Here, Yang et al. report that TEX15, a testis-specific protein, is required for transposable element (TE) silencing. They show that loss of Tex15 causes TE desilencing with intact piRNA production, and their findings identify TEX15 as a new essential epigenetic regulator that may function as a nuclear effector of MILI to silence TEs by DNA methylation.
Keywords: DNA methylation, epigenetics, MILI, spermatogenesis, TEX15, transposable element
Abstract
DNA methylation is a major silencing mechanism of transposable elements (TEs). Here we report that TEX15, a testis-specific protein, is required for TE silencing. TEX15 is expressed in embryonic germ cells and functions during genome-wide epigenetic reprogramming. The Tex15 mutant exhibits DNA hypomethylation in TEs at a level similar to Mili and Dnmt3c but not Miwi2 mutants. TEX15 is associated with MILI in testis. As loss of Tex15 causes TE desilencing with intact piRNA production, our results identify TEX15 as a new essential epigenetic regulator that may function as a nuclear effector of MILI to silence TEs by DNA methylation.
Transposable elements (TEs), including LINEs, SINEs, and endogenous retroviruses, occupy ∼40% of the mammalian genome (Goodier and Kazazian 2008). Retrotransposons amplify in the genome by a “copy and paste” mechanism and retrotransposition of TE sequences into new genomic sites can change the expression level or pattern of neighboring genes or generate new genes. While TEs play an important role in genome evolution at the species level, integration of TEs can potentially disrupt the function of targeted genes and thus be deleterious at the individual level. Indeed, TEs are culprits in >60 human genetic diseases (Goodier and Kazazian 2008).
Given the dramatic impact of TEs on genome integrity, cells have evolved multiple robust silencing mechanisms. DNA methylation is the major silencing mechanism for TEs in mammals (Bourc'his and Bestor 2004). Maintenance (DNMT1) and de novo (DNMT3A and DNMT3B) DNA methyltransferases are required for methylation of IAP (intracisternal A particle) retrotransposons (Walsh et al. 1998; Kato et al. 2007). Inactivation of DNMT3L, a noncatalytic germ cell-specific paralog of DNMT3A/3B, causes reactivation of TEs in male germ cells and loss of maternal genomic imprints, resulting in sterility (Bourc'his et al. 2001; Bourc'his and Bestor 2004). Loss of DNMT3C, a rodent and germ cell-specific de novo DNA methyltransferase, leads to reactivation of LINE1 and IAP retrotransposons (Barau et al. 2016). In addition to DNA methylation, repressive histone modifications (H3K9me2 and H3K9me3) are important for silencing of transposable elements (Di Giacomo et al. 2013; Liu et al. 2014; Pezic et al. 2014; Zamudio et al. 2015).
The piRNA (Piwi-associated small noncoding RNAs) pathway is an evolutionarily conserved RNA-based TE silencing mechanism in the germline (Siomi et al. 2011). piRNAs are mostly derived from TEs in early germ cells (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2010). Genome-wide demethylation in primordial germ cells (PGCs) causes a burst of TE expression (Bourc'his and Bestor 2004). During PGC epigenetic reprogramming, the piRNA pathway is activated and the increased expression of TE transcripts drives piRNA production, which serve two purposes (Aravin et al. 2008). First, piRNAs direct Piwi proteins to specifically cleave TE transcripts to enforce posttranscriptional silencing. Second, piRNAs derived from activated TEs instruct de novo methylation of TE promoters, thereby establishing transcriptional silencing through a mechanism that is yet to be completely elucidated in mammals.
Our previous cDNA subtraction genomic screen identified 36 germ cell-specific genes from mouse spermatogonia (Wang et al. 2001). Five genes from this genomic screen (Mili, Vasa, Tdrd1, Mov10l1, and Rnf17) were subsequently found to encode key protein components of the piRNA pathway. MOV10L1, a bona fide RNA helicase, forms a complex with MILI (PIWIL2) and TDRD1, binds specifically to piRNA precursors to initiate piRNA biogenesis, and is required for silencing retrotransposons in male germ cells (Zheng et al. 2010; Vourekas et al. 2015). The hallmarks of most piRNA pathway mouse mutants are activation of TEs in germ cells, meiotic arrest, and male sterility. Notably, Tex15 (testis-specific gene 15) was also first identified in this genomic screen (Wang et al. 2001). TEX15 is a large protein of 3059 residues with no known domain or binding partner. As previously reported, the Tex15−/− males are sterile due to meiotic arrest but females are fertile (Yang et al. 2008). Tex15−/− spermatocytes exhibit a failure in chromosomal synapsis and meiotic recombination (Yang et al. 2008). Because the Tex15−/− mice mimic mouse mutants with desilencing of TEs, we asked whether TEs are desilenced in male germ cells from Tex15−/− mice. In addition, mutations in human TEX15 cause infertility and meiotic arrest in men (Okutman et al. 2015). Here we report that TEX15 associates with MILI and is a new and essential factor in the epigenetic silencing of TEs in male germ cells.
Results and Discussion
TEX15 represses evolutionarily young transposable elements in embryonic male germ cells
We examined the expression of Tex15 in perinatal male germ cells. RNA-seq analysis showed that Tex15 transcript abundance was high at embryonic day 16.5 (E16.5) and E18.5 and increased at postnatal day 2.5 (P2.5). In contrast, Mili (Piwil2) and Miwi2 (Piwil4) levels were high at E16.5 and decreased at P2.5 (Supplemental Fig. S1A). By immunofluorescence, we found that TEX15 protein localized to both nucleus and cytoplasm in embryonic male germ cells—gonocytes at E17.5 (Fig. 1A). The TEX15 localization signal was specific, as it was absent in the Tex15−/− germ cells (Fig. 1A). The expression in embryonic testis suggests a functional role for TEX15 in early germ cells.
Figure 1.
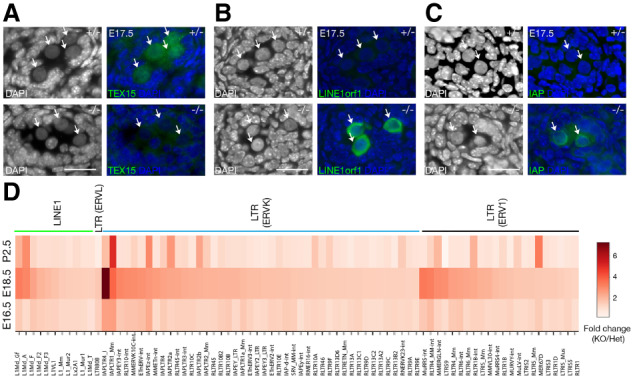
Derepression of transposable elements in Tex15-deficient gonocytes. (A) Cytoplasmic and nuclear localization of TEX15 in E17.5 Tex15+/− gonocytes (indicated by arrows). (B,C) Derepression of LINE1 and IAP in E17.5 Tex15−/− gonocytes. Scale bars, 25 µm. (D) Heat map of differentially expressed transposable elements at E16.5, E18.5, and postnatal day 2.5 (P2.5). The expression level cutoff is ≥1 cpm (counts per million reads).
Tex15−/− male mice were sterile with meiotic arrest, while females were fertile (Yang et al. 2008). We noticed the presence of a unique class of spermatocytes with enlarged nuclei and loose chromatin (Yang et al. 2008), a phenotype commonly observed in mutant testes, such as Miwi2−/− and Mov10l1−/− mutants, in which TEs are derepressed (Carmell et al. 2007; Zheng et al. 2010). To assess the potential for TE derepression, we focused on the expression of LINE1 (non-LTR retrotransposons) and IAP (LTR retrotransposons). LINE1 is the most abundant retrotransposon and, while most retrotransposons have degenerated due to mutations, ∼100 copies of LINE1 in the human genome and ∼3000 copies in the mouse genome remain competent for retrotransposition (Mandal and Kazazian 2008). To assess LINE1 expression, we monitored the expression of LINE1orf1, an abundantly expressed LINE1-encoded structural protein. As expected, LINE1 expression was not detected in E17.5 Tex15+/− control embryonic germ cells (Fig. 1B). In contrast, LINE1 was highly expressed in Tex15−/− embryonic male germ cells (Fig. 1B). Assessment of IAP, an LTR retrotransposon that is active in the mouse genome, was monitored by expression of its GAG protein. IAP expression was also up-regulated in E17.5 Tex15−/− embryonic male germ cells in comparison with Tex15+/− control (Fig. 1C). These results show that TEX15 is required for silencing of both LINE1 and IAP retrotransposons in male embryonic germ cells.
Shortly after sex determination, embryonic male germ cells cease dividing and undergo genome-wide epigenetic reprogramming, which includes genome-wide de novo DNA methylation and establishment of paternal imprinting (Hackett et al. 2012). At E16.5, genome methylation levels in male germ cells are low, become higher at E18.5, and at postnatal day 2.5 (P2.5), genome-wide de novo methylation is largely complete. To systematically profile TE expression, we performed RNA-seq analysis of FACS-sorted Tex15−/− and Tex15+/− male germ cells at E16.5, E18.5, and P2.5 (Fig. 1D; Supplemental Table S1). Germ cells were sorted from Oct4-GFP-positive testes. Three biological replicates were performed per genotype per time point. Two-thousand to 6000 germ cells from each embryo or pup were used per RNA-seq library. At E16.5, most TE families exhibited less than twofold change in Tex15−/− germ cells, although a few ERVK families (IAPLTR4_I, IAPLTR1_Mm, IAPEz-Int) and one ERV1 family (RLTR4_MM-int) were up-regulated by greater than or equal to twofold in Tex15−/− germ cells (Fig. 1D; Supplemental Table S2). At E18.5, a large number of transposon families in three classes of retrotransposons (LINE1, ERVK, and ERV1) exhibited more pronounced up-regulation in Tex15−/− germ cells. Compared with E18.5, the up-regulation level of most TEs in Tex15−/− germ cells at P2.5 was reduced, although some (L1Md_A, IAPEz-int, IAPLTR2a, IAPLTR2b, and RLTR1B-int) remained comparable, and two (IAPLTR1_Mm and MER67D) were more highly expressed. The different patterns of TE desilencing might be attributed to the dynamic nature of epigenetic reprogramming in male germ cells during this period and the diverse silencing mechanisms of different TE families (Yang and Wang 2016). Notably, the up-regulated LINE1s, including L1Md_A, L1Md_Gf, and L1Md_T are evolutionarily young. Young LINE1s are full-length intact copies and their desilencing is deleterious to the genome integrity. In conclusion, TEX15 is essential for silencing of a large number of TE families in male germ cells.
Binary activation of LINE1 and IAP retrotransposons in Tex15−/− spermatocytes and spermatogonia
We next examined activation of TEs in adult testes by immunofluorescence. In control seminiferous tubules from adult Tex15+/− testes, LINE1 and IAP were barely detectable (Fig. 2A). However, in Tex15−/− testes, LINE1orf1 was highly expressed in spermatocytes but not in spermatogonia (Fig. 2B), whereas the IAP-encoded GAG protein was highly expressed in spermatogonia but not in spermatocytes (Fig. 2C). The binary derepression of these two classes of retrotransposons suggests that additional mechanisms are involved in silencing of different retrotransposon classes. LINE1 silencing requires both DNA methylation and H3K9me2 in spermatogonia, but as H3K9me2 is absent in spermatocytes, DNA methylation is sufficient for LINE1 silencing in spermatocytes (Di Giacomo et al. 2013; Zamudio et al. 2015). qRT-PCR showed an up-regulated expression of LINE1 and IAP in P10 and P14 Tex15−/− testes (Fig. 2D). Correspondingly, Western blotting analysis revealed the increased abundance of LINE1orf1 and IAP GAG proteins in Tex15−/− testes (Fig. 2E). We then asked whether LINE1 promoter regions were hypomethylated in Tex15-deficient testes. Methylation-sensitive Southern blotting analysis showed that LINE1 hypomethylation was detected in P10 and P14 Tex15−/− testes, but not discernable in Tex15+/− testes (Fig. 2F). Bisulfite sequencing analysis of LINE1-Gf, a young LINE1, showed that CpG methylation decreased from 79% in Tex15+/− testes to 41% in Tex15−/− testes at P10 (Fig. 2G). Together, these results demonstrate that TEX15 is required for methylation and silencing of a subset of LINE1 retrotransposons in postnatal male germ cells.
Figure 2.
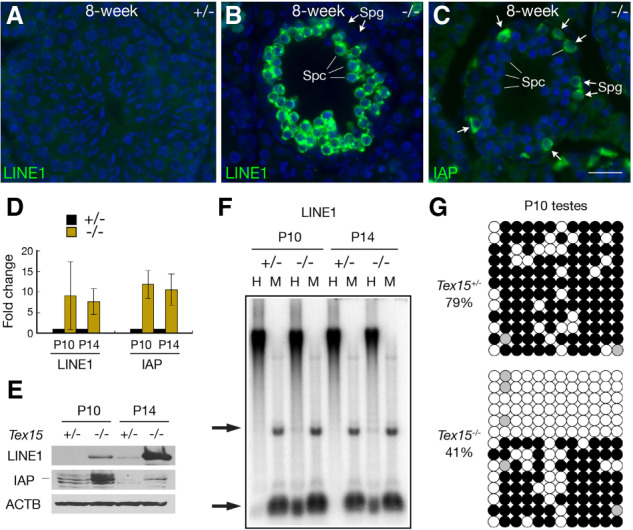
Binary desilencing of LINE1 and IAP retrotransposons in adult Tex15−/− testes. (A,B) Desilencing of LINE1 in spermatocytes (spc) from 8-wk-old Tex15−/− testis but not Tex15+/− testis. (C) Desilencing of IAP in spermatogonia (spg) from 8-wk-old Tex15−/− testis. Scale bar, 25 µm. (D) qRT-PCR analysis of LINE1 and IAP transcripts in P10 and P14 testes. (E) Western blot analysis of LINE1 and IAP in P10 and P14 testes. (F) Methylation-sensitive Southern blot analysis of LINE1 in P10 and P14 testis genomic DNA. (H) HpaII (methylation-sensitive); (M) MspI (methylation-insensitive). Arrows indicate the position of restriction fragments. (G) Bisulfite sequencing of LINE1 (L1_Gf) in P10 testes. CpGs are shown as circles. (Filled) Methylated; (open) unmethylated; (gray) unknown.
piRNA biogenesis is intact in Tex15-deficient testes
Our Tex15 mutant mice exhibit a meiotic phenotype that is similar to a number of piRNA pathway mutant mice: activation of retrotransposons, meiotic arrest, and male sterility. Therefore, we next asked whether TEX15 functions in the piRNA pathway. Among the three Piwi proteins in mice, MILI and MIWI2 are expressed in embryonic germ cells (Supplemental Fig. S1B,C; Carmell et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). While MILI is cytoplasmic, MIWI2 localizes to the nucleus in embryonic germ cells, where it binds to retrotransposon-derived antisense piRNAs. Nuclear MIWI2–piRNA complexes are required for de novo methylation of retrotransposons, but the underlying mechanism is unknown. In piRNA pathway mutants in which biogenesis of piRNAs is affected, unloaded MIWI2 is mislocalized to the cytoplasm (Reuter et al. 2009; Kuramochi-Miyagawa et al. 2010; Zheng et al. 2010; Pandey et al. 2013; Yang et al. 2016). Interestingly, we found that MIWI2 remained localized to the nucleus in Tex15-deficient gonocytes (Supplemental Fig. S1B), suggesting that piRNA biogenesis is intact in Tex15−/− mice.
To further assess whether piRNA biogenesis is intact, we performed MILI and MIWI2 immunoprecipitations from neonatal (P0) testes and found that both MILI- and MIWI2-bound piRNAs were present in Tex15−/− testes (Supplemental Fig. S2A). We also sequenced total small RNAs from E16.5 Tex15+/− and Tex15−/− testes and confirmed that piRNAs were present in Tex15−/− testes (Supplemental Fig. S2B,C; Supplemental Table S3). Length distribution of genome mapped reads was compared between Tex15−/− and Tex15+/−. Abundance of both miRNAs (22 nt long reads) and piRNAs (24- to 31-nt-long reads) was similar in Tex15+/− and Tex15−/− samples (Supplemental Fig. S2B,C). Sequencing of MILI- and MIWI2-bound small RNAs revealed slightly increased production of TE (repeat)-derived piRNAs associated with MILI and, to a lesser extent with MIWI2 (Supplemental Fig. S2D). The repeats with increased abundance of piRNAs belonged to LTR and LINE retrotransposons (Supplemental Fig. S2E). The increased production of piRNAs from retrotransposons in Tex15−/− testes can most likely be attributed to the increased abundance of retrotransposon transcripts, which were presumably fed into the piRNA biogenesis machinery. Such an increase in repeat-derived piRNA production has also been observed in other mutants such as Morc1−/− and Dnmt3l−/− testes with desilencing of retrotransposons (Aravin et al. 2008; Pastor et al. 2014). We next analyzed the ping–pong amplification signature of MILI-bound piRNA pairs on L1 and IAP repeats by measuring the 5′–5′ end distance. Like in Tex15+/− testis, the 10-nt 5′–5′ distance of piRNA pairs was apparent in Tex15−/− testis, showing normal ping–pong piRNA amplification in the absence of TEX15 (Supplemental Fig. S2F). As TEX15 still localized to the nucleus in Miwi2−/− germ cells (Supplemental Fig. S1C), these results suggest that TEX15 either functions independently of piRNA production or serves as a nuclear effector protein downstream of the piRNA pathway in silencing of TEs in male germ cells.
Hypomethylation of transposable elements in Tex15−/− germ cells
Methylation-sensitive Southern blot analysis revealed hypomethylation of LINE1 elements in Tex15−/− testes (Fig. 2F), showing that desilencing of retrotransposons in Tex15−/− germ cells is likely due to loss of DNA methylation. To address whether TEX15 affects global DNA methylation levels, we performed whole-genome bisulfite sequencing (WGBS) on flow-sorted Tex15−/−, Tex15+/−, and Tex15+/+ germ cells from P2.5 male pups (two biological replicates each) using the previously described protocol (Pastor et al. 2014). CpG methylation in genomic domains was unaltered: CpG islands remained unmethylated, whereas intergenic, intragenic, satellite, and transposon regions were hypermethylated in the presence or absence of TEX15 (Fig. 3A). CpG methylation levels were also similar among transposon families (Fig. 3B). Together, these results indicate that TEX15 does not affect de novo DNA methylation at the genome-wide level (Supplemental Table S4).
Figure 3.
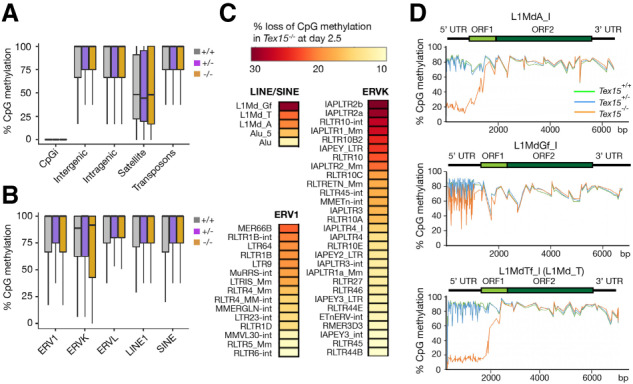
DNA hypomethylation of transposable elements in Tex15−/− male germ cells. WGBS was performed with P2.5 Tex15+/+, Tex15+/−, and Tex15−/− gonocytes. (A,B) Box and whisker plots of CpG methylation among genomic compartments (A) and classes of transposable elements (B). (CpGi) CpG islands. (C) Percentage of CpG DNA methylation loss in individual retrotransposon families in Tex15−/− gonocytes. Only members with >10% loss of CpG methylation in Tex15−/− germ cells are shown. (D) Metaplots of DNA methylation over full-length LINE1 classes.
DNA methylation is a key silencing mechanism of TEs in perinatal germ cells in mammals. Given the derepression of TEs (Fig. 1), we analyzed the CpG methylation levels in individual TE families. LINE1, ERVK, and ERV1 families were all hypomethylated (Fig. 3C) and transcriptionally derepressed in Tex15−/− germ cells (Fig. 1D). Hypomethylation preferentially affected evolutionarily young LINE1s (Fig. 3D; Supplemental Fig. S3). LINE1 5′ UTRs function as promoters and young LINE1s (L1Md_A, L1Md_Gf, and L1Md_T) were specifically hypomethylated at their promoters (5′ UTRs) but not in the ORFs and 3′ UTRs in Tex15−/− male germ cells (Fig. 3D). The CpG methylation level of the 5′ UTR in L1MdA_I and L1MdTf_I was 80% in Tex15+/− germ cells but only 20% in Tex15−/− germ cells (Fig. 3D). Focused analysis of L1MdA families showed hypomethylation in young LINE1s (L1MdA_I to L1MdA_III) but not in ancient LINE1s (L1MdA_VI and L1MdA_VII) in Tex15−/− germ cells (Supplemental Fig. S3). Deep sequencing of each WGBS library yielded sixfold genome coverage. Genome-wide DNA methylation levels were similar among Tex15−/−, Tex15+/−, and wild-type germ cells at P2.5 (Supplemental Fig. S4A; Supplemental Table S4). Statistical analysis of DMRs (differentially methylated regions) in Tex15−/− versus Tex15+/− germ cells revealed that there were more hypomethylated DMRs (430) than hypermethylated DMRs (116) in the absence of TEX15 (Supplemental Fig. S4B). The majority of hypomethylated DMRs overlapped with LINE1, ERVK, and ERV1, in comparison with the control regions (Supplemental Fig. S4C). Therefore, retrotransposon hypomethylation is associated with transcriptional derepression in Tex15−/− male germ cells.
In addition to TE desilencing, RNA-seq analysis identified differentially expressed protein-coding genes (DEGs) in Tex15−/− germ cells (Supplemental Fig. S5; Supplemental Table S5). Comparison of Tex15+/− versus Tex15+/+ germ cells at P2.5 revealed a similar small number of up-regulated and down-regulated lowly expressed genes (Supplemental Fig. S5A). While only eight genes were up-regulated in Tex15−/− germ cells at E16.5, 659 genes were up-regulated and 75 genes were down-regulated in Tex15−/− germ cells at E18.5. At P2.5, the number of up-regulated and down-regulated DEGs decreased to 135 and 81, respectively (Supplemental Fig. S5B). Notably, E18.5 Tex15−/− germ cells showed the highest number of both DEGs and up-regulated TEs (Fig. 1D; Supplemental Fig. S5). Gene ontology analysis of up-regulated DEGs at E18.5 and P2.5 revealed the following two top enriched biological processes: G-protein-coupled receptor signaling pathway and sensory perception of smell, because a large number of olfactory receptor genes were up-regulated in Tex15−/− germ cells (Supplemental Fig. S5C). However, while hypomethylated DMRs at P2.5 substantially overlapped with TEs (Supplemental Fig. S4C), we did not find a correlation of DMRs with DEGs at this time point (Supplemental Fig. S6). Except for Gtsf1, none of the known factors (Dnmt3l, Dnmt3c, Morc1, Mili, Miwi2, Mov10l1, Ddx4, Mael, and other piRNA factors) required for TE silencing were differentially expressed in Tex15−/− versus Tex15+/− germ cells. GTSF1 is required for secondary piRNA biogenesis (Yoshimura et al. 2018) and Gtsf1 was down-regulated in Tex15−/− germ cells but only at E18.5 (Supplemental Table S5). Of the other known TE-derepressing mutants, the Mili mutant was reported to exhibit a high number of DEGs, with more than 100 DEGs at P10 (Manakov et al. 2015).
TEX15 associates with MILI in testis
To investigate the mechanism of TEX15 function, we sought to identify TEX15-interacting proteins. We generated 3×Flag-TEX15 knock-in mice, in which Flag is fused to the N terminus of TEX15 (Fig. 4A; Supplemental Fig. S7A). Immunoprecipitation (IP) and Western blot analysis with anti-Flag antibody detected the full-length TEX15 protein (>300 kDa) and a shorter TEX15 isoform (∼150 kDa) (Fig. 4B). The 150-kDa isoform could be generated through endolytic cleavage of the full-length protein and its physiological importance is unknown. By immunofluorescence, Flag-TEX15 was both cytoplasmic and nuclear in E17.5 male germ cells (Supplemental Fig. S7B), confirming the localization pattern revealed with anti-TEX15 antibody (Fig. 1A). Of three mouse Piwi proteins, only MILI, like TEX15, is expressed in both embryonic and postnatal germ cells (Kuramochi-Miyagawa et al. 2004; Aravin et al. 2007). MILI was detected in the protein complexes immunoprecipitated from testes with anti-Flag (TEX15) antibody (Fig. 4C). Importantly, reciprocal IP confirmed this association (Fig. 4D). These results demonstrate that TEX15 is associated with MILI in testis.
Figure 4.
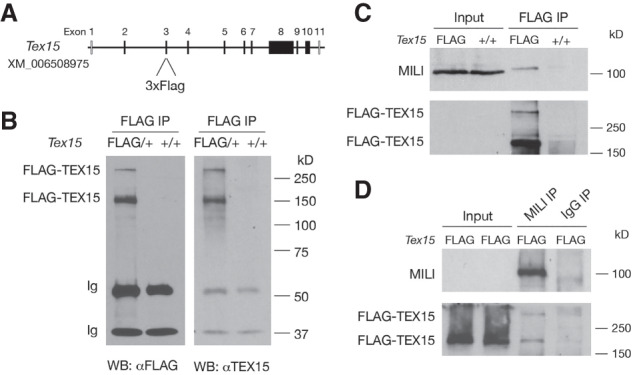
TEX15 forms a complex with MILI in testis. (A) Design of the Flag-Tex15 knock-in allele. The coding sequence of 3xFlag epitope tag is inserted into exon 3 in-frame by CRISPR/Cas9 genome editing. (B) Western blot analysis of Flag-TEX15 proteins in postnatal day 20 testes. Note the presence of two isoforms of TEX15 proteins. (C) Coimmunoprecipitation of MILI with Flag-TEX15 in P20 testes. Input was 1.5% of the extract used for immunoprecipitation. (D) Reciprocal coimmunoprecipitation of Flag-TEX15 with MILI in P20 testes.
In conclusion, TEX15 is required for DNA methylation of TEs during genome-wide epigenetic reprogramming and thus silencing of TEs in male germ cells. In comparison with Miwi2−/− germ cells, Mili−/− or Dnmt3c−/− germ cells show derepression of a greater number of TEs and more severe hypomethylation at TEs (Supplemental Fig. S8; Kuramochi-Miyagawa et al. 2008; Manakov et al. 2015; Nagamori et al. 2015; Barau et al. 2016). Mili−/− germ cells show a more dramatic reduction in piRNA production than Miwi2−/− germ cells. The Tex15−/− mutant (Fig. 3D) more closely resembles the Mili−/− mutant or the Dnmt3c mutant than the Miwi2−/− mutant in terms of the degree of DNA hypomethylation at TEs (for example, L1MdA_I, L1MdGf_I, and L1MdTf_I in Supplemental Fig. S9) and the number of derepressed TEs (Molaro et al. 2014; Manakov et al. 2015; Barau et al. 2016). In contrast to Miwi2−/− germ cells, both Tex15−/− and Mili−/− germ cells showed a substantial number of differentially expressed genes. The mechanism underlying the differential epigenetic defects observed in Mili and Miwi2 mutants could be partly explained by the TEX15–MILI association.
In Drosophila, Silencio/Panoramix bridges Piwi protein to the epigenetic silencing machinery in the nucleus to silence transposable elements (Sienski et al. 2015; Yu et al. 2015); however, the Drosophila Silencio/Panoramix has no sequence orthologs in vertebrates. Consistently, TEX15 is vertebrate-specific. Thus, our finding on the association of TEX15 with MILI supports that TEX15 might be the vertebrate equivalent (functional homolog) of the fly Silencio/Panoramix. TEX15 is both cytoplasmic and nuclear, and although MILI is abundantly expressed in the cytoplasm, it is also present in the nucleus of germ cells (Beyret and Lin 2011). Our working model is that MILI/piRNA binds to nascent TE transcripts and interacts with TEX15, which in turn recruits the epigenetic silencing machinery to the transposon loci. Loss of TEX15 does not affect piRNA production. Therefore, we propose that TEX15 functions as a molecular bridge between piRNA complex and the downstream effectors in vertebrates.
Materials and methods
Tex15, Miwi2, and Oct4-GFP mice
Tex15−/−, Miwi2−/−, and Oct4-GFP mice were generated previously (Szabó et al. 2002; Carmell et al. 2007; Yang et al. 2008). In Oct4-GFP mice, the expression of GFP is under the control of the Oct4 promoter and ΔPE enhancer (Szabó et al. 2002). Intercrossing produced Tex15+/− Oct4-GFPHom mice, in which Oct4-GFP is homozygous. Timed interbreeding of Tex15+/− Oct4-GFPHom mice was performed to generate Tex15+/+ Oct4-GFP, Tex15+/− Oct4-GFP, and Tex15−/− Oct4-GFP embryos at E16.5 and E18.5 or pups at P2.5.
Generation of Flag-Tex15 knock-in mice
To generate 3xFlag-Tex15 knock-in mice, the guide RNA mapping to Tex15 exon 3 was cloned into the px330 vector (Addgene) and transcribed in vitro (Supplemental Fig. S7A). The ssDNA repair template contains the 3xFlag-coding sequence (Supplemental Fig. S7A). A mixture of 50 ng/µL guide RNA, 100 ng/µL ssDNA template, and 100 ng/µL Cas9 mRNA (Trilink L-7206) was microinjected into ∼50 mouse zygotes (1 to 2 pL per zygote). The zygotes were transferred into three recipient females. Five founder mice were obtained. Sanger sequencing of tail genomic DNA identified one founder mouse with the intact 3xFlag insertion, which transmitted the Tex15Flag allele through germline. The wild-type (242-bp) and Tex15Flag (323-bp) alleles were genotyped by PCR with primers 5′-TGAGGACTTTCAGGTTGCTA-3′ and 5′-CTTTTTCAGTCGTCCTCCTGA-3′. All experiments were approved by the Institute of Animal Care and Use Committee (IACUC) of University of Pennsylvania.
Supplementary Material
Acknowledgments
We thank N.A. Leu for microinjection of zygotes; S.L. Martin and A. Bortvin for L1 ORF1p antibody; A. Bortvin for L1 Southern blot probe; B.R. Cullen for IAP antibody; G. Hannon for MIWI2 antibody; J. Schug at Next-Generation Sequencing Core for sequencing; L.B. King for help with the manuscript; and Y. Guan, R. Liu, R. Guo, and H. Lin for critical reading of the manuscript. This work was supported by the National Institute of Health grant P50HD068157 (P.J.W. and M.S.B.) and the Swiss National Science Foundation ERC Transfer Grant GermMethylation CRETP3_166923 (to R.S.P.) and Project Grant 310030B_185386 (to R.S.P.).
Author contributions: F.Y. and P.J.W. conceptualized the study. F.Y. performed the experiments. Y.L. performed bioinformatic analyses of RNA-seq and WGBS-seq data with contributions from S.L.B. and M.S.B. R.R.P., D.H., and R.S.P. contributed the piRNA data. F.Y., S.L.B., M.S.B., R.S.P., and P.J.W. analyzed data. P.J.W., D.H., R.S.P., and F.Y. wrote the manuscript with contributions from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.335489.119.
References
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. 10.1126/science.1142612 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, Guillou F, Bourc'his D. 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354: 909–912. 10.1126/science.aah5143 [DOI] [PubMed] [Google Scholar]
- Beyret E, Lin H. 2011. Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev Biol 355: 215–226. 10.1016/j.ydbio.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99. 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539. 10.1126/science.1065848 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D. 2013. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50: 601–608. 10.1016/j.molcel.2013.04.026 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH Jr. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35. 10.1016/j.cell.2008.09.022 [DOI] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, Surani MA. 2012. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet 28: 164–174. 10.1016/j.tig.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280. 10.1093/hmg/ddm179 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. 10.1242/dev.00973 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917. 10.1101/gad.1640708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. 2010. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 24: 887–892. 10.1101/gad.1902110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brind'Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. 2014. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev 28: 2041–2055. 10.1101/gad.244848.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. 2015. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep 12: 1234–1243. 10.1016/j.celrep.2015.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Kazazian HH Jr. 2008. SnapShot: vertebrate transposons. Cell 135: 192–192.e1. 10.1016/j.cell.2008.09.028 [DOI] [PubMed] [Google Scholar]
- Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, Hannon GJ. 2014. Two waves of de novo methylation during mouse germ cell development. Genes Dev 28: 1544–1549. 10.1101/gad.244350.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamori I, Kobayashi H, Shiromoto Y, Nishimura T, Kuramochi-Miyagawa S, Kono T, Nakano T. 2015. Comprehensive DNA methylation analysis of retrotransposons in male germ cells. Cell Rep 12: 1541–1547. 10.1016/j.celrep.2015.07.060 [DOI] [PubMed] [Google Scholar]
- Okutman O, Muller J, Baert Y, Serdarogullari M, Gultomruk M, Piton A, Rombaut C, Benkhalifa M, Teletin M, Skory V, et al. 2015. Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet 24: 5581–5588. 10.1093/hmg/ddv290 [DOI] [PubMed] [Google Scholar]
- Pandey RR, Tokuzawa Y, Yang Z, Hayashi E, Ichisaka T, Kajita S, Asano Y, Kunieda T, Sachidanandam R, Chuma S, et al. 2013. Tudor domain containing 12 (TDRD12) is essential for secondary PIWI interacting RNA biogenesis in mice. Proc Natl Acad Sci 110: 16492–16497. 10.1073/pnas.1316316110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc'his D, et al. 2014. MORC1 represses transposable elements in the mouse male germline. Nat Commun 5: 5795 10.1038/ncomms6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezic D, Manakov SA, Sachidanandam R, Aravin AA. 2014. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 28: 1410–1428. 10.1101/gad.240895.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646. 10.1038/nsmb.1615 [DOI] [PubMed] [Google Scholar]
- Sienski G, Batki J, Senti KA, Dönertas D, Tirian L, Meixner K, Brennecke J. 2015. Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev 29: 2258–2271. 10.1101/gad.271908.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Szabó PE, Hübner K, Schöler H, Mann JR. 2002. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev 115: 157–160. 10.1016/S0925-4773(02)00087-4 [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, Pillai RS, Mourelatos Z, Wang PJ. 2015. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev 29: 617–629. 10.1101/gad.254631.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117. 10.1038/2413 [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. 2001. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27: 422–426. 10.1038/86927 [DOI] [PubMed] [Google Scholar]
- Yang F, Wang PJ. 2016. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin Cell Dev Biol 59: 118–125. 10.1016/j.semcdb.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. 2008. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol 180: 673–679. 10.1083/jcb.200709057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Chen KM, Pandey RR, Homolka D, Reuter M, Janeiro BK, Sachidanandam R, Fauvarque MO, McCarthy AA, Pillai RS. 2016. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol Cell 61: 138–152. 10.1016/j.molcel.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Watanabe T, Kuramochi-Miyagawa S, Takemoto N, Shiromoto Y, Kudo A, Kanai-Azuma M, Tashiro F, Miyazaki S, Katanaya A, et al. 2018. Mouse GTSF1 is an essential factor for secondary piRNA biogenesis. EMBO Rep 19: e42054 10.15252/embr.201642054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J, Czech B, Hannon GJ. 2015. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350: 339–342. 10.1126/science.aab0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio N, Barau J, Teissandier A, Walter M, Borsos M, Servant N, Bourc'his D. 2015. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev 29: 1256–1270. 10.1101/gad.257840.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ. 2010. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci 107: 11841–11846. 10.1073/pnas.1003953107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


