Abstract
Introduction:
Acute exposure to ionizing radiation (IR) is hazardous or even lethal. Accurate estimation of the doses of IR exposure is critical to wisely determining the following treatments. Exosomes are nanoscale vesicles harboring biomolecules and mediate the communications among cells and tissues to influence biological processes. Screening out the microRNAs (miRNAs) contained in exosomes as biomarkers can be useful for estimating the IR exposure doses and exploring the correlation between these miRNAs and the occurrence of disease.
Methods:
We treated mice with 2.0, 6.5, and 8.0 Gy doses of IR and collected the mice sera at 0, 24, 48, and 72 hours after exposure. Then, the serum exosomes were isolated by ultracentrifuge and the small RNA portion was extracted for sequencing and the following bioinformatics analysis. Qualitative polymerase chain reaction was performed to validate the potential dose-specific markers.
Results:
Fifty-six miRNAs (31 upregulated, 25 downregulated) were differentially expressed after exposure of the above 3 IR doses and may act as common IR exposure miRNA markers. Bioinformatic analysis also identified several dosage-specific responsive miRNAs. Importantly, IR-induced miR-151-3p and miR-128-3p were significantly and stably increased at 24 hours in different mouse strains with distinct genetic background after exposed to 8.0 Gy of IR.
Conclusion:
Our study shows that miR-151-3p and miR-128-3p can be used as dose-specific biomarkers of 8.0 Gy IR exposure, which can be used to determine the exposure dose by detecting the amount of the 2 miRNAs in serum exosomes.
Keywords: ionizing radiation, exosomes, serum, miRNA, biomarkers
Introduction
Exosomes are nanoscale vesicles actively secreted by different types of cells and are abundantly in the body fluids.1 Exosomes contain proteins, DNAs and RNAs, and are important for material transportation and signal transduction. Previous reports indicated that microRNAs (miRNAs) in exosomes are very important for common biological processes in organisms.2 Circulating miRNAs in exosomes are useful tools for detecting many diseases. It was reported that serum exosomal miRNAs might represent potential diagnostic biomarkers in clear cell renal cell carcinoma.3 Serum exosomal miR-9 and miR-124 are promising biomarkers for diagnosing acute ischemic stroke and evaluating the degree of damage caused by ischemic injury.4 Expression levels of miR-125b in exosomes obtained from serum are low and associated with advanced melanoma disease, probably reflecting the tumor cell dysregulation.5
Since Roentgen discovered X-ray, the history of research and application of ionizing radiation (IR) has been more than a century. Acute exposure to IR is hazardous or even lethal. Radiation sickness includes acute radiation sickness and chronic radiation sickness. Acute radiation sickness are systemic diseases caused by a large dose (>1 Gy) of IR in a short time (several seconds to days). Acute radiation sickness includes acute radiation sickness of bone marrow (1-10 Gy), acute radiation sickness of intestinal type (>10 Gy), and acute radiation sickness of brain (>50 Gy). Ionizing radiation exposure affects the abundance of exosomes and, in particular, changes their molecular composition, which promotes the migration phenotype upon absorption.6 After X-ray irradiation, proteins and RNAs of exosomes can induce changes in telomeric metabolism, which can lead to genomic instability in epithelial cancer cells.7 The bystander effect and genomic instability are at least partially mediated by exosomal RNA.8 The exosomal miR-7-5p is a crucial mediator of bystander autophagy and associated with EGFR/Akt/mTOR signaling pathway.9
Biomarkers can be used to diagnose diseases, determine disease stages, or to evaluate the safety and effectiveness of new drugs or therapies in target populations. Studies have evaluated the efficacy of acute-phase protein serum amyloid A (SAA) as a biomarker for IR exposure in mice. A 24-hour total body irradiation (TBI) dose prediction model was generated using SAA levels. The model successfully divided TBI mice into the control group, 1.0 Gy group, and ≥2.0 Gy group with high accuracy.10 People exposed to irradiation had radiation nephropathy in the late stage. The expression of miR-1224 and miR-21 are significantly upregulated in mice urine exposed to IR. MiR-1224 is a good dose-dependent responder after 2.0, 4.0, 6.0, and 8.0 Gy of TBI, indicating that it could be used as a biomarker for IR exposure.11 The expression of adhesion molecules and their related functions of adhesion and migration can be used to identify the IR-induced and dose-dependent effects in peripheral blood mononuclear cells (PBMCs). Compared with the control group, the adherence ability of the irradiated PBMC to the immunoglobulin G (IgG) substrate was significantly increased at 6 hours after irradiation exposure. With the increase in IR dose, the migration ability of MIP-1α chemokines in rats toward PBMCs was significantly decreased. Therefore, cell-surface molecules and proteins associated with cellular functions may be potential biomarkers that identify radiation exposure doses.12 One study found that miR-133b, miR-215, and miR-375 can be used to distinguish whether or not it has been irradiated, based on 2 miRNAs, miR-30a and miR-126 can be used to predict radiation-induced mortality.13
The purpose of this study was to analyze exosome miRNA signatures in serum in response to different doses of IR. We found that miRNA signatures can effectively distinguish 0, 2.0, 6.5, and 8.0 Gy whole-body irradiation groups at different time points (0, 24, 48, and 72 hours) after IR exposure. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to investigate the effects of different doses of IR on signal pathways. Importantly, miR-128-3p and miR-151-3p were identified as dose-specific and stable exosomal miRNA markers for 8.0 Gy IR exposure after 24 hours. The differentially expressed miRNA markers found in this study not only provide a series of information markers for identifying radiation exposure doses but also lay the foundation for future clinical interventions.
Materials and Methods
Animals
This study was approved by the Institutional Animal Care and Use Committee of Southern Medical University (approval code L2016103, September 13, 2016), which is following the guidelines of the Asian Federation of Laboratory Animal Science Associations and the National Regulations for the Administration of Affairs Concerning Experimental Animals (January 8, 2011). All animals were purchased from the Guangdong Medical Laboratory Animal Center and treated following standard guidelines for the care and use of laboratory animals. Male C57BJ/L or BALB/c mice were separately housed at 25 °C and 50% to 60% relative humidity in cages under a 12/12-hour light/dark cycle, with free access to food and water.
Ionizing Radiation Treatment and Serum Collection
Ionizing radiation treatment was performed using X-ray at dose rate of 110 cGy/min. A total of 130 male C57BJ/L or BALB/c mice were separated into sham control group and 4 IR-treated groups. (Both mice are 6-8 weeks old, and the weight of C57BL/6 mice is 17.5-19.0 g while 19.5-21.0 g for the BALB/c mice.) As indicated in Supplementary Table S1, besides for the sham control group (10 mice without IR treatment), the remaining mice (120 mice) were equally separated into 4 groups (30 mice per group) and exposed to 4 different doses (0, 2.0, 6.5, and 8.0 Gy) of IR exposure. The serum samples were collected at 24, 48, and 72 hours after IR exposure with 10 mice per dose time point. The blood samples were incubated at 4 °C for 4 to 5 hours to clot and then centrifuged at 4000 rpm for 10 minutes at 4 °C. In every group, 200 µL of translucent blood plasma per mice was collected from the upper layer into a sterilized tube. The serum samples were preserved at −80 °C for further analysis.
Exosome Isolation by Ultracentrifugation
Add 250-µL serum to 10 mL of phosphate-buffered serum (PBS) to mix and pass through a 0.22-µm filter. After collecting the filtrate, it was centrifuged at 10 000g for 60 minutes. Then the supernatant was centrifuged at 110 000g for 70 minutes. After resuspended the precipitate with 10 mL PBS, it was centrifuged at 110 000g for 70 minutes. The final precipitated exosomes were resuspended by adding 100-µL PBS and store at −80 °C for further analysis.
Nanoparticle Tracking Analysis of Exosomes
We used nanoparticle tracking analysis (NTA) to quantify the number and size of exosomes isolated from serum samples using Nanosight NS300 (Malvern, Worcestershire, United Kingdom). Based on Brownian motion, NTA can visualize and analyze particles. When the nanoparticles are scattered under laser irradiation, the size of the nanoparticles in the sample can be detected in the range of 10 to 2000 nm. Exosome samples were measured 5-fold for 5 × 30 seconds each after diluted at 1:500.
Transmission Electron Microscopy
Exosomes examined with a scanning electron microscope (SEM) were loaded onto a carbon-coated electron microscope grid as mentioned previously.14 The samples were fixed with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 mol/L sodium cacodylate buffer at pH 7.3 for 3 hours at room temperature. The sample was dried at the critical point, mounted on the sample stub, spray-coated, and observed with a Hitachi S3400 SEM.
Western Blot Analysis
Twenty microgram of protein was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel and then transferred to a polyvinylidenedifluoride membrane (Millipore). Membranes were blocked and incubated with anti-CD63 (1:2000, Santa Cruz Biotechnology) and anti-TSG101 (1:2000, Abcam) for 2 hours. Anti-mouse or anti-rabbit IgG labeled with horseradish peroxidase was used as the secondary antibody (TBST 1:500 dilution). Bound antibodies were visualized with the Luminata forte Western HRP substrate (Millipore).
MicroRNA Library Construction and Sequencing
Total RNA was extracted from exosomes and used to prepare small RNA libraries and sequences (RiboBio). In short, total RNA samples were fractionated using 15% Tris-borate-EDTA polyacrylamide gel (Invitrogen), and small RNAs of 18 to 30 nucleotides (nt) were used for library preparation. Small RNAs were reverse transcribed and amplified by polymerase chain reaction (PCR). The PCR products were sequenced using the Illumina HiSeq 3000 platform.
Kyoto Encyclopedia of Genes and Genomes Analysis
Kyoto Encyclopedia of Genes and Genomes signaling pathway enrichment analysis was performed by differentially expressed miRNAs at various doses. The target genes corresponding to each miRNA were analyzed by miRDB tool, and they were enriched by KEGG pathway by DAVID tool.
Isolation of RNA and Quantitative Real-Time PCR
The total RNA of exosomes was extracted with Trizol (Invitrogen) according to the manufacturer’s protocol. RNA was quantified using Nanodrop 2000 at OD260. TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) was used to perform miRNA reverse transcription for miR-128-3p, miR-151-5p, and U6. TaqMan miRNA assays (Applied Biosystems) for miR-128-3p, miR-151-5p, and U6 were performed using TaqMan Universal Master Mix II, no UNG (Applied Biosystems) on a Roche LightCycler 96 instrument. Cycling parameters were 95 °C for 10 minutes, 40 cycles of 95 °C (15 seconds), and annealed/extended at 60 °C for 40 seconds. The gene expression ΔΔCt values of miR-128-3p and miR-151-5p from each sample were calculated by normalizing with internal control of U6, respectively. Fold-change was calculated using the equation 2−ΔΔCt. All experiments were performed in triplicates.
Statistical Analysis
All of data were used IBM SPSS Statistics (version 17) to analyze. We used Student t test to compare relative expression levels between individual groups. A P < 0.05 indicates a statistically significant difference. We used receiver operating characteristic (ROC) analysis and Youden index to evaluate the power of each biomarker.
Results
8.0 Gy Is Lethal to Experimental Mice
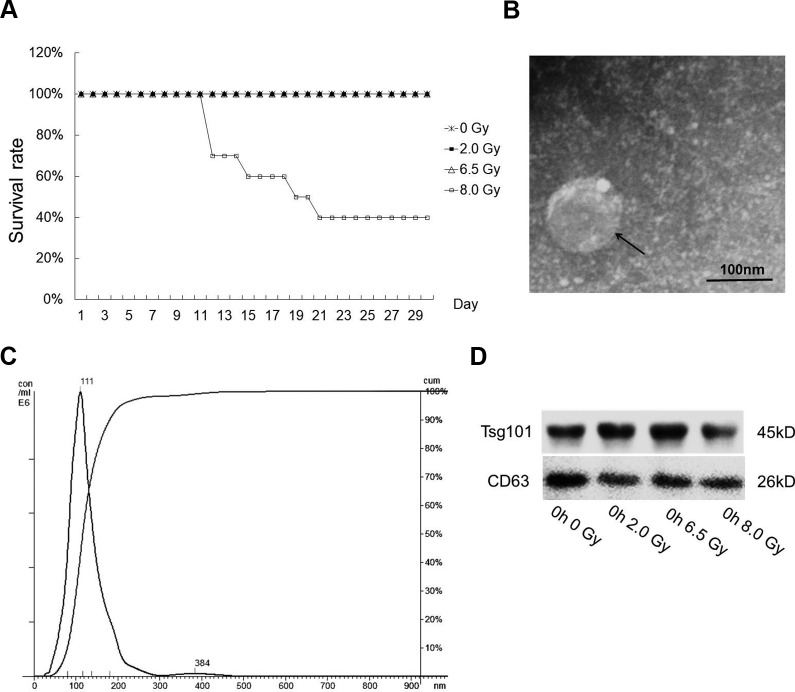
To systematically analyze the impact of IR on C57BJ/L mice, except a sham control group (10 mice without treatment), the mice were separated into 4 groups treated with different IR doses (0, 2.0, 6.5, and 8.0 Gy). We first analyzed the survival rates of mice irradiated with the above doses of X-ray. Within 1-month observation period after IR exposure, no death has been observed in the 0, 2.0, and 6.5 Gy groups within 30 days. While the mice in the 8.0 Gy group began to die from the 12th day, only 40% of mice in the same group were still alive at the end of the observation period (Figure 1A). No significant differences appeared among mice in the first 3 days after IR exposure. Because our primary goal is to find dose-specific markers for early diagnosis, we choose 24, 48, and 72 hours important time points to explore.
Figure 1.
Characterization of serum exosomes isolated by ultracentrifugation. A, Survival curves of C57BJ/L male mice exposed to 0 (control), 2.0, 6.5, or 8.0 Gy of total body irradiation (n = 10 per group). B, Exosomes were analyzed under electron microscopy. Bar 100 nm. C, The size distribution of exosomes analyzed by the qNano particle counter. D, Western immunoblotting of exosomal markers TSG101 and CD63 from different dose groups.
We also evaluated the impact of IR exposure on the body weight of mice (Supplementary Table S2). There is no significant difference in body weight among different groups before IR treatment. Yet, the body weight was influenced immediately after IR exposure, as shown by significantly low body weight in 2.0 and 6.5 Gy groups compared with 0 Gy group (Supplementary Table S2A, 0 hour). This situation continued to be more obvious until 2.0, 6.5, and 8.0 Gy groups all showed significantly low body weight compared with 0 Gy group (Supplementary Table S2A, 48 and 72 hours). We have compared body weights in the same group at different times after radiation exposure and found significant weight gain in the control group at 48 and 72 hours after exposure. There was no significant difference in body weight between 2.0 and 6.5 Gy groups before and after exposure. In the 8.0 Gy group, body weight decreased significantly at 48 and 72 hours after exposure (Supplementary Table S2B). It is suggested the IR treatment significantly influences the body weight in experimental animals.
Isolation and Identification of Exosomes From IR-Treated Serum Samples
After IR treatments, the serum samples were collected from all groups (1 control group and 4 IR-treated groups) at different time points (0, 24, 48, and 72 hours). We collected 10 serum samples (about 300 µL each) at every specific the dose time point and combined the 10 serum samples with the equal 200 µL volume for that dosage time point for the following exosome isolation, miRNA extraction, and small RNA sequencing.
The quality of serum exosomes was examined by SEM and NTA (Figure 1B and C). Isolated exosomes are particles ranging from 50 to 150 nm in diameter (Figure 1B). By NTA, it has been determined the median size was 111 nm (Figure 1C). The presence of known exosomal markers Tsg101 and CD63 in all isolated exosomes was confirmed by Western blot (Figure 1D). The above analysis confirmed the isolated vesicles from sera were exosomes. The expression of Tsg101 and CD63 in exosomes of other groups is shown in Supplementary Figure S1.
Differentially Expressed Exosomal MiRNAs Were Identified From IR-Treated Serum Samples by Small RNA Sequencing
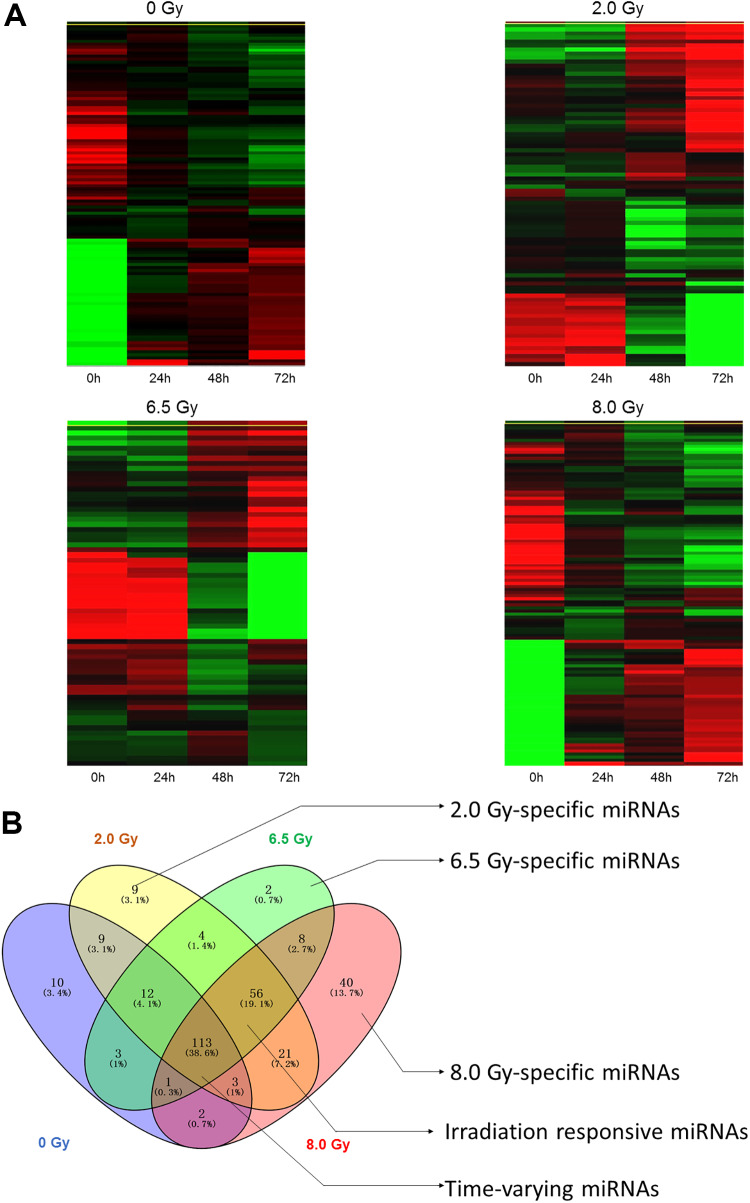
To investigate the expression profiles of exosomal miRNAs isolated from serum of control and IR-treated mice, the RNA samples were extracted from serum exosomes and small RNA libraries were constructed and sequenced using Illumina HiSeq 3000. To identify the conserved miRNAs, all miRNA reads from control and IR-treated group libraries were compared with the known mouse miRNAs in miRBase version 21. To focus on the highly represented miRNAs, we removed miRNAs which sequences less than 10 reads. A total of 461 miRNAs was detected from all the exosomal samples. To identify differential expressed miRNAs, we grouped exosomal miRNA samples (24, 48, and 72 hours) according to exposure dosage (0, 2.0, 6.5, and 8.0 Gy) and compared them with the 0 hour samples. It was thought that miRNAs with more than 2-fold expression changes (P < . 05) were significantly differentially expressed and these miRNAs were used for hierarchical clustering, which indicates the differentially expressed miRNAs in response to IR exposure (Figure 2A).
Figure 2.
Differential expression of microRNAs in serum exosomes between control, 2.0 Gy, 6.5 Gy, and 8.0 Gy groups. A, The hierarchical clustering of differentially expressed microRNAs. B, Venn diagrams indicated the numbers of overlapping and nonoverlapping differentially expressed microRNAs in serum exosomes after 0, 2.0, 6.5, and 8.0 Gy exposure.
We compared the 3 dose groups with the control group (0 hour sample) and found 227 miRNAs (53 upregulated, 174 downregulated) were differentially expressed in the 2 Gy group (24, 48, and 72 hours) and 199 miRNAs (38 upregulated, 161 downregulated) were differentially expressed in the 6.5 Gy group (24, 48, and 72 hours). When compared 8.0 Gy group (24, 48, and 72 hours) with the control group (0 hour sample), there were 244 mioRNAs (84 upregulated, 160 downregulated) that were differentially expressed. The top 30 differentially expressed miRNAs from 2.0, 6.5, and 8.0 Gy group are listed in Table 1. To screen common markers of IR exposure and dose-specific markers, Venn analysis of significantly varied miRNAs were performed using Venny version 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny; Figure 2B). 9 (5 downregulated, 4 upregulated), 2 (downregulated), and 40 (33 upregulated, 7 downregulated) miRNAs were identified as dose-specific miRNAs response to 2.0, 6.5, and 8.0 Gy IR exposure, respectively (Figure 2B, Table 2), whereas 56 miRNAs (31 upregulated, 25 downregulated) are common IR exposure-responsive miRNAs shared by the above 3 IR doses (Figure 2B, Table 3). We also found that 113 miRNAs were time dependently varied miRNAs, indicating their changing is not caused by IR exposure and thus were excluded (Figure 2B). The common and dose-specific IR responsive exosomal miRNAs were selected for further bioinformatic and experimental evaluations.
Table 1.
Top 30 Differentially Expressed MicroRNAs Between the Irradiation Exposure Group and the Non-Irradiation Exposure Group.a
| 0 Gy 0 hours versus (0 Gy 24 hours, 0 Gy 48 hours, 0 Gy 72 hours common) | 0 Gy 0 hours versus (2.0 Gy 24 hours, 2.0 Gy 48 hours, 2.0 Gy 72 hours common) | 0 Gy 0 hours versus (6.5 Gy 24 hours, 6.5 Gy 48 hours, 6.5 Gy 72 hours common) | 0 Gy 0 hours versus (8.0 Gy 24 hours, 8.0 Gy 48 hours, 8.0 Gy 72 hours common) | ||||
|---|---|---|---|---|---|---|---|
| miRNA_ID | Fold change | miRNA_ID | Fold change | miRNA_ID | Fold change | miRNA_ID | Fold change |
| miR-5100 | −500.66 | miR-15a-5p | −3192.79 | miR-18a-5p | −1010.74 | miR-124-3p | 4327.45 |
| miR-331-5p | −500.66 | let-7i-3p | −1057.98 | miR-1191a | −680.15 | miR-15a-5p | −3192.79 |
| miR-17-3p | −491.21 | miR-18a-5p | −1010.74 | miR-181c-5p | −614.00 | miR-130a-5p | −632.89 |
| miR-10a-3p | −396.75 | miR-130a-5p | −632.89 | miR-17-3p | −491.21 | miR-495-3p | 608.45 |
| miR-8097 | −387.30 | miR-15a-3p | −623.44 | miR-872-3p | −443.96 | miR-129-2-3p | 525.05 |
| miR-3473b | −358.97 | miR-181c-5p | −614.00 | miR-3470b | −425.08 | miR-17-3p | −491.21 |
| miR-30d-3p | −358.97 | miR-17-3p | −491.21 | miR-3473b | −358.97 | miR-200b-5p | 427.74 |
| miR-1945 | −330.61 | miR-495-3p | 422.67 | miR-30d-3p | −358.97 | miR-409-3p | 405.17 |
| miR-331-3p | −321.17 | miR-30d-3p | −358.97 | miR-16-2-3p | −321.17 | miR-485-5p | 387.43 |
| miR-3535 | 260.29 | miR-500-3p | −340.05 | miR-331-3p | −321.17 | miR-30d-3p | −358.97 |
| miR-6967-3p | −255.04 | miR-16-2-3p | −321.17 | miR-3535 | 305.09 | miR-96-5p | 350.07 |
| miR-7653-5p | −236.16 | miR-331-3p | −321.17 | miR-450a-5p | −302.27 | miR-3535 | 347.36 |
| miR-449a-5p | −236.16 | miR-409-3p | 306.70 | miR-190b-5p | −292.83 | miR-760-3p | 338.99 |
| miR-6960-5p | −236.16 | miR-190b-5p | −292.83 | miR-1983 | −283.38 | miR-331-3p | −321.17 |
| miR-25-5p | −236.16 | miR-202-5p | −283.38 | miR-202-5p | −283.38 | miR-433-3p | 307.11 |
| miR-3061-5p | −226.71 | miR-200b-5p | 275.36 | miR-324-5p | −273.94 | miR-770-3p | 299.58 |
| let-7f-2-3p | −217.26 | miR-324-5p | −273.94 | miR-411-5p | −264.50 | miR-202-5p | −283.38 |
| miR-1960 | −207.82 | miR-322-5p | −255.04 | miR-200b-5p | 257.67 | miR-511-5p | 270.20 |
| miR-7669-5p | −198.38 | miR-25-5p | −236.16 | miR-322-5p | −255.04 | miR-322-5p | −255.04 |
| let-7g-3p | −188.92 | miR-449a-5p | −236.16 | miR-449a-5p | −236.16 | miR-7043-3p | 254.57 |
| miR-1957a | −188.92 | miR-3061-5p | −226.71 | miR-3061-5p | −226.71 | miR-204-3p | 245.03 |
| miR-1950 | −188.92 | miR-3535 | 221.23 | miR-98-3p | −217.26 | miR-449a-5p | −236.16 |
| miR-495-3p | 180.12 | let-7f-2-3p | −217.26 | miR-1960 | −207.82 | miR-543-3p | 235.57 |
| miR-33-3p | −179.48 | miR-98-3p | −217.26 | miR-495-3p | 203.84 | miR-3061-5p | −226.71 |
| miR-7065-3p | −179.48 | miR-485-5p | 217.11 | miR-7669-5p | −198.38 | miR-98-3p | −217.26 |
| miR-5126 | −179.48 | miR-136-5p | −207.82 | miR-147-3p | −188.92 | let-7f-2-3p | −217.26 |
| miR-1947-5p | −179.48 | miR-1960 | −207.82 | let-7g-3p | −188.92 | miR-5114 | 200.27 |
| miR-6538 | −170.02 | miR-511-5p | 205.12 | miR-1950 | −188.92 | miR-7669-5p | −198.38 |
| miR-30b-3p | −160.59 | miR-7669-5p | −198.38 | miR-33-3p | −179.48 | miR-1950 | −188.92 |
| miR-7036b-5p | −160.59 | miR-7043-3p | 193.41 | miR-1947-5p | −179.48 | miR-130b-5p | 182.14 |
Abbreviation: miRNA, microRNA.
a Positive value and negative of fold-change indicated upregulation and downregulation, respectively. P value calculated from t test and statistical significance was defined as P < .05.
Table 2.
Dosage-Specific MicroRNAs to 2.0 and 6.5 Gy and Top 10 Dosage-Specific MicroRNAs to 8.0 Gy.
| 2.0 Gy (24, 48, 72 hours common) | 6.5 Gy (24, 48, 72 hours common) | 8.0 Gy (24, 48, 72 hours common) | |||
|---|---|---|---|---|---|
| miRNA_ID | Fold change | miRNA_ID | Fold change | miRNA_ID | Fold change |
| miR-152-5p | −188.92 | miR-411-5p | −264.50 | miR-129-2-3p | 525.05 |
| miR-3071-5p | −141.69 | miR-9-5p | −4.94 | miR-96-5p | 350.07 |
| miR-344-3p | −113.35 | miR-129b-5p | 116.69 | ||
| miR-122-5p | 3.00 | miR-10b-3p | 116.47 | ||
| miR-101c | −6.81 | miR-211-5p | 113.45 | ||
| miR-2137 | 2.32 | miR-3099-3p | 84.28 | ||
| miR-139-3p | 77.63 | miR-666-5p | 74.76 | ||
| miR-8116 | 65.23 | miR-34c-3p | 68.92 | ||
| miR-341-3p | 2.05 | miR-151-5p | 68.28 | ||
| miR-30c-2-3p | 65.82 | ||||
Abbreviation: miRNA, microRNA.
Table 3.
56 MicroRNAs Are Shared Irradiation Exposure–Responsive miRNA.
| miRNA_ID | miRNA_ID | miRNA_ID | miRNA_ID | ||||
|---|---|---|---|---|---|---|---|
| 1 | miR-98-3p | 15 | miR-297b-3p | 29 | miR-149-5p | 43 | miR-27a-5p |
| 2 | miR-363-3p | 16 | miR-29c-5p | 30 | miR-5114 | 44 | miR-543-3p |
| 3 | miR-138-5p | 17 | miR-297c-3p | 31 | miR-193a-5p | 45 | miR-338-5p |
| 4 | miR-200b-5p | 18 | miR-297a-3p | 32 | miR-30a-5p | 46 | miR-21a-5p |
| 5 | miR-151-3p | 19 | miR-125b-1-3p | 33 | miR-1839-5p | 47 | miR-29a-3p |
| 6 | miR-700-5p | 20 | miR-433-3p | 34 | miR-195a-3p | 48 | miR-1843b-5p |
| 7 | miR-450b-5p | 21 | miR-455-3p | 35 | miR-541-5p | 49 | miR-1843a-5p |
| 8 | miR-199b-5p | 22 | miR-770-3p | 36 | miR-144-5p | 50 | miR-6979-3p |
| 9 | miR-362-3p | 23 | miR-30b-5p | 37 | miR-615-3p | 51 | miR-669n |
| 10 | miR-182-5p | 24 | miR-127-3p | 38 | miR-322-3p | 52 | miR-125a-3p |
| 11 | miR-330-3p | 25 | miR-191-3p | 39 | miR-1843b-3p | 53 | miR-126b-5p |
| 12 | miR-1191a | 26 | miR-1a-1-5p | 40 | miR-128-3p | 54 | miR-760-3p |
| 13 | miR-485-5p | 27 | miR-1943-5p | 41 | miR-103-3p | 55 | miR-7010-5p |
| 14 | miR-7043-3p | 28 | miR-130b-5p | 42 | miR-196b-5p | 56 | miR-155-5p |
Abbreviation: miRNA, microRNA.
Kyoto Encyclopedia of Genes and Genomes Analysis of Predicted Downstream Target Genes Indicates the Potential Biological Pathways Regulated by Exosomal MiRNA Markers
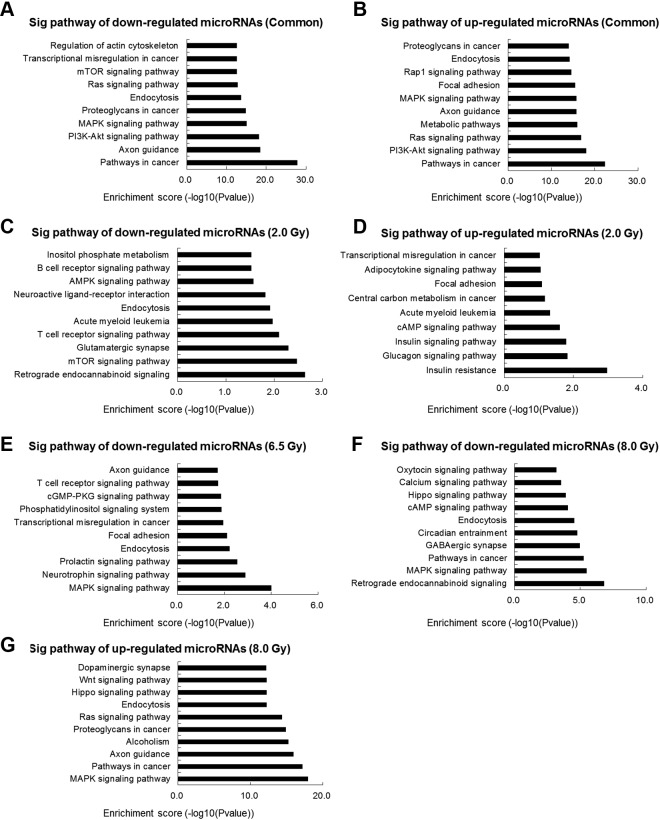
To investigate the involved biological pathways of downstream target genes of the differentially expressed miRNAs, we performed KEGG pathway analysis. Interestingly, the top 10 KEGG pathways predicted from different groups are quite different. When the pathways were predicted by common IR exposure-responsive miRNAs, the target genes of downregulated common IR-responsive exosomal miRNAs are mainly enriched in “pathways in cancer,” “axon guidance,” “PI3K-Akt signaling pathway,” “MAPK signaling pathway,” and “proteoglycans in cancer” (Figure 3A), while signal pathways target genes of upregulated common IR-responsive exosomal miRNAs are involved mainly in “pathways in cancer,” “PI3K-Akt signaling pathway,” “Ras signaling pathway,” “metabolic pathways,” and “axon guidance” (Figure 3B). The pathways predicted by target genes in 2.0-Gy downregulated group are significantly enriched in critical pathways, including “retrograde endocannabinoid signaling,” “mTOR signaling pathway,” “glutamatergic synapse,” “T-cell receptor signaling pathway,” and “acute myeloid leukemia” (Figure 3C). Compared with 2.0-Gy downregulated group, the target genes of upregulated miRNAs are mainly enriched in “transcriptional misregulation in cancer,” “glucagon signaling pathway,” “insulin signaling pathway,” “cAMP signaling pathway,” and “insulin resistance” (Figure 3D). In the 6.5 Gy group, the target genes of downregulated miRNAs are involved in “MAPK signaling pathway,” “neurotrophin signaling pathway,” “prolactin signaling pathway,” and “endocytosis and focal adhesion” (Figure 3E). In 8.0 Gy group, the target genes of downregulated miRNAs are involved mainly in “retrograde endocannabinoid signaling,” “MAPK signaling pathway,” “pathways in cancer,” “GABAergic synapse,” and “circadian entrainment” (Figure 3F), while target genes of upregulated miRNAs are involved in mainly include “dopaminergic synapse,” “Wnt signaling pathway,” “Hippo signaling pathway,” “endocytosis,” and “Ras signaling pathway” (Figure 3G). Based on the above analysis, the potential signaling pathways influenced by the target genes of exosomal miRNA markers are quite different, which may indicate the differential pathophysiological impacts by different IR doses.
Figure 3.
Kyoto Encyclopedia of Genes and Genomes pathways analysis of differentially expressed microRNAs. The most significant pathways for common downregulated genes (A) and commonly upregulated genes (B) in serum exosomes after irradiation exposure. The most significant pathways for downregulated genes (C) and upregulated genes (D) in serum exosomes after 2.0 Gy irradiation exposure. The most significant pathways for downregulated genes (E) in serum exosomes after 6.5 Gy irradiation exposure. The most significant pathways for downregulated genes (F) and upregulated genes (G) in serum exosomes after 8.0 Gy irradiation exposure.
Identification of MiR-128-3p and MiR-151-3p as Biomarkers for IR Exposure
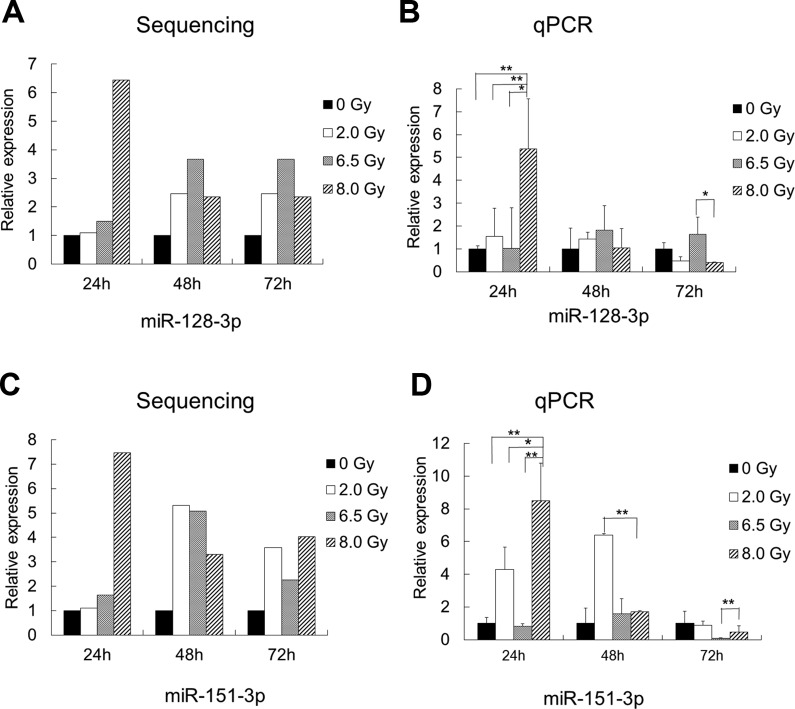
The development of more effective diagnostic methods to determine IR doses is significant and can help to control the hazards of radiation exposure. All of the differentially expressed miRNAs were analyzed to screen miRNAs that can be used to distinguish the lethal dose 8.0 Gy within 24 hours. First, we arranged 2 sets of comparisons: one set is 0 Gy compared with 2.0, 6.5, and 8.0 Gy, while the other set is 8.0 Gy compared with 0, 2.0, and 6.5 Gy. The 2 comparisons generated a list of 46 differentially expressed miRNAs. Next, by excluding miRNAs presenting in 0 Gy and restrict the time point to 24 hours, 6 miRNAs are significantly varied, including miR-129-5p, miR-151-3p, miR-195a-3p, miR-128-3p, miR-99b-3p, and miR-501-3p. Further, considering miRNA significantly upregulated would be more suitable to act as diagnostic biomarkers, we selected the upregulated exosomal miRNAs, miR-128-3p and miR-151-3p, for further validation (Figure 4A and C). The qualitative real-time PCR of 8.0-Gy group clearly showed that these 2 miRNA markers reached the significance criteria at 24 hours (P <.05; Figure 4B and D). We also performed the same validation in BALB/c mice and obtained similar results (Supplementary Figure S2).
Figure 4.
Differential expression results of sampling microRNAs from microarray analysis (A, C) and expression profiles of exosomal miR-128-3p and miR-151-3p from C57BJ/L mice exposed to ionizing radiation (IR) were validated by qualitative polymerase chain reaction (B, D). * P < .05. Assessment of miR-128-3p and miR-151-3p as biomarkers for IR exposure.
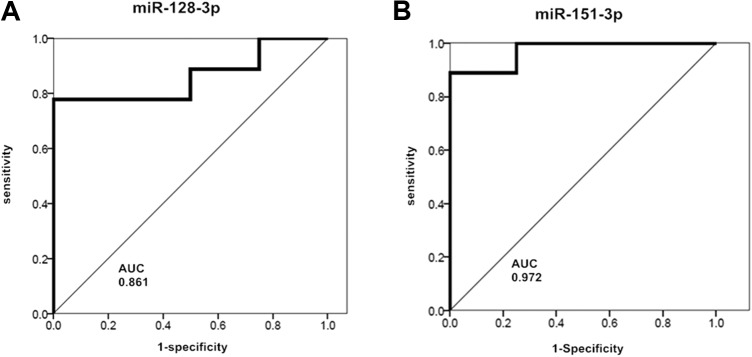
To test the sensitivity and specificity of our results, we performed the ROC curve analysis to evaluate the predictive power of miR-128-3p and miR-151-3p. The IR exposure diagnostic efficacy of miR-128-3p and miR-151-3p was evaluated by comparing none-IR exposure and IR exposure serum exosomal miRNA expression profiles. The area under the curve (AUC) of miR-128-3p was 0.861 (P = .045), and AUC of miR-151-3p was 0.972 (P = .009; Figure 5A and B), which shows good specificity and sensitivity to IR exposure. Youden index is a kind of method to evaluate the authenticity of the screening test method (Youden index = sensitivity + specificity − 1). The larger the Youden index, the better the results of the screening experiment, and the higher its authenticity. According to calculations, Youden index of miR-128-3p as an IR marker is 0.778 and that of miR-151-3p as an IR marker is 0.750. Therefore, based on the above evaluation, miR-128-3p and miR-151-3p are valuable molecular markers for identifying 8.0 Gy IR exposure.
Figure 5.
Receiver operating characteristic curve for miR-128-3p (A) and miR-151-3p (B). * P < .05. AUC indicates area under the curve.
Discussion
In this study, we selected 2.0 Gy as a low dose, 6.5 Gy as a sublethal dose, and 8.0 Gy as a lethal dose. Thirty days after IR treatments, there is no death in 0, 2.0, and 6.5 Gy groups, while the survival rate of 8.0 Gy group is only 40%. Our results are consistent with previous reports15 and set a solid foundation for identifying dose-specific markers.
There were a lot of methods that can be used to evaluate the radiometric sensitivity of normal and tumor cells with specific dosages. Chromosome aberration analysis is a widely used method, but the workload is heavy and the experiment cycle is time-consuming. Researchers have to collect peripheral blood samples, incubate lymphocytes in vitro, determine the first mitotic region of the double chromosome and the ring frequency (dic + r), and then calculate the IR exposure dose.16,17 Early maturation chromosome agglutination analysis is also a commonly used radiation biological dose index. Electron spin resonance analysis can predict wide range of doses, while the biological samples are the teeth or bones and these materials are difficult to obtain.18 Therefore, novel diagnostic methods and biomarkers with stable, fast, and accurate features are urgently necessary for identifying IR exposure doses.
As encapsulated by lipid bilayers of exosomes, exosomal miRNAs in serum have been considered to be stable biomarkers or indicators for many diseases. Many reports have suggested exosomal miRNAs from the body fluids potentially act as diagnostic markers. MiR-92a in serum exosomes can be used to reflect the body’s Brown adipose tissue (BAT) activity. There was a negative correlation between the expression level of miR-92a and the activity of BAT.19 The expression levels of 4 muscle-specific miRNAs (including miR-1, miR-133a, miR-133b, and miR-206) in serum exosomes are related to the degree of muscle wasting in patients with myotonic dystrophy type 1 (DM1), which can be used as biomarkers for monitoring the progress of muscle wasting in patients with DM1.20 The expression level of miR-17-92a cluster in human exosomes is related to the progression of human colorectal cancer (CRC), and patients with low expression of miR-19a in exosomes had significantly better prognoses.21 Thus, the above studies indicated the selection of serum exosomes miRNAs as markers for IR dose identification has been widely validated.
The expression of mir-151-3p and mir-128-3p are also changes in other diseases. There are many reports about the role of these 2 miRNAs in tumorigenesis and development. For example, it has been shown that miR-151-3p directly regulates TWIST1 expression to inhibiting migration and invasion of human breast cancer cells by enhancing E-cadherin expression.22 Studies have shown that miR-450b-3p and miR-151-3p in exosomes secreted by pancreatic cancer cells are expected to become new targets for the treatment of pancreatic cancer. Because they can significantly downregulate the expression of IRS1 and P110α in skeletal muscle, which leads to increased lipid deposition and decreased glucose uptake in skeletal muscle cells, eventually promoting insulin resistance.23 Serum hsa-miR-122-5p and hsa-miR-151-3p may function as new biomarkers in significant liver injury with persistently normal alanine transaminase levels patients.24 Overexpression of miR-151-3p upregulates myoblast proliferation and reduces slow muscle gene expression in C2C12 myocardium and primary cultures. And miR-151-3p directly targets ATP2a2 encoding slow bone and cardiac-specific Ca2+ ATPase genes and SERCA2 to downregulate slow muscle gene expression.25 Hydroxyurea in patients with hematological disorders is treated with induced hemoglobin (HbF) to treat sickle cell anemia, whereas the increase in miR-151-3p expression is associated with hydroxyurea-mediated HbF induction.26
Studies have shown that miR-128-3p expression level is an independent prognostic indicator and plays an important role in cell proliferation, metabolism, and metastasis (lung, breast, glioblastoma (GBM), gastric cancer, and CRC). It was found that miR-128-3p induces mesenchymal and stemness-like properties through downregulating multiple inhibitors of Wnt/β-catenin and tumor growth factor (TGF)-β pathways and leading to their overactivation. In patients with non-small cell lung cancer (NSCLC), miR-128-3p levels were related to activated β-catenin and TGF-β signaling and pro-epithelial-to-mesenchymal transition/pro-metastatic protein levels. Therefore, miR-128-3p might be a potential target against metastasis and chemoresistance in NSCLC.27 Previous studies have shown that IR exposure potentially induces brain tumors. It was reported that miR-128-3p was downregulated in glioma tissues in comparison with paracarcinoma tissues. Overexpression of miR-128-3p inhibited glioma cell proliferation and affected cell cycle and apoptosis by regulating GREM1.28 MiR-128-3p was significantly higher in cerebrospinal fluid of stroke patients compared to controls.29 Statistical analysis showed that the expression of miR-128a was negatively correlated with the expression of mesenchymal markers in GBM, it can increase the expression of mesenchymal markers in GBM cell lines and mesenchymal signaling in GBM may be negatively regulated by miR-128a.30 MiR-128-3p can promote the sensitivity of radiotherapy. BMI1 (multiple comb ring oncogene) is an oncogene associated with radiation resistance in tumor cells. When miR-128-3p is overexpressed in laryngeal cancer cells, it can target BMI1 to reduce its expression. MiR-128-3p causes chemotherapeutic sensitivity.31 When U-87MG glioma cells were exposed to IR, the expression level of miR-128-3p was upregulated in the 1 and 2.0 Gy dose groups and downregulated in the 8.0 Gy dose group, while the mRNA expression level of BMI1 was downregulated in the 2.0 Gy group and upregulated in the 6 and 8.0 Gy groups. Therefore, miR-128-3p and its downstream target gene BMI1 may play an important role in the radioresistance of U87MG glioma cells.32 Prions can cause infectious neurological diseases, and some miRNAs are dysregulated in prion-infected brain tissue. Studies have found that expression of miR-128-3p is upregulated in exosomes released by prion-infected nerve cells. The results showed that circulating exosomes released during prion infection have unique miRNA characteristics and can be used to diagnose and understand the pathogenesis of prion disease.33 PHF6 is a tumor suppressor gene for T-cell acute lymphoblastic leukemia. It has been reported that miR-128-3p can target PHF6 in T-cell acute lymphoblastic leukemia cell lines. The results of in vivo experiments showed that overexpression of miR-128-3p in a Notch1-induced T-cell acute lymphoblastic leukemia mouse model can promote the development of leukemia, thus obtaining evidence for the carcinogenic effect of the miRNA in T-cell acute lymphoblastic leukemia. Therefore, miR-128-3p is a cancer-causing miRNA that targets PHF6 tumor suppressor gene in T-cell acute lymphoblastic leukemia.34
We think that these miRNAs shall involve in acute injury and immediate recovery through their downstream target genes, such as Sirt1 and Dclre1c, as miR-128-3p targets and Atg5 as miR-151-3p target. Sirt1 is a conserved NAD+-dependent deacetylase that is involved in regulating cell survival and stress response, and its downregulation increases radiation sensitivity. It is involved in the pathway Sirt1/NF-κB/Smac associated with radiation damage.35 Dclre1c (DNA cross-link repair 1C) is also a target gene involved in the repair of DNA damage caused by radiation. ATG5 is an autophagy-related gene. It increases radiation sensitivity and is associated with radiation-induced cell death.36 Other miRNAs may also be involved in acute injury and immediate recovery through their downstream targets. In the future, these predicted injury- and/or recovery-related roles will be validated by functional study.
In this work, using high-throughput small RNA sequencing, we have identified plenty of dose-specific and common IR exposure-responsive exosomal miRNAs. These exosomal miRNAs are involved in a great number of biological and pathophysiological processes through several critical pathways, including PI3K-Akt signaling pathway, MAPK signaling pathway, mTOR signaling pathway, and so on. With rigorous validations, miR-128-3p and miR-151-3p were determined to be 8.0-Gy IR exposure markers. The findings presented here are only the preliminary results, and more exosomal miRNA markers will be discovered with further validations. In the future, studies can be focused on targeting tissues or organs of serum exosomes by tracing techniques to better explore the mechanism of the biological effects of radiation.
Conclusion
In this study, we treated mice with 0, 2.0, 6.5, and 8.0 Gy of IR for exploring dose-specific and common IR exposure markers. Through high-throughput sequencing of serum exosome samples and bioinformatic analysis, we identified miR-128-3p and miR-151-3p to be 8.0-Gy-specific IR exposure markers from serum exosomes at 24 hours’ time point, which were validated in 2 mouse strains by qualitative PCR. Our findings provide a novel and quick method for determining the 8.0-Gy exposure and possess great potential to be used in clinical applications.
Supplemental Material
Supplementary_Figure_S1 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response
Supplementary_Figure_S2 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response
Supplementary_Table_S1 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response
Supplementary_Table_S2 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response
Footnotes
Authors’ Note: Ying Zhang and Jiabin Liu contributed equally to this work. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The research was supported by the National Natural Science Foundation of China (grant no. 81573076) and grants from the School of Public Health of Southern Medical University, China (grant no. GW201716).
ORCID iD: Meijuan Zhou  https://orcid.org/0000-0003-4962-3164
https://orcid.org/0000-0003-4962-3164
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Qin J, Xu Q. Functions and application of exosomes. Acta Pol Pharm. 2014;71(4):537–543. [PubMed] [Google Scholar]
- 2. Yu DD, Wu Y, Shen HY, et al. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015;106(8):959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang W, Ni M, Su Y, et al. MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur Urol Focus. 2016:8. [DOI] [PubMed] [Google Scholar]
- 4. Ji Q, Ji Y, Peng J, et al. Increased brain-specific miR-9 and miR-124 in the serum exosomes of acute ischemic stroke patients. PLoS One. 2016;11(9):e163645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alegre E, Sanmamed MF, Rodriguez C, Carranza O, Martin-Algarra S, Gonzalez A. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch Pathol Lab Med. 2014;138(6):828–832. [DOI] [PubMed] [Google Scholar]
- 6. Arscott WT, Tandle AT, Zhao S, et al. Ionizing radiation and glioblastoma exosomes: implications in tumor biology and cell migration. Transl Oncol. 2013;6(6):638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Mayah AH, Bright SJ, Bowler DA, Slijepcevic P, Goodwin E, Kadhim MA. Exosome-mediated telomere instability in human breast epithelial cancer cells after X irradiation. Radiat Res. 2017;187(1):98–106. [DOI] [PubMed] [Google Scholar]
- 8. Al-Mayah AH, Irons SL, Pink RC, Carter DR, Kadhim MA. Possible role of exosomes containing RNA in mediating nontargeted effect of ionizing radiation. Radiat Res. 2012;177(5):539–545. [DOI] [PubMed] [Google Scholar]
- 9. Song M, Wang Y, Shang ZF, et al. Bystander autophagy mediated by radiation-induced exosomal miR-7-5p in non-targeted human bronchial epithelial cells. Sci Rep. 2016;6:30165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K. Serum amyloid A as a biomarker for radiation exposure. Radiat Res. 2015;184(1):14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhayana S, Song F, Jacob J, et al. Urinary miRNAs as biomarkers for noninvasive evaluation of radiation-induced renal tubular injury. Radiat Res. 2017;188(6):626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li MJ, Cui FM, Cheng Y, Sun D, Zhou PK, Min R. Changes in the adhesion and migration ability of peripheral blood cells: potential biomarkers indicating exposure dose. Health Phys. 2014;107(3):242–247. [DOI] [PubMed] [Google Scholar]
- 13. Fendler W, Malachowska B, Meghani K, et al. Evolutionarily conserved serum microRNAs predict radiation-induced fatality in nonhuman primates. Sci Transl Med. 2017;9(379):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer. 2014;13:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Acharya SS, Fendler W, Watson J, et al. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med. 2015;7(287):269r–287r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Q, Cao J, Wang ZQ, et al. Dose estimation by chromosome aberration analysis and micronucleus assays in victims accidentally exposed to (60)Co radiation. Br J Radiol. 2009;82(984):1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamadrid BA, Romero AI, Terzoudi GI, Gonzalez MJ, Pantelias G, Garcia O. Rapid assessment of high-dose radiation exposures through scoring of cell-fusion-induced premature chromosome condensation and ring chromosomes. Mutat Res. 2013;757(1):45–51. [DOI] [PubMed] [Google Scholar]
- 18. Chiaravalle AE, Mangiacotti M, Marchesani G, Vegliante G. Electron spin resonance (ESR) detection of irradiated fish containing bone (gilthead sea bream, cod, and swordfish). Vet Res Commun. 2010;34(suppl 1):S149–S152. [DOI] [PubMed] [Google Scholar]
- 19. Chen Y, Buyel JJ, Hanssen MJ, et al. Exosomal microRNA miR-92a concentration in serum reflects human brown fat activity. Nat Commun. 2016;7:11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koutsoulidou A, Photiades M, Kyriakides TC, et al. Identification of exosomal muscle-specific miRNAs in serum of myotonic dystrophy patients relating to muscle disease progress. Hum Mol Genet. 2017;26(17):3285–3302. [DOI] [PubMed] [Google Scholar]
- 21. Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeh TC, Huang TT, Yeh TS, et al. MiR-151-3p targets TWIST1 to repress migration of human breast cancer cells. PLoS One. 2016;11(12):e168171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L, Zhang B, Zheng W, et al. Exosomes derived from pancreatic cancer cells induce insulin resistance in C2C12 myotube cells through the PI3K/Akt/FoxO1 pathway. Sci Rep. 2017;7(1):5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng JL, Zhao H, Yang SG, Chen EM, Chen WQ, Li LJ. Plasma miRNA-122-5p and miRNA-151a-3p identified as potential biomarkers for liver injury among CHB patients with PNALT. Hepatol Int. 2018;12(3):277–287. [DOI] [PubMed] [Google Scholar]
- 25. Wei H, Li Z, Wang X, et al. MicroRNA-151-3p regulates slow muscle gene expression by targeting ATP2a2 in skeletal muscle cells. J Cell Physiol. 2015;230(5):1003–1012. [DOI] [PubMed] [Google Scholar]
- 26. Walker AL, Steward S, Howard TA, et al. Epigenetic and molecular profiles of erythroid cells after hydroxyurea treatment in sickle cell anemia. Blood. 2011;118(20):5664–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai J, Fang L, Huang Y, et al. Simultaneous overactivation of WNT/beta-catenin and TGF beta signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. 2017;8:15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of miR-128-3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–1157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Sorensen SS, Nygaard AB, Carlsen AL, Heegaard N, Bak M, Christensen T. Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Biomark Res. 2017;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma X, Yoshimoto K, Guan Y, et al. Associations between microRNA expression and mesenchymal marker gene expression in glioblastoma. Neuro Oncol. 2012;14(9):1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen H, Jin L, Zhou L, Huang JM. Overexpressed miR-128a enhances chemoradiotherapy to laryngeal cancer cells and its correlation with BMI1. Future Oncol. 2018;14(7):611–620. [DOI] [PubMed] [Google Scholar]
- 32. Ye L, Yu G, Wang C, et al. MicroRNA128a, BMI1 polycomb ring finger oncogene, and reactive oxygen species inhibit the growth of U87 MG glioblastoma cells following exposure to X-ray radiation. Mol Med Rep. 2015;12(4):6247–6254. [DOI] [PubMed] [Google Scholar]
- 33. Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mets E, Van Peer G, Van der Meulen J, et al. MicroRNA-128-3p is a novel oncomiR targeting PHF6 in T-cell acute lymphoblastic leukemia. Haematologica. 2014;99(8):1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ji K, Sun X, Liu Y, et al. Regulation of apoptosis and radiation sensitization in lung cancer cells via the Sirt1/NF-kappaB/Smac pathway. Cell Physiol Biochem. 2018;48(1):304–316. [DOI] [PubMed] [Google Scholar]
- 36. Qiang L, Sample A, Shea CR, Soltani K, Macleod KF, He YY. Autophagy gene ATG7 regulates ultraviolet radiation-induced inflammation and skin tumorigenesis. Autophagy. 2017;13(12):2086–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Figure_S1 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response
Supplementary_Figure_S2 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response
Supplementary_Table_S1 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response
Supplementary_Table_S2 for Exosomal Small RNA Sequencing Uncovers Dose-Specific MiRNA Markers for Ionizing Radiation Exposure by Ying Zhang, Jiabin Liu, Liang Zhou, Shuai Hao, Zhenhua Ding, Lin Xiao and Meijuan Zhou in Dose-Response