Abstract
Background:
In burn patients, wound healing is often accompanied by hypertrophic scar (HS) development, resulting in both functional and aesthetic problems. HSs are characterised by abundant presence of myofibroblasts that contribute to overproduction of extracellular matrix (ECM) that is regulated by the TGF-β signalling pathway. Studies have shown that inhibition of TGF-β receptors in fibrotic diseases reduces the fibrotic load. In the present study, we aim to inactivate ALK5, also known as TGF-β receptor I, in human HS fibroblasts by exon skipping using antisense oligonucleotides (AONs).
Methods:
HS biopsies were used to isolate and set up fibroblast monocultures. AONs targeting ALK5 were supplemented to the fibroblast cultures to induce exon skipping, while pharmacological ALK5 inhibition was induced using SB431542. AON delivery in HS fibroblasts was examined using immunofluorescence (IF), while TGF-β signalling downstream targets, such as Smad2/3, PAI-1, ACTA2, COL1A1 and COL3A1, were analysed using touchdown polymerase chain reaction (PCR), quantitative PCR (qPCR), IF or western blotting.
Results:
Our data clearly demonstrate that AONs were successfully delivered in the nuclei of HS fibroblasts and that functional exon skipping of ALK5 took place as confirmed with touchdown PCR and qPCR. In addition, exon skipping affected the expression of ECM-related genes, such as type I/III collagens, PAI-1 and CCN2. Moreover, AON treatment did not affect the migration of HS fibroblasts in a model for wound healing.
Conclusion:
Exon skipping is a promising tool to modulate the TGF-β signalling pathway in HS. This would open a therapeutic window for the treatment of patients suffering from HSs.
Keywords: Hypertrophic scars, (myo)fibroblasts, TGF-β, ALK5, exon skipping, antisense oligonucleotides
Lay Summary
In this research article, we describe the effect of exon skipping, a type of gene therapy, on fibroblasts that were isolated from hypertrophic scar biopsies. In these fibroblasts, we targeted the overactivated molecular pathway involved in the development of hypertrophic scars. Furthermore, this approach has been successfully studied in different diseases that show the same overactive molecular pathway. It is therefore of great interest to assess the effects of exon skipping in hypertrophic scars.
The biopsies were retrieved from excisions performed on patients that developed post-burn hypertrophic scars. Upon arrival, the hypertrophic scar biopsies were prepared and the fibroblasts were isolated and put in culture on a Petri dish. All treatments were then added to the culture medium of the fibroblasts.
In this study, we found that exon skipping was able to affect the main molecular pathway that is involved in the development of hypertrophic scars. Further, since this specific molecular pathway is of great importance in normal wound healing, we determined the effect of exon skipping on the migration of these fibroblasts. By simulating wound healing in a so-called scratch assay, we found that exon skipping does not affect fibroblast migration.
Altogether, the present study shows that exon skipping has great potential as a novel treatment option in the management of hypertrophic scars. Additional research is warranted before we can start clinical trials.
Background
The healing of burn wounds is often followed by the development of hypertrophic scars (HSs), with a prevalence of 70%, resulting in functional and aesthetic problems.1,2 HSs are characterised by excessive deposition of extracellular matrix (ECM) components (e.g. type I and III collagens) produced by myofibroblasts (MFs), which are abundantly present in HSs and other fibrotic diseases.3 The transforming growth factor (TGF)-β signalling pathway plays a central role in the activity of MFs. The TGF-β signalling pathway regulates multiple processes, such as embryonic development, homeostasis, wound healing, chemotaxis and cell cycle control.4,5 In a full thickness wound, inflicted by burn injury, fibroblasts migrate to the wound and differentiate into activated MFs and express α-smooth muscle actin (αSMA), enabling contraction and closure of the wound.3
The differentiation of MFs is largely regulated by TGF-β signalling.6 In short, under normal wound-healing conditions, activated TGF-β binds to the TGF-β receptor type II (TGFβRII), which then forms a heteromeric complex and activates TGF-β receptor type I (TGFβRI, also known as activin like kinase 5, ALK5) by phosphorylation. The signal is then propagated by Smad2/3/4 complex and induces subsequent transcription of ECM-related genes such as type I and III collagens, plasminogen activator inhibitor-1 (PAI-1) and connective tissue growth factor (CTGF). Once the wound is closed and remodelling of the collagen fibres is completed, the MFs enter apoptosis and a scar is formed. However, under pathological conditions the MFs become insensitive to the apoptotic signals and continue to proliferate, producing excessive amounts of ECM components and αSMA. This results in a thickened scar showing disorganised collagen alignment and severe contraction, hence a HS.7,8
To date, several treatment options are available, such as surgery, corticoid steroid injections, laser therapy, pressure therapy, cryotherapy and radiation.9 However, in most cases, the HSs become resistant to therapy and treatment has to be repeated.10 New developments in the treatment of HS have been reported (e.g. interferon, 5-Fluorouracil and SB431542); however, these new treatment options showed to have adverse effects or unstable pharmacokinetics in vivo.11,12 Therefore, it is warranted to search for new treatments options in order to treat and/or prevent the development of HSs in patients with burns.
Previous studies have shown that targeting ALK5 in fibrotic diseases (e.g. renal fibrosis and Dupuytren’s disease [DD]), using exon skipping, is able to reduce the fibrotic load. In these studies, the ligand-binding domain coding exon 2 of ALK5 was specifically targeted using antisense oligonucleotides (AONs). This approach ensures that a specific exon is excluded from mature messenger RNA (mRNA). The intact open reading frame is translated, which results in a protein lacking a specific peptide sequence.13,14 The aim of this study was to investigate modulation of TGF-β signalling in HS fibroblasts, targeting ALK5 by exon skipping. In the present study, we have induced exon skipping of ALK5 in HS-derived monocultures and assessed the anti-fibrotic effect. We show that ALK5 exon 2 is successfully skipped in HS-derived monocultures, and that this inhibition affects the expression of ECM-related genes and downstream targets of the TGF-β signalling pathway as assessed on mRNA and protein level. Furthermore, exon skipping does not affect fibroblast migration. In conclusion, exon skipping is a promising tool in the treatment and management of HS and could open a new window for the treatment of scars in burn patients.
Materials and methods
Isolation, cell culture
Excised HS biopsies were provided by the Red Cross Hospital, Beverwijk, The Netherlands (Table 1). Upon arrival, the biopsies were cut in equal pieces of 0.5 × 0.5 mm and incubated overnight in dispase II at 4 °C and subsequently incubated for 30 min at 37 °C. Next, tissue samples were treated with Collagenase (Invitrogen, Breda, The Netherlands) and Dispase (Roche Diagnostics, Almere, The Netherlands), mixed in a 3:1 ratio for 2 h at 37 °C, and poured through a cell strainer (Falcon® 70 µm cell strainer; Corning Inc., Corning, NY, USA). The isolated fibroblasts were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco/Invitrogen, Breda, The Netherlands) supplemented with 5% fetal bovine serum (FBS; HyClone, Thermo Scientific, Etten-Leur, The Netherlands) and 1% penicillin–streptomycin (Invitrogen). Next, the fibroblast cultures (n = 3) were tested for myofibroblast abundance, since this fibroblast subtype is target cell of interest. The percentage of myofibroblasts was in the range of 13%–30% as assessed using αSMA staining (Figure 1).3 The fibroblasts were kept at 37 °C at 5% CO2 and used for the following experiments between the third and fifth passage.
Table 1.
Patient information.
| Donor no. | Gender | Age (years) | Ethnicity |
|---|---|---|---|
| 1 | M | 8 | Caucasian |
| 2 | F | 48 | Caucasian |
| 3 | M | 20 | Caucasian |
| 4 | F | 10 | Caucasian |
| 5 | M | 13 | Caucasian |
Information on the HS biopsies obtained from five different patients used in this study.
F, female; M, male.
Figure 1.
Isolation and immunofluorescence staining of HS-derived fibroblasts. (a) H&E staining on HT-scar formalin-fixed sections, displaying the epidermis (I) and dermis (II) in HS tissue. (b) Fibroblasts were isolated from the dermis and cultured on glass slides. (c) Immunostaining for αSMA (red) positive cells and nuclear counterstaining were performed with DAPI (blue). Scale bars: 100 μm. n = 3. H&E, hematoxylin and eosin; HS, hypertrophic scar.
Based on the declaration of Helsinki principles, patient consent was not required since the excised scar tissue was considered waste material. Experiments were conducted in accordance with article 7:467 of the Dutch Law on Medical Treatment Agreement and the Code for proper Use of Human Tissue of the Dutch Federation of Biomedical Scientific Societies (https://www.federa.org/codes-conduct). As of this national legislation, coded HS tissue can be used for scientific research purposes when no written objection is made by the informed donor. Therefore, additional approval of an ethics committee regarding scientific use of excised HS tissue was not required. Thus, no specific approval by institutional ethics committee was requested.
Compounds
In order to apply exon skipping, we used vivo-morpholinos (ViM), containing the following AON sequences (5′-3′): ALK5ViM: GCAGTGGTCCTGATTGCAGCAATAT (to target ALK5 exon 2) and ScrViM (scrambled sequence) : CCTCTTACCTCAGTTAC AATTTATA (GeneTools, LLC, OR, USA). Both AONs were used at a concentration of 2 µM and added to the culture medium as described in the manufacturer’s protocol. As a positive control, the pharmacological ALK5 inhibitor SB431542 (10 µM) (TOCRIS Bioscience, Bristol, UK), a small molecule inhibitor which interacts with the ATP-binding site of ALK5, ALK4 and ALK7 (these ALKs show high similarity in their kinase domains compared with ALK5), was supplemented to the culture medium.15
AON delivery assay
Since the AONs assert their function in the nucleus of the cell, we determined whether AONs were present in the nucleus by conducting a delivery assay. For the AON delivery assay, three different HS donors were cultured on a six-well plate (2.5 × 105 cells per well) (Greiner Bio-One, Kremsmünster, Austria), serum-deprived overnight (O/N) (DMEM, containing 0.5% FBS; 1% P/S) and treated for 24 h with an ScrViM (1 µM, 5 µM and 10 µM) containing a red-emitting fluorescent tag (3’-Lissamine tag; excitation peak: 575 nm; emission peak 539 nm; GeneTools, LLC). For visualisation of the nuclei, Hoechst 33342 at a dilution of 1:2000 (Thermo Fisher Scientific, Waltham, MA, USA) was used. Successful oligo delivery was indicated by dim and diffuse fluorescence throughout the cytosol and nuclear compartment according to the manufacturer’s protocol. Imaging of ViM delivery was performed using an inverted microscope (IX53; Olympus Corporation, Shinjuku, Tokio, Japan) and CellScan software (Imstar, Paris, France).
Polymerase chain reaction (PCR)
To assess whether exon skipping induces an anti-fibrotic effect at gene expression level, we conducted a quantitative polymerase chain reaction (qPCR) and calculated the mRNA expression levels of a panel of genes (Table 2) that are up- or downregulated during a fibrotic response.16–22 Fibroblasts were plated on 6-cm dishes (1.0 × 105 cells per well) (Greiner Bio-One). Before treatment, the fibroblasts were serum-deprived in culture medium containing 0.5% serum O/N. Next, the fibroblasts were treated with AONs (2 µM) (GeneTools, LLC) treatment or SB431542 (10 µM) (TOCRIS Bioscience) for 24 h and followed by 6 h hTGF-β1 (5 ng/mL) (Cell Signaling, MA, USA) treatment for half of the samples. The treated fibroblasts were processed for RNA extraction using an RNA extraction kit (Favorgen®, Biotech Corp., Ping-Tung, Taiwan). RNA extraction was conducted according to the manufacturer’s protocol. Next, the total RNA was used for complementary DNA (cDNA) synthesis, using a cDNA Synthesis Kit (iScript™, Bio-Rad Laboratories Inc., Hercules, CA, USA). For qPCR analysis, the cDNA was amplified in a Real-Time PCR Detection System (CFX Connect Real-Time PCR Detection System, Bio-Rad Laboratories Inc.) using SYBR Green Supermix reagent (Bio-Rad Laboratories Inc.). The list of forward and reverse primers are depicted in Table 2. The expression levels were normalised to reference genes (SND, SDHA, EI24 and Golga1) and the relative normalised expression levels were calculated using Bio-Rad CFX Manager™ Version 3.1 (Bio-Rad Laboratories Inc.). In order to calculate fold changes after exon skipping, we have set ScrViM-treated cells as control samples in order to calculate the fold change in mRNA expression after exon skipping. As for TGF-β1 treated cells, the samples treated with TGF-β1 and ScrViM were set as control for comparison for exon skipping. For SB431542 treatment, we have set the untreated (ctrl) condition as control in CFX Manager™ and for TGF-β1-treated cells TGF-β1.
Table 2.
Primer list.
| Gene | Forward primer (5’ → 3’) | Reverse primer (5’ → 3’) |
|---|---|---|
| MMP1 | TTGGCCACAAAGTTGATGCAG | CTGGAGAGTCAAAATTCTCTTCGT |
| MMP2 | CTCATCGCAGATGCCTGGAA | AGGCACCCTTGAAGAAGTAGC |
| TIMP2 | CCTTATACCAGAGTTATGAGATCAA | AGTGATGTGCAAGAGTCCATCC |
| ACTA2 | TTGCCTGATGGGCAAGTGAT | GTGGTTTCATGGATGCCAGC |
| SERPINE1 | AGTGAAGATCGAGGTGAACGA | CCACAAAGAGGAAGGGTCTGT |
| CCN2 | CACCCGGGTTACCAATGACA | TACGGATGCACTTTTTGCCC |
| TGF-β1 | CACCGGAGTTGTGCGGCAGT | GGCCGGTAGTGAACCCGTTGATG |
| ALK5E2/3 | CAGTGTTTCTGCCACCTCTG | TGAATGACAGTGCGGTTGTGG |
| ALK5E5/6 | AGCGGTCTTGCCCATCTTC | TGCAATACAGCAAGTTCCATTCT |
| JUN | ATGAAGAACATCTCCTTCTGCGG | AAAGACGTGGCAGGCGAAG |
| ERK1/2 | GTAGAACAGGCTCTGGCCC | AACTTGAATGGTGCTTCGGC |
| STAT-1 | TTGATGGCCCTAAAGGAACTGG | ACATGGGGAGCAGGTTGTCT |
| COL1A1 | GAGGCATGTCTGGTTCGGC | TGACCTTCCTGCGCCTGATG |
| COL3A1 | ACGGAAACACTGGTGGACAG | GCCGTTAGGACTTGAAGGAC |
| EI24 | TTCACCGCATCCGTCGCCTG | GAGCGGGTCCTGCCTTCCCT |
| SDHA | AACCAAACGCTGGGGAAGAA | GGAACACGGCAGCATGATTT |
| GOLGA1 | ATGCCCGCGAGATCAACTTT | GTTCAGCAACACTGACACAGC |
| SND1 | CGTGCAGCGGGGCATCATCA | TGCCCAGGGCTCATCAGGGG |
ACTA2, actin alpha 2; ALK5E2/3, activin-like kinase 5 exon 2 and 3; ALK5E5/6, activin-like kinase 5 exon 5 and 6; CCN2, cellular communication network factor 2; COL1A1, collagen type 1 alpha 1; COL3A1, collagen type 3 alpha 1; EI24, etoposide-induced 2.4; ERK1/2, extracellular signal-regulated kinase 1/2; GOLGA1, golgin A1; JUN, Jun proto-oncogene; MMP1, matrix metalloproteinase-1; MMP2, metalloproteinase-2; SDHA, succinate dehydrogenase complex flavoprotein subunit A; SERPINE1, serpin family E member 1; SND1, staphylococcal nuclease and tudor domain containing 1; STAT-1, signal transducer and activator of transcription 1; TGF-β1, transforming growth factor beta 1; TIMP2, tissue inhibitor of metalloproteinases 2.
In parallel with executing qRT-PCR, touch down PCR (TD-PCR) was performed using a DNA polymerase kit (KOD Hot Start DNA Polymerase, Novagen®, Merck, Darmstadt, Germany). The mastermix was prepared according to the manufacturer’s protocol, using 20–30 µg of the synthesised cDNA. The PCR product was then loaded on a 2% agarose gel and sequential visualisation was performed using a detection system for agarose gels (ChemiDoc™ MP imager, Bio-Rad Laboratories Inc.).
Immunofluorescence
To further elucidate the modulatory effect of exon skipping of TGF-β signalling on protein level, we conducted immunofluorescence staining for TGFβRI (ALK5) activation, nuclear translocation of activated SMAD2 (pSMAD2) and expression of αSMA. For immunofluorescence analyses, cells were plated on chamber slides (3.0 × 104 cells per slide) (Thermo Fisher Scientific). For Phospho-Smad2 and ALK5 detection, HS fibroblasts were serum-deprived O/N and treated with AONs or SB431542 for 24 h. After ALK5 inhibition, half of the slides were treated with hTGF-β1 (5 ng/mL) (Cell Signaling) for 1 h before being washed in phosphate buffered saline (PBS) and fixed with 4% formaldehyde. As for αSMA detection, half of the samples were pre-treated with hTGF-β1 (5 ng/mL) (Cell Signaling) for 24 h followed by AON treatment (2 µM) (GeneTools, LLC) treatment or SB431542 (10 µM) (TOCRIS Bioscience) treatment. Next, cells were washed in PBS and fixed in 4% formaldehyde. The fixed cells were incubated with the primary antibody O/N at 4 °C. Primary antibodies, used for immunofluorescence, consisted of phospho-Smad2 (1:250; Bioss antibodies, Woburn, MA, USA), TGF-β receptor I (1:500; Abcam, Cambridge, UK), ACTA2 (1:500; Sigma-Aldrich, MO, USA). Primary staining was then followed by secondary antibody incubation (1:250; Alexa Flour® Dyes, Thermo Fisher Scientific) for 1.5 h at room temperature. After antibody incubation, the cells were washed with PBS and counterstained the nuclei using 4’,6-diamidino-2-phenylindole (DAPI; 1:4000; Carlsbad, CA, USA) and mounted for visualisation with mounting medium that preserves fluorescence (VECTASHIELD, VECTOR Laboratories, Burlingame, CA, USA). Analysis was performed with ImageJ. The positive area fraction was calculated by setting an upper and lower threshold, in order to count the pixels exhibiting a positive fluorescent signal. Next, fold induction was calculated (area fraction samples divided by area fraction control samples) for every condition over their putative control condition.
Western blot
In the present study, we used western blot analysis to determine MAPK/Erk kinase (MEK) activation and αSMA protein expression. For assessing MEK activation, the HS fibroblasts were plated on 6-cm dishes (1.0 × 105 cells per well) (Greiner Bio-One). Next, the fibroblasts were serum-deprived O/N followed by AON (2 µM) (GeneTools, LLC) or SB431542 (10 µM) (TOCRIS Bioscience ) treatment for 24 h. Followed by hTGF-β1 (5 ng/mL) (Cell Signaling) for 1 h, before harvesting.
For αSMA protein analysis, fibroblasts were plated on 6-cm dishes (1.0 × 105 cells per well) (Greiner Bio-One). Next, the HS fibroblasts were pre-treated with hTGF-β1 (5 ng/mL) (Cell Signaling) for 24 h followed by ALK5 inhibition for 24 h, consisting of AON (2 µM) (GeneTools, LLC) treatment or SB431542 (10 µM) (TOCRIS Bioscience) treatment.
For gel electrophoresis, 5 μg of protein was loaded. After electrophoresis, the protein was transferred to a polyvinylidene difluoride (PVDF) membrane and sequential incubation with primary antibodies for αSMA primary antibody (1:500; Merck, Darmstadt, Germany). MEK (1:1000; Cell Signaling) and p(hospho)-MEK (1:1000; Cell Signaling) were performed O/N at 4 °C. As a loading control, GAPDH (1:1500, Cell Signaling) or β-actin (1:2000, Cell Signaling) was used. The membranes were then incubated with a secondary antibody (Pierce, anti-mouse IgG and anti-rabbit IgG HRP-conjugated, 1:2500 diluted in blocking buffer) for 60 min at room temperature and prepared for visualisation using enhanced chemiluminescence (ECL) substrate solution (Pierce, SuperSignal West Femto Maximum Sensitivity Substrate) in a dark environment. Visualisation was performed with the ChemiDoc™ MP System (Bio-Rad Laboratories B.V., Veenendaal).
For quantification of protein expression, ImageJ was used. As for quantification of MEK and pMEK, we first calculated the relative band intensity per sample by dividing the band density of MEK and pMEK by its corresponding GAPDH band intensity. Next, MEK activation was calculated by dividing the relative density of pMEK by the relative density of total MEK. For αSMA expression, the band intensity of αSMA and β-actin was determined for every condition. Next, the αSMA band intensity was divided by its corresponding β-actin band intensity in order to calculate the relative band intensity of αSMA.
Scratch assay
In order to assess whether exon skipping would affect the migration of fibroblasts during wound healing, a scratch assay was performed using HS-derived fibroblasts. The fibroblasts were plated on a six-well plate (1.0 × 105 cells per well) until a confluence of 70%–80% was reached. Next, the cells were serum-deprived for 12 h and subsequently treated with AONs (2 µM) or SB431542 (10 µM), with or without 4 h TGF-β (5 ng/mL) pre-treatment. After 6 h of ViM or SB431542 treatment, a scratch was made, using a 200-μL pipet tip, and culture medium was carefully refreshed. Images were taken at t(0), t(4), t(8) and t(20). Open area percentages of the scratch assay images were assessed using the software tool TScratch (www.cse-lab.ethz.ch/software.html). The migration capacity of the treated fibroblasts was expressed by the migration rate (%), from which the calculation was derived from Zhou et al., 2012 : [percentage open area at t0 – open area at tx]/ percentage open area at t0, where t0 is the starting point directly after inducing a scratch and tx any other timepoint with a 4-h interval. In the present study, the migration rates (in percentages) for the different conditions were calculated after 20 h with respect to 0 h. For further reading considering the algorithm behind the software and manual, we refer to the report of Gebäck et al.23
Statistics
For statistical analysis, we have used GraphPad Prism version 6.00 for Windows (GraphPad Software, CA, USA). Statistical analysis for gene expression analysis was conducted using the multiple t-test. For correction of multiple comparisons, we used the Holm–Šídák method. For the scratch assay and immunofluorescence for ALK5 phosphorylation, we performed a one-way ANOVA, followed by Tukey’s multiple comparisons test. For pSmad2 nuclear translocation, we performed one-way ANOVA with the Holm–Šídák post-test. The differences were noted as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Results
TGF-β1 elevated gene expression of pro-fibrotic genes
Since we treated half of the samples with hTGF-β1 in all experiments, we first assessed the effect TGF-β1 treatment compared to untreated HS fibroblasts (Figure 2).
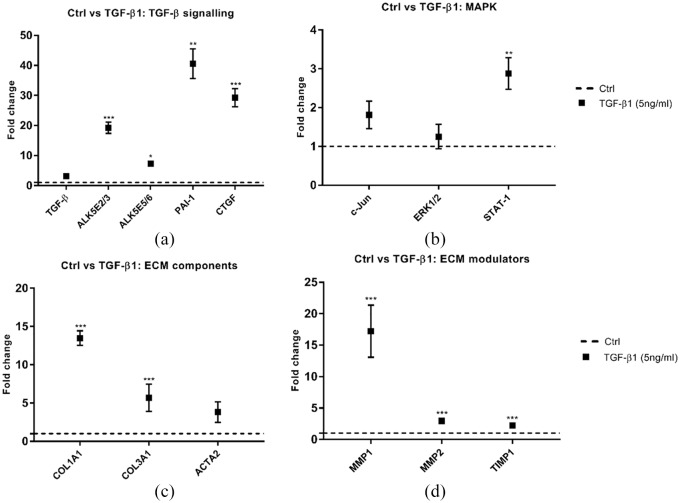
Figure 2.
The effect of TGF-β1 treatment in HS-derived fibroblast monocultures. (a–d) Relative mRNA expression of TGF-β signalling pathway downstream targets (a), MAPK pathway downstream targets (b), ECM components (c) and ECM modulators (d) treated with TGF-β1 for 6 h. The graph represents the relative normalised mRNA expression values of the treated samples over the untreated (Ctrl) samples (set at 1; dashed line). n = 3; *P < 0.05; **P < 0.01; ***P < 0.001; multiple t-test, error bars indicate standard of the mean. ECM, extracellular matrix; HS, hypertrophic scar.
Gene expression analysis showed a substantial induction of TGF-β downstream targets PAI-1 (P < 0.01) and CTGF (P < 0.001) (Figure 2a). A similar result was found for ECM components COL1A1 and COL3A1 (P < 0.001) (Figure 2c). ACTA2 mRNA levels showed a slight elevation compared to basal condition; however, this upregulation was not statistically significant (P < 0.06) (Figure 2c). Further, expression levels of ECM modulators also showed a substantial elevation. Besides the canonical pathway, we also assessed the gene expression levels of three downstream targets of the non-canonical MAPK pathway: c-Jun; ERK1/2: and STAT1 (Figure 2b). Here, we found an induction in STAT1 gene expression levels after TGF-β1 treatment (P < 0.01).
Vivo-morpholino AONs are successfully delivered into the nucleus and are functionally active
Next, we determined whether ViMs could be successfully delivered in HS-derived fibroblasts. To this end, we treated these fibroblasts isolated from HS biopsies obtained from three different burn patients for 24 h with a 3’-Lissamine-tagged (red emitting tag) non-targeting AON sequence (ScrViM). At a concentration of 10 μM, there was clear delivery of the AON, compared to the untreated (Ctrl) condition. In Figure 3a, the images are presented of the delivery of the tagged ScrViM.
Figure 3.
Successful AON delivery of a 3’-Lissamine tagged ScrViM and assessment of ViM functionality. (a) Untreated HS-derived fibroblasts. Fibroblasts were treated with an oligo concentration of 10 µM showed successful AON delivery by representing a dim and diffuse fluorescence. n = 3. Scale bar: 50 μm. (b) Validation and confirmation of exon 2 skipping using TD-PCR. n = 3. AON, antisense oligonucleotides; HS, hypertrophic scar.
Next, we assessed whether the delivered ViMs were functionally active, hence, able to induce skipping of ALK5 exon 2. Therefore, we performed a nested PCR on fibroblasts treated with ViMs (2 μM) for 24 h and loaded the PCR product on an agarose gel (Figure 3b). Here we show that the control sample (Ctrl), ScrViM (non-targeting AON sequence) treated and SB431542 (positive control) samples only exhibit the full-length (exon 1-3) product, whereas ALK5ViM treated samples clearly show the exon-skipping product, containing only exon 1 and 3, confirming skipping of exon 2. At higher concentrations, we observed a cytotoxic effect after ViM treatment.
Exon skipping modulates TGF-β signalling and reduces fibrotic gene signature
First, we assessed the effect of SB431542 on HS- derived fibroblasts at basal levels and after TGF-β1 treatment (Additional file 1). In order to determine the effect of SB431542 on ALK5 activity and ALK5 exon 2, we designed ALK5 exon 2/3 primers sequences (ALK5E2/3) and as an internal positive control we determined relative expression levels of ALK5 exon 5/6 (ALK5E5/6). Here, we show at baseline levels and after hTGF-β1 treatment that ALK5 exon 2/3 and ALK5 exon 5/6 show concomitant changes in expression levels, confirming the inhibitory effect of SB4311542 on ALK5 activity.15 In both groups, we found that SB431542 lowered the expression of PAI-1 (baseline: P < 0.01) and CTGF (baseline: P < 0.001; TGF-β1 post-treatment: P < 0.01), TGF-β downstream targets known to be overexpressed in fibrotic diseases.19,24 The expression levels in the ECM components ACTA2, COL1A1 and COL3A1 showed in both groups a significant reduction upon ALK5 inhibition (Figure S1c and S1g). As for the ECM modulators (Figure S1e and S1h) that were taken into account in this assay, we found in both groups a twofold induction of MMP1 transcript (P < 0.01) compared to control samples. Chemical inhibition showed no substantial effect on MMP2 and TIMP1 expression. As for the non-canonical MAPK pathway, no statistically significant changes were found.
We then continued assessing the effect of exon skipping on TGF-β signalling and its downstream targets in HS-derived fibroblast cultures. The fibroblasts of three different patients with HSs were treated in duplicate with the ScrViM, ALK5ViM sequence or SB431542 for 24 h. The relative mRNA expression levels are displayed in Figure 4.
Figure 4.
Validation and anti-fibrotic effect of exon skipping in HS-derived fibroblasts after 24 h. Half of the samples were treated with hTGF-β1 for 6 h and, thereby, establishing two groups: baseline and TGF-β1 post-treatment. (a–d) Fold change in mRNA expression of TGF-β1, ALK5 exon 2/3 and ALK5 exon 5/6, downstream targets PAI-1 and CTGF, MAPK downstream targets c-Jun, ERK1/2 and STAT1, ECM components ACTA2, COL1A1 and COL3A1 and ECM modulators MMP1, MMP2 and TIMP1 at baseline and TGF-β1 post-treatment (6 h). These graphs represent the relative normalised expression of ALK5ViM treatment (24 h) over ScrViM as control (set at 1, dashed line). n = 3; *P < 0.05; **P < 0.01; ***P < 0.001; multiple t-test, error bars indicate standard error of the mean. ECM, extracellular matrix; HS, hypertrophic scar.
First, we assessed the effect and specificity of exon skipping on ALK5 exon 2 by evaluating the relative mRNA expression levels of ALK5E2/3 and ALK5E5/6 (internal positive control) (Figure 4a and 4e). Here, we found in both groups an almost complete loss of ALK5E2/3 expression (P < 0.001), thus confirming successful skipping of exon 2. Furthermore, this finding suggests that TGF-β1 is not able to overcome the inhibitory effect of ALK5ViM. Moreover, in both groups ALK5E5/6 showed similar expression levels compared to control samples and, thereby, confirming the specificity of ALK5ViM targeting exon 2. Next, exon skipping showed a similar effect on TGF-β1 signalling downstream targets PAI-1 and CTGF relative mRNA expression levels compared to control samples (Figure 4a and 4e). MAPK pathway downstream targets c-Jun and STAT1 showed a slight reduction after exon skipping (Figure 4b). As for the ECM components (ACTA2, COL1A1 and COL3A1) (Figure 4c and 4g) and ECM modulator MMP1 (Figure 4d and 4h), we found that exon skipping and SB431542 treatment showed a similar effect in both groups as expected.
ALK5 activation is reduced after exon skipping
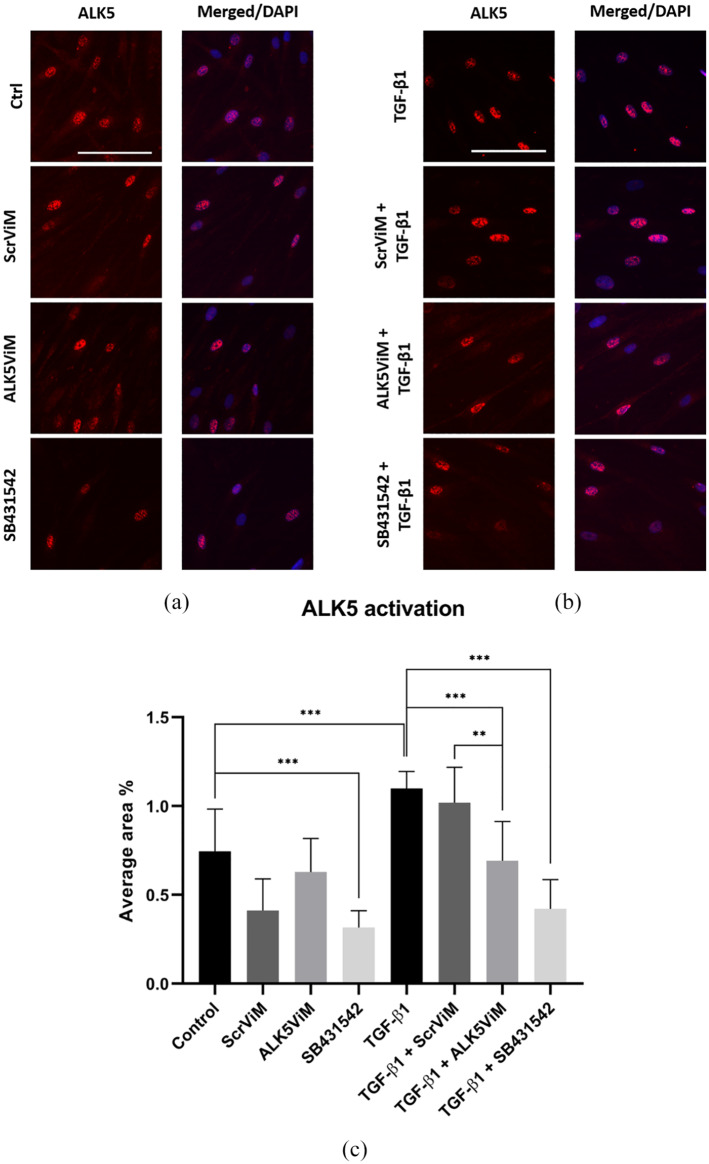
Next, we assessed the effect of exon skipping on ALK5 activity by investigating activated ALK5 which is primarily located around the nucleus (Figure 5). Here, activated ALK5 is indicated in red and the nuclear staining in blue (merged images). Figure 5a represents images of samples at basal level. Here, only SB431542 was able to substantially reduce ALK5 activation (P < 0.001) (Figure 5a and 5c). However, after TGF-β1 treatment, ALK5 activation was found to be elevated compared to the control condition (P < 0.001) (Figure 5b and 5c).
Figure 5.
Immunofluorescence and quantification of activated ALK5 in HS-derived fibroblasts. Fibroblasts were pre-treated with ViMs (2 μM) or SB431542 (10 μM) for 24 h. Next, fibroblasts were treated with hTGF-β1 (5 ng/mL) for 1 h. (a) ALK5 IF staining (red) for the following conditions: TGF-β1, ScrViM+TGF-β1, ALK5ViM+TGF-β1 and SB431542+TGF-β1. Nuclear staining was performed with DAPI, displayed in corresponding merged images. Scale bar: 100 µm. (b) Quantification of ALK5 activation presented as average area fraction. Fluorescent signal was calculated as area fraction for every donor in eight different conditions (Control [untreated], ScrViM, ALK5ViM, SB431542, TGF-β1, ScrViM+TGF-β1, ALK5ViM+TGF-β1 and SB431542+TGF-β1) and was calculated with ImageJ. n = 3; *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA, error bars indicate standard error of the mean.
Exon skipping reduces p(hospho)-Smad2 nuclear translocation and MEK activation
Since translocation of activated Smad2 to the nucleus is a crucial step in TGF-β signal transduction and activates expression of other TGF-β downstream targets, we analysed pSmad2 nuclear translocation in HS-derived fibroblasts in order to determine to what extent exon skipping reduces TGF-β signalling at basal and TGF-β1 treated samples (Figure 6). In Figure 6a and 6b, the images are shown that represent the graphs (Figure 6c–e). At basal levels, nuclear pSmad2 showed a reduction in positive fluorescent signal after ALK5 inhibition by exon skipping and pharmacological inhibition compared to the untreated (Ctrl) condition (Figure 6a and 6c). One hour after treatment with TGF-β1 showed even a slight reduction in pSmad2 nuclear translocation compared to the untreated samples (Figure 6b and 6d). Further, TGF-β1 post-treatment samples showed a similar effect as on basal levels. Here we found that all three donors collectively showed lower nuclear pSmad2 compared to their control condition (TGF-β1 treatment) (Figure 6e). Next, we continued assessing the effect of exon skipping on the activation of the non-canonical MAPK signalling pathway by performing an immunoblot staining for (p)MEK (Figure 6f). After ALK5 inhibition, we found a trend which showed a reduction in MEK activation. (Figure 6g). Although not significant, the trend in these data suggests that ALK5 inhibition is able to reduce pSmad2 nuclear translocation and MEK action.
Figure 6.
pSmad2 immunofluorescence together with (p)MEK immunoblot staining performed on HS-derived fibroblasts. (a–e) Fibroblasts were pre-treated with ViMs (2 μM) or SB431542 (10 μM) for 24 h. Next, fibroblasts were treated with hTGF-β1 (5 ng/mL) for 1 h. (a) pSmad2 IF (red) images of ScrViM or ALK5ViM treated HS-derived fibroblasts. Nuclear staining was performed with DAPI. Scale bar: 50 μm. (b–d) Quantification of fold induction of pSmad2 nuclear translocation. One dot represents one donor. Fluorescent signal was calculated as area fraction for every donor in eight different conditions (Ctrl [untreated], ScrViM, ALK5ViM, SB431542, TGF-β1, ScrViM+TGF-β1, ALK5ViM+TGF-β1 and SB431542+TGF-β1) with ImageJ. Positive values for pSmad2 are represented as fold induction over their control condition (at baseline level: Ctrl and TGF-β1 for hTGF-β1-post treated samples) indicated by the grey line. n = 3, error bars represent standard error of the mean. (f, g) Fibroblasts were pre-treated with ViMs (2 μM) or SB431542 (10 μM) for 24 h. Next, fibroblasts were treated with hTGF-β1 (5 ng/mL) for 1 h. The relative band intensity of total MEK and pMEK was calculated by dividing the relative band intensity of each sample by its corresponding relative GAPDH (loading control) band intensity. For quantification of pMEK activation, pMEK:total MEK ratio was calculated for each sample. n = 2.
Exon skipping shows a decline in αSMA protein expression
We continued investigating the anti-fibrotic potential by inferring whether exon skipping could reduce αSMA expression (Figure 7). Here, we found that exon skipping was able to reduce αSMA protein expression levels after 24 h compared to samples treated with the scrambled sequence (ScrViM), although not significant (P < 0.07; Student’s t-test) (Figure 7a). As for pharmacological ALK5 inhibition, after TGF-β1 treatment, we found similar αSMA expression levels compared to TGF-β1 treated fibroblasts (Figure 7b).
Figure 7.
Western blot analysis and immunofluorescence of αSMA expression in HS-derived fibroblasts. HS- derived fibroblasts were pre-treated with hTGF-β1 (5 ng/mL) followed by ALK5 inhibition, subdivided into exon skipping (ALK5ViM [2 µM]) (a) and SB431542 (10 µM) (b). After 24 h, samples were harvested for protein analysis or fixed for immunofluorescence (c). (a, b) The mean relative band intensity of αSMA was calculated by dividing the relative αSMA band intensity by its corresponding relative β-actin (loading control) band intensity. n = 3, error bars represent standard error of the mean. (c) Immunofluorescence for αSMA (red) and merged images show αSMA DAPI nuclear staining (blue). These images are representative for two separate experiments. Scalebar: 100 µm. HS, hypertrophic scar.
Thus, ALK5 inhibition by means of exon skipping, as opposed to pharmacological ALK5 inhibition, is able to substantially reduce αSMA protein expression levels.
Exon skipping does not affect fibroblast migration rate
Next, we determined whether exon skipping affects fibroblast migration and for this purpose we conducted a scratch wound-healing assay (Figure 8). We found that exon skipping (ALK5ViM) showed a small reduction in cell migration when monitored after 20 h; however, this alteration was not significant over the five donors. Further, ScrViM did not show any change in fibroblast migration compared to control, while ScrViM and SB431542 treatments showed an equal migration rate after 20 h. Moreover, TGF-β supplementation showed an inhibitory effect on fibroblast migration rate (P < 0.05). When fibroblasts were treated with TGF-β1 before exon skipping or SB431542 treatment, we found that ALK5 inhibition rescued the inhibitory effect of TGF-β1 to some extent, hence, reversing fibroblast migration rate to control condition.
Figure 8.
Effect of exon skipping on HS-derived fibroblast migration rate. Fibroblasts were treated with ViMs, SB431542 (for 24 h) or TGF-β1 (for 4 h). (a) Images of the scratches were taken at 0 h and 20 h and show visible wound closure over time. Scale bar: 200 μm. (b) The calculated migration rates for the different conditions 20 h post-scratch. Images were analysed using TScratch software. n = 5; *P < 0.05; one-way ANOVA, error bars represent standard error of the mean.
Discussion
In the present study, we have investigated the effect of exon skipping in a HS-derived fibroblast two-dimensional (2D) monoculture model. Here we showed for the first time that we were able to successfully deliver AONs into the nucleus of HS-derived fibroblasts and confirmed its functionality regarding loss of exon 2 and its anti-fibrotic effect by modulation of the TGF-β signalling pathway. Due to a high rate of recurrence with all its consequences on the quality of life of burn victims, it is warranted to search for a permanent solution in HS treatment.10 In previous studies, performed by Kemaladewi et al.13 and Karkampouna et al.,14 it has been reported that ALK5 exon 2 was successfully spliced out using exon skipping and reduced activity of TGF-β signalling downstream targets. Therefore, we used these studies as a scaffold to show for the first time the ECM modulating effect of exon skipping in HS 2D models. In order to be able to specifically skip an exon of interest, one can use AONs. AONs are short fragments of nucleic acid, consisting of 13–25 nucleotides, and are able to hybridise a specific target sequence of an exon, ensuring the target exon to be spliced out. In the present study, we make use of ViMs.13,25,26 On the backbone of a morpholino oligonucleotide, each subunit comprises a nucleic acid base, a morpholine ring and a non-ionic phosphorodiamidate intersubunit linkage. Furthermore, ViMs produced by GeneTools provide ViMs containing an octaguadinine moiety for optimal uptake by cells through endocytosis. Moreover, ViMs do not induce an immunological response. Therefore, ViMs are an excellent tool to induce exon skipping HS-derived fibroblasts.26–28
During the present study, the chemical ALK5 inhibitor SB431542 was used as a positive control. Exon skipping using AONs has three major advantages over pharmacological ALK5 inhibition. First, ALK5 SB431542 is also an inhibitor of the transforming growth factor- superfamily type I Activin Receptor-Like Kinase (ALK) Receptors ALK4 and ALK7. Second, SB431542 works properly in in vitro models and shows a significant anti-fibrotic effect. However, in vivo studies showed that this compound is pharmacokinetically unstable.12 Third, in fibrotic diseases, TGF-β signalling is conducted through Smad-dependent and Smad-independent pathways (such as a MAPK pathway), whereas SB431542 only inhibits TGF-β signalling through the Smad-dependent pathway.29,30
Before we conducted experiments to investigate the effects of exon skipping in HS fibroblasts, we first determined delivery of AONs into the nucleus. Although the backbone of morpholinos are considered to be non-toxic, some sequences can be toxic due to base-pairing interactions.15 Furthermore, the arginine-rich delivery moiety causes the ViMs to be toxic as the number of morpholino is increased.15 Therefore, the optimal concentration was determined. Since the typical concentration range of ViMs in cell cultures is between 1 µM and 10 µM, we tested the delivery of AONs at 5 µM and 10 µM.15 Here, we found that a concentration of 10 μM showed clear delivery of the AON, compared to untreated (Ctrl) condition. Further, according to the manufacturer’s protocol, the oligo concentration for subsequent knockdown studies is lower (up to 10 times) than the optimal concentration for delivery studies. Therefore, in the present study, we decided to use a ViM concentration of 2 µM for all experiments.
During our gene study, we analysed mRNA expression levels of different target genes involved in the TGF-β signalling pathway, ECM components and modulators by qPCR. Here we found that ALK5ViM was able to mimic the anti-fibrotic effect of SB431542 after 24 h by significant downregulation of TGF-β downstream targets and ECM component mRNA expression levels. Furthermore, MMP1 showed an almost fourfold increase upon ALK5 inhibition together with substantial downregulation of COL1A1 and COL3A1 mRNA expression. This is in agreement with other studies showing that collagen type 1 and 3 are mainly digested by MMP-1.31,32 Besides the canonical Smad-dependent pathway, we have assessed downstream targets of the non-canonical MAPK pathway. In contrast to previous studies, we did not find a significant effect of ALK5 inhibition on MAPK signalling pathway at mRNA level. However, this discrepancy could be explained by: (1) pharmacological agents; (2) the time of treatment, which could have been too short in order to detect a substantial effect; (3) the initial fibroblast type (e.g. HS-derived versus TGF-β activated fibroblasts). As expected, TGF-β1 treatment showed a significant elevation of STAT1 (P < 0.01), demonstrating bilateral activation of the canonical and non-canonical pathways. Another explanation why exon skipping did not affect MAPK signalling could be addressed to kinase activity upstream in the signalling cascade. Here we found that exon skipping was able to slightly reduce MEK phosphorylation. This observation could explain why the downstream targets were unaffected (Figure 6f and 6g). In order to observe an elevation in other MAPK downstream targets, one should vary the time intervals of the various treatments. Altogether, these data suggest that exon skipping induces an anti-fibrotic effect at gene expression level, which is most profound through Smad-dependent signalling.
Next, we investigated whether exon skipping affected ALK5 activity. Upon TGF-β binding TGFβRII phosphorylates ALK5 . Next, activated ALK5 phosphorylates receptor-associated Smad2 and 3, which in turn then bind to common mediator Smad4 . The complex is translocated to the nucleus in order to activate pro-fibrotic transcription factors.33,34 It has been reported that proper intracellular propagation of Smad signalling requires internalisation of ALK5 by endocytic vesicle formation.35 In the present study, we found ALK5 to be primarily expressed around the nucleus where numerous foci were found, which showed to be highly positive for ALK5. Based on findings in previous studies, these foci could represent ALK5 in vesicles due to receptor internalisation.35,36 Therefore, we could consider these foci to represent the portion of activated ALK5. Thus, one would expect elevated levels of activated ALK5 after TGF-β1 activation. Indeed, TGF-β1 treatment significantly increased ALK5 activation. In a previous study performed by Bock and colleagues,22 fibroblasts derived from normal skin, keloids and HSs were isolated and treated. Here, researchers found that ALK5 mRNA expression were elevated, although not significantly. In the present study, we were able to substantially reduce it by exon skipping. As stated before, several studies have reported new insights on internalisation of TGFβR . However, it is warranted to further elucidate the exact mechanisms. New information on this topic could provide in the development of novel anti-fibrotic therapeutics regarding internalisation of ALK5.
Since Smad2/3 are the first downstream targets to be activated upon ALK5 activation, we assessed the nuclear translocation of pSmad2 before and after treatment. Here we found that basal nuclear pSmad2 levels exhibited similar (and in some cases even higher) fold induction of nuclear pSmad2, which was not significantly increased by TGF-β1 activation. One possible explanation could be due to the high turnover rate of pSmad2.
Altogether, our findings show that exon skipping is able to reduce pSmad2 nuclear translocation, suggesting that this therapy indeed could balance the activation of TGF-β downstream targets, such as αSMA expression: the main cause of severe contraction in HSs.3,37 In order to determine the effect αSMA expression upon exon skipping, we have treated the HS-derived fibroblasts with h TGF-β1 in order to push fibroblasts to differentiate into MFs. Sequential AON treatment was able to reduce αSMA expression after 24 h, although not significant, indicate the (partial) release of contractile forces by αSMA. Thus, myofibroblast inhibition. However, in contrast to previous studies, SB431542 did not show any reduction of αSMA.14,38,39 Therefore, we assessed the effect of ALK5 inhibition by staining cells for αSMA. Indeed, here we found that exon skipping, as well as SB431542 treatment reduced αSMA expression in HS fibroblasts. Moreover, we found that lowered ALK5 activity was followed by a reduction in pSmad2 nuclear translocation. Consequently, αSMA showed reduced expression after exon skipping. Altogether, demonstrating the inhibitory effects of ALK5 exon skipping on TGF-β signalling. Furthermore, we found that exon skipping was able to significantly reduce TGF-β1-induced contraction in collagen lattices (data not shown).
The focus of the present study was to modulate the TGF-β signalling pathway in HS-derived fibroblasts. However, since TGF-β signalling has a pleiotropic effect, we assessed the effect of exon skipping on wound healing simulated in a scratch assay. Here we found a slight decrease in migration rate after exon skipping; however, it was not significant. Further, TGF-β1 before treatment showed a significant inhibitory effect on the migration rate of the fibroblasts, suggesting a delayed migration of fibroblasts upon TGF-β1 treatment. This finding is in agreement with previous in vitro studies reporting the effect of TGF-β on fibroblasts in which TGF-β1 treatment showed an inhibitory effect on cell motility.40,41 In addition, sequential exon skipping showed the rescuing of the inhibitory effect of TGF-β to some extent. Moreover, SB431542 after treatment restored the migration rate back to control conditions (P < 0.05). A possible explanation for this improvement could be due to the fact that SB431542 also inhibits ALK4 and ALK7 and could therefore be responsible for the improvement in the fibroblast migration rate.15 Altogether, this finding suggests that ALK5 inhibition does not affect the migratory rate of HS-derived fibroblasts and could rescue the inhibitory effect of TGF-β1 on the migratory capabilities of the fibroblasts.
With this approach in which we downregulate TGF-β signalling through exon skipping of ALK5 ligand binding domain, one should consider that upon treatment a compensatory mechanism could be induced. A possible mechanism that could induce take-over is facilitated by TGFβRII and/or the assessor TGF-β receptor associated proteins betaglycan and endoglin, also known as TGF-β type III receptors (TGFβRIII).42 It has been reported by Zhang et al.43 that TGFβRII is able to establish homodimers with itself and thereby being able to activate through its intracellular domain Smad2/3 upon TGF-β ligand binding in a TGFβRI-independent manner. TGFβRIII, comprising betaglycan and endoglin, that share sequence similarity in amino acid sequence of the transmembrane domain, is a transmembrane receptor that possess an extracellular domain which allows binding of TGF-β with high affinity.44,45 However, TGFβRIII has a cytoplasmic signalling domain without kinase activity. Upon TGF-β binding, TGFβRIII presents the ligand to TGFβRII and/or TGFβRI and induces sequential activation of the TGF-β signalling pathway, hence, acting as a co-receptor.46 Both assessor proteins are expressed on fibroblasts and could activate the TGF-β signalling cascade in a TGFβRI (ALK5) independent manner and may elicit a pro-fibrotic effect.45,47–49 It would be of great interest to elucidate the role of TGFβRIII in the development of HS, the anti-fibrotic potential of TGFβRIII downregulation and to what extent TGFβRIII contributes to a compensatory mechanism upon downregulation of TGFβRI activity.
Further investigation should include the comparison on the effect of exon skipping in HS fibroblasts versus normal dermal fibroblasts (NDF), since there exists heterogeneity between HS fibroblasts and NDFs, in terms of proliferation, responsiveness towards hTGF-β1 treatment and anti-TGF-β treatment.50 Furthermore, regarding isolating fibroblasts from HS biopsies, the age of the biopsy has to be taken into account. A previous study has reported that proliferation and regression of HSs is closely correlated with fibroblast biology.51 These dynamic changes emphasise one of the challenges in the present study, in terms of the clinical application of this therapeutic approach and when to apply it. Therefore, timing will be an important question that needs to be answered before exon skipping can enter the clinic. Moreover, one would preferably prevent development of HSs in burn patients. In the updated international clinical recommendations on scar management, good surgery and wound management are necessary in order reduce the risk of HS development.52 At this point during treatment, one should step in and treat fresh burn trauma with exon skipping.
Conclusions
In the present study, we have shown for the first time that exon skipping is able modulate the TGF-β signalling cascade and is able to reduce ECM-related components involved in the development of HS. Furthermore, several lines of evidence point to a central role of the TGF-β signalling pathway in coordinating the biological actions that constitute the development of other fibrotic diseases, such as Dupuytren’s disease and scleroderma.53 Therefore, exon skipping could be a promising novel treatment option beyond HSs. Altogether, exon skipping is a promising tool in the treatment of HSs and could open a new therapeutic window in the treatment of patients with HSs.
Supplemental Material
Supplemental material, Supplementary for Exon skipping of TGFβRI affects signalling and ECM expression in hypertrophic scar-derived fibroblasts by Rajiv S Raktoe, Marion H Rietveld, Jacoba J Out-Luiting, Marianna Kruithof-de Julio, Paul PM van Zuijlen, Remco van Doorn and Abdoelwaheb El Ghalbzouri in Scars, Burns & Healing
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the Dutch Burn Foundation (NO. 14.105) located at Beverwijk.
ORCID iD: Rajiv S Raktoe  https://orcid.org/0000-0003-4168-6443
https://orcid.org/0000-0003-4168-6443
Supplemental material: Supplemental material for this article is available online.
References
- 1. Brusselaers N, Monstrey S, Vogelaers D, et al. Severe burn injury in europe: a systematic review of the incidence, etiology, morbidity, and mortality. Crit Care 2010; 14(5): R188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bombaro KM, Engrav LH, Carrougher GJ, et al. What is the prevalence of hypertrophic scarring following burns? Burns 2003; 29(4): 299–302. [DOI] [PubMed] [Google Scholar]
- 3. Tredget EE, Nedelec B, Scott PG, et al. Hypertrophic scars, Keloids and Contractures. Surg Clin North Am 1997; 77(3): 701–730. [DOI] [PubMed] [Google Scholar]
- 4. Schmid P, Itin P, Cherry G, et al. Enhanced expression of transforming growth factor-beta type I and type II receptors in wound granulation tissue and hypertrophic scar. Am J Pathol 1998; 152(2): 485–493. [PMC free article] [PubMed] [Google Scholar]
- 5. Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol 2000; 1(3): 169–178. [DOI] [PubMed] [Google Scholar]
- 6. Desmoulière A, Geinoz A, Gabbiani F. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol 1993; 122(1): 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Penn JW, Grobbelaar AO, Rolfe KJ. The role of the TGF-β family in wound healing, burns and scarring: a review. Int J Burns Trauma 2012; 2(1): 18–28. [PMC free article] [PubMed] [Google Scholar]
- 8. Klass BR, Grobbelaar AO, Rolfe KJ. Transforming growth factor β1 signalling, wound healing and repair: a multifunctional cytokine with clinical implications for wound repair, a delicate balance. Postgrad Med J 2009; 85(999): 9–14. [DOI] [PubMed] [Google Scholar]
- 9. Bloemen MCT, van der Veer WM, Ulrich MMW, et al. Prevention and curative management of hypertrophic scar formation. Burns 2009; 35(4): 463–475. [DOI] [PubMed] [Google Scholar]
- 10. Del Toro D, Dedhia R, Tollefson TT. Advances in scar management. Curr Opin Otolaryngol Head Neck Surg 2016; 24: 322–329. [DOI] [PubMed] [Google Scholar]
- 11. Friedstat JS, Hultman CS. Hypertrophic Burn Scar Management. Ann Plast Surg 2014; 72(2): S198–201. [DOI] [PubMed] [Google Scholar]
- 12. Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov 2012; 11(11): 886–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kemaladewi DU, Pasteuning S, van der Meulen JW, et al. Targeting TGF-β Signaling by Antisense Oligonucleotide-mediated Knockdown of TGF-β Type I Receptor. Mol Ther - Nucleic Acids 2014; 3(February): e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karkampouna S, Kruithof BP, Kloen P, et al. Novel Ex Vivo Culture Method for the Study of Dupuytren’s Disease: Effects of TGFβ Type 1 Receptor Modulation by Antisense Oligonucleotides. Mol Ther - Nucleic Acids 2014; 3(June 2013): e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inman GJ, Nicolás FJ, Callahan JF, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 2002; 62(1): 65–74. [DOI] [PubMed] [Google Scholar]
- 16. Murray JC. Scars and Keloids. Dermatol Clin 1993; 11(4): 697–708. [PubMed] [Google Scholar]
- 17. Neely AN, Clendening CE, Gardner J, et al. Gelatinase activity in keloids and hypertrophic scars. Wound Repair Regen 1999; 7(3): 166–171. [DOI] [PubMed] [Google Scholar]
- 18. Ulrich D, Ulrich F, Unglaub F, et al. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in patients with different types of scars and keloids. J Plast Reconstr Aesthet Surg 2010; 63(6): 1015–1021. [DOI] [PubMed] [Google Scholar]
- 19. Ghosh AK, Vaughan DE. PAI-1 in tissue fibrosis. J Cell Physiol 2012; 227(2): 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Colwell AS, Phan T-T, Kong W, et al. Hypertrophic scar fibroblasts have increased connective tissue growth factor expression after transforming growth factor-beta stimulation. Plast Reconstr Surg 2005; 116(5): 1382–1387. [DOI] [PubMed] [Google Scholar]
- 21. Hu X, Wang H, Liu J, et al. The role of ERK and JNK signaling in connective tissue growth factor induced extracellular matrix protein production and scar formation. Arch Dermatol Res 2013; 305(5): 433–445. [DOI] [PubMed] [Google Scholar]
- 22. Bock O, Yu H, Zitron S, et al. Studies of transforming growth factors beta 1-3 and their receptors I and II in fibroblast of keloids and hypertrophic scars. Acta Derm Venereol 2005; 85(3): 216–220. [DOI] [PubMed] [Google Scholar]
- 23. Gebäck T, Schulz MMP, Koumoutsakos P, et al. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 2009; 46(4): 265–274. [DOI] [PubMed] [Google Scholar]
- 24. Grotendorst GR. Connective tissue growth factor: a mediator of TGF-β action on fibroblasts. Cytokine Growth Factor Rev 1997; 8(3): 171–179. [DOI] [PubMed] [Google Scholar]
- 25. Sazani P, Kole R. Therapeutic potential of antisense oligonucleotides as modulators of alternative splicing. J Clin Invest 2003; 112(4): 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: A non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques 2008; 45(6): 613–623. [DOI] [PubMed] [Google Scholar]
- 27. Partridge M, Vincent A, Matthews P, et al. A simple method for delivering morpholino antisense oligos into the cytoplasm of cells. Antisense Nucleic Acid Drug Dev 1996; 6(3): 169–175. [DOI] [PubMed] [Google Scholar]
- 28. Moulton JD. Guide for morpholino users: toward therapeutics. J Drug Discov Dev Deliv 2016; 3(2): 1023. [Google Scholar]
- 29. Sorrentino A, Thakur N, Grimsby S, et al. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol 2008; 10(10): 1199–1207. [DOI] [PubMed] [Google Scholar]
- 30. Kim S, Il Kwak JH, Na H-J, et al. Transforming Growth Factor-β (TGF-β1) Activates TAK1 via TAB1-mediated Autophosphorylation, Independent of TGF-β Receptor Kinase Activity in Mesangial Cells. J Biol Chem 2009; 284(33): 22285–22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mizuno S, Matsumoto K, Kurosawa T, et al. Reciprocal balance of hepatocyte growth factor and transforming growth factor-β1 in renal fibrosis in mice. Kidney Int 2000; 57(3): 937–948. Available from: http://www.sciencedirect.com/science/article/pii/S0085253815468267 [DOI] [PubMed] [Google Scholar]
- 32. Jinnin M, Ihn H, Mimura Y, et al. Effects of Hepatocyte Growth Factor on the Expression of Type I Collagen and Matrix Metalloproteinase-1 in Normal and Scleroderma Dermal Fibroblasts. J Invest Dermatol 2005; 124(2): 324–330. [DOI] [PubMed] [Google Scholar]
- 33. Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003; 425(6958): 577–584. [DOI] [PubMed] [Google Scholar]
- 34. Walton KL, Johnson KE, Harrison CA. Targeting TGF-β Mediated SMAD Signaling for the Prevention of Fibrosis. Front Pharmacol 2017; 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Penheiter SG, Mitchell H, Garamszegi N, et al. Internalization-dependent and -independent requirements for transforming growth factor beta receptor signaling via the Smad pathway. Mol Cell Biol 2002; 22(13): 4750–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayes S, Chawla A, Corvera S. TGF beta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J Cell Biol 2002; 158(7): 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi Y, Massagué J. Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell 2003; 113(6): 685–700. [DOI] [PubMed] [Google Scholar]
- 38. Sanders YY, Cui Z, Le Saux CJ, et al. SMAD-independent down-regulation of caveolin-1 by TGF-β: effects on proliferation and survival of myofibroblasts. PLoS One 2015; 10(2): e0116995–e0116995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mori Y, Ishida W, Bhattacharyya S, et al. Selective inhibition of activin receptor-like kinase 5 signaling blocks profibrotic transforming growth factor beta responses in skin fibroblasts. Arthritis Rheum 2004; 50(12): 4008–4021. [DOI] [PubMed] [Google Scholar]
- 40. Allolio C, Baxova K, Vazdar M, et al. Guanidinium Pairing Facilitates Membrane Translocation. J Phys Chem B 2016; 120(1): 143–153. [DOI] [PubMed] [Google Scholar]
- 41. Mii S, Ware JA, Kent KC. Transforming growth factor-beta inhibits human vascular smooth muscle cell growth and migration. Surgery 1993; 114(2): 464–470. [PubMed] [Google Scholar]
- 42. Nakerakanti S. The Role of TGF-β Receptors in Fibrosis. Open Rheumatol J 2012; 6(1): 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang W, Jiang Y, Wang Q, et al. Single-molecule imaging reveals transforming growth factor-beta-induced type II receptor dimerization. Proc Natl Acad Sci U S A 2009; 106(37): 15679–15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang XF, Lin HY, Ng-Eaton E, et al. Expression cloning and characterization of the TGF-β type III receptor. Cell 1991; 67(4): 797–805. [DOI] [PubMed] [Google Scholar]
- 45. López-Casillas F, Wrana JL, Massagué J. Betaglycan presents ligand to the TGF-β signaling receptor. Cell 1993; 73(7): 1435–1444. [DOI] [PubMed] [Google Scholar]
- 46. You HJ, Bruinsma MW, How T, et al. The type III TGF-β receptor signals through both Smad3 and the p38 MAP kinase pathways to contribute to inhibition of cell proliferation. Carcinogenesis 2007; 28(12): 2491–2500. [DOI] [PubMed] [Google Scholar]
- 47. Conley BA, Smith JD, Guerrero-Esteo M, et al. Endoglin, a TGF-beta receptor-associated protein, is expressed by smooth muscle cells in human atherosclerotic plaques. Atherosclerosis 2000; 153(2): 323–335. [DOI] [PubMed] [Google Scholar]
- 48. Hermida N, López B, González A, et al. A synthetic peptide from transforming growth factor-β1 type III receptor prevents myocardial fibrosis in spontaneously hypertensive rats. Cardiovasc Res 2009; 81(3): 601–609. [DOI] [PubMed] [Google Scholar]
- 49. Holmes AM, Ponticos M, Shi-wen X, et al. Elevated CCN2 expression in scleroderma: a putative role for the TGFβ accessory receptors TGFβRIII and endoglin. J Cell Commun Signal 2011; 5(3): 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou L-J, Ono I, Kaneko F. Role of transforming growth factor-β1 in fibroblasts derived from normal and hypertrophic scarred skin. Arch Dermatol Res 1997; 289(11): 646–652. [DOI] [PubMed] [Google Scholar]
- 51. Chun Q, ZhiYong W, Fei S, et al. Dynamic biological changes in fibroblasts during hypertrophic scar formation and regression. Int Wound J 2016; 13(2): 257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gold MH, McGuire M, Mustoe TA, et al. Updated international clinical recommendations on scar management: Part 2 - Algorithms for scar prevention and treatment. Dermatologic Surg 2014; 40(8): 825–831. [DOI] [PubMed] [Google Scholar]
- 53. Leask A, Abraham DJ. TGF-β signaling and the fibrotic response. FASEB J 2004; 18(7): 816–827. [DOI] [PubMed] [Google Scholar]
How to cite this article
- Raktoe RS, Rietveld MH, Out-Luiting JJ, Julio M, van Zuijlen PPM, van Doorn R, El Ghalbzouri A. Exon skipping of TGFβRI affects signalling and ECM expression in hypertrophic scar-derived fibroblasts. Scars, Burns & Healing, Volume 6, 2020. DOI: 10.1177/2059513118908857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary for Exon skipping of TGFβRI affects signalling and ECM expression in hypertrophic scar-derived fibroblasts by Rajiv S Raktoe, Marion H Rietveld, Jacoba J Out-Luiting, Marianna Kruithof-de Julio, Paul PM van Zuijlen, Remco van Doorn and Abdoelwaheb El Ghalbzouri in Scars, Burns & Healing