Abstract
RT-PCR analysis of gar pituitary and brain indicated that different combinations of gar melanocortin receptor mRNAs are present in the same tissues with mRNAs for gar mrap1 and gar mrap2. Against this background, an objective of this study was to determine whether the ligand sensitivity for either ACTH or α-MSH was affected when gar (g) melanocortin receptors (Mcrs) were co-expressed with either of the accessory proteins gMrap1 or gMrap2 in Chinese Hamster Ovary cells. The results indicated that gMc2r has an obligatory requirement for co-expression with gMrap1 in order for the receptor to be activated by hACTH(1–24). In addition, activation of gMc2r did not occur when the receptor was expressed alone or co-expressed with gMrap2. Furthermore, coexpression of gMc2r with gMrap1 followed by stimulation with NDP-MSH resulted in a low level of activation (only at 10−7M and 10−6M). However, gMc1r, gMc3r, gMc4r, and gMc5r responded to stimulation by NDP-MSH in a more robust manner. Co-expression of gMc1r, gMc3r, gMc4r, and gMc5r with gMRAP1 had no effect on sensitivity to stimulation by NDP-MSH or hACTH(1–24). Co-expression with gMRAP2 had no negative or positive effect on ligand sensitivity for gMc1r, gMc3r, and gMc5r, however this treatment did increase the activation of CHO cells transfected with gMc4r following stimulation with both hACTH(1–24) (p < 0.001), and NDP-MSH (p < 0.001). Co-expression of gMC5R with either gMRAP1 or gMRAP2 increased trafficking of gMC5R to the plasma membrane. These pharmacological observations are compared to the response of melanocortin receptors from other neopterygian fishes, cartilaginous fishes, and tetrapods to stimulation by ACTH (1–24) and forms of α-MSH.
Keywords: Lepisosteus oculatus, Melanocortin receptors, ACTH, α-MSH, Evolution
1. Introduction
Subclass Neopterygii is an assemblage of ray-finned bony fishes in class Actinopterygii that includes order Lepisosteiformes (the gars), order Amiformes (the bowfin) and division Teleostei (the modern bony fishes) (Nelson, 1994). The fossil record indicates that this assemblage emerged during the late Permian, and the Lepisosteiformes may be the oldest of the three lineages (Carroll, 1988s). Analyses of the morphology of extant and fossil neopterygygians have alternatively placed the Lepiosteiformes as the sister group to a clade consisting of the Semionotiformes (a group of extinct neopterygians), the Amiformes, and the Teleostei, or have placed the Lepisosteiformes as the sister group to a clade consisting of the Amiformes and the Teleostei, as summarized by Nelson (1994). Subsequent sequencing (Amores et al., 2011) and annotation of the genome of Lepisosteus oculatus (Braasch et al., 2016) favors the latter hypothesis. In addition, analysis of the slowly evolving gar genome has provided perspectives on the evolution of several gene families, (e.g., immunity, mineralization, and development) for not only their sister group, the teleosts, but also for the corresponding human genes (Braasch et al., 2016). Hence the gar genome appears to occupy a unique phylogenetic position within the bony vertebrates for analyzing the radiation of gene families (Braasch et al., 2016). In this regard, an analysis of the pharmacological properties of gar melanocortin receptors (gMcrs) may be informative with respect to the functional radiation of this gene family in teleosts and tetrapods.
Melanocortin receptors are unique to the chordates (Vastermark and Schiöth, 2011), and are members of the rhodopsin family of G protein-coupled receptors (GPCR; Yang, 2011). This family of receptors usually includes five single-exon members (i.e., Mc1r, Mc2r, Mc3r, Mc4r, and Mc5r) located on autosomal chromosomes, and have evolved novel functions as reviewed in Cone (2006) and Metz et al. (2006). Melanocortin receptors (Mcrs) are activated by the melanocortin peptides ACTH, and the MSH-sized ligands (i.e., α-MSH, β-MSH, γ-MSH and δ-MSH; Takahashi et al., 2001; Cone, 2006) which are in turn, derived from the common precursor protein gene, Pomc (Nakanishi et al., 1979; Amemiya et al., 2000). The Mcr genes and the Pomc gene have co-evolved during the radiation of the chordates with two accessory protein genes (for review see Dores et al., 2016), Mrap1 (melanocortin-2 accessory protein 1; Metherell et al., 2005), and Mrap2 (melanocortin-2 accessory protein 2; Chan et al., 2009). For some of the melanocortin receptors, the interplay between the receptor and one of these accessory proteins can influence ligand selectivity, ligand sensitivity, and trafficking of the receptor to the plasma membrane.
For example, the functional expression of human (h) Mc2r is completely dependent on co-expression with the Mrap1 gene (Metherell et al., 2005; Sebag and Hinkle, 2007). When hMc2r forms a heterodimer with Mrap1, the complex will traffic from the endoplasmic reticulum to the plasma membrane. As a result, hMc2r will be in the correct configuration to be stimulated by ACTH, but the receptor cannot be activated by any of the MSH-sized ligands at physiological concentrations (Metherell et al., 2005; Sebag and Hinkle, 2007). In addition, in the absence of this interaction, hMc2r is unable to move to the plasma membrane. These same trafficking and ligand selectivity properties have also been observed for three teleost MC2R orthologs; zebrafish (Agulleiro et al., 2010), sea bass (Agulleiro et al., 2013), and rainbow trout (Liang et al., 2015). With respect to the gMc2r ortholog, the prediction would be that this Mc2r ortholog should also have an obligatory requirement for interaction with gMrap1.
While Mrap1 interaction with hMc2r facilitates trafficking to the plasma membrane, Mrap1 interaction with hMc5r has the opposite effect (Sebag and Hinkle, 2009). If the hMc5r/Mrap1 interaction is an ancestral feature for bony vertebrate Mc5r orthologs, then the prediction would be that formation of a gMc5r/gMrap1 heterodimer will decrease trafficking of the receptor to the plasma membrane.
Finally, Mrap2 co-expression with either mouse Mc4r (Asai et al., 2013) or zebrafish Mc4r (Sebag et al., 2013) enhances ligand sensitivity of the receptor to activation by NDP-MSH, and in the case of the zebrafish Mc4r, sensitivity for activation by ACTH increases as well (Josep Agulleiro et al., 2013). For the gMc4r ortholog, the prediction would be that the ligand selectivity and sensitivity of gMc4r would also be affected by co-expression with gMrap2.
To evaluate these predictions and to determine whether the ligand selectivity and sensitivity properties of gMc1r, gMc2r, gMc3r, gMc4r, and gMc5r are influenced by co-expression with gMrap1 or gMrap2, individual gar melanocortin receptors were transiently transfected into Chinese Hamster Ovary cells, and stimulated with either hACTH(1–24) or NDP-MSH, and the effects of these manipulations on activation of the respective receptor were evaluated by using a cAMP-reporter gene assay. In addition, an RT-PCR analysis was also done to identify gar tissues that co-express gmcr and gmrap mRNAs.
2. Materials and methods
2.1. DNA constructs
The cDNA sequences of gMc1r, gMc2r, gMc3r, gMc4r, gMc5r, gMrap1, and gMrap2 were obtained from the spotted gar genome database (http://uswest.ensembl.org/Lepisosteus_oculatus/Info/Index) and the identification codes for each cDNA were: gMc1r, ENSLOCT00000001382; gMc2r, ENSLOCG00000009532; gMc3r, ENSLOCT00000022499; gMc4r, ENSLOCG00000018161; gMc5r, ENSLOCG00000018340), gMrap1, ENSLOCG00000009184; and gMrap2, ENSLOCG00000016569). Each cDNA was synthesized by GenScript (Piscataway, NJ) and inserted into a pcDNA3.1 + vector. The cAMP reporter gene construct CRE-Luciferase (Chepurny and Holz, 2007) was provided by Dr. Patricia Hinkle (University of Rochester, NY). An alignment of the gmer amino acid sequences is presented in Supplemental Fig. S1, and the amino acid sequences of gMrap1 and gMrap2 are presented in Supplemental Fig. S2.
2.2. RT-PCR protocol
Spotted gar (Lepisosteus oculatus) were obtained as embryos from hormone-induced spawns of wild-caught broodstock from bayous near Thibodaux, LA, USA and then raised in 150-300-gallon tanks in accordance with approved Institutional Animal Care and Use Committee (IACUC) protocols at Michigan State University (protocol number AUF 10/16-179-00). Gar tissues (pituitary, hypothalamus, brain minus hypothalamus, liver, heart, immature gonad, and muscle) were dissected from four sub-adults following euthanasia in 300 mg/L MS-222 (Sigma) from fish at the age of 20 months with total/standard lengths between 25.8/20.6 cm and 31.6/25.1 cm.
Total RNA was obtained from the RNAlater-preserved tissues using Isogen RNA extraction system (Nippongene, Tokyo, Japan) according to manufacturer’s protocol. One microgram of total RNA of each tissue was treated with DNase I (ThermoFisher Scientific, Waltham, MA) to remove genomic DNA, and reverse transcribed using High Capacity cDNA Reverse Transcription Kit with the random hexamer protocol. Negative controls for reverse transcription were performed without reverse transcriptase. The PCR was performed using an ABI 9700 thermal cycler (ThermoFisher Scientific, Waltham, MA) with KAPA Taq PCR Kit (KAPABIOSYSTEMS, Wilmington, MA). Primer information for each target gene was provided in Supplemental Table 1. PCR was performed in a 10 μL reaction volume for 40 cycles with the profile of 20 sec at 95 °C for denaturation, 30 s at 55 °C for annealing, and 30 s at 72 °C for extension. A final 5 min extension at 72 °C was given. PCR products were electrophoresed on 2.0% agarose gels stained by ethidium bromide and photographed using a gel-doc system.
2.3. Tissue culture
Chinese Hamster Ovary (CHO) cells (ATCC, Manassas, VA) were grown in Kaighn’s Modification of Ham’s F12K media (ATCC) supplemented with 10% fetal bovine serum, 100unit/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml normocin, and maintained in a humidified incubator with 95% air and 5% CO2 at 37 °C. When reaching 80% confluence, CHO cells were split into subcultures using 0.05% trypsin/0.53 mM EDTA.
2.4. ACTH analog peptides
Human (h) ACTH(1–24) and NDP-MSH (α-melanocyte-stimulating hormone) were purchased from Sigma-Aldrich Inc.(St. Louis, MO). For the cAMP-reporter gene assay, hACTH(1–24) was used to stimulate transfected cells at concentrations ranging from 10−13M to 10−6M. NDP-MSH was used at concentrations ranging from 10−12M to 10−6M.
2.5. cAMP-reporter gene assay
For the cAMP-reporter gene assay (Liang et al., 2011), 3.0 × 106 CHO cells were transiently co-transfected with a gMcr cDNA construct and either a gMrap1 or a gMrap2 cDNA construct, and the CRE-Luciferase cDNA construct (2 μg each) using the Amaxa Cell Line Nucleofector II system (Lonza, Portsmouth, NH) utilizing the Solution T transfection kit and program U-23 as recommended by the company. After a 10-minute post-transfection recovery period, cells were then seeded in white 96-well plate at a final density of 1 × 105 cells/well. 48 h after transfection, cells were stimulated with concentrations of hACTH(1–24) or NDP-MSH diluted in serum-free CHO media for 4 h at 37 °C. Following the incubation period, the stimulating media was removed, and the luciferase substrate reagent Bright GLO (Promega, Madison, WI) was added to the wells for a 5 min incubation period at room temperature. Luminescence was immediately measured using a Bio-Tek Synergy HTX plate reader (Winooski, VT). To determine the background levels of cAMP production, a set of transfected CHO cells were stimulated with serum free CHO media for the four hour incubation period, and the average background luminescence reading for these control wells was subtracted from the ligand-stimulated luminescence readings. The dose response curves for the stimulated cells were analyzed using the Michaelis-Menten equation to obtain EC50 values. All assays were done in triplicate. The data were plotted using the Kaleidograph software (www.synergy.com).
2.6. Cell surface ELISA assay
The Cell Surface ELISA assay was done following the protocol outline in Barney et al., 2019 with the following modifications. CHO cells were plated in a 24-well culture dish (0.75 × 105 cells/well), and grown overnight. Cells were transfected with cDNAs encoding gMcrs in combination with either gMrap1 or gMrap2 using jetPRIME transfection reagents (Polyplus transfection, Illkirch, France). After 48 hr, cells were fixed in 4% Paraformaldehyde, washed and then incubated with polyclonal V5-epitope antibody (Genetex, Irvine, CA, USA), followed by secondary HRP-conjugated goat anti-rabbit antibody. Cells were washed and treated with one-step 2,2′azinobis-3ethylbenzthiazoline-6-sulfonic acid (one-step ABTS) (Thermo Fisher Scientific, Waltham, MA, USA). An aliquot of supernatant was removed and absorbance at 405 nm was determined using a Spectramax i3 plate reader (Molecular Devices, San Jose, CA, USA). For this assay, data was analyzed using either the Student’s T-test or a one-way ANOVA with Tukey’s multi-comparison post-test using GraphPad Prism software (GraphPad Inc, La Jolla, CA, USA) and the threshold for significance was set at p < 0.05.
2.7. Statistical analysis of cAMP-Reporter gene assay
Data points are expressed as the mean ± standard error of the mean (n = 3). Statistical differences between the EC50 value of the gMcr positive control and the gMcr co-expressed with either gMrap1 or gMrap2 were evaluated using either the Student’s t-test for equal variance or one-way ANOVA followed by Tukey’s multi-comparison test using GraphPad Prism 2 (GraphPad Software Inc, La Jolla, CA, USA) for equal variance as specified in the figure legend for the dose response curve. Significance was set at P ≤ 0.05.
3. Results
3.1. RT-PCR of gar tissues
RT-PCR analysis of mrap1 and mrap2 indicated that they are ubiquitously expressed in all tissues examined (Fig. 1). In many samples, higher molecular weight bands were observed long with the expected bands, and sequencing results confirmed that they are the un-spliced transcripts of mrap1 and mrap2 (Supplementary Figs. S3–S5). In whole pituitary (i.e., Pars Distalis, Pars Intermedia, Pars Nervosa), the expression of mc1r, mc2r, mc3r, mc4r, and mc5r was detected (Fig. 1). While the mc1r was not expressed in the pituitary of two individuals, mc3r and mc4r were not expressed in one individual (Supplemental Fig. 3). RT-PCR analysis of the hypothalamus also indicated the presence of mRNAs corresponding to mc1r, mc2r, mc4r, and mc5r. However, in this tissue the mc3r band was either very weak or not detectable (Fig. 1 and Supplemental Fig. S3). Furthermore, the analysis of the rest of the brain (i.e., forebrain minus hypothalamus, midbrain, hindbrain), revealed an mRNA profile nearly identical to that of the hypothalamus.
Fig. 1.
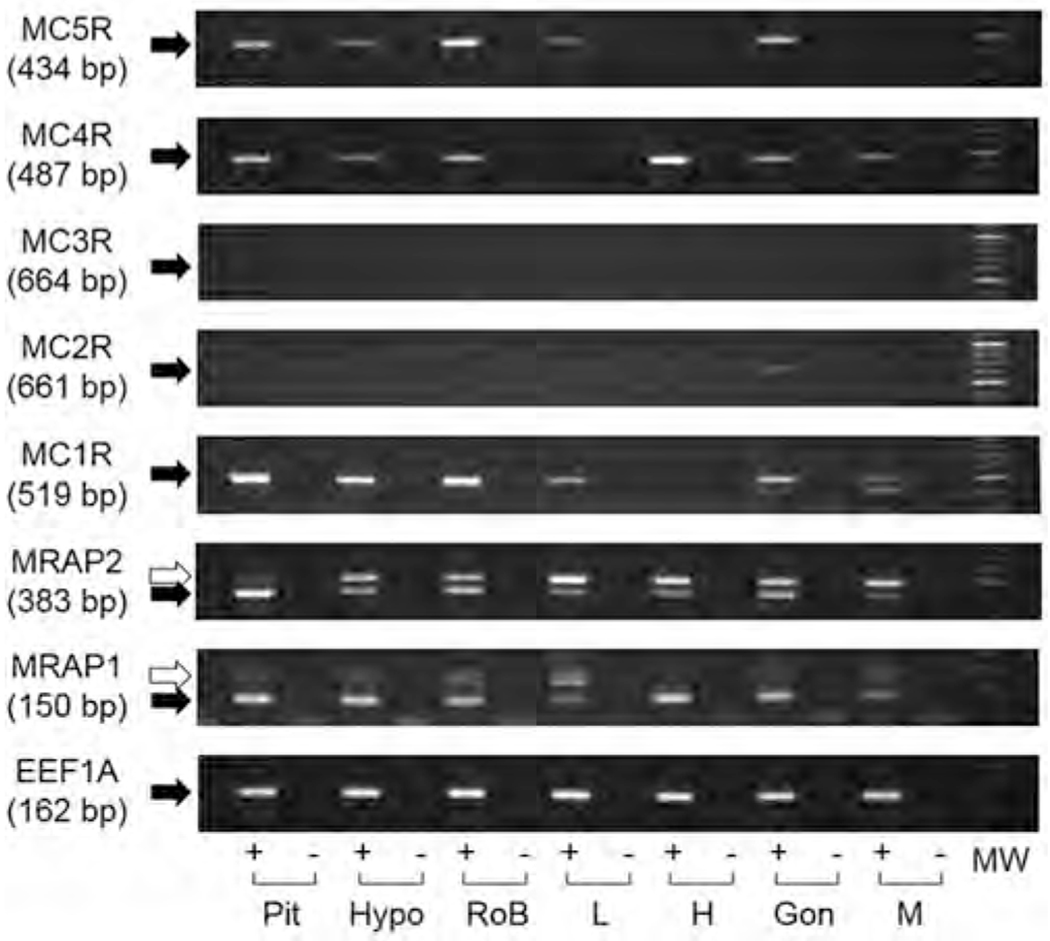
mRNA Distribution of Mcrs and Mraps various tissues of spotted gar. All the PCR amplifications were carried out for 40 cycles. Black arrows indicate the sizes of the expected PCR products, while white arrows indicate the unspliced transcripts found in Mraps. Elongation factor 1 α was amplified to indicate the integrity of cDNA. RNA sample was reverse transcribed with (+) or without (−) reverse transcriptase. Pit, pituitary; Hypo, hypothalamus; RoB, rest of brain; L, liver; H, heart; Gon, gonad; M, muscle; MW, molecular marker.
By contrast, mc1r and mc5r mRNAs were present in liver (Fig. 1), mc4r mRNA was detected in heart (Fig. 1, Supplemental Fig. S3), mc1r mRNAs were detected in gonad (Supplemental Fig. 3), and mc1r mRNAs were detected in muscle (Fig. 1 and Supplemental Fig. S3). It was unfortunate that our repeated attempts to identify the glucocorticoid-synthesizing tissue associate with the head kidney of the gar were unsuccessful (data not shown).
To verify the identity of each cDNA band, sequencing analysis was performed on the RT-PCR products, and for gMc1r, gMc2r, gMc3r, gMc4r, gMc5r, gMrap1, and gMrap2 published in the spotted gar genome database (http://uswest.ensembl.org/Lepisosteus_oculatus/Info/Index). As noted splice variants of gmrap1 and gmrap2 were detected (Supplemental Figs. 4 and 5).
3.2. Co-expression of gar Mcrs and gMraps in CHO cells -cAMP reporter gene assay
Since the RT-PCR analysis indicated tissues in which gar melanocortin receptor mRNAs were also detected with gar mrapl, and gar mrap2 mRNAs, we conducted a pharmacological study to evaluate whether co-expression of individual gar Mcrs with either gar Mrap1 or gar Mrap2 had an effect on ligand sensitivity. To this end, each gar melanocortin receptor was separately expressed in CHO cells either in the presence or absence of the respective gar Mrap paralog, and stimulated with either human (h) ACTH(1–24) or NDP-MSH. For these experiments hACTH (1–24) was used because this peptide has 96% primary sequence identity with gACTH(1–24) (gPOMC: XP_015206930; Supplemental Fig. S6). NDP-MSH (Nle4, D-Phe7-α-MSH) was used in place of α-MSH (N-acetyl-SYSMEHFRWGKPV-NH2) because this melanocortin peptide was used in previous studies on the ligand selectivity of the melanocortin receptors of a teleost (Klovins et al., 2004), and human Mc2r (Mountjoy et al., 1992).
3.3. Melanocortin-2 receptor
As predicted, expression of gMc2r alone in CHO cells did not result in activation of the receptor following stimulation with hACTH(1–24) (Fig. 2A). However, activation of the receptor was recovered when gMc2r was co-expressed with gMrap1 (EC50 = 5.2 × 10−11 M ± 1.9 × 10−11; Fig. 2A). In addition, activation did not occur when gMc2r was co-expressed with gMrap2 (Fig. 2A). Finally, as shown in Fig. 2B, when gMc2r was co-expressed with gMrap1 and stimulated with NDP-MSH, activation was observed, but only at concentrations of 10−7 M and 10−6M. The degree of stimulation by NDP-MSH at 10−6 M was roughly 40% of the stimulation observed when activation of hMc2r reached saturation at 10−9M hACTH(1–24).
Fig. 2.
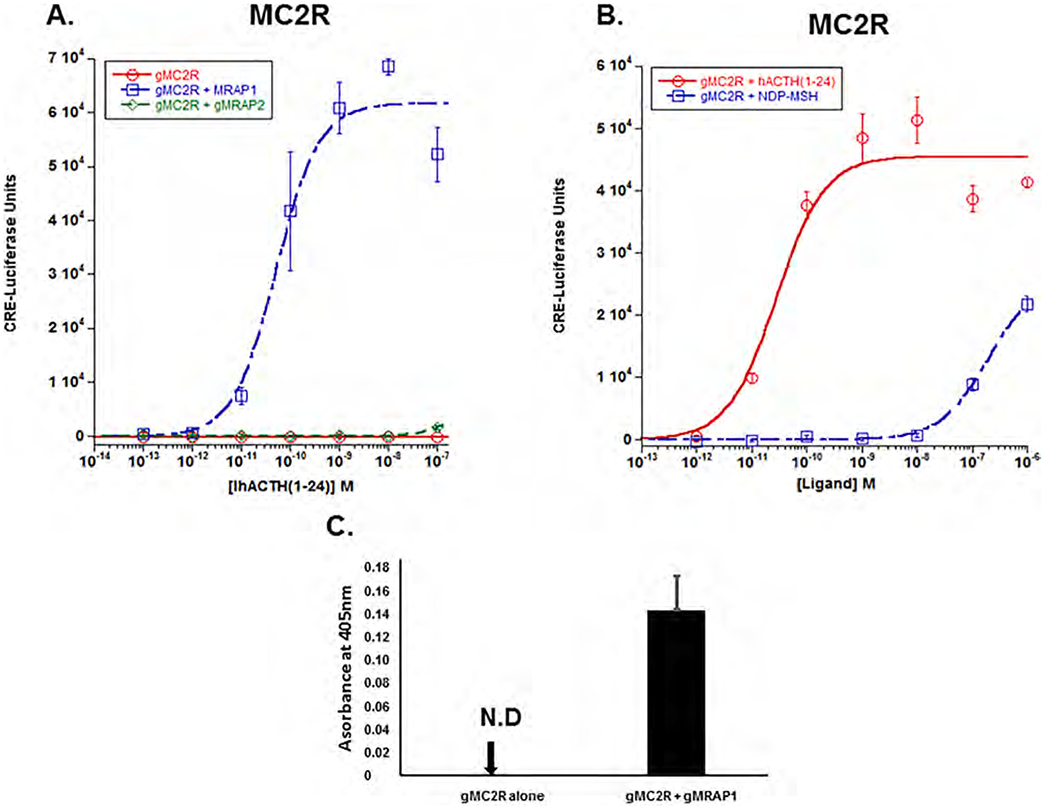
Co-expression of gMc2r with either gMrap1 or gMrap2. A. Gar Mc2r was expressed in CHO cells in the presence of absence of gMrap1 and gMrap2, and the transfected cells were stimulated with hACTH(1–24) as described in Methods. Following stimulation with hACTH(1–24), gMc2r had no activation when expressed alone, and an EC50 value of 5.6 × 10−11M ± 1.9 × 10−13 when co-expressed with gMrap1. When gMc2r was co-expressed with gMrap2 and stimulated with hACTH (1–24) there was no activation. B) Gar Mc2r was co-expressed with gMrap1, and separate sets of transfected cells were stimulated with either hACTH(1–24) or NDP-MSH. Following stimulation with hACTH(1–24), the gMc2r/gMrap1 transfected cells had an EC50 value of 4.3 × 10 11M ± 3.9 × 10 11, and when gMc2r/gMrap1 transfected cells stimulated with NDP-MSH which resulted in weak activation only at 10 7M and 10−6M NDP-MSH. C) Cell surface ELISA analysis of gMc2r and gMc2r/gMrap1. n = 3.
A cell surface ELISA assay was used to evaluate the role that gMrap1 plays in the trafficking of gMc2r to the plasma membrane. As shown in Fig. 2C, when expressed alone, gMc2r could not be detected on the plasma membrane. However, when co-expressed with gMrap1, the receptor clearly trafficked to the plasma membrane.
3.4. Melanocortin 1 receptor
As shown in Fig. 3A, stimulation of gMc1r with hACTH(1–24) resulted in an EC50 value of 1.4 × 10−10M ± 2.5 × 10−11. When the receptor was co-expressed with gMrap1, there was no positive or negative effect on the sensitivity of the receptor for stimulation by hACTH (1–24) (EC50 value = 1.0 × 10−10M ± 2.3 × 10−11; p = 0.90). However, co-expression with gMrap1 resulted in a decrease in the Vmax value when stimulating with hACTH(1–24) (p < 0.001; T-test). In addition, when gMc1r was expressed alone and stimulated with NDP-MSH (Fig. 2A), the EC50 value was 4.2 × 10−12M ± 1.6 × 10−12; whereas, co-expression with gMrap1 resulted in an EC50 value of 6.0 × 10−12M ± 1.7 × 10−12 (p = 0.92). Clearly, co-expression with gMrap1 had no effect on the sensitivity of gMc1r for stimulation by NDP-MSH, however, there was a decrease in the Vmax value for the gMc1r/gMrap1 transfected cells (p = 0.02; T-test), when the receptor was stimulated with NDP-MSH. Finally, a comparison of the EC50 values for gMc1r expressed alone and stimulated with either hACTH (1–24) or NDP-MSH indicated that this receptor is much more sensitive to stimulation by NDP-MSH than to stimulation by hACTH(1–24) (p < 0.001).
Fig. 3.
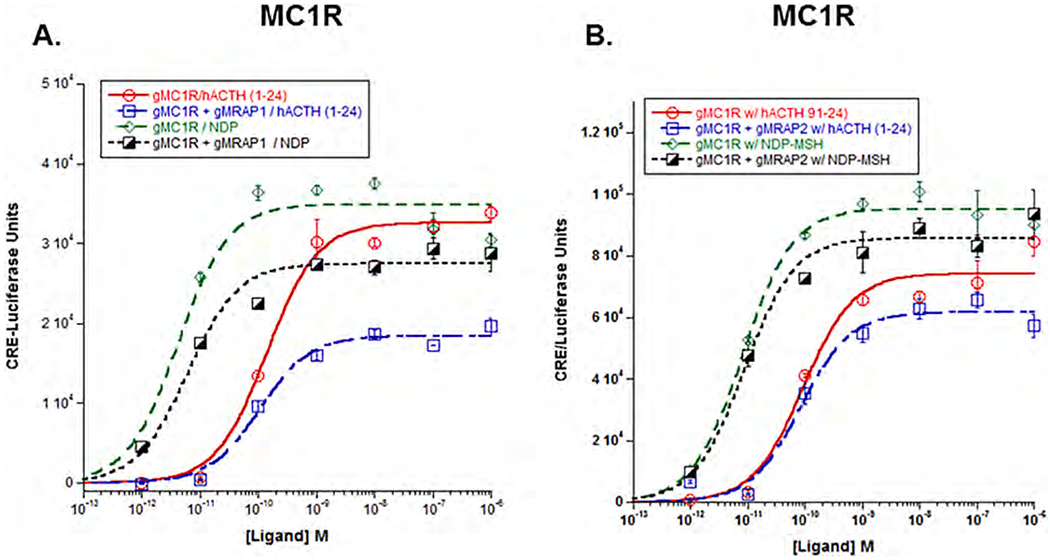
Co-expression of gMc1r with either gMrap1 or gMrap2. A) Gar Mclr was expressed in CHO cells in the presence of absence of gMrap1, and the transfected cells were stimulated with either hACTH(1–24) or NDP-MSH as described in METHODS. The EC50 value for each dose response curve is presented in Results Section 3b. Statistical analysis was done using one-way ANOVA as described in METHODS. B) Gar Mclr was expressed in the presence of absence of gMrap2, and the transfected cells were stimulated with hACTH(1–24) or NDP-MSH as described in METHODS. Statistical analysis was done using one-way ANOVA as described in METHODS, n = 3.
Similar experiments were done to evaluate the response of gMc1r co-expressed with gMrap2 (Fig. 3B). In this experiment, when gMc1r was expressed alone and stimulated with hACTH(1–24), the EC50 value was 9.1 × 10−11M ± 3.0 × 10−11 as compared to an EC50 value of 8.4 × 10−11M ± 2.4 × 10−11 when the receptor was co-expressed with gMrap2 (p = 0.12). In addition, when gMc1r was co-expressed with gMRAP2 and stimulated with NDP-MSH, the EC50 value was 8.6 × 10−12M ± 2.0 × 10−12; whereas, when gMc1r was expressed alone and stimulated with NDP-MSH the EC50 value was 8.1 × 10−12M ± 1.3 × 10−12 (p = 0.28). Apparently, co-expression with gMrap2 had no effect, in either a positive or negative manner, on the sensitivity of gMc1r for stimulation by either hACTH(1–24) or NDP-MSH. In addition, co-expression with gMrap2 had no effect on the Vmax value for hACTH(1–24) stimulated cells (P = 0.43; T-test), or the NDP-MSH stimulated cells (p = 0.42; T-test).
3.5. Melanocortln 3 receptor
The results for gMc3r (Fig. 4A&B) were similar, but not identical to the results observed for gMc1r. Co-expression with gMrap1 had no effect either positive or negative on sensitivity to stimulation by hACTH (1–24); the EC50 value for the receptor expressed alone was of 2.2 × 10−10M ± 4.6 × 10−11, and when co-expressed with gMrap1 the EC50 value was 1.7 × 10−10M ± 2.5 × 10−11 (p = 0.49). Co-expression with gMrap1 also had no effect on the Vmax value for the transfected cells. Stimulation of the receptor expressed alone with NDP-MSH resulted in an EC50 value of 2.6 × 10−11 ± 5.8 × 10−12, and when the receptor was co-expressed with gMrap1 and stimulated with NDP-MSH, the EC50 value was 2.5 × 10−11M ± 4.9 × 10−12 (p = 0.98). Once again, co-expression with gMrap1 had no effect on the Vmax value for the transfected cells. Finally, in terms of ligand efficacy, gMc3r has a higher sensitivity for stimulation by NDP-MSH than by stimulation with hACTH(1–24) (p = 0.03).
Fig. 4.
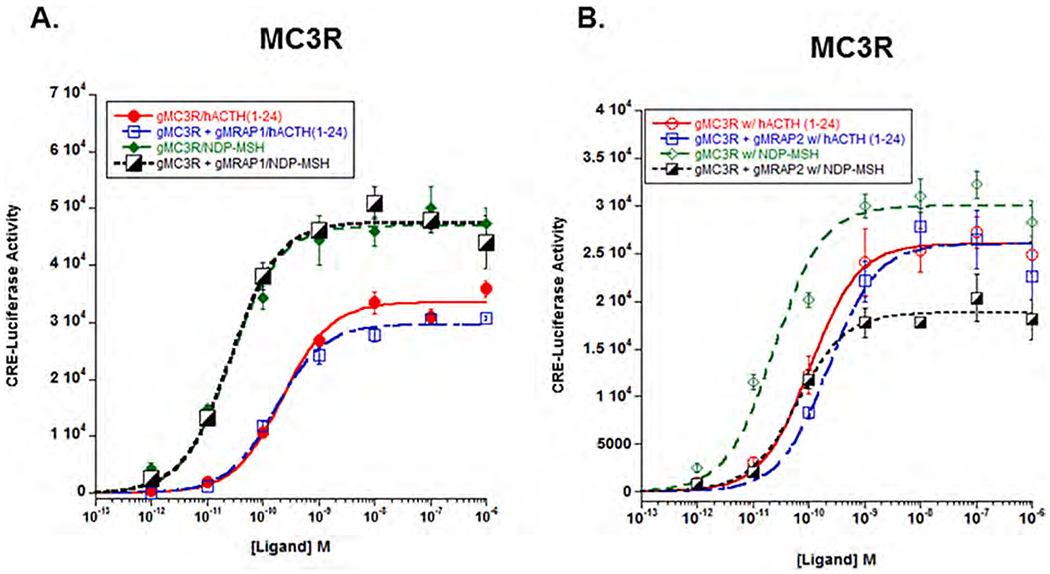
Co-expression of gMc3r with either gMrap1 or gMrap2. A) Gar Mc3r was expressed in CHO cells in the presence of absence of gMrap1, and the transfected cells were stimulated with hACTH(1–24) or NDP-MSH as described in Methods. The EC50 value for each dose response curve is presented in Results Section 3c. Statistical analysis was done using one-way ANOVA as described in METHODS. Following stimulation with hACTH(1–24) for Mc3r alone vs. gMc3r/gMrap1, p = 0.49. Following stimulation with NDP-MSH for Mc3r alone vs. gMc3r/gMrap1, p = 0.98. B) Gar Mc3r was expressed in CHO cells in the presence or absence of gMrap2, and the transfected cells were stimulated with hACTH(1–24) or NDP-MSH. Statistical analysis was done using one-way ANOVA as described in METHODS. Following stimulation with hACTH(1–24), gMc3r expressed alone had an EC50 value of 1.0 × 10−10 ± 1.5 e−11, and when the receptor was co-expressed with gMrap2 the EC50 value was 2.2 × 10−10 ± 1.2 × 10−10 (p = 0.98). When gMc3r was stimulated with NDP-MSH and expressed alone, the EC50 value was 2.4 × 10−11 ± 8.6 × 10−12, and when the receptor was co-expressed with gMrap2 the EC50 value was 6.3 × 10−11 ± 1.3 × 10−11 (p = 0.98). n = 3.
Co-expression with gMrap2 also had no effect either positive or negative on the sensitivity of gMc3r for stimulation by either hACTH (1–24) or NDP-MSH. The EC50 values and the result of the one-way ANOVA analysis appear in the legend for Fig. 4B. In addition, co-expression with gMRAP2 had no effect on the Vmax value for the transfected cells stimulated with hACTH(1–24) (p – 0.99; T-test); however, there was a decrease in the Vmax value for the transfected cells stimulated with NDP-MSH (p > 0.001; T-test).
3.6. Melanocortin 4 receptor
Co-expression with gMrap1 had no statistical effect on the sensitivity of gMc4r to stimulation by either hACTH(1–24) or NDP-MSH (Fig. 5A). The EC50 values and the results of the statistical analyses are presented in the legend for Fig. 4. In addition, co-expression with gMrap1 had no effect on the Vmax value for the transfected cells stimulated with either hACTH(1–24) or NDP-MSH. Finally, gMc4r can be activated with equal efficacy by either hACTH(1–24) or NDP-MSH (p = 0.11).
Fig. 5.
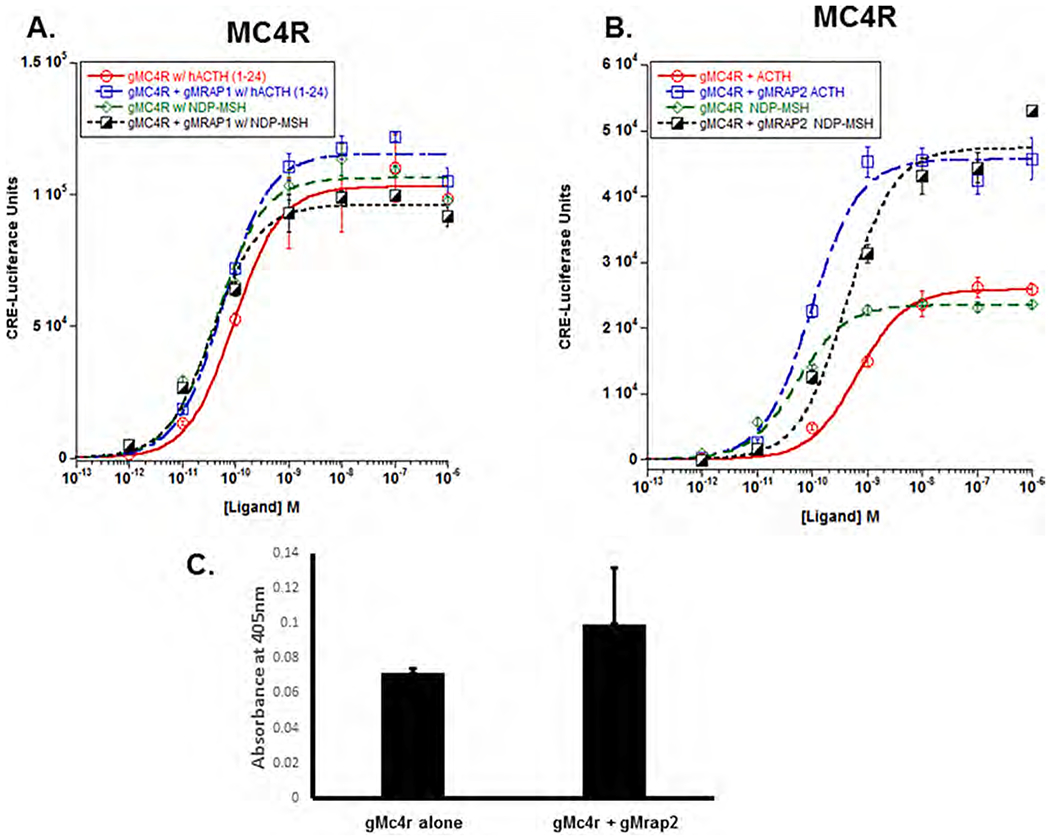
Co-expression of gMc4r with either gMrap1 or gMrap2. A) Gar MC4R was expressed in the presence of absence of gMRAP1, and the transfected cells were stimulated with hACTH(1–24) or NDP-MSH as described in Methods. Statistical analysis was done using one-way ANOVA as described in METHODS. Following stimulation with hACTH(1–24), gMc4r had an EC50 value of 9.3 × 10 11M ± 1.5 × 10− when expressed alone, and an EC50 value of 5.7 × 10−11M ± 1.2 × 10−11 when co-expressed with gMrap1 (p = 0.49). When gMc4r was stimulated with NDP-MSH, the EC50 value was 3.8 × 10−11M ± 8.0 × 10−12 when expressed alone, and the EC50 value was 2.8 × 10e−11M ± 2.6 × 10−11 when co-expressed with gMrap1 (p = 0.98). B) Gar Mc4r was expressed in the presence or absence of gMrap2, and the transfected cells were stimulated with either hACTH(1–24) or NDP-MSH. Statistical analysis was done using one-way ANOVA as described in METHODS. The EC50 values and the results of the statistical analyses are presented in Results section 3.d. n = 3. C) Cell surface ELISA analysis of gMc4r and gMc4r/gMrap2. n = 3.
However, the response of gMc4r to co-expression with gMrap2 was quite distinct relative to the other gar Mcrs (Fig. 5B). When gMc4r was expressed alone and stimulated with hACTH(1–24), the EC50 value was 6.6 × 10−10M ± 9.7 × 10−10. However, co-expression with gMrap2 resulted in an EC50 value of 9.6 × 10−11M + 1.9 × 10−11. The gMc2r/gMrap2 enhanced the sensitivity of the receptor for activation by hACTH(1–24) (p < 0.001; One-Way ANOVA), and the Vmax for the gMc4r/gMrap2 dose response curve was statistically higher than the Vmax for the dose response curve when gMc4r was expressed alone (p < 0.001; T-Test). In addition, when gMc4r was expressed alone and stimulated with NDP-MSH, the EC50 value was 5.3 × 10−11M ± 1.0 × 10−11. Co-expression of the receptor with gMrap2 resulted in a lowering of sensitivity to stimulation by NDP-MSH (EC50 value of 4.2 × 10−10M ± 1.3 × 10−10; p = 0.003; One-Way ANOVA), yet the Vmax for the gMc4r/gMrap2 dose response curve was statistically higher than the Vmax for the dose response curve when gMc4r was expressed alone (p < 0.001; T-test), and comparable in magnitude to the Vmax value for the gMc4r/gMrap2 cells stimulated hACTH(1–24) (Fig. 4B). When the co-expression of gMc4r and gMrap2 was analyzed using a cell surface ELISA protocol (Fig. 5C), there was no apparent effect on the trafficking of gMc4r in either a positive or negative manner.
3.7. Melanocortin 5 receptor
For the analysis of gMc5r, we elected to look first at the effect of coexpression of the receptor with either gMrap1 or gMrap2 on sensitivity to stimulation with hACTH(1–24) (Fig. 6A). In this experiment, when gMc5r was expressed alone, the EC50 value was 2.5 × 10−10M ± 2.0 × 10−11. Co-expression with gMrap1 resulted in a negligible shift in sensitivity (EC50 value = 1.1 × 10−10M ± 2.0 × 10−11; p = 0.5), and no change in the Vmax value (p = 0.44; T-test). Co-expression with gMrap2 also did not affect sensitivity to stimulation with hACTH(1–24) (EC50 value = 1.6 × 10−10M ± 1.9 × 10−11; p = 0.5), however, there was an increase in the Vmax value (p = 0.001; T-test).
Fig. 6.
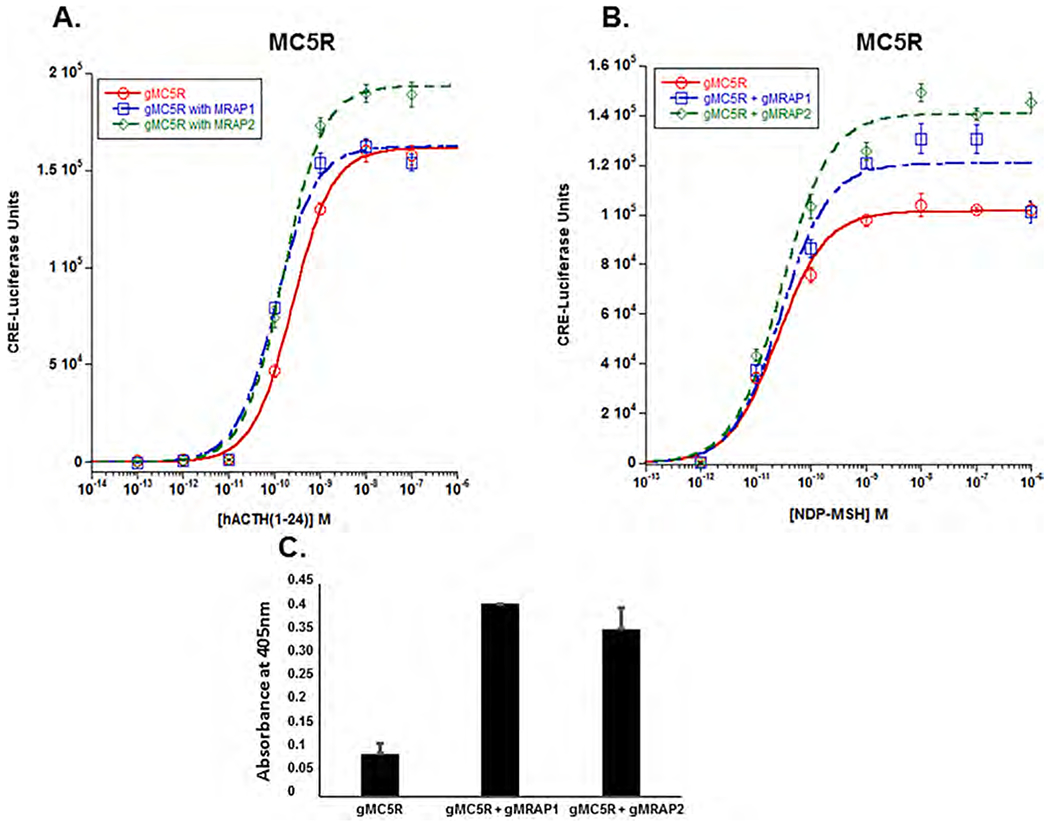
Co-expression of gMc5r with either gMrap1 or gMrap2. A) Gar Mc5r was expressed alone or in the presence of either gMrap1 or gMrap 2, and each set of transfected cells was stimulated with hACTH(1–24). The EC50 value for each dose response curve, and the results of the statistical analysis of the EC50 values (one-way ANOVA) are presented in Results Section 3e. B) Gar Mc5r was expressed alone or co-expressed with either gMrap1 or gMrap2 and the sets of transfected cells were stimulated with NDP-MSH. The EC50 values for each dose response curve was analyzed by one-way ANOVA as described in Methods. The EC50 values and the results of the statistical analyses are presented in Results Section 3e. n = 3. C) Cell surface ELISA analysis of gMc5r, gMc5r + gMrap1, and gMc5r + gMrap2. n = 3.
When gMc5r was expressed alone and stimulated with NDP-MSH (Fig. 5B), the EC50 value was 2.5 × 10−11M ± 3.9 × 10−12. This value was not statistically different from stimulation with hACTH (1–24) (Fig. 6B), (p = 0.07; T-test). While co-expression of gMc5r with gMrap1 had no effect on sensitivity to stimulation by NDP-MSH (EC50 value = 2.8 × 10−11M ± 1.2 × 10−11; p = 0.64), there was an increase in Vmax (p = 0.01; T-test). In a similar manner, co-expression with gMrap2 did not increase sensitivity to stimulation by NDP-MSH (EC50 value = 2.9 × 10−11 M ± 6.9 × 10−12), however, there was an increase in Vmax (p = 0.007; T-test).
To determine whether interaction between gMc5r and gMrap1 or gMc5r and gMrap2 has an effect on trafficking, a cell surface ELISA analysis was done (Fig. 6C). gMc5r will move to plasma membrane when expressed alone in CHO cells. However, there was a statistically significant increase in trafficking when the receptor was co-expressed with gMrap1 (p = 0.009; One-way ANOVA). However, when co-expression with gMrap2 appeared to increase the trafficking of gMc5r to the plasma membrane, yet One-way ANOVA analysis indicated that the apparent increase in trafficking was not statistically significant (p = 0.09).
4. Discussion
The rationale for selecting the spotted gar for this study on melanocortin receptor/Mrap interactions was based on the phylogenetic position of the gar lineage relative to the teleost and the tetrapod lineages (Carroll, 1988). Since the number of melanocortin receptors in this gene family was the result of at least two genome duplication events and one local gene duplication event that occurred prior to the diversification of the ancestral gnathostomes (Schiöth et al., 2003; Vastermark and Schiöth, 2011), the eventual dichotomy of the ancestral gnathostomes into ancestral cartilaginous fishes and ancestral bony fishes has led to parallel yet distinct trajectories for at least Mc2r orthologs and Mc5r orthologs. This is a topic that has been recently addressed from the perspective of the cartilaginous fishes (Dores et al., 2018; Barney et al., 2019). Hence, this study analyzed the pharmacological properties of melanocortin receptors from a bony vertebrate perspective. Within the bony vertebrate radiation, the spotted gar lineage (order Lepisosteiformes, subclass Neopterygii) is one of the older lineages in which the genome has been sequenced, and the gar lineage is at a position that precedes the emergence of the teleost and tetrapod lineages. Hence, studies on this species may reveal traits that are ancestral for both teleosts and tetrapods. This type of relationship has been shown for genes associated with immunity, mineralization, and development (Braasch et al., 2016). Against this background, one of the objectives of this study was to see if gar melanocortin receptor networks have retained features that can be considered ancestral for both teleosts and tetrapods with a particular focus on the interactions between gar melanocortin receptors and the gar accessory proteins, Mrap1 and Mrap2.
To this end, an RT-PCR analysis of selected tissues was done to identify tissues that co-express melanocortin receptor mRNAs and mrap mRNAs. Previous studies on elephant shark interrenal tissue and chicken adrenal tissue indicated that multiple melanocortin receptors are present in these tissues along with Mrap1. In addition, pharmacological studies indicated that co-expression of a species specific Mrap1 with the corresponding Mc2r and Mc5r orthologs of these species enhanced sensitivity to stimulation by ACTH (Thomas et al., 2018; Dores et al., 2018; Barney et al., 2019). We attempted to analyze the glucocorticoid tissues (head kidney) of the gar, but were not successful in obtaining the correct tissues. However, an analysis of the spotted gar pituitary and hypothalamus did reveal the presence of multiple melanocortm receptor mRNAs, as well as mRNAs for mrapl and mrap2 in the respective tissues (Fig. 1). These observations provided a rationale for the pharmacological studies on gar Mcr/Mrap interactions presented in Figs. 2–6. In addition, the detection of multiple mcr and mrap mRNAs in the pituitary and hypothalamus of the spotted gar raised some interesting issues with respect to the extent of melanocortin signaling networks in this species.
RT-PCR analysis of the spotted gar pituitary (Fig. 1) detected mc1r, mc2r, mc4r, and mc5r mRNAs as well as mRNAs for mrap1 and mrap2. Since the analysis was done on whole pituitary (i.e., Pars Distalis, Pars Intermedia, Pars Nervosa) it is not possible to determine which pituitary cell types are making these mRNAs. For example, melanocortin receptors may be playing an autocrine/paracrine role in spotted gar corticotropes (Pars Distalis; ACTH synthesizing cells) or melanotropes (Pars Intermedia; MSH synthesizing cells). A recent study on the pituitary of the elephant shark yielded similar results (Barney et al, 2019). Collectively, these observations raise the possibility of a melanocortin peptide hypophysiotropic network for both these species. This possibility is not unprecedented. In a recent study on the hypothalamus/pituitary network of the zebrafish, melanocortin peptides immunoreactivity was detected in neurosecretory cell terminals located in close proximity to Pars Distalis cells expressing zebrafish MC4R (Zhang et al., 2012). Thus, the presence of a melanocortin hypophysiotropic system may be more widespread in vertebrates than previously suspected.
The RT-PCR analysis of the spotted gar hypothalamus also detected mRNAs for mc1r; mc2r, mc4r, and mc5r as well as mrap1 and mrap2 (Fig. 1). Multiple melanocortin receptor mRNAs (i.e., mc3r, mc4r, and mc5r) and mrap2 mRNA have also been detected in the hypothalamus of the elephant shark (Barney et al., 2019), and several species of teleosts (Cerdá-Reverter et al., 2011). Among tetrapods, multiple melanocortin receptor mRNAs (i.e., mc1r, mc3r, mc4r) and mrap2 mRNA have been detected in an avian hypothalamus (Thomas et al., 2018), and mc3r, mc4r, and mrap2 mRNA have been detected in adult mammalian brain (Mountjoy, 2010). Interestingly, during stages of mammalian brain development other melanocortin receptors, such as MC2R have also been detected in the hypothalamus (Mountjoy, 2010). In zebrafish and mouse hypothalamus, Mc4r and Mrap2 are playing a role in feeding behavior (Asai et al., 2013; Sebag et al., 2013; Zhang and Tao, 2017), whether the melanocortin network in the hypothalamus of the spotted gar plays a role in feeding behavior has yet to be experimentally determined. In addition, the physiological role of Mc5r in gar liver cells, Mcr4 in heart cells, and Mclr in muscles is also unknown (Fig. 1). Finally, a recurrent feature of the RT-PCR analysis was the detection of mrap1 and mrap2 mRNA in every tissue analyzed. In mammals, mrap1 mRNA and mrap2 mRNA tend to have more restrictive expression patterns. In the hypothalamus, mrap2 mRNA is present but not mrap1 mRNA is not detected (Chan et al., 2009). Conversely, in the adrenal cortex mrap1 mRNA is, but not mrap2 mRNA is not detected (Metherell et al., 2005; Mountjoy, 2010). That said, in the tissues of the teleost, Oncorrhynchus mykis (rainbow trout) a similar expression pattern to the spotted gar for mrap1 and mrap2 mRNAs has been observed (Mathilakath M. Vijayan, University of Calgary, personal communication).
The pharmacological experiments presented in this study set out to address the predictions made in the INTRODUCTION with respect to the interaction between Mc2r and Mrap1, Mc5r and Mrap1, and Mc4r and Mrap2, respectively. As shown in Fig. 2A, gMc2r has an obligatory requirement for co-expression with gMrap1 in order for the receptor to be activated by hACTH(1–24). This interaction includes the trafficking of gMc2r to the plasma membrane (Fig. 2C). In addition, gMrap2 did not facilitate activation of gMc2r. These same properties have been observed for Mc2r/Mrap1 interactions of teleosts (Agulleiro et al., 2010; Liang et al., 2015; Zhang and Tao, 2017) and tetrapods (Sebag and Hinkle, 2007; Davis et al., 2013; Thomas et al., 2018). Hence, it would be reasonable to assume that these properties for Mc2r/Mrap1 interaction were established relatively early in the radiation of the ancestral bony vertebrates. Hence, it would be reasonable to predict that when the glucocorticoid tissue of the spotted gar is identified, this species will have a Hypothalamus/Pituitary/Interrenal axis that functions in the same manner as the HPI/A axes of other bony vertebrates.
A surprising observation was that gMC2R/gMrap1 interaction could be activated to a limited degree by NDP-MSH at concentrations of 10−7M and 10−6M. While the stimulation by NDP-MSH is not physiologically relevant, the Mc2r ortholog of the teleost, Oncorhynchus mykis (rainbow trout) was not activated under the same experimental conditions (Supplemental Fig. S7), and in our hands, incubation of hMc2r/mMrap1 with NDP-MSH also did not evoke a response (Liang et al., 2011). Perhaps stimulation of Mc2r orthologs by α-MSH-related peptides is an ancestral gnathostome trait. Studies on cartilaginous fishes indicate that these Mc2r orthologs when co-expressed with a cartilaginous fish Mrap1 do respond to stimulation by a-MSH-related peptides at concentrations in a physiologically relevant range (Reinick et al., 2012; Dores et al., 2018;, Barney et al., 2019).
While the interaction between bony vertebrate Mc2r orthologs and their corresponding Mrap1 orthologs is fairly uniform, the same cannot be said for the interaction between Mc5r orthologs and Mrap1 orthologs. As shown in Fig. 6A and 6B, co-expression of gMc5r with gMrap1 gMrap2 had no effect, either positive or negative, on the sensitivity of the receptor to stimulation by either hACTH(1–24) or NDP-MSH. However Cell Surface ELISA analysis, indicated that co-expression with gMrap1 increased trafficking of gMc5r to the plasma membrane (Fig. 6C). These results are in sharp contrast to what has been observed for hMc5r. Co-expression of this receptor with mouse Mrap1 resulted in a decrease in trafficking of the receptor to the plasma membrane (Sebag and Hinkle, 2009). In addition, co-expression of Gallus gallus (chicken; c) Mc5r with cMrap1 significantly increased the sensitivity of gMc5r to stimulation by hACTH(1–24) (Thomas et al., 2018). Currently, data is not available for the effects of co-expression of a teleost Mc5r ortholog with a teleost Mrap1 on ligand sensitivity or trafficking. In any event, a comparison of the amino acid sequences of gar, mouse, and chicken Mrap1 orthologs (Fig. 7) does not reveal distinctive amino acid motifs in the Mrap1 orthologs that might explain these observations, and a uniform mechanism to explain Mc5r/Mrap1 interaction may not exist for the bony fishes and tetrapods.
Fig. 7.

Amino Acid Alignment of Mrap1 and Mrap2 Sequences. The amino acid sequences of gar Mrap1 (gMrap1), mouse Mrap1 (mMrap1; NP 084120), chicken Mrap1 (cMrap1; NP 416703.1), gar Mrap2 (gMrap2) chicken Mrap2 (cMrap2; NP 001346884.1), and human Mrap2 (hMrap2; NP 001333473.1) were aligned. [A] is the location of the activation motif in Mrap1 orthologs. [B] is the location of the reverse topology motif in Mrap1 and Mrap2 paralogs. [C] is the location of the transmembrane domain in Mrap1 and Mrap2 paralogs. Positions that are underlined are identical in either the Mrap1 orthologs or the Mrap2 orthologs. (*) indicates a putative N-linked glycosylation site. Note that the transmembrane domain for the three Mrap2 orthologs have identical amino acid sequences.
Co-expression of gMc5r with gMrap2 also did not affect ligand sensitivity (Fig. 6A & B); however, this interaction did enhance the trafficking of gMc5r to the plasma membrane (Fig. 6C). The same outcome was not seen in mammals. For example, co-expression of human (h) Mc5R with hMrap2 decreased trafficking of the receptor to the plasma membrane (Chan et al., 2009). Since the transmembrane domain of Mrap2 is responsible for trafficking, and these domains are identical in gMrap2 and hMrap2 (Fig. 7), it is not clear why the two accessory proteins produce different effects with their respective Mc5r ortholog.
Finally, orthologs of Mrap2 have been shown to influence the ligand selectivity and trafficking of Mc4r in a teleost (zebrafish; Agulleiro et al., 2013; Sebag et al., 2013; Zhang and Tao, 2017) and a mammal (mouse; Asai et al., 2013), and this interaction in the hypothalamus affects feeding behavior for both species. Gar mc4r and gmrap2 mRNAs were both detected in the hypothalamus of the spotted gar (Fig. 1). Coexpression of gMc4r with gMrap2 statistically increased sensitivity to stimulation by both hACTH(1–24) and NDP-MSH (Fig. 5B), and the amplitude of stimulation (Vmax) also increased for both ligands (Fig. 5B). In this regard, the ability of Mrap2 to influence the functionality of Mc4r orthologs appears to be a feature common to bony vertebrates. For gMc4r, the increase in stimulation does not appear to be due to more efficient trafficking of the receptor to the plasma membrane (Fig. 5C).
In summary, gar melanocortin receptors display properties with respect to ligand selectivity that are nearly identical to the properties of melanocortin receptors in teleosts and tetrapods. In this regard, the strict requirement for interaction with Mrap1 makes this receptor very selective for activation by ACTH, and is a common feature of bony vertebrate melanocortin receptors. In addition, the need for interaction with Mrap1 for the trafficking of Mc2r orthologs to the plasma membrane is also a feature common to all bony vertebrate Mc2r orthologs that have been studied. Finally, the interaction between gMc4r and gMrap2 also appears to be a feature common to bony vertebrates. Thus, at least for the Mc2r/Mrap1 mediated network and the Mc4r/Mrap2 mediated network, the studies on the spotted gar did reveal common evolutionary features in both teleosts and tetrapods.
Supplementary Material
Acknowledgements
This research was partially supported by the Long Research Fund (R.M.D.; University of Denver), and by JSPS Grant-in-Aid 16K18575 awarded to MW. We wish to thank Prof. Susumu Hydodo (University of Tokyo) for his comments on this project. Gar research in the Braasch Lab is supported by NIH R01OD011116; we thank Brett Racicot for gar husbandry and Andrew W. Thompson for assistance with gar dissections.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://doi.org/10.1016/j.ygcen.2019.113215.
References
- Agulleiro MJ, Roy S, Sanchez E, Puchol S, Gallo-Payet N, Cerda-Reverter JM, 2010. Role of melanocortin receptor accessory proteins in the function of zebrafish melanocortin receptor type 2. Mol. Cell. Endocrinol 320, 145–152. [DOI] [PubMed] [Google Scholar]
- Agulleiro MA, Sánchez E, Leal E, Cortés R, Fernández-Durán B, Guillot R, Gallo-Payet N, Dores RM, Davis P, Cerdá-Reverter JM, 2013. Molecular characterization and functional regulation of melanocortin 2 receptor (MC2R) in the Sea Bass. A putative role in the adaptation to stress. PLoS One 27;8(5):e65450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya Y, Takahashi A, Suzuki N, Sasayama Y, Kawauchi H, 2000. Molecular cloning of proopiomelanocortin cDNA from an Elasmobranch, Dasyatis akajei . Gen. Comp. Endocrinol 118, 105–112. [DOI] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH, 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188, 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai M, Ramachandrappa S, Joachim M, Shen Y, Zhang R, Nuthalapati N, Ramanathan V, Strochlic DE, Ferket P, Linhart K, Ho C, Novoselova TV, Garg S, Ridderstrale M, Marcus C, Hirschhorn JN, Keogh JM, O’Rahilly S, Chan LF, Clark AJ, Farooqi S, Majzoub JA, 2013. Loss of function of the melanocortin 2 receptor accessory protein 2 is associated with mammalian obesity. Science 34, 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barney Emily, Dores Michael R., McAvoy Danielle, Davis Perry, Racareanu Rona Cristina, Iki Ayuko, Hyodo Susumu, Dores Robert M., 2019. Elephant shark melanocortin receptors: novel interactions with MRAP1 and implication for the HPI axis. Gen. Comp. Endocrinol 272, 42–51. [DOI] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi B, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Canestro C, Sydes J, Beaudry FEG, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SMJ, Aken B, Yandell M, Schneider I , Yoder JA, Volff J-N, Meyer A, Amemiya CT, Venkatesh B, Holland PWH, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alfoldi J, Lindblad-Toh K, Postlethwait JH, 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet 48, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RI, 1988. In: Vertebrate Paleontology and Evolution. W.H. Freeman and Company, New York, pp. 102–109. [Google Scholar]
- Cerda-Reverter JM, Josep Agulleiro M, Guillot R, Sanchez E, Ceinos R, Rotllant J, 2010. Fish melanocortin system. Eur. J. Pharm 600, 53–60. [DOI] [PubMed] [Google Scholar]
- Chan LF, Webb TR, Chung T-T, Meimaridou E, Cooray SN, Guasti L, Chappie JP, Egertova M, Elphick MR, Cheethamc ME, Metherell LA, Clark AJL, 2009. MRAP and MRAP2 are bidirectional regulators of the melanocortin receptor family. Proc. Natl. Acad. Sci. U.S.A 106, 6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepurny OG, Holz GG, 2007. A novel cyclic adenosine monophosphate responsive luciferase reporter incorporating a nonpalindromic cyclic adenosine monophosphate response element provides optimal performance for use in G protein coupled receptor drug discovery efforts. J. Biomol. Screening 12, 740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone RD, 2006. Studies on the physiological functions of the melanocortin system. Endocr. Rev 27, 736–749. [DOI] [PubMed] [Google Scholar]
- Davis P, Franquemont S, Liang L, Angleson JK, Dores RM, 2013. Review: volution of the melanocortin-2 receptor in tetrapods: studies on Xenopus tropicalis MC2R and Anolis carolinensis MC2R. Gen. Comp. Endocrinol 188, 75–84. [DOI] [PubMed] [Google Scholar]
- Dores RM, Liang L, Davis P, Thomas AL, Petko B, 2016. Melanocortin receptors: evolution of ligand selectivity for melanocortin peptides. J. Mol. Endocrinol 56, T1–T16. [DOI] [PubMed] [Google Scholar]
- Dores Robert M., Scuba-Gray Michelle, McNally Bridgette, Davis Perry, Takahashi Akiyoshi, 2018. Evaluating the interactions between red stingray (Dasyatis akajei) melanocortin receptors and elephant shark (Callorhinchus milii) MRAP1 and MRAP2 following stimulation with either stingray ACTH(l-24) or stingray Des-Acetyl-aMSH: a pharmacological study in Chinese Hamster Ovary Cells. Gen. Comp. Endocrinol 265, 133–140. [DOI] [PubMed] [Google Scholar]
- Josep Agulleiro M, Cortes R, Fernandez-Duran B, Navarro S, Guillot R, Meimaridon E, Clark AJ, Cerda-Reverter JM, 2013. Melanocortin 4 receptor becomes an ACTH receptor by coexpression of melanocortin receptor accessory protein 2. Mol. Endocrinol 27, 1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins J, Haitina T, Fridmanis D, Kilianova Z, Kapa K, Fredriksson R, Gallo-Payet N, Schioth HB, 2004. The melanocortin system in Fugu: determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs. Mol. Biol. Evol 21, 563–579. [DOI] [PubMed] [Google Scholar]
- Liang L, Sebag JA, Eagelston L, Serasinghe MN, Veo K, Reinick C, Angleson J, Hinkle PM, Dores RM, 2011. Functional Expression of frog and rainbow trout melanocortin 2 receptors using heterologous MRAPls. Gen. Comp. Endocrinol 174, 5–14. [DOI] [PubMed] [Google Scholar]
- Liang L, Schmid K, Sandhu N, Angleson JK, Vijayan MM, Dores RM, 2015. Structure/function studies on the activation of the rainbow trout melanocortin-2 receptor. Gen. Comp. Endocrinol 210, 145–151. [DOI] [PubMed] [Google Scholar]
- Metherell LA, Chappie JP, Cooray S, David A, Becker C, Ruschendorf F, Naville D, Begeot M, Khoo B, Nurnberg P, Huebner A, Cheethan ME, Clark AJ, 2005. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nat. Genet 37, 160–170. [DOI] [PubMed] [Google Scholar]
- Metz JR, Peters JJ, Flik G, 2006. Molecular biology and physiology of the melanocortin system in fish: a review. Gen. Comp. Endocrinol 148, 150–162. [DOI] [PubMed] [Google Scholar]
- Mountjoy KG, 2010. Distribution and function of melanocortin receptors within the brain In: Catania A (Ed.), Melanocortins: Multiple Actions and Therapeutic Potential. Advances in Experimental Medicine and Biology. Springer, New York, NY, pp. 48–58. [DOI] [PubMed] [Google Scholar]
- Mountjoy KG, Robbins LS, Mortrud MT, Cone RD, 1992. The cloning of a family of genes that encode the melanocortin receptors. Science 257, 1248–1251. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Inoue A, Kita T, Nakamura M, Chung ACY, Cohen SN, Numa S, 1979. Nucleotide sequence of cloned cDNA for bovine corticotropin-beta-lipotropin precursor. Nature 278, 423–427. [DOI] [PubMed] [Google Scholar]
- Nelson JS, 1994. In: Fishes of the World, third ed Wiley Press, New York, pp. 83–87. [Google Scholar]
- Reinick CL, Liang L, Angleson JK, Dores RM, 2012. Functional expression of Squalus acanthias melanocortin-5 receptor in CHO cells: ligand selectivity and interaction with MRAP. Eur. J. Pharmacol 680 (1–3), 1–7. [DOI] [PubMed] [Google Scholar]
- Schiöth HB, Raudsepp T, Ringholm A, Fredriksson R, Takeuchi S, Larhammar D, Chowdhary BP, 2003. Remarkable synteny conservation of melanocortin receptors in chicken, human, and other vertebrates. Genomics 81, 504–509. [DOI] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM, 2007. Melanocortin-2 receptor accessory protein MRAP forms antiparallel homodimers. Proc. Natl. Acad. Sci. U.S.A 104, 20244–20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM, 2009. Opposite effects of the melanocortin-2 (MC2) receptor accessory protein MRAP on MC2 and MC5 receptor dimerization and trafficking. J. Biol. Chem 284, 22641–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebag JA, Zhang C, Hinkle PM, Bradshaw AM, Cone RD, 2013. Developmental control of the melanocortin-4 receptor by MRAP2 proteins in zebrafish. Science 341, 278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Amemiya Y, Nozaki M, Sower SA, Kawauchi H, 2001. Evolutionary significance of proopiomelanocortin in agnathan andchondrorichthyes. Comp. Biochem. Physiol. B Biochem Mol. Biol 129, 283–289. [DOI] [PubMed] [Google Scholar]
- Thomas AL, Maekawa F, Kawashima T, Sakamoto H, Sakamoto T, Davis P, Dores RM, 2018. Analyzing the effects of co-expression of chick (Gallus gallus) melanocortin receptors with either chick MRAP1 or MRAP2 in CHO cells on sensitivity to ACTH(1–24) or ACTH(1–13)NH2: implications for the HPA axis and melanocortin circuits in the hypothalamus of the chick. Gen. Comp. Endocrinol 256, 50–56. [DOI] [PubMed] [Google Scholar]
- Vastermark A, Schiöth HB, 2011. The early origin of melanocortin receptors, agouti-related peptide, agouti signaling peptide, and melanocortin receptor-accessory proteins, with emphasis on pufferfishes, elephant shark, lampreys, and amphioxus. Eur. Jr. Pharm 660, 61–69. [DOI] [PubMed] [Google Scholar]
- Yang Y, 2011. Structure, function and regulation of the melanocortin receptors. Eur. J. Pharmacol 660, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Forlano PM, Cone RD, 2012. AgRP and POMC neurons are hypophysiotropic and coordinately regulate multiple endocrine axes in a larval teleost. Cell Metabol. 15, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Tao Y-X, 2017. Melanocortin Receptor Accessory Proteins (MRAPs): functions in the melanocortin system and beyond. Biochim. Biophys. Acta 1863 (10ptA). 10.1016/j.bdais.2017.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


