaining probes may be subsequently amplified by helicase-dependent amplification, generating high fluorescence within 30 minutes. This assay yielded a low detection limit of 12.8 fM and exhibited a large dynamic range from 100 fM to 10 nM. Moreover, it was able to discriminate different microRNA family members and was applied to quantify the endogenous microRNAs extracted from cancer cells.
3.1.2. Aptamers
Different than the complete hybridization used in the above examples, aptamers acquire their affinity to the targets through folding into well-defined three-dimensional structures. Such a conformational change is taken advantage of in bioassays to design highly sensitive and selective methods for detection of non-nucleic acid targets. In addition to the understanding of the aptamer critical sequence for target binding, it is possible to further modify the aptamers with conjugation species, such as fluorophores, nanoparticles, or enzymes without affecting its target affinity. In effect, this can greatly reduce the difficulty in labeling the aptamers with the signaling units in the design of fluorescence-based assays. Such a feature becomes extremely beneficial in the cases of coupling aptamer-based target recognition with nanomaterial-based signaling. This is because nanomaterials, such as QDs, silver nanoclusters, upconversion nanomaterials, cationic conjugated polymers, etc., are much bulkier than organic fluorophores but usually provide higher signal intensity, better photostability, a wider, size-tunable fluorescence spectrum, and larger versatility in assay platform designs.
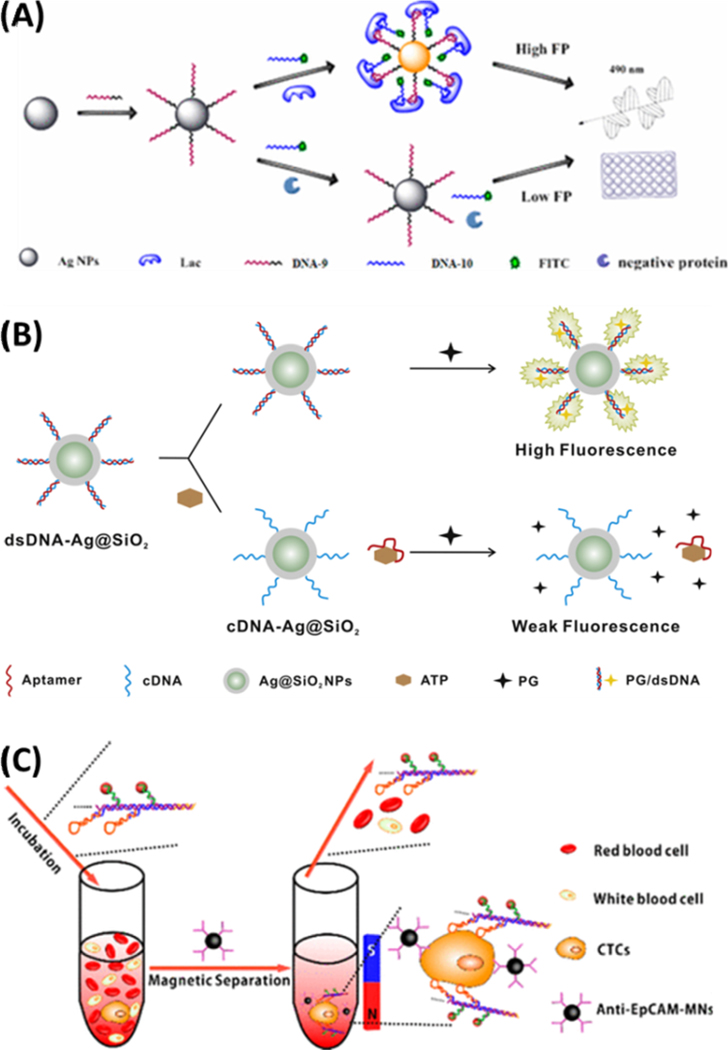
A silver-enhanced bivalent aptasensor, reported by Chen et al., employs the two split parts of an aptamer and the metal-enhanced fluorescence (MEF) effect of silver decahedral nanoparticles (Ag10NPs) to enable rapid detection of lactoferrin (Lac) in milk.139 As shown in Figure 5A, the bivalent aptamers were modified with fluorescein and the fluorescence enhancer, the Ag10NPs. The split aptamers could bind to different sites of Lac and assemble into an aptamer-target complex, narrowing the distance between Ag10NPs and FITC dye and resulting in MEF. The authors proved that the sensitivity of this assay was about 3 orders of magnitude higher than that obtained with traditional aptamer-based homogeneous assays, providing a detection limit of 1.25 pM.
Figure 5.
(A) Schematic illustration of the aptasensor produced from the bivalent aptamers and the Ag10NPs for dual signal amplification. Reproduced from Chen, Z.; Li, H.; Jia, W.; Liu, X.; Li, Z.; Wen, F.; Zheng, N.; Jiang, J.; Xu, D. Anal. Chem. 2017, 89, 5900–5908 (ref 139). Copyright 2017 American Chemical Society (B) The sensing scheme for ATP detection assisted by the Ag@SiO2 NPs. Reprinted from Biosens. Bioelectron., vol. 77, Song, Q.; Peng, M.; Wang, L.; He, D.; Ouyang, J. A Fluorescent Aptasensor for Amplified Label-free Detection of Adenosine Triphosphate Based on Core-shell Ag@SiO2 Nanoparticles (ref. 140). Copyright 2015, with permission from Elsevier. (C) Helispecific recognition and isolation of CTCs by the Ag2S nanoassembly and the anti-EpCAM-MNPs. Reproduced from Ding, C.; Zhang, C.; Yin, X.; Cao, X.; Cai, M.; Xian, Y. Anal. Chem. 2018, 90, 6702–6709 (ref 146). Copyright 2018 American Chemical Society.
Song and co-workers presented a label-free aptasensor for adenosine triphosphate (ATP) detection in aqueous solutions by using an ultra-sensitive nucleic acid stain PicoGreen (PG) as a fluorescent indicator and the core–shell Ag@SiO2 NPs as the MEF effector.140 The complementary DNA (cDNA)/aptamer duplex stained with PG was originally confined onto the Ag@SiO2 NPs’ surface (Figure 5B). Binding to ATP released the aptamers into the solution, causing a significant reduction in PG fluorescence with the MEF effect. This aptasensor achieved a detection limit of 14.2 nM for ATP with a wide linear range and exhibited a good assay performance in complex biological samples.
Furthermore, lv et al. developed a simple, rapid, low cost, and highly sensitive platform for detection of ochratoxin A (OTA) based on AuNPs.141 The dispersed AuNPs exhibited a better quenching effect to the fluorescein label on the anti-OTA aptamer than the aggregated AuNPs. In the absence of OTA, the aptamer was adsorbed onto the surface of AuNPs, which not only quenched fluorescein but also helped to enhance the stability of AuNPs in salty solution. Upon binding to OTA, the conformation of the aptamer changed, which removed the protection effect from the aptamer, induced AuNP aggregation, and recovered the probe’s fluorescence. The assay showed a linear response toward OTA concentration in the range of 25 nM to 300 nM. The limit of detection for OTA was determined to be 22.7 nM. Similarly, to obtain high detection sensitivity, much complex platforms were also developed based on the fluorescence quenching mechanism, such as tuning the aggregation/disaggregation behavior of graphene quantum dots by structure-switching aptamers142 and modulating the inner filter effect of AuNPs on the fluorescence of CdTe QDs via aptamers to detect acetamiprid143.
Conformational change in aptamers can also trigger signal amplification events. Gao et al. developed an upconversion fluorescent aptasensor based on hybridization chain reaction (HCR) and nicking endonuclease for detection of polychlorinated biphenyls (PCBs).144 Target recognition probes were designed to partially hybridize with the aptamers for the PCBs, i.e., PCB72/106, which would release the cDNA from the magnetic microspheres (MMPs) upon target binding to initiate HCR and open the stem of hairpins. Since the stem brought the quencher (BHQ-1) close to the upconversion nanoparticles (UCNPs), stem opening by HCR could turn on the UCNPs to emit strong fluorescence. In addition, a nicking enzyme was employed to further cleave the hairpins to stabilize the fluorescence, enabling PCB detection within the linearity range of 0.004 to 800 ng/mL and a detection limit of 0.0035 ng/mL. Similarly, Deng et al. developed a FRET-based ratiometric fluorescent biosensor for a highly sensitive and selective detection of kanamycin by using the aptamer-recognition event to trigger signal amplification.145 In this system, kanamycin-aptamer binding would initiate polymerase-catalyzed amplification (PCA) to produce single-strand DNA products, which could catalyze hairpin assembly (CHA) to enhance FRET between the donor molecule of Cy3 and the acceptor molecule of Cy5. The constructed biosensors result in highly sensitive detection of kanamycin down to 0.29 nM within a linear range from 1.0 nM to 80.0 nM.
Near-Infrared (NIR) fluorescent Ag2S nanodots (NDs) were developed for efficient detection of circulating tumor cells through HCR-based signal amplification.146 As shown in Figure 5C, the Ag2S NDs were linked through HCR using the aptamers conjugated on the ND surface, forming the DNA1-Ag2S tags with each tag carrying multiple Ag2S NDs to enhance detection sensitivity. The NIR fluorescence greatly enhanced the background to facilitate detection in blood samples. The DNA1-Ag2S tags were bound to the MCF7 cells through the anti-MUC1 aptamers that were assembled onto the tags via the hairpins used in HCR. Since each of the DNA1-Ag2S-aptamer assemblies contained multiple aptamers to attain multivalency in target recognition, the binding ability with tumor cells was enhanced in comparison with the single-aptamer methods. Moreover, the anti-epithelial cell adhesion molecule (EpCAM) antibody-labeled magnetic nanospheres were used to capture the rare tumor cells from whole blood, which were also tagged by the DNA1-Ag2S-aptamer assemblies. Both the signal amplification approach and the magnetic pull-down step enabled highly efficient detection of the circulating tumor cells, showing good potential in cancer diagnostics.
Aptamer-containing probes can also be designed to serve for both purposes of target capture and signaling in fluorescence-based assays. Wang et al. employed the anti-Zearalenone (ZEN) aptamer to label the magnetic nanoparticles (MNPs), which could bind to ZEN and release a region for complementary hybridization with the detection DNAs tagged with the time-resolved fluorescence (TRFL) nanoparticles.147 After magnetic separation, the TRFL intensity at 544 nm was used to measure ZEN in the linearity range of 0.001–10 ng mL−1 and a limit of detection down to 0.21 pg mL−1.
3.1.3. DNAzyme
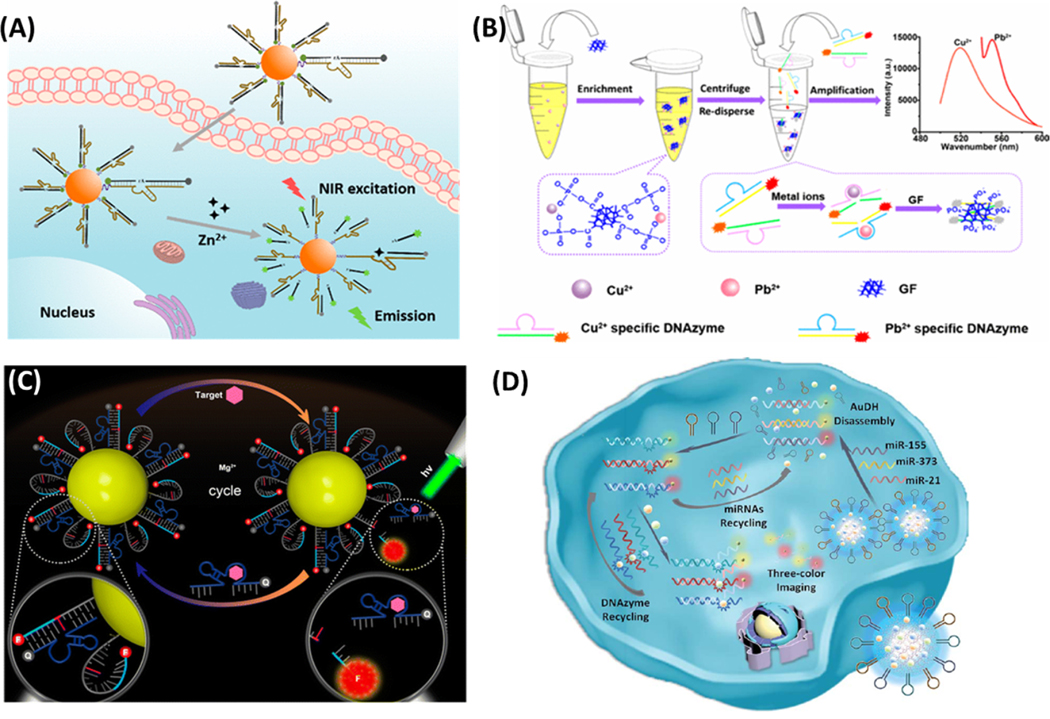
DNAzyme is a nucleic acid sequence that has catalytic activity, and thus, can act as both the target recognition unit and the signaling unit. All known DNAzymes have been discovered via in vitro selection, and most of them exhibit catalytic activity in the presence of metal cofactors. Their high selectivity to the type of metal ions makes them effective tools for metal detection. For instance, Yang et al. developed an RNA-cleaving DNAzyme-based two-photon (TP) imaging probe (TP-8−17ES−AuNP) for Zn2+ detection in living cells (Figure 6A).148 They modified a Zn2+-specific DNAzyme (8−17) with a TP fluorophore (TP-8−17ES) and coupled it onto the AuNPs for intracellular delivery. The modified TP-8−17ES exhibited good two-photon properties and excellent photostability. In the absence of Zn2+, the TP fluorophore was quenched by the AuNPs. In the presence of Zn2+, the DNAzyme cleaved the TP fluorophore-labeled substrate strand, resulting in fluorescence enhancement and TP imaging. Very recently, our group combined the high selectivity of the metal-responsive DNAzymes and the strong absorptivity of nanomaterials to design a sensor for ultrasensitive and multiplexed metal detection.149 As shown in Figure 6B, the graphene foam (GF) was employed to enrich divalent metal ions, like Pb2+ and Cu2+, that activated the DNAzymes to produce the fluorescently labeled single-stranded DNAs. The ssDNA had high affinity to the GF so that the fluorophore was quenched. GF enrichment and DNA cleavage, catalyzed by the DNAzymes, worked in synchronization to greatly improve detection sensitivity, resulting in metal quantitation as low as 50 pM and 0.6 nM for Pb2+ and Cu2+, respectively. In both cases, the DNAzymes not only specifically recognize the presence of the target metals but also help with signal amplification by its catalytic activity.
Figure 6.
(A) A fluorescent sensor for intracellular Zn2+ prepared from immobilizing the Zn-sensitive DNAzyme carrying a TP fluorescent dye onto the AuNPs. Reproduced from Yang, C.; Yin, X.; Huan, S.-Y.; Chen, L.; Hu, X.-X.; Xiong, M.-Y.; Chen, K.; Zhang, X.-B. Anal. Chem. 2018, 90, 3118–3123 (ref 148). Copyright 2018 American Chemical Society (B) A metal sensor prepared by coupling the GF with metal-sensitive DNAzymes. Reproduced from Fang, X.; Liu, Y.; Jimenez, L.; Duan, Y.; Adkins, G. B.; Qiao, L.; Liu, B.; Zhong, W. Anal. Chem. 2017, 89, 11758–11764 (ref 149). Copyright 2017 American Chemical Society. (C) The working mechanism of the aptazyme–AuNP sensor for fluorescent detection of intracellular ATP. Reproduced from Yang, Y.; Huang, J.; Yang, X.; Quan, K.; Wang, H.; Ying, L.; Xie, N.; Ou, M.; Wang, K. Anal. Chem. 2016, 88, 5981–5987 (ref 152). Copyright 2016 American Chemical Society. (D) Detection of miRNAs in living cells using AuDH/Mn+/H-mediated dual signal amplification. Reproduced from Meng, X.; Zhang, K.; Dai, W.; Cao, Y.; Yang, F.; Dong, H.; Zhang, X. Chem. Sci. 2018, 9, 7419–7425 (ref 153), with permission of The Royal Society of Chemistry
Alternatively, DNAzyme can be combined with other target recognition units and contribute only to signal amplification in fluorescence-based assays. Such strategies have been utilized to detect microRNA, ATP, bacterium, etc.150,151 As shown in Figure 6C, Yang and co-workers reported an aptazyme sensor that coupled the aptamer for ATP to the Mg2+-dependent 10−23 DNAzyme.152 The AuNP was modified with the substrate strand hybridized to the aptazyme. Target binding activated the aptazyme to cleave and release the fluorophore-labeled substrate from the AuNP, resulting in fluorescence enhancement. Since each target molecule could cleave multiple fluorophore-labeled substrates, the fluorescence signal was amplified, resulting in a limit of detection 2 or 3 orders of magnitude lower than that using just the aptamer for ATP detection.
Meng et al. successfully developed an AuNP - DNA hydrogel (AuDH) network for detection of intracellular miRNAs. The AuDH was constructed from three different DNA-capped AuNPs and their complementary fluorophore-modified DNA probes.153 Three hairpin-locked DNAzyme strands (H1, H2 and H3) and their specific metal ions (Cu2+, Mg2+ and Zn2+) were also loaded into the AuDH (Figure 6D). Once delivered into the cells, the intracellular miRNAs could trigger strand displacement reactions with the corresponding DNA probes to release the fluorophores from the AuNPs and unlock the DNAzymes for target recycling, enabling dual signal amplification. The assay was able to detect multiple miRNAs simultaneously down to 1 fM in different cells. Although the application was demonstrated for intracellular detection, this sensor can be easily adapted for fluorescence-based HTS of miRNAs.
3.2. Protein-based recognition units
3.2.1. Antibodies
Antibody is the most widely used target recognition unit because of its high specificity, strong affinity, wide availability and extensively studied structure and binding features. In a typical immunoassay, the antibody is conjugated onto a solid surface for target capture, and another target-binding molecule, probably with relatively lower specificity, such as a secondary antibody, can bind to the target and emit signal. For instance, a sensitive immunoassay was developed to employ multifunctional probes for detection of cancer cells.154 A fluorescent dye (Cy3) and the UCNPs were coupled to the secondary antibody to tag the anti-EpCAM antibody bound to cancer cells. The Cy3 and the UCNPs enabled the dual-modal fluorescence (FL)/upconversion luminescence (UCL) imaging in the confocal microscope under excitation wavelengths of 528 nm and 980 nm, respectively. Coupling the UCNPs with the downshifting luminescent Ln3+-doped NaREF4 nanocrystals (NCs), in vitro detection of the prostate-specific antigen with a limit of detection down to 1.8 ng mL−1 was achieved. Additionally, in vivo imaging in the near-infrared range was also attained with a high signal-to-noise ratio of 12.
The dual-antibody recognition format also facilitates the design of energy-transfer signaling systems in immunoassays to improve accuracy in quantification. For example, a ratiometric sensor was reported by Remcho et al. that used a capture antibody labeled with carbon dot-doped silica nanoparticles (Ab1-CD-SNPs) and a fluorescein-conjugated secondary antibody (Ab2-FITC) for detection of alphafetoprotein (AFP), a potential cancer marker.155 In this design, the CD-SNPs functioned as both the solid support to facilitate washing and target separation, as well as the built-in signal reference to correct for inconsistent environmental effects. The target protein acted as the bridge to connect the CD-SNPs and fluorescein to induce energy transfer between these two signaling molecules and realize ratiometric detection. A similar design but employing two fluorophores was developed by Wang et al., which detected both fluorescence and scattering light cross-correlation spectroscopy (FSCCS) using silver nanoparticle (SNPs) and Alexa Fluor 488 (Alexa).156 The assay could distinguish the interaction between the two fluorescent-labeled biomolecules with the target molecules. It was applied to detect AFP, yielding a linear range of AFP from 5 pM to 580 pM and a detection limit of 3.1 pM.
Similarly, a FRET pair was generated by linking the FRET acceptor – semiconductor quantum dots (QDs) – with the long-lived lanthanide donors.157 An antibody that selectively recognized ADP but not ATP was labeled with terbium (Tb). An ADP modified His6-peptide was conjugated to the QD surface via metal-affinity coordination. Time-resolved FRET (TR-FRET) was measured to minimize the background. With the addition of free ADP, the antibody was competitively separated from the QD and the change in the ratiometric emission could be correlated with the free ADP concentration.
3.2.2. Protein receptors
Besides antibodies, protein receptors are another important group of the protein-based target recognition units that provide high affinity and specificity in ligand binding. Such receptors span on a cell surface and can establish a preferred intermembrane spacing as well as favorable orientation for proper ligand acceptance based on molecular dimensions and structures. Recombinant protein receptors have been employed to detect their native ligands in fluorescence-based assays. For example, Liu et al. reported an estrogen receptor (ER)-based biosensor for facile screening of estrogenic compounds in the environment, based on the competitive binding between the xenoestrogens and 17β-estradiol (E2) for the recombinant human estrogen receptor α (hERα).158 The competition freed up E2 to bind to the fluorophore-labeled anti–E2 monoclonal antibody; the unbound anti–E2 antibody then interacted with the immobilized E2–protein conjugate on the fiber surface, yielding fluorescence emission induced by the evanescent field. Protein receptors often require the membrane environment to remain stable in aqueous solutions and function properly. Biswas et al. then incorporated the EphA2 receptor in the membrane structure supported by the silica microbeads, together with E-cadherin.159 Epifluorescence was used to precisely determine the intermembrane receptor−ligand dimension through receptor−ligand binding with the EphrinA1 and anti-E-cadherin antibody, respectively, offering a space resolution down to 1 nm in estimation of the intermembrane receptor−ligand dimension.
The target recognition functions of protein receptors have also been exploited to assist target enrichment. For example, T-cell immunoglobulin domain protein 4 (Tim4) is a type of transmembrane protein expressed on macrophages, which strongly binds phosphatidylserine, specifically on apoptotic cells, exosomes and microvesicles.160 This protein has been utilized to isolate and detect the extracellular vesicles (EVs),161 in which the isolated cells and EVs were easily released by a chelating reagent for Ca2+ because Tim4-phosphatidylserine binding is Ca2+-dependent. In addition, a two-stage microfluidic platform (ExoPCD-chip) that integrated on-chip isolation and in situ electrochemical analysis of EVs from serum was also reported to use the Tim4 modified magnetic beads to capture exosomes in a Y-shaped micropillars mixing pattern.162
3.3. Supramolecular receptors
As non-biological target recognition units, supramolecular strategies have become more and more popular in solving problems in analyte detection and imaging. They have been developed by chemically assembling macrocycles for interaction/accommodation with multiple guests.163 Compared to the biological target recognition units mentioned above, supramolecular receptors offer several advantages. Their structures can be tailored to improve specificity in target recognition. In addition, the binding affinity and selectivity can be tuned through the cooperation between different structural units within the receptor and by the solution components in the sensing environment. Moreover, they form dynamic molecular interactions with the targets and the “on-and-off” binding format produces less interference to the function of target molecules during in vitro or in vivo applications. These features, along with the feasibility of large-scale production with lower costs than biomolecules, make synthetic receptors excellent target recognition units in bioassays.
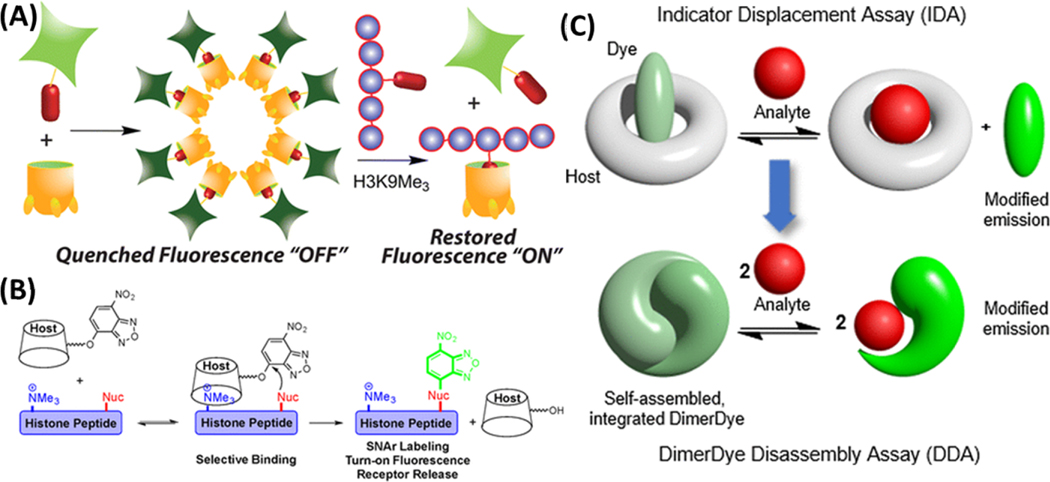
By taking advantage of the electrostatic interaction between a cationic histone peptide and an anionic cavitand, a fluorescent indicator displacement sensing system has been created for the detection of trimethylated peptides and determination of histone demethylase activity.164 Selective recognition of suitably sized trimethylammonium salts by the anionic cavitand can induce reversible lipophilic aggregation, providing a unique quenching mechanism for production of the supramolecular receptor-based sensor for methylated histone peptides (Figure 7A). This sensor demonstrated excellent capability in sensing the activity of the methylation-related enzymes and is suitable for HTS of enzyme inhibitors owing to its high simplicity in operation. Sensitive discrimination concerning the state and position of methylation on the protein is necessary to further understand how different situations of the modifications could impact protein function. Therefore, built upon the work of the aggregation-based sensor, we further developed a dual-mode synthetic receptor array for discrimination of histone peptides carrying various situations of methylations using variably functionalized cavitands.165 The cavitands not only bind specifically to the trimethyllysine (KMe3) groups, but also have secondary upper rim structures to provide sensitive discrimination of targets with identical KMe3 binding handles, i.e. identical lysine modifications at different sites of histone peptides. The synergistic application of multiple variables allows the synthetic receptors to approach high levels of recognition selectivity.
Figure 7.
(A) Self-aggregation based host-guest sensing of histone methylation. (B) Supramolecular affinity labeling of the histone peptides containing trimethyllysine. Reproduced from Gober, I. N.; Waters, M. L. J. Am. Chem. Soc. 2016, 138 (30), 9452–9459 (ref 169). Copyright 2016 American Chemical Society. (C) Self-assembled dimer-dye based fluorescent sensing in biological media. Reproduced from Beatty, M. A.; Borges-Gonzalez, J.; Sinclair, N. J.; Pye, A. T.; Hof, F. J. Am. Chem. Soc. 2018, 140 (10), 3500–3504 (ref 171). Copyright 2018 American Chemical Society.
In addition, to further understand epigenetic regulation, the deep cavitand array was applied to monitor site-specific modification catalyzed by the lysine methyltransferase PRDM9, or the lysine demethylase JMJD2E, with interference from the non-substrate histone peptides.166 Moreover, inclusion of a fluorogenic guest, trans-4-[4-(dimethylamino)styryl]-1-methylpyridinium iodide (DSMI), added the ability of phosphorylation recognition to our sensor arrays, which enabled in situ monitoring of the influence from the adjacent methylation on the reaction rate of a protein kinase.167 Our works on supramolecular receptor-based assays have broaden the scope of peptide modification detection by chemical means and may facilitate the development of more versatile in vitro enzymatic assays.
As a side application, we also applied our cavitand array to detect metals, relying on the coordination of the metal to the fluorescent guests or to the cavitands which induced fluorescence change. Variable host–metal and host–guest–metal interactions led to both turn-on and turn-off fluorescence, providing the discriminatory properties of our array towards a series of transition and heavy metals.168 The limit of detection for certain metals was as low as 70 nM, and highly similar metals such as lanthanides and actinides were distinguished at low micromolar concentrations in environmental water samples.
An interesting fluorescent labeling approach for lysine methylation detection was developed by the Waters group (Figure 7B).169 Calix-[4]-arene (CX4-Ar), a trimethyl lysine receptor, was used to direct a reactive fluorophore to the trimethylation site, which could subsequently label the nearby lysine on the same peptide. The labeling efficiency was proved to be highly sensitive to the distance between the trimethylation lysine and target lysine. This fluorescence turn-on approach was applied to monitor the histone deacetylase activity in vitro. The same group also recently developed an indicator displacement sensor array containing four synthetic receptors to detect a large number of PTMs, including methylation, phosphorylation and acetylation.170
A self-assembled dimer-dye based sensor was reported by the Hof group, utilizing a strategy different from the well-known indicator displacement assays (IDAs) (Figure 7C).171 The fluorophore employed formed dimers due to strong hydrophobic attraction and electrostatic repulsion and self-quenched, until target addition, which disassembled the fluorophore and resumed the fluorescence. Another approach done by the Hof group involves using the cucurbit[7]uril–dye (Rhodamine B) pair for monitoring acetyl amantadine, a biomarker for aggressive cancer.172 The detection limit was 0.087 μM with a linear range up to 1 μM. Switching the rhodamine dye with a berberine fluorophore further reduced the detection limit to 0.077 μM.
Recently, exciting results for the differentiation of steroids with the commercially available synthetic receptors - cucurbit[n]urils (CB7 and CB8) were reported by Nau’s group.173 A total of 21 steroids were tested, thanks to the nanomolar binding affinity of the CBs to the steroids. Comprehensive studies using NMR, X-ray crystal diffraction, ITC, and quantum chemical calculations were also conducted in this work to gain better understanding of the binding behavior. It was observed that the binding between steroid and CBs were stable in gastric acid and serum; binding to CBs greatly enhanced the aqueous solubility of the steroids by more than 100 times. Additionally, the assay was employed to monitor the destruction of nandrolone 17-propionate to nandrolone by the pig liver esterase. Additional work by this group produced a simple fluorescence-based assay to continuously monitor the activity of ornithine decarboxylase.174 The enzyme catalyzed the transformation of l-ornithine to putrescine, which could be sensed by the combination of CB6 and the fluorogenic guest DSMI. This design is suitable for determination of inhibition constants, and for inhibitor HTS.
By conjugation of the synthetic host cucurbit[7]uril (CB7) to the fluorescent dye tetramethylrhodamine (TMR), Hirani et al. were able to analyze the cellular uptake of the synthetic host.175 The host-guest conjugate was found primarily in the cytoplasm but not organelles, imposing no perturbation to cell morphology or viability after 4-day incubation at a concentration of 2.2 μM. Very recently, the same group reported for the first time the recognition of the N-terminal methionine peptide by CB8 in biological solution.176 They screened > 100 peptide sequences to select four showing strong affinity to CB8, demonstrating that non-aromatic peptides could be suitable guests for CBs. However, the affinity requires the presence of hydrophobic or cationic residues near the methionine site.
Supramolecular sensors have been exploited to sense drug molecules, amino acids, nitrate, etc.163 Shcherbakova et al. used acyclic cucurbituril (aCBs) to sense morphine, heroin, and oxycodone as well as their metabolites.177 The sensor was comprised of four glycouril units terminated on both ends with naphthalene fluorophore walls. The recognition event between the aCBs and guests resulted in changes in the fluorescence of the naphthalene fluorophore walls, which was exploited to discriminate the target analytes.
One interesting development in this field are the applications of synthetic receptors in cell imaging. Rood et al. applied the host- guest complex between β-cyclodextrin (CD) and adamantane (Ad) to study overexpression of the membrane protein chemokine receptor 4 (CXCR4) in cells.178 The specific ligand to CXCR4, the Ac-TZ14011 peptide, was functionalized with Ad to specifically target CXCR4. A multivalent host molecule, the fluorescent β-CD-Poly(isobutylene-alt-maleic-anhydride)-polymer, could then be used to image the presence of CXCR4 on the cell surface through interaction between β-CD and Ad.178 Alternatively, Pal et al. utilized the supramolecular host−guest chemistry to functionalize nanoparticles for cell labeling.179 Cyclodextrin was attached to QDs to facilitate folate and riboflavin conjugation, where cyclodextrin acted as a host for the folate/riboflavin guest. The QDs could then selectively label the cells with over-expressed folate/riboflavin receptors and induce endocytosis, just as well as the particles covalently conjugated with folate/riboflavin.179
4. ASSAY FORMAT
Besides the choices of proper signaling and target recognition units, assay platforms that strategically connect these two parts to realize sensitive and specific target detection are extremely important for the success of fluorescence-based assays. The most convenient design is to generate difference in fluorescence signal, i.e. “fluorescence-on” or “fluorescence-off”, upon target recognition in homogeneous solutions. Otherwise, heterogeneous assay formats are designed to remove the fluorophores without target recognition. In addition, platform design is the key to improve assay performance in sample throughput, reproducibility, and quantitation accuracy, as well as to reduce overall operation costs, simplify operation, and shorten assay duration. In this review, we will discuss the progress made in the past few years in designing lateral flow devices, microfluidics, and microarray platforms for fluorescence-based assays.
4.1. Lateral flow device
The lateral flow assay (LFA) is a membrane-based platform for detection and quantification of analytes in complex mixtures. In an LFA device, the liquid sample containing the analytes of interest is placed on a membrane strip and moves under capillary action through various zones to interact with the immobilized target recognition and signaling units.180–182 Low cost, high portability, fast testing, and ease of production are the prevalently attractive features of LFAs that have resulted in their wide applications in multiple fields, like biomedicine, agriculture, food and environmental sciences. LFAs offer quick monitoring of analytes of interest, in particular in hospitals, physician’s offices and clinical laboratories for quick survey of pathogens, specific antigen and antibodies, as well as testing gene signatures.180–182
To improve analyte detectability, new nanomaterials have been developed to visualize the presence of analytes with enhanced sensitivity in LFAs, substituting the common labels of colored microbeads,183 Au NPs,184–186 and fluorescent NPs.187 For instance, Fe3O4-Pt/core-shell NPs (MPt/CS NPs) with various compositions were synthesized and integrated in lateral flow immunoassay (LFIA) strips to achieve 100-fold improvement in sensitivity in comparison to Au NPs.188 The Fe3O4 core of this nanocomposite acted as a nanozyme with catalytic activity superior to the natural enzymes; and its catalytic activity was further improved by incorporating Pt to the shell when building the heterogeneous nanostructures. In addition, Fe3O4 had magnetic properties to enable analyte enrichment to increase sensitivity of the LFA (Figure 8A).185 On the other hand, an Au core coated with a europium(III)-chelate fluorophore-doped silica shell (AuNPs@SiO2–Eu3+) was synthesized and conjugated to antibodies for the determination of human thyroid stimulating hormone (hTSH) in LFIA devices. The AuNPs@SiO2–Eu3+ nanocomposites allowed for both colorimetric and fluorometric detection on the LFIA device, because the Au core could adsorb visible light due to the localized surface plasmon resonance and the Eu(III)-chelate-modified shell could emit fluorescence under irradiation with a hand-held UV lamp (365 nm). The limits of detection (LOD) for naked eye observation could reach 5 μIU mL−1 and 0.1 μIU mL−1 for the colorimetric and fluorometric detection, respectively. This LFIA assay platform was combined with a smartphone and digital color analysis to record the fluorescence signals, giving a linear relationship between the fluorescence intensity at 625 nm and the logarithmic concentration of hTSH (R2 = 0.988) and an LOD of 0.02 μIU mL−1. In addition, to improve analyte quantification, Shah et al. coupled the low-cost ultraviolet light-emitting diodes with the long Stokes-shift QDs to enable ratiometric fluorescence measurements on a lateral flow device using a smart phone without optical filters.187 Ratiometric imaging with unmodified smartphone cameras improved the contrast and attenuated the impact of excitation intensity variability by 15×. This platform was applied to detect influenza A with the nucleoproteins spiked into the simulated nasal matrix, achieving LODs of 1.5 and 2.6 fmol using two mobile phones, which were comparable to that attained with a gel imager, and about 100 times better than using Au NPs.
Figure 8.
(A) Design of the magnetic nanozyme-based LFA strips for target detection. Reproduced from Kim, M. S.; Kweon, S. H.; Cho, S.; An, S. S. A.; Kim, M. I.; Doh, J. and Lee, J. ACS Appl. Mater. Interfaces 2017, 9, pp35133–35140 (ref 185). Copyright 2017 American Chemical Society. (B) Schematic illustration of the preconcentration unit of a LFA kit. Reproduced from Kim, C.; Yoo, Y. K.; Han, S. I.; Lee, J.; Lee, D.; Lee, K.; Hwang, K. S.; Lee, K. H.; Chung, S.; Lee, J. H. Lab Chip, 2017, 17, pp 2451–2458 (ref 188), with permission of The Royal Society of Chemistry
Another approach to improve detection performance in LFAs is to couple it with sample enrichment steps. For instance, a paper-based preconcentration unit was developed by Kim et al. to be used with a commercial human chorionic gonadotropin (β-hCG)-based LFA test (Figure 8B).188 For the preconcentration unit, we designed an ICP-based pre-concentrator by combining a paper channel layer with the pre-cut Nafion-pattern sheets, and attaching the cellulose paper on a supporting substrate, in which substantial sensitivity was enhanced and the limit of detection was as low as 150 ng/mL β-hCG. Such a low target concentration cannot be monitored without the preconcentration step.
4.2. Microfluidics
Microfluidic systems integrate microscale channels (tens to hundreds of micrometers) to control and manipulate small volumes of fluid (usually 10−9 to 10−18 L) to flow in designated configurations.189,190 They provide the advantages of small reagent consumption, fast analysis, miniaturization, high-throughput, and high degree of automation.189,190 Microfluidic devices coupled with fluorescence-based assays to detect protein or nucleic acid targets have been widely used in surveillance of highly prevalent diseases like human immunodeficiency virus (HIV), hepatitis and tuberculosis, or in other biomedical applications.189,190 The materials used in fabrication of the microfluidic devices could offer unique properties to help with target detection. For instance, Pivetal et al. reported a sensitive colorimetric and fluorescence detection method for clinically relevant protein biomarkers using a Teflon®-FEP microfluidic device.191 The inert Teflon®-FEP microcapillaries were functionalized with a permanent coating of the high-molecular weight polyvinyl alcohol (PVOH). While such a surface permits antibody coupling with a variety of chemical reactions, the authors used a bi-functional crosslinker and the (3-aminopropyl)triethoxysilane (APTES)-coated PVOH to enhance surface coverage with the capture antibody without increasing background binding.191 Paper-based microfluidics have also been employed by Kong et al.192 for fast, inexpensive and direct detection of biological macromolecules. In this work, the Cy3/FAM labeled ssDNA was employed as the reporter and the graphene oxide as the quencher in FRET, with the glass fiber paper being the supporting surface. The chip was designed and fabricated by a cutting machine, forming a miniature biosensor that could monitor fluorescence recovery from FRET. The hybridization assay and fluorescence detection were both simplified, and the surface of the chip did not require immobilization or washing. The biosensing capability of this design met many potential requirements for disease diagnosis and biological analysis.
In recent years, development and applications of one important subcategory of microfluidics, droplet microfluidics190,193–196, in HTS have become more and more popular. Such devices generate and manipulate discrete droplets through multiphase flows inside various microchannels, with each droplet acting as a reactor or a detection well. Since the volume of each droplet is very small (in the range of subnanoliters), the droplets show enhanced mixing and mass transfer within short diffusion distances.190 One representative development was carried by He et al.197 which fabricated the fluorescent magnetic multifunctional microparticles by a facile droplet microfluidic strategy. Two sodium alginate streams, one doped with the Fe3O4 NPs and the other with the CdSe/ZnS QDs, were introduced into a flow-focusing channel as a type of parallel laminar flow to form the droplets containing two distinct parts. Then, in the serpentine channel, the Ca2+ in the oil phase diffused into the droplets, causing droplet solidification. The as-prepared fluorescent magnetic Janus particles were successfully employed for DNA assay, which demonstrated the potential of multifunctional microbeads in biological applications. 197
Chen et al.193 reported a series of interesting works in droplet microfluidics for fluorescence bioassays. For instance, they used the microfluidic technique to fabricate the bifunctional copolymers into thousands of mono-disperse, droplet-templated hydrogel microparticles for controlled encapsulation and triggered release of mammalian cells. Importantly, the hydrogel scaffolds were readily fabricated via quick, on-chip gelation by triggered release of Ca2+ from the Ca-EDTA complex. Very mild release of the microencapsulated cells was achieved via controlled degradation of the hydrogel scaffolds taking advantage of competitive binding with Ca2+ between Alginate and EDTA (Figure 9A). Moreover, they developed another droplet-based microfluidic approach to fabricate a large number monodisperse, portable microtissues. Using water–water– oil double emulsions as templates and spatially they assembled hepatocytes in the core and fibroblasts in the shell, forming a 3D liver model in a drop to co-culture cells.193 They labeled the alginate in the shell with fluorescein and imaged the 3D core-shell structure under confocal microscopy, and used the LIVE/DEAD® Viability/Cytotoxicity Kit the hepatocytes (HepG2 cells) in the core surrounded by fibroblasts (NIH-3T3 cells) in the shell were clearly observed (Figure 9B).
Figure 9.
(A) Construction of the 3D core-shell scaffold: a - Crosslink of the alginate network by release of Ca2+ from the Ca-EDTA complex; b - Schematic diagram of the PDMS device; c - Fabrication of the core–shell droplets using w/w/o double emulsion as the template, in which shell algination was triggered by release of Ca2+; d - Monodisperse core–shell droplets generated using the droplet-based microfluidics. (B) Spatial assembly of different cells in the 3D core–shell scaffold: a - HepG2 cells confined in the core by the hydrogel shell; b - NIH-3T3 fibroblasts immobilized by the crosslinked alginate network in the shell; c - Simultaneous assembly of hepatocytes in the core and fibroblasts in the shell, forming an artificial liver in a drop. The scale bars are 100 μm. Caption cited and figures produced from Chen, Q.; Utech, S.; Chen, D.; Prodanovic, R.; Lin, J. M.; Weitz, D. A. Lab Chip, 2016, 16, 1346–1349 (ref 193), with permission of The Royal Society of Chemistry.
4.3. Microarrays
Microarray is a two-dimensional array printed on a solid substrate such as a glass slide or a silicon thin-film that can simultaneously test multiple analytes in a miniaturized and parallel processing format, an ideal platform fluorescence-based assays. The solid substrate can be made with materials to enhance fluorescence detection. For example, an array of photonic crystal beads (PCBs) encoded by their characteristic reflection spectrum (CRS) was developed by Zhao et al. along with a hyperspectral imaging system and algorithms for high throughput decoding of the PCB array with accuracy up to 98.56% for ∼500 PCBs, and excellent ability to extract low-intensity fluorescence intensities (Figure 10).198 This technique can simultaneously obtain both spatial and spectral information from the PCBs, making this array a powerful platform for multiplex fluorescence assays. In addition, a localized surface plasmon resonance chip (LSPR chip) having the nanostructured arrays marked on the LSPR substrate was combined with a microfluidic chip by Zhou et al.199 These two chips were integrated with micropumps, a light source for fluorescence excitation, and a camera system, forming a point-of-care device for automatic sample analysis and data acquisition. To enhance detection sensitivity, a plasmonic field was used to excite the fluorescence dyes conjugated to the analyte instead of direct detection of the LSPR wavelength shift induced by analyte binding. Their method could enhance the biomarker detection sensitivity by 10 to 100 times upon careful design of the metal nanostructures and the location of the fluorescence dyes.
Figure 10.
Schematic illustration of the developed hyperspectral imaging system. Reproduced from Zhao, X.; Ma, T.; Zeng, Z.; Zheng, S.; Gu, Z. Analyst, 2016, 141, 6549–6556 (ref 198), with permission of The Royal Society of Chemistry.
OUTLOOK AND OPPORTUNITIES
The present review summarizes the recent progress in designing fluorescence-based assays applicable in HTS and beneficial for disease diagnosis and marker/drug discovery. Miscellaneous strategies were employed to improve target recognition, enhance signal intensity, and simplify assay processes. While great progresses have been made to move the field forward, many challenges exist in detection of low abundance targets with diverse properties in complex environment using rapid and easy procedures, calling for further technical advancements.
Detection of low-abundance targets relies on signaling units that can yield a high signal-to-noise ratio per target recognition event. Rapid development of nanotechnology produces large numbers of excellent nanomaterial-based fluorescent labels spanning wide spectral ranges. Still, issues exist in stability, biocompatibility, and reproducibility in bioconjugation and large-scale production. Continuous improvements in these aspects are needed in future works to further revolutionize the applications of fluorescent nanomaterials in medical diagnostic, imaging, and therapeutics. On the other hand, signal can be improved via amplification approaches like isothermal amplification, which does not require sophisticated equipment and is suitable for point-of-care applications. Still, problems like capability of target discrimination, limitation in detection of multiple targets, high rates of false positives etc., need to be overcome for current isothermal amplification strategies in order for them to replace the gold standard, PCR.
One interesting direction in development of new signaling molecules deserved more future research efforts is those responsive to external “stimuli”. As the simplest design in fluorescence-based assays, such signaling units like the fluorogenic dyes and the ET pairs permit direct measurement in homogeneous solutions, requiring no additional assay platforms. With proper design, target specificity can be greatly increased and signal from interfering molecules be dramatically reduced, enabling quick and sensitive response to targets and eliminating the sample preparation step. Another attractive feature of such designs, in particular, with the ET pairs, is the possibility of carrying out ratiometric measurement based on the relative fluorescence intensities between the donor and the acceptor. However, responsivity to diverse target molecules is not easy to be attained. For example, most of the fluorogenic dyes respond to reactive small molecules like ROS, RNS, HS, metals, etc.,44 or to certain environmental conditions such as hydrophobicity, limited by the types of chemical or physical changes experienced by the dye molecules.17,40 They could be employed to label certain functional groups on macromolecules but cannot be used to reveal the identity of proteins or nucleic acids. Ensuring fast and complete reaction is another difficulty in designing such probes. To broaden the scope of analytes targeted by the “stimuli-responsive” signaling units, ET pairs cab be formed, the distance between which can be controlled by the target binding events mediate by specific target recognition units. But the background signal is heavily dependent on the strength and stability of the interaction. One promising approach to solve these problems is the nanomaterial-based enzymes, nanozymes,200–203 that can catalyze the productions of multiple fluorescent molecules from each target molecule. Most of the nanozymes reported in literature are peroxidase mimicking, and “stimuli-responsive” signaling units derived from the nanozymes system require cooperation of biological enzymes.204–206 Improvements in the design of the cascade enzymatic systems as well as discovery of more nanozymes against diverse analytes are desired.
The challenge of target recognition in complex environment calls for the discovery of more, and highly specific probes. With the great success of the macromolecular affinity probes like antibodies and protein receptors, target recognition units that can be rationally designed or discovered in a procedure involved no animals, and chemically produced with lower cost and in a larger scale become more and more popular. One excellent category of such target recognition units is the synthetic receptors. Tremendous progress has been achieved in the past few years in employing synthetic receptors in fluorescence assays. Still, much more improvements are desired in target binding affinity and specificity, as well as in signal intensity. Detection in complex biological environments remains a big challenge for synthetic receptors. While rationale design of the receptor molecule is the most important approach to solve these difficulties, proper employment of statistical analysis and machine learning tools holds great promise for improving the performance of synthetic receptors in molecular detection.207,208 These tools, coupled with a good numbers of sensor elements that target various properties of the analytes, are expected to reduce the noise from interfering molecules and enlarge the target-specific signals by multivariate data analysis. Another potential solution is to couple specific target enrichment with synthetic receptors, i.e. relying on analytical separation, to improve specificity and sensitivity.209
With the multi-well plates being the common HTS platforms, microarrays and microfluidics have been the popular solutions for assay miniaturization and automation. The platform that can remove the interfering molecules and enrich the trace analytes is the key to move fluorescence-based assays from research labs to medical clinics. The present work only highlights few developments that are the most relevant to fluorescence-based assays, because insights of the recent progress in the design, fabrication, and specific applications of such platforms have been reviewed recently.210–215
In summary, opportunities exist in development of novel fluorescence-based assays by expanding the scope of selections of both the signaling and the target recognition units, and by integrating them on judiciously designed platforms. All three elements play important roles in improving assay accuracy, specificity and sensitivity, as well as in automating the detection procedure, facilitating adaptation in HTS and real-world applications. The recent developments highlighted in this Review provide excellent examples on how careful consideration on choosing or designing these elements can greatly enhance assay performance, paving the way for future works on further improvements in the specificity, sensitivity, simplicity, and speed of analysis of fluorescence-based assays.
Acknowledgments
The authors gratefully acknowledge funding from the National Institutes of Health (Grant R01-CA188991) and the National Science Foundation (Grant CHE-1707347).
Biographies
Xiaoni Fang earned her Ph.D. in 2016 from the Department of Chemistry at Fudan University under the supervision of Prof. Baohong Liu, then began her postdoctoral research at Prof. Wenwan Zhong’s lab at University of California, Riverside. Her research mainly focuses on development of novel nanomaterials and applied in bioassays for study of biomolecules.
Yongzan Zheng obtained his Ph.D. in 2017 from the Department of Chemistry at Tsinghua University under the supervision of Prof. Jin-Ming Lin, working on nanomaterial-enhanced chemiluminescence. Currently he is working in Prof. Wenwan Zhong’s lab at University of California, Riverside, as a postdoctoral researcher, developing isolation methods for circulating biomarkers.
Yaokai Duan is an assistant project scientist in the Department of Chemistry at University of California, Riverside supervised by Prof. Wenwan Zhong. He obtained his M.S. in Medicinal Chemistry at Peking University Health Science Center in 2013, and Ph.D. in Chemistry from University of California, Riverside in 2018. His research focuses on development of analytical methods to characterize protein corona of nanomaterials.
Yang Liu obtained his Ph.D. in 2017 from the Environmental Toxicology program at the University of California Riverside with the guidance of Prof. Wenwan Zhong. His research mainly focused on developing supramolecular sensors for posttranslational modifications. He moved to Yale University to continue his postdoctoral research after graduation. His research now focused on developing a microfluidic platform for single-cell transcriptome.
Wenwan Zhong is a Professor in Department of Chemistry and the Director of the Environmental Toxicology Graduate Program at the University of California, Riverside (UCR). She earned her Ph.D. in Chemistry at Iowa State University with Prof. Edward S. Yeung (2003). Before beginning her independent career at UCR, she was a Postdoctoral Research Associate at Los Alamos National Laboratory (2003−2006). Prof. Zhong and her research team focus their efforts on development and applications of microscale separation and bioassays for discovery, functional study, and detection of epigenetic markers such as circulating non-coding RNAs and post-translational modifications in protein, employing chemical tools like functional nucleic acids and synthetic receptors, as well as analytical technologies like flow field flow fractionation, capillary electrophoresis, mass spectrometry, and optical spectroscopy.
REFERENCES
- (1).Arnold M; Sierra MS; Laversanne M; Soerjomataram I; Jemal A; Bray F Gut 2017, 66, 683. [DOI] [PubMed] [Google Scholar]
- (2).Hall MD; Yasgar A; Peryea T; Braisted JC; Jadhav A; Simeonov A; Coussens NP Methods Appl fluores 2016, 4, 022001–022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kolanowski JL; Liu F; New EJ Chem. Soc. Rev. 2018, 47, 195–208. [DOI] [PubMed] [Google Scholar]
- (4).Ma F; Li Y; Tang B; Zhang C.-y. Acc Chem Res 2016, 49, 1722–1730. [DOI] [PubMed] [Google Scholar]
- (5).Pastoriza-Santos I; Kinnear C; Pérez-Juste J; Mulvaney P; Liz-Marzán LM Nat Rev Mater 2018, 3, 375–391. [Google Scholar]
- (6).Wu D; Sedgwick AC; Gunnlaugsson T; Akkaya EU; Yoon J; James TD Chem. Soc. Rev. 2017, 46, 7105–7123. [DOI] [PubMed] [Google Scholar]
- (7).Yang Z; Mao Z; Xie Z; Zhang Y; Liu S; Zhao J; Xu J; Chi Z; Aldred MP Chem. Soc. Rev. 2017, 46, 915–1016. [DOI] [PubMed] [Google Scholar]
- (8).Yang Z; Sharma A; Qi J; Peng X; Lee DY; Hu R; Lin D; Qu J; Kim JS Chem. Soc. Rev. 2016, 45, 4651–4667. [DOI] [PubMed] [Google Scholar]
- (9).Yoo SM; Lee SY Trends Biotech 2016, 34, 7–25. [DOI] [PubMed] [Google Scholar]
- (10).Zhu H; Fan J; Du J; Peng X Acc Chem Res 2016, 49, 2115–2126. [DOI] [PubMed] [Google Scholar]
- (11).Specht EA; Braselmann E; Palmer AE Annu Rev Physiol 2017, 79, 93–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Huang X; Song J; Yung BC; Huang X; Xiong Y; Chen X Chem. Soc. Rev. 2018, 47, 2873–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Qian L; Li L; Yao SQ Acc Chem Res 2016, 49, 626–634. [DOI] [PubMed] [Google Scholar]
- (14).Chi Z; Zhang X; Xu B; Zhou X; Ma C; Zhang Y; Liu S; Xu J Chem. Soc. Rev. 2012, 41, 3878–3896. [DOI] [PubMed] [Google Scholar]
- (15).Mu B; Zhang J; McNicholas TP; Reuel NF; Kruss S; Strano MS Acc Chem Res 2014, 47, 979–988. [DOI] [PubMed] [Google Scholar]
- (16).Fenneteau J; Chauvin D; Griffiths AD; Nizak C; Cossy J Chem Commun 2017, 53, 5437–5440. [DOI] [PubMed] [Google Scholar]
- (17).Klymchenko AS Acc Chem Res 2017, 50, 366–375. [DOI] [PubMed] [Google Scholar]
- (18).Liu Y; Qu Z; Cao H; Sun H; Gao Y; Jiang X ACS Nano 2017, 11, 12446–12452. [DOI] [PubMed] [Google Scholar]
- (19).Miao L; Zhu C; Jiao L; Li H; Du D; Lin Y; Wei Q Anal. Chem. 2018, 90, 1976–1982. [DOI] [PubMed] [Google Scholar]
- (20).Gupta R; Goddard NJ Sensor Actuat B: Chem 2017, 244, 549–558. [Google Scholar]
- (21).Lu Y-J; Deng Q; Hou J-Q; Hu D-P; Wang Z-Y; Zhang K; Luyt LG; Wong W-L; Chow C-F ACS Chemical Biology 2016, 11, 1019–1029. [DOI] [PubMed] [Google Scholar]
- (22).Niu W; Chen X; Tan W; Veige AS Angew Chem Int Ed Engl 2016, 55, 8889–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Farrukh A; Paez JI; Salierno M; del Campo A Angew Chem Int Ed Engl 2016, 55, 2092–2096. [DOI] [PubMed] [Google Scholar]
- (24).Li Y; Bai Y; Zheng N; Liu Y; Vincil GA; Pedretti BJ; Cheng J; Zimmerman SC Chem Commun 2016, 52, 3781–3784. [DOI] [PubMed] [Google Scholar]
- (25).Liu W; Samanta SK; Smith BD; Isaacs L Chem. Soc. Rev. 2017, 46, 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tateishi H; Monde K; Anraku K; Koga R; Hayashi Y; Ciftci HI; DeMirci H; Higashi T; Motoyama K; Arima H; Otsuka M; Fujita M Sci. Rep 2017, 7, 8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wu C; Deng Z; Shang B; Ikkala O; Peng B Chem Commun 2018, 54, 12726–12729. [DOI] [PubMed] [Google Scholar]
- (28).Peltomaa R; Amaro-Torres F; Carrasco S; Orellana G; Benito-Peña E; Moreno-Bondi MC ACS Nano 2018. [DOI] [PubMed] [Google Scholar]
- (29).He L; Dong B; Liu Y; Lin W Chem. Soc. Rev. 2016, 45, 6449–6461. [DOI] [PubMed] [Google Scholar]
- (30).Wang Y; Shi H; Cui K; Zhang L; Ge S; Yan M; Yu J Biosens Bioelectron 2018, 117, 515–521. [DOI] [PubMed] [Google Scholar]
- (31).Wang S-Y; Wang C-F; Lv Y-K; Shen S-G Trac-Trend Anal Chem 2018, 106, 53–61. [Google Scholar]
- (32).Yang D; Liu X; Zhou Y; Luo L; Zhang J; Huang A; Mao Q; Chen X; Tang L Anal. Methods 2017, 9, 1976–1990. [Google Scholar]
- (33).Ranallo S; Amodio A; Idili A; Porchetta A; Ricci F Chem. Sci. 2016, 7, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Aw SS; Tang MXM; Teo YN; Cohen SM Nucleic Acids Res 2016, 44, e92–e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Becherer L; Bakheit M; Frischmann S; Stinco S; Borst N; Zengerle R; von Stetten F Anal. Chem. 2018, 90, 4741–4748. [DOI] [PubMed] [Google Scholar]
- (36).Liao R; He K; Chen C; Chen X; Cai C Anal. Chem. 2016, 88, 4254–4258. [DOI] [PubMed] [Google Scholar]
- (37).Zhang X; Cheng R; Shi Z; Jin Y Biosens Bioelectron 2016, 75, 101–107. [DOI] [PubMed] [Google Scholar]
- (38).Linz TH; Hampton Henley W; Michael Ramsey J Lab Chip 2017, 17, 1076–1082. [DOI] [PubMed] [Google Scholar]
- (39).Zhao J; Bao X; Wang S; Lu S; Sun J; Yang X Anal. Chem. 2017, 89, 10529–10536. [DOI] [PubMed] [Google Scholar]
- (40).Lavis LD Annual Rev Biochem 2017, 86, 825–843. [DOI] [PubMed] [Google Scholar]
- (41).Hövelmann F; Seitz O Acc Chem Res 2016, 49, 714–723. [DOI] [PubMed] [Google Scholar]
- (42).Yang Z; Cao J; He Y; Yang JH; Kim T; Peng X; Kim JS Chem. Soc. Rev. 2014, 43, 4563–4601. [DOI] [PubMed] [Google Scholar]
- (43).Dube S; Dube H; Green NB; Larsen EM; White A; Johnson RJ; Kowalski JR ChemBioChem 2017, 18, 1807–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Jiao X; Li Y; Niu J; Xie X; Wang X; Tang B Anal. Chem. 2018, 90, 533–555. [DOI] [PubMed] [Google Scholar]
- (45).Pelaz B; del Pino P; Maffre P; Hartmann R; Gallego M; Rivera-Fernández S; de la Fuente JM; Nienhaus GU; Parak WJ ACS Nano 2015, 9, 6996–7008. [DOI] [PubMed] [Google Scholar]
- (46).Corbo C; Molinaro R; Parodi A; Toledano Furman NE; Salvatore F; Tasciotti E Nanomedicine 2015, 11, 81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).von Roemeling C; Jiang W; Chan CK; Weissman IL; Kim BY S. Trends Biotech 2017, 35, 159–171. [DOI] [PubMed] [Google Scholar]
- (48).Boutureira O; Bernardes GJ L. Chem. Rev. 2015, 115, 2174–2195. [DOI] [PubMed] [Google Scholar]
- (49).Ashby J; Duan Y; Ligans E; Tamsi M; Zhong W Anal. Chem. 2015, 87, 2213–2219. [DOI] [PubMed] [Google Scholar]
- (50).Duan Y; Liu Y; Shen W; Zhong W Anal. Chem. 2017, 89, 12160–12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lincoln R; Greene LE; Zhang W; Louisia S; Cosa GJ Am. Chem. Soc. 2017, 139, 16273–16281. [DOI] [PubMed] [Google Scholar]
- (52).Backus KM; Correia BE; Lum KM; Forli S; Horning BD; Gonzálezpáez GE; Chatterjee S; Lanning BR; Teijaro JR; Olson AJ Nature 2016, 534, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Pan S; Jang SY; Liew SS; Fu J; Wang D; Lee JS; Yao SQ Angew Chem Int Ed Engl 2018, 57, 579. [DOI] [PubMed] [Google Scholar]
- (54).Jiang X; Yu Y; Chen J; Zhao M; Chen H; Song X; Matzuk AJ; Carroll SL; Tan X; Sizovs A; Cheng N; Wang MC; Wang J ACS Chemi Bio 2015, 10, 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Umezawa K; Yoshida M; Kamiya M; Yamasoba T; Urano Y Nat Chem 2017, 9, 279–286. [DOI] [PubMed] [Google Scholar]
- (56).Liu Z; Zhou X; Miao Y; Hu Y; Kwon N; Wu X; Yoon J Angew Chem Int Ed Engl 2017, 129, 5812. [DOI] [PubMed] [Google Scholar]
- (57).Chen J; Jiang X; Zhang C; MacKenzie KR; Stossi F; Palzkill T; Wang MC; Wang J ACS Sensors 2017, 2, 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Gao M; Tang BZ Acs Sens 2017, 2. [DOI] [PubMed] [Google Scholar]
- (59).Su D; Teoh CL; Wang L; Liu X; Chang YT Chem. Soc. Rev. 2017, 46. [DOI] [PubMed] [Google Scholar]
- (60).Lukinavičius G; Reymond L; Umezawa K; Sallin O; D’Este E; Göttfert F; Ta H; Hell SW; Urano Y; Johnsson KJ Am. Chem. Soc.2016, 138, 9365–9368. [DOI] [PubMed] [Google Scholar]
- (61).Uno SN; Kamiya M; Yoshihara T; Sugawara K; Okabe K; Tarhan MC; Fujita H; Funatsu T; Okada Y; Tobita S Nat Chem 2014, 6, 681. [DOI] [PubMed] [Google Scholar]
- (62).Karch S; Broichhagen J; Schneider J; Böning D; Hartmann S; Schmid B; Tripal P; Palmisano R; Alzheimer C; Johnsson K; Huth TJ Med. Chem. 2018, 61, 6121–6139. [DOI] [PubMed] [Google Scholar]
- (63).Collot M; Fam TK; Ashokkumar P; Faklaris O; Galli T; Danglot L; Klymchenko AS J. Am. Chem. Soc. 2018, 140, 5401–5411. [DOI] [PubMed] [Google Scholar]
- (64).Yoshida W; Baba Y; Karube I Anal. Chem. 2016, 88, 9264–9268. [DOI] [PubMed] [Google Scholar]
- (65).Hildebrandt N; Spillmann CM; Algar WR; Pons T; Stewart MH; Oh E; Susumu K; Díaz SA; Delehanty JB; Medintz IL Chem.Rev. 2017, 117, 536–711. [DOI] [PubMed] [Google Scholar]
- (66).Chansuvarn W; Tuntulani T; Imyim A Trac-Trend Anal Chem 2015, 65, 83–96. [Google Scholar]
- (67).Du X; Jiang D; Li H; Hao N; You T; Wang K Sensor Actuat B: Chem 2018, 259, 316–324. [Google Scholar]
- (68).Elbaz J; Yin P; Voigt CA Nat Commun 2016, 7, 11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Ikbal J; Lim GS; Gao Z Trac-Trend Anal Chem 2015, 64, 86–99. [Google Scholar]
- (70).Goryacheva IY; Speranskaya ES; Goftman VV; Tang D; De Saeger S Trac-Trend Anal Chem 2015, 66, 53–62. [Google Scholar]
- (71).Chen Q; Zhu L; Chen J; Jiang T; Ye H; Ji H; Tsang S; Zhao Z; Yi T; Chen H Food Chem 2019, 277, 162–178. [DOI] [PubMed] [Google Scholar]
- (72).Bazin I; Tria SA; Hayat A; Marty J-L Biosens Bioelectron 2017, 87, 285–298. [DOI] [PubMed] [Google Scholar]
- (73).Gong S; Xia Y Chem Commun 2016, 52, 9660–9663. [DOI] [PubMed] [Google Scholar]
- (74).Gu W; Yan Y; Pei X; Zhang C; Ding C; Xian Y Sensor Actuat B: Chem 2017, 250, 601–607. [Google Scholar]
- (75).Zheng XT; Ananthanarayanan A; Luo KQ; Chen P Small 2014, 11, 1620–1636. [DOI] [PubMed] [Google Scholar]
- (76).Matea CT; Mocan T; Tabaran F; Pop T; Mosteanu O; Puia C; Iancu C; Mocan L Int J. Nanomed 2017, 12, 5421–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Bala R; Swami A; Tabujew I; Peneva K; Wangoo N; Sharma RK Biosens Bioelectron 2018, 104, 45–49. [DOI] [PubMed] [Google Scholar]
- (78).Huang L; Liao T; Wang J; Ao L; Su W; Hu J Adv Funct Mater 2017, 28, 1705380. [Google Scholar]
- (79).Justin R; Tao K; Román S; Chen D; Xu Y; Geng X; Ross IM; Grant RT; Pearson A; Zhou G; MacNeil S; Sun K; Chen B Carbon 2016, 97, 54–70. [Google Scholar]
- (80).Zuo P; Lu X; Sun Z; Guo Y; He H Microchim Acta 2016, 183, 519–542. [Google Scholar]
- (81).Meziani MJ; Dong X; Zhu L; Jones LP; LeCroy GE; Yang F; Wang S; Wang P; Zhao Y; Yang L; Tripp RA; Sun Y-P ACS Appl. Mater. Interfaces 2016, 8, 10761–10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Zhang J; Cheng F; Li J; Zhu J-J; Lu Y Nano Today 2016, 11, 309–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gu W; Pei X; Cheng Y; Zhang C; Zhang J; Yan Y; Ding C; Xian Y ACS Sensors 2017, 2, 576–582. [DOI] [PubMed] [Google Scholar]
- (84).Jin M; Liu X; Zhang X; Wang L; Bing T; Zhang N; Zhang Y; Shangguan D ACS Appl. Mater. Interfaces 2018, 10, 25166–25173. [DOI] [PubMed] [Google Scholar]
- (85).Xu M; Gao Z; Zhou Q; Lin Y; Lu M; Tang D Biosens Bioelectron 2016, 86, 978–984. [DOI] [PubMed] [Google Scholar]
- (86).Ji X; Wang H; Song B; Chu B; He Y Frontiers in Chemistry 2018, 6, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Peng F; Su Y; Zhong Y; Fan C; Lee S-T; He Y Acc Chem Res 2014, 47, 612–623. [DOI] [PubMed] [Google Scholar]
- (88).Sonawane MD; Nimse SB J. Chem. 2016, 2016, 19. [Google Scholar]
- (89).Das S; Batuta S; Alam MN; Fouzder C; Kundu R; Mandal D; Begum NA Colloid Surfaces B 2017, 157, 286–296. [DOI] [PubMed] [Google Scholar]
- (90).Reisch A; Klymchenko AS Small 2016, 12, 1968–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Yan J; Estévez MC; Smith JE; Wang K; He X; Wang L; Tan W Nano Today 2007, 2, 44–50. [Google Scholar]
- (92).Wu C; Chiu DT Angew Chem Int Ed Engl 2013, 52, 3086–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Yang X; Grailer JJ; Rowland IJ; Javadi A; Hurley SA; Matson VZ; Steeber DA; Gong S ACS Nano 2010, 4, 6805–6817. [DOI] [PubMed] [Google Scholar]
- (94).Sonawane SL; Asha SK ACS Appl. Mater. Interfaces 2016, 8, 10590–10599. [DOI] [PubMed] [Google Scholar]
- (95).Huth K; Glaeske M; Achazi K; Gordeev G; Kumar S; Arenal R; Sharma SK; Adeli M; Setaro A; Reich S; Haag R Small 2018, 14, 1800796. [DOI] [PubMed] [Google Scholar]
- (96).Luby BM; Charron DM; MacLaughlin CM; Zheng G Adv Drug Deliver Rev 2017, 113, 97–121. [DOI] [PubMed] [Google Scholar]
- (97).Cheng R; Ge C; Qi L; Zhang Z; Ma J; Huang H; Pan T; Dai Q; Dai LJ Phy. Chem. C 2018, 122, 13314–13321. [Google Scholar]
- (98).Sun X; Fan J; Zhang Y; Chen H; Zhao Y; Xiao J Biosens Bioelectron 2016, 79, 15–21. [DOI] [PubMed] [Google Scholar]
- (99).Yu M; Wang H; Fu F; Li L; Li J; Li G; Song Y; Swihart MT; Song E Anal. Chem. 2017, 89, 4085–4090. [DOI] [PubMed] [Google Scholar]
- (100).Li J; Li D; Yuan R; Xiang Y ACS Appl. Mater. Interfaces 2017, 9, 5717–5724. [DOI] [PubMed] [Google Scholar]
- (101).Cai Q-Y; Li J; Ge J; Zhang L; Hu Y-L; Li Z-H; Qu L-B Biosens Bioelectron 2015, 72, 31–36. [DOI] [PubMed] [Google Scholar]
- (102).Yan X; Song Y; Zhu C; Li H; Du D; Su X; Lin Y Anal. Chem. 2018, 90, 2618–2624. [DOI] [PubMed] [Google Scholar]
- (103).Jin B; Wang S; Lin M; Jin Y; Zhang S; Cui X; Gong Y; Li A; Xu F; Lu TJ Biosens Bioelectron 2017, 90, 525–533. [DOI] [PubMed] [Google Scholar]
- (104).Liu H; Yang Y; Wang A; Han M; Cui W; Li J Advanced Functional Materials 2016, 26, 2561–2570. [Google Scholar]
- (105).Liu Y; Wang Y-M; Sedano S; Jiang Q; Duan Y; Shen W; Jiang J-H; Zhong W Chem Commun 2018, 54, 4329–4332. [DOI] [PubMed] [Google Scholar]
- (106).Tan B; Zhao H; Du L; Gan X; Quan X Biosens Bioelectron 2016, 83, 267–273. [DOI] [PubMed] [Google Scholar]
- (107).Martynov VI; Pakhomov AA; Deyev IE; Petrenko AG Biochimica et Biophysica Acta (BBA) - General Subjects 2018, 1862, 2924–2939. [DOI] [PubMed] [Google Scholar]
- (108).Zlobovskaya OA; Sergeeva TF; Shirmanova MV; Dudenkova VV; Sharonov GV; Zagaynova EV; Lukyanov KA BioTechniques 2016, 60, 62–68. [DOI] [PubMed] [Google Scholar]
- (109).Bozhanova NG; Baranov MS; Sarkisyan KS; Gritcenko R; Mineev KS; Golodukhina SV; Baleeva NS; Lukyanov KA; Mishin AS ACS Chemical Biology 2017, 12, 1867–1873. [DOI] [PubMed] [Google Scholar]
- (110).David JSB Methods and Applications in Fluorescence 2018, 6, 020201. [DOI] [PubMed] [Google Scholar]
- (111).Choi J-W; Jo B-G; deMello AJ; Choo J; Kim HY Analyst 2016, 141, 6499–6502. [DOI] [PubMed] [Google Scholar]
- (112).Zhang J; Wang M; Tang R; Liu Y; Lei C; Huang Y; Nie Z; Yao S Anal. Chem. 2018, 90, 3245–3252. [DOI] [PubMed] [Google Scholar]
- (113).Nasu Y; Asaoka Y; Namae M; Nishina H; Yoshimura H; Ozawa T Anal. Chem. 2016, 88, 838–844. [DOI] [PubMed] [Google Scholar]
- (114).Wang A; Feng J; Li Y; Zou P ACS Chemical Neuroscience 2018, 9, 639–650. [DOI] [PubMed] [Google Scholar]
- (115).Chernov KG; Redchuk TA; Omelina ES; Verkhusha VV Chem. Soc. Rev. 2017, 117, 6423–6446. [DOI] [PubMed] [Google Scholar]
- (116).Lei C; Wang Z; Nie Z; Deng H; Hu H; Huang Y; Yao S Anal. Chem. 2015, 87, 1974–1980. [DOI] [PubMed] [Google Scholar]
- (117).Petrova AS; Lukonina AA; Dementyev DV; Ya. Bolsunovsky A; Popov AV; Kudryasheva NS Analytical and Bioanalytical Chemistry 2018, 410, 6837–6844. [DOI] [PubMed] [Google Scholar]
- (118).Jang Y-J; Lee K-H; Yoo TH; Kim D-M Anal. Chem. 2017, 89, 9638–9642. [DOI] [PubMed] [Google Scholar]
- (119).Ma Y; Mao Y; An Y; Tian T; Zhang H; Yan I; Zhu Z; Yang CJ Analyst 2018, 143, 1679–1684. [DOI] [PubMed] [Google Scholar]
- (120).Pecht I European Biophysics Journal 2018, 47, 363–371. [DOI] [PubMed] [Google Scholar]
- (121).Munke A; Persson J; Weiffert T; De Genst E; Meisl G; Arosio P; Carnerup A; Dobson CM; Vendruscolo M; Knowles TPJ; Linse S Proceedings of the National Academy of Sciences 2017, 114, 6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Brinton LT; Bauknight DK; Dasa SSK; Kelly KA PLOS ONE 2016, 11, e0155244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Fang X; Tan W Acc Chem Res 2010, 43, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Poolsup S; Kim C-Y Current Opinion in Biotechnology 2017, 48, 180–186. [DOI] [PubMed] [Google Scholar]
- (125).Huang B-T; Lai W-Y; Chang Y-C; Wang J-W; Yeh S-D; Lin EP-Y; Yang P-C Molecular Therapy - Nucleic Acids 2017, 8, 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Zhou J; Rossi J Nature Reviews Drug Discovery 2016, 16, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Erdőssy J; Horváth V; Yarman A; Scheller FW; Gyurcsányi RE Trac-Trend Anal Chem 2016, 79, 179–190. [Google Scholar]
- (128).Figueiredo L; Erny GL; Santos L; Alves A Talanta 2016, 146, 754–765. [DOI] [PubMed] [Google Scholar]
- (129).Eddy SR Nature Reviews Genetics 2001, 2, 919. [DOI] [PubMed] [Google Scholar]
- (130).Venkatesan N; Jun Seo Y; Hyean Kim B Chem. Soc. Rev. 2008, 37, 648–663. [DOI] [PubMed] [Google Scholar]
- (131).Dai N; Kool ET Chem. Soc. Rev. 2011, 40, 5756–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Miao X; Cheng Z; Ma H; Li Z; Xue N; Wang P Anal. Chem. 2018, 90, 1098–1103. [DOI] [PubMed] [Google Scholar]
- (133).Peng H; Wang J; Li J; Zhao M; Huang S.-k.; Gu Y.-y.; Li Y; Sun X.-j.; Yang L; Luo Q; Huang C.-z. Life Sciences 2016, 151, 235–242. [DOI] [PubMed] [Google Scholar]
- (134).Li W; Li N; Kang X; Shi K Clinica Chimica Acta 2017, 475, 152–156. [DOI] [PubMed] [Google Scholar]
- (135).Deng R; Zhang K; Li J Acc Chem Res 2017, 50, 1059–1068. [DOI] [PubMed] [Google Scholar]
- (136).Kong JE; Wei Q; Tseng D; Zhang J; Pan E; Lewinski M; Garner OB; Ozcan A; Di Carlo D ACS Nano 2017, 11, 2934–2943. [DOI] [PubMed] [Google Scholar]
- (137).Ball CS; Light YK; Koh C-Y; Wheeler SS; Coffey LL; Meagher RJ Anal. Chem. 2016, 88, 3562–3568. [DOI] [PubMed] [Google Scholar]
- (138).Ma F; Liu M; Tang B; Zhang C.-y. Anal. Chem. 2017, 89, 6182–6187. [DOI] [PubMed] [Google Scholar]
- (139).Chen Z; Li H; Jia W; Liu X; Li Z; Wen F; Zheng N; Jiang J; Xu D Anal. Chem. 2017, 89, 5900–5908. [DOI] [PubMed] [Google Scholar]
- (140).Song Q; Peng M; Wang L; He D; Ouyang J Biosens Bioelectron 2016, 77, 237–241. [DOI] [PubMed] [Google Scholar]
- (141).Lv X; Zhang Y; Liu G; Du L; Wang S RSC Advances 2017, 7, 16290–16294. [Google Scholar]
- (142).Wang S; Zhang Y; Pang G; Zhang Y; Guo S Anal. Chem. 2017, 89, 1704–1709. [DOI] [PubMed] [Google Scholar]
- (143).Guo J; Li Y; Wang L; Xu J; Huang Y; Luo Y; Shen F; Sun C; Meng R Anal Bioanal Chem 2016, 408, 557–566. [DOI] [PubMed] [Google Scholar]
- (144).Wang Y; Bai J; Huo B; Yuan S; Zhang M; Sun X; Peng Y; Li S; Wang J; Ning B; Gao Z Anal. Chem. 2018, 90, 9936–9942. [DOI] [PubMed] [Google Scholar]
- (145).Deng J; Liu Y; Lin X; Lyu Y; Qian P; Wang S Sensors & Actuators: B. Chemical 2018, 273, 1495–1500. [Google Scholar]
- (146).Ding C; Zhang C; Yin X; Cao X; Cai M; Xian Y Anal. Chem. 2018, 90, 6702–6709. [DOI] [PubMed] [Google Scholar]
- (147).Niazi S; Wang X; Pasha I; Khan IM; Zhao S; Shoaib M; Wu S; Wang Z Talanta 2018, 186, 97–103. [DOI] [PubMed] [Google Scholar]
- (148).Yang C; Yin X; Huan S-Y; Chen L; Hu X-X; Xiong M-Y; Chen K; Zhang X-B Anal. Chem. 2018, 90, 3118–3123. [DOI] [PubMed] [Google Scholar]
- (149).Fang X; Liu Y; Jimenez L; Duan Y; Adkins GB; Qiao L; Liu B; Zhong W Anal. Chem. 2017, 89, 11758–11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Wang L; Zhou H; Liu B; Zhao C; Fan J; Wang W; Tong C Anal. Chem. 2017, 89, 11014–11020. [DOI] [PubMed] [Google Scholar]
- (151).Zou M; Li D; Yuan R; Xiang Y Biosens Bioelectron 2017, 92, 624–629. [DOI] [PubMed] [Google Scholar]
- (152).Yang Y; Huang J; Yang X; Quan K; Wang H; Ying L; Xie N; Ou M; Wang K Anal. Chem. 2016, 88, 5981–5987. [DOI] [PubMed] [Google Scholar]
- (153).Meng X; Zhang K; Dai W; Cao Y; Yang F; Dong H; Zhang X Chem. Sci. 2018, 9, 7419–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Yang B; Zhang Y; Chen B; He M; Yin X; Wang H; Li X; Hu B Biosens Bioelectron 2017, 96, 77–83. [DOI] [PubMed] [Google Scholar]
- (155).Wu Y; Wei P; Pengpumkiat S; Schumacher EA; Remcho VT Analytical Methods 2016, 8, 5398–5406. [Google Scholar]
- (156).Wang J; Huang X; Liu H; Dong C; Ren J Anal. Chem. 2017, 89, 5230–5237. [DOI] [PubMed] [Google Scholar]
- (157).Díaz SA; Lasarte-Aragones G; Lowery RG; Aniket; Vranish JN; Klein WP; Susumu K Medintz IL. ACS Applied Nano Materials 2018, 1, 3006–3014. [Google Scholar]
- (158).Liu L; Zhou X; Lu Y; Shan D; Xu B; He M; Shi H; Qian Y Biosen. Bioelectron. 2017, 97, 16–20. [DOI] [PubMed] [Google Scholar]
- (159).Biswas KH; Groves JT Langmuir 2016, 32, 6775–6780. [DOI] [PubMed] [Google Scholar]
- (160).Kobayashi N; Karisola P; Peña-Cruz V; Dorfman DM; Jinushi M; Umetsu SE; Butte MJ; Nagumo H; Chernova I; Zhu B; Sharpe AH; Ito S; Dranoff G; Kaplan GG; Casasnovas JM; Umetsu DT; Dekruyff RH; Freeman GJ Immunity 2007, 27, 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Nakai W; Yoshida T; Diez D; Miyatake Y; Nishibu T; Imawaka N; Naruse K; Sadamura Y; Hanayama R Sci. Rep 2016, 6, 33935–33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Xu H; Liao C; Zuo P; Liu Z; Ye B-C Anal. Chem.2018. [DOI] [PubMed] [Google Scholar]
- (163).Elacqua E; Zheng X; Shillingford C; Liu M; Weck M Acc Chem Res 2017, 50, 2756–2766. [DOI] [PubMed] [Google Scholar]
- (164).Liu Y; Perez L; Mettry M; Easley CJ; Hooley RJ; Zhong WJ Am. Chem. Soc. 2016, 138, 10746–10749. [DOI] [PubMed] [Google Scholar]
- (165).Liu Y; Perez L; Mettry M; Gill AD; Byers SR; Easley CJ; Bardeen CJ; Zhong W; Hooley RJ Chem. Sci. 2017, 8, 3960–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Liu Y; Perez L; Gill AD; Mettry M; Li L; Wang Y; Hooley RJ; Zhong WJ Am. Chem. Soc. 2017, 139, 10964–10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Liu Y; Lee J; Perez L; Gill AD; Hooley RJ; Zhong WJ Am. Chem. Soc. 2018, 140, 13869–13877. [DOI] [PubMed] [Google Scholar]
- (168).Liu Y; Mettry M; Gill AD; Perez L; Zhong W; Hooley RJ Anal. Chem. 2017, 89, 11113–11121. [DOI] [PubMed] [Google Scholar]
- (169).Gober IN; Waters ML J. Am. Chem. Soc. 2016, 138, 9452–9459. [DOI] [PubMed] [Google Scholar]
- (170).Peacor BC; Ramsay CM; Waters ML Chem. Sci. 2017, 8, 1422–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Beatty MA; Borges-Gonzalez J; Sinclair NJ; Pye AT; Hof FJ Am. Chem. Soc. 2018, 140, 3500–3504. [DOI] [PubMed] [Google Scholar]
- (172).Li W; Kuehne NW; Dallin E; Gordon R; Hof F Can. J. Chem. 2016, 94, 969–975. [Google Scholar]
- (173).Lazar AI; Biedermann F; Mustafina KR; Assaf KI; Hennig A; Nau WM J. Am. Chem. Soc. 2016, 138, 13022–13029. [DOI] [PubMed] [Google Scholar]
- (174).Nilam M; Gribbon P; Reinshagen J; Cordts K; Schwedhelm E; Nau WM; Hennig A SLAS Discovery 2017, 22, 906–914. [DOI] [PubMed] [Google Scholar]
- (175).Hirani Z; Taylor HF; Babcock EF; Bockus AT; Urbach AR; Varnado CD Jr.; Bielawski CW J. Am. Chem. Soc. 2018, 140, 12263–12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Bockus AT; Smith LC; Grice AG; Ali OA; Young CC; Mobley W; Leek A; Roberts JL; Urbach AR; Vinciguerra B; Isaacs LJ Am. Chem. Soc. 2016, 138, 16549–16552. [DOI] [PubMed] [Google Scholar]
- (177).Shcherbakova EG; Zhang B; Gozem S; Minami T; Zavalij PY; Pushina M; Isaacs LD; Anzenbacher PJ Am. Chem. Soc. 2017, 139, 14954–14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Rood MT; Spa SJ; Welling MM; Ten Hove JB; van Willigen DM; Buckle T; Velders AH; van Leeuwen FW Sci. Rep. 2017, 7, 39908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Pal S; Dalal C; Jana NR ACS omega 2017, 2, 8948–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Heldt S; Hoenigl M Current fungal infection reports 2017, 11, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (181).Koczula KM; Gallotta A Essays in biochemistry 2016, 60, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Raeisossadati MJ; Danesh NM; Borna F; Gholamzad M; Ramezani M; Abnous K; Taghdisi SM Biosens Bioelectron 2016, 86, 235–246. [DOI] [PubMed] [Google Scholar]
- (183).Hu LM; Luo K; Xia J; Xu GM; Wu CH; Han JJ; Zhang GG; Liu M; Lai WH Biosens Bioelectron 2017, 91, 95–103. [DOI] [PubMed] [Google Scholar]
- (184).Foo PC; Chan YY; Mohamed M; Wong WK; Nurul Najian AB; Lim BH Analytica chimica acta 2017, 966, 71–80. [DOI] [PubMed] [Google Scholar]
- (185).Kim MS; Kweon SH; Cho S; An SSA; Kim MI; Doh J and Lee J ACS Appl. Mater. Interfaces 2017, 9, pp 35133–35140. [DOI] [PubMed] [Google Scholar]
- (186).Oh HK; Kim JW; Kim JM; Kim MG Analyst 2018, 143, 3883–3889. [DOI] [PubMed] [Google Scholar]
- (187).Shah KG; Yager P Anal. Chem. 2017, 89, 12023–12029. [DOI] [PubMed] [Google Scholar]
- (188).Kim C; Yoo YK; Han SI; Lee J; Lee D; Lee K; Hwang KS; Lee KH; Chung S; Lee JH Lab Chip 2017, 17, 2451–2458. [DOI] [PubMed] [Google Scholar]
- (189).Gong MM; Sinton D Chem. Rev. 2017, 117, 8447–8480. [DOI] [PubMed] [Google Scholar]
- (190).Shang L; Cheng Y; Zhao Y Chem. Rev. 2017, 117, 7964–8040. [DOI] [PubMed] [Google Scholar]
- (191).Pivetal J; Pereira FM; Barbosa AI; Castanheira AP; Reis NM; Edwards AD Analyst 2017, 142, 959–968. [DOI] [PubMed] [Google Scholar]
- (192).Chen Q; Chen D; Wu J; Lin JM Biomicrofluidics 2016, 10, 064115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Chen Q; Utech S; Chen D; Prodanovic R; Lin JM; Weitz DA Lab Chip 2016, 16, 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Cole RH; Gartner ZJ; Abate AR Journal of visualized experiments : JoVE 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Rhee M; Liu P; Meagher RJ; Light YK; Singh AK Biomicrofluidics 2014, 8, 034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Srinivas RL; Johnson SD; Doyle PS Anal. Chem. 2013, 85, 12099–12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (197).Lan J; Chen J; Li N; Ji X; Yu M; He Z Talanta 2016, 151, 126–131. [DOI] [PubMed] [Google Scholar]
- (198).Zhao X; Ma T; Zeng Z; Zheng S; Gu Z Analyst 2016, 141, 6549–6556. [DOI] [PubMed] [Google Scholar]
- (199).Zhou X; Wong TI; Song HY; Wu L; Wang Y; Bai P; Kim D-H; Ng SH; Tse MS; Knoll W Plasmonics 2014, 9, 835–844. [Google Scholar]
- (200).Korschelt K; Tahir MN; Tremel W Chemistry - A European Journal 2018, 24, 9703–9713. [DOI] [PubMed] [Google Scholar]
- (201).Sun H; Zhou Y; Ren J; Qu X Angew Chem Int Ed Engl 2018, 57, 9224–9237. [DOI] [PubMed] [Google Scholar]
- (202).Wang Q; Wei H; Zhang Z; Wang E; Dong S Trac-Trend Anal Chem 2018, 105, 218–224. [Google Scholar]
- (203).Wu J; Li S; Wei H Chem. Commun. 2018, 54, 6520–6530. [DOI] [PubMed] [Google Scholar]
- (204).Liu J-W; Luo Y; Wang Y-M; Duan L-Y; Jiang J-H; Yu R-Q ACS Appl. Mater. Interfaces 2016, 8, 33439–33445. [DOI] [PubMed] [Google Scholar]
- (205).Wang Y-M; Liu J-W; Adkins GB; Shen W; Trinh MP; Duan L-Y; Jiang J-H; Zhong W Anal. Chem. 2017, 89, 12327–12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Wang Y-M; Liu J-W; Jiang J-H; Zhong W Anal. Bioanal. Chem. 2017, 409, 4225–4232. [DOI] [PubMed] [Google Scholar]
- (207).Abraham Y; Zhang X; Parker Christian N Journal of biomolecular screening 2014, 19, 628–639. [DOI] [PubMed] [Google Scholar]
- (208).Braun R Advances in Experimental Medicine and Biology 2015, 844, 153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Lee J; Perez L; Liu Y; Wang H; Hooley RJ; Zhong W Anal. Chem. 2018, 90, 1881–1888. [DOI] [PubMed] [Google Scholar]
- (210).Cohen L; Walt DR Chemical Reviews (Washington, DC, United States) 2018, Ahead of Print. [Google Scholar]
- (211).Jauset-Rubio M; El-Shahawi MS; Bashammakh AS; Alyoubi AO; O’Sullivan CK Trac-Trend Anal Chem 2017, 97, 385–398. [Google Scholar]
- (212).Cho H; Kim J; Song H; Sohn KY; Jeon M; Han K-H Analyst (Cambridge, United Kingdom) 2018, 143, 2936–2970. [DOI] [PubMed] [Google Scholar]
- (213).Kaminski TS; Garstecki P Chem. Soc. Rev. 2017, 46, 6210–6226. [DOI] [PubMed] [Google Scholar]
- (214).Zhu Z; Yang CJ Acc. Chem. Res. 2017, 50, 22–31. [DOI] [PubMed] [Google Scholar]
- (215).Serra M; Ferraro D; Pereiro I; Viovy JL; Descroix S Lab Chip 2017, 17, 3979–3999. [DOI] [PubMed] [Google Scholar]