Abstract
Brain disorders, from neurodegenerative to psychiatric disorders, are among the most challenging conditions to study because of the intricate nature of the human brain and the limitations of existing model systems in recapitulating all these intricacies. However, innovations in stem cell technologies now allow us to reprogram patient somatic cells to induced pluripotent stem cells (iPSCs), which can then be differentiated to disease-relevant neural and glial cells. iPSCs are a valuable tool to model brain disorders, as they can be derived from patients with known symptom histories, genetics, and drug-response profiles. Here, we discuss the premise and validity of the iPSC-based in vitro model system and highlight key findings from the most commonly studied neurodegenerative and psychiatric disorders.
Neuropsychiatric and neurodegenerative disorders constitute a bulk of brain disorders, contributing significantly to the global burden of disease. The uniquely human abilities of higher-order thought, symbolic abstract thought, and the ability to generate and share complex ideas emerged along with a large brain. Disorders of the brain are therefore challenging to study as no single model system fully encapsulates the complexity of the human brain, one of the least accessible organs within the body. While postmortem data can provide information regarding the end-stage pathology of a disease, its causes and consequences remain difficult to parse. While animal models provide a complex and live system that enables us to study specific endophenotypes of disorders, they do not fully recapitulate the complex human-specific pathology. Therefore, more model systems are required.
Human-induced pluripotent stem cells (iPSCs) as a model system address many of the issues associated with animal models and postmortem tissue. The reprogramming of somatic cells to a pluripotent state—that is a state capable of generating cells from all germ layers—was first achieved by the overexpression of the transcription factors OCT4, SOX2, KLF4, NANOG, and MYC using integrating viral vectors (Takahashi and Yamanaka 2006). Currently, the field has moved to using a variety of integration-free methods for delivery of these so-called Yamanaka factors for reprogramming to avoid potential host cell genome mutations as a result of transgene integration (Kim et al. 2008; Okita et al. 2008). These integration-free reprogramming methods include nonintegrating viral vectors such as Sendai virus, adenovirus, and nonviral vectors such as episomal DNAs, PiggyBac transposons, synthetic modified messenger RNAs (mRNAs), microRNAs, and recombinant proteins (Shi et al. 2017). Of these, Sendai virus, synthetic mRNAs, and episomal DNAs are most commonly used for generating iPSCs because of their high reprogramming efficiency and wide application to different cell types (Takahashi and Yamanaka 2006). iPSCs were first generated from dermal fibroblasts, which are easily accessible and grow readily in culture. Fibroblasts and blood cells remain the most popular somatic cell types for reprogramming, but several other cells from different lineages have also been used to generate iPSCs (Sohn et al. 2012): keratinocytes, cord blood cells, hepatocytes, mesenchymal stromal cells, urine cells, amniotic/yolk-sac cells, neural stem cells, melanocytes, lymphocytes, adipose tissue-derived cells, and pancreatic B cells as well as malignant cancer cell lines (Sohn et al. 2012).
iPSCs provide a renewable resource for generating disease-relevant cell types, screening drugs and potentially generating cells and tissues to replace damaged tissues. Cellular reprogramming is evolving rapidly in the context of disease modeling, drug screening, and regenerative medicine (Shi et al. 2017). Over the last decade, iPSC technology and relevant phenotypic assays have been used for neurological “disease-in a-dish” modeling, using tissue from patients with the idiopathic disease or with known genetics that could serve as tools for screening therapeutic compounds (Yamanaka 2012). With the right set of signaling cues, iPSCs have been differentiated into countless cell types representing nearly all organ systems. Neural differentiation and patterning protocols facilitate the study of complex neurodegenerative disorders on relatively pure populations of impacted cell types from affected patients, which allows for a powerful reductionist approach to perform detailed analyses on cell-intrinsic properties of monogenic and idiopathic disorders. Further, as a patient's genetic background is controlled for, common cellular phenotypes can be parsed from polygenic disorders (Vadodaria et al. 2018).
Great progress has been made in generating various methods and tools to direct the differentiation of iPSCs to diverse neuronal subtypes. Differentiation protocols involve exposing iPSCs to neural patterning cues and developmental signaling molecules discovered in rodent models. Modulating the combination, concentration, and temporal exposure of key signaling molecules has yielded protocols for generating enriched subtypes of neurons and glia from iPSCs (Liu and Zhang 2011). Thus far, researchers have developed protocols for reliably generating several neural and glial cell types (Fig. 1), including glutamatergic, GABAergic, dopaminergic, cholinergic, serotonergic, motor, astrocytes, microglia, and more (Han et al. 2011; Williams et al. 2012). Recent technologies for examining transcriptional signatures in single cells have led to the discovery of previously unknown subpopulations of cells, highlighting the functional diversity of cells within the brain, and the importance of continuing efforts to generate more disease-relevant cell subpopulations (Lake et al. 2016). A complementary approach for generating cell subpopulations is to transdifferentiate somatic cells to other differentiated cells by overexpressing fate-driving transcription factors. This method does not require reprogramming of somatic cells to a pluripotent state and enables the generation of terminally differentiated cells from somatic cells (Wernig et al. 2010).
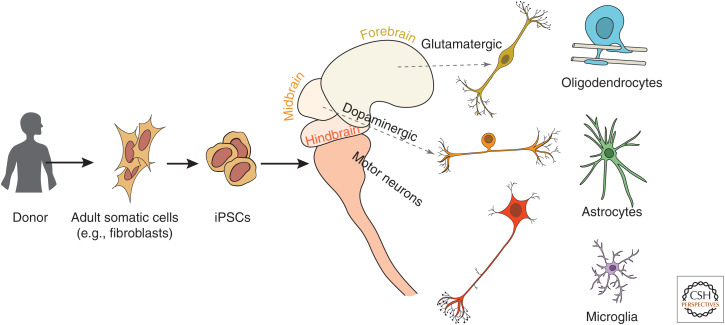
Figure 1.
Schematic depicts the general process of generating differentiated brain cell types from adult humans. (Left to right) Biopsied somatic cells (such as dermal fibroblasts) from patients are reprogrammed to induced pluripotent stem cells (iPSCs) and differentiated to generate cells from different regions of the brain. Neuronal subtypes such as cortical, sensory, and motor neurons can be generated as well as glial cells such as oligodendrocytes, astrocytes, and microglia.
One advantage of patient-derived stem cell modeling is the ability to capture the full genetic component of complex polygenic brain disorders. Although many neuropsychiatric and neurodegenerative diseases are highly heritable suggesting a strong genetic component, large-scale human genome-wide association studies (GWAS) have led to the discovery of multiple disease-associated variants with only low penetrance (Martin et al. 2019). Through iPSC modeling, the entire genetic component can be captured without prior knowledge of the causal variants. Conversely, in the case of highly penetrant disease-associated alleles, iPSCs represent a clear tool for genetic engineering via CRISPR/Cas9, as they are a renewable source of cells. One can directly induce a mutation(s) in a wild-type (WT) genomic background and/or correct a mutation in a disease background to generate isogenic lines. These genetically modified iPSCs can be used for parallel differentiations into multiple neuronal subtypes to understand the contribution of different cell types in disease pathology. Furthermore, they can be used in complex multi-culture systems where only one subtype harbors the mutation to interrogate cell- and non-cell-autonomous effects of cell types on disease phenotypes.
To summarize, stem-cell-based brain disorder modeling encompasses (1) generating iPSCs from individuals with a disorder with or without disease-causing mutation(s) or generating isogenic iPSC lines with the specific mutation introduced, (2) differentiating iPSCs into relevant brain cells and subtypes for (3) examination using assays to probe pathological phenotypes relevant to the disease (Fig. 2). iPSC-based disease modeling enables the study of neurological disorders at the cellular and molecular level and at a resolution not previously possible. Here we discuss the validity, advantages, and limitations of iPSC-based assay systems used in the field and provide an overview of how iPSCs have been utilized to study neurodegenerative and psychiatric disorders. We highlight here recent and mostly consistent findings for Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), amyotrophic lateral sclerosis (ALS), schizophrenia (SCZ), autism spectrum disorders (ASDs), and bipolar disorder (BD).
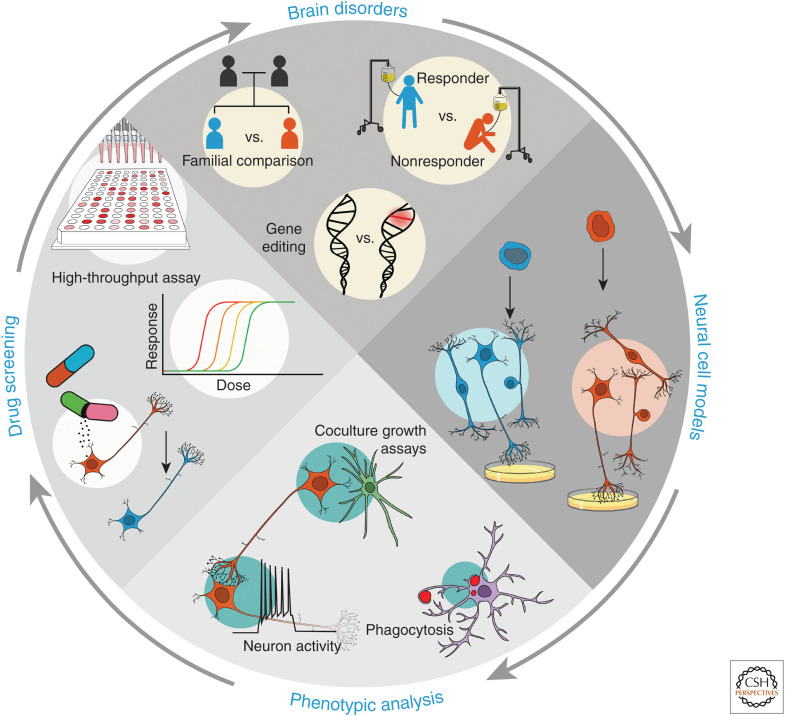
Figure 2.
Key elements in the process of modeling brain disorders using induced pluripotent stem cells (iPSCs). Clockwise from top of the circle (arrows): patient cohorts, generating iPSCs and differentiating neural cells in vitro, studying disease-relevant cellular phenotypes, and high-throughput assays for drug discovery. (Top panel) For comparing healthy versus diseased groups, shown are examples of patient cohorts, such as those within families, or those that are treatment-responsive or -resistant, or comparing cohorts of gene-edited cells containing disease-associated variants. (Right panel) Differentiating iPSCs into diverse disease-relevant neural cell types from different patient cohorts enables in vitro analysis of cellular phenotypes. (Bottom panel) Various cellular assays can be used to examine cellular functions and phenotypes in vitro. For example, neuron–astrocyte interactions via coculture, neuronal activity patterns, and phagocytic properties of microglia can be examined in vitro. (Left panel) Drug-response profiles of patient-cells can be measured in vitro. Patient iPSC-derived cells can be scaled up for high-throughput assays and drug screening based on phenotypic rescue, eventually contributing to therapeutics, cell replacement, and precision medicine for the patient.
VALIDITY OF THE iPSC MODEL SYSTEM
iPSC-derived neurons are a model to study in vivo development. As such, the aphorism, all models are wrong but some are useful, becomes relevant to the discussion. A major goal of modeling is to create a reduced system that simulates the real-world phenomenon. This system should enable the user to manipulate individual components and to test how the distinct parts contribute to the overall complexity of the system. To confidently interpret the results from manipulating such a model system, we first need to understand which aspects of the real-world phenomenon are being accurately represented in the model. In short, how useful is our model?
Over the years, many assays have been utilized to examine whether iPSC-derived cells are similar to developmentally derived neuronal populations of interest. These assays segregate into three main categories: molecular markers, morphology, and function. Staining for well-described molecular markers of specific populations is useful for determining the accuracy and efficiency of a protocol (Tao and Zhang 2016). Importantly, the proportional enrichment of cell-type-specific marker genes has been found to be highly consistent across laboratories, which speaks to the reproducibility of these differentiation methods for generating cell types of interest.
Although marker genes have been relied on for decades to validate the presence of a given cell type both in culture and in vivo, basing validation on the expression of a select set of markers is not infallible. Inaccuracies from the species specificity of marker expression, batch differences in antibodies, functional changes associated with fixation, and even the ubiquity of marker expression across cell types can all negatively impact the precision of marker assays (Fritschy 2008; Zeisel et al. 2018; Hodge et al. 2019). Furthermore, because in practice it is common to use only a few protein markers to distinguish a cell type, it is worth considering whether an experiment should be deemed a failure if the derived cell type has failed to express a single key marker gene. This raises a question: is the presence of defined markers sufficient to determine a cell type? Answering this question requires more information about the functional utility of the marker gene and the reason for its failure. However, we do not always have direct access to this information. Perhaps because of the limitation of previous techniques, biological classification of cell types has been dependent on the expression of a few known markers. With the advent of modern technologies, we can expand our classification to include a higher dimensionality of marker expression. Transcriptome-wide approaches, including microarray, bulk RNA sequencing, and single-cell RNA sequencing, can all be used to perform a high-dimensional, unbiased validation of iPSC-derived cellular models. For example, single-cell RNA sequencing can be used to validate neurons that give rise to heterogeneous, but region-specific cell types, such as hippocampal neurons (Illes et al. 2019).
When assessing the validity of iPSC models, it is important not only to examine the presence of discrete cell types but also to understand their developmental state. Many approaches have been developed to assess the spatiotemporal state of neurons through machine-learning based approaches such as ABAEnrichment, cerebroViz, BrainImageR, and CoNTExT (Stein et al. 2014; Bahl et al. 2016; Grote et al. 2016; Linker et al. 2019). Interestingly, temporal analyses of iPSC-derived neurons consistently show a similarity with neurons during prenatal development, most commonly the second trimester. Indeed, there are multiple signs that iPSC-derived neurons are transcriptionally similar to those in midgestation, including the maintenance of early developmental genes like doublecortin, electrophysiological properties such as a resting membrane potential above −65 mV (Prè et al. 2014), neurons with abortive or single-action potentials (Bardy et al. 2016), and synchronized bursting patterns that mimic early developmental neurons (Khazipov and Luhmann 2006). In an attempt to mature 2D neuronal cultures, advances have been made including coculture with astrocytes (Johnson et al. 2007; Gunhanlar et al. 2018) and modifications of the differentiation media (Bardy et al. 2015). Despite these advances, iPSC-derived neurons continue to exhibit a relatively early developmental state, suggesting an important role for the extracellular matrix and actual time in regulating maturation. Interestingly, while the temporal state should be taken into consideration when interpreting results, iPSC-derived neurons have been found to exhibit disease-associated characteristics even when adult-onset disorders are examined. This finding indicates that, despite the time of disease onset, iPSC-derived neurons may be useful for modeling postnatal or adult neuronal phenotypes (Mertens et al. 2015; Stern et al. 2018; Vadodaria et al. 2019). iPSC-derived neurons may display specific features of mature neurons than either the transcriptomic or physiological signatures indicate; another interpretation is that adult-onset disorders impact neuronal physiology long before behavioral changes are first detected. This possibility underscores the importance of iPSC-disease modeling across neurological disorders independent of the age of onset and speaks to the utility of these models for early diagnostic capabilities.
Cellular assays probing the properties and functions of neural and glial cells are central to discovering phenotypes associated with disease pathology, which may be key to developing better therapeutics in the future. Several cellular functions have been studied with assays, including neural progenitor migration assessed with the scratch assay and neurosphere assays, neuronal activity assessed by imaging, patch-clamp or multielectrode array, dendritic morphology assessed by tracing, and phagocytosis to assess microglial function. Through these assays, iPSC-derived cells have proven to be a valid model system across a range of functional metrics. Importantly, while iPSC models may not completely resemble the in vivo brain, they provide a much-needed simplified and accessible system that can be manipulated to assess downstream phenotypic changes, which are detailed in the sections below. More importantly, iPSC-derived cells serve as a complementary model system to the in vivo rodent model. While rodent studies have provided a wealth of information about basic biology in the setting of a living animal, species-specific differences in development and disease progression confound interpretation and can even send researchers down a path of study that is not applicable to humans. For example, clinical trials based on mouse studies have a notoriously low success rate, with some reports estimating <8% (Mak et al. 2014). To increase these success rates and to better understand the cellular nature of complex human neuronal diseases, human models must be studied. IPSC-models are an attractive system to do so and have the added benefit that they can be derived directly from patients, thereby capturing the complex genetic landscape without the need for prior genetic information of disease-associated loci. In summary, the model is useful.
NEURODEGENERATIVE DISORDERS
Though the field is still young, iPSC-based disease modeling has already demonstrated utility in modeling some of the most widespread neurodegenerative diseases: AD, PD, and ALS. Pathological hallmarks associated with AD, PD, and ALS are present in the neurons generated from iPSCs, despite the reprogramming process and a more fetal transcriptional profile of the resulting neuronal cells. iPSCs generated from familial and sporadic AD patients are most often differentiated to forebrain glutamatergic neurons (85%–90%), with a small proportion of GABAergic neurons (10%–15%) (Israel et al. 2012). Neurons generated from AD iPSCs are electrically active and display pathology consistent with both animal models and human clinical data. Aβ accumulation and increases in toxic Aβ species (Yagi et al. 2011; Israel et al. 2012), tau hyperphosphorylation (Ochalek et al. 2017; Wang et al. 2017), and proteinaceous inclusions (Marchetto et al. 2010; Ross and Akimov 2014) have been commonly noted across multiple studies. In addition to the major pathological hallmarks of AD, multiple studies have noted reactive oxygen species (ROS) and GSK3B activation in AD neurons (Israel et al. 2012; Kondo et al. 2013; Ochalek et al. 2017). To date, over 99% of therapeutics identified as potentially effective for treating AD in preclinical studies have failed in clinical trials (Becker et al. 2008). Therefore, it is crucial to explore new avenues for disease modeling, target discovery, and drug screening. iPSC modeling of AD allows for the generation of large-scale, patient-specific neurons that can serve as tools for high-throughput drug screens. Further, multiple neuronal types can be derived to create an AD profile of a wide range of human neurons. (For a detailed review of AD iPSC modeling, see Mungenast et al. 2016.)
Loss of dopaminergic neurons in the substantia nigra pars compacta underlies the initial onset of the motor dysfunction common to PD (Kalia and Lang 2015). Despite several autosomal-dominant mutations linked to PD, like AD, over 90% of PD cases are idiopathic and lack clear etiology (Li et al. 2018) but have a large known genetic component, making PD an ideal disease to study with iPSC modeling. Common pathological features between idiopathic and genetic cases include intracellular proteinaceous inclusions, termed Lewy bodies, which lead to progressive loss of dopaminergic neurons. Several studies have used PD patient-derived iPSCs to generate midbrain dopaminergic neurons at ∼80% purity (Soldner et al. 2011; Ambasudhan et al. 2014). Numerous studies have reported increased expression/aggregation of the protein α-synuclein (Byers et al. 2011; Chung et al. 2016), ROS (Nguyen et al. 2011; Ryan et al. 2013), and other metabolic and mitochondrial abnormalities (Morais et al. 2014; Sanders et al. 2014; Chung et al. 2016). Interestingly, PD pathology impacts neuronal morphology and neurite outgrowth, a common assay for in vitro disease modeling (Jiang et al. 2012; Flierl et al. 2014; Skibinski et al. 2014). (For a detailed review of iPSC work in PD, see Li et al. 2018.)
Cortical glutamatergic neurons and midbrain dopaminergic neurons are not the only neuronal subtypes susceptible to degenerative processes. Motor neurons in the motor cortex and spinal cord are impacted in several neurodegenerative disorders. One of the most common is ALS (Brown and Al-Chalabi 2017). Upon onset, the disease rapidly progresses, resulting in paralysis and eventually death usually within 4 years of onset (Sances et al. 2016). The most common genetic mutations associated with ALS susceptibility and motor neuron death are C9ORF72, SOD1, TARDP, and FUS. Highly pure motor neurons (>95% HB9+) can be differentiated from iPSCs (Du et al. 2015) with high homeobox domain resolution (Lippmann et al. 2015) from ALS patient iPSCs containing these mutations (Sances et al. 2016). Using this disease-in-a-dish approach, iPSC-based ALS models have recapitulated the pathological hallmarks associated with ALS, including aggregation of SOD1 (Chen et al. 2014), TDP-43 (Fujimori et al. 2018), changes in RNA processing (Donnelly et al. 2013; Sareen et al. 2013), and hyperexcitability (Wainger et al. 2014; Devlin et al. 2015). As with the other proteinaceous neurodegenerative diseases discussed in this review, neuronal morphological changes are often noted. With pathological and functional readouts present in ALS patient motor neurons, drug screening is now possible on bona fide human astrocytes (for a detailed review, see Guo et al. 2017). While the cell-intrinsic properties of motor neurons in ALS represent an important avenue for the development of potential therapeutics, non-cell-autonomous disease progression has been demonstrated in animal models (Boillée et al. 2006). Non-cell-autonomous contributions have also been implicated in AD, PD, and ALS (Lobsiger and Cleveland 2007; Streit et al. 2009; Chai and Kohyama 2019). Moving forward, an important consideration for future iPSC research will be how to appropriately model both cell-autonomous and nonautonomous factors to capture the complexities of neurodegenerative disease etiology.
NEUROPSYCHIATRIC DISORDERS
Unlike neurodegenerative disorders that often display region- or cell-type-specific pathology, neuropsychiatric disorders do not present clear regional etiology. Furthermore, with the significant heterogeneity in symptomatology, neuropsychiatric disorders have been notoriously challenging to study and the lack of novel therapeutics is a testament to this fact. Diagnostic and Statistical Manual of Mental Disorders (DSM)-based diagnoses of psychiatric disorders rely primarily on behavioral, cognitive, and emotional symptoms without the incorporation of biological data. iPSC-based disease modeling may be especially promising for studying psychiatric disorders, allowing us to probe deeper into the human-specific cellular phenotypes. So far, diverse study designs have discovered in vitro phenotypes associated with SCZ, BD, and ASD. Here we discuss consistent findings from studies utilizing iPSC-derived neural and glial cells that give insight into disease pathology.
SCZ is a severe disorder characterized by positive (hallucinations, thought pattern disruption, delusions, movement disorder), negative (affective and daily behavioral abnormalities), and cognitive (memory, attention, executive functioning deficits) symptoms with a strong genetic component. Studies using idiopathic patient iPSC-derived neurons show consistent findings of decreased activity of pan-cortical glutamatergic neurons, as well as hippocampal neurons. This decreased activity coincides with a decrease in neuronal connectivity and increased levels of serotonin, dopamine, and norepinephrine from the same patient cohort-derived neurons (Brennand et al. 2011; Hook et al. 2014; Yu et al. 2014; Sarkar et al. 2018). Similarly, Shao et al. (2019) found SCZ patient-derived cortical interneurons to have reduced neurite arborization and reduced synaptic density, which may further contribute to circuit activity alterations (Shao et al. 2019). While these reports reveal cell-intrinsic differences in patient neurons, it is possible that some of these deficits are the result of non-cell-autonomous differences in patient-derived glial cells, which are also present in neural cultures in vitro. Recent studies with SCZ patient glia uncovered non-cell-autonomous effects as well. Using humanized glia chimeric mice, Windrem et al. (2017) showed premature migration, delayed differentiation and abnormal morphology of patient astrocytes. Another study using in vitro models of synaptic pruning revealed increased synapse elimination in SCZ patient-derived microglia (Sellgren et al. 2019). In genetic studies, deletion of 22q11.2 and 15q11.2 copy-number variants has been found to be prominent risk factors for SCZ (Rees et al. 2014; Marshall et al. 2017). 22q11.2-deleted iPSC-derived cells display decreased capacity for neural differentiation, abnormal neurite growth, and migration as well as abnormal neuron-to-glia ratios. 15q11.2 microdeletion iPSC-derived neural progenitor cells (NPCs) display deficits in adherens junctions and apical polarity (Toyoshima et al. 2016). Similarly, DISC1 (disrupted in schizophrenia 1) and DISC2 are relevant genes discovered in the study of a family where 29 of 69 members were diagnosed with psychiatric illness and were found to have a translocation between these two genes (Blackwood et al. 2001). iPSCs from patients with DISC1 mutations exhibited impaired synaptic vesicle release (Wen et al. 2014) and TALEN- and CRISPR-Cas9-edited iPSCs with DISC1 mutations display increased Wnt signaling and abnormal neural differentiation (Srikanth et al. 2015). These studies indicate that multiple cellular phenotypes are associated with SCZ and potentially lie downstream from diverse genetic causes. Furthermore, unlike in neurodegenerative disorders, disease-associated phenotypes are more difficult to assess in the postmortem brains of psychiatry patients, making iPSC technology a powerful method to study functional and developmental cellular dysfunctions in these diseases.
Another devastating psychiatric disorder, BD, affects 1% of the population worldwide, and is characterized by extreme mood states going from high-energy states known as manic episodes to low mood/energy states known as depressive episodes. Given the complex genetic etiology underlying BD, a majority of iPSC-based studies on BD have utilized cells from idiopathic patients with diverse strategies for patient stratification. In a family-based paradigm, Madison et al. (2015) derived iPSCs from brothers with BD and unaffected parents. A comparison of the two BD children versus unaffected parents revealed decreased progenitor proliferation and neurogenesis associated with altered expression of WNT pathway genes and increased CXCR4 (Madison et al. 2015). Another strategy for patient stratification is based on pharmacological responsiveness. Typically, BD patients are prescribed mood stabilizers such as lithium (Li), but a subset of patients are nonresponsive to treatment. Strikingly, from two separate cohorts of Li-responsive and nonresponsive patients, Mertens et al. (2015) and Stern et al. (2018) showed that BD patient-derived dentate granule-like neurons were hyperexcitable, with alterations in mitochondrial function. This hyperexcitability was rescued by treatment with Li only in Li-responder patient-derived neurons (Mertens et al. 2015). Similarly, Chen et al. (2014) found alterations in calcium signaling pathways in BD patient-derived cortical neurons, which may tie in with hyperexcitability phenotypes. These results highlight common BD-associated phenotypes found across multiple patient cohorts in different study designs. The observed phenotypes may lie downstream from alterations in several molecular pathways. Gene expression studies revealed altered regulation of miRNA-134 which is predicted to target multiple genes known to be risk factors for BD. miRNA-134 was found to be up-regulated in BD-patient-derived neurons and NPCs with enhanced miR-34a expression displayed defects in neuronal differentiation, expression of synaptic proteins, and neuronal morphology (Bavamian et al. 2015). Together, these studies suggest that alterations in neuronal differentiation and activity downstream from diverse signaling pathways may contribute to BD disease pathology.
Autism is called a spectrum disorder because of the immense heterogeneity in the symptoms and severity of patient experience. It is categorized as a neurodevelopmental disorder affecting more than 1% of children in the United States. Patients display impaired social interaction with limited interests and repetitive behaviors. Family and twin studies show a strong genetic component in the etiology of ASD, with an impact both from the common genetic burden and elevated de novo genetic variation (Sanders et al. 2015). Rett syndrome, a monogenic disorder with ASD-like symptoms, is associated with mutations in genes such as MECP2 and CDKL5. Rett syndrome patient iPSC-derived NPCs in a 3D culture system exhibits deficits in NPC migration, and decreased neurite length, spines, synapses, and activity have been observed in both 2D and 3D culture systems (Marchetto et al. 2010; Zhang et al. 2016). Copy-number variations in the 16p11.2 region are associated with neurodevelopmental disorders such as ASD and SCZ. Studies with iPSC-derived neurons revealed increased neuronal soma size and dendrite length in 16p11.2 deletion carriers and opposite phenotypes with 16p11.2 duplication. These phenotypes are in agreement with in vivo phenotypes of patients with 16p11.2 copy-number variants (Deshpande et al. 2017). Similarly, CACNA1C mutations have been associated with Timothy syndrome and cortical neurons derived from patients exhibit abnormal migration and saltation in a 3D iPSC model system (Birey et al. 2017). Interestingly, increased tyrosine hydroxylase and downstream neurotransmitters norepinephrine and dopamine were also found to be higher in Timothy syndrome patient-derived neurons, suggesting potentially divergent cellular mechanisms underlying the syndrome (Paşca et al. 2011). Idiopathic cases of ASD with unknown genetic causes but biological features such as transient megaloencephaly (MEG) also reveal an interesting pattern of correlation between in vitro and in vivo phenotypes (Marchetto et al. 2017). MEG-ASD patient-derived NPCs display increased proliferation, accelerated cell cycle, mildly correlating with patient brain size, accelerated early growth transcriptional networks and subsequent reduction in neuronal synapses, and a shift in differentiation to GABAergic neurons (Mariani et al. 2015; Stern et al. 2018; Schafer et al. 2019).
Taken together, these results show key cellular phenotypes associated with specific genetic and biological features of SCZ, BD, and ASD, indicating that iPSC-based model systems may help uncover convergent mechanisms underlying neuropsychiatric pathology.
CHALLENGES AND FUTURE DIRECTIONS
iPSCs represent a powerful model system for studying human disease. However, it is clear that no single model system is sufficient to fully encapsulate the complex nature of human central nervous system (CNS) disorders. As is so often the case, contributions made by various models over the years are required to make progress and ease patient suffering. With this perspective, it is important to juxtapose the exciting new avenues of research with some of the challenges currently facing the iPSC disease modeling field. In the following, we highlight some of the underinvestigated scientific areas relevant to iPSC disease modeling and point out some limitations to the field as we ask big-picture questions that can move the field forward.
One of the underinvestigated areas of investigation is the role of glial cells in all of the human maladies discussed here. The majority of work on “disease-in-a-dish” modeling has logically focused on the impacted neuronal population for any given disease. Though iPSCs provide access to the cell-intrinsic properties of neurons, disease-associated phenotypes do not always manifest in a clear cell-autonomous manner. The three major glial cell types, astrocytes, microglia, and oligodendrocytes, constitute a large number of the total cells in the human CNS. It is a reflection of the state of glia biology that the relative proportion of glia to neurons is still controversial (Kettenmann and Verkhratsky 2008). Likewise, the relative contributions of glia to neurodegenerative and neuropsychiatric diseases represent a growing interest for researchers and for iPSC-based disease modeling, as some glial cells are the primary source of some neurological disorders (Jones et al. 2018). Recent advances in differentiation protocols have demonstrated that pure populations of astrocytes (Krencik et al. 2011; Santos et al. 2017), microglia (Abud et al. 2017), and oligodendrocytes (Wang et al. 2013) can be generated from iPSCs. Aside from disease contributions, iPSC-derived glia will be instructive for answering fundamental biological questions such as regional heterogeneity among glia populations and for developing complex coculture systems for studying neuron–glia interactions. The combinatorial role and contribution of each cell type can be studied for the first time, opening new avenues for understanding disease progression and developing therapies to target underlying biological causes. Because iPSC cell-type generation is scalable, new and complex screening platforms are starting to show promise for novel therapies to treat the complex disorders of the human CNS.
There is a strong connection between neuronal structure and function and alterations in neuronal morphology have been implicated in most neurodegenerative and neuropsychiatric disorders discussed here. Current 2D substrates are limited and, without more organ-specific plating substrates, it is difficult to conclude whether these disease states cause the observed morphological differences or alternatively make them less adaptable to a 2D in vitro substrate. In general, more complex culture systems will contribute to progress and complement iPSC disease modeling. Currently, efforts are underway to generate 3D organoids and in vitro blood–brain barrier models to better resemble an in vivo extracellular matrix. Advances in microfluidic devices and 3D printing with compatible biomaterials also represent future avenues for research for modeling complex neural circuitry.
In the context of age-associated diseases, how can we study the impact of biological aging on a cell in a dish and its relevance to pathologic changes associated with diseases? Even though the somatic cell type of origin can be identified in an unbiased fashion at early passages after reprogramming cells to a pluripotent state, the reprogramming procedure causes drastic epigenetic changes. Reports indicate a significant loss of epigenetic memory in iPSCs, and epigenetic marks associated with age are generally lost (Mertens et al. 2018). Transdifferentiation or direct conversion technologies enable the conversion of aged somatic cells directly to neurons, thereby maintaining elements of age in transdifferentiated neurons. It is likely that the combination of iPSC and transdifferentiation technologies may provide insights into the contribution of age to neurodegenerative phenotypes helping parse potential differences between biological aging and disease.
CONCLUDING REMARKS
iPSC models provide a much-needed simplified, accessible, and flexible system that can be manipulated to assess the genetic basis of disease-associated phenotypic changes (Fig. 2). Though they may not completely resemble the in vivo brain, iPSCs complement large-scale genetic studies, and other animal- and cell-based model systems to help unravel the complex disorders of the human brain. Furthermore, iPSC-based disease models are scalable, with obvious implications for target discovery, drug screening, and cell-replacement therapies. For the first time, human brain disorders can be studied with patient genetic background controlled for, providing a powerful platform for the study of cellular functions and disease phenotypes.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health (NIH) (P30 CA014195, U01 MH106882, National Cooperative Reprogrammed Cell Research Groups (NCRCRG), Grant Nos. U19 MH106434, R01 AG057706, R01 AG056511, R01 MH-114030, and R01 AG056306), the American Heart Association/Paul G. Allen Frontiers Group Initiative in Brain Health and Cognitive Impairment (19PABHI34610000), the Leona M. and Harry B. Helmsley Charitable Trust Grant #2017-PG-MED001, the Grace Foundation, the JPB Foundation, Annette C. Merle-Smith, the Robert and Mary Jane Engman Foundation, Lynn and Edward Streim, the Takeda-Sanford Consortium Innovation Alliance grant program (Takeda Pharmaceutical Company), and the Ray and Dagmar Dolby Family Fund. K.C.V. was supported by a Swiss National Science Foundation Outgoing Fellowship. We thank M.L. Gage for editorial comments on the manuscript.
Footnotes
Editors: Cristina Lo Celso, Kristy Red-Horse, and Fiona M. Watt
Additional Perspectives on Stem Cells: From Biological Principles to Regenerative Medicine available at www.cshperspectives.org
REFERENCES
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, et al. 2017. IPSC-derived human microglia-like cells to study neurological diseases. Neuron 94: 278–293.e9. 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R, Dolatabadi N, Nutter A, Masliah E, Mckercher SR, Lipton SA. 2014. Potential for cell therapy in Parkinson's disease using genetically programmed human embryonic stem cell-derived neural progenitor cells. J Comp Neurol 522: 2845–2856. 10.1002/cne.23617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl E, Koomar T, Michaelson JJ. 2016. CerebroViz: an R package for anatomical visualization of spatiotemporal brain data. Bioinformatics 33: btw726 10.1093/bioinformatics/btw726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, Gorris M, Galet B, Palomares V, Brown J, et al. 2015. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci 112: E2725–E2734. 10.1073/pnas.1504393112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy C, van den Hurk M, Kakaradov B, Erwin JA, Jaeger BN, Hernandez RV, Eames T, Paucar AA, Gorris M, Marchand C, et al. 2016. Predicting the functional states of human IPSC-derived neurons with single-cell RNA-Seq and electrophysiology. Mol Psychiatry 21: 1573–1588. 10.1038/mp.2016.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, Madison JM, Zhou F, Rueckert EH, Barker D, et al. 2015. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol Psychiatry 20: 573–584. 10.1038/mp.2014.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RE, Greig NH, Giacobini E. 2008. Why do so many drugs for Alzheimer's disease fail in development? Time for new methods and new practices? J Alzheimers Dis 15: 303–325. 10.3233/JAD-2008-15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, et al. 2017. Assembly of functionally integrated human forebrain spheroids. Nature 545: 54–59. 10.1038/nature22330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St. Clair DM, Porteous DJ, Muir WJ. 2001. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet 69: 428–433. 10.1086/321969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillée S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. 2006. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312: 1389–1392. 10.1126/science.1123511 [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. 2011. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473: 221–225. 10.1038/nature09915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RH, Al-Chalabi A. 2017. Amyotrophic lateral sclerosis. New Engl J Med 377: 162–172. 10.1056/NEJMra1603471 [DOI] [PubMed] [Google Scholar]
- Byers B, Cord B, Nguyen HN, Schüle B, Fenno L, Lee PC, Deisseroth K, Langston JW, Pera RR, Palmer TD. 2011. SNCA triplication Parkinson's patient's IPSC-derived DA neurons accumulate α-synuclein and are susceptible to oxidative stress. PLoS ONE 6: e26159 10.1371/journal.pone.0026159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai MC, Kohyama J. 2019. Non-cell-autonomous neurotoxicity in Parkinson's disease mediated by astroglial α-synuclein. Stem Cell Rep 12: 183–185. 10.1016/j.stemcr.2019.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, DeLong CJ, Bame M, Rajapakse I, Herron TJ, McInnis MG, O'Shea KS. 2014. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiatry 4: e375 10.1038/tp.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SY, Kishinevsky S, Mazzulli JR, Graziotto J, Mrejeru A, Mosharov EV, Puspita L, Valiulahi P, Sulzer D, Milner TA, et al. 2016. Parkin and PINK1 patient IPSC-derived midbrain dopamine neurons exhibit mitochondrial dysfunction and α-synuclein accumulation. Stem Cell Rep 7: 664–677. 10.1016/j.stemcr.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A, Yadav S, Dao DQ, Wu ZY, Hokanson KC, Cahill MK, Wiita AP, Jan YN, Ullian EM, Weiss LA. 2017. Cellular phenotypes in human IPSC-derived neurons from a genetic model of autism spectrum disorder. Cell Rep 21: 2678–2687. 10.1016/j.celrep.2017.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AC, Burr K, Borooah S, Foster JD, Cleary EM, Geti I, Vallier L, Shaw CE, Chandran S, Miles GB. 2015. Human IPSC-derived motoneurons harbouring TARDBP or C9ORF72 ALS mutations are dysfunctional despite maintaining viability. Nat Commun 6: 5999 10.1038/ncomms6999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. 2013. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80: 415–428. 10.1016/j.neuron.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZW, Chen H, Liu H, Lu J, Qian K, Huang CL, Zhong X, Fan F, Zhang SC. 2015. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun 6: 6626 10.1038/ncomms7626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierl A, Oliveira LM, Falomir-Lockhart LJ, Mak SK, Hesley J, Soldner F, Arndt-Jovin DJ, Jaenisch R, Langston JW, Jovin TM, et al. 2014. Higher vulnerability and stress sensitivity of neuronal precursor cells carrying an α-synuclein gene triplication. PLoS ONE 9: e112413 10.1371/journal.pone.0112413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy J. 2008. Is my antibody-staining specific? How to deal with pitfalls of immunohistochemistry. Eur J Neurosci 28: 2365–2370. 10.1111/j.1460-9568.2008.06552.x [DOI] [PubMed] [Google Scholar]
- Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, Hadano S, Aoki M, Saya H, Sobue G, et al. 2018. Modeling sporadic ALS in IPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med 24: 1579–1589. 10.1038/s41591-018-0140-5 [DOI] [PubMed] [Google Scholar]
- Grote S, Prüfer K, Kelso J, Dannemann M. 2016. ABAEnrichment: an R package to test for gene set expression enrichment in the adult and developing human brain. Bioinformatics 32: 3201–3203. 10.1093/bioinformatics/btw392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunhanlar N, Shpak G, van der Kroeg M, Gouty-Colomer LA, Munshi ST, Lendemeijer B, Ghazvini M, Dupont C, Hoogendijk WJG, Gribnau J, et al. 2018. A simplified protocol for differentiation of electrophysiologically mature neuronal networks from human induced pluripotent stem cells. Mol Psychiatry 23: 1336–1344. 10.1038/mp.2017.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fumagalli L, Prior R, van den Bosch L. 2017. Current advances and limitations in modeling ALS/FTD in a dish using induced pluripotent stem cells. Front Neurosci 11: 671 10.3389/fnins.2017.00671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SSW, Williams LA, Eggan KC. 2011. Constructing and deconstructing stem cell models of neurological disease. Neuron 70: 626–644. 10.1016/j.neuron.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. 2019. Conserved cell types with divergent features in human versus mouse cortex. Nature 573: 61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook V, Brennand KJ, Kim Y, Toneff T, Funkelstein L, Lee KC, Ziegler M, Gage FH. 2014. Human IPSC neurons display activity-dependent neurotransmitter secretion: aberrant catecholamine levels in schizophrenia neurons. Stem Cell Rep 3: 531–538. 10.1016/j.stemcr.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illes J, Weiss S, Bains J, Chandler JA, Conrod P, De Koninck Y, Fellows LK, Groetzinger D, Racine E, Robillard JM, et al. 2019. A neuroethics backbone for the evolving Canadian brain research strategy. Neuron 101: 370–374. 10.1016/j.neuron.2018.12.021 [DOI] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, et al. 2012. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature 482: 216–220. 10.1038/nature10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Ren Y, Yuen EY, Zhong P, Ghaedi M, Hu Z, Azabdaftari G, Nakaso K, Yan Z, Feng J. 2012. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat Commun 3: 668 10.1038/ncomms1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Weick JP, Pearce RA, Zhang SC. 2007. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J Neurosci 27: 3069–3077. 10.1523/jneurosci.4562-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JR, Kong L, Hanna MG 4th, Hoffman B, Krencik R, Bradley R, Hagemann T, Choi J, Doers M, Dubovis M, et al. 2018. Mutations in GFAP disrupt the distribution and function of organelles in human astrocytes. Cell Rep 25: 947–958.e4. 10.1016/j.celrep.2018.09.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, Lang AE. 2015. Parkinson's disease. Lancet 386: 896–912. 10.1016/S0140-6736(14)61393-3 [DOI] [PubMed] [Google Scholar]
- Kettenmann H, Verkhratsky A. 2008. Neuroglia: the 150 years after. Trends Neurosci 31: 653–659. 10.1016/j.tins.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Khazipov R, Luhmann HJ. 2006. Early patterns of electrical activity in the developing cerebral cortex of humans and rodents. Trends Neurosci 29: 414–418. 10.1016/j.tins.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. 2008. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454: 646–650. 10.1038/nature07061 [DOI] [PubMed] [Google Scholar]
- Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, et al. 2013. Modeling Alzheimer's disease with IPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell 12: 487–496. 10.1016/j.stem.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Krencik R, Weick JP, Liu Y, Zhang ZJ, Zhang SC. 2011. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol 29: 528–534. 10.1038/nbt.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. 2016. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352: 1586–1590. 10.1126/science.aaf1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jiang H, Zhang B, Feng J. 2018. Modeling Parkinson's disease using patient-specific induced pluripotent stem cells. J Parkinsons Dis 8: 479–493. 10.3233/JPD-181353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker SB, Hsu JY, Pfaff A, Amatya D, Ko SM, Voter S, Wong Q, Gage FH. 2019. BrainImageR: spatiotemporal gene set analysis referencing the human brain. Bioinformatics 35: 343–345. 10.1093/bioinformatics/bty618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann ES, Williams CE, Ruhl DA, Estevez-Silva MC, Chapman ER, Coon JJ, Ashton RS. 2015. Deterministic HOX patterning in human pluripotent stem cell-derived neuroectoderm. Stem Cell Rep 4: 632–644. 10.1016/j.stemcr.2015.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang SC. 2011. Specification of neuronal and glial subtypes from human pluripotent stem cells. Cell Mol Life Sci 68: 3995–4008. 10.1007/s00018-011-0770-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobsiger CS, Cleveland DW. 2007. Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat Neurosci 10: 1355–1360. 10.1038/nn1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison JM, Zhou F, Nigam A, Hussain A, Barker DD, Nehme R, van der Ven K, Hsu J, Wolf P, Fleishman M, et al. 2015. Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and MRNA expression abnormalities. Mol Psychiatry 20: 703–717. 10.1038/mp.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak IW, Evaniew N, Ghert M. 2014. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res 6: 114–118. [PMC free article] [PubMed] [Google Scholar]
- Marchetto MCN, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. 2010. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143: 527–539. 10.1016/j.cell.2010.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria K, Beltrao-Braga P, Trujillo CA, Mendes APD, Padmanabhan K, et al. 2017. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry 22: 820–835. 10.1038/mp.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, et al. 2015. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162: 375–390. 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, et al. 2017. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49: 27–35. 10.1038/ng.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Daly MJ, Robinson EB, Hyman SE, Neale BM. 2019. Predicting polygenic risk of psychiatric disorders. Biol Psychiatry 86: 97–109. 10.1016/j.biopsych.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, Zheng Y, Diffenderfer KE, Zhang J, Soltani S, et al. 2015. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527: 95–99. 10.1038/nature15526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Reid D, Lau S, Kim Y, Gage FH. 2018. Aging in a dish: IPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu Rev Genet 52: 271–293. 10.1146/annurev-genet-120417-031534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais VA, Haddad D, Craessaerts K, De Bock PJ, Swerts J, Vilain S, Aerts L, Overbergh L, Grünewald A, Seibler P, et al. 2014. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 344: 203–207. 10.1126/science.1249161 [DOI] [PubMed] [Google Scholar]
- Mungenast AE, Siegert S, Tsai LH. 2016. Modeling Alzheimer's disease with human induced pluripotent stem (IPS) cells. Mol Cell Neurosci 73: 13–31. 10.1016/j.mcn.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schüle B, Dolmetsch RE, Langston W, et al. 2011. LRRK2 mutant IPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell 8: 267–280. 10.1016/j.stem.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochalek A, Mihalik B, Avci HX, Chandrasekaran A, Téglási A, Bock I, Giudice ML, Táncos Z, Molnár K, László L, et al. 2017. Neurons derived from sporadic Alzheimer's disease IPSCs reveal elevated TAU hyperphosphorylation, increased amyloid levels, and GSK3B activation. Alzheimers Res Ther 9: 90 10.1186/s13195-017-0317-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. 2008. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322: 949–953. 10.1126/science.1164270 [DOI] [PubMed] [Google Scholar]
- Paşca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Paşca AM, Cord B, Palmer TD, Chikahisa S, Nishino S, et al. 2011. Using IPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med 17: 1657–1662. 10.1038/nm.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prè D, Nestor MW, Sproul AA, Jacob S, Koppensteiner P, Chinchalongporn V, Zimmer M, Yamamoto A, Noggle SA, Arancio O. 2014. A time course analysis of the electrophysiological properties of neurons differentiated from human induced pluripotent stem cells (IPSCs). PLoS ONE 9: e103418 10.1371/journal.pone.0103418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Kirov G, Sanders A, Walters JT, Chambert KD, Shi J, Szatkiewicz J, O'Dushlaine C, Richards AL, Green EK, et al. 2014. Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry 19: 37–40. 10.1038/mp.2013.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Akimov SS. 2014. Human-induced pluripotent stem cells: potential for neurodegenerative diseases. Hum Mol Genet 23: R17–R26. 10.1093/hmg/ddu204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, et al. 2013. Isogenic human IPSC Parkinson's model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell 155: 1351–1364. 10.1016/j.cell.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances S, Bruijn LI, Chandran S, Eggan K, Ho R, Klim JR, Livesey MR, Lowry E, Macklis JD, Rushton D, et al. 2016. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci 19: 542–553. 10.1038/nn.4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders LH, Laganière J, Cooper O, Mak SK, Vu BJ, Huang YA, Paschon DE, Vangipuram M, Sundararajan R, Urnov FD, et al. 2014. LRRK2 mutations cause mitochondrial DNA damage in IPSC-derived neural cells from Parkinson's disease patients: reversal by gene correction. Neurobiol Dis 62: 381–386. 10.1016/j.nbd.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. 2015. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87: 1215–1233. 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Vadodaria KC, Jaeger BN, Mei A, Lefcochilos-Fogelquist S, Mendes APD, Erikson G, Shokhirev M, Randolph-Moore L, Fredlender C, et al. 2017. Differentiation of inflammation-responsive astrocytes from glial progenitors generated from human induced pluripotent stem cells. Stem Cell Rep 8: 1757–1769. 10.1016/j.stemcr.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. 2013. Targeting RNA foci in IPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 5: 208ra149 10.1126/scitranslmed.3007529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Mei A, Paquola ACM, Stern S, Bardy C, Klug JR, Kim S, Neshat N, Kim HJ, Ku M, et al. 2018. Efficient generation of CA3 neurons from human pluripotent stem cells enables modeling of hippocampal connectivity in vitro. Cell Stem Cell 22: 684–697.e9. 10.1016/j.stem.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ST, Paquola ACM, Stern S, Gosselin D, Ku M, Pena M, Kuret TJM, Liyanage M, Mansour AA, Jaeger BN, et al. 2019. Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat Neurosci 22: 243–255. 10.1038/s41593-018-0295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, Fu T, Worringer K, Brown HE, Wang J, et al. 2019. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci 22: 374–385. 10.1038/s41593-018-0334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Noh H, Bin Kim W, Ni P, Nguyen C, Cote SE, Noyes E, Zhao J, Parsons T, Park JM, et al. 2019. Dysregulated protocadherin-pathway activity as an intrinsic defect in induced pluripotent stem cell–derived cortical interneurons from subjects with schizophrenia. Nat Neurosci 22: 229–242. 10.1038/s41593-018-0313-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, Yamanaka S. 2017. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16: 115–130. 10.1038/nrd.2016.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G, Nakamura K, Cookson MR, Finkbeiner S. 2014. Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J Neurosci 34: 418–433. 10.1523/jneurosci.2712-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn YD, Han JW, Yoon Ys. 2012. Generation of induced pluripotent stem cells from somatic cells. Prog Mol Biol Transl Sci 111: 1–26. 10.1016/B978-0-12-398459-3.00001-0 [DOI] [PubMed] [Google Scholar]
- Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. 2011. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146: 318–331. 10.1016/j.cell.2011.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth P, Han K, Callahan DG, Makovkina E, Muratore CR, Lalli MA, Zhou H, Boyd JD, Kosik KS, Selkoe DJ, et al. 2015. Genomic DISC1 disruption in HiPSCs alters wnt signaling and neural cell fate. Cell Rep 12: 1414–1429. 10.1016/j.celrep.2015.07.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernández IA, Marchetto MC, Baker DK, Lu D, Hinman CR, Lowe JK, et al. 2014. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron 83: 69–86. 10.1016/j.neuron.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S, Santos R, Marchetto MC, Mendes APD, Rouleau GA, Biesmans S, Wang QW, Yao J, Charnay P, Bang AG, et al. 2018. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol Psychiatry 23: 1453–1465. 10.1038/mp.2016.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit WJ, Braak H, Xue QS, Bechmann I. 2009. Dystrophic (senescent) rather than activated microglial cells are associated with tau pathology and likely precede neurodegeneration in Alzheimer's disease. Acta Neuropathol 118: 475–485. 10.1007/s00401-009-0556-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tao Y, Zhang SC. 2016. Neural subtype specification from human pluripotent stem cells. Cell Stem Cell 19: 573–586. 10.1016/j.stem.2016.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima M, Akamatsu W, Okada Y, Ohnishi T, Balan S, Hisano Y, Iwayama Y, Toyota T, Matsumoto T, Itasaka N, et al. 2016. Analysis of induced pluripotent stem cells carrying 22q11.2 deletion. Transl Psychiatry 6: e934 10.1038/tp.2016.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadodaria KC, Amatya DN, Marchetto MC, Gage FH. 2018. Modeling psychiatric disorders using patient stem cell-derived neurons: a way forward. Genome Med 10: 1 10.1186/s13073-017-0512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadodaria KC, Ji Y, Skime M, Paquola A, Nelson T, Hall-Flavin D, Fredlender C, Heard KJ, Deng Y, Le AT, et al. 2019. Serotonin-induced hyperactivity in SSRI-resistant major depressive disorder patient-derived neurons. Mol Psychiatry 24: 795–807. 10.1038/s41380-019-0363-y [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041. 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, et al. 2014. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep 7: 1–11. 10.1016/j.celrep.2014.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Bates J, Li X, Schanz S, Chandler-Militello D, Levine C, Maherali N, Studer L, Hochedlinger K, Windrem M, et al. 2013. Human IPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell 12: 252–264. 10.1016/j.stem.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ward ME, Chen R, Liu K, Tracy TE, Chen X, Xie M, Sohn PD, Ludwig C, Meyer-Franke A, et al. 2017. Scalable production of IPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep 9: 1221–1233. 10.1016/j.stemcr.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, et al. 2014. Synaptic dysregulation in a human IPS cell model of mental disorders. Nature 515: 414–418. 10.1038/nature13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LA, Davis-Dusenbery BN, Eggan KC. 2012. SnapShot: directed differentiation of pluripotent stem cells. Cell 149: 1174–1174.e1. 10.1016/j.cell.2012.05.015 [DOI] [PubMed] [Google Scholar]
- Windrem MS, Osipovitch M, Liu Z, Bates J, Chandler-Militello D, Zou L, Munir J, Schanz S, McCoy K, Miller RH, et al. 2017. Human IPSC glial mouse chimeras reveal glial contributions to schizophrenia. Cell Stem Cell 21: 195–208.e6. 10.1016/j.stem.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. 2011. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum Mol Genet 20: 4530–4539. 10.1093/hmg/ddr394 [DOI] [PubMed] [Google Scholar]
- Yamanaka S. 2012. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10: 678–684. 10.1016/j.stem.2012.05.005 [DOI] [PubMed] [Google Scholar]
- Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, Mei A, McHenry L, Lisuk D, Grasmick JM, et al. 2014. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Rep 2: 295–310. 10.1016/j.stemcr.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. 2018. Molecular architecture of the mouse nervous system. Cell 174: 999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZN, Freitas BC, Qian H, Lux J, Acab A, Trujillo CA, Herai RH, Nguyen Huu VA, Wen JH, Joshi-Barr S, et al. 2016. Layered hydrogels accelerate IPSC-derived neuronal maturation and reveal migration defects caused by MeCP2 dysfunction. Proc Natl Acad Sci 113: 3185–3190. 10.1073/pnas.1521255113 [DOI] [PMC free article] [PubMed] [Google Scholar]