Abstract
The mononuclear phagocyte system in liver is a frequent target for nanoparticles (NPs). We performed a toxicological profiling of metal-based NPs in Kupffer and hepatocyte cell lines. Sixteen NPs were provided by the Nanomaterial Health Implications Research Consortium of the National Institute of Environmental Health Sciences to study the toxicological effects in KUP5 (KC) and Hepa 1-6 cells. Five NPs (Ag, CuO, ZnO, SiO2, and V2O5) exhibited cytotoxicity in both cell types, while SiO2 and V2O5 induced IL-1β production in KC. Ag, CuO, and ZnO induced caspase-3 generated apoptosis in both cell types was accompanied by ion shedding and generation of mitochondrial ROS in both cell types. However, the cell death response to SiO2 in KC differed by inducing pyroptosis as a result of potassium efflux, caspase-1 activation, NLRP3 inflammasome assembly, IL-1β release and cleavage of gasdermin-D. This releases pore-performing peptide fragments responsible for pyroptotic cell swelling. Interestingly, although V2O5 induced IL-1β release and delayed caspase-1 activation by vanadium ion interference in membrane Na+/K+ ATPase activity, the major cell death mechanism in KC (and Hepa 1-6) was caspase-3 mediated apoptosis. These findings improve our understanding of the mechanisms of metal-based ENM toxicity in liver cells towards comprehensive safety evaluation.
Keywords: Nanoparticles, Kupffer Cells, Hepatocytes, Macrophages, NLRP3 Inflammasome Activation, Potassium (K+) Efflux, Pyroptosis
1. Introduction
It has long been appreciated that the mononuclear phagocyte system (MPS, a.k.a. the reticuloendothelial system or RES) is frequently targeted by nanoparticles (NPs) during inadvertent or therapeutic exposures.[1, 2] An important phagocytic component of the liver is the Kupffer cell (KC), which constitutes 15% of liver cells or 80–90% of all the tissue macrophages in the body.[3-5] KCs play a major role in phagocytosis of foreign materials, endotoxin removal, and modulation of innate immune responses and also serves as the first line of defense to NPs by phagocytic removal in the liver.[1, 3, 6] In contrast, hepatocytes, which comprises 60-80% of liver cells,[1, 3, 6, 7] are involved in activities such as protein synthesis and storage, carbohydrate metabolism, synthesis of cholesterol, phospholipids, and bile salts, detoxification and excretion of chemical substances in the bile.[8-10] Importantly, there is also extensive cell–cell communications between KCs and hepatocytes, including for the transformation of drugs and xenobiotics that may be released from nanocarriers in capturing KCs.[5, 8, 11] ENM-induced liver toxicity is often evaluated by quantifying the release of liver enzymes such as aspartate aminotransferase (AST), alkaline phosphatase (ALP), and alanine aminotransferase (ALT) to the systemic circulation, as well as determining serum levels of bilirubin, albumin, and total protein.[2, 8] Since most of the screening is focused on the evaluation of hepatocyte function, much less attention has been given to the potential adverse impact on KCs.[2, 7, 12-16]
The Nanomaterial Health Implications Research (NHIR) Consortium at the National Institute of Environmental Health Sciences (NIEHS) aims at developing harmonized approaches to nanomaterial safety assessment through the provision of characterized NPs to consortium members, including release of a library of 16 metal and metal oxide nanoparticles for analysis. The availability of these materials presented us with a unique opportunity to elucidate response differences between KCs and hepatocytes for materials that are widely used in food products (such as confectionery, chewing gums, sauces, cakes, and pastries), drug formulations, dental and cosmetic materials, etc.[17-19] For instance, SiO2 and TiO2 exposure have been estimated to be around 100 mg/day. Van der Zande et al. documented the occurrence of liver fibrosis in rats fed with food-grade pyrogenic fumed silica.[20] Comparatively little information is available for the impact of metal-based NPs on Kupffer cells as compared to the effect on hepatocytes.[2, 7, 12-14] This includes previous research demonstrating that TiO2 and ZnO NPs induce toxicity in human hepatoblastoma C3A cells based on their ability to generate oxidative stress,[2] while Ag and ZnO NPs induced higher toxicity in 3D liver microtissues (comprised of human hepatocytes and Kupffer cells) compared to TiO2 NPs and multiwalled carbon nanotubes.[13] Moreover, our own studies have demonstrated that rare earth oxide NPs can induce a pyroptotic cell death in primary and transformed KCs through a unique effect that differs from the pro-oxidative effects of transition metal oxides NPs in liver cells.[7]
In this communication, we used the metal and metal oxide NPs provided by the NHIR Consortium for comprehensive toxicological profiling in transformed KCs and hepatocytes. This material panel included Ag (20 nm), Al2O3 (20 nm), CeO2 (10 nm and 30 nm), CuO (60 nm), Fe2O3 (10 and 100 nm), MgO (25 nm), TiO2 (30 nm and 100 nm), WO3 (20 nm), pyrolytic (fumed) silica (20 nm), V2O5 (400 nm), ZnO (50 nm) as well as the metal sulfides, CdS (10 nm) and ZnS (200 nm).[21, 22] The strategy for cellular screening was premised on the properties of metal and metal oxide nanomaterials that can lead to a compromise of cell viability as a result of ENM dissolution, metal ion shedding, surface membrane perturbation, NLRP3 inflammasome activation and the generation of hierarchical oxidative stress responses. We demonstrate similarity and differences in KC versus hepatocyte responses, including unique involvement of the NLRP3 inflammasome in terms of the cell viability features of fumed silica and V2O5 versus pro-oxidative NPs.
2. Results
2.1. Physical Characterization of Metal and Metal Oxide Nanoparticles
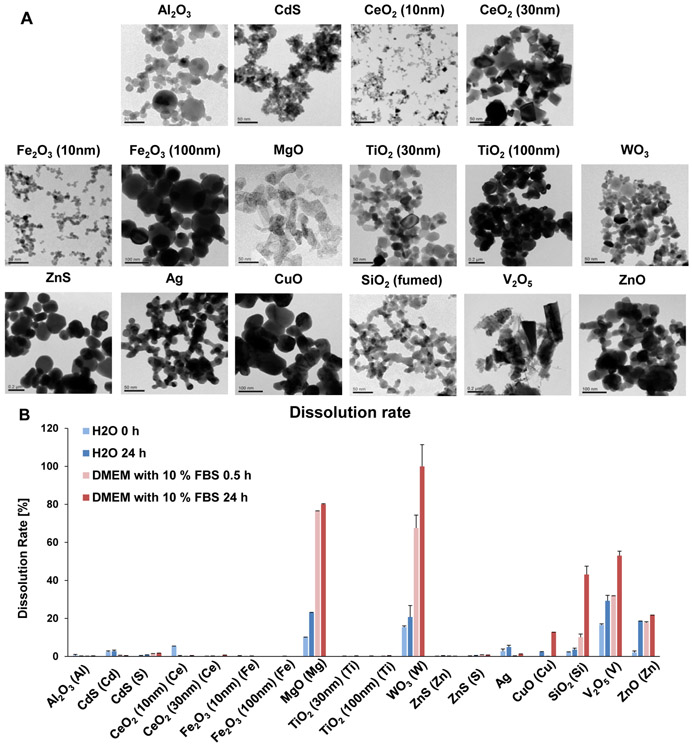
Safety assessment of metal, metal oxide (MOx) and metal sulfide nanoparticles was performed on materials provided by the Nanomaterials Health Implications Research (NHIR) Consortium at the NIEHS (Table 1). All the materials were thoroughly characterized for primary size, shape, hydrodynamic diameter, zeta (ζ)-potential (Table 1 and Figure 1A) and dissolution rate (Figure 1B). Transition electron microscopy (TEM) was used to assess primary particle sizes and shapes (Table 1). ENM sizes varied from 9.5 to 209 nm, except for V2O5 that was ~395 nm. Most nanoparticles displayed spherical shapes during TEM analysis, except for the irregular shapes of MgO and V2O5 and the chain-like structure of fumed SiO2 (Figure 1A). A Powder X-ray diffraction (XRD) spectrum (using a Panalytical X'Pert Pro diffractometer) was obtained to determine the crystal structure and oxidation status of the V2O5 NPs. The results (data not shown) demonstrate that all the characteristic peaks in our V2O5 sample matched the orthorhombic crystal structure of V2O5, indicating that V5+ was the associated vanadium ion species. NP characterization in aqueous and cell culture media showed that the majority of the NPs formed agglomerates with hydrodynamic sizes of 100–1000 nm, except for Fe2O3 (10 nm) and WO3, which formed smaller agglomerates of 72.1 and 79.4 nm, respectively (Table 1). The majority of the NPs exhibited a positive ζ potential in deionized (DI) water, except for Ag, CdS, CuO, SiO2, V2O5, WO3 and ZnO, which displayed negative surface charges (Table 1). The ζ potential changed to negative values of −10 to −20 mV in the presence of complete Dulbecco's Modified Eagle Medium (DMEM) culture medium, which contained 10% fetal calf serum. This likely reflects the formation of a protein corona (Table 1).
Table 1. Metal, metal oxide and metal sulfite nanoparticle library, depicting the primary size, hydrodynamic size and zeta potential.
The hydrodynamic size and zeta potential were determined in deionized water and complete DMEM (supplemented with 10% fetal bovine serum).
| Nanoparticles | Primary Size (nm) | Hydrodynamic Size (nm) |
z-Potential (mV) | Hydrodynamic Size (nm) | z-Potential (mV) |
|---|---|---|---|---|---|
| DI Water | DMEM | ||||
| Al2O3 | 19.9 ± 12.5 | 109.8 ± 2.5 | 47.9 ± 0.9 | 153.3 ± 2.4 | −20.2 ± 3.1 |
| CdS | 10.8 ± 2.8 | 436.3 ± 66.1 | −15.4 ± 0.9 | 406.4 ± 5.8 | −11.6 ± 2.8 |
| CeO2 (10 nm) | 9.7 ± 3.7 | 103.9 ± 2.4 | 37.9 ± 1.0 | 198.0 ± 7.1 | −10.9 ± 0.9 |
| CeO2 (30 nm) | 35.3 ± 21.9 | 143.2 ± 2.2 | 39.1 ± 1.4 | 225.3 ± 1.9 | −13.9 ± 3.8 |
| Fe2O3 (10 nm) | 9.5 ± 4.0 | 72.1 ± 1.3 | 40.4 ± 0.7 | 250.1 ± 6.3 | −14.0 ± 1.4 |
| Fe2O3 (100 nm) | 97.9 ± 52.1 | 997.4 ± 72.2 | 26.6 ± 0.5 | 1531.4 ± 88.8 | −11.8 ± 1.9 |
| MgO | 25.5 ± 9.1 | 544.3 ± 8.7 | 18.7 ± 0.8 | 896.9 ± 70.2 | −14.9 ± 1.5 |
| TiO2 (30 nm) | 27.0 ± 9.7 | 202.0 ± 1.6 | 32.7 ± 1.5 | 564.9 ± 54.9 | −16.0 ± 1.6 |
| TiO2 (100 nm) | 123.7 ± 46.3 | 181.3 ± 5.2 | 34.3 ± 2.3 | 445.1 ± 4.6 | −14.6 ± 1.6 |
| WO3 | 21.6 ± 8.9 | 79.4 ± 0.8 | −47.2 ± 2.3 | 186.4 ± 1.5 | −12.2 ± 0.8 |
| ZnS | 209.4 ± 63.9 | 271.1 ± 9.85 | 15.6 ± 1.4 | 506.7 ± 23.0 | −13.5 ± 1.4 |
| Ag | 20.0 ± 7.2 | 114.4 ± 1.7 | −28.6 ± 1.6 | 196.1 ± 3.4 | −15.6 ± 2.4 |
| CuO | 62.2 ± 14.6 | 290.9 ± 5.8 | −12.1 ± 1.9 | 362.1 ± 7.1 | −13.1 ± 1.6 |
| SiO2 | 16.1 ± 4.9 | 153.8 ± 4.9 | −25.6 ± 0.6 | 221.6 ± 5.8 | −13.8 ± 3.2 |
| V2O5 | 395.0 ± 230.5 | 610.4 ± 22.1 | −22.0 ± 2.0 | 1079.6 ± 12.4 | −11.3 ± 2.1 |
| ZnO | 47.7 ± 17.4 | 209.9 ± 5.9 | −36.5 ± 1.7 | 498.1 ± 12.1 | −14.1 ± 1.7 |
Figure 1. Characterization of nanoparticle morphology, dissolution characteristics, and cellular uptake.
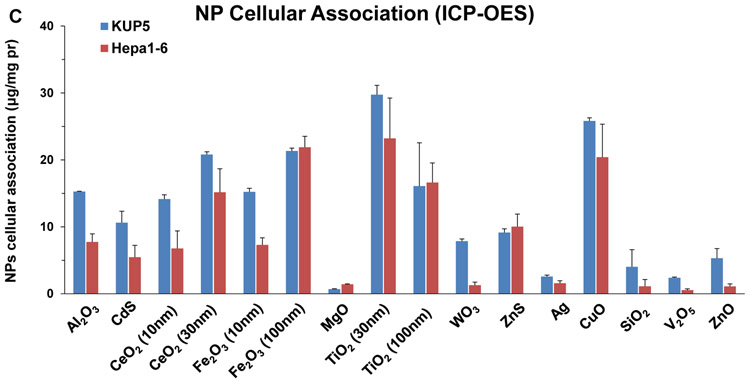
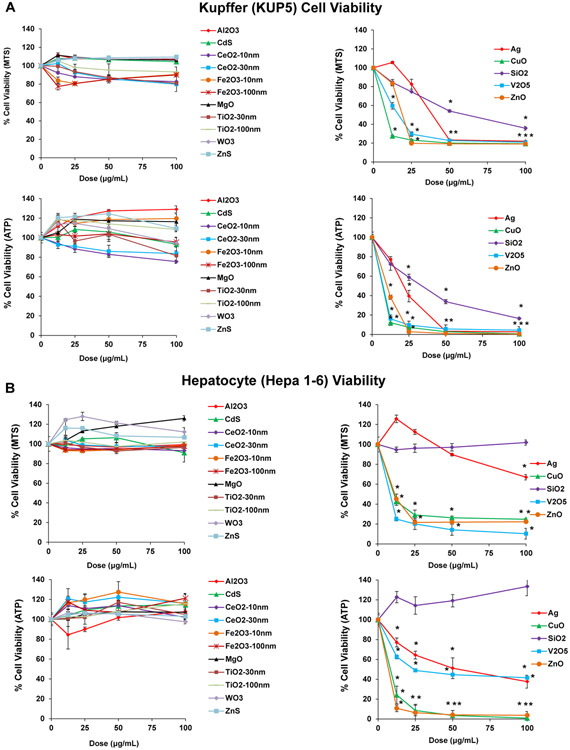
(A) TEM images of 16 nanoparticles provided by the consortium. The images were captured using a JEOL 1200-EX TEM with accelerating voltage of 80 kV. (B) ICP-OES analysis of nanoparticle dissolution. The graph shows the % metal dissolution for the individual nanoparticles in water and complete DMEM (DMEM with 10 % FBS) at two time points (0 and 24 h). The analysis was performed by suspending 100 μg/mL of each material in deionized H2O, followed by incubation at 37 °C for 0 or 24 h. The supernatants were collected by centrifugation at 15, 000 g for 30 min and digested by concentrated nitric acid at 90 °C for 3 h. The digested solutions were dried by evaporation at 120 °C and dissolved in 3 mL of 5% nitric acid for ICP-OES analysis. (C) Cellular uptake of nanoparticles. After 4 h treatment with 12.5 μg/mL nanoparticles, KUP5 and Hepa 1–6 cell pellets were collected and acid digested for assessment of their metal content by ICP-OES.
An additional characteristic of high relevance to metal and metal oxide toxicity was to assess the dissolution rate of the NPs in DI water and DMEM (Figure 1B). This demonstrated quantifiable dissolution for 7 materials (SiO2, Ag, CuO, ZnO, WO3, MgO and V2O5) in water, which was amplified by the presence of salt and protein in complete DMEM. WO3 and MgO showed the highest dissolution rates (>80% of total NPs in suspensions), while SiO2, ZnO, CuO and V2O5 showed higher dissolution (>10%) than Ag (< 5%) in complete medium (Figure 1B). Particle dissolution presents one mechanism by which ion shedding from nanoparticles can increase the cellular metal and metal oxide content. A second mechanism involves ion release inside the cell, following uptake of intact or partially dissolved NPs. Inductively coupled plasma optical emission spectrometry (ICP-OES) was used to assess the intracellular metal content following exposure to NPs (Figure 1C). While the uptake of low solubility NPs (e.g., Al2O3, CeO2, Fe2O3, TiO2, CdS, and ZnS) were generally higher in KUP5 cells than Hepa 1-6 cells, more dissolvable NPs (e.g., MgO, WO3, Ag, SiO2, V2O5, CuO, ZnO) exhibited lesser uptake in both cell types, except for CuO (Figure 1C). The lower cellular uptake of fumed SiO2 can be explained by its chain-like structure and reactive surface silanol groups, which promotes strong binding derivatives with surface membrane components, precluding cellular uptake. [21-23] We will discuss the significance of cell membrane perturbation in the activation of the NLRP3 inflammasome by fumed silica.
2.2. Nanoparticles Induce Differential Cytotoxic Responses in KUP5 and Hepa 1–6 Cells
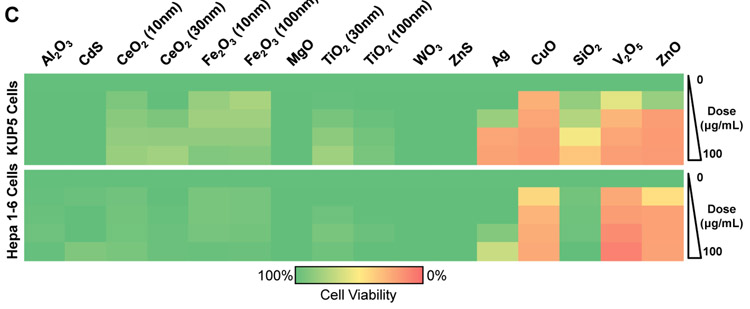
Cell viability studies were undertaken to obtain a provisional toxicological profiling of the particles in transformed KC (KUP5) and hepatocyte (Hepa 1–6) cell lines, which represent two of the principal liver cell types impacted by nanomaterials. Moreover, it is also known that there are differences in the toxicological responses of phagocytic cells (such as KC) and hepatocytes.[21-23] Our first analysis was to assess cell viability by MTS and adenosine triphosphate (ATP) assays over a particle dose range of 12.5–100 μg/mL (Figure 2A and B). The MTS results demonstrated that there were essentially two response profiles as a reflection of particle composition in both cell types (Figure 2A and B). Thus, while Al2O3, CeO2 (10 nm), CeO2 (30 nm), Fe2O3 (10 nm), Fe2O3 (100 nm), MgO, TiO2 (30 nm), and TiO2 (100 nm) failed to interfere in cell viability in either cell type, Ag, CuO, V2O5 and ZnO were toxic in both cell lines. In contrast, fumed SiO2 was toxic in KUP5 but not in Hepa 1–6 cells (Figure 2A and B). The MTS results were confirmed by a luminescence-based viability assay that determines ATP content (Figure 2A and B). Please notice that although we did not observe size-dependent differences, the material library was not constructed with the intention to make an independent assessment of this parameter.
Figure 2. Cytotoxicity screening and cellular uptake of nanoparticles in KUP5 and Hepa 1–6 cells.
MTS and ATP assays were used to assess the viability of (A) KUP5 and (B) Hepa 1–6 cells after particle exposure at a dose range of 12.5–100 μg/mL for 24 h. The results are reported according to the material response characteristics. The left-hand panel shows profiling of nontoxic materials, while the right-hand panel shows the response to particles with significant toxicity. The viability of non-treated control cells was regarded as 100%. (C) Heat map display for KUP5 and Hepa 1–6 cells according to the color scale at the bottom. The heat map was generated by using Excel, based on the MTS viability data in (A) and (B). The viability of non-treated control cells was regarded as 100% and assigned green, while the 0% viability was assigned red.
2.3. Ag, CuO, SiO2, V2O5 and ZnO induce Cellular Oxidative Stress Responses
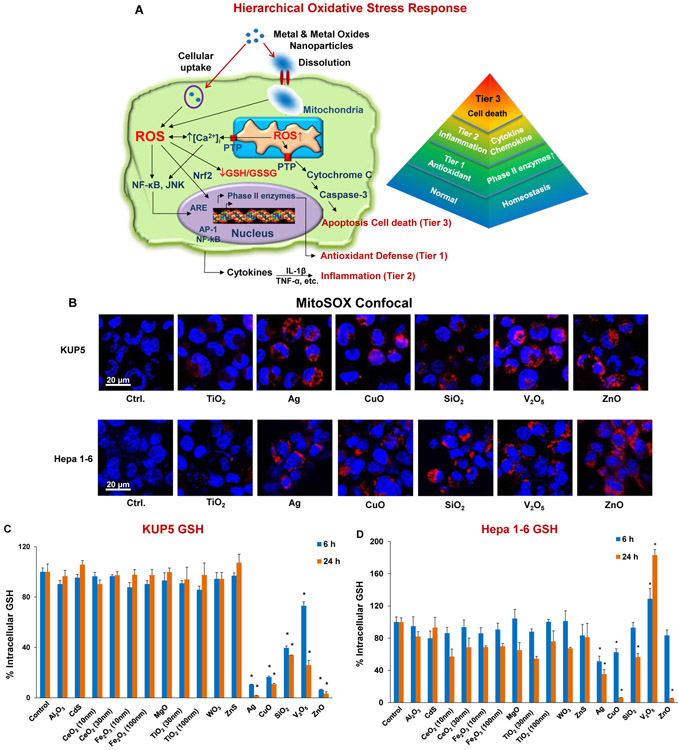
It has been shown that metal oxides are capable of inducing a tiered or hierarchical oxidative stress response in a variety of non-hepatic cells, as outlined in Figure 3A.[24, 25] Particle-induced generation of reactive oxygen species (ROS) production is capable of triggering a tier 1 antioxidant defense at the lowest level of oxidative stress.[26, 27] This response tier is dependent on thiol crosslinking of the transcription factor, erythroid 2-related factor (Nrf2), which leads to activation of the promoter of Phase II enzymes such as heme oxygenase 1 (HO-1) and glutathione peroxidase (GPx).[26, 27] Further increase in oxidative stress can lead to transcriptional activation of pro-inflammatory cytokines and chemokines (e.g., TNF-α, IL-1β, etc.), through the activation of the NF-κB, JNK/AP-1 signaling cascades.[26, 27] Tier 3 constitutes toxic oxidative stress, where triggering of the mitochondrial permeability transition pore and caspase activation can lead to cell death.[21, 24, 25, 28-30]
Figure 3. Cellular mitochondrial ROS generation and oxidative stress in KUP5 and Hepa 1–6 cells post-exposed to nanoparticles.
(A) Schematic to depict the hierarchical oxidative stress response to metals and metal oxide nanoparticles as previously elucidated for profiled nanomaterials.[23] Abbreviations in the schematic: JNK (c-Jun N-terminal kinases); ARE (Antioxidant Response Element); GSH (glutathione); AP-1 (activating protein-1); PTP (permeability transition pore). (B) Mitochondrial ROS generation was identified by MitoSOX red confocal microscopy following exposure to nanoparticles at 25 μg/mL for 5 h. Cells were stained with 5 × 10–6 M of MitoSOX for 20 min. The scale bar is 20 μm. (C and D) Impact on GSH levels as determined by the lmninescence-based GSH-Glo assay. KUP5 (C) and Hepa 1–6 (D) cells were treated with 50 μg/mL nanoparticles for 6 and 24 h. GSH levels were expressed as a percentage of the luminescence intensity compared to control cells (100%). (*) p < 0.05, compared to control. (E) HO-1 expression determined by immunoblotting. Both cell lines were treated with 12.5 μg/mL NPs for 6 h. (F) Dose-dependent IL-1β release by the full range of nanomaterials in KUP5 cells. LPS-primed (1 μg/mL, 4 h) KUP5 cells were exposed to nanoparticles (12.5–100 μg/mL) for 24 h. Supernatants were collected to measure IL-1β production by ELISA; *p < 0.05 compared to control cells. (G) Confocal microscopy images showing nanoparticle-induced caspases 3/7 expression in KUP5 and Hepa 1–6 cells. LPS-primed (1 μg/mL, 4 h) KUP5 and Hepa 1–6 cells were exposed to 25 μg/mL of chosen NPs for 5 h and stained with the FAM-FLICA caspase reagent for 1 h. Cell nuclei were stained with Hoechst 33342. The scale bar is 20 μm.
The initial assessment of the particles was to evaluate ROS production and cellular glutathione (GSH) levels, which reflect the cellular redox equilibrium as a trigger of diverse response outcomes (Figure 2). Mitochondrial ROS production was assessed by the following MitoSOX red fluorescence in KUP5 and Hepa 1–6 cells during viewing under a confocal microscope (Figure 3B). All particles induced fluorogenic oxidation of the dye (by superoxide), except non-toxic TiO2 ENM that was used as a control. In this study, we did not observe toxic effects of TiO2 NPs in either cell line, which is consistent with previous findings in other cell lines, e.g., BEAS-2B, THP-1, and RAW264.7.[29, 30] Measurement of GSH levels by a luminescence-based GSH-Glo assay demonstrated that 50 μg/mL Ag, CuO, SiO2, V2O5 and ZnO NPs, for 6 and 24 h, induced a significant reduction of GSH levels in KUP5 cells (Figure 3C). Although Ag, CuO, SiO2, and ZnO NPs significantly reduced GSH levels, V2O5 triggered an increase in the levels of this antioxidant in Hepa 1-6 cells (Figure 3D). In order to determine if the V2O5 effect could reflect and anti-oxidant protective response by Nrf2 activation, Western blotting was performed to assess the expression of HO-1, a frequently used Tier 1 oxidative stress biomarker (Figure 3E). Curiously, while Ag, CuO and ZnO NPs induced strong HO-1 responses in both cell lines there was no evidence that SiO2 and V2O5 NPs acted similarly. A possible explanation is that vanadium has been shown to specifically interfere with HO-1 expression by reducing mRNA levels.[31] SiO2 will be discussed later.
In order to assess pro-inflammatory effects that could be related to Tier 2 oxidative stress effects, we assessed TNF-α release in the supernatant of KUP5 and Hepa 1-6 cells in response to the full range of NPs (Figure S1A and B). This demonstrated that while CuO, SiO2 and ZnO NPs could induce significant TNF-α production in KUP5 cells, no response could be detected in Hepa 1-6 cells (Figure S1A and B). We also assessed ENM effects on IL-1β production, premised on the previous demonstration that the induction of this cytokine reflects activation of the NLRP3 inflammasome by nanomaterials.[7, 32-35] Importantly, only SiO2 and V2O5 NPs could be seen to induce IL-1β production in KUP5 cells (Figure 3F). In contrast, none of the materials induced IL-1β production in Hepa 1-6 cells (Figure S2A), likely due to the absence of pro-IL-1β expression in Hepa 1-6 cells (Figure S2B).
To assess the possibility that the NPs could induce apoptotic cell death as a consequence of toxic oxidative stress (Tier 3), confocal microscopy was used to assess caspase-3 and 7 activation through cleavage of the specific fluorescent FAM-FLICA substrate (Figure 3G). This demonstrated robust activation of these protease enzymes in KUP5 as well as Hepa 1–6 cells by all the materials leading to the cytotoxic cell responses (Figure 2). Please notice that SiO2 had a relatively weak effect on caspase activation in comparison to V2O5 NPs, a finding that will be discussed later (Figure 3G). TiO2 had no effect.
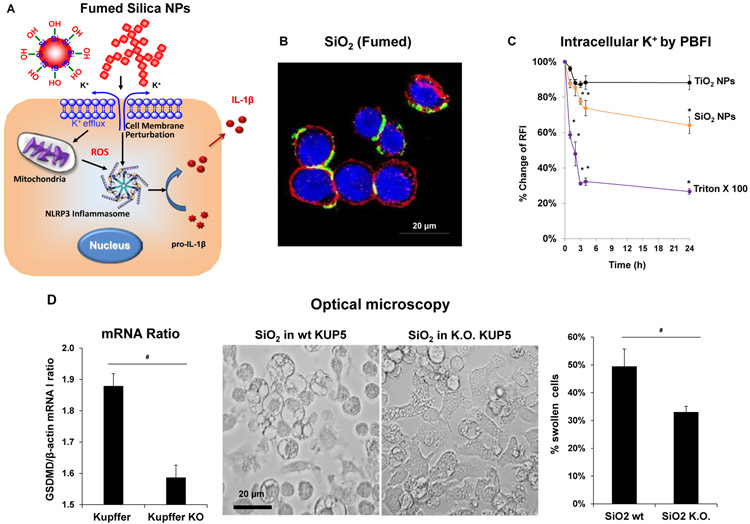
2.4. SiO2 Nanoparticles Induce Pyroptosis in KUP5 Cells Through Triggering Cell Membrane Depolarization and Inflammasome Activation
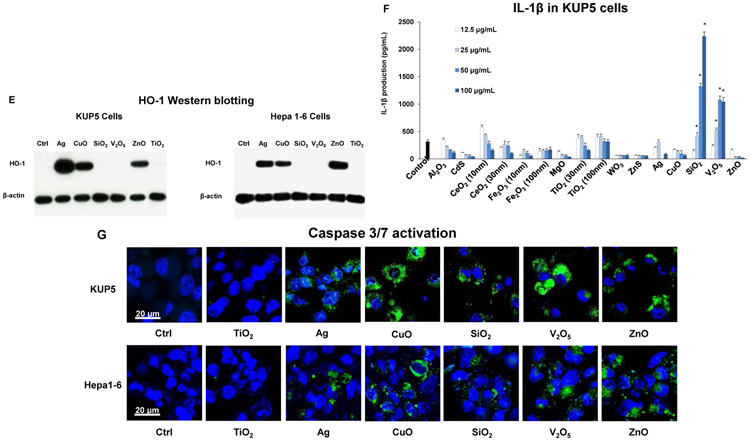
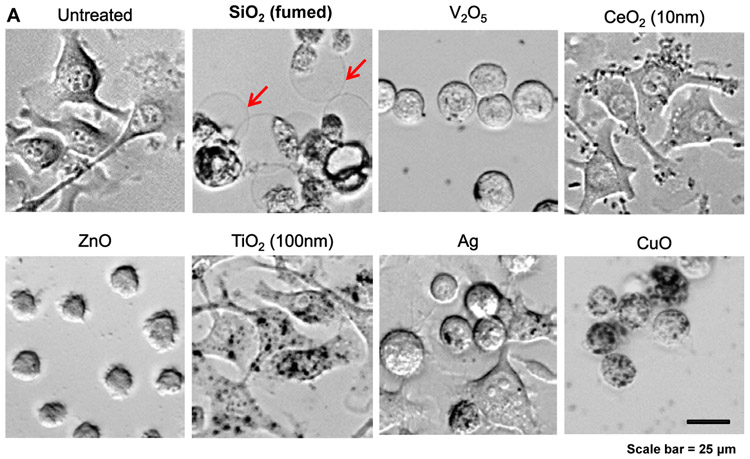
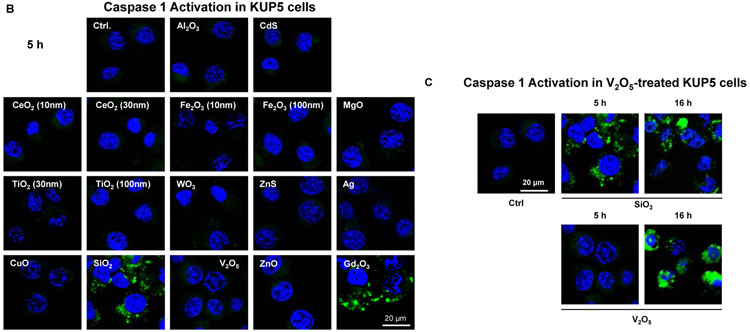
In the process of performing cell death profiling, the use of light optic microscopy revealed contrasting morphological features for fumed SiO2 compared to other NPs in KUP5 cells (Figures 4A and S3). This manifested as the appearance of swollen KUP5 cells, exhibiting giant surface blebs as compared to cellular shrinkage without surface blebbing during treatment with Ag, CuO, V2O5, and ZnO (Figure 4A). Also, there was no change in cell morphology for non-toxic NPs (Figures 4A and S3). Since the morphological changes to fumed SiO2 was accompanied by IL-1β production (Figure 3F), the possibility was that this could represent a pyroptosis response in KUP5 cells.[7, 36-38] Pyroptosis is characterized by cellular swelling and surface blebbing as a result of the formation of surface membrane pores by gasdermin D (GSDMD) upon its cleavage by caspase-1.[7, 36-45] Caspase-1 activation was assessed by confocal microscopy looking at the cleavage of the substrate, FAM-YVAD-FMK. Confocal imaging demonstrated caspase-1 activation in KUP5 cells, treated for 5 h with fumed SiO2 (Figure 4B). The rare earth oxide, Gd2CO3, was used as positive control. Noteworthy, there was no activation of this caspase by other NPs, including during treatment with V2O5 for 5h (Figure 4C). However, it was possible to demonstrate caspase-1 activation by V2O5 after a longer incubation period (16 h), suggesting differences in the response kinetics and mechanism of caspase activation, in comparison to SiO2 (Figure 4C). No evidence of caspase-1 activation was obtained in Hepa 1-6 cells (Figure S4).
Figure 4. Assessment of nanoparticles to induce Caspase 1 and SiO2 induced pyroptotic cell death in KUP5 cells.
(A) Optical microscopy images of ENM-treated KUP5 cells. Optical microscopy images showing the morphology of KUP5 cells exposed to seven of the NPs (fumed SiO2, Ag, CeO2, CuO, V2O5, TiO2 and ZnO) at 12.5 μg/mL for 6 h. The scale bar is 25 μm. The rest of the optical images for other particles appear in Figure S3. (B) Assessment of caspase 1 activation in ENM-treated KUP5 cells. The LPS-primed KUP5 cells were incubated with nanoparticles at 25 μg/mL for 5 h. Cells were washed with PBS and stained with FAM-FLICA caspase substrate for 1 h at 37 °C. Cells were then stained with Hoechst 33342. (C) Time-dependent activation of caspase 1 in V2O5-treated KUP5 cells. The LPS-primed KUP5 cells were incubated with fumed SiO2 and V2O5 for 5 and 16 h, and the staining process was repeated. The cells were imaged using Leica Confocal SP8-SMD microscope. The scale bar is 20 μm.
We have previously demonstrated that a variety of high aspect ratio nanomaterials are capable of generating IL-1β production in the myeloid cell line, THP-1, as a consequence of lysosomal damage and assembly of the NLRP3 inflammasome (Figure 5A).[32, 35, 46-55] In contrast, inflammasome activation by fumed SiO2 have been shown to proceed independent of lysosome damage, involving an pathway premised on K+ efflux from the surface membrane.[21-23] This pathway is dependent on the morphology of fumed silica ENM, allowing reactive surface silanol groups, situated in its chain-linked structure, to tightly adhere to the surface membrane of its cellular target. This triggers K+ efflux, which is one of the major stimuli for assembly of the NLRP3 inflammasome, leading to caspase-1 activation (Figure 5A).[22, 33, 34, 56, 57] Thus, as the first order of business, we assessed the effect of SiO2 NPs on lysosome damage and K+ efflux in KUP5 cells. Lysosomal damage was assessed by using the Magic Red-labeled substrate for cathepsin B to assess enzyme release from KUP5 lysosomes during confocal microscopy (Figure S5). In contrast to the lysosome disrupting properties of a positive control metal oxide ENM, Gd2CO3, which results in the disappearance of the punctate red staining pattern of lysosomal cathepsin B,[7, 58, 59] none of the consortium materials (including SiO2) had an impact on lysosome integrity (Figure S5). To investigate the impact on K+ efflux, FITC labeling of the SiO2 ENM were used to study particle association with KUP5 cells. Confocal microscopy demonstrated robust SiO2 ENM association with the cell surface membrane (Figure 5B). The next assessment was to study the impact of SiO2 on the intracellular [K+], using potassium-binding benzofuran isophthalate (PBFI) staining and confocal microscopy (Figure 5C). This demonstrated that SiO2 NPs could trigger a significant drop in the cellular [K+] (Figure 5C).
Figure 5. Generation of pyroptosis by fumed SiO2 in KUP5 cells, including the effect of GSDMD knockout.
(A) Schematic of the mechanism of fumed SiO2 NPs toxicity following surface membrane perturbation, potassimn efflux and the assembly of the NLRP3 inflammasome. (B) SiO2-induced plasma membrane perturbation. Confocal image of KUP5 cells treated with FITC-labeled fumed SiO2 nanoparticles. KUP5 cells were exposed to fumed SiO2 (100 μg/mL) for 12 h. The scale bar is 20 μm. The cell membrane was stained with Alexa Fluor 594 WGA; fumed SiO2 was tagged with FITC, and the nucleus stained with Hoechst 33342 (blue). (C) Intracellular K+ efflux in KUP5 cells, which were exposed to fumed SiO2 NPs before cellular staining with 5 μM PBFI-AM for 1 h. Triton X-100 (0.2 %) was used as positive control. Data were expressed as % change of relative fluorescence intensity (RFI), which is defined as the percentage change of fluorescence intensity of treated cells normalized to the intensity of non-treated control cells; *p < 0.05 compared to control cells. (D) Optical microscope images to compare the morphological changes by fumed SiO2 NPs at 12.5 μg/mL for 6 h in wildtype and GSDMD−/− KUP5 cells. Following image acquisition with an optical microscope, the % of cells (right panel) expressing at least two giant blebs were determined. The scale bar is 20 μm. GSDMD/β-actin mRNA ratio (left panel) to confirm that the knockdown of GSDMD in KUP5 cells. # p < 0.05 between wildtype and GSDMD knockout KUP5 cells.
While surface membrane perturbation by SiO2 can explain the particle effect on NLRP3 inflammasome and caspase-1 activation, the induction of pyroptosis in KUP5 cells also need to consider the role of gasdermin D (GSDMD).[7, 36-45] Cleavage of this protein by caspase-1 releases an N-terminal GSDMD fragments that assemble into pore-forming surface membrane structures; the pores are responsible for the lytic cell death response, a.k.a. pyroptosis.[7, 36-45] In order to assess the role of GSDMD in SiO2-induced KUP5 death, siRNA knockdown was performed. RT PCR analysis showed a significant decrease of the GSDMD/β-actin mRNA ratio in GSDMD−/− compared to wild type KUP5 cells (left panel in Figure 5D). Subsequent exposure to SiO2 NPs demonstrated a significant reduction in cytotoxicity (Figure S6A) as well as IL-1β production (Figure S6B) in GSDMD−/− compared to wildtype KUP5 cells. In addition, optical microscopy confirmed that there were less cell swelling and surface blebbing in GSDMD−/− compared to wildtype KUP5 cells (Figure 4A and 5D). All considered, these data show that SiO2-induced pyroptosis in KUP5 requires surface membrane perturbation, K+ efflux, NLRP3 inflammasome and caspase-1 activation, GSDMD cleavage and the formation of membrane pores.
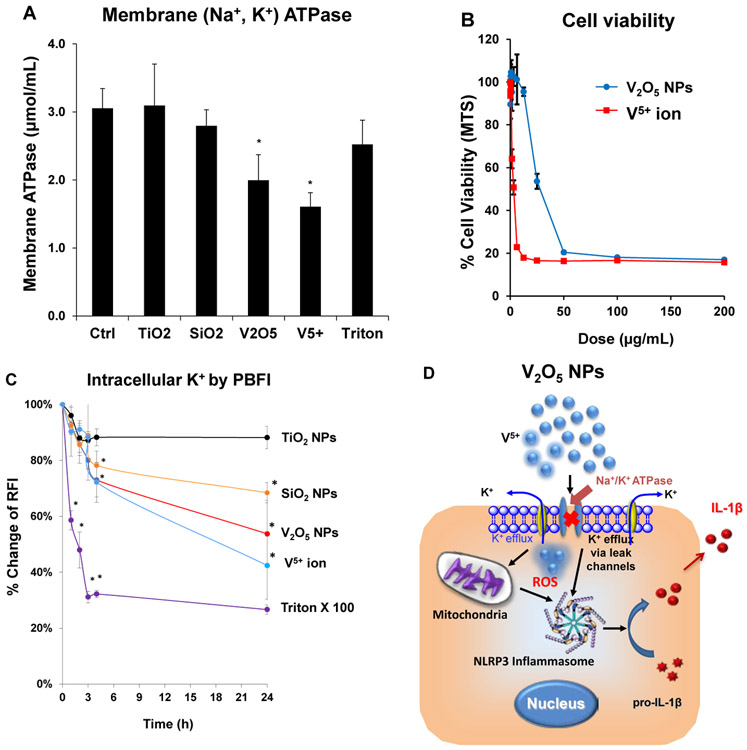
2.5. V2O5 Nanoparticles Induce NLRP3 Activation through perturbation of the Na+-K+-ATPase, leading to K+ efflux in KUP5 cells
Different from the pyroptosis response to SiO2 in KUP5 cells, V2O5 NPs promoted apoptotic cell death that is accompanied by later-onset caspase-1 activation (Figure 4C) and IL-1β production (Figure 3F). V2O5 does not exhibit the chain-like structure of fumed SiO2 NPs and has a relatively high dissolution rate that does not permit robust cellular uptake or lysosomal damage.[21-23] The key question therefore becomes how to reconcile these findings with the cell death response in KUP5 cells that are also associated with caspase-1 activation? It is known that vanadium ions (V5+) are capable of inhibiting Na+/K+ ATPase in the surface membrane of cells,[60-63] with the capacity to lower cellular [K+] levels as a result of cation leakage from the surface membrane.[61] We hypothesized, therefore, that perturbation of intracellular [K+] could serve as the basis for V2O5 triggering of NLRP3 inflammasome assembly, caspase-1 activation and IL-1β production.[23, 34, 56] First, we conducted an assay measuring Na+/K+ ATPase activity in KUP5 cells exposed to vanadium ions (V5+) and V2O5 NPs. Both the particles and vanadium ions were capable of inhibiting Na+/K+ ATPase activity (Figure 6A), leading to a reduction in cell viability (Figure 6B). In contrast, SiO2 and TiO2 NPs had no effect on ATPase activity (Figure 6A). A second experiment demonstrated that treatment with V5+ or V2O5 NPs could induce significant and time-dependent decline in intracellular [K+] levels in KUP5 cells (Figure 6C), which is in agreement with caspase-1 activation and IL-1β production by V2O5 in KUP5 cells (Figures 4C and 3F). However, while inflammasome activation and caspase-1 activation is also seen for SiO2, the absence of pyroptosis in response to V2O5 likely reflects the robust activation of caspases 3 and 7 by the latter particle type (Figure 3G). Caspases-3 and −7 utilize alternative cleavage GSDMD cleavage sites that interfere in the generation of pore-forming GSDMD fragments by caspase-1.[39] This aspect will be further discussed later.
Figure 6. The impact of V2O5 ENM on NLRP3 inflammasome activation by inhibiting membrane Na+/K+ ATPase.
(A) Membrane Na+/K+ ATPase activity in NPs-induced KUP5 cells. KUP5 cells, exposed to V2O5, SiO2, TiO2 NPs and V5+ ions, were sonicated to collect the lysed cell pellets in a centrifuge tube. The pellets were used to assess Na+/K+ ATPase activity with a commercial kit (MyBioSource). *p < 0.05 compared to control cells. (B) Cell viability. In response to treatment with V2O5 NPs and V5+ ions in KUP5 cells. An MTS assay was used to assess the viability of LPS-primed (1 μg/mL for 4 h) KUP5 cells that were subsequently exposed to V2O5 NPs and V5+ ions for 18 h. The viability of nontreated control cells was regarded as 100%. *p < 0.05 compared to control cells. (C) Intracellular K+ leakage in KUP5 cells. Same experiment as in Figure 5C, using KUP5 cells exposed to V2O5, SiO2, TiO2 NPs and V5+ ion. TiO2 NPs were used as a negative control. Triton X-100 (0.2 %) was used as positive control. Data were expressed as % change in RFI, which is defined as the percentage change of fluorescence intensity of treated cells normalized to the intensity of non-treated control cells. *p < 0.05 compared to control cells. (D) Schematic to explain the mechanism of V2O5 NPs toxicity through their effect on interference in Na+/K+ ATPase activity, potassium efflux, NLRP3 inflammasome assembly and IL-1β release.
In summary, Figure 6D summarizes the proposed pathway for V2O5 _induced apoptosis in KUP5 cells. We propose that particle dissolution generates toxic V5+ ions, capable of activating the NLRP3 inflammasome by decreasing the cellular [K+] content through interference in membrane Na+/K+ ATPase activity. However, V2O5 also generates ROS production and activation of caspases-3 and −7, which is responsible for apoptotic cell death and interference in caspase-1 mediated pyroptosis in KUP5 cells.
3. Discussion
In this study, we conducted a comprehensive analysis of the mechanisms of cytotoxicity of a panel of 16 metal, metal oxide, and metal sulfide nanoparticles, provided by the NIEHS Consortium, in transformed KCs and hepatocytes. Interestingly, five of these materials (Ag, CuO, ZnO, SiO2 and V2O5) exhibited cytotoxic potential in both liver cell types through different mechanisms of cytotoxicity, which reflects the involvement of oxidative stress, caspase activation, assembly of the NLRP3 inflammasome and GSDMD cleavage. Noteworthy, Ag, CuO, and ZnO NPs could induce caspase-3/7 (Figure 3G) mediated apoptotic cell death in KUP5 and Hepa 1-6 cells that can be explained by particle dissolution and ROS production. In contrast to the apoptosis response in hepatocytes, SiO2 NPs induced GSDMD dependent pyroptotic cell death in KUP5 cells. The pyroptosis effect reflects interaction of fumed SiO2 with the surface membrane, triggering NLRP3 activation, which, in turn, leads to GSDMD cleavage by caspase-1 and IL-1β production. In contrast, V2O5 activated caspases-3 and −7 promoted apoptotic cell death in KUP5 and Hepa 1-6 cells without resulting in pyroptosis in KUP5 cells, in spite of caspase-1 activation. The mechanism of caspase-1 activation by V2O5 NPs could be explained by particle dissolution and interference in membrane Na+/K+ ATPase activity by V5+ ions. This leads to a significant reduction in intracellular [K+] and triggering of the NLRP3 inflammasome in KUP5 cells. However, this event does not lead to pyroptosis, presumably due to the robust activation of caspases 3 and 7, which interfere in the generation of pore forming GSDMD fragments by caspase-1. In summary, the study provides important insight into the pathways of toxicity of metal and metal oxide nanoparticles in phagocytic liver cells versus hepatocytes.
The KUP5 and Hepa 1-6 cells were chosen in this work because they are widely used and well-characterized mouse cell lines for liver research. Although we did not include other hepatic cells to demonstrate the molecular mechanisms of pyroptosis or apoptosis in this work, we have previously shown[7] that KUP5 cells exhibit the same toxicological profiling in response to metal oxide nanoparticles (e.g., oxidative stress and pyroptosis) as primary human KCs, primary bone-marrow-derived macrophage (BMDM), as well as other phagocytic cell lines including J774A.1 and RAW 264.7 cells. In addition, the results obtained from Hepa 1-6 cells were similar to that of primary human hepatocytes in the same study.[7] An important finding of the current study is the demonstration of the role different toxicological pathways for metal and metal oxide NPs in KCs and hepatocytes. Overall, five materials (Ag, CuO, ZnO, SiO2 and V2O5 NPs) showed cytotoxic effects in liver cells, with eleven other NPs being exempt from effects on cell viability. The shedding of toxic ions by Ag, CuO and ZnO NPs played a role in the generation of oxidative stress, including by triggering ROS production in mitochondria. The generation of oxidative stress in KUP5 and Hepa 1-6 could be linked to GSH depletion, HO-1 expression and the initiation of caspase-3 and 7 mediated cell death. Moreover, CuO and ZnO NPs were associated with the release of TNF-α in KC (Figure S1A), which could reflect the generation of oxidative stress. However, there was no TNF-α release from Hepa 1-6 cells, which is in agreement with the observation that under in vitro conditions hepatocytes (Hepa 1-6 cells) can only produce TNF-α production during co-culture with macrophages.[64]
Of what relevance are the data for cellular studies to the potential toxicity of NPs in vivo and in humans? Among the 16 materials, the most obvious impact to consumers would be the potential toxic effect of fumed silica on the liver. Fumed silica has been estimated to be ingested as a food additive in human, leading to exposure of 0.28 and 4.53 mg/kg bw/day.[18] Van der Zande et al. documented the occurrence of liver fibrosis in rats fed with food-grade pyrogenic fumed silica.[17] It is also important to clarify that the absence of cellular toxicity is not necessary indicative of the absence of harmful effects during in vivo exposures. Take TiO2 for example. While we have not seen any adverse impact of these particles in liver cells, the use of TiO2 as a food additive (e.g., in confectionery and chewing gums, etc.)[17] has been associated in rat studies with preneoplastic lesions in the colon.[20] These in vivo outcomes could be further impacted by exposure duration, dose levels, systemic immune responses, and the involvement of microbiome etc. It is worth noting that France banned the use of titanium dioxide in all food products since January 1st, 2020. Other types of nanoparticles in this library have exposure potentials via oral pathway as well. These facts warrant the assessment of their toxicological potential and mechanisms of toxicity in liver.
SiO2 NPs could induce pyroptosis in KUP5 but not Hepa 1-6 cells. Pyroptosis differs significantly from other forms of cell death in its morphological and biochemical features.[36, 38, 39, 44] While pyroptosis may participate in immune defense against intracellular bacterial infections, excessive inflammasome activation can induce aseptic inflammation and liver disease such as acute or chronic hepatitis or liver fibrosis.[36, 38, 39, 44, 65] We have previously shown that rare earth oxide NPs induce pyroptosis in Kupffer cells via a pathway that involves intracellular uptake and lysosome damage, leading to NLRP3 and caspase-1 activation.[7] Caspase-1 is responsible for cleavage of GSDMD, resulting in the release of N-terminal peptide fragments from the autoinhibitory C-terminal domain of this protein.[7, 40, 41, 43] These fragments integrated in the cell surface membrane, where peptide oligomerization leads to pore formation.[7, 40, 41, 43] This leads to ion and water flux influx across the membrane, resulting in cellular swelling and the formation of giant cell blebs.[7, 38, 40, 41, 43, 44, 65] In contrast, the mechanism of SiO2-induced pyroptosis is different from the rare earth oxides insofar as the interaction of these reactive chain-link structures with the surface membrane is concerned.[21, 23] We have previously shown that the fumed silica toxicity derives from strained three-member siloxane rings (3MRs) that leads to the display of highly reactive silanol groups, acting in concert with hydroxyl groups and electrostatic interactions to cause surface membrane damage in cells coming in contact with these particles.[21] The accompanying surface membrane perturbation triggers K+ efflux, NLRP3 inflammasome assembly, caspase-1 activation and cleavage of GSDMD. The role of GSDMD in SiO2-induced pyroptosis was demonstrated by siRNA knockdown of GSDMD in KUP5 cells. In contrast to fumed silica, colloidal or Stober silica do not display 3MRs and are less prone to cause membrane damage of hemolysis.[21] However, there is one recent report showing Stober silica aggregates inducing pyroptosis in N9 microglial cells, a type of residential macrophage in the central nervous system.[66] Interestingly, in these studies the authors reported that the 50 nm particles to exhibit a high surface silanol density, different from the colloidal silica that we used. This toxicity could be attenuated by decreasing the surface silanol display through calcination and metal doping.[23, 66]
V2O5 NPs undergoes rapid dissolution, leading to shedding of >50% of the pentavalent vanadium ions with a +5 oxidation state. These particles were capable of inducing caspase-3 and 7 mediated apoptosis in KUP5 and Hepa 1-6 cells. Moreover, the particles also induced caspase-1 activation and IL-1β production without pyroptosis in KUP5 (Figure 5A). However, different from SiO2 NPs, activation of the NLRP3 inflammasome by V2O5 was not accompanied by the surface membrane perturbation (of SiO2) or the lysosome damage (of rare earth oxide NPs) in KUP5 cells (Figure S4). Instead, vanadate [in its orthovanadate (VO43−) form] is structurally similar to phosphate, and known to be a potent inhibitor of the intracellular hydrolysis domain of Na+/K+ ATPase in the cell surface membrane.[61, 67] Physiologically, this ion pump allows mammalian cells to maintain a high K+-to-Na+ ratio by ATP hydrolysis, allowing 3 Na+ to be pumped out of the cell in exchange of 2 K+ entering the cell.[61, 67] Thus, inhibition of the enzyme activity can lead to lowering of the intracellular [K+], which is a potent stimulus for activating the NLRP3 inflammasome and IL-1β production in KUP5 cells.[34, 56, 57] In addition, orthovanadate could also inhibit Ca2+ ATPase in the cell membrane and endoplasmic reticulum (ER), generating an increase in intracellular calcium.[68] This divalent cation is a potent inducer of mitochondrial ROS production.
An interesting question is why V2O5 NPs did not induce pyroptosis in KUP5 cells in spite of NLRP3 and caspase-1 activation? A likely explanation is the crosstalk between caspase-1 and caspase-3 and 7 in the cleavage of GSDMD (Figure S7).[39] In this regard, it has been shown that during apoptosis in macrophages and monocytes (as seen with V2O5 in KUP5 cells) caspases-3 and -7 can block pyroptosis by cleavage of GSDMD at sites that differ from the proteolysis by inflammatory caspases (caspase-1, -4, and -5).[39] This leads to the generation of inactive GSDMD fragments that are incapable of inducing surface membrane pores and pyroptotic cell death.[39] This notion is compatible with the finding that V2O5 is a robust inducer of caspases-3 and 7 in KUP5 cells, where the activation of caspase-1 is delayed (Figure 5C). This likely reflects the slower kinetics of Na+/K+ ATPase inhibition and inflammasome activation by V2O5 (Figure 6A). In contrast, SiO2 NPs has a more rapid rate of caspase-1 activation, with early GSDMD cleavage resulting in pyroptosis (Figures 5C and S7). It has also been demonstrated that in the absence of the GSDMD, caspase-1 could also activate caspases-3 and 7 to induce apoptosis.
4. Conclusion
We demonstrate that among 16 metal, metal oxide, and metal sulfide nanoparticles, five materials elicit toxicity in KCs and hepatocytes. Ag, CuO, and ZnO NPs induced caspase-3 and 7 mediated apoptotic cell death in KCs and hepatocytes. While SiO2 induced pyroptosis in KCs cells, the same outcome was not observed in hepatocytes. This pyroptosis response in KC is premised on surface membrane perturbation, K+ efflux, NLRP3 inflammasome activation and gasdermin D (GSDMD) cleavage by SiO2. In contrast, V2O5-induced apoptosis in the face of caspase-1 activation can be explained by inhibition of Na+/K+ ATPase, which leads to late-onset NLRP3 activation in KUP5 cells (at which point, caspases-3 and 7 already degraded the GSDMD pore-forming protein). Our study is of considerable importance for the screening and analysis of the potentially hazardous effects of metal based nanoparticles in the liver. The mechanistic understanding will help to establish a knowledge base for the safety assessment and risk analysis of NPs.
5. Experimental Section
Reagents and Materials:
A particle library of 16 materials was obtained from The National Institute of Environmental Health Sciences (NIEHS) Nanomaterials Health Implications Research (NHIR) Consortium. The CellTiter 96 AQueous MTS Assay and GSH-Glo Glutathione Assay kits were purchased from Promega (Madison, WI). The FAM-FLICA Caspase-1, Caspase-3/7 and Magic Red Cathepsin B Assay Kits were purchased from ImmunoChemistry Technologies, LLC (Bloomington, MN). The lipopolysaccharide (LPS) and GSDMD, NLRP3 and caspase 1 siRNAs were obtained from Sigma (St. Louis, MO). The ELISA kit for mouse IL-1β was purchased from R&D Systems (Minneapolis, MN). The ELISA kit for the mouse IL-1β pro-form was purchased from Thermo Fisher Scientific (Waltham, MA). The mouse Kupffer line, KUP5, was purchased from RIKEN CELL BANK (Japan). The hepatocyte cell line, Hepa 1–6, was purchased from ATCC.
Physical Characterizations of Nanoparticles:
The nanoparticles were characterized as received without further purification or modification. Their morphologies and primary sizes were assessed by TEM (JEOL 1200 EX transmission electron microscope). For particle characterization in suspension, the stock solutions were prepared at a concentration of 5 mg/mL in filtered deionized (DI) water, followed by sonication for 15 min in a water bath sonicator. These suspensions were then diluted in DI water or cell culture media to final concentrations of 50 μg/mL, followed by further sonication for 15 min. The hydrodynamic diameters and ζ potentials of particles in DI water and DMEM were determined using a ZetaPALS instrument (Brookhaven Instrument, Holtsville, NY). The particles were collected and thoroughly washed with deionized water before their morphology was characterized by TEM.
Fluorescence labeling of fumed silica nanoparticles:
2.5 mg of fluorescein isothiocyanate (FITC) was dissolved in 1.5 mL of dry toluene and 1.87 μL of 3-aminopropyltri ethoxy silane (APTES) was added and allowed to react under dry N2 for 2 h. The fluorescein/APTES mixture was added to 20 mL of 5 mg/mL fumed silica nanoparticles suspension in dry toluene, and allowed to react overnight over dry N2 at 80°C. The FITC-labeled fumed silica was collected through centrifugation (7000 rpm for 2 min), washed twice with toluene and twice with methanol. Fluorescein attachment was verified through IR spectroscopy and UV-Vis spectroscopy.
Cell Culture:
KUP5 cells were grown in high-glucose DMEM media supplemented with 10% fetal bovine serum (FBS), 10 μg/mL bovine insulin, and 250 μM 1-thioglycerol. Hepa 1–6 cells were grown in high-glucose DMEM media supplemented with 10% FBS and 100 U/mL-100 μg/mL penicillin streptomycin.
Assessment of cell viability:
The viability of KUP5 and Hepa 1–6 cells were assessed by the MTS assay. Cells were exposed to the nanoparticles at the indicated concentrations for 24 h in a 96-well plate, followed by removal of the medium and replacement with 120 μL of complete cell culture media containing 16.7 % of an MTS stock solution for 0.5 h at 37 °C. The plates were centrifuged at 2000g for 10 min in an Eppendorf 5430 microcentrifuge, and then 100 μL of supernatant was collected from each well and transferred into a new 96-well plate. The absorbance was read at 490 nm on a SpectraMax M5e microplate reader (Molecular Devices, Sunnyvale, CA).
Determining Intracellular GSH Levels:
The intracellular GSH level for KUP5 and Hepa 1–6 cells was assessed by the GSH-Glo Glutathione Assay. Cells were exposed to the nanoparticles at 50 μg/mL for 6 or 24 h in a 96-well plate, followed by removal of the medium and replacement with 100 μL of GSH-Glo reagent. After 30 min incubation, 100 μL of reconstituted luciferin detection reagent was added to the plate. Following 15 min incubation, luminescence was measured using a SpectraMax M5e microplate reader (Molecular Devices, Sunnyvale, CA).
Western Blot Analysis for HO-1 Expression:
1.6 × 105 cells from each line were seeded into the wells of six-well plates (Costar, Corning, NY). After overnight culture, each well received 1.6 mL of the appropriate culture medium containing 12.5 μg/mL NPs for an additional 6 h. Untreated cells were used as a blank control, while TiO2 NPs were used as a negative control. Cells were washed with PBS three times and harvested by scraping. The cell pellets were resuspended in cell lysis buffer containing Triton X-100 and protease inhibitors. After sonication and centrifugation, the protein content of the supernatants was measured by the Bradford method and 30 μg protein from each sample was electrophoresed by 10% SDS-PAGE and transferred to a PVDF membrane. After blocking, the membranes were incubated with anti-HO-1 monoclonal antibody (1:1000) (ENZO Life Sciences, Plymouth Meeting, PA) or anti-metallothionein monoclonal antibody (1:1000) (Abcam, Cambridge, MA). The membranes were overlaid with biotinylated secondary antibody (1:1000) before the addition of the HRP-conjugated avidin-biotin complex (1:10 000). The proteins were detected using the ECL reagent according to the manufacturer's instructions.
Determination of Mitochondrial ROS Generation:
KUP5 or Hepa 1-6 cells were exposed to nanoparticles at 37 °C and 5% CO2. After 3 h exposure to nanoparticles, the cells were washed twice with PBS and treated with 5 μM of MitoSOX (Invitrogen, Carlsbad, CA) in HBSS at 37 °C and 5% CO2 for 20 min. The cells were fixed with 4 % paraformaldehyde in PBS, stained with Hoechst 33342 (5 μg/mL), and imaged using a Leica Confocal SP8-SMD microscope.
Assessment of Pyroptotic Cell Death under Optical Microscopy:
Optical microscopy was used to monitor the morphology of wild type and GSDMD knockdown KUP5 cells, pretreated with 1 μg/mL LPS for 4 h. Cells were exposed to 12.5 μg/mL nanoparticles for 6 h at 37 °C and 5% CO2 in an 8-well Lab-Tek chamber slide. The cells were examined using a Zeiss Optical Microscope. In order to monitor the time kinetics of pyroptosis in LPS primed cells, 10 × 104 KUP5 cells/mL were exposed to 50 μg/mL nanoparticles for 6 h in a 35 mm glass bottom dish (In Vitro Scientific) at 37 °C and 5% CO2. Light optic images were obtained using a Leica Confocal SP5 Blue microscope.
Determination of IL-1β Production:
KUP5 and Hepa 1–6 cells were plated overnight at a density of 2 × 104 cells per well in a 96-well plate. The cells were primed by replacing the tissue culture medium with fresh medium containing 1 μg/mL LPS for 4 h. The primed cells were exposed to nanoparticles at the indicated concentrations and time periods as shown in the figure legends. The cellular supernatants were collected for IL-1β quantification by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions.
Determination of Pro-IL-1β Activity in LPS-Primed Cells:
KUP5 and Hepa 1–6 cells were plated overnight in 96-well plates at 2 × 104 cells per well. Following priming of the cells with 1 μg/mL LPS for 4 h, cells were lysed by using three freeze–thaw cycles in 100 μL of lysis buffer (10 μM 2-ME, 9 mM MgC12, and 0.1% triton X-100 in PBS). The cellular lysates were obtained after centrifugation and used for the quantification of pro-IL-1β by ELISA (Thermo Fisher Scientific).
Assessment of Lysosomal Damage by Cathepsin B Staining:
After KUP5 cells were cultured overnight in an 8-well Lab-Tek chamber slide at 5 × 104 cells per well, the cells were primed with LPS (1 μg/mL) for 4 h and exposed to particles (50 μg/mL) for 1 h at 37 °C. The cells were washed with PBS and incubated with Magic Red working solution for 30 min at 37 °C. Cells were washed with PBS and fixed with 4% paraformaldehyde in PBS. Finally, the cells were stained with Hoechst 33342 (5 μg/mL), washed with PBS, and imaged under a Leica Confocal SP8-SMD microscope.
Determination of Caspases 1, 3 and 7 Activation by Confocal Microscopy:
KUP5 or Hepa 1-6 cells were cultured in an 8-well Lab-Tek chamber slide at 5 × 104 cells/400 μL medium at 37 °C and 5% CO2. The KUP5 cells were primed with LPS (1 μg/mL) for 4 h and incubated with nanoparticles at 50 μg/mL for 3 h. The treated cells were washed with PBS and stained with FAM-FLICA Caspase-1 or Caspase 3/7 substrates for 1 h at 37 °C according to the manufacturer’s protocol. Finally, the cells were fixed with 4% paraformaldehyde in PBS, stained with Hoechst 33342 (5 μg/mL), and imaged using a Leica Confocal SP8-SMD microscope.
Evaluation of Intracellular Potassium Efflux and Na+/K+ ATPase activity:
NPs-induced potassium efflux in KUP5 cells was determined using PBFI AM (Molecular Probes, Carlsbad, CA). The KUP5 cells were primed with LPS (1 μg/mL) for 4 h and then incubated with NPs at 50 μg/mL. PBFI AM was added to the cells at 5 μM and cells were incubated at 37 °C and 5% CO2 for 1 h. Triton X-100 (0.2%) treated cells were used as controls. The fluorescence of PBFI AM was measured at excitation/emission at 340/505 nm, using a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA). For determination of Na+/K+ ATPase activity, ENM-treated KUP5 cells were incubated on ice for 30 min and then sonicated (with power 25%, sonication 3 s, pause 10 s, repeat 30 times) to collect lysates in a centrifuge tube. Following centrifugation, the supernatants were removed and placed in a new centrifuge tubes on ice to assess the Na+/K+ ATPase activity by a commercial kit (MyBioSource).
Determining Particle Dissolution in Cell Culture Medium:
Particle suspensions were prepared at 50 μg/mL in 1.5 mL of DI water or DMEM. The suspensions were incubated at 37 °C for 24 h, followed by centrifugation at 15,000 rpm for 30 min. Dissolution rate at 0 h indicated that data collected after centrifugation of particle suspension for 30 min. The supernatants were collected and subjected to acid digestion by using 10 mL mixture of concentrated HNO3 (65–70%, trace metal grade, Fisher Scientific) and HCl (35–38%, trace metal grade, Fisher Scientific) in a ratio of 1:3 at 95 °C for 2 days in a HotBlock (SC100, Environmental Express). ICP-OES analysis was performed to determine the metal content. The metal content was quantified by an ICP-OES (ICPE-9000, Shimadzu, Japan), using triplicate analysis of each sample and standard in the presence of 2% (v/v) nitric acid. Digested DMEM and DI water, which do not contain nanoparticles, served as blank reagent.
siRNA Knockdown in KUP5 Cells:
Knockdown of GSDMD gene was performed in KUP5 cells using electroporation at the Integrated Molecular Technologies Core Facility (University of California, Los Angeles). Briefly, 6 μg of siRNA in 100 μL of media was electroporated into 1 × 106 KUP5 cells. After electroporation, cells were maintained in complete media for another 48 h before cytotoxicity and IL-1β production analysis.
Statistical Analysis:
All the Statistical analysis represent mean±SD. Statistical significance was determined by two-tailed Student's t test for two-group analysis or one-way ANOVA for multiple group comparisons.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the Nanotechnology Health Implications Research (NHIR) Consortium of the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number (U01ES027237). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The engineered nanomaterials used in the research presented in this publication have been procured/developed, characterized, and provided by the Engineered Nanomaterials Resource and Coordination Core established at Harvard T. H. Chan School of Public Health (NIH grant # U24ES026946) as part of the Nanotechnology Health Implications Research Consortium. The authors thank the CNSI Advanced Light Microscopy/Spectroscopy and Electron Imaging Center for NanoMachines Core Facilities, the Flow Cytometry Core Facility of Jonsson Comprehensive Cancer Center, the Microscopic Techniques and Electron Microscope Core Facility of Brain Research Institute, and the Integrated Molecular Technologies Core (CURE/P30 DK041301) at UCLA. The authors would also like to thank Sydney Kusumoputro, Candice Lau, Shannon Tseng and Marissa Huang for their technical support on sample preparation, cell culture and paper editing.
Footnotes
Conflict of Interest
Andre E. Nel is co-founder and equity holders in Westwood Biosciences Inc and NAMMI Therapeutics. The remaining authors declare no conflict of interest.
Reference
- [1].Hume DA, Curr Opin Immunol 2006, 18, 49–53. [DOI] [PubMed] [Google Scholar]
- [2].Kermanizadeh A, Gaiser BK, Johnston H, Brown DM, Stone V, Brit J Pharmacol 2014, 171, 3980–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bilzer M, Roggel F, Gerbes AL, Liver Int 2006, 26, 1175–1186. [DOI] [PubMed] [Google Scholar]
- [4].Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger J, Braeuning A, Budinsky RA, Burkhardt B, Cameron NR, Camussi G, Cho CS, Choi YJ, Rowlands JC, Dahmen U, Damm G, Dirsch O, Donato MT, Dong J, Dooley S, Drasdo D, Eakins R, Ferreira KS, Fonsato V, Fraczek J, Gebhardt R, Gibson A, Glanemann M, Goldring CEP, Gomez-Lechon MJ, Groothuis GMM, Gustavsson L, Guyot C, Hallifax D, Hammad S, Hayward A, Haussinger D, Hellerbrand C, Hewitt P, Hoehme S, Holzhutter HG, Houston JB, Hrach J, Ito K, Jaeschke H, Keitel V, Kelm JM, Park BK, Kordes C, Kullak-Ublick GA, LeCluyse EL, Lu P, Luebke-Wheeler J, Lutz A, Maltman DJ, Matz-Soja M, McMullen P, Merfort I, Messner S, Meyer C, Mwinyi J, Naisbitt DJ, Nussler AK, Olinga P, Pampaloni F, Pi JB, Pluta L, Przyborski SA, Ramachandran A, Rogiers V, Rowe C, Schelcher C, Schmich K, Schwarz M, Singh B, Stelzer EHK, Stieger B, Stober R, Sugiyama Y, Tetta C, Thasler WE, Vanhaecke T, Vinken M, Weiss TS, Widera A, Woods CG, Xu JJ, Yarborough KM, Hengstler JG, Arch Toxicol 2013, 87, 1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Harbrecht BG, Billiar TR, Shock 1995, 3, 79–87. [PubMed] [Google Scholar]
- [6].Kodali V, Littke MH, Tilton SC, Teeguarden JG, Shi L, Frevert CW, Wang W, Pounds JG, Thrall BD, ACS nano 2013, 7, 6997–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mirshafiee V, Sun BB, Chang CH, Liao YP, Jiang W, Jiang JH, Liu XS, Wang X, Xia T, Nel AE, ACS nano 2018, 12, 3836–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gissen P, Arias IM, J Hepatol 2015, 63, 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gramignoli R, Tahan V, Dorko K, Venkataramanan R, Fox IJ, Ellis ECS, Vosough M, Strom SC, Cell Transplant 2014, 23, 1545–1556. [DOI] [PubMed] [Google Scholar]
- [10].Zinchenko YS, Culberson CR, Coger RN, Tissue Eng 2006, 12, 2241–2251. [DOI] [PubMed] [Google Scholar]
- [11].Billiar TR, Curran RD, Jpen-Parenter Enter 1990, 14, S175–S180. [DOI] [PubMed] [Google Scholar]
- [12].Kermanizadeh A, Gaiser BK, Hutchison GR, Stone V, Particle and fibre toxicology 2012, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kermanizadeh A, Lohr M, Roursgaard M, Messner S, Gunness P, Kelm JM, Moller P, Stone V, Loft S, Particle and fibre toxicology 2014, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hussain SM, Hess KL, Gearhart JM, Geiss KT, Schlager JJ, Toxicology in vitro : an international journal published in association with BIBRA 2005, 19, 975–83. [DOI] [PubMed] [Google Scholar]
- [15].Zhu SS, Zhang JQ, Zhang L, Ma WT, Man N, Liu YM, Zhou W, Lin J, Wei PF, Jin PP, Zhang YJ, Hu Y, Gu EW, Lu XF, Yang ZL, Liu XS, Bai L, Wen LP, Adv Healthc Mater 2017, 6. [DOI] [PubMed] [Google Scholar]
- [16].Kojima S, Negishi Y, Tsukimoto M, Takenouchi T, Kitani H, Takeda K, Toxicology 2014, 321, 13–20. [DOI] [PubMed] [Google Scholar]
- [17].Winkler HC, Notter T, Meyer U, Naegeli H, J Nanobiotechnol 2018, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fruijtier-Polloth C, Arch Toxicol 2016, 90, 2885–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen Z, Han S, Zheng P, Zhou D, Zhou S, Jia G, Nanoscale 2020. [DOI] [PubMed] [Google Scholar]
- [20].van der Zande M, Vandebriel RJ, Groot MJ, Kramer E, Rivera ZEH, Rasmussen K, Ossenkoppele JS, Tromp P, Gremmer ER, Peters RJB, Hendriksen PJ, Marvin HJP, Hoogenboom RLAP, Peijnenburg AACM, Bouwmeester H, Particle and fibre toxicology 2014, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang HY, Dunphy DR, Jiang XM, Meng H, Sun BB, Tarn D, Xue M, Wang X, Lin SJ, Ji ZX, Li RB, Garcia FL, Yang J, Kirk ML, Xia T, Zink JI, Nel A, Brinker CJ, J Am Chem Soc 2012, 134, 15790–15804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun BB, Wang X, Liao YP, Ji ZX, Chang CH, Pokhrel S, Ku J, Liu XS, Wang M, Dunphy DR, Li RB, Meng H, Maedler L, Brinker CJ, Nel AE, Xia T, ACS nano 2016, 10, 8054–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sun B, Pokhrel S, Dunphy DR, Zhang H, Ji Z, Wang X, Wang M, Liao Y, Chang CH, Dong J, Li R, Mädler L, Brinker J, Nel A, Xia T, ACS nano 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M, Nat Mater 2009, 8, 543–557. [DOI] [PubMed] [Google Scholar]
- [25].Nel A, Xia T, Madler L, Li N, Science 2006, 311, 622–627. [DOI] [PubMed] [Google Scholar]
- [26].Li N, Alam J, Venkatesan MI, Eiguren-Fernandez A, Schmitz D, Di Stefano E, Slaughter N, Killeen E, Wang XR, Huang A, Wang MY, Miguel AH, Cho A, Sioutas C, Nel AE, J Immunol 2004, 173, 3467–3481. [DOI] [PubMed] [Google Scholar]
- [27].Wang MY, Xiao GG, Li N, Xie YM, Loo JA, Nel AE, Electrophoresis 2005, 26, 2092–2108. [DOI] [PubMed] [Google Scholar]
- [28].Zhang HY, Pokhrel S, Ji ZX, Meng H, Wang X, Lin SJ, Chang CH, Li LJ, Li RB, Sun BB, Wang MY, Liao YP, Liu R, Xia T, Madler L, Nel AE, J Am Chem Soc 2014, 136, 6406–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang HY, Ji ZX, Xia T, Meng H, Low-Kam C, Liu R, Pokhrel S, Lin SJ, Wang X, Liao YP, Wang MY, Li LJ, Rallo R, Damoiseaux R, Telesca D, Madler L, Cohen Y, Zink JI, Nel AE, ACS nano 2012, 6, 4349–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].George S, Pokhrel S, Ji ZX, Henderson BL, Xia T, Li LJ, Zink JI, Nel AE, Madler L, J Am Chem Soc 2011, 133, 11270–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang J, Yuan Z, Zhang K, Ding X, Bai S, Zeng Q, Peng H, Celi P, Poultry science 2018, 97, 3109–3118. [DOI] [PubMed] [Google Scholar]
- [32].Wang X, Xia T, Duch MC, Ji ZX, Zhang HY, Li RB, Sun BB, Lin SJ, Meng H, Liao YP, Wang MY, Song TB, Yang Y, Hersam MC, Nel AE, Nano Lett 2012, 12, 3050–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang X, Sun BB, Liu SJ, Xia T, Nanoimpact 2017, 6, 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sun BB, Wang X, Ji ZX, Li RB, Xia T, Small 2013, 9, 1595–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang X, Chang CH, Jiang JH, Liu Q, Liao YP, Lu JQ, Li LJ, Liu XS, Kim J, Ahmed A, Nel AE, Xia T, Small 2019, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bergsbaken T, Fink SL, Cookson BT, Nat Rev Microbiol 2009, 7, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cunha LD, Zamboni DS, Frontiers in cellular and infection microbiology 2013, 3, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fink SL, Cookson BT, Cell Microbiol 2007, 9, 2562–2570. [DOI] [PubMed] [Google Scholar]
- [39].Taabazuing CY, Okondo MC, Bachovchin DA, Cell Chem Biol 2017, 24, 507–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].He WT, Wan HQ, Hu LC, Chen PD, Wang X, Huang Z, Yang ZH, Zhong CQ, Han JH, Cell Res 2015, 25, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu JS, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang YF, Bertram EM, Goodnow CC, Dixit VM, Nature 2015, 526, 666–671. [DOI] [PubMed] [Google Scholar]
- [42].Liu X, Zhang ZB, Ruan JB, Pan YD, Magupalli VG, Wu H, Lieberman J, Nature 2016, 535, 153–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shi JJ, Zhao Y, Wang K, Shi XY, Wang Y, Huang HW, Zhuang YH, Cai T, Wang FC, Shao F, Nature 2015, 526, 660–665. [DOI] [PubMed] [Google Scholar]
- [44].Vande Walle L, Lamkanfi M, Curr Biol 2016, 26, R568–R572. [DOI] [PubMed] [Google Scholar]
- [45].Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Dueber EC, P Natl Acad Sci USA 2016, 113, 7858–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xia T, Hamilton RF, Bonner JC, Crandall ED, Elder A, Fazlollahi F, Girtsman TA, Kim K, Mitra S, Ntim SA, Orr G, Tagmount M, Taylor AJ, Telesca D, Tolic A, Vulpe CD, Walker AJ, Wang X, Witzmann FA, Wu NQ, Xie YM, Zink JI, Nel A, Holian A, Environ Health Persp 2013, 121, 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang X, Mansukhani ND, Guiney LM, Lee JH, Li RB, Sun BB, Liao YP, Chang CH, Ji ZX, Xia T, Hersam MC, Nel AE, ACS nano 2016, 10, 6008–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang X, Lee JH, Li RB, Liao YP, Kang J, Chang CH, Guiney LM, Mirshofiee V, Li LJ, Lu JQ, Xia T, Hersam MC, Nel AE, Small 2018, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang X, Duch MC, Mansukhani N, Ji Z, Liao YP, Wang MY, Zhang HY, Sun BB, Chang CH, Li RB, Lin SJ, Meng H, Xia T, Hersam MC, Nel AE, ACS nano 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sun BB, Wang X, Ji ZX, Wang MY, Liao YP, Chang CH, Li RB, Zhang HY, Nel AE, Xia T, Small 2015, 11, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sun BB, Ji ZX, Liao YP, Wang MY, Wang X, Dong JY, Chang CH, Li RB, Zhang HY, Nel AE, Xia T, ACS nano 2013, 7, 10834–10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sun BB, Ji ZX, Liao YP, Chang CH, Wang X, Ku J, Xue CY, Mirshafiee V, Xia T, Acs Appl Mater Inter 2017, 9, 21697–21705. [DOI] [PubMed] [Google Scholar]
- [53].Lin SJ, Wang X, Ji ZX, Chang CH, Dong Y, Meng H, Liao YP, Wang MY, Song TB, Kohan S, Xia T, Zink JI, Lin S, Nel AE, Aspect Ratio Plays a Role in the Hazard Potential of CeO2 Nanoparticles in Mouse Lung and Zebrafish Gastrointestinal Tract. In ACS nano, 2014; Vol. 8, pp 4450–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li RB, Wang X, Ji ZX, Sun BB, Zhang HY, Chang CH, Lin SJ, Meng H, Liao YP, Wang MY, Li ZX, Hwang AA, Song TB, Xu R, Yang Y, Zink JI, Nel AE, Xia T, ACS nano 2013, 7, 2352–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ji Z, Wang X, Zhang H, Lin S, Meng H, Sun B, George S, Xia T, Nel AE, Zink JI, ACS nano 2012, 6, 5366–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Walev I, Reske K, Palmer M, Valeva A, Bhakdi S, The EMBO journal 1995, 14, 1607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].El Kebir D, Jozsef L, Khreiss T, Filep JG, Cell Signal 2006, 18, 2302–2313. [DOI] [PubMed] [Google Scholar]
- [58].Li RB, Ji ZX, Qin HQ, Kang XD, Sun BB, Wang MY, Chang CH, Wang X, Zhang HY, Zou HF, Nel AE, Xia T, ACS nano 2014, 8, 10280–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Li RB, Ji ZX, Dong JY, Chang CH, Wang X, Sun BB, Wang MY, Liao YP, Zink JI, Nel AE, Xia T, ACS nano 2015, 9, 3293–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Higashino H, Bogden JD, Lavenhar MA, Bauman JW, Hirotsu T, Aviv A, Am J Physiol 1983, 244, F105–F111. [DOI] [PubMed] [Google Scholar]
- [61].Searle BM, Higashino H, Khalil F, Bogden JD, Tokushige A, Tamura H, Kino M, Aviv A, Circ Res 1983, 53, 186–191. [DOI] [PubMed] [Google Scholar]
- [62].North P, Post RL, J Biol Chem 1984, 259, 4971–4978. [PubMed] [Google Scholar]
- [63].Cantley LC Jr., Resh MD, Guidotti G, Nature 1978, 272, 552–4. [DOI] [PubMed] [Google Scholar]
- [64].Takano M, Sugano N, Mochizuki S, Koshi RN, Narukawa TS, Sawamoto Y, Ito K, J Periodontal Res 2012, 47, 89–94. [DOI] [PubMed] [Google Scholar]
- [65].Wu JL, Lin S, Wan B, Velani B, Zhu YY, Aging Dis 2019, 10, 1094–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Du QQ, Ge D, Mirshafiee V, Chen C, Li M, Xue CY, Ma XH, Sun BB, Nanoscale 2019, 11, 12965–12972. [DOI] [PubMed] [Google Scholar]
- [67].Cantley LC, Josephson L, Warner R, Yanagisawa M, Lechene C, Guidotti G, J Biol Chem 1977, 252, 7421–7423. [PubMed] [Google Scholar]
- [68].Clausen JD, Bublitz M, Arnou B, Olesen C, Andersen JP, Moller JV, Nissen P, Structure 2016, 24, 617–623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.