In this study, Rickman et al. set out to map BRCA2 DNA-binding domain (DBD) mutants in patients presenting with atypical Fanconi anemia phenotypes. Using BRCA2 DBD mutants discovered in FA patients that were determined to confer loss of replication fork protection but only moderate HR deficiency, the authors show that BRCA2 DBD is required for replication fork protection and that BRCA2 fork protection at HU-induced and BRCA2 fork protection at ICL-induced stalled forks are distinct processes.
Keywords: BRCA2, Fanconi anemia, homologous recombination, DNA interstrand cross-link repair, ICL, replication fork protection, RAD51, DNA2, WRN, MRE11
Abstract
DNA interstrand cross-links (ICLs) are a form of DNA damage that requires the interplay of a number of repair proteins including those of the Fanconi anemia (FA) and the homologous recombination (HR) pathways. Pathogenic variants in the essential gene BRCA2/FANCD1, when monoallelic, predispose to breast and ovarian cancer, and when biallelic, result in a severe subtype of Fanconi anemia. BRCA2 function in the FA pathway is attributed to its role as a mediator of the RAD51 recombinase in HR repair of programmed DNA double-strand breaks (DSB). BRCA2 and RAD51 functions are also required to protect stalled replication forks from nucleolytic degradation during response to hydroxyurea (HU). While RAD51 has been shown to be necessary in the early steps of ICL repair to prevent aberrant nuclease resection, the role of BRCA2 in this process has not been described. Here, based on the analysis of BRCA2 DNA-binding domain (DBD) mutants (c.8488-1G>A and c.8524C>T) discovered in FA patients presenting with atypical FA-like phenotypes, we establish that BRCA2 is necessary for the protection of DNA at ICLs. Cells carrying BRCA2 DBD mutations are sensitive to ICL-inducing agents but resistant to HU treatment consistent with relatively high HR repair in these cells. BRCA2 function at an ICL protects against DNA2–WRN nuclease–helicase complex and not the MRE11 nuclease that is implicated in the resection of HU-induced stalled replication forks. Our results also indicate that unlike the processing at HU-induced stalled forks, the function of the SNF2 translocases (SMARCAL1, ZRANB3, or HLTF), implicated in fork reversal, are not an integral component of the ICL repair, pointing to a different mechanism of fork protection at different DNA lesions.
DNA interstrand cross-links (ICLs) are a deleterious form of DNA damage that covalently link the Watson and Crick strands of DNA. ICLs can be produced by exogenous compounds such as mitomycin C (MMC), diepoxybutane (DEB), cisplatin, psoralen, and nitrogen mustards, or by naturally occurring biological metabolites such as aldehydes (Langevin et al. 2011; Kottemann and Smogorzewska 2013; Garaycoechea and Patel 2014).
The importance of the proper repair of ICLs is emphasized by the rare genetic disorder, Fanconi anemia (FA). FA is characterized by developmental abnormalities, bone marrow failure (BMF), predisposition to solid tumors and leukemia, and cellular hypersensitivity to cross-linking agents (Auerbach 2009). FA results from pathogenic variants in one of the 22 FANC genes (FANCA-W) whose protein products are required for proper ICL repair (Kottemann and Smogorzewska 2013; Wang and Smogorzewska 2015; Ceccaldi et al. 2016; Niraj et al. 2019).
When an ICL is encountered during DNA replication, it causes fork stalling and FA pathway activation (Garcia-Higuera et al. 2001; Knipscheer et al. 2009). The removal of an ICL is a multistep process requiring activation of the FA core complex and monoubiqutination of FANCD2 and FANCI (Garcia-Higuera et al. 2001; Timmers et al. 2001; Smogorzewska et al. 2007). Monoubiquitinated FANCD2 and FANCI form a heterodimer that is recruited to chromatin and is required for ICL processing, which entails nucleolytic unhooking of the cross-linked DNA (Niedernhofer et al. 2004; Kim et al. 2011, 2013; Klein Douwel et al. 2014; Alcón et al. 2020; Tan et al. 2020; Wang et al. 2020). Unhooking of the ICL enables translesion bypass on one-strand and double-strand break (DSB) repair by homologous recombination (HR) on the second strand (Howlett et al. 2002; Litman et al. 2005; Xia et al. 2007; Long et al. 2011).
A number of FA proteins, BRCA2/FANCD1, PALB2/FANCN, FANCJ/BRIP1, RAD51C/FANCO, RAD51/FANCR, and BRCA1/FANCS are known for facilitating HR (Howlett et al. 2002; Litman et al. 2005; Rahman et al. 2007; Xia et al. 2007; Vaz et al. 2010; Sawyer et al. 2015). BRCA2/FANCD1 is an essential gene and single allele pathogenic variants predispose to breast and ovarian cancer and biallelic pathogenic variants result in a subtype of Fanconi anemia, FA-D1 (Howlett et al. 2002). FA is a heterogeneous disease, but even within the disease spectrum, patients with biallelic pathogenic variants in BRCA2/FANCD1 are phenotypically distinct from the most common complementation groups, FA-A, FA-C, and FA-G. A higher proportion of FA-D1 patients have developmental abnormalities and nearly one hundred percent have a malignancy by 5 yr of age (Alter et al. 2007), which is most likely due to HR deficiency.
Functional analysis of BRCA2 has largely focused on canonical HR, and the role of BRCA2 in ICL repair has been associated with the repair of DSBs generated by programmed incisions at the ICL. Outside of their role in HR and ICL repair, BRCA2 and RAD51, along with a number of other recently described proteins, function in replication fork protection (Rickman and Smogorzewska 2019). In the absence of replication fork protection, newly synthesized DNA is degraded at replication forks stalled due to dNTP imbalance secondary to hydroxyurea (HU) treatment, and a number of nucleases including MRE11, CTIP, and EXO1 have been implicated in the process (Lemaçon et al. 2017; Przetocka et al. 2018; Rickman and Smogorzewska 2019).
Another nuclease, DNA2, has also been shown to resect DNA at ICLs in cells expressing the RAD51/FANCR separation of function mutant, p.T131P, identified in an individual with FA-like syndrome. The mutant RAD51 p.T131P has a dominant-negative effect on RAD51 function that does not seem to affect HR at cellular levels but disrupts the function of RAD51 at ICLs, suggesting a fork protection role for RAD51 in ICL repair. The requirement for BRCA2 in the early steps of ICL repair to prevent aberrant resection has not previously been determined. Here we investigated the requirements of BRCA2 with RAD51 in fork protection at ICLs and demonstrate that the two proteins are both required to prevent hyperresection by the DNA2-WRN nuclease-helicase complex, but not MRE11. These studies were performed using BRCA2 DNA-binding domain (DBD) mutants discovered in FA patients and these variants were determined to confer loss of replication fork protection but only moderate HR deficiency. Our results indicate that the BRCA2 DBD is required for replication fork protection and that BRCA2 fork protection at HU-induced and ICL-induced stalled forks are distinct processes.
Results
Atypical presentation of Fanconi anemia in individuals with BRCA2/FANCD1 DNA-binding domain variants
Two female siblings, enrolled in the International Fanconi Anemia Registry (IFAR), with unknown causative gene mutations, were born with a multitude of congenital abnormalities and had mildly elevated levels of chromosomal breakage at birth (see Supplemental Table S1 for clinical presentation). Biallelic BRCA2/FANCD1 variants (c.2330dupA and c.8524C>T) were identified by whole-exome sequencing (WES) and no other likely pathogenic FA gene variants were observed. These results were surprising since neither sibling displayed the typical clinical findings of the FA-D1 complementation group, with no history of malignancy or bone marrow failure at the ages of 20 and 23. There is no reported family history of FA, but there are cases of breast cancer that were diagnosed later in life (above 60 yr of age), individuals with skin cancer in the family, and early onset colorectal cancer in the father (40 yr old) (Fig. 1A).
Figure 1.
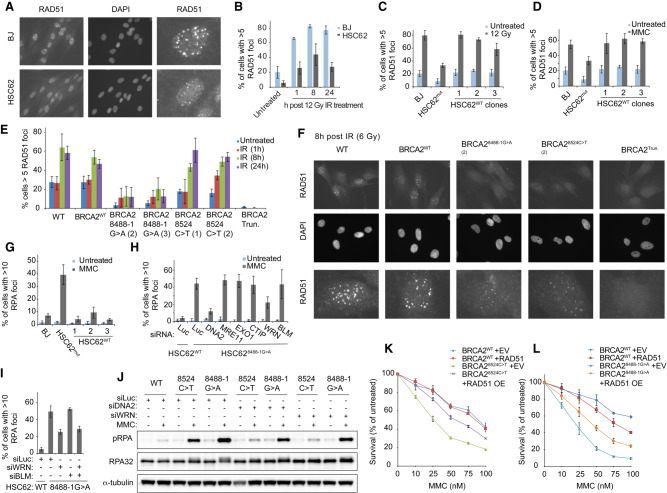
BRCA2 variants identified in individuals with atypical Fanconi anemia. (A) Family pedigree showing a sibling pair with Fanconi anemia (red circles) who are compound heterozygous for BRCA2 variants c.2330dupA (maternal inheritance) and c.8524C>T (paternal inheritance). Family history of breast cancer (purple, all diagnosed in 60s and 70s), skin cancer (gray), and colon cancer (green; diagnosed at 40 yr old). (B) Schematic of BRCA2 domain structure and key interacting proteins. (C) Alignment of exon 20 BRCA2 DBD peptide sequence demonstrating that it is evolutionary conserved across many species. In green are the amino acid residues modified by the patient variants, p.W2830_K2833del (c.8488-1G>A) and p.R2842C (c.8524C>T). Purple arrows indicate amino acid residues that contact DNA (Yang et al. 2005). (D) Immunoblot showing BRCA2 levels in WT (RA2985) control, FA-D1 (RA2525), and patient RA3105 and RA3106 LCLs. (E) Quantification of chromosome breaks following DEB treatment of WT (RA2985), FA-A (RA2939), and patient RA3105 and RA3106 LCLs. (F,G) Cell survival assays of patient-derived lymphoblast cell lines (LCLs) RA3105, FA-A (RA2939), WT (RA2985), and FA-D1 (RA2525) after MMC and PARP inhibitor olaparib (PARPi) treatment. Relative cell survival was normalized to untreated controls to give percent survival. Error bars indicate SD. (H) Quantification of chromosome breaks following MMC treatment of BJ wild-type fibroblasts, FA-A patient fibroblasts, and HSC62 fibroblasts. (I) Cell survival of HSC62 (c.8488-1G>A) fibroblasts compared with BJ WT fibroblast and complemented FA-A patient cells (RA3087) expressing wild-type FANCA (FA-A+A) or empty vector (FA-A+EV). Cells were treated with increasing concentrations of MMC. Relative cell survival was normalized to untreated controls to give the percent survival. Error bars indicate SD. (J) Cell survival of MMC-treated HSC62 uncorrected patient cell line (HSC62mut) compared with BJ WT fibroblast and CRISPR/Cas9 corrected wild-type HSC62 (HSC62WT) clones 1-3. (K,L) Cell survival of BJ WT fibroblasts, and CRISPR/Cas9-targeted BJ fibroblasts: BJ WT fibroblast clone (BRCA2WT), c.8488-1G>A BJ clones (BRCA28488-1G>A), c.8524C>T BJ clones (BRCA28524C>T), and exon 20 BRCA2 frameshift mutant (BRCA2Trun.). Cells were treated with increasing concentrations of MMC or PARPi. Error bars indicate SD. Kruskal-Wallis ANOVA, with Dunn's post-test. (***) P < 0.001; (****) P < 0.0001.
The frameshift c.2330dupA variant of exon 11 (maternal origin) results in premature truncation of BRCA2 (p.Asp777Glufs*11) and has been described previously in hereditary breast and ovarian cancer (HBOC) (Supplemental Fig. S1A). The c.8524C>T missense variant of exon 20 (paternal origin) results in an p.Arg2842Cys residue change in the highly conserved DNA-binding domain (DBD) of BRCA2 and has previously been identified as a variant of unknown significance (VUS) in HBOC (Fig. 1B,C; Supplemental Fig. S1B,C). At the protein level, the missense variant results in the p.Arg2842Cys change at a highly conserved residue at the base of the BRCA2 Tower domain of the DBD (Supplemental Fig. S1C). Sequencing of peripheral blood and lymphocytes demonstrated the presence of both variants and no evidence of somatic mosaicism.
A third individual with FA, biallelic BRCA2 variants, and an atypical presentation, was identified in the literature (Howlett et al. 2002). This individual was homozygous for the c.8488-1G>A variant (alias “IVS19-1G>A”) that alters the splice acceptor site of exon 20. cDNA analysis demonstrated the use of an alternate splice acceptor that results in the loss of 12 bp of exon 20 and translates into p.Trp2830_Lys2833del (Fig. 1B,C; Howlett et al. 2002). Amino acid residues 2830–2833 are located within the DBD at the transition of the OB2 fold and the base of the Tower domain (Fig. 1B,C; Supplemental Fig. S1C). This individual was 30 yr of age at last follow up, was born with a thumb malformation, but had no history of bone marrow failure or malignancy. Similar to the sibling pair, chromosomal breakage was modest (Howlett et al. 2002).
BRCA2 DNA-binding domain variants identified in FA patients confer defects in the response to replication stress
Lymphoblastoid cell lines (LCL) (RA3105 and RA3106) were derived from the sibling pair with compound heterozygous BRCA2 variants, c.2330dupA and c.8524C>T. FA pathway activation requires monoubiqutination of FANCI, which was observed in patient-derived LCLs (Supplemental Fig. S1D). Analysis of BRCA2 expression by Western blot demonstrated a full-length (∼390-kDa) band, the presumed product of the c.8524C>T allele, for both patient cell lines (Fig. 1D). DEB-induced breakage analysis confirmed previous clinical data that breakage was elevated, but not to levels of the typical FANCA-deficient (FA-A) LCLs (RA2939) (Fig. 1E). RA3105 LCL displayed hypersensitivity to the cross-linking agents MMC and DEB, but to a lesser degree than RA2939 (Fig. 1F; Supplemental Fig. S1F). RA3105 was also hypersensitive to replication stress-inducing agents including olaparib, a PARP inhibitor (PARPi), and camptothecin (CPT), a topoisomerase I inhibitor (Fig. 1G; Supplemental Fig. S1G).
Similarly, analysis of patient-derived fibroblasts, HSC62 (Howlett et al. 2002), from the individual with homozygous c.8488-1G>A variant also revealed more moderate chromosomal breakage to DEB and MMC and cellular hypersensitivity to cross-linking agents (Fig. 1H,I; Supplemental Fig. S1O). The cells were not hypersensitive to ionizing radiation (IR), but were sensitive to replication stress induced by CPT and PARPi (Supplemental Fig. S1J–L). In contrast, the cells were not sensitive to replication stress produced by the agents aphidicolin and HU (Supplemental Fig. S1M,N).
We corrected the pathogenic variants in the HSC62 patient fibroblast cell line to demonstrate that the c.8488-1G>A variant caused the observed defects. The homozygous c.8488-1G>A variant was corrected to wild type at the endogenous locus using CRISPR/Cas9 gene targeting. Both heterozygous and homozygous clones were recovered (HSC62WT/MUT or HSC62WT/WT) (Supplemental Fig. S1P). cDNA analysis demonstrated that restoration of the splice acceptor base (A>G) in HSC62WT/MUT or HSC62WT/WT clones restored the cDNA exon 19-20 junction (Supplemental Fig. S1Q). Both HSC62WT/MUT and HSC62WT/WT clones rescued hypersensitivity to replication stress-inducing agents MMC, CPT, and PARPi (Fig. 1J; Supplemental Fig. S1S,T).
For a direct comparison of the BRCA2 DNA-binding domain variants, we generated isogenic cell lines by introducing the variants, c.8524C>T (p.R2842C) and c.8488-1G>A (p.Trp2830_Lys2833del), into wild-type BJ fibroblasts with CRISPR/Cas9 gene editing (Supplemental Fig. S2A,B). Knock-in of the BRCA2 c.8488-1G>A variant in BJ fibroblasts conferred the same splicing defect observed in HSC62 cells (Supplemental Fig. S2A). Western blot analysis of BRCA2 demonstrated an ∼390-kDa band for all mutants except for BRCA2 clones containing exon 20 frameshift variants obtained in parallel using CRISPR/Cas9 gene targeting (Supplemental Fig. S2C). The BRCA2 frameshift mutant is homozygous c.8531dupA with a predicted p.R2845Kfs*22 truncation (BRCA2Trun.). Analysis of cellular sensitivity of the BRCA2 DBD mutants revealed that presence of both DBD variants sensitize cells to MMC, PARPi, and CPT but not aphidicolin, recapitulating phenotypes of patient HSC62 fibroblasts (Fig.1K,L; Supplemental Fig. S2D,E).
BRCA2 DNA-binding domain variants confer defects in RAD51 recruitment after IR and MMC
To determine the impact of DBD variants on the ability of BRCA2 to load RAD51 onto ssDNA following DNA damage, we analyzed RAD51 foci formation after IR and MMC. Levels of RAD51 foci and focus size were reduced after IR and MMC treatment in HSC62 cells, which was rescued by the CRISPR/Cas9 gene editing (Fig. 2A–D; Supplemental Fig. S2H,I). Analysis of isogenic BJ cell lines with DBD mutations also demonstrated defects in RAD51 foci formation following IR and MMC (Fig. 2E,F; Supplemental Fig. S2J,K). The c.8488-1G>A variant had a stronger impact on RAD51 foci formation, resulting in fewer cells with RAD51 foci and reduced focus size. The c.8524C>T mutant did not show a significant reduction in the number of cells with RAD51 foci; however, the foci were smaller in size (Fig. 2F; Supplemental Fig. S2K). By comparison, the BRCA2Trun. mutant had complete loss of observable RAD51 foci. These data indicate that the BRCA2 DBD mutants are hypomorphic in their mediator function.
Figure 2.
Defective ICL repair in BRCA2 DBD mutants results in increased ssDNA that is WRN and DNA2 dependent. (A) Immunofluorescence images of RAD51 foci, 8 h following 12 Gy ionizing radiation (IR) of BJ WT fibroblast and patient derived HSC62 fibroblast, detected with anti-RAD51 antibody. Third row images are individual cells enlarged to better demonstrate differences in RAD51 focus size. (B) Quantification of RAD51 foci 1 h, 8 h, and 24 h following 12 Gy IR of BJ WT fibroblast and HSC62 fibroblast. Error bars indicate SD of two independent experiments (≥200 cells per experiment). (C) Quantification of RAD51 foci 8 h after 12 Gy IR of BJ WT fibroblast, wild-type HSC62 (HSC62WT) clones 1–3, and HSC62 uncorrected patient cell line (HSC62mut). (D) Quantification of RAD51 foci 24 h following 1-h treatment with 3 µM MMC. Error bars indicate SD of three independent experiments (≥200 cells per experiment). (E) Quantification of RAD51 foci in isogenic BJ fibroblasts clones at 1 h, 8 h, and 24 h following 6 Gy IR of BJ WT fibroblasts, BJ WT fibroblast clone (BRCA2WT), BRCA28488-1G>A BJ clones 2–3, BRCA28524C>T BJ clones 1–2, and a BRCA2 homozygous truncation mutant, c.8531dupA (BRCA2Trun). Error bars indicate SD of three independent experiments (≥200 cells per experiment). (F) Representative images of RAD51 foci in isogenic BJ fibroblasts clones, 8 h after 6 Gy IR, detected by immunofluorescence with anti-RAD51 antibody. Third row images are individual cells enlarged to better demonstrate differences in RAD51 focus size. (G) Quantification of RPA foci 24 h following 1-h treatment with 3 μM MMC of BJ WT fibroblast, CRISPR/Cas9 corrected wild-type HSC62 clones (HSC62WT), and HSC62 uncorrected patient cell line (HSC62mut). (H) Quantification of RPA foci 24 h following 1-h treatment with 3 μM MMC in HSC62mut cells depleted of DNA2, MRE11, EXO1, CTIP, WRN, or BLM by siRNA compared with luciferase control (Luc). Error bars indicate SD of four independent experiments. (I) Quantification of RPA foci 24 h following 1 h treatment with 3 μM MMC in HSC62mut cells depleted of DNA2, WRN, BLM, or codepleted of WRN and BLM by siRNA compared with luciferase control (Luc). Error bars indicate SD of three independent experiments. (J) Immunoblot analysis of RPA phosphorylation in isogenic BJ fibroblasts clones 24 h after 1-h treatment with 3 μM MMC. BRCA2WT, BRCA28524C>T, and BRCA28488-1G>A BJ fibroblast cells were transfected with siRNA control luciferase (Luc) or siRNAs targeting DNA2 or WRN. (K,L) MMC cell survival of BJ BRCA2WT, BRCA28488-1G>A, and BRCA28524C>T fibroblasts overexpressing (OE) WT RAD51 or empty vector (EV) control. Relative cell survival was normalized to untreated controls to give percent survival. Error bars indicate SD.
Increased ssDNA in BRCA2 DBD variants is dependent on DNA2 and WRN
The previously described RAD51/FANCR p.T131P patient-derived cell line that is proficient for HR but defective in ICL repair displays increased RPA phosphorylation and foci formation indicating an increase in ssDNA upon MMC treatment (Wang et al. 2015). Given that the interaction of BRCA2 and RAD51 is required for their canonical function in HR and their noncanonical function in replication fork protection at HU-induced stalled forks, we investigated whether BRCA2 also functions in preventing increased ssDNA generation at ICLs (Schlacher et al. 2011; Wang et al. 2015; Mijic et al. 2017; Bhat et al. 2018). We observed an increase in RPA foci formation in HSC62MUT cells compared with wild-type fibroblasts upon MMC treatment (Fig. 2G; Supplemental Fig. S3A). Similar to RAD51/FANCR p.T131P-expressing patient cells, the increased RPA foci formation in HSC62 cells was also dependent on DNA2 and WRN activity, but not MRE11, EXO1, CTIP, or BLM (Fig. 2H; Supplemental Fig. S3B–D). Codepletion of WRN with BLM did not further rescue the increased RPA foci in HSC62MUT cells after MMC (Fig. 2I; Supplemental Fig. S3E). Increased RPA foci and phosphorylation following MMC was also observed for c.8524C>T and c.8488-1G>A mutants, with a greater increase for the c.8488-1G>A mutants (Fig. 2J; Supplemental Fig. S3F–J). These results suggest that BRCA2 is functioning with RAD51 to protect against aberrant processing by DNA2 and WRN at ICLs, but not against the other effectors of DSB end resection such as MRE11, EXO1, or CTIP. Overexpression of RAD51 in the BRCA2 c.8524C>T- and c.8488-1G>A-expressing cells partially rescued cellular sensitivity to MMC and RPA foci formation after MMC (Fig. 2K,L; Supplemental Fig. S3K,L). This data supports that RAD51 and BRCA2 function interdependently at ICLs.
To determine whether blocking ICL unhooking or nuclease mediated fork collapse (McPherson et al. 2004; Niedernhofer et al. 2004; Dendouga et al. 2005; Kim et al. 2011; Bogliolo et al. 2013) would rescue RPA foci formation in BRCA2 DBD mutant cells following MMC, we depleted SLX4 and MUS81. Depletion of either SLX4 or MUS81 did not rescue the increase in chromatin bound RPA in the BRCA28524C>T and BRCA28488-1G>A- expressing cells (Supplemental Fig. S3M–O). SLX4 depletion further increased the RPA foci formation, indicating further defects in ICL repair in its absence, which may be the result of loss of function of the associated nucleases.
ICLs are a substrate of nucleolytic processing in the absence of a functioning FA pathway
Having demonstrated that BRCA2 and RAD51 share a role in protecting ICLs from overresection by DNA2 and WRN, we investigated whether other FA proteins are also required for protection against DNA hyperresection at ICLs. Analysis of a panel of FA patient-derived cells with mutations in FANCA, FANCL, FANCD2, FANCI, FANCJ, and SLX4/FANCP demonstrated increased RPA foci formation following MMC treatment for all complementation groups (Fig. 3A). To determine whether the genetic requirement for RPA suppression was the same as in BRCA2 and RAD51 mutant cells, DNA2 and WRN were depleted in a complemented pair of FANCA (Fig. 3B,C; Supplemental Fig. S3P,Q) and FANCG patient-derived cells (Fig. 3D). Interestingly, the dependence on DNA2 was the same, but the helicase dependency was different, as WRN did not rescue RPA levels but BLM depletion did (Fig. 3C,D; Supplemental Fig. S3R–T). These data demonstrate a dependence on the FA core complex and pathway associated proteins to prevent resection of ICLs by DNA2 and BLM. They also suggest that different nuclease–helicase pairs engage when ICL repair is halted at different stages of the process.
Figure 3.

Proper ICL repair is required to prevent aberrant nuclease processing. (A) Quantification of RPA foci 8 h, 24 h, and 48 h following 1-h treatment with 3 μM MMC of FA patient-derived fibroblasts compared with BJ wild-type fibroblasts. Patient cells lines from FA complementation group FA-R (RAD51/FANCR), FA-A (FANCA), FA-L (FANCL), FA-D2 (FANCD2), FA-I (FANCI), FA-J (FANCJ), and FA-P (SLX4/FANCP). FA-A patient complemented cell lines were generated by transducing WT FANCA cDNA or EV. Error bars indicate SD of two independent experiments. (B) FA-A patient cells expressing WT FANCA (FA-A+FANCA) or empty vector (FA-A+EV) were transfected with siRNA control luciferase (Luc) or siRNAs targeting DNA2 and WRN. Quantification of RPA foci 24 h following 1-h treatment with 3 μM MMC. Error bars indicate SD of two independent experiments. (C) FA-A+EV were transfected with siRNA Luc or siRNAs targeting DNA2 and BLM. Quantification of RPA foci 24 h following 1-h treatment with 3 μM MMC. Error bars indicate SD of two independent experiments. (D) FA-G patient cells expressing WT FANCG (FA-G+FANCG) or empty vector (FA-G+EV) were transfected with siRNA control luciferase (Luc) or siRNAs targeting DNA2, WRN, and BLM. Quantification of RPA foci 24 h following 1-h treatment with 3 μM MMC. Error bars indicate SD of three independent experiments.
Determination of homologous recombination efficiency in DNA-binding domain mutants
To determine the HR proficiency of BRCA28488-1G>A and BRCA28524C>T -expressing cells, we used a HDR assay that targets DSBs at the LMNA locus (Pinder et al. 2015; Arnoult et al. 2017). The assay was performed in HEK293T cells after CRISPR-Cas9 gene editing to engineer either BRCA2 DBD variants or the exon 27 p.S3291A variant, previously reported to have an effect on replication fork protection but not on HR (Supplemental Fig. S4A,B; Schlacher et al. 2011; Kim et al. 2014). Compared with wild-type cells, HR in all BRCA2 clones, including the S3291A mutant, was moderately decreased (Fig. 4A). Cells with DBD BRCA28488-1G>A and BRCA28524C>T variants showed similar decreases in HR levels to approximately half that of wild-type cells but retained significantly more HR activity than cells depleted of RAD51 and BRCA2 or BRCA2Trun cells, consistent with a previous study (Siaud et al. 2011).
Figure 4.
BRCA2 DBD and C-terminal domain variants confer a moderate defect in HR and disrupt replication fork protection function. (A) Levels of mClover-positive cells were normalized to WT HEK293T (siLuc). Error bars indicate SD of three independent experiments performed in triplicate. P-values were determined by ANOVA and Tukey's multiple comparison. (****) P < 0.0001. (B) Sister chromatid exchange (SCE) assay in BJ WT fibroblast and HSC62 patient derived fibroblast following treatment with 0.1 μg/mL or 0.2 μg/mL MMC. (C) Isogenic BJ fibroblast BRCA2 mutants, BRCA2Trun., BRCA28524C>T, and BRCA28488-1G>A were analyzed for replication fork resection. Cells were labeled with DNA analogs, IdU for 20 min, and then CldU for 20 min. Cells were then incubated in 6 mM HU with and without MRE11 inhibitor mirin (50 µM) for 4 h before being harvested. DNA fibers were prepared and visualized by immunofluorescence detection of IdU and CldU and measured. Error bars indicate SD. (D) Isogenic BJ fibroblast BRCA2 mutants, BRCA2Trun., BRCA28524C>T, BRCA28488-1G>A, and BRCA2S3291A were transfected with siRNA control luciferase (Luc) or siRNAs targeting DNA2 or MRE11. Cells were treated and labeled with DNA analogs as above. Error bars indicate SD. (E) BJ fibroblast with BRCA2 variants, BRCA28524C>T and BRCA28488-1G>A, were analyzed for replication fork resection when depleted of RADX by shRNA or transduced with shRNA control (shCONT.). Cells were treated and labeled with DNA analogs as above. Data of two replicates plotted. Error bars indicate SD. (F) Quantification of chromosome breaks in isogenic BJ fibroblast BRCA2 mutants following 5 h of 6 mM HU and released into colcemid. Breakage was not significantly increased in BRCA28524C>T and BRCA28488-1G>A compared with BRCA2WT. Kruskal-Wallis ANOVA, with Dunn's post-test. (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001.
Given the normal resistance to IR in HSC62 fibroblasts, we assessed sister chromatid exchange (SCEs) levels as a readout of HR (Sonoda et al. 1999). SCEs were induced by increasing concentrations of MMC or depletion of BLM. There was no significant difference in SCE levels observed in wild-type BJ fibroblasts and HSC62 cells (Fig. 4B; Supplemental Fig. S4E–G); however, SCE levels were suppressed in BRCA2Trun fibroblasts (Supplemental Fig. S4H). These observations suggest that the DNA-binding domain defect in HSC62 cells, while decreasing RAD51 foci formation, does not significantly reduce HR as observed by normal resistance to IR and SCE levels in these cells. Taken together, the variants moderately reduce HR at Cas-9 targeted DSBs but do not impact cellular HR readouts, which is similar to the behavior of cells carrying the RAD51 p.T131P mutation (Wang et al. 2015).
The BRCA2 DNA-binding domain is required for replication fork protection at HU-induced stalled forks
To determine the requirement for the BRCA2 DBD in replication fork protection after HU treatment, BRCA28524C>T- and BRCA28488-1G>A-expressing cells were examined by DNA fiber analysis. Replication fork protection by BRCA2 has largely been attributed to the C-terminal RAD51 interacting domain by analysis of the BRCA2 p.S3291A variant (Schlacher et al. 2011). Analysis of BRCA2Trun-, BRCA28524C>T-, and BRCA28488-1G>A-expressing cells demonstrated defects in replication fork protection of HU-induced stalled forks as measured by the degradation of nascent DNA tracks labeled with nucleotide analogs, IdU and CldU. As previously reported, nascent strand degradation in the absence of BRCA2 was rescued by the MRE11 inhibitor mirin and MRE11 depletion (Fig. 4C,D). These data demonstrate that the BRCA2Trun-, BRCA28524C>T-, and BRCA28488-1G>A-expressing cells are all similarly defective for replication fork protection and that the DBD is required for protection of replication forks from MRE11 processing. Depletion of DNA2 also rescues resection after HU in cells expressing all of the BRCA2 mutants including BRCA2Trun., BRCA28524C>T, BRCA28488-1G>A, and BRCA2S3291A (Fig. 4D). RADX depletion has been shown to rescue nascent strand degradation at HU-induced stalled replication forks in BRCA2-deficient cells without restoring HR function (Dungrawala et al. 2017). Consistent with these studies, depletion of RADX in the BRCA2 DBD mutant-expressing cells did not rescue HR defects (Supplemental Fig. SF5A–D) but did rescue nascent strand degradation (Fig. 4E). Taken together, these data demonstrate that both the DBD and C-terminal domain of BRCA2 are required for proper replication fork protection at HU-induced stalled forks, and that both domains are required to protect against degradation by the nucleases MRE11 and DNA2.
Although all of the BRCA2 mutants showed similar levels of nascent strand resection as measured by DNA fibers, the levels of chromosomal breakage differed (Fig. 4F; Supplemental Fig. S5E,F). Metaphases were analyzed after 5 h of 6 mM HU and release into colcemid. BRCA2Trun.-expressing cells showed a large increase in genomic instability upon stalling with HU in comparison with WT and the other BRCA2 mutants. Cells with BRCA28524C>T and BRCA2S3291A variants did not show an elevation in breakage and BRCA28488-1G>A-expressing cells had a mild increase. The elevated chromosomal breakage in BRCA2Trun. cells were reduced by MRE11 depletion, but exacerbated by DNA2 depletion (Fig. 4F; Supplemental Fig. S5E). DNA2 depletion resulted in a mild increase in breakage for all mutants but resulted in a synergistic increase in cells with BRCA2Trun. Previous studies have reported elevated breakage resulting from replication fork degradation in p.S3291A expressing cells and BRCA2 deficient cells (Schlacher et al. 2011; Mijic et al. 2017). In contrast, the newly characterized BRCA28524C>T- or BRCA2S3291A-expressing cells in this study do not have a significant increase in breakage after HU, despite having levels of fork degradation similar to BRCA2Trun (Fig. 4F; Supplemental Fig. S5F). Our data demonstrate that different levels of BRCA2 function impairment have different consequences on HU-induced stalled forks and that replication fork resection at HU-induced stalled forks does not always manifest in chromosomal breakage. How this breakage occurs in BRCA2 depleted or LOF cells needs to be investigated further, but like nascent DNA degradation, it is partially dependent on MRE11.
SMARCAL1, ZRANB3, and HLTF function is not required for ICL repair
Replication fork reversal has been observed as a response to replication stress induced by a number of different classes of genotoxic agents including MMC (Zellweger et al. 2015). SMARCAL1, ZRANB3, and HLTF are ATPase-dependent DNA translocases of the SNF2 family of chromatin remodelers that have recently been shown to promote replication fork reversal in vitro and in vivo. Depletion of any of the three translocases rescues nascent strand resection at HU-induced stalled forks in BRCA2-deficient cells (Mijic et al. 2017; Taglialatela et al. 2017). Similarly, depletion of the translocases in the BRCA28524C>T- and BRCA28488-1G>A-expressing cells rescued nascent strand degradation (Fig. 5A). However, depletion of SMARCAL1, ZRANB3, or HLTF did not rescue the increased RPA phosphorylation and foci formation after MMC (Fig. 5B,C; Supplemental Fig. S5G–I). Codepletion of SMARCAL1 and ZRANB3 also had no effect on decreasing RPA foci formation after MMC (Supplemental Fig. S5J,K). To determine whether the proteins implicated in replication fork reversal are important for the repair of ICLs, wild-type cells were depleted of SMARCAL1 or ZRANB3 and tested for sensitization to MMC. Cells depleted of either translocase were not significantly sensitized to MMC (Fig. 5D). Additionally, depletion of either translocase did not rescue cellular hypersensitivity to MMC or CPT in BRCA28488-1G>A cells (Fig. 5E,F). These data suggest that the function of these translocases is not required during MMC-induced ICL repair.
Figure 5.
SNF2 translocases are not required for ICL repair. (A) BJ fibroblast mutants BRCA28524C>T and BRCA28488-1G>A were analyzed for replication fork resection when depleted of either SMARCAL1 or ZRANB3 by shRNA or transduced with control shRNA (shLuc). Cells were labeled with DNA analogs, IdU for 20 min and then CldU for 20 min. Cells were then incubated in 6 mM HU for 4 h before being harvested. DNA fibers were prepared and visualized by immunofluorescence detection of IdU and CldU and measured. Error bars indicate SD (B) Quantification of RPA foci in isogenic BJ fibroblasts clones 24 h following 1-h treatment with 3 µM MMC in cells depleted of SMARCAL1 or ZRANB3. Error bars indicate SD of two independent experiments. (C) Quantification of RPA foci in BJ fibroblasts clones 24 h following 1-h treatment with 3 µM MMC in cells depleted of HLTF. Error bars indicate SD of two independent experiments. (D) MMC cell survival of isogenic BJ BRCA2WT fibroblasts depleted of SMARCAL1 or ZRANB3 by shRNA or transduced with shRNA luciferase control (shLuc). Relative cell survival was normalized to untreated controls to give percent survival. Error bars indicate SD. (E,F) MMC and CPT cell survival assay of isogenic BJ BRCA28488-1G>A or BRCA28524C>T clones depleted of either SMARCAL1 or ZRANB3 by shRNA or transduced with shRNA luciferase control (shLuc). Relative cell survival was normalized to untreated controls to give percent survival. Error bars indicate SD. Kruskal-Wallis ANOVA, with Dunn's post-test. (***) P < 0.001; (****) P < 0.0001.
Discussion
BRCA2 and RAD51 function at the ICL
Here we studied the functional consequences of pathogenic BRCA2 variants in the DNA-binding domain in the context of homologous recombination, and protection of stalled replication forks due to dNTP depletion or DNA interstrand cross-link lesions. The DBD variants did not affect IR sensitivity, SCE levels, or HU sensitivity, suggesting that the HR levels in cells carrying the DBD variants is sufficiently intact. We also saw only a moderate reduction in HR using an HR reporter assay. Similar to the previously described patient cell line with RAD51/FANCR p.T131P mutation (Wang et al. 2015), the cells with BRCA2 DBD variants were sensitive to ICL-inducing agents and showed increased RPA foci formation after MMC that was DNA2-WRN dependent. These data suggest that like the well-described interdependence of BRCA2 and RAD51 in HR, BRCA2, and RAD51 function together in the early steps of ICL repair to prevent DNA resection and that the function of the BRCA2 DBD is important for this role. This expands the role of BRCA2 in ICL repair beyond HR to include protection of DNA at the ICL stalled replication fork from aberrant nucleolytic processing (Fig. 6).
Figure 6.

The role of BRCA2 in response to replication stress produced by hydroxyurea and DNA interstrand cross-links is distinct. Schematic representing the different roles of BRCA2 in replication fork protection and homologous recombination. (A) During homologous recombination repair of DSBs, BRCA2 assembles RAD51 nucleofilaments onto ssDNA overhangs, which is important for the RAD51-mediated homology search of the sister chromatid. (B) During DNA interstrand cross-link repair, homologous recombination is required to repair the programmed DSBs. BRCA2 has a role in two distinct types of replication fork protection. (C) At HU stalled forks, replication fork remodeling depends on RAD51 and the SNF2 translocases, SMARCAL1, ZRANB3, and HLTF. BRCA2 and RAD51 protect the reversed replication fork from degradation by nucleases. The MRE11 nuclease has been reported numerous times to be responsible for the degradation of HU stalled forks in the absence of fork protection. More recently, other nucleases have been described in nascent strand degradation including EXO1 and DNA2. (D) At ICLs, BRCA2 and RAD51 protect the fork from resection by the DNA2-WRN nuclease helicase complex. The ssDNA generated after MMC is not dependent on MRE11, CTIP, or EXO1, as described for HU stalled forks. ICL repair does not require the function of the SNF2 translocases, suggesting that reversed forks present in MMC treated cells are likely the result of a more global cellular response to replication stress. We also propose that an additional role of the FA core complex and associated proteins at the ICL is to prevent aberrant resection by the DNA2–BLM nuclease helicase complex.
Depletion of the replication fork remodelers SMARCAL1, ZRANB3, and HLTF and the RAD51 modulator RADX rescued nascent strand degradation at HU-induced stalled forks in cells carrying DBD variants consistent with the previous data on the role of BRCA2 in this process (Dungrawala et al. 2017; Lemaçon et al. 2017; Mijic et al. 2017; Taglialatela et al. 2017). However, depletion of the translocases did not mitigate cellular sensitivity or increased RPA after MMC in the BRCA2 DBD mutant-expressing cells. Our study demonstrates that remodeling by the translocases is not a major step in the repair of ICLs and suggests that the MMC-induced replication fork reversal may be a more general response to replication stress but not specifically at the fork that is stalled at an ICL (Zellweger et al. 2015; Mutreja et al. 2018). These data further support that the protection by BRCA2 and RAD51 at a HU-induced stalled fork is different from protection at an ICL (Fig. 6).
The mechanism of DNA protection at the ICL by the BRCA2 DBD domain remains to be explored. However, the location of the variants at the transition of the OB2 fold and base of the Tower domain suggests a plausible mechanism of protection at an ICL stalled fork. The OB2 fold binds to ssDNA and the Tower domain contains a 3HB domain at the apex that is capable of binding to dsDNA (Yang et al. 2002). We speculate that the mutations in this region of the DBD may preclude efficient binding/bridging at ssDNA–dsDNA junctions, which is a structure expected at stalled forks, and lack of this binding would lead to the deprotection phenotype. Lack of proper placement of BRCA2 may also preclude proper RAD51 loading, which may lead to inappropriate DNA resection. Biochemical analysis of the BRCA2 variants we have identified in atypical Fanconi anemia patients will further our understanding of how BRCA2 interacts with different replication fork structures.
FA protein function at the ICL
FA proteins have been shown previously to be important for protection at HU-induced stalled replication forks (Schlacher et al. 2012). Here we show that FA patient cell lines from various complementation groups also demonstrate increased ssDNA and RPA foci formation after MMC. However, in FANCA-deficient cells, the increase in RPA foci is dependent on DNA2 and BLM, but not WRN. This suggests that the fork protection of BRCA2–RAD51 is not redundant with the FA core complex, but further investigation will be needed to determine the genetic dependency of increased ssDNA in the absence of the other FA proteins. DNA2 has previously been reported to interact with FANCD2 and be recruited to ICLs where it is required for repair, but is deleterious in the absence of FANCD2 (Karanja et al. 2012, 2014). BLM has been reported to interact with a number of FA proteins and colocalize with FANCD2 at ICLs (Meetei et al. 2003; Pichierri et al. 2004; Suhasini and Brosh 2012). Consistent with BLM depletion rescuing increased ssDNA at the fork in the absence of FANCA, BLM knockout was also recently reported to rescue ICL sensitivity and reduce DNA damage in FA-deficient cells (Moder et al. 2017). It is possible that DNA2, WRN, and BLM are recruited to ICLs for normal functions, but in the absence of key FA/BRCA pathway components are left unregulated, resulting in aberrant processing of the fork.
BRCA2 function at the HU-induced stalled replication fork
Our analysis of BRCA2 DBD mutants engineered into the endogenous BRCA2 locus demonstrates that the function of the DBD is also required for protection at HU-induced stalled replication forks to prevent nuclease degradation. This is in contrast to a previous report that the DBD was dispensable for replication fork protection at HU-induced stalled forks, a disparity that is most likely due to a difference in experimental set-up (Schlacher et al. 2011). Our findings extend the replication fork protection role of BRCA2 at HU-induced stalled replication forks beyond the C-terminal domain and show that fork protection likely requires the DBD to bind DNA at the stalled replication fork. It remains to be determined whether the DBD variants have an effect on replication fork reversal after HU treatment, but the dependency on the resection phenotype on the translocases suggest that they will.
While the role of MRE11 in nascent strand degradation of BRCA2 deficient cells has been widely shown, there is conflicting data about resection mediated by DNA2 (Ray Chaudhuri et al. 2016; Lemaçon et al. 2017; Przetocka et al. 2018). A role for DNA2 with WRN in replication fork restart has been described, and it has also been reported that DNA2 degrades nascent DNA at stalled forks in the setting of RECQ1, BOD1L, or Abro1 deficiency (Higgs et al. 2015; Thangavel et al. 2015; Xu et al. 2017; Rickman and Smogorzewska 2019). Here we show in isogenic cell lines that BRCA2 function is required to also prevent DNA2 resection at HU-induced stalled forks.
The observation that genomic instability results from the absence of proper replication fork protection after HU treatment has largely been studied by RNAi depletion of BRCA2 (Lemaçon et al. 2017; Mijic et al. 2017; Taglialatela et al. 2017). By studying BRCA2 mutants, we show that a significant increase in chromosomal breakage after HU does not correlate with replication fork resection. For some of the BRCA2 mutants (c.8524C>T and p.S3291A) replication fork protection at HU-induced stalled forks is defective, but there is no significant increase in chromosomal breakage after HU. We observed increased chromosomal breakage in cells expressing BRCA2 LOF truncation variant, which is consistent with many previous reports that BRCA2 knockdown results in increased chromosomal breakage (Schlacher et al. 2011; Lemaçon et al. 2017; Mijic et al. 2017; Taglialatela et al. 2017). The DNA damage in cells with BRCA2 LOF variants was similarly rescued by MRE11 depletion/inhibition. However, all of the BRCA2 mutants in our analysis that undergo MRE11-dependent fork resection at HU-induced stalled replication forks do not have significantly elevated chromosomal breakage. These results also correlate with the cellular sensitivity observed in the BRCA2 mutants; LOF mutants show sensitivity to replication stress induced by HU and aphidicolin, whereas the DBD mutants did not. These results demonstrate the importance of using BRCA2 mutants that permit separation between different BRCA2 functions as opposed to RNAi depletion or LOF mutants that remove all protein functions. It is possible that in studies using BRCA2 depletion or LOF mutants, the loss of BRCA2 HR function contributes to the breakage phenotype at the unprotected and degraded replication forks.
We show that DNA2 depletion in cells with BRCA2 mutations also rescues resection at HU-induced stalled forks, but at the same time we observe that DNA2 depletion exacerbates chromosomal breakage after HU treatment. This observation suggests that in the setting of BRCA2 deficiency, DNA2 depletion is deleterious, which may be due to its requirement in replication-coupled repair or modulation of reversed forks (Hu et al. 2012; Karanja et al. 2012; Thangavel et al. 2015). Recent reports have also implicated EXO1 and CTIP as degrading HU-induced stalled forks in the absence of BRCA2 (Lemaçon et al. 2017). Conversely, CTIP has been reported to be required to restrain DNA2 activity at stalled replication forks in the absence of BRCA1/2 (Przetocka et al. 2018). Taken together, resection of the regressed fork in the absence of BRCA2 is now reported to involve all of the DSB end-resection nucleases. MRE11 and DNA2 are already reported to be required for replication fork restart (Bryant et al. 2009; Thangavel et al. 2015). However, further investigation is required to determine whether all of these factors have a normal function in processing stalled forks or restoring reversed forks under wild-type genetic conditions. These results are also interesting in that all of the nucleases are implicated at HU-induced stalled forks, but only DNA2 has activity at the ICL in BRCA2-deficient cells.
Clinical implications
The identification of BRCA2 DBD variants in conjunction with atypical disease presentation gives the opportunity to investigate how defects in the DBD impact BRCA2 function and gives insight into how these defects may give rise to the developmental defects characteristic of FA but not the early childhood malignancies seen in other patients with biallelic FANCD1/BRCA2 variants. The disease presentation of these individuals resembles the phenotype of FA-like patients described for FA-R (FANCR/RAD51) and FA-O (FANCO/RAD51C) complementation groups (Vaz et al. 2010; Wang et al. 2015). Due to the moderate impact that these DBD variants have on HR, we hypothesize that the retention of ∼50% of HR function that we observe is sufficient enough to safeguard against early tumor development. Diagnosis and classification as FA-D1 (BRCA2/FANCD1) complementation group should also be considered for patients presenting with FA-like syndrome.
Furthermore, this study has implications for how we think about BRCA2 variants of unknown significance (VUS) in human disease, including HBOC and FA. Evaluation of BRCA2 VUS relies on multifactorial probability models (Guidugli et al. 2014) or functional assays assessing HR (Guidugli et al. 2013) to estimate whether a variant is pathogenic. Some VUS can be easily classified as pathogenic if HR is dramatically reduced; however, a number of VUS show intermediate phenotypes, making it difficult to interpret their role in HBOC (Guidugli et al. 2013; Shimelis et al. 2017). The contribution of other BRCA2 functions, including replication fork protection, to cellular function and tumorigenesis requires further investigation. Here we demonstrated that the BRCA2 DBD mutations that we studied are pathogenic. It is possible that some DBD mutations carry only low to moderate risk for HBOC related to their preservation of HR function, but still result in FA when biallelic BRCA2 mutations are inherited due to a predominant defect in ICL repair.
Materials and methods
Study subjects
DNA samples and cell lines were derived from subjects enrolled in the International Fanconi Anemia Registry (IFAR) after obtaining informed written consent. The Institutional Review Board of The Rockefeller University approved these studies.
Cell lines
Patient-derived fibroblast cell lines (Supplemental Table S2) and BJ foreskin normal control fibroblasts (ATCC) were transformed by expression of HPV16 E6E7 and immortalized with the catalytic subunit of human telomerase (hTERT). Fibroblasts were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 15% FBS, 100 U/mL penicillin, 0.1 mg of streptomycin/mL, nonessential amino acids, and glutamax (Invitrogen). Fibroblasts cell lines were incubated at 37°C, 5% CO2, and 3% O2. Lymphoblast cell lines (Supplemental Table S1) were established from patient peripheral blood mononuclear cells by Epstein-Barr virus (EBV) transformation and grown in Roswell Park Memorial Institute medium (RPMI) with 20% FBS and further supplemented as above. HEK293T (ATCC) cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin and glutamax as indicated above. Lymphoblast and HEK293T cell lines were incubated at 37°C, 5% CO2, and ambient O2.
Viral transfection/transduction
HA-RAD51 cDNA was delivered by retroviral transduction after packaging in HEK293T cells (TransIT-293 transfection reagent, Mirus). HEK293T cells were plated at 4.5 × 106 the evening before transfection of DNA and viral packaging vectors. Transfection was performed according to the manufacturer's instructions. The day after transfection, cell medium was replaced and 2 d after transfection, viral supernatants were harvested and used to infect target cells in the presence of 4 mg/mL polybrene. Stably expressing cells were selected with 2 µg/mL puromycin.
RNAi
Cells were transfected with pools of three siRNAs against MRE11, DNA2, EXO1, CTIP, WRN, BLM, BRCA2, RAD51, MUS81, XPF, and SLX4. For RADX and HLTF depletion, a single previously published siRNA was used (Supplemental Table S3; Dungrawala et al. 2017; Taglialatela et al. 2017). Cells were transfected using Lipofectamine RNAiMAX (Invitorgen) according to the manufacturer's instructions. For shRNA depletion, virus was packaged in HEK293T cells and used to infect target cells and cells with stable integration were selected. shRNA constructs for SMARCAL1 and ZRANB3 were a gift from Alberto Ciccia (Supplemental Table S4). shRNAs to RADX were purchased from Transomics and used in the pZIP_hCMV_Puro vector or pMSCV-PM-mir30. shRNAs were PCR amplified and cloned into pMSCV-PM-mir30 by digestion with XhoI and MluI and vector ligation. See Supplemental Table S5 for PCR primers for amplification of shRNA from UltramiRs of pZIP_hCMV vector. RNAi knockdown was measured by RT-qPCR or Western blot.
PCR, reverse transcription, and RT-qPCR
PCR reactions were performed using Taq DNA Polymerase (Qiagen), Phusion high-fidelity PCR master mix with GC buffer (Thermo Scientific), and PCR SuperMix high fidelity (Invitrogen) according to manufacturer's protocols and primers are listed in Supplemental Table S6. Total messenger RNA was extracted using RNeasy plus kit (Qiagen). RNA was reverse transcribed to cDNA using the SuperScript III reverse transcriptase (Invitrogen). Platinum SYBR Green SuperMix-UDG (Invitrogen) was used according to manufacturer's protocol to determine relative transcript levels, which were normalized against GAPDH levels (see Supplemental Table S7 for RT-qPCR primers).
Gene targeting
To correct the BRCA2 c.8488-1G>A variants in HSC62 fibroblasts, cells were transduced with the pCW-Cas9-Puro (Addgene 50661) vector, which contains a doxycycline-inducible Cas9. Subsequently, HSC62 cells were transduced with plentiGuide-Hygro (derived from Addgene 52963) that expresses a single guide RNA (sgRNA) (see Supplemental Table S8 for sgRNA sequence) that targets DNA in proximity to the c.8488-1G>A variant. sgRNAs were designed using the online CRISPR design tool from the Zhang laboratory (http://www.crispr.mit.edu). Cells (1 × 106) were electroporated with a 100-bp template oligonucleotide (see Supplemental Table S9 for sequence) using Lonza 2b-Nucleofector. Cells were cultured in 500 ng/mL doxycycline for 48 h to induce Cas9 expression and then incubated in fresh doxycycline-free medium for another 48 h before being single-cell-cloned into 96-well plates. Clones were expanded and screened by sequencing of genomic DNA. For clones HSC62mut/WT-1 and HSC62WT/WT-2, cells were selected in low dose MMC (50 ng/mL) once a week for 3 wk before seeding in 96-wells. Clone 3 (HSC62WT/WT) was not selected for.
The rest of the gene targeting was performed by electroporation of Cas9/gRNA ribonucleoprotein (RNP) complexes with 100 nt oligonucleotide donor templates, with phosphorothioate-protected ends. sgRNA was prepared by combining crRNA (designed using crispr.mit.edu) and universal tracrRNA as per manufactures guidelines (IDT). To form RNP complexes, gRNA duplex and Cas9-3NLS (IDT) were combined, incubated for 10–15 min at room temperature, and then placed on ice until used. RNP complexes and 10 µg of 100-nt donor template oligonucleotide were electroporated into 2 × 105 fibroblasts or 3.5 × 105 HEK293T cells using Lonza 4D-Nucleofector. Cells were plated in a 12-well for 48–72 h to recover before single-cell plating in 96-wells. Clones were expanded and screened by sequencing of genomic DNA. No selection was used.
Chromosomal breakage
Cells were treated with 0.1 µg DEB/mL of media for 48–72 h or 45–100 nM of MMC for 24 h. HU treatments were as indicated. LCLs were arrested with colcemid (0.17 µg/mL) for 20 min and fibroblasts for 90 min. Cells were harvested and incubated in 0.075 M KCL for 10 min before being fixed in methanol and acetic acid (3:1). Cells were dropped onto wet slides and dried for at least 1 h at 40°C before staining with Karyomax Giemsa (Invitrogen) for 3 min. Dry slides were then imaged on the Metasystems Metafer slide scanning platform.
Cell survival studies
Fibroblasts were seeded overnight in triplicate and treated the next day with DNA damaging agents at indicated concentrations. Cells were grown for 4–6 d and passaged once at appropriate ratios. Once cells reached near confluence (7–9 d), they were counted using Z2 Coulter counter (Beckman Coulter). In the case of cisplatin treatment, drug was removed after 1 h and cells were washed with PBS and given fresh drug-free media. For aphidicolin treatment, after 48 h cells were washed with PBS and given fresh drug-free media. For PARPi treatment, cells were given fresh media with olaparib daily. For ionizing, radiation cells were treated with the indicated IR dose in Falcon tubes prior to being plated. LCLs were treated at the time of seeding, agitated daily, and counted on the seventh day. HEK293T cells were seeded overnight, treated with MMC, passaged after 3 d, and counted on the fifth day.
Western blot
Whole-cell extracts were prepared by lysing cell pellets in Laemmli sample buffer (Bio-Rad or 4% SDS, 20% glycerol, 125 mM Tris-HCl at pH 6.8). Samples were either sonicated or vortexed at highest speed for 30 sec. Samples were boiled for 5 min. For pRPA and BRCA2 Western blots, samples were instead heated for 10 min at 50°C. Proteins were separated on 4%–12% or 3%–8% gradient gels (Invitrogen) by SDS-PAGE. Immunoblotting was performed using the antibodies indicated in Supplemental Table S10.
Immunofluorescence
Cells were seeded on coverslips the day before. For FANCD2 foci, cells were treated with 1 µM MMC for 24 h. For RAD51 foci, cells were irradiated for indicated dose or treated with 3 µM MMC for 1 h and harvested at indicated times. For RPA foci, cells were treated with 3 µM MMC for 1 h and harvested at indicated times. Cells were washed with PBS twice, fixed in 3.7% formaldehyde for 10 min, washed twice with PBS, and permeablized with 0.5% Triton in PBS for 10 min. Cells were blocked in 5% (v/v) FBS in PBS and incubated with primary antibodies in blocking buffer for 2 h at room temperature or overnight at 4°C (for antibodies see Supplemental Table S9). Cells were washed three times for 5 min with blocking buffer and then incubated with secondary antibody (Alexa fluor; 1:1000). Cells were washed again three times with blocking buffer, rinsed quickly with water, air dried, and then embedded on glass slides with DAPI Fluoromount-G (SouthernBiotech).
Sister chromatid exchange
For MMC-induced SCEs, fibroblasts were cultured for 24 h in 10 µg/mL BrdU and then treated with 0.1 or 0.2 µg/mL MMC for 1 h. Cells were washed and put into fresh media with 10 µg/mL BrdU for another 24 h. For cells depleted of BLM, siRNA transfection was performed twice as described. For the second siRNA transfection, 10 µg/mL BrdU was added to media and cells were cultured in BrdU for a total of 48 h before harvest. Cells were collected, fixed, and dropped on glass slides for metaphases as previously described. Slides were dried overnight at 42°C and then stained in 20 µg/mL Hoechst 33342 for 30 min. Slides were treated with 254 nM UV light for 3 h. Slides were incubated for 2 h at 65°C in 2× SCC, then rinsed in 1× GURR buffer, and stained in 8% Giemsa Karyomax for 3 min. Metaphases were scanned and imaged on Metasystems Metafer slide scanning platform.
mClover homologous recombination assay
Cells were plated in a 24-well plate the day before and transfected with 0.25 µg of pCMV-Cas9-sgLMNA-BFP and 0.4 µg of pDONR-LMNA using TransIT-293 Transfection Reagent (Mirus) according to manufacturer's instructions (plasmids were a gift from Jan Karlseder) (Arnoult et al. 2017). Twenty-four hours after transfection cell media was replaced. Cells were incubated for another 48 h and were then harvested and analyzed on BD LSRII to determine the proportion of mClover positive cells and data was analyzed with FlowJo.
DNA fibers
For DNA fibers, cells were plated the evening before and labeled with nucleotide analogs and treated with 6 mM HU for 5 h. Cells were harvested and cell pellets were washed once in cold PBS. Cells were resuspended at a concentration of 1 × 106 cells/mL in cold PBS. On a clean glass coverslip, 10 µL of droplets of spreading buffer (0.5% SDS, 200 mM Tris-HCl at pH 7.4, and 50 mM EDTA at pH 8) was placed. Cell suspension (2.5 µL) was pipetted into the spreading buffer, stirred, and pipetted up and down three times. Coverslips were incubated horizontally for 9 min at room temperature before gently being tilted vertically to allow the buffer to run down the slide. Coverslips were dried at room temperature at an angle and then heated for 30 min at 65°C. Coverslips were fixed in methanol/acetic acid 3:1 overnight at 4°C. The next day coverslips were washed in PBS three times at room temperature and then incubated in 2.5 M HCl for 1 h. Coverslips were then washed five times for 5 min with PBS and after the final wash they were blocked in 5% FBS in PBS for 30 min. For immunostaining, coverslips were incubated with primary antibodies for 2.5 h at room temperature. Rat anti-BrdU antibody (1:40) was used to detect CldU and mouse anti-BrdU antibody (1:20) was used to detect ldU. Coverslips were washed five times with PBS with 0.2% Tween and then blocked for 30 min in 5% FBS in PBS. Coverslips were incubated with secondary (Alexa Fluor) anti-rat (594) and anti-mouse (488) at a dilution of 1:300 for 1 h at room temperature. Coverslips were washed five times with with PBS with 0.2% Tween and rinsed with water and air-dried. Dry coverslips were mounted on glass slides using Fluoromount-G (SouthernBiotech). DNA tracks were all imaged on the DeltaVision Image Restoration microscope and measured using ImageJ. For each experimental condition, ≥200 fibers were analyzed for each experiment.
Whole-exome sequencing
The libraries for whole-exome sequencing (WES) were constructed and sequenced on Illumina HiSeq 2000 or Illumina GA-IIX using 76-bp paired-end reads at the Broad Institute or by using Agilent SureSelect Human All Exon V4 capture kit and 100-bp paired-end sequencing on Illumina HiSeq 2500. Sequence was aligned to human genome build GRCh37 using BWA (Burrows-Wheeler aligner) (Li and Durbin 2009). Duplicate reads were marked using Picard (http://picard.sourceforge.net). Genome Analysis Toolkit (GATK) was used for base quality score recalibration (BQSR), and local realignment around indels (DePristo et al. 2011). Variant discovery was performed in part by variant calling with GATK HaplotypeCaller and then joint genotyping with GATK GenotypeGVCFs. The variant call sets were then refined with variant quality score recalibration (VQSR) and VQSR scores helped discriminate low-quality variants. Variant annotation was performed using SnpEff, VCFtools, and in-house software (NYGC) (Danecek et al. 2011; Cingolani et al. 2012). All WES was analyzed with the NYGC sequence analysis pipeline.
NCBI references are BRCA2/FANCD1, RefSeq NM_000059.3, and protein NP_000050.2
Supplementary Material
Acknowledgments
We thank the family and patients for their participation in the International Fanconi Anemia Registry (IFAR) and this study. We thank Alberto Ciccia for sharing shRNAs to SMARCAL1 and ZRANB3. We thank Alan D'Andrea for sharing HSC62 cells. We thank the A.S. laboratory members for their input and discussion of the manuscript. Initial BRCA2 studies were funded by Starr Foundation grant I8-A8-101. RAD51 and BRCA2 studies in the laboratory are supported by grant RO1 CA204127 from National Institutes of Health (NIH) and grant UL1TR001866 from the National Center for Advancing Translational Sciences, NIH Clinical and Translational Science Award program. K.A.R. was supported by the Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program and the William Randolph Hearst Foundation Fellowship at the Rockefeller University. A.S. is a Howard Hughes Medical Institute Faculty Scholar.
Author contributions: K.A.R. and A.S. conceived the ideas and designed experiments for this study. K.A.R., R.J.N., F.P.L., S.S., A.T.W., and A.H. performed the experiments. A.A. and K.A.R. performed bioinformatic analysis of the exomes. A.D.A. directed clinical testing in the proband at the time of IFAR enrollment and excluded number of complementation groups in the patient. M.K. provided clinical information. K.A.R. and A.S. wrote the manuscript with essential input from other authors. A.S. acquired funding and supervised all studies.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.336446.120.
References
- Alcón P, Shakeel S, Chen ZA, Rappsilber J, Patel KJ, Passmore LA. 2020. FANCD2–FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair. Nat Struct Mol Biol 27: 240–248. 10.1038/s41594-020-0380-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Rosenberg PS, Brody LC. 2007. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet 44: 1–9. 10.1136/jmg.2006.043257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Correia A, Ma J, Merlo A, Garcia-Gomez S, Maric M, Tognetti M, Benner CW, Boulton SJ, Saghatelian A, et al. 2017. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549: 548–552. 10.1038/nature24023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. 2009. Fanconi anemia and its diagnosis. Mutat Res 668: 4–10. 10.1016/j.mrfmmm.2009.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Krishnamoorthy A, Dungrawala H, Garcin EB, Modesti M, Cortez D. 2018. RADX modulates RAD51 activity to control replication fork protection. Cell Rep 24: 538–545. 10.1016/j.celrep.2018.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillón J, Ramírez MJ, Pujol R, et al. 2013. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet 92: 800–806. 10.1016/j.ajhg.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. 2009. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 28: 2601–2615. 10.1038/emboj.2009.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D'Andrea AD. 2016. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol 17: 337–349. 10.1038/nrm.2016.48 [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendouga N, Gao H, Moechars D, Janicot M, Vialard J, McGowan CH. 2005. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol Cell Biol 25: 7569–7579. 10.1128/MCB.25.17.7569-7579.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Bhat KP, Le Meur R, Chazin WJ, Ding X, Sharan SK, Wessel SR, Sathe AA, Zhao R, Cortez D. 2017. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol Cell 67: 374–386.e5. 10.1016/j.molcel.2017.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Patel KJ. 2014. Why does the bone marrow fail in Fanconi anemia? Blood 123: 26–34. 10.1182/blood-2013-09-427740 [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7: 249–262. 10.1016/S1097-2765(01)00173-3 [DOI] [PubMed] [Google Scholar]
- Guidugli L, Pankratz VS, Singh N, Thompson J, Erding CA, Engel C, Schmutzler R, Domchek S, Nathanson K, Radice P, et al. 2013. A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res 73: 265–275. 10.1158/0008-5472.CAN-12-2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidugli L, Carreira A, Caputo SM, Ehlen A, Galli A, Monteiro AN, Neuhausen SL, Hansen TV, Couch FJ, Vreeswijk MP, et al. 2014. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum Mutat 35: 151–164. 10.1002/humu.22478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel V, Miller ES, Zlatanou A, Nieminuszczy J, Ryan EL, Davies NJ, et al. 2015. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol Cell 59: 462–477. 10.1016/j.molcel.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609. 10.1126/science.1073834 [DOI] [PubMed] [Google Scholar]
- Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W, et al. 2012. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149: 1221–1232. 10.1016/j.cell.2012.04.030 [DOI] [PubMed] [Google Scholar]
- Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL. 2012. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle 11: 3983–3996. 10.4161/cc.22215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja KK, Lee EH, Hendrickson EA, Campbell JL. 2014. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle 13: 1540–1550. 10.4161/cc.28476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. 2011. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 43: 142–146. 10.1038/ng.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. 2013. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood 121: 54–63. 10.1182/blood-2012-07-441212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TM, Son MY, Dodds S, Hu L, Hasty P. 2014. Deletion of BRCA2 exon 27 causes defects in response to both stalled and collapsed replication forks. Mutat Res 766-767: 66–72. 10.1016/j.mrfmmm.2014.06.003 [DOI] [PubMed] [Google Scholar]
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Räschle M, Walter JC, Knipscheer P. 2014. XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54: 460–471. 10.1016/j.molcel.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701. 10.1126/science.1182372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493: 356–363. 10.1038/nature11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475: 53–58. 10.1038/nature10192 [DOI] [PubMed] [Google Scholar]
- Lemaçon D, Jackson J, Quinet A, Brickner JR, Li S, Yazinski S, You Z, Ira G, Zou L, Mosammaparast N, et al. 2017. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat Commun 8: 860 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8: 255–265. 10.1016/j.ccr.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. 2011. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333: 84–87. 10.1126/science.1204258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, et al. 2004. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science 304: 1822–1826. 10.1126/science.1094557 [DOI] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol 23: 3417–3426. 10.1128/MCB.23.10.3417-3426.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Ray Chaudhuri A, Nussenzweig A, Janscak P, et al. 2017. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun 8: 859 10.1038/s41467-017-01164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moder M, Velimezi G, Owusu M, Mazouzi A, Wiedner M, Ferreira da Silva J, Robinson-Garcia L, Schischlik F, Slavkovsky R, Kralovics R, et al. 2017. Parallel genome-wide screens identify synthetic viable interactions between the BLM helicase complex and Fanconi anemia. Nat Commun 8: 1238 10.1038/s41467-017-01439-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutreja K, Krietsch J, Hess J, Ursich S, Berti M, Roessler FK, Zellweger R, Patra M, Gasser G, Lopes M. 2018. ATR-mediated global fork slowing and reversal assist fork traverse and prevent chromosomal breakage at DNA interstrand cross-links. Cell Rep 24: 2629–2642.e5. 10.1016/j.celrep.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, et al. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol 24: 5776–5787. 10.1128/MCB.24.13.5776-5787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niraj J, Färkkilä A, D'Andrea AD. 2019. The Fanconi anemia pathway in cancer. Annu Rev Cancer Biol 3: 457–478. 10.1146/annurev-cancerbio-030617-050422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Rosselli F. 2004. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J 23: 3154–3163. 10.1038/sj.emboj.7600277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder J, Salsman J, Dellaire G. 2015. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res 43: 9379–9392. 10.1093/nar/gkv993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przetocka S, Porro A, Bolck HA, Walker C, Lezaja A, Trenner A, von Aesch C, Himmels SF, D'Andrea AD, Ceccaldi R, et al. 2018. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol Cell 72: 568–582.e6. 10.1016/j.molcel.2018.09.014r [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, et al. 2007. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 39: 165–167. 10.1038/ng1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535: 382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman K, Smogorzewska A. 2019. Advances in understanding DNA processing and protection at stalled replication forks. J Cell Biol 218: 1096–1107. 10.1083/jcb.201809012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kähkonen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian Genomics, FORGE Canada Consortium, Majewski J, Dyment DA, Innes AM, et al. 2015. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov 5: 135–142. 10.1158/2159-8290.CD-14-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51–BRCA1/2. Cancer Cell 22: 106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimelis H, Mesman RLS, Von Nicolai C, Ehlen A, Guidugli L, Martin C, Calléja F, Meeks H, Hallberg E, Hinton J, et al. 2017. BRCA2 hypomorphic missense variants confer moderate risks of breast cancer. Cancer Res 77: 2789–2799. 10.1158/0008-5472.CAN-16-2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N, Barbera MA, Egashira A, Lam I, Christ N, Schlacher K, Xia B, Jasin M. 2011. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet 7: e1002409 10.1371/journal.pgen.1002409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129: 289–301. 10.1016/j.cell.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol 19: 5166–5169. 10.1128/MCB.19.7.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhasini AN, Brosh RM Jr. 2012. Fanconi anemia and Bloom's syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet 28: 7–13. 10.1016/j.tig.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R, et al. 2017. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol Cell 68: 414–430.e8. 10.1016/j.molcel.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, van Twest S, Leis A, Bythell-Douglas R, Murphy VJ, Sharp M, Parker MW, Crismani WM, Deans AJ. 2020. Monoubiquitination by the human Fanconi anemia core complex clamps FANCI:FANCD2 on DNA in filamentous arrays. Elife 9: e54128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, et al. 2015. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol 208: 545–562. 10.1083/jcb.201406100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D'Andrea AD, et al. 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell 7: 241–248. 10.1016/S1097-2765(01)00172-1 [DOI] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. 2010. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet 42: 406–409. 10.1038/ng.570 [DOI] [PubMed] [Google Scholar]
- Wang AT, Smogorzewska A. 2015. SnapShot: Fanconi anemia and associated proteins. Cell 160: 354–354.e1. 10.1016/j.cell.2014.12.031 [DOI] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. 2015. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol Cell 59: 478–490. 10.1016/j.molcel.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Wang S, Dhar A, Peralta C, Pavletich NP. 2020. DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex. Nature 580: 278–282. 10.1038/s41586-020-2110-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. 2007. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39: 159–161. 10.1038/ng1942 [DOI] [PubMed] [Google Scholar]
- Xu S, Wu X, Wu L, Castillo A, Liu J, Atkinson E, Paul A, Su D, Schlacher K, Komatsu Y, et al. 2017. Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev 31: 1469–1482. 10.1101/gad.299172.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–1848. 10.1126/science.297.5588.1837 [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. 2005. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433: 653–657. 10.1038/nature03234 [DOI] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol 208: 563–579. 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.