Abstract
Xenopus has been used to study a wide array of developmental processes, benefiting from vast quantities of relatively large, externally developing eggs. Xenopus is particularly amenable to examining the cardiac system because many of the developmental processes and genes involved in cardiac specification, differentiation, and growth are conserved between Xenopus and human and have been characterized in detail. Furthermore, compared with other higher vertebrate models, Xenopus embryos can survive longer without a properly functioning heart or circulatory system, enabling investigation of later consequences of early embryological manipulations. This biology is complemented by experimental technology, such as embryonic explants to study the heart, microinjection of overexpression constructs, and, most recently, the generation of genetic mutations through gene-editing technologies. Recent investigations highlight Xenopus as a powerful experimental system for studying injury/repair and regeneration and for congenital heart disease (CHD) modeling, which reinforces why this model system remains ideal for studying heart development.
Studies in amphibians have formed the basis of cardiac biology in vertebrates for >80 years, yielding many of our most important insights (Taylor 1931; Nieuwkoop 1947; Chuang and Tseng 1957; Jacobson 1960, 1961; Monnickendam and Balls 1973). The frog has several advantages over other species for studying heart development. For example, mice are genetically tractable but are difficult for live imaging or studying biochemistry of the heart. Fish are an outstanding system for live imaging, but their small size and clutch numbers make systems-level proteomic approaches difficult. Xenopus has an advantage over these species in that a suite of novel tools exists that will allow—in a single organism—integration of systems-level genomic and proteomic analyses with quantitative live imaging of cardiac cell behaviors.
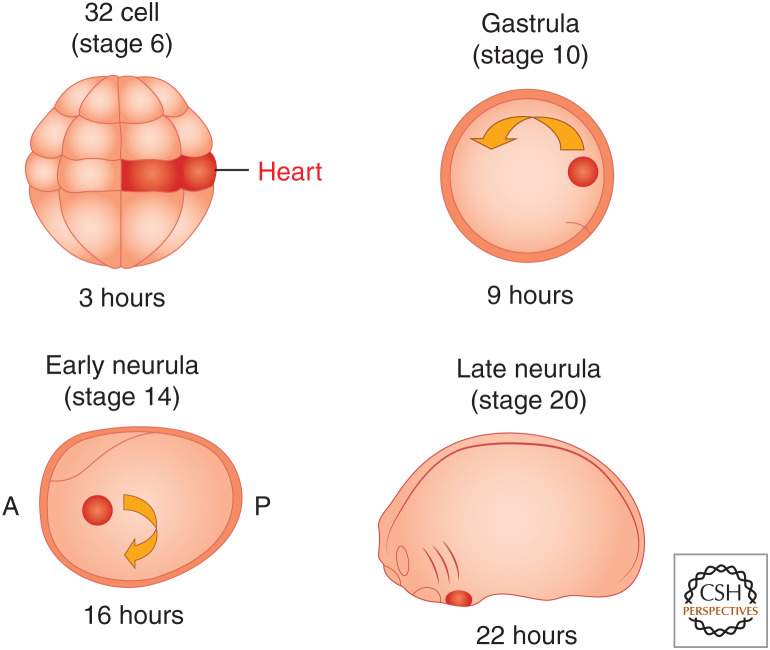
Classical fate-mapping studies have shown that at a mere 3 hours after fertilization, at the 32-cell stage, four blastomeres in the dorsal equatorial region of Xenopus are fated to become the adult heart (Fig. 1; Dale and Slack 1987; Moody 1987a,b). As in all embryos with yolk-filled blastomeres, cells do not undergo extensive mixing before gastrulation. Instead, the descendants of the four blastomeres remain as a coherent group of cells that lie juxtaposed between the organizer and the underling endoderm; these two tissue types induce the cardiac lineage (Fig. 1). During gastrulation, the cardiac cells are the first cells to become specified and determined (Symes et al. 1994; Nascone and Mercola 1995; Mercola 1999; Mohun and Leong 1999; Zhu et al. 1999; Kolker et al. 2000; Mohun et al. 2000).
Figure 1.
Schematic of early Xenopus laevis heart development. Stages are shown above embryos, and hours of development at room temperature are shown beneath. Blastomeres at the 32-cell stage fated to become heart tissue are labeled in red and their decedents labeled in red at gastrula, early neurula, and late neurula. (A) Anterior, (P) posterior.
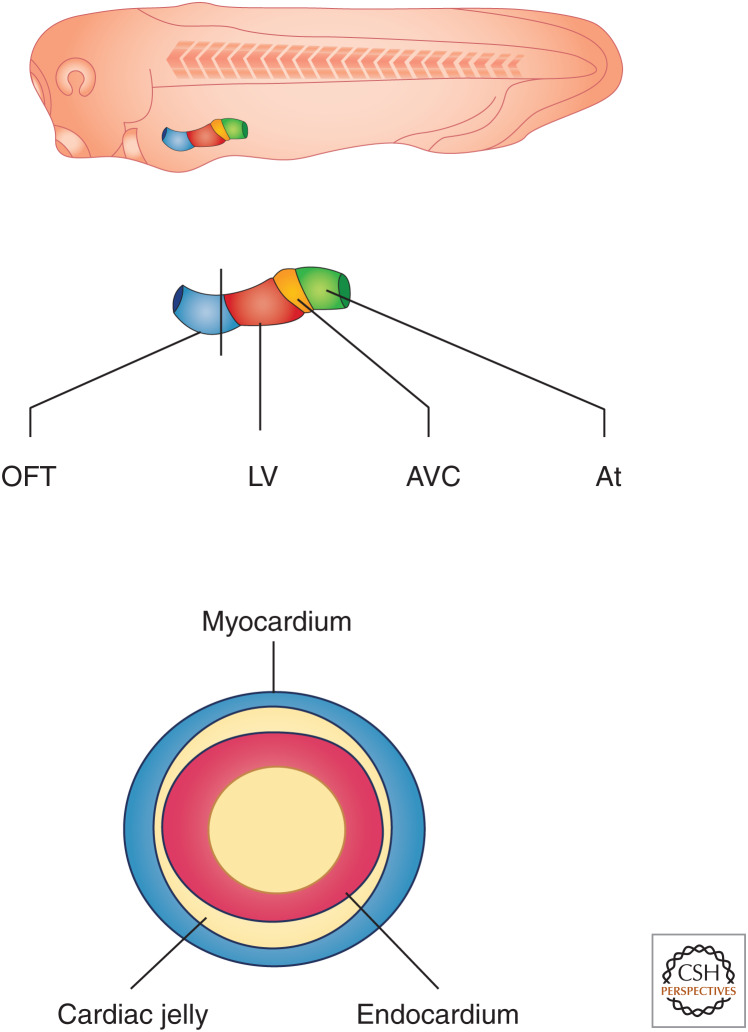
During neurulation, the cardiac cells move as two distinct populations on either side of the embryo toward the forward end of the mesoderm sheet as it engulfs the yolk and encompasses the embryo (Fig. 1). The cells converge to form a single sheet at the ventral anterior midline, where they form a tube positioned along the anteroposterior axis (Figs. 1 and 2). The linear heart tube is bilaminar, composed of an outer myocardium and an inner endocardium (Fig. 2; Mohun and Leong 1999; Kolker et al. 2000; Mohun et al. 2000).
Figure 2.
(Top) A schematic of Xenopus laevis heart development at the 32-cell stage showing the position of the linear heart tube. (Middle) Specific relative positions of cell type along the heart tube outflow tract (OFT), left ventricle (LV), atrioventricular canal (AVC), and atrium (At). (Bottom) Section of the heart tube with black solid lines in the middle of the schematic showing that the heart is comprised of a bilaminar heart tube with an inner endocardium and an outer myocardium, which are separated by extracellular cardiac jelly.
As with all vertebrates, the blood in Xenopus flows from the tail to the head. As development proceeds, the cardiac tube begins to undergo looping. Collectively, this activity folds the heart, bringing the inflow and outflow tracts near each other to make the proper connections with the developing vascular system and to place the two developing atria on top of the single ventricle. The atria will then undergo a slow septation process, leading to the formation of a three-chambered heart (Mohun and Leong 1999; Kolker et al. 2000; Mohun et al. 2000).
UNDERPINNING CLASSICAL EMBRYOLOGY AND EXPERIMENTAL BIOLOGY
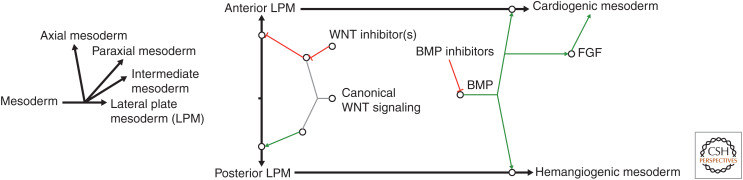
The large amphibian embryos introduced above provide a distinct experimental advantage. The large embryos allow for observation of the conserved vertebrate body as it develops, and the associated large embryonic cells (called blastomeres) allow the experimental researcher to use lineage tracing and targeted tissue- and organ-specific delivery of experimental reagents by microinjection (e.g., target the red blastomeres in the 32-blastomere embryo in Fig. 1). The large embryos also lend themselves to powerful, so-called “cut-and-paste” experiments to study the role of tissue interactions, particularly in heart development, and “just cut” embryonic explant experiments to study, for instance, the development of heart tissue in relative isolation. Such approaches can be used to separate cardiogenic from noncardiogenic mesoderm (i.e., dorsal marginal zone [DMZ], ventral marginal zone [VMZ]) (reviewed by Afouda 2012) to study the function of inducers and repressors of heart development (e.g., Fig. 3; Foley and Mercola 2005). Many of the fundamental discoveries made in Xenopus are a result of these intrinsic advantages of the experimental model. For instance, the important role of Wnt signaling in cardiogenesis, both canonical β-catenin-mediated (Schneider and Mercola 2001) and noncanonical (Pandur et al. 2002), was discovered in Xenopus, as well as bone morphogenetic protein (BMP) (Breckenridge et al. 2001) and later fibroblast growth factor (FGF) signaling (Deimling and Drysdale 2011). These discoveries, together with findings in other models such as chick (Marvin et al. 2001; Wittig and Münsterberg 2019), gave rise to an often reproduced figure in textbooks (Fig. 4) regarding signaling pathways, and also inhibition of signaling pathways, specifying anterior and posterior lateral plate mesoderm and subsequently cardiogenic versus hemangiogenic tissue differentiation. Combined with state-of-the art molecular approaches, Xenopus research continues to provide unique opportunities for current and future pioneering discoveries relevant to cardiovascular biology and medicine.
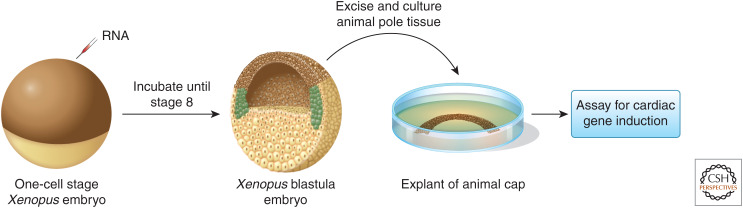
Figure 3.
Schematic of Xenopus animal cap assay. (Left-hand panel) a one-cell-stage embryo injected with mRNA or DNA is then allowed to develop to stage 8 (pre-gastrula) stage embryo, at which time point the ectodermal or animal cap is removed and placed in isolation, cultured, and assayed for the presence, absence, or type of cardiac tissue.
Figure 4.
Schematic outlining role of signaling pathways and inhibition of signaling pathways in regulating cell fate specification in the mesoderm toward cardiac differentiation. (BMP) Bone morphogenetic protein, (FGF) fibroblast growth factor. (Figure based on data in Gilbert and Barresi 2016.)
WIDENING ACCESS TO STEM-CELL(-LIKE) EXPERIMENTS
One further type of Xenopus embryonic explant deserves further mention: the so-called “animal cap” and—specifically for the field of heart development—its use as an “activin cap.” The animal cap is the tissue from the animal pole of the Xenopus embryo, which is removed from the rest of the embryo before gastrulation by straightforward microdissection (Fig. 3). Importantly, this extracted tissue retains pluripotency and can be used in stem-cell approaches inexpensively (Furue and Asashima 2004). Remarkably, treatment of this pluripotent animal cap with activin (resulting in “activin caps”) leads to differentiation of autonomously beating heart tissue (Fig. 3; Kinoshita et al. 2010; Afouda 2012). This heterologous heart tissue differentiation stem-cell(-like) system can be combined with microinjection experiments and transcriptomics analysis to study the regulatory mechanisms driving vertebrate heart development (Afouda et al. 2018).
COMPLEMENTARY APPROACHES USING XENOPUS LAEVIS AND XENOPUS TROPICALIS
The traditional species associated with Xenopus research is X. laevis. It has particularly large embryos, even among Xenopus, and is robust in its husbandry and recovery of normal embryonic development after quite rough experimental manipulation. Its cousin, X. tropicalis, has a simpler genome organization, which benefitted many high-throughput genomic studies and transcriptome analyses (Table 1; Fig. 5; Hellsten et al. 2010; Kashiwagi et al. 2010; Owens et al. 2016). Genome sequencing and analysis has shown that X. laevis is not a tetraploid but rather is an allotetraploid frog possessing two diploid genomes, the L and S genomes, derived from the mating of two distinct ancestral species 17–18 million years ago (Session et al. 2016). Approximately 56% of genes in the present-day X. laevis genome are duplicated between the L and S genomes as a result of the allotetraploidy event (Session et al. 2016). Thus, the two species are very closely related and often can be used interchangeably because most probes work in both species. Possible subtle but consequential differences in regenerative potential in the heart, which are currently debated (see below) may further benefit from this two-species approach (Fig. 4).
Table 1.
High-throughput genomic studies and transcriptome analyses of Xenopus laevis and Xenopus tropicalis
| X. laevis | X. tropicalis | |
|---|---|---|
| Size of adults (♂; ♀) | ∼7 cm; ∼10 cm | ∼3 cm; ∼5 cm |
| Size of embryo (diameter; volume) | ∼1–1.4 mm; ∼1–2 mL | ∼0.6–0.8 mm; ∼0.2–0.5 mL |
| No. of chromosomes | 18 chromosomes | 10 chromosomes |
| Size of genome | 3.1 × 109 bp | 1.7 × 109 bp |
| Generation time | ∼13 months | ∼4 months |
| Brood size | ≥1000 embryos | ≥300 embryos |
X. laevis is allotetraploid, believed to have emerged after hybridization of two X. tropicalis–like diploid ancestors (but not directly X. tropicalis itself). The 18 chromosomes of X. laevis can still be clearly ordered into two subgenomes (chromosomes 1L to 9L and 1S to 9S, respectively, presumably derived from either prehybridization diploid ancestor). Chromosomes 1L to 8L and 1S to 8S in X. laevis correspond to X. tropicalis chromosomes 1 to 8, whereas sequences on X. tropicalis chromosomes 9 and 10 correlate with the ninth chromosomes from either subgenome in X. laevis (therefore now often called chromosomes 9_10L and 9_10S, respectively) (Session et al. 2016). Under ideal laboratory conditions, the generation time of X. laevis has been reduced to 8 months and that of X. tropicalis to 3 months (Hirsch et al. 2002).
Figure 5.
Xenopus laevis flanked by smaller cousin Xenopus tropicalis. (Photo by Atsushi Suzuki.)
KNOCKDOWN, KNOCKOUT, KNOCKIN, AND TRANSGENICS
The use of Xenopus for gain- and loss-of-function experiments has been legendary because of its ease of experimental manipulation. This traditional strength has been complemented with efficient transgenesis approaches (see recent review in Horb et al. 2019) and gene editing (see recent reviews in Tandon et al. 2017; Deniz et al. 2018), including knockin (Aslan et al. 2017). These approaches have already proven particularly powerful in Xenopus, not least because analysis can begin in transheterozygotes of the F0 generation only hours after CRISPR-Cas9 technology application (Blitz et al. 2013) and in targeted mosaics (Naert et al. 2016).
REGENERATION
In their early stages, both X. tropicalis and X. laevis appear able to regenerate heart tissue. However, in adults, X. tropicalis but not X. laevis reportedly undergo cardiac regeneration on injury. Because side-by-side studies have not been conducted, the differences in adults may simply represent differences in protocols or the relative age of the adult frogs. However, they might instead be the result of genetic differences between the two species in abundance of proteins associated with antigen-specific adaptive immunity (Liao et al. 2017, 2018; Marshall et al. 2017, 2018, 2019).
Recently, Federspiel et al. (2019) conducted a direct comparison of the adult cardiac proteomes of four model vertebrates with dual circulatory systems: the pig (Sus scrofa), the mouse (Mus musculus), X. laevis, and X. tropicalis, which were all compared with human. Surprisingly, a significant increase in protein abundance in a vast array of cell-cycle proteins was observed in X. laevis versus X. tropicalis in age-matched female hearts. Thus, one alternative explanation for the ability of adult X. tropicalis to regenerate is their ability to induce cardiac cell-cycle genes on injury. Also noteworthy, the investigators further observed a significant increase in proteins that control metabolic growth including the TOR pathway in X. tropicalis versus X. laevis (Fig. 5). It will be exciting to learn whether these differences underlie the observed differences in the ability of the two species to undergo adult regeneration.
HEART DISEASE MODELING
Xenopus has proven to be a great asset for defining the molecular and cellular underpinnings of several human congenital heart disease (CHD) states, including Holt–Oram disease (Horb and Thomsen 1999; Garg et al. 2003; Brown et al. 2005; Goetz et al. 2006; Puskaric et al. 2010; Herrmann et al. 2011; Steimle et al. 2018), Tbx20-related CHD (Brown et al. 2003, 2005; Stennard et al. 2003; Showell et al. 2006; Mandel et al. 2010; Kaltenbrun et al. 2013), and Nkx2.5-related heart disease (Biben and Harvey 1997; Jiang et al. 1999; Raffin et al. 2000; Small et al. 2000; Jamali et al. 2001a,b; Shiratori et al. 2001; Kasahara et al. 2003; Small and Krieg 2003; Stennard et al. 2003; Bartlett et al. 2007). In addition, it has recently proven to be a powerful tool in defining the molecular source of CHD. One of the central issues facing clinicians in treating CHD is to try to decipher from massive patient data sets which human differences in DNA or RNA sequence represent naturally occurring single-nucleotide polymorphisms (SNPs) and which are somatic mutations in coding regions of potential disease-causing genes (Musunuru et al. 2018). One approach in addressing this issue has come from pioneering work by the Khokha laboratory, which has used a high-throughput CRISPR-Cas9 system to screen potential human mutations in Xenopus for those that affect essential cardiac genes (Fakhro et al. 2011; Boskovski et al. 2013; Duncan and Khokha 2016; Garfinkel and Khokha 2017; Griffin et al. 2018; Kulkarni and Khokha 2018; Kulkarni et al. 2018; Sempou et al. 2018; Robson et al. 2019; Sempou and Khokha 2019). This unbiased, patient-driven gene discovery approach has led to the identification of new genes and protein pathways that may have been missed through other approaches, as well as providing a genetic resource for studying the normal and disease states (Fakhro et al. 2011; Boskovski et al. 2013; Duncan and Khokha 2016; Garfinkel and Khokha 2017; Griffin et al. 2018; Kulkarni and Khokha 2018; Kulkarni et al. 2018; Sempou et al. 2018; Robson et al. 2019; Sempou and Khokha 2019).
CONCLUSION
There is no single best model for studying heart development and disease. The accessibility of embryonic tissues and the bounty of experimental sample material available for state-of-the-art analysis methods undoubtedly make Xenopus an important part of an approach that complements other model systems. Moreover, Xenopus has the unique advantage of combining a matchless suite of novel tools that will allow—in a single organism—integrating systems-level genomic and proteomic analyses with quantitative live imaging of cardiac cell behaviors. These studies can be conducted at the level of single cells, in stem-cell-like and organoid-like explants, or even in whole embryos and animals. By applying this innovative experimental toolbox, Xenopus will continue its impressive track record as an important experimental system for groundbreaking novel discoveries in vertebrate heart development, regeneration, and disease.
ACKNOWLEDGMENTS
We thank Atsushi Suzuki for Figure 5. F.L.C. is supported by a grant from the National Heart, Lung and Blood Institute (HL135007). S.H. is supported by a British Heart Foundation Programme Grant (RG/18/8/33673).
Footnotes
Editors: Benoit G. Bruneau and Paul R. Riley
Additional Perspectives on Heart Development and Disease available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Afouda BA. 2012. Stem-cell-like embryonic explants to study cardiac development, pp. 515–523. Humana, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Afouda BA, Lynch AT, de Paiva Alves E, Hoppler S. 2018. Genome-wide transcriptomics analysis identifies sox7 and sox18 as specifically regulated by gata4 in cardiomyogenesis. Dev Biol 434: 108–120. 10.1016/j.ydbio.2017.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan Y, Tadjuidje E, Zorn AM, Cha SW. 2017. High-efficiency non-mosaic CRISPR-mediated knock-in and indel mutation in F0 Xenopus. Development 144: 2852–2858. 10.1242/dev.152967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett HL, Sutherland L, Kolker SJ, Welp C, Tajchman U, Desmarais V, Weeks DL. 2007. Transient early embryonic expression of Nkx2-5 mutations linked to congenital heart defects in human causes heart defects in Xenopus laevis. Dev Dyn 236: 2475–2484. 10.1002/dvdy.21244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Harvey RP. 1997. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev 11: 1357–1369. 10.1101/gad.11.11.1357 [DOI] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, Cho KW. 2013. Biallelic genome modification in F0 Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51: 827–834. 10.1002/dvg.22719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. 2013. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 504: 456–459. 10.1038/nature12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge RA, Mohun TJ, Amaya E. 2001. A role for BMP signalling in heart looping morphogenesis in Xenopus. Dev Biol 232: 191–203. 10.1006/dbio.2001.0164 [DOI] [PubMed] [Google Scholar]
- Brown DD, Binder O, Pagratis M, Parr BA, Conlon FL. 2003. Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev Genes Evol 212: 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Martz SN, Binder O, Goetz SC, Price BM, Smith JC, Conlon FL. 2005. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development 132: 553–563. 10.1242/dev.01596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HH, Tseng MP. 1957. An experimental analysis of the determination and differentiation of the mesodermal structures of neurula in urodeles. Sci Sin 6: 669–708. [PubMed] [Google Scholar]
- Dale L, Slack JM. 1987. Fate map for the 32-cell stage of Xenopus laevis. Development 99: 527–551. [DOI] [PubMed] [Google Scholar]
- Deimling SJ, Drysdale TA. 2011. Fgf is required to regulate anterior–posterior patterning in the Xenopus lateral plate mesoderm. Mech Dev 128: 327–341. 10.1016/j.mod.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Deniz E, Mis EK, Lane M, Khokha MK. 2018. CRISPR/Cas9 F0 screening of congenital heart disease genes in Xenopus tropicalis. Methods Mol Biol 1865: 163–174. 10.1007/978-1-4939-8784-9_12 [DOI] [PubMed] [Google Scholar]
- Duncan AR, Khokha MK. 2016. Xenopus as a model organism for birth defects—congenital heart disease and heterotaxy. Semin Cell Dev Biol 51: 73–79. 10.1016/j.semcdb.2016.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton JA, Lifton RP, Khokha MK, Brueckner M. 2011. Rare copy number variations in congenital heart disease patients identify unique genes in left–right patterning. Proc Natl Acad Sci 108: 2915–2920. 10.1073/pnas.1019645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federspiel JD, Tandon P, Wilczewski CM, Wasson L, Herring LE, Venkatesh SS, Cristea IM, Conlon FL. 2019. Conservation and divergence of protein pathways in the vertebrate heart. PLoS Biol 17: e3000437 10.1371/journal.pbio.3000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley AC, Mercola M. 2005. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev 19: 387–396. 10.1101/gad.1279405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furue M, Asashima M. 2004. Isolation of pluripotential stem cells from Xenopus embryos. In Handbook of stem cells (ed. Lanza, R), Vol. 1, pp. 483–492. Academic, San Diego. [Google Scholar]
- Garfinkel AM, Khokha MK. 2017. An interspecies heart-to-heart: using Xenopus to uncover the genetic basis of congenital heart disease. Curr Pathobiol Rep 5: 187–196. 10.1007/s40139-017-0142-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. 2003. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424: 443–447. 10.1038/nature01827 [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Barresi MJF. 2016. Developmental biology, 11th ed Sinauer, Sunderland, MA. [Google Scholar]
- Goetz SC, Brown DD, Conlon FL. 2006. TBX5 is required for embryonic cardiac cell cycle progression. Development 133: 2575–2584. 10.1242/dev.02420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JN, Del Viso F, Duncan AR, Robson A, Hwang W, Kulkarni S, Liu KJ, Khokha MK. 2018. RAPGEF5 regulates nuclear translocation of β-catenin. Dev Cell 44: 248–260.e4. 10.1016/j.devcel.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636. 10.1126/science.1183670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann F, Bundschu K, Kühl SJ, Kühl M. 2011. Tbx5 overexpression favors a first heart field lineage in murine embryonic stem cells and in Xenopus laevis embryos. Dev Dyn 240: 2634–2645. 10.1002/dvdy.22776 [DOI] [PubMed] [Google Scholar]
- Hirsch N, Zimmerman LB, Gray J, Chae J, Curran KL, Fisher M, Ogino H, Grainger RM. 2002. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev Dyn. 225: 522–535. 10.1002/dvdy.10188 [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. 1999. Tbx5 is essential for heart development. Development 126: 1739–1751. [DOI] [PubMed] [Google Scholar]
- Horb M, Wlizla M, Abu-Daya A, McNamara S, Gajdasik D, Igawa T, Suzuki A, Ogino H, Noble A; Centre de Ressource Biologique Xenope team in France, et al. 2019. Xenopus resources: transgenic, inbred and mutant animals, training opportunities, and web-based support. Front Physiol 10: 387 10.3389/fphys.2019.00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson AG. 1960. Influences of ectoderm and endoderm on heart differentiation in the newt. Dev Biol 2: 138–154. 10.1016/0012-1606(60)90003-8 [DOI] [PubMed] [Google Scholar]
- Jacobson AG. 1961. Heart determination in the newt. J Exp Zool 146: 139–151. 10.1002/jez.1401460204 [DOI] [PubMed] [Google Scholar]
- Jamali M, Karamboulas C, Rogerson PJ, Skerjanc IS. 2001a. BMP signaling regulates Nkx2-5 activity during cardiomyogenesis. FEBS Lett 509: 126–130. 10.1016/S0014-5793(01)03151-9 [DOI] [PubMed] [Google Scholar]
- Jamali M, Rogerson PJ, Wilton S, Skerjanc IS. 2001b. Nkx2-5 activity is essential for cardiomyogenesis. J Biol Chem 276: 42252–42258. 10.1074/jbc.M107814200 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Drysdale TA, Evans T. 1999. A role for GATA-4/5/6 in the regulation of Nkx2.5 expression with implications for patterning of the precardiac field. Dev Biol 216: 57–71. 10.1006/dbio.1999.9469 [DOI] [PubMed] [Google Scholar]
- Kaltenbrun E, Greco TM, Slagle CE, Kennedy LM, Li T, Cristea IM, Conlon FL. 2013. A Gro/TLE-NuRD corepressor complex facilitatesTbx20-dependent transcriptional repression. J Proteome Res 12: 5395–5409. 10.1021/pr400818c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Ueyama T, Wakimoto H, Liu MK, Maguire CT, Converso KL, Kang PM, Manning WJ, Lawitts J, Paul DL, et al. 2003. Nkx2.5 homeoprotein regulates expression of gap junction protein connexin 43 and sarcomere organization in postnatal cardiomyocytes. J Mol Cell Cardiol 35: 243–256. 10.1016/S0022-2828(03)00002-6 [DOI] [PubMed] [Google Scholar]
- Kashiwagi K, Kashiwagi A, Kurabayashi A, Hanada H, Nakajima K, Okada M, Takase M, Yaoita Y. 2010. Xenopus tropicalis: an ideal experimental animal in amphibia. Exp Anim 59: 395–405. 10.1538/expanim.59.395 [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Ariizumi T, Yuasa S, Miyoshi S, Komazaki S, Fukuda K, Asashima M. 2010. Creating frog heart as an organ: in vitro-induced heart functions as a circulatory organ in vivo. Int J Dev Biol 54: 851–856. 10.1387/ijdb.093036mk [DOI] [PubMed] [Google Scholar]
- Kolker S, Tajchman U, Weeks DL. 2000. Confocal imaging of early heat development in Xenopus laevis. Dev Biol 218: 64–73. 10.1006/dbio.1999.9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SS, Khokha MK. 2018. WDR5 regulates left-right patterning via chromatin-dependent and -independent functions. Development 145, dev159889 10.1242/dev.159889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SS, Griffin JN, Date PP, Liem KF Jr, Khokha MK. 2018. WDR5 stabilizes actin architecture to promote multiciliated cell formation. Dev Cell 46: 595–610.e3. 10.1016/j.devcel.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Dong W, Lv L, Guo H, Yang J, Zhao H, Huang R, Yuan Z, Chen Y, Feng S, et al. 2017. Heart regeneration in adult Xenopus tropicalis after apical resection. Cell Biosci 7: 70 10.1186/s13578-017-0199-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Dong W, Zhao H, Huang R, Qi X, Cai D. 2018. Cardiac regeneration in Xenopus tropicalis and Xenopus laevis: discrepancies and problems. Cell Biosci 8: 32 10.1186/s13578-018-0230-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel EM, Kaltenbrun E, Callis TE, Zeng XX, Marques SR, Yelon D, Wang DZ, Conlon FL. 2010. The BMP pathway acts to directly regulate Tbx20 in the developing heart. Development 137: 1919–1929. 10.1242/dev.043588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Vivien C, Girardot F, Péricard L, Demeneix BA, Coen L, Chai N. 2017. Persistent fibrosis, hypertrophy and sarcomere disorganisation after endoscopy-guided heart resection in adult Xenopus. PLoS ONE 12: e0173418 10.1371/journal.pone.0173418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Girardot F, Demeneix BA, Coen L. 2018. Is adult cardiac regeneration absent in Xenopus laevis yet present in Xenopus tropicalis? Cell Biosci 8: 31 10.1186/s13578-018-0231-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall LN, Vivien CJ, Girardot F, Péricard L, Scerbo P, Palmier K, Demeneix BA, Coen L. 2019. Stage-dependent cardiac regeneration in Xenopus is regulated by thyroid hormone availability. Proc Natl Acad Sci 116: 3614–3623. 10.1073/pnas.1803794116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. 2001. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev 15: 316–327. 10.1101/gad.855501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola M. 1999. Embryological basis for cardiac left–right asymmetry. Semin Cell Dev Biol 10: 109–116. 10.1006/scdb.1998.0280 [DOI] [PubMed] [Google Scholar]
- Mohun TJ, Leong LM. 1999. Heart formation and the heart field in amphibian embryos. In Heart development (ed. Rosenthal HA), pp. 37–49. Academic, San Diego. [Google Scholar]
- Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. 2000. The morphology of heart development in Xenopus laevis. Dev Biol 218: 74–88. 10.1006/dbio.1999.9559 [DOI] [PubMed] [Google Scholar]
- Monnickendam MA, Balls M. 1973. Amphibian organ culture. Experientia 29: 1–17. 10.1007/BF01913222 [DOI] [PubMed] [Google Scholar]
- Moody SA. 1987a. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev Biol 119: 560–578. 10.1016/0012-1606(87)90059-5 [DOI] [PubMed] [Google Scholar]
- Moody SA. 1987b. Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev Biol 122: 300–319. 10.1016/0012-1606(87)90296-X [DOI] [PubMed] [Google Scholar]
- Musunuru K, Bernstein D, Cole FS, Khokha MK, Lee FS, Lin S, McDonald TV, Moskowitz IP, Quertermous T, Sankaran VG, et al. 2018. Functional assays to screen and dissect genomic hits: doubling down on the national investment in genomic research. Circ Genom Precis Med 11: e002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naert T, Colpaert R, Van Nieuwenhuysen T, Dimitrakopoulou D, Leoen J, Haustraete J, Boel A, Steyaert W, Lepez T, Deforce D, et al. 2016. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci Rep 6: 35264 10.1038/srep35264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascone N, Mercola M. 1995. An inductive role for endoderm in Xenopus cardiogenesis. Development 121: 515–523. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD. 1947. Investigations on the regional determination of the central nervous system. J Exp Biol 24: 145–183. [DOI] [PubMed] [Google Scholar]
- Owens NDL, Blitz IL, Lane MA, Patrushev I, Overton JD, Gilchrist MJ, Cho KWY, Khokha MK. 2016. Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep 14: 632–647. 10.1016/j.celrep.2015.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur P, Lasche M, Eisenberg LM, Kuhl M. 2002. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature 418: 636–641. 10.1038/nature00921 [DOI] [PubMed] [Google Scholar]
- Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H, Rappold G. 2010. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum Mol Genet 19: 4625–4633. 10.1093/hmg/ddq393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffin M, Leong LM, Rones MS, Sparrow D, Mohun T, Mercola M. 2000. Subdivision of the cardiac Nkx2.5 expression domain into myogenic and nonmyogenic compartments. Dev Biol 218: 326–340. 10.1006/dbio.1999.9579 [DOI] [PubMed] [Google Scholar]
- Robson A, Makova SZ, Barish S, Zaidi S, Mehta S, Drozd J, Jin SC, Gelb BD, Seidman CE, Chung WK, et al. 2019. Histone H2B monoubiquitination regulates heart development via epigenetic control of cilia motility. Proc Natl Acad Sci 116: 14049–14054. 10.1073/pnas.1808341116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Mercola M. 2001. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev 15: 304–315. 10.1101/gad.855601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempou E, Khokha MK. 2019. Genes and mechanisms of heterotaxy: patients drive the search. Curr Opin Genet Dev 56: 34–40. 10.1016/j.gde.2019.05.003 [DOI] [PubMed] [Google Scholar]
- Sempou E, Lakhani OA, Amalraj S, Khokha, MK. 2018. Candidate heterotaxy gene FGFR4 is essential for patterning of the left-right organizer in Xenopus. Front Physiol 9: 1705 10.3389/fphys.2018.01705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. 2001. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol Cell 7: 137–149. 10.1016/S1097-2765(01)00162-9 [DOI] [PubMed] [Google Scholar]
- Showell C, Christine KS, Mandel EM, Conlon FL. 2006. Developmental expression patterns of Tbx1, Tbx2, Tbx5, and Tbx20 in Xenopus tropicalis. Dev Dyn 235: 1623–1630. 10.1002/dvdy.20714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EM, Krieg PA. 2003. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol 261: 116–131. 10.1016/S0012-1606(03)00306-3 [DOI] [PubMed] [Google Scholar]
- Small EM, Vokes SA, Garriock RJ, Li D, Krieg PA. 2000. Developmental expression of the Xenopus Nkx2-1 and Nkx2-4 genes. Mech Dev 96: 259–262. 10.1016/S0925-4773(00)00400-7 [DOI] [PubMed] [Google Scholar]
- Steimle JD, Rankin SA, Slagle CE, Bekeny J, Rydeen AB, Chan SS, Kweon J, Yang XH, Ikegami K, Nadadur RD, et al. 2018. Evolutionarily conserved Tbx5-Wnt2/2b pathway orchestrates cardiopulmonary development. Proc Natl Acad Sci 115: E10615–E10624. 10.1073/pnas.1811624115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, McDonald LP, Niederreither K, Dolle P, Bruneau BG, et al. 2003. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol 262: 206–224. 10.1016/S0012-1606(03)00385-3 [DOI] [PubMed] [Google Scholar]
- Symes K, Yordán C, Mercola M. 1994. Morphological differences in Xenopus embryonic mesodermal cells are specified as an early response to distinct threshold concentrations of activin. Development 120: 2339–2346. [DOI] [PubMed] [Google Scholar]
- Tandon P, Conlon F, Furlow JD, Horb ME. 2017. Expanding the genetic toolkit in Xenopus: approaches and opportunities for human disease modeling. Dev Biol 426: 325–335. 10.1016/j.ydbio.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NB. 1931. The relation of temperature to the heart rate of the South African frog (Xenopus dactylethra). J Physiol 71: 156–168. 10.1113/jphysiol.1931.sp002723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wittig JG, Münsterberg A. 2019. The chicken as a model organism to study heart development. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Marvin MJ, Gardiner A, Lassar AB, Mercola M, Stern CD, Levin M. 1999. Cerberus regulates left–right asymmetry of the embryonic head and heart. Curr Biol 9: 931–938. 10.1016/S0960-9822(99)80419-9 [DOI] [PubMed] [Google Scholar]