In this review, Brockdorff et al. discuss progress toward determining the role of newly defined factors in establishing features of Xi chromatin and in gene silencing by Xist RNA, including a summary of the current understanding of their relative contribution and interplay.
Keywords: X chromosome inactivation, Xist, SPEN, NCoR–HDAC3, LBR, Polycomb, RBM15, m6A RNA methylation, chromatin
Abstract
The X inactive-specific transcript (Xist) gene is the master regulator of X chromosome inactivation in mammals. Xist produces a long noncoding (lnc)RNA that accumulates over the entire length of the chromosome from which it is transcribed, recruiting factors to modify underlying chromatin and silence X-linked genes in cis. Recent years have seen significant progress in identifying important functional elements in Xist RNA, their associated RNA-binding proteins (RBPs), and the downstream pathways for chromatin modification and gene silencing. In this review, we summarize progress in understanding both how these pathways function in Xist-mediated silencing and the complex interplay between them.
X chromosome inactivation, the process that evolved in mammals to balance levels of X-linked gene expression in XX females relative to XY males, is controlled by a master regulator locus, Xist (X inactive-specific transcript), that is located on the X chromosome and functions in cis (for review, see Heard et al. 1997). Xist produces a 15- to 17-kb-long noncoding RNA (lncRNA) that accumulates over the chromosome from which it is transcribed, recruiting factors/complexes that act to modify the underlying chromatin environment and repress X-linked gene expression. Gene knockout and transgenic experiments demonstrated that Xist is both necessary and sufficient for chromosome inactivation (Penny et al. 1996; Lee and Jaenisch 1997).
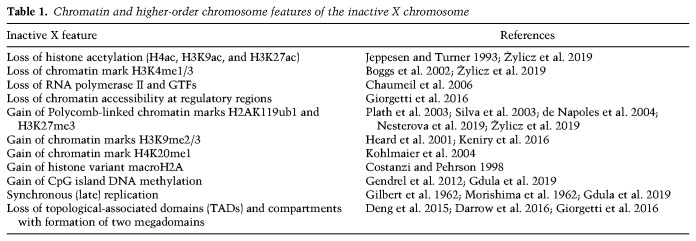
Studies over many years have defined chromatin features, chromatin-associated factors, and higher-order chromosome organization associated with the inactive X chromosome (Xi) (Fig. 1; Table 1). An important challenge for the field is to understand cause and effect. Which features of Xi are required to establish gene silencing and depend on factors recruited directly by Xist RNA and which are secondary but possibly still important for long-term chromosome silencing through development and into adulthood? To address this issue, studies on early mouse embryos and using mouse embryonic stem cell (mESC) models in vitro defined Xi features that are established early following the onset of Xist RNA expression, interpreted as indicating a role in establishment of X inactivation. Important examples include exclusion of RNA Polymerase II (RNAPII), loss of histone modifications associated with gene activity, and acquisition of histone modifications catalyzed by the Polycomb complexes PRC1 and PRC2. Conversely, Xi features that are established after a delay following Xist RNA expression, for example, CpG island DNA methylation and deposition of the histone variant macroH2A, are thought to function in maintenance rather than establishment of silencing.
Figure 1.
Features of the inactive X chromosome. (A) Comparison of chromatin features on the active X chromosome (Xa) and the inactive X chromosome (Xi) at the nucleosomal scale. Xi is characterized by depletion of histone modifications associated with gene activity, shown here histone acetylation, normally enriched at promoters and enhancers, H3K4me3, enriched at promoters and H3K36me3, high levels of which occur within the bodies of active genes. Additionally, Xi chromatin has enhanced levels of histone modifications associated with gene repression; for example, H3K27me3, H2AK119ub1, H3K9me2/3, and high levels of the histone variant macroH2A. DNA methylation is acquired at CpG islands of Xi genes. RNA polymerase II (RNAPII) and associated general transcription factors (GTFs) are depleted from Xi, as are enhancer-bound transcription factors (TFs), as evidenced by reduced chromatin accessibility seen using ATAC-seq. Similarly, there are reduced levels of the insulator protein CTCF and the cohesin complex at insulator/boundary elements. (B) Changes in higher order structure of the Xi chromosome. Key examples are an approximately twofold chromatin compaction relative to Xa, association of Xi with the nuclear and/or nucleolar periphery, reduced TAD and compartment structure (related to reduced CTCF/cohesin binding), with interactions instead occurring within one of two large megadomains, and synchronous replication across most of the chromosome, usually in late S phase.
Table 1.
Chromatin and higher-order chromosome features of the inactive X chromosome
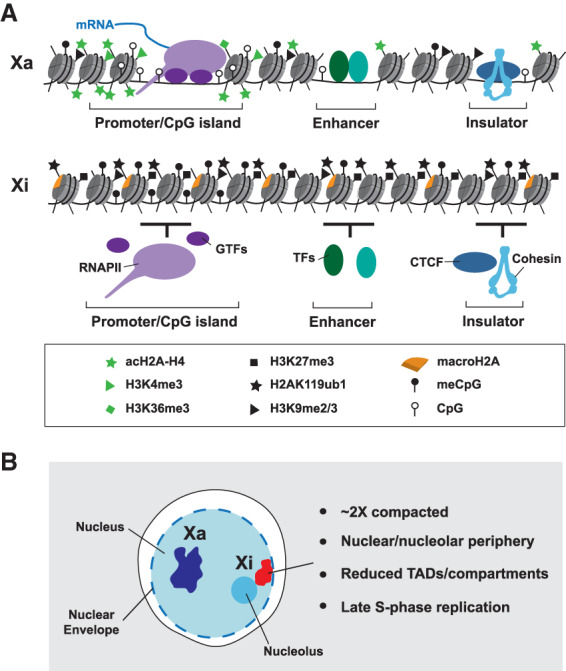
Significant progress toward defining the critical pathways for establishment of gene silencing has come from the identification of functional sequence elements in Xist RNA and the RNA-binding proteins (RBPs) that they bind. Thus, deletion analysis defined the A repeat, one of a number of conserved tandem repeat blocks (repeated X7.5 in mice and X8 in humans) present in Xist RNA sequence (Fig. 2A), as being critical for silencing, while multiple regions of the transcript were defined as functioning redundantly to mediate local accumulation of Xist RNA (Wutz et al. 2002). An important breakthrough toward identifying the factors that interact with the A repeat (and other regions of Xist RNA), came from complementary studies that applied either proteomic or genetic screening strategies (Chu et al. 2015; McHugh et al. 2015; Minajigi et al. 2015; Moindrot et al. 2015; Monfort et al. 2015). Leading candidates that emerged from this work are summarized in Figure 2A,B. The RBP SPEN, which functions through interaction with the histone deacetylase complex NCoR-HDAC3 (and possibly other factors), was shown to bind Xist A repeat directly (Monfort et al. 2015). The closely related RBP RBM15 also binds the A-repeat, and functions at least in part by recruiting the METTL3/14 complex responsible for catalyzing N6-methyladenosine (m6A) on mRNA (Patil et al. 2016). Additionally, the Lamin B receptor (LBR), was identified as binding Xist RNA (McHugh et al. 2015). Although not a known RBP, subsequent work reported that LBR has an unstructured SR domain that mediates interaction with Xist RNA (Chen et al. 2016). Finally the RBP hnRNPK was identified as an abundant Xist-binding protein (Chu et al. 2015), with subsequent studies revealing that it recognizes the Xist B/C-repeat region and directs recruitment of PRC1 and PRC2 Polycomb complexes to Xi (Cooper et al. 2016; Almeida et al. 2017; Pintacuda et al. 2017). It should be noted that the latter studies overturned the prior model for Polycomb recruitment to Xi, which invoked PRC2 interaction with Xist A repeat as the initiating event (Zhao et al. 2008). In this review, we discuss progress toward determining the role of these newly defined factors in establishing features of Xi chromatin and in gene silencing by Xist RNA, including a summary of our understanding of their relative contribution and interplay.
Figure 2.
Xist RNA-binding proteins (RBPs) with a role in chromosome silencing. (A) Binding profiles of key RBPs discussed in this article. iCLIP/eCLIP data obtained from various sources (Chen et al. 2016; Lu et al. 2016; Cirillo et al. 2017; Nesterova et al. 2019) is shown against a map of the Xist gene with tandem repeat elements A–F indicated below. A-repeat and B/C-repeat regions are highlighted with pale-blue shading. The m6A track illustrates sites of m6A deposition as determined by m6A-seq (Coker et al. 2020). Gray bar indicates the location of LBS on Xist. (B) Features of the RBPs SPEN, RBM15, LBR, and hnRNPK. Schematics indicate domain architecture with key interacting partners shown at the right. Validated direct interactors are shown in black. (RRM) RNA recognition motif; (RID) RBPJ interaction domain; (SPOC) Spen paralog and ortholog C-terminal; (RS) arginine/serine; (PEMT) phosphatidylethanolamine N-methyltransferase; (KH) K homology; (KI) K interaction.
Assaying Xist-mediated chromosome silencing
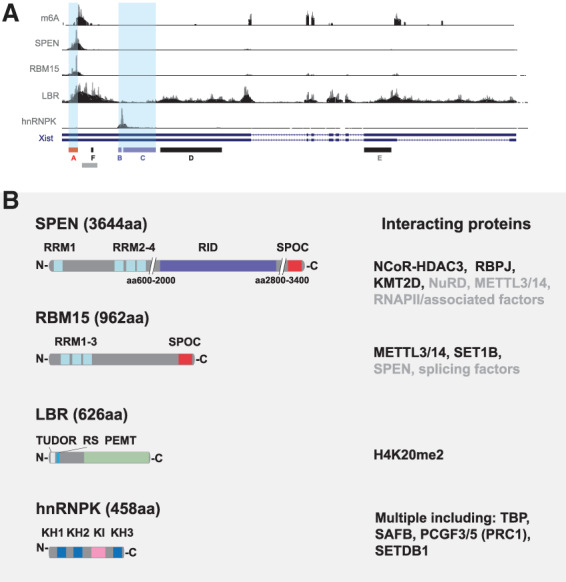
In considering recent advances in this field it is important to appreciate the varied models and assays that have been used in analyzing the contribution of different factors to Xist-mediated chromosome silencing, not least because reported findings may otherwise appear contradictory. As summarized in Figure 3A, the principal models that provide access to the developmental stage when X inactivation is established are early mouse embryos and pluripotent embryonic stem cells (mESCs). The latter include XX mESCs, which express Xist RNA from a single X chromosome when differentiated in vitro, XY mESCs with Xist expression driven by a doxycycline-activated promoter, either from the single X chromosome allele, or from an autosomally integrated Xist transgene, and XX mESCs with a doxycycline inducible promoter engineered into one of the two Xist alleles. Models using inducible promoters offer the advantage of regulatable, synchronous, and homogeneous Xist RNA expression within cell populations, albeit at levels not necessarily equivalent to expression from the physiological Xist promoter. Analysis of X inactivation in embryos is important to verify observations in a physiological context. In addition to the aforementioned models, some studies have analyzed XX somatic cells in which X inactivation is already established. However, maintenance of X inactivation in somatic cells has been reported to be Xist-independent (Csankovszki et al. 1999), albeit using relatively crude gene silencing assays, and as such these models may not be ideally suited to studying the establishment of Xist-mediated silencing.
Figure 3.
Models and assays for X inactivation studies. (A) Early mouse embryos. X inactivation (indicated as condensed bar with Xist RNA represented as green dashed line) proceeds at the two- to four-cell stage (E1.5–E2.5), initially on the paternally inherited X chromosome (Xp), referred to as imprinted X chromosome inactivation (iXCI). iXCI is maintained in trophectoderm (orange) and extraembryonic endoderm (green) derived tissues through development (shown here E3.5–E10.5), whereas in cells of the embryo proper (blue), initial reactivation of Xp at E3.5 is followed by random X inactivation (rXCI) of Xp or the maternal X chromosome (Xm) at around E5.5. (B) In XX mouse embryonic stem cells (mESCs) both X chromosomes are active. Random X inactivation proceeds following the onset of cellular differentiation in vitro. (C,D) X inactivation in mESCs can be uncoupled from differentiation by using an inducible promoter (red line) to drive Xist expression, illustrated on the single X chromosome in XY mESCs (C) or one of two X chromosome in XX mESCs (D). Silencing can be induced with or without cell differentiation in these models. Assays used to assess Xist-mediated silencing in wild-type (+/+) versus silencing factor mutant (−/−) include the following: (E) Phenotype of female versus male embryos/adults. (F,G) Imaging-based assays, notably nascent RNA FISH where abrogated silencing in mutants is indicated by the detection of nascent RNA (large red dot) signal within the Xist RNA domain (gray; F), and single-molecule (sm) FISH to quantify mRNA from an X-linked gene in the presence or absence of Xist RNA induction (G). smFISH illustration shows Xist RNA (gray) induced on the single X chromosome in an XY mESC line with X-linked gene mRNA (small red dots) being strongly reduced in wild-type, but not in silencing factor mutant cells. Finally, molecular assays have been used to directly quantify levels of X-linked mRNAs. (H) RNA-seq analysis using cells in which divergent X chromosomes with high single-nucleotide polymorphism density (orange and black bars), allow precise determination of the Xa:Xi ratio for RNAs (orange and black dots) for most X-linked genes when used in conjunction with nonrandom X inactivation/Xist induction.
Assays for Xist-mediated silencing can be grouped into three categories; indirect phenotype/viability assays, direct imaging based assays and direct molecular assays, for example, allelic RNA-seq, illustrated in Figure 3B. Each of the approaches affords distinct advantages, but equally has specific limitations. Phenotypic assays provide evidence for the importance of a given factor in a physiological setting, but do not quantify the underlying deficiency in gene silencing. Imaging-based assays provide a good basis for measuring silencing in individual cells, within a population, for example, in early embryos, but provide limited quantitation and throughput (the number of X-linked genes that is practical to assay). Molecular assays such as allelic RNA-seq provide highly quantitative measurements of silencing for multiple genes, but the data are in most cases averaged values for large and often heterogeneous populations of cells. The increasing application of single-cell RNA-seq technologies has the potential to overcome this latter limitation.
A central role for SPEN in Xist-mediated silencing
The RBP SPEN was identified as a key factor for establishment of Xist-mediated silencing in several independent studies (Chu et al. 2015; McHugh et al. 2015; Moindrot et al. 2015; Monfort et al. 2015). Recent work has validated this conclusion in a physiological model, preimplantation mouse embryos (Dossin et al. 2020). SPEN (synonyms: MINT and SHARP), >400 kDa, is a conserved RNA-binding protein containing four canonical RRM (RNA recognition motif) domains at the N terminus and a C-terminal SPOC (SPEN paralog and ortholog C-terminal) domain (Fig. 2B). The RRM and SPOC domains are responsible for SPEN's protein–RNA interactions and protein–protein interactions, respectively. EMSA experiments confirmed that a direct interaction occurs between SPEN RRM2–4 and Xist A repeat, with comparable binding affinity to SRA (steroid receptor RNA activator) RNA, previously identified as a SPEN target (Arieti et al. 2014; Monfort et al. 2015). PARIS (psoralen analysis of RNA interactions and structures), together with SPEN RRM iCLIP-seq, suggest that SPEN binding to the Xist A repeat likely occurs at the single-stranded nucleotides 3–5 nt upstream of the interrepeat duplex (Lu et al. 2016). Analysis of the interaction of SPEN with SRA RNA revealed that RRM3 is the principal domain mediating protein–RNA interaction (Arieti et al. 2014).
SPEN was originally identified as having a role in the Notch/RBP-J signaling pathway in Drosophila (Oswald et al. 2002), functioning through an interaction with the SMRT/NCoR–HDAC3 histone deacetylase complex, which represses gene transcription (Shi et al. 2001). HDAC3 is a stoichiometric component of the SMRT/NCoR complex whose core enzymatic activity and thus repressive function depends on this interaction (Emmett and Lazar 2019). Similar to SPEN, HDAC3 loss of function strongly abrogates Xist-mediated silencing (McHugh et al. 2015; Żylicz et al. 2019). These findings indicate that SPEN is directly recruited to Xist RNA through binding to the A repeat by its RRM domain, with interaction with SMRT/NCOR–HDAC3 through the SPEN SPOC domain (Ariyoshi and Schwabe 2003; Mikami et al. 2014), resulting in histone deacetylation and repression of X-linked genes. Consistent with this model, a recent study reported that deacetylation of H4 and H3K27 are among the earliest changes in chromatin modification following the onset of Xist RNA expression (Żylicz et al. 2019).
While recruitment of NCoR–HDAC3 by SPEN is a key mechanism in Xist-mediated silencing, emerging evidence indicates that SPEN has additional functions in X inactivation. Specifically, deletion of the SPEN RNA-binding domains and/or the entire protein leads to a more dramatic loss of silencing than HDAC3 loss of function (Nesterova et al. 2019; Żylicz et al. 2019; Dossin et al. 2020). Interestingly, deletion of the C-terminal SPOC domain alone results in an intermediate silencing deficit (Dossin et al. 2020). Together, these findings suggest that there are other interacting factors, in addition to HDAC3, important for SPEN function in Xist-mediated silencing. One candidate is the H3K4 methyltransferase coactivator complex KMT2D, which has been reported to interact with the SPEN SPOC domain (Oswald et al. 2016). SPEN and KMT2D also physically interact in Drosophila and permit the chromatin regulation of Notch targets in vivo (Oswald et al. 2016). Sequestration of KMT2D by SPEN could conceivably be linked to the loss of H3K4me3 on Xi. A recent analysis of the SPEN SPOC domain interactome in mESCs (Dossin et al. 2020), has highlighted other candidate factors, notably the NuRD complex, a finding that is consistent with a prior study (Shi et al. 2001), and also RNAPII and associated cofactors. The latter may indicate directed RNAPII inactivation by SPEN, possibly accounting for the reduced RNAPII levels over the Xi chromosome. Further studies are required to determine whether these additional SPEN-linked functions contribute to Xist-mediated silencing.
Besides its interactions with chromatin modifying factors, SPEN loss of function has been reported to reduce Xist RNA levels and to disrupt local accumulation over the Xi domain, as has deletion of the Xist A-repeat region (Nesterova et al. 2019), and this also could be a contributory factor in abrogated silencing following SPEN loss of function.
A role for RBM15 and the METTL3/14 complex in Xist-mediated chromosome silencing
RNA modification has emerged as a new layer in controlling a variety of processes in RNA metabolism (Roundtree et al. 2017). Xist RNA shows high levels of m6A (Ke et al. 2015; Linder et al. 2015; Patil et al. 2016; Coker et al. 2019; Nesterova et al. 2019), an abundant RNA modification in mRNA and noncoding RNA (Linder et al. 2015; Patil et al. 2016). Major m6A peaks in Xist RNA lie immediately downstream from the A repeat in Xist exon I and at downstream locations in Xist exons III/IV and VII (Fig. 2A). The m6A modification is deposited on mRNA by the METTL3/14 complex consisting of a catalytic heterodimer of METTL3–METTL14 and regulatory subunit WTAP, together with accessory proteins including RBM15 and KIAA1429 (Meyer and Jaffrey 2017). YTH-domain-containing proteins YTHDC1/2 and YTHDF1/2/3 serve as m6A readers in vivo (Meyer and Jaffrey 2017).
RBM15/15B (synonyms: OTT1/3) derive from the same protein family as SPEN and share a conserved arrangement of N-terminal RRM domains and a C-terminal SPOC domain. RBM15 interacts directly with the Xist A-repeat region as determined by CLIP-seq (Fig. 2A,B; Patil et al. 2016). As noted above, RBM15 (and its direct homolog, RBM15B), have been implicated in targeting the METTL3/14 complex to specific RNAs, including Xist (Patil et al. 2016). However, like SPEN, RBM15 also interacts with other factors/complexes. Examples include components of the RNA export pathway (Zolotukhin et al. 2009), and SET1B (synonym: KMT2G), a subunit of an H3K4 methyltransferase complex that interacts directly with the RBM15 SPOC domain (Lee and Skalnik 2012). Similar to the argument above for SPEN and KMT2D, sequestration of SET1B by RBM15 could contribute to reduced H3K4me3 on Xi. A recent analysis of the RBM15 interactome in mESCs confirmed association with the m6A and the SET1B complexes, and in addition, reported several other potentially interesting links, including RNA splicing factors and, intriguingly, SPEN (Coker et al. 2020). Whether or not these associations involve direct interaction with RBM15 remains to be determined.
Perturbation of RBM15/METTL3/14 complex subunits affects Xist-mediated silencing to varying degrees, ranging from little or no silencing deficiency following RNAi-mediated knockdown of the regulatory subunit WTAP (Chu et al. 2015; Moindrot et al. 2015) through to a major silencing deficiency following knockdown of the catalytic subunit METTL3 (Patil et al. 2016). Loss of function of the m6A reader YTHDC1 was also reported to strongly abrogate silencing, and, moreover, tethering YTHDC1 to the 3′ end of XIST was sufficient to rescue its silencing function in the absence of the m6A methylation complex (Patil et al. 2016). Set against these findings, more recent studies reported a relatively modest effect on Xist-mediated silencing following CRISPR/Cas9-mediated deletion of genes encoding RBM15 and subunits of the METTL3/14 complex (Nesterova et al. 2019). This conclusion was supported by analysis of deletions encompassing the major m6A peak located immediately downstream from the Xist A repeat (Nesterova et al. 2019; Coker et al. 2020). The functional role of other Xist m6A sites and how they are established have not been analyzed to date. Gene knockout of RBM15 in mice results in midgestation embryo lethality but with no apparent female bias (Raffel et al. 2009). This finding supports that RBM15 plays only a modest role in Xist-mediated silencing, although possible redundancy with RBM15B may be important in this context.
Several factors likely underlie the disparate findings on the role of RBM15/METTL3/14 complex in Xist-meditated silencing. The METTL3/14 complex has a global influence on ∼7000 mRNAs, affecting pre-mRNA processing, RNA stability, and translation (for review, see Yue et al. 2015), so loss of function likely generates significant secondary effects. The manifestation of secondary effects will in turn be influenced by the means for abrogating function (knockdown vs. chronic knockout vs. acute knockout). Additional confounding factors are functional redundancy of METTL3/14 complex subunits (for example, RBM15 and RBM15B) (Patil et al. 2016), and possibly nonessentiality of regulatory subunits (for example, WTAP) in m6A catalysis. Finally, as noted above, use of different models (XY vs. XX mESCs) and silencing assays (single-molecule FISH vs. allelic RNA-seq), is likely a contributory factor for the aforementioned studies coming to different conclusions (see also comments below).
While the emerging consensus is that RBM15 and m6A play a relatively minor role in Xist-mediated gene silencing, at least compared with other factors such as SPEN and the Polycomb system, more work is needed to corroborate this conclusion and also to understand the mechanistic basis for the effect that is seen. In the latter case, possibilities include that m6A readers such as YTHDC1 mediate gene silencing, or that m6A modification impacts on Xist RNA folding and/or binding of other RBPs linked to silencing (so-called “m6A switch”) (Liu et al. 2015), for example, SPEN. With this in mind it is interesting to consider how RBM15 and SPEN may affect one another's interaction with A repeat, given both proteins likely compete for common binding sites. A final consideration, given that m6A has been reported to regulate RNA stability (Wang et al. 2014), is that RBM15/m6A may affect Xist RNA turnover, and thereby modify Xist-mediated silencing indirectly.
A reassessment of the role of LBR in Xist-mediated silencing
Proteomic screens identified Lamin B receptor (LBR), an integral component of the nuclear lamina, as a direct Xist interactor (McHugh et al. 2015; Minajigi et al. 2015). Consistent with this finding LBR was identified as a candidate mRNA binder in the mRNA-bound proteome (Baltz et al. 2012). More recent work has reported that LBR binds to three sites across Xist RNA as determined by CLIP-seq, with the most prominent, referred to as LBS, located downstream from the major 5′ m6A peak in and encompassing the entire F repeat (Fig. 2A; Chen et al. 2016). The interaction of LBR with Xist RNA was shown to be through an arginine–serine tract (RS) ( Fig. 2B). In addition to major sites of enrichment there is a widespread distribution of LBR across Xist RNA (Chen et al. 2016; Cirillo et al. 2017), in contrast to other well-characterized Xist RBPs such as SPEN, RBM15, and hnRNPK (see Fig. 2A). This may indicate a relatively low sequence specificity in the interaction of LBR with RNA.
In initial studies, LBR was reported to be critically important for Xist-mediated silencing, as determined using smFISH (see Fig. 3) following LBR depletion by RNAi (McHugh et al. 2015; Chen et al. 2016). Similar loss of silencing was seen upon deletion of LBS, and moreover, this deficit was complemented by tethering LBR synthetically to the 3′ end of Xist RNA. Further analysis suggested that LBR facilitates silencing by recruiting the inactive X chromosome to the nuclear lamina to enable Xist RNPs to spread to actively transcribed genes (Chen et al. 2016). A more recent study reported contrasting conclusions with only a minor effect on silencing following CRISPR/Cas9-mediated deletion of Lbr or LBS following Xist induction in XX mESCs (Nesterova et al. 2019). This study also analyzed Lbr deletion in an autosomal Xist transgenic XY mESC line and found no detectable effects on Xist-mediated silencing. The minor silencing deficiency observed in XX mESCs disappeared after extended Xist RNA induction (6 d), suggesting that LBR may enhance the rate of Xist-mediated silencing, possibly, as reported by Chen et al. (2016), by facilitating association of Xi with the nuclear periphery. The conclusion that LBR plays a relatively minor role in Xist-mediated silencing is supported by analysis of Lbr gene mutations in mice, for which there are no reported female-specific phenotypes (Shultz et al. 2003; Cohen et al. 2008).
The contrasting findings in different studies on the role of LBR in X inactivation are likely attributable to the use of different models and assay systems. In particular, silencing deficiencies determined using the smFISH assay (Fig. 3) appear to be considerably greater than those seen using allelic RNA-seq, also evident in relation to the disparate findings on the role of RBM15 and the METTL3/14 complex discussed above. The quantitative power of allelic RNA-seq, together with the greater coverage of X-linked genes, suggests this assay likely provides a more reliable measure, although this point is open to debate. An additional consideration in relation to studies on LBR is the use of different perturbation strategies, specifically RNAi-mediated knockdown, which is relatively acute, compared with chronic effects in null mESCs generated using CRISPR/Cas9 gene deletion. Again, further studies should address whether this is relevant.
Recruitment of hnRNPK and the Polycomb system through B/C repeat contributes to Xist-mediated silencing
The Polycomb system, which comprises several multiprotein complexes that catalyze the histone modifications H2AK119ub1 (canonical and variant PRC1 complexes), and H3K27me3 (PRC2 complexes), is recruited to Xi in an Xist RNA-dependent manner (for review, see Brockdorff 2017). Briefly, the RBP hnRNPK bound to the Xist RNA B/C repeat initiates Polycomb recruitment via direct interaction of the hnRNPK KI domain (Fig. 2B), and the PCGF3/5 subunit of PCGF3/5–PRC1. PCGF3/5–PRC1-mediated H2AK119ub1 further directs concentration of other variant PRC1 complexes, PRC2, and ultimately, canonical PRC1 via binding to PRC2-mediated H3K27me3 (da Rocha et al. 2014; Chu et al. 2015; Cooper et al. 2016; Almeida et al. 2017; Pintacuda et al. 2017). hnRNPK has three RNA-binding KH domains (Fig. 2B) that function cooperatively by each recognizing a C-rich RNA motif (Paziewska et al. 2004). The B/C-repeat region in both humans and mice comprises ∼30 tandem repeats (WGCCC), which could theoretically bind up to 10 hnRNPK molecules. Occupancy of this element by multiple hnRNPK molecules is supported by CLIP-seq (Fig. 2A; Cirillo et al. 2017), and EMSA analyses (Colognori et al. 2019). Perturbation of hnRNPK strongly abrogates Polycomb recruitment, phenocopying deletion of the B/C repeat (Chu et al. 2015; Pintacuda et al. 2017; Colognori et al. 2019), while tethering of hnRNPK to XistΔB/C can rescue recruitment of Polycomb (Pintacuda et al. 2017).
Early studies reported that perturbation of PRC2 results in a weak effect on Xist-mediated silencing (Wang et al. 2001; Silva et al. 2003; Kalantry and Magnuson 2006; Kalantry et al. 2006;). More recent studies addressed the role of PRC1 and PRC2 together (deletion of PCGF3/5 or the B/C repeat region), and reported a substantive reduction in silencing efficiency, using both inducible autosomal Xist in XY mESCs and XX mESC models (Almeida et al. 2017; Pintacuda et al. 2017; Nesterova et al. 2019). It was further shown that complete PRC1 loss of function as opposed to PRC2 loss (and as a consequence, canonical but not variant PRC1) is the principal contributor to abrogated silencing (Nesterova et al. 2019). Presumably H2AK119ub1 catalysis is important for this effect, although this remains to be formally proven. A primary role for PRC1 was substantiated through phenotype analysis of Pcgf3/5 null embryos, with female lethality occurring at an earlier stage than in male embryos (E7.5–E9.5 compared with E9.5–E12.5).
Other recent studies broadly support that the B/C-repeat element, hnRNPK, and PRC1 mediate the recruitment of Polycomb to Xi (Chu et al. 2015; Bousard et al. 2019; Colognori et al. 2019), although there are some differences in interpretation. Colognori et al. (2019) found that a low level of PRC2-mediated H3K27me3 is retained over Xi following deletion of the catalytic subunits of PRC1, and this was suggested to indicate an independent pathway for PRC2 recruitment by Xist RNA. However, because in this instance deletion of PRC1/Xist B repeat was performed in XX somatic cells (MEFs), after establishment of X inactivation, retention of H3K27me3 could be linked to PRC2 self-templating, as has been described previously (Hansen et al. 2008; Margueron et al. 2009). The study by Bousard et al. (2019), also reported low levels of H3K27me3 enrichment on Xi after induction of Xist-RNA lacking the B/C repeat region, although this was only within the bodies of silenced genes and is likely due to loss of active gene-associated histone modifications, specifically H3K4me3 and H3K36me3, that directly inhibit PRC2 catalytic activity (Schmitges et al. 2011).
Bousard et al. (2019) concluded that the Polycomb pathway makes a relatively small contribution to Xist-mediated silencing, in contrast to the conclusions of Pintacuda et al. (2017), Nesterova et al. (2019), and Colognori et al. (2019). This difference could relate to the use of different cell models as Bousard et al. (2019) analyzed XY mESCs with inducible Xist expression on the single X chromosome. There were also differences in the extent of the B/C-repeat region deletions, and the time points following onset of Xist RNA expression. Indeed, Nesterova et al. (2019) reported a more pronounced effect on Xist-mediated silencing following deletion of the B/C repeat region after 6 d (with mESC differentiation) compared with 1 d of Xist RNA induction. Bousard et al. (2019) were not able to analyze later time points as Xist expression triggers rapid cell death in the XY mESC model.
The mechanism by which Polycomb represses genes is not well understood, and this applies also in the case of Xist-mediated silencing. As noted above, both H2AK119ub1 and H3K27me3 enrichment occur over gene-rich domains covering much of the Xi chromosome, encompassing regulatory elements (REs), gene bodies, and intergenic regions. Thus, silencing could result from the activity of reader proteins that bind these histone modifications, or alternatively, through effects on chromatin structure/accessibility, for example chromatin compaction limiting TF and RNAPII accessibility at REs.
In relation to Polycomb reader proteins, recent work has shown that recruitment of the chromosomal architecture protein SmcHD1 to Xi, which occurs several days after the onset of Xist expression (Gendrel et al. 2012), is dependent on PRC1 activity (Jansz et al. 2018b). SmcHD1 is thought to function in long-term maintenance of X inactivation, being required for DNA methylation at the majority of Xi CpG island promoters, and also for Xi-specific higher-order chromosome structure (Gendrel et al. 2013; Mould et al. 2013; Jansz et al. 2018a; Sakakibara et al. 2018; Wang et al. 2018; Gdula et al. 2019). This pathway may therefore account for the more pronounced contribution of Polycomb to Xist-mediated silencing at later stages of mESC differentiation.
A final consideration in how Polycomb impacts Xist-mediated silencing comes from a recent report showing abnormal Xist RNA localization following deletion of the Xist B repeat, hnRNPK, or PRC1 (Colognori et al. 2019). Thus, similar to SPEN and the A repeat, at least some of the observed silencing deficiency in the absence of the Polycomb system may result from perturbation of Xist–RNA localization rather than Xi chromatin modification.
The interplay of silencing pathways in X inactivation
A key question leading on from the identification of pathways for Xist-mediated silencing is their relative contribution and relationship to one another. In addressing this it is important to consider that not all genes on the X chromosome, or for that matter on autosomes silenced by Xist transgenes, respond equivalently. Several studies have reported variation in the silencing rate of individual genes and gene subsets (Okamoto et al. 2004; Lin et al. 2007; Berletch et al. 2015; Marks et al. 2015; Nesterova et al. 2019), and a number of genes show either partial or complete escape from X inactivation, in some cases varying depending on developmental stage and cell type (Berletch et al. 2015).
As discussed above, the use of different models, time points, and assays in analyzing key factors means that it is difficult to make meaningful quantitative comparisons between studies. More recently this has been addressed in a systematic analysis of the major pathways, analyzing common time points following the onset of Xist RNA expression (Nesterova et al. 2019). A-repeat/SPEN was found to be the predominant pathway, with only low levels of silencing detected in its absence. The B-repeat/hnRNPK/PRC1 pathway was the second most significant in terms of overall contribution to silencing, with both the RBM15/METTL3/14 complex and LBS/LBR contributing relatively little. A simple interpretation of these observations is that the A-repeat/SPEN pathway establishes silencing of X-linked genes, and that other pathways function downstream, augmenting A-repeat/SPEN-mediated repression and/or stabilizing gene silencing for long-term maintenance of X inactivation. In support of this view, Żylicz et al. (2019) reported that HDAC3 loss of function delays the acquisition of Polycomb-mediated H2AK119ub1/H3K27me3 on Xi. This interpretation nevertheless needs to be tempered taking into consideration that A-repeat/SPEN and B-repeat/hnRNPK/PRC1 have more than one downstream effector and, moreover, that both pathways contribute to correct localization and/or maintaining high levels of Xist RNA on the chromosome.
In relation to the importance of different pathways for silencing specific genes or gene subsets, two recent studies applied machine learning to define which features affect the rate or efficiency of silencing of X-linked genes. In both cases, the principal determinants that were identified were 2D/3D proximity relative to the Xist transcription site and pre-existing chromatin features linked to gene activity (reducing silencing efficiency) or gene repression (enhancing silencing efficiency) (Barros de Andrade et al. 2019; Nesterova et al. 2019). The results of the chromatin feature analysis suggest that highly expressed genes are silenced less well and vice versa. Accordingly Nesterova et al. (2019), reported that the SPEN/NCoR-HDAC3 pathway is especially important for silencing highly transcribed genes, a finding that accords with a prior study that analyzed allelic silencing in trophoblast tissues of female embryos following deletion of the A repeat (Sakata et al. 2017). Consistent with these observations, initial deacetylation events linked to SPEN-HDAC3 occur preferentially at transcriptionally active regions (Żylicz et al. 2019). Moreover, a recent study reported that SPEN-binding sites on Xi chromatin correspond to promoters and enhancers of active genes, correlating closely with sites of RNAPII enrichment (Dossin et al. 2020). As silencing progresses, the level of SPEN enrichment reduces proportionately, suggesting that active transcription is required for the ongoing recruitment of SPEN.
The B/C-repeat/hnRNPK/PRC1 pathway results in increased deposition of H2AK119ub1 and H3K27me3 chromosome wide, notably over gene-rich domains where Xist RNA is concentrated (Marks et al. 2009; Calabrese et al. 2012; Pinter et al. 2012; Nesterova et al. 2019; Żylicz et al. 2019). Accordingly, recent analyses using allelic RNA-seq indicate that the contribution of Polycomb-mediated silencing is equivalent for most, if not all, of the genes located in cis with expressed Xist RNA (Almeida et al. 2017; Pintacuda et al. 2017; Colognori et al. 2019; Nesterova et al. 2019). Nesterova et al. (2019) did find, however, that genes with pre-existing polycomb marks, which in general are expressed at a relatively low level, tend to be more dependent on PCGF3/5 than those with preactive marks. Consistent with these findings, the placement of the repressive marks H2AK119ub1 and H3K27me3 occurs preferentially at pre-existing repressive environments (Nesterova et al. 2019). The likely reason for this is that positive feedback facilitates amplification of existing Polycomb-mediated chromatin modifications. Conversely, the small contribution made by PRC2 in silencing X-linked genes was found to occur predominantly at highly active genes during later progression of X inactivation, suggesting a role in maintaining or reinforcing depletion of histone modifications associated with gene activity, for example H3K27ac.
The differential effects on repression of gene subsets by the two major silencing pathways are relatively subtle, with most genes showing equivalent dependence on both pathways. Similarly, for the RBM15/METTL3/14 and LBR/LBS pathways there were no significant differential effects on gene subsets (Nesterova et al. 2019). Thus, the overall picture is that the different silencing pathways function synergistically, contributing to the silencing of all X-linked genes rather than through division of labor, to distinct gene subsets.
Future perspectives
Recent years have seen significant progress toward understanding how Xist RNA establishes chromosome-wide silencing. Chromatin modifications linked to SPEN/HDAC3 and hnRNPK/PRC1 provide a compelling explanation for classical features of Xi chromatin, namely, histone hypoacetylation and deposition of H2AK119ub1 and H3K27me3 histone modifications, all of which occur commensurate with the onset of Xist RNA expression (Nesterova et al. 2019; Żylicz et al. 2019). The basis for other classical Xi features linked to establishment of Xist-mediated silencing, specifically depletion of RNAPII and loss of H3K4me2/3 (Chaumeil et al. 2006), remain to be fully resolved. One possibility is that these are indirect consequences of a repressive chromatin state linked to the aforementioned histone modifications, and negative feedback mechanisms such as inhibition of H3K4me3 by PRC2-mediated H3K27me3 (Schmitges et al. 2011). Alternatively, more direct mechanisms may play a role, supported by the findings that the SPEN SPOC domain interacts with RNAPII/associated cofactors (Dossin et al. 2020), and both SPEN and RBM15 have direct interactions with complexes that catalyze H3K4me3 (Lee and Skalnik 2012; Oswald et al. 2016; Coker et al. 2020). The mechanistic basis for the relatively minor contribution of RBM15/m6A and LBR in establishment of silencing is at present a matter of debate but may relate to known roles for the RBM15/m6A pathway in regulating mRNA dynamics, and the role of LBR in associating the Xi with the nuclear periphery. Figure 4 summarizes key findings discussed in this review.
Figure 4.
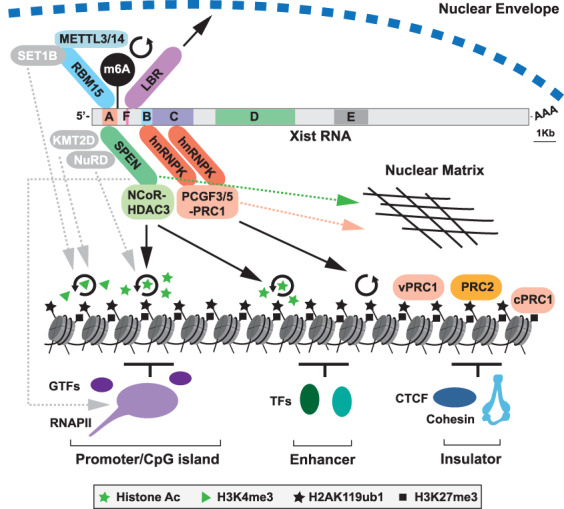
Pathways for Xist-mediated silencing. Schematic illustrates Xist RNA with tandem repeats A-F indicated together with associated RBPs and downstream effectors implicated in chromosome silencing. SPEN is linked to promoter/enhancer histone deacetylation through NCoR–HDAC3. hnRNPK recruits PCGF3/5–PRC1, which catalyzes widespread deposition of H2AK119ub1. H2AK119ub1 provides a binding platform for other variant (v) PRC1 complexes and PRC2, with PRC2-mediated H3K27me3, then recruiting canonical (c) PRC1. RBM15 is linked to m6A deposition on Xist RNA via recruitment of the METTL3/14 complex, facilitating silencing through yet to be determined mechanisms. LBR links Xist/Xi with the nuclear envelope and the repressive Lamin-associated environment. Other putative pathways, indicated in gray with dashed gray arrows, are RBM15-SET1B and/or SPEN–KMT2D potentially contributing to loss of H3K4me3 on Xi, SPEN–NuRD, potentially contributing to Xi histone deacetylation, and direct inhibitory interaction of SPEN with RNAPII/cofactors. Circular arrows denote catalytic activity. Both SPEN and PRC1 also contribute to anchoring Xist RNA to the nuclear matrix (dashed pink/green arrows), which may in turn contribute to their role in silencing.
As the field has progressed toward defining the pathways for Xist-mediated silencing, new frontiers and challenges have begun to open up. A notable example is the possible role of liquid-liquid phase separation driven by intrinsically disordered proteins and associated nucleic acids, recently recognized as a fundamental mechanism in a wide range of molecular interactions in biology (for review, see Hyman et al. 2014). Several of the RBPs that interact with Xist RNA have intrinsically disordered regions, and thus have the potential to drive formation of liquid-liquid phase separated condensates. This mechanism has been proposed to be important for Xist RNA function, potentially impacting on Xist-mediated silencing or Xist RNA localization (Cerase et al. 2019). A second important challenge is to define the role of key factors/pathways in Xist-mediated silencing versus local accumulation of Xist RNA in cis. Previously these were considered to be mechanistically separable processes (Wutz et al. 2002), but as indicated above (and see Fig. 4), there are emerging examples of factors that have dual roles both in silencing and Xist RNA localization. A third important challenge will be to better understand how pathways that function in the establishment of silencing link to pathways for maintenance of X inactivation, for example, CpG island methylation and deposition of macroH2A, and similarly how silencing and maintenance pathways determine the unique topological organization of Xi. Finally, it has been shown that Xist-dependent silencing occurs only within a restricted window of opportunity in early development (Wutz and Jaenisch 2000), and yet the major silencing pathways discussed herein apparently function throughout development and in adult tissues. Determining why Xist RNA functions, in large part only in early developmental cell types, presents an intriguing and important future challenge.
Inspired by the progress toward understanding chromosome silencing by Xist RNA it will be interesting to determine whether there are other examples of genetic regulation using similar mechanisms. In a recent report Airn and Kcnq1ot1 loci, which are required for parental imprinting of gene clusters on mouse chromosome 17 and chromosome 7, respectively, were found to function in long-range recruitment of Polycomb complexes through a mechanism that is reminiscent of Xist (Schertzer et al. 2019). Specifically, lncRNAs derived from the Airn and Kcnq1ot1 loci induce Polycomb-dependent chromatin modifications in cis over domains spanning several mega-bases in trophoblast stem cells in an hnRNPK-dependent pathway. The extent of gene silencing and Polycomb modifications correlates with topological proximity to the lncRNA locus, pre-existing genome architecture, and the abundance of the lncRNA itself. These examples may represent only the tip of the iceberg as many lncRNAs have been implicated in gene silencing by associating with repressive histone-modifying complexes (Khalil et al. 2009; Guttman et al. 2011; Guttman and Rinn 2012).
Acknowledgments
We thank Heather Coker and Mafalda Almeida for critical reading of the manuscript. Work in the Brockdorff laboratory is supported by the Wellcome Trust (grant no. 215513).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.337196.120.
Freely available online through the Genes & Development Open Access option.
References
- Almeida M, Pintacuda G, Masui O, Koseki Y, Gdula M, Cerase A, Brown D, Mould A, Innocent C, Nakayama M, et al. 2017. PCGF3/5–PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356: 1081–1084. 10.1126/science.aal2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieti F, Gabus C, Tambalo M, Huet T, Round A, Thore S. 2014. The crystal structure of the split end protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs. Nucleic Acids Res 42: 6742–6752. 10.1093/nar/gku277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi M, Schwabe JW. 2003. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev 17: 1909–1920. 10.1101/gad.266203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Barros de Andrade ESL, Jonkers I, Syx L, Dunkel I, Chaumeil J, Picard C, Foret B, Chen CJ, Lis JT, Heard E, et al. 2019. Kinetics of Xist-induced gene silencing can be predicted from combinations of epigenetic and genomic features. Genome Res 29: 1087–1099. 10.1101/gr.245027.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, Deng X. 2015. Escape from X inactivation varies in mouse tissues. PLoS Genet 11: e1005079 10.1371/journal.pgen.1005079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet 30: 73–76. 10.1038/ng787 [DOI] [PubMed] [Google Scholar]
- Bousard A, Raposo AC, Zylicz JJ, Picard C, Pires VB, Qi Y, Gil C, Syx L, Chang HY, Heard E, et al. 2019. The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep 20: e48019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. 2017. Polycomb complexes in X chromosome inactivation. Philos Trans R Soc Lond Biol Sci 372: 20170021 10.1098/rstb.2017.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T. 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963. 10.1016/j.cell.2012.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerase A, Armaos A, Neumayer C, Avner P, Guttman M, Tartaglia GG. 2019. Phase separation drives X-chromosome inactivation: a hypothesis. Nat Struct Mol Biol 26: 331–334. 10.1038/s41594-019-0223-0 [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20: 2223–2237. 10.1101/gad.380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, Cerase A, McDonel P, Guttman M. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354: 468–472. 10.1126/science.aae0047 [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo D, Blanco M, Armaos A, Buness A, Avner P, Guttman M, Cerase A, Tartaglia GG. 2017. Quantitative predictions of protein interactions with long noncoding RNAs. Nat Methods 14: 5–6. 10.1038/nmeth.4100 [DOI] [PubMed] [Google Scholar]
- Cohen TV, Klarmann KD, Sakchaisri K, Cooper JP, Kuhns D, Anver M, Johnson PF, Williams SC, Keller JR, Stewart CL. 2008. The lamin B receptor under transcriptional control of C/EBPε is required for morphological but not functional maturation of neutrophils. Hum Mol Genet 17: 2921–2933. 10.1093/hmg/ddn191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker H, Wei G, Brockdorff N. 2019. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochimi Biophys Acta Gene Regul Mech 1862: 310–318. 10.1016/j.bbagrm.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Coker H, Wei G, Moindrot B, Mohammed S, Nesterova T, Brockdorff N. 2020. The role of the Xist 5′ m6A region and RBM15 in X chromosome inactivation. Wellcome Open Res 5: 31 10.12688/wellcomeopenres.15711.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognori D, Sunwoo H, Kriz AJ, Wang CY, Lee JT. 2019. Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol Cell 74: 101–117.e10. 10.1016/j.molcel.2019.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Grijzenhout A, Underwood E, Ancelin K, Zhang T, Nesterova TB, Anil-Kirmizitas B, Bassett A, Kooistra SM, Agger K, et al. 2016. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun 7: 13661 10.1038/ncomms13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393: 599–601. 10.1038/31275 [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. 1999. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet 22: 323–324. 10.1038/11887 [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. 2014. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell 53: 301–316. 10.1016/j.molcel.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, Sun Z, Huang SC, Sanborn AL, Machol I, Shamim M, et al. 2016. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci 113: E4504–E4512. 10.1073/pnas.1609643113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676. 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Deng X, Ma W, Ramani V, Hill A, Yang F, Ay F, Berletch JB, Blau CA, Shendure J, Duan Z, et al. 2015. Bipartite structure of the inactive mouse X chromosome. Genome Biol 16: 152 10.1186/s13059-015-0728-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossin F, Pinheiro I, Żylicz JJ, Roensch J, Collombet S, Le Saux A, Chelmicki T, Attia M, Kapoor V, Zhan Y, et al. 2020. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578: 455–460. 10.1038/s41586-020-1974-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmett MJ, Lazar MA. 2019. Integrative regulation of physiology by histone deacetylase 3. Nat Rev Mol Cell Biol 20: 102–115. 10.1038/s41580-018-0076-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula MR, Nesterova TB, Pintacuda G, Godwin J, Zhan Y, Ozadam H, McClellan M, Moralli D, Krueger F, Green CM, et al. 2019. The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat Commun 10: 30 10.1038/s41467-018-07907-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Apedaile A, Coker H, Termanis A, Zvetkova I, Godwin J, Tang YA, Huntley D, Montana G, Taylor S, et al. 2012. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell 23: 265–279. 10.1016/j.devcel.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Tang YA, Suzuki M, Godwin J, Nesterova TB, Greally JM, Heard E, Brockdorff N. 2013. Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol Cell Biol 33: 3150–3165. 10.1128/MCB.00145-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert CW, Muldal S, Lajtha LG, Rowley J. 1962. Time-sequence of human chromosome duplication. Nature 195: 869–873. 10.1038/195869a0 [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535: 575–579. 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346. 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300. 10.1038/ncb1787 [DOI] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P. 1997. X-chromosome inactivation in mammals. Annu Rev Genet 31: 571–610. 10.1146/annurev.genet.31.1.571 [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107: 727–738. 10.1016/S0092-8674(01)00598-0 [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. 2014. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Jansz N, Keniry A, Trussart M, Bildsoe H, Beck T, Tonks ID, Mould AW, Hickey P, Breslin K, Iminitoff M, et al. 2018a. Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat Struct Mol Biol 25: 766–777. 10.1038/s41594-018-0111-z [DOI] [PubMed] [Google Scholar]
- Jansz N, Nesterova T, Keniry A, Iminitoff M, Hickey PF, Pintacuda G, Masui O, Kobelke S, Geoghegan N, Breslin KA, et al. 2018b. Smchd1 targeting to the inactive X is dependent on the Xist-HnrnpK-PRC1 pathway. Cell Rep 25: 1912–1923.e9. 10.1016/j.celrep.2018.10.044 [DOI] [PubMed] [Google Scholar]
- Jeppesen P, Turner BM. 1993. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74: 281–289. 10.1016/0092-8674(93)90419-Q [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T. 2006. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet 2: e66 10.1371/journal.pgen.0020066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. 2006. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol 8: 195–202. 10.1038/ncb1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. 2015. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 29: 2037–2053. 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A, Gearing LJ, Jansz N, Liu J, Holik AZ, Hickey PF, Kinkel SA, Moore DL, Breslin K, Chen K, et al. 2016. Setdb1-mediated H3K9 methylation is enriched on the inactive X and plays a role in its epigenetic silencing. Epigenetics Chromatin 9: 16 10.1186/s13072-016-0064-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672. 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: E171 10.1371/journal.pbio.0020171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. 1997. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386: 275–279. 10.1038/386275a0 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. 2012. Rbm15-Mkl1 interacts with the Setd1b histone H3-Lys4 methyltransferase via a SPOC domain that is required for cytokine-independent proliferation. PLoS One 7: e42965 10.1371/journal.pone.0042965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O'Neill LP, Turner BM. 2007. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol 5: e326 10.1371/journal.pbio.0050326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. 2015. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518: 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. 10.1038/nature08398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. 2009. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res 19: 1361–1373. 10.1101/gr.092643.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kerstens HH, Barakat TS, Splinter E, Dirks RA, van Mierlo G, Joshi O, Wang SY, Babak T, Albers CA, et al. 2015. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol 16: 149 10.1186/s13059-015-0698-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. 2017. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol 33: 319–342. 10.1146/annurev-cellbio-100616-060758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami S, Kanaba T, Takizawa N, Kobayashi A, Maesaki R, Fujiwara T, Ito Y, Mishima M. 2014. Structural insights into the recruitment of SMRT by the corepressor SHARP under phosphorylative regulation. Structure 22: 35–46. 10.1016/j.str.2013.10.007 [DOI] [PubMed] [Google Scholar]
- Minajigi A, Froberg J, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al. 2015. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349: aab2276 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep 12: 562–572. 10.1016/j.celrep.2015.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep 12: 554–561. 10.1016/j.celrep.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima A, Grumbach MM, Taylor JH. 1962. Asynchronous duplication of human chromosomes and the origin of sex chromatin. Proc Natl Acad Sci 48: 756–763. 10.1073/pnas.48.5.756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould AW, Pang Z, Pakusch M, Tonks ID, Stark M, Carrie D, Mukhopadhyay P, Seidel A, Ellis JJ, Deakin J, et al. 2013. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin 6: 19 10.1186/1756-8935-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Wei G, Coker H, Pintacuda G, Bowness JS, Zhang T, Almeida M, Bloechl B, Moindrot B, Carter EJ, et al. 2019. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun 10: 3129 10.1038/s41467-019-11171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649. 10.1126/science.1092727 [DOI] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, et al. 2002. SHARP is a novel component of the Notch/RBP-Jκ signalling pathway. EMBO J 21: 5417–5426. 10.1093/emboj/cdf549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Rodriguez P, Giaimo BD, Antonello ZA, Mira L, Mittler G, Thiel VN, Collins KJ, Tabaja N, Cizelsky W, et al. 2016. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res 44: 4703–4720. 10.1093/nar/gkw105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paziewska A, Wyrwicz LS, Bujnicki JM, Bomsztyk K, Ostrowski J. 2004. Cooperative binding of the hnRNP K three KH domains to mRNA targets. FEBS Lett 577: 134–140. 10.1016/j.febslet.2004.08.086 [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. 10.1038/379131a0 [DOI] [PubMed] [Google Scholar]
- Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, et al. 2017. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell 68: 955–969.e10. 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. 2012. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res 22: 1864–1876. 10.1101/gr.133751.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. 10.1126/science.1084274 [DOI] [PubMed] [Google Scholar]
- Raffel GD, Chu GC, Jesneck JL, Cullen DE, Bronson RT, Bernard OA, Gilliland DG. 2009. Ott1 (Rbm15) is essential for placental vascular branching morphogenesis and embryonic development of the heart and spleen. Mol Cell Biol 29: 333–341. 10.1128/MCB.00370-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y, Nagao K, Blewitt M, Sasaki H, Obuse C, Sado T. 2018. Role of SmcHD1 in establishment of epigenetic states required for the maintenance of the X-inactivated state in mice. Development 145: dev166462 10.1242/dev.166462 [DOI] [PubMed] [Google Scholar]
- Sakata Y, Nagao K, Hoki Y, Sasaki H, Obuse C, Sado T. 2017. Defects in dosage compensation impact global gene regulation in the mouse trophoblast. Development 144: 2784–2797. 10.1242/dev.149138 [DOI] [PubMed] [Google Scholar]
- Schertzer MD, Braceros KCA, Starmer J, Cherney RE, Lee DM, Salazar G, Justice M, Bischoff SR, Cowley DO, Ariel P, et al. 2019. lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG island DNA. Mol Cell 75: 523–537.e10. 10.1016/j.molcel.2019.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev 15: 1140–1151. 10.1101/gad.871201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, Dreger C, Herrmann H, Kalscheuer V, Olins AL, et al. 2003. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huet anomaly. Hum Mol Genet 12: 61–69. 10.1093/hmg/ddg003 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4: 481–495. 10.1016/S1534-5807(03)00068-6 [DOI] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. 2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet 28: 371–375. 10.1038/ng574 [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Jégu T, Chu HP, Oh HJ, Lee JT. 2018. SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174: 406–421.e25. 10.1016/j.cell.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705. 10.1016/S1097-2765(00)80248-8 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C. 2015. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 29: 1343–1355. 10.1101/gad.262766.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin AS, Uranishi H, Lindtner S, Bear J, Pavlakis GN, Felber BK. 2009. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res 37: 7151–7162. 10.1093/nar/gkp782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żylicz JJ, Bousard A, Zumer K, Dossin F, Mohammad E, da Rocha ST, Schwalb B, Syx L, Dingli F, Loew D, et al. 2019. The implication of early chromatin changes in X chromosome inactivation. Cell 176: 182–197.e23. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]