Abstract
Shine-Dalgarno sequences (SD) in prokaryotic mRNA facilitate protein translation by pairing with rRNA in ribosomes. Although conventionally defined as AG-rich motifs, recent genomic surveys reveal great sequence diversity, questioning how SD functions. Here, we determined the molecular fitness (i.e., translation efficiency) of 49 synthetic 9-nt SD genotypes in three distinct mRNA contexts in Escherichia coli. We uncovered generic principles governing the SD fitness landscapes: (1) Guanine contents, rather than canonical SD motifs, best predict the fitness of both synthetic and endogenous SD; (2) the genotype-fitness correlation of SD promotes its evolvability by steadily supplying beneficial mutations across fitness landscapes; and (3) the frequency and magnitude of deleterious mutations increase with background fitness, and adjacent nucleotides in SD show stronger epistasis. Epistasis results from disruption of the continuous base pairing between SD and rRNA. This “chain-breaking” epistasis creates sinkholes in SD fitness landscapes and may profoundly impact the evolution and function of prokaryotic translation initiation and other RNA-mediated processes. Collectively, our work yields functional insights into the SD sequence variation in prokaryotic genomes, identifies a simple design principle to guide bioengineering and bioinformatic analysis of SD, and illuminates the fundamentals of fitness landscapes and molecular evolution.
Translation initiation is often the rate-limiting step in protein synthesis and fundamental to gene regulation (Laursen et al. 2005). In bacteria and archaea, it begins with attachment of 30S ribosome subunits to ribosome binding sites (RBS) immediately upstream of the protein-coding sequence in mRNA (Fig. 1A). Subsequent recruitment of initiator tRNA and 50S subunits leads to formation of the complete 70S translation machinery. The Shine-Dalgarno sequence (SD), typically an AG-rich region in RBS, has been thought to play a key role in this process (Shine and Dalgarno 1975). SD facilitates 30S subunit-mRNA binding by base-pairing interaction with the conserved CU-rich anti-SD sequence (aSD) at the 3′ tail of the 16S rRNA in 30S subunits. Stable SD:aSD base pairing promotes the translation efficiency, which in turn determines the protein yield. The mechanism of translation initiation is thought to be well-understood, but recent surveys of thousands of bacterial genomes raise questions on this presumption (Nakagawa et al. 2010; Omotajo et al. 2015; Hockenberry et al. 2018). Although these studies detect AG-rich motifs in the majority of RBS, they also identify a significant proportion of AG-less RBS. This finding implies that either the AG-rich rule is not absolute, or there exists unidentified mechanisms besides SD:aSD pairing and the 70S ribosome-mediated translation specifically for leaderless mRNA (Laursen et al. 2005).
Figure 1.
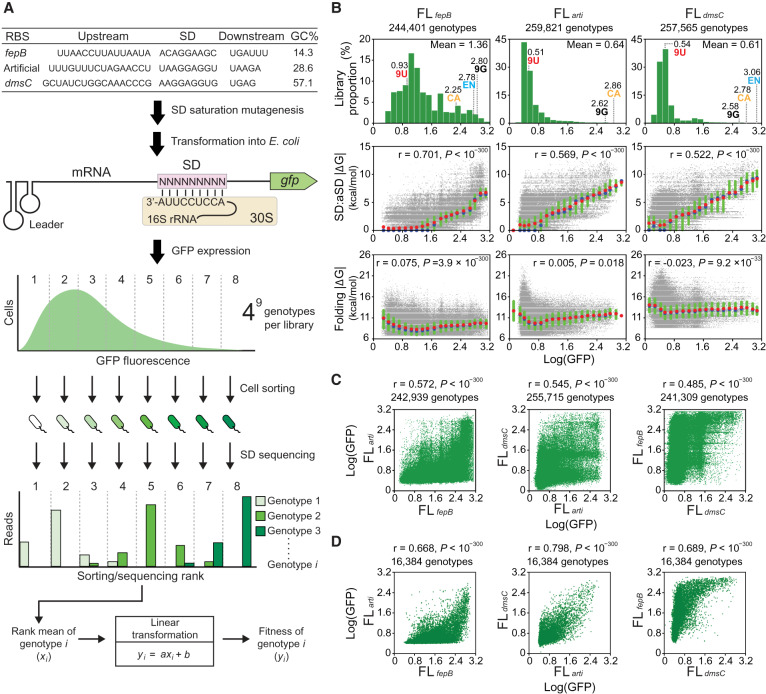
G-P associations of the SD fitness landscapes. (A) Determining SD G-P associations. SD libraries under three RBS contexts are generated and assayed in E. coli. Cell sorting divides a library into multiple ranks based on GFP expression. The genotypic composition of each rank is revealed by Illumina sequencing. The fitness (Log[GFP]) of a genotype is determined by its read count distribution transformed through a linear equation describing the correlation between the rank means and GFP fluorescence. (B) Distribution of the fitness of genotypes (upper) and its correlations (Pearson's r, t-test) with SD:aSD base-pairing energy (ΔG, middle) and mRNA folding energy (ΔG, lower). ΔG (predicted values ≤0) is shown as absolute values for simplicity. Genotypes are ranked by fitness and grouped into 20 equal-sized bins. Group means, medians, and interquartile ranges of ΔG are shown as red circles, blue circles, and green bars, respectively. The fitness of canonical SD (CA), endogenous SD (EN), and genotypes made of nine guanines (9G) and nine uracils (9U) is shown. (C) Phenotypic correlation (Pearson's r, t-test) of 9-nt SD genotypes between fitness landscapes. (D) Phenotypic correlation (Pearson's r, t-test) of 7-nt SD genotypes. The 9-nt SD genotypes of each fitness landscape are divided into 47 7-nt genotypic subsets according to the sequence located 7–13 nt upstream of the start codon. The mean fitness of 9-nt genotypes in each 7-nt subset is computed and compared between fitness landscapes.
A comprehensive understanding of the SD genotype-phenotype (G-P) associations is necessary to clarify the processes of translation initiation, illuminate the patterns of mRNA-rRNA interaction, uncover factors shaping the RBS diversity among prokaryotes, and guide the design of RBS in synthetic circuits (Salis et al. 2009). Prior experimental and bioinformatic studies both contributed greatly to this subject. Experimental dissection of molecular mechanisms revealed significant functional dependence of SD on its surrounding RBS context: Besides SD:aSD pairing strength, translation initiation was also influenced by the SD:aSD pairing location and the accessibility of SD caused by local mRNA folding (de Smit and van Duin 1990; Chen et al. 1994; Osterman et al. 2013; Espah Borujeni et al. 2014). However, with limited experimental throughput, former works were unable to investigate the global architecture of SD G-P mapping, such as its phenotype distribution (Blanco et al. 2019), the distribution of mutational effects (Eyre-Walker and Keightley 2007), the causes and consequences of epistasis (i.e., functional dependence of mutations on genetic backgrounds) (Domingo et al. 2019), robustness and evolvability (Wagner 2008), and how RBS contexts impacted the aforementioned properties. Alternatively, bioinformatic analyses scanned endogenous RBS of diverse prokaryotic species for motifs capable of pairing tightly with aSD (Chang et al. 2006; Nakagawa et al. 2010; Scharff et al. 2011; Omotajo et al. 2015; Hockenberry et al. 2018). By assigning RBS into “SD-led” or “non-SD-led” categories (i.e., with or without AG-rich motifs, respectively), these studies identified significant associations between the AG-richness of RBS and features like local mRNA folding, synonymous codon usage, and gene function. Nevertheless, because the observed correlation suggests selective pressure acting on gene expression, inspecting endogenous SD likely samples a biased subset of genotypes preserved by adaptive evolution. Moreover, as most species included in these studies lack genome-wide measurement of protein synthesis rates, research relying on just sequence information cannot assess the quantitative contribution of the SD genotypic composition on protein translation.
Here, we performed high-throughput experiments to quantify the molecular fitness (i.e., efficiency to initiate protein translation) of 262,144 SD genotypes under distinct RBS contexts in vivo. Our study illuminated the global architecture of SD fitness landscapes and the distribution of mutational effects, yielded novel biochemical insights into SD G-P mapping, and uncovered a new form of epistasis stemming from the nature of RNA base pairing.
Results
Developing a sort-seq platform for SD G-P mapping
We assembled a green fluorescent protein (GFP) expression cassette on plasmids to report the fitness of SD (Supplemental Fig. S1). To enhance the resolution of quantification, the plasmid system was designed to maximize GFP expression while minimizing its impact on cellular growth (Supplemental Fig. S2A). Hereafter fitness referred strictly to the translation efficiency of SD but not the reproductive success of host cells. Transcription of the cassette produced mRNA bearing a translation insulator leader sequence (self-folding RNA fragment designed to avoid interaction with RBS), a 30-nt RBS region, and the gfp coding sequence (Fig. 1A; Davis et al. 2011). As RNA secondary structure formed by RBS also affected translation initiation, our experimental design brought in two endogenous (RBSfepB and RBSdmsC of E. coli fepB and dmsC genes, respectively) or one artificial RBS (termed RBSarti) to control for this context effect. The GC contents of RBSfepB, RBSarti, and RBSdmsC differed greatly (14.3%, 28.6%, and 57.1%, respectively) and gave low to high structure-forming potential. Under each RBS context, we used saturation mutagenesis to synthesize all possible 262,144 (=49) 9-nt genotypes within an 11-nt SD region (5–15 nt upstream of the start codon) where the sequence composition strongly influenced 30S subunit-mRNA binding (Gao et al. 2016). We restricted our mutagenesis scope to 9 nt for each library (5–13, 6–14, and 7–15 nt upstream of the start codon for RBSfepB, RBSarti, and RBSdmsC, respectively) to achieve high genotypic coverage but meanwhile jointly scan the entire 11-nt SD region. The resulting 9-nt genotype libraries, FLfepB, FLarti, and FLdmsC, represented three SD fitness landscapes under distinct RBS contexts. Additionally, we built a FLarti-Y library, identical to FLarti except using the yellow fluorescent protein reporter, to evaluate the credibility of GFP-based fitness quantification. As the properties of FLarti-Y turn out to be very similar to FLarti, the results of its characterization are not discussed but shown in Supplemental Figure S3. Once constructed, the SD libraries were separately transformed into E. coli and grown in liquid culture until its optical density (OD) reached 0.55–0.65 (Supplemental Fig. S2). Based on GFP expression, each SD library was divided into multiple ranks through fluorescence-activated cell sorting (FACS), and the genotypic composition of each rank was determined by Illumina sequencing. The fitness of a genotype was estimated based on its sequencing read distribution across ranks (Fig. 1A). To quantify the effects of mutations (additive) and epistasis (nonadditive), we defined the logarithm of GFP expression (Log[GFP]) as the fitness of SD because the molecular basis of its G-P mapping, from changes in the SD sequence, the SD:aSD duplex length, base-pairing energy (ΔG), to the logarithm of protein production, showed an overall additive relationship (for justification, see Supplemental Text and Supplemental Fig. S4; for experimental support, see Fig. 1B). Details of our experimental design and implementation are described in Methods and Supplemental Methods.
High-throughput experiments reliably determine SD G-P associations
The fitness of SD genotypes in the FLfepB, FLarti, and FLdmsC libraries ranged between 0.19 and 3.14, 0.11 and 3.10, and 0.03 and 3.12, respectively (Fig. 1B). Three replicates of sort-seq experiments were performed for each library. Through accurate FACS (76.80%–99.84% purity per rank) and deep sequencing (100.02–218.49 average reads per genotype), we detected 99.641%–99.995% of all possible 262,144 genotypes in each library (Supplemental Figs. S5, S6; Supplemental File S1). The measurement error, defined as the standard deviation of fitness measured in three replicates, generally declined with an increasing read count of a genotype (Supplemental Fig. S7A). To ensure the measurement accuracy and genotypic coverage, we considered only genotypes with 25 or more reads and combined the data of three replicate experiments for analysis (Supplemental Files S2–S4). In the replicate-combined data sets of FLfepB, FLarti, and FLdmsC, the measurement errors on average were 0.168, 0.108, and 0.198 (or 15.8%, 19.5%, and 36.6% in terms of the coefficient of variation) (Supplemental Fig. S7B), respectively. The Pearson's correlations of fitness measured by replicate experiments were 0.687–0.937 (P < 10−300) (Supplemental Fig. S7C,D). The high measurement error of FLdmsC was partly attributed to its highly right-skewed fitness distribution, which disproportionally inflated the coefficient of variation (Fig. 1B).
Our quantification method assumed a direct correspondence between cellular fluorescence and GFP expression, and GFP expression mainly determined by the influence of SD on protein translation rather than mRNA stability. To validate these assumptions, we performed dot blot experiments for six representative genotypes (Log[GFP] = 0.43–3.01) and showed a significant linear relationship between the absolute GFP production and cellular fluorescence (Pearson's r = 0.997, P < 0.05) (Supplemental Fig. S8). Then we performed qPCR experiments for 24 representative genotypes and found that the mRNA level only increased significantly above a fitness threshold (Log[GFP] ≥ 2.8) (Supplemental Fig. S9A). This elevated mRNA level likely results from increased mRNA stability attributable to ribosome shielding and has been noted in another high-throughput study (Kosuri et al. 2013). We chose not to normalize fitness measurements with respect to the mRNA level because this could greatly underestimate the fitness of genotypes (Log[GFP] ≥ 2.8) whose mRNA level increased by three- to fivefold. Additionally, ribosome shielding affected just a small fraction of genotypes based on the observed fitness threshold (FLfepB: 15,744 [6.44%], FLarti: 1424 [0.55%], FLdmsC: 1064 [0.41%]), which had a marginal effect on our conclusion (Supplemental Fig. S9B,C).
SD G-P associations depend strongly on the RBS contexts
To elucidate the relationship between fitness and other molecular features, we assigned genotypes of each fitness landscape into 20 fitness-ranked groups and reported group characteristics. Examining the fitness of genotypes in FLfepB, FLarti, and FLdmsC revealed distinct distributions (Fig. 1B, upper panels). Although most genotypes in FLarti and FLdmsC had fitness below 0.8, the majority of genotypes in FLfepB was above this value and spread across the fitness spectrum. In both FLfepB and FLdmsC, the endogenous SD of RBSfepB and RBSdmsC had fitness higher than most genotypes and the canonical SD (i.e., reverse complement of aSD), suggesting it as the product of natural selection. Besides the overall fitness distribution, the fitness of individual genotypes across the three fitness landscapes showed just moderate correlations (Pearson's r = 0.485–0.572) (Fig. 1C). One potential cause of lower correlations was the difference in the physical location of SD genotypes (relative to the start codon) in RBSfepB, RBSarti, and RBSdmsC (Fig. 1A). To remove this confounding factor from cross-library G-P comparison, we assigned the 9-nt SD genotypes to 47 genotypic subsets based on their sequence identity in the 7-nt overlapping region (i.e., 6–12 nt upstream of the gfp start codon). Then we compared the mean fitness of each 7-nt genotypic subset across FLfepB, FLarti, and FLdmsC. Although the correlations increased significantly after eliminating the location effect (Pearson's r = 0.668–0.798) (Fig. 1D), part of this increment was also attributed to averaging variables (i.e., comparing subsets rather than individual genotypes) (for extensive analysis, see Supplemental Fig. S10).
We used a RNA folding algorithm to predict the influence of SD genotypes on local mRNA folding and SD:aSD base-pairing interaction (Lorenz et al. 2011). The mRNA folding energy of genotypes in FLfepB, FLarti, and FLdmsC was −8.88, −10.66, and −12.74 kcal/mol, respectively, on average (Fig. 1B, lower panels). This difference correlated with the GC contents of the three RBS contexts and corroborated with the observed fitness distribution. In contrast, analysis within each fitness landscape showed fitness correlating strongly with SD:aSD base-pairing energy (Pearson's r = 0.522–0.701) (Fig. 1B, middle panels) but hardly with mRNA folding energy (Pearson's r = −0.023 to 0.075). These trends were in agreement with multiple linear regression, which suggested SD:aSD base-pairing strength having a higher impact on fitness than the SD:aSD base-pairing location and mRNA folding energy in our experimental system (Supplemental Text). Examining the characteristics of fitness-ranked groups showed less fitness dependence of FLfepB on SD:aSD base-pairing energy than FLarti and FLdmsC. In FLfepB more than 50% of genotypes with fitness as 0.64–1.60 showed detectable GFP expression (the fitness of a GFP-negative control was 0.45 ± 0.04) but was not predicted to form SD:aSD base pairing (i.e., ΔG = ; blue circles in Fig. 1B indicate group medians), suggesting SD:aSD base pairing may be nonessential to translation initiation of weakly structured RBS. Collectively, results manifested strong RBS context effects on SD G-P mapping and indicated that SD:aSD base pairing played a prominent role in each fitness landscape.
G-P correlations increase both mutational robustness and evolvability
A central question of evolutionary genetics has been how G-P mapping structure affects the distribution of mutational effects, in other words, the probability of genotypes to acquire beneficial, neutral, or deleterious mutations (Orr 2005). Prior studies of fitness landscapes addressed this by exploring mostly the G-P associations surrounding wild-type RNA and proteins or variants showing superior molecular function (Podgornaia and Laub 2015; Li et al. 2016; Sarkisyan et al. 2016; Domingo et al. 2018). Our comprehensive G-P mapping of SD in three RBS contexts provided an unprecedented opportunity to reveal general features throughout entire fitness landscapes. Taking each genotype as the reference, we inspected the relationship between its fitness and the individual or mean fitness of its 27 mutation neighbors in FLfepB, FLarti, and FLdmsC (Fig. 2A, “Authentic”). We found modest (Pearson's r = 0.549–0.663) and strong (Pearson's r = 0.883–0.929) correlations for the former and latter cases, respectively, indicating that overall similar genotypes showed similar phenotypes, but fitness variation between close genotypes was substantial. The observed G-P correlations were biologically significant because such trend was absent among 10,000 G-P–shuffled landscapes generated by randomly pairing the genotypic and fitness data of the three fitness landscape while keeping their overall fitness distribution identical (Fig. 2A, “G-P–shuffled”).
Figure 2.
G-P correlation and the distribution of mutational effects. (A) Correlations (Pearson's r, permutation test) between the fitness of a genotype and the individual fitness (blue) or the mean fitness (red) of its 27 single-mutation neighbors in the authentic fitness landscapes or in a representative of 10,000 G-P–shuffled fitness landscapes. (B) Relationship between the fitness of a genotype and the amount of beneficial, neutral, and deleterious mutations in this genetic background in the authentic or the representative G-P–shuffled fitness landscapes. The 27 point mutations a genotype would acquire are assigned into beneficial, neutral, and deleterious categories based on an operational cutoff defined by sort-seq measurement errors (FLfepB = 0.168, FLarti = 0.108, and FLdmsC = 0.198). Genotypes are ranked by fitness and grouped into 20 equal-sized bins. Lines and bars indicate group means and standard deviations, respectively. (A,B) Only genotypes with their 27 single-mutation neighbors fully characterized are considered.
To assess the influence of G-P correlations on the distribution of mutational effects, we classified the 27 point mutations each genotype would gain as beneficial, neutral, or deleterious (with respect to the translation efficiency) using one- or twofold sort-seq measurement errors as the operational cutoff (Fig. 2B; Supplemental Fig. S11A). Regardless of differences in fitness distribution and measurement errors, FLfepB, FLarti, and FLdmsC showed similar trends: From low-fitness to high-fitness genetic backgrounds, we observed a decline of beneficial mutations, an elevation of deleterious mutations, and neutral mutations peaking at the fitness range occupied by most genotypes. As expected, high-fitness genotypes like endogenous SD were more likely to gain deleterious mutations than beneficial mutations. This result confirmed the decreasing availability of beneficial mutations as a cause of diminishing returns in adaptive evolution (Orr 2005). It also reflected a restricted view of the distribution of mutational effects generated by prior wild-type centered mutagenesis studies (Eyre-Walker and Keightley 2007; Li et al. 2016; Sarkisyan et al. 2016). To investigate the influence of G-P mapping structure on this distribution, we applied the same analysis to 10,000 G-P–shuffled fitness landscapes (Fig. 2B; Supplemental Fig. S11A,B). Although the overall trends seemed similar between the authentic and G-P–shuffled fitness landscapes, two unique features emerged: The authentic fitness landscapes had lower and higher input of deleterious and neutral mutations, respectively, throughout the fitness spectrum of genetic backgrounds. Moreover, authentic fitness landscapes showed a steady and higher supply of beneficial mutations across broad fitness altitudes (around Log[GFP] = 1.0–3.0). Relative to FLfepB, the stable input of beneficial mutations in FLarti and FLdmsC at similar fitness altitudes seemed unexpected given their highly right-skewed fitness distribution (Fig. 1B). Together, the two properties would make SD both mutationally robust and evolvable, granting a better chance to evolve into high-fitness genotypes.
Guanine contents predict the fitness of synthetic and endogenous SD
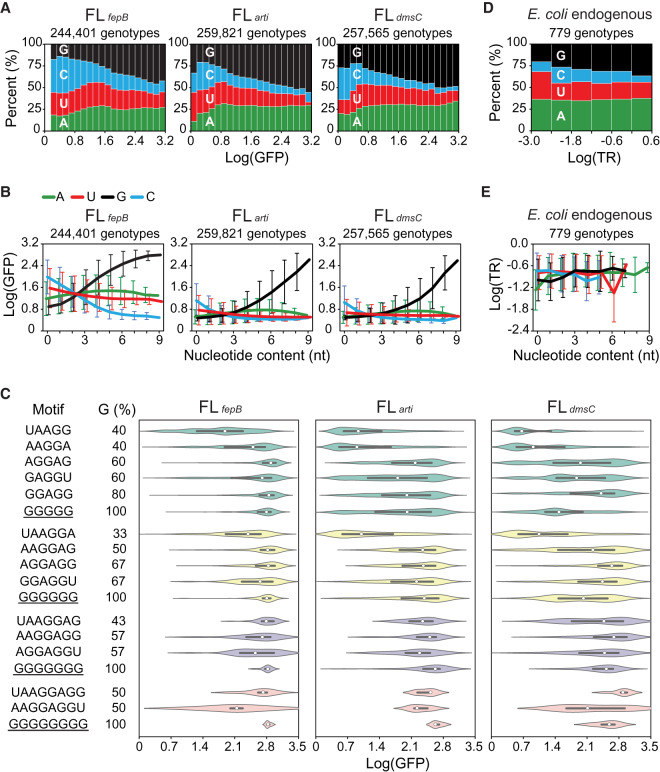
To elucidate the biochemical basis of SD fitness landscapes, we examined the nucleotide composition of genotypes across the 20 fitness-ranked groups in FLfepB, FLarti, and FLdmsC (Fig. 3A; Supplemental Figs. S12, S13). From low-fitness to high-fitness groups, we observed a huge and consistent increase of guanine, a significant and continuous decrease of cytosine, a moderate decrease of uracil, and a slight increase of adenine in all the three fitness landscapes. Although the preceding trends were consistent with the positive and negative roles of guanine and cytosine, respectively, in SD:aSD base pairing, the conventional view of adenine being critical for SD function was questionable because its ratio only changed slightly at low fitness (Log[GFP] = 0–0.8) and remained stable beyond this range. To gauge the functional significance of adenine, uracil, guanine, and cytosine, we categorized genotypes in terms of the amount of each nucleotide type and inspected the relationship between the nucleotide content and the mean fitness of genotypes in each category. In all the three fitness landscapes, we discovered a strong positive correlation between the guanine content and fitness (Fig. 3B; Supplemental Fig. S14). Even genotypes made of nine guanines conferred high fitness (Log[GFP] = 2.58–2.80) (Figs. 1B, 3B). On the contrary, the cytosine content showed a negative correlation with fitness, and the influences of adenine and uracil were both minor. We compared poly(G) tracts and canonical SD motifs with respect to their influence on the fitness of genotypes. Relative to the whole library, genotypes bearing both types of sequence motifs showed strong left-skewed fitness distribution (Fig. 3C). It was noteworthy that the skewness of distribution appeared to correlate with the guanine content of these motifs, consistent with the result of genotype-level analysis.
Figure 3.
Biochemical properties of the SD sequence. (A) Relationship between fitness (Log[GFP]) and nucleotide composition. Genotypes are ranked by fitness and grouped into 20 equal-sized bins. (B) Relationship between the nucleotide content and fitness. Genotypes are ranked by the amount of each nucleotide type and grouped into 10 bins (i.e., 0–9 nt). Lines and bars indicate group means and standard deviations of fitness, respectively. (C) Influence of sequence motifs on fitness. The fitness distribution of genotypes bearing 5- to 8-nt poly(G) tracts (underlined) or canonical SD motifs is shown as violin plots, where medians and interquartile ranges are indicated by white circles and black lines, respectively. (D) Relationship between the translation rate (TR; ribosomes/s) and nucleotide composition of endogenous SD sequences in E. coli (Gorochowski et al. 2019). Genotypes are ranked by TR and grouped into six equal-sized bins. (E) Relationship between the nucleotide content and TR of endogenous SD. Genotypes are ranked by the amount of each nucleotide type and grouped into 12 bins (i.e., 0–11 nt). Lines and bars indicate group means and standard deviations of TR, respectively. (D,E) Endogenous SD is defined as the 11-nt region 5–15 nt upstream of the start codon of a protein-coding sequence. TR is shown in the logarithmic scale to allow a direct comparison with fitness quantified in this study.
To see if observation of our synthetic SD libraries predicted the biochemical properties of endogenous SD in E. coli, we applied the same analysis to a published data set reporting the precisely measured translation rates of 779 endogenous SD (Gorochowski et al. 2019). We observed a similar relationship between the translation rate of endogenous SD and its nucleotide composition despite that each endogenous SD was situated in a unique RBS context (Fig. 3D). Endogenous SD had an overall elevated adenine content, which compressed the variation of other nucleotides. In contrast, the correlation between the nucleotide content and the translation rate of endogenous SD was less obvious (Fig. 3E). From low to high translation rates, endogenous SD still showed slight increases and decreases in its guanine and cytosine contents, respectively, but the variation was pronounced. The less significant trends may be attributed to the analytic method and data composition: Relative to synthetic libraries, poly(G) and poly(C) tracts were underrepresented in endogenous SD. Moreover, endogenous SD was enriched for adenine (36.02% and 34.51% for the 779 and total 4566 endogenous SD, respectively) and guanine (31.06% and 32.29% for the 779 and total 4566 endogenous SD, respectively), hence compressing the variation of cytosine and uracil (Hayashi et al. 2006). One distinguishable feature of endogenous SD was its high adenine content regardless of the translation rate (Fig. 3D), suggesting certain functional constraints on sequence evolution. Collectively, our analysis of both synthetic and endogenous SD showed the guanine content as an indicator of the translation efficiency. To our knowledge, this simple relation and efficient translation initiation by guanine-only SD have not been proposed or experimentally shown before.
Increasing the guanine content of SD enhances mRNA-ribosome binding
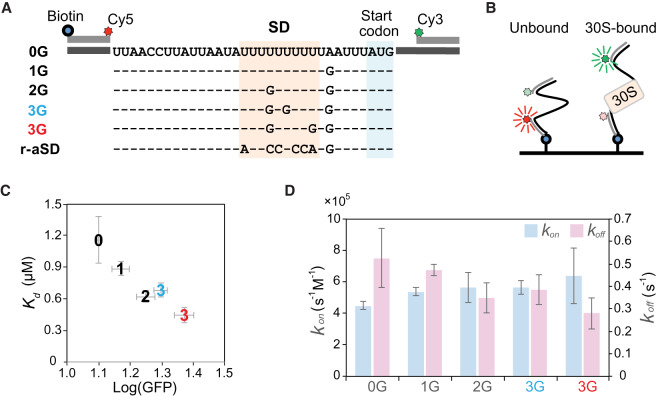
In addition to revealing a positive correlation between the guanine content and fitness, inspecting FLfepB, FLarti, and FLdmsC identified significant GFP expression by poly(U) SD in weakly structured RBSfepB (Figs. 1B, 3B; Supplemental Fig. S9A). Although the poor SD:aSD pairing capacity of poly(U) SD seemed odd to E. coli (none of its endogenous SD bore poly(U) tracts longer than five nucleotides) (Hayashi et al. 2006), it resembled the abundant non-SD-led RBS in bacteroides, cyanobacteria, and plastids (Nakagawa et al. 2010; Scharff et al. 2011; Omotajo et al. 2015). To investigate the translation mechanism of non-SD-led RBS and validate the additivity of guanine contents on mRNA-ribosome binding and translation initiation, we modified RBSfepB to create six synthetic RBS bearing 0–3 guanines in the SD region and quantified their in vivo fitness and in vitro binding kinetics with 30S subunits in the absence of initiation factors and initiator tRNA (Fig. 4A,B; Supplemental Fig. S15). Under the latter condition, the measured kinetics would reflect the intrinsic 30S-binding activity to mRNA. Synthetic RBS was designed to minimize the difference in RNA folding energy (predicted as −6.8 to −9.3 kcal/mol). The fitness and dissociation constants (Kd) of synthetic RBS were negatively correlated (Pearson's r = −0.962, P < 0.05) (Fig. 4C). Except the negative control, all of them, including synthetic RBS devoid of guanine, bound to the 30S subunit, and their guanine contents correlated positively with fitness (Pearson's r = 0.969, P < 0.05) and negatively with Kd (Pearsons’ r = −0.923, P < 0.05). Changes in Kd (= koff/kon) were caused by a larger difference in the RBS-30S subunit dissociation rate (koff) than the association rate (kon) (Fig. 4D). This result supported the main role of SD:aSD base pairing in stabilization of the mRNA-ribosome complex (Rodnina 2016). It also suggested that the non-SD-led RBS of bacteroides, cyanobacteria, and plastids might bind to ribosomes through the regular means as SD-led RBS, but their ribosomes may have evolved a SD:aSD pairing-independent way to stabilize the mRNA-ribosome complex. As such, there seems no need to evoke alternative mechanisms to explain the translation initiation of non-SD-led RBS.
Figure 4.
In vivo and in vitro characterization of synthetic RBS. (A,B) Determining the RBS-30S subunit binding kinetics by single-molecule fluorescence resonance energy transfer (FRET). RBSfepB is modified to generate six synthetic RBS bearing 0–3 guanines in the 11-nt SD region. Each RBS in A is tagged with biotin, Cy3, and Cy5 dyes and attached to a slide surface (B). The sequence of RBS devoid of guanine (0G) is shown as the consensus. Upon binding with the 30S subunit, the distance between Cy3 and Cy5 dyes is increased, causing reduced FRET signals. (C) Correlation (Pearson's r = −0.962, P < 0.05) between the in vivo fitness (Log[GFP]) and the in vitro dissociation constants (Kd). (D) The RBS-30S subunit association (kon) and dissociation rates (koff). (C,D) RBS is indicated according to the abbreviations in A, and bars show the standard deviations of three independent measurements. The SD region of the negative control (r-aSD) contains the reverse sequence of aSD. Since no apparent binding event is detected, the Kd, kon, and koff of r-aSD cannot be determined and are not shown here.
RNA thermodynamics explains the biochemical principles of G-P mapping
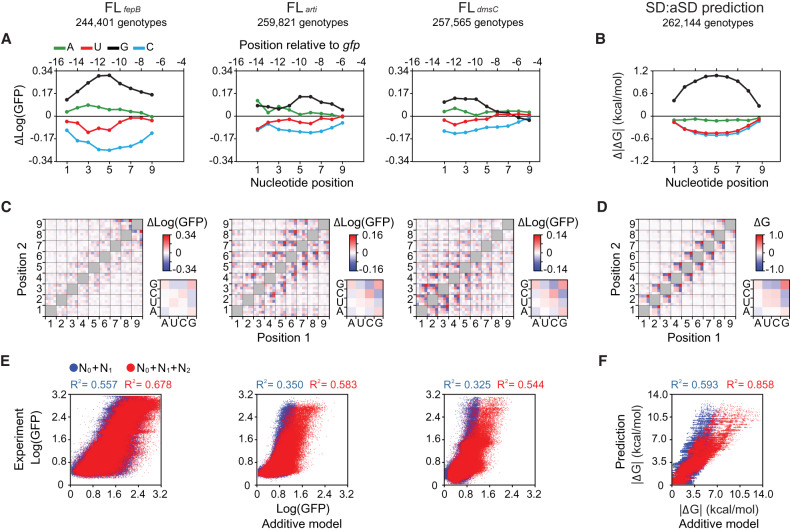
Following investigation of the RBS-ribosome binding rules, we studied how these biochemical properties would transform into the fitness contribution of adenine, uracil, guanine, cytosine, and their epistatic interactions at each nucleotide position in SD. Because our experimental data covered the great majority of genotypes in FLfepB, FLarti, and FLdmsC (Fig. 1B), we applied a variance partition method to estimating the mean fitness effect of the RBS context (1 term), the mean fitness effects of single nucleotides (36 terms: four nucleotide types at nine positions), and those of pairwise epistasis (576 terms: 36 paired combinations of nine positions where each position had four nucleotide types) across all genotypes (Eqs. 4–6 in Supplemental Text). The mean fitness effects of single nucleotides and pairwise epistasis in FLfepB, FLarti, and FLdmsC were qualitatively similar. For single nucleotides, general ranking of their mean fitness effects was G > A > U > C across positions (Fig. 5A). Concerning pairwise epistasis, nucleotides interacted strongly with those in adjacent positions, and pairwise interactions between four nucleotide types were largely similar regardless of positions (Fig. 5C). Would this pervasive pattern of epistasis stem from SD:aSD base pairing? We applied the same analysis to the free energy of SD:aSD base pairing predicted by a RNA folding algorithm (Lorenz et al. 2011). Indeed, results revealed great similarities between in silico predictions and our in vivo data at both the single-nucleotide and pairwise epistasis levels (Fig. 5B,D). This congruence highlights the predominant role of SD:aSD base pairing in translation initiation despite significant RBS context effects (Fig. 1B,C). Additionally, it shows the capability of our high-throughput experiments to elucidate RNA thermodynamics despite measuring phenotypic variation at the protein level.
Figure 5.
Fitness contribution of single nucleotides and pairwise epistasis. (A,B) Mean effects of single nucleotides on fitness (Log[GFP]) (A) and SD:aSD base-pairing energy (ΔG) (B). (C,D) Mean effects of pairwise epistasis on fitness (C) and SD:aSD base-pairing energy (D). Color bars, large heatmaps, and small heatmaps show measurement scales, nucleotide–nucleotide epistasis across the SD sequence, and the averaged patterns of epistasis between each nucleotide position and its two upstream and two downstream neighbors, respectively. (E,F) Explanatory power (R2) of the RBS context (N0), single nucleotides (N1), and pairwise epistasis (N2) on fitness (E) and SD:aSD base-pairing energy (F). (B,D,F) ΔG (predicted values ≤0) is shown as absolute values for simplicity.
Epistasis contributes greatly to fitness variation
Using an additive model (Eq. 7 in Supplemental Text), we evaluated the overall contribution of the RBS context, single nucleotides, and pairwise epistasis to fitness. The additive model predicted the fitness of each genotype in FLfepB, FLarti, and FLdmsC by summing up either the mean fitness effects of the RBS context plus 9 single nucleotides, or those of the RBS context plus 9 single nucleotides and the 36 types of pairwise epistasis. Then we correlated these predictions with experimental data to calculate the explanatory power (R2) of the additive model (Fig. 5E). The contribution of single nucleotides and pairwise epistasis to SD:aSD binding energy was analyzed similarly except that the RBS context was not considered (Fig. 5F). The additive model could explain 32.5%–55.7% and 54.4%–67.8% of experimental data, respectively, when considering the RBS context plus single nucleotides, or the RBS context plus single nucleotides and pairwise epistasis. Likewise, the additive model explained 59.3% and 85.8% of predicted SD:aSD binding energy, respectively, when considering single nucleotides or single nucleotides plus pairwise epistasis. In both the in vivo and in silico cases, single nucleotides exerted a greater influence than pairwise epistasis on fitness. Among the three experimentally characterized fitness landscapes, the magnitude of explanatory power of FLfepB resembled that of the in silico predictions most, reaffirming the weak structured nature of the RBSfepB context. Conversely, even after taking the proportion of measurement errors into consideration (Supplemental Fig. S7B), the additive model was unable to explain 9%–22.3% of fitness variation in FLfepB, FLarti, and FLdmsC, suggesting the influence of higher-order epistasis (i.e., interaction involving more than two nucleotides) or other uncaptured factors.
Global impacts of higher-order epistasis and the constraint of RNA duplex stability on mutational effects
To explore the source of unexplained fitness variation, we performed a complementary analysis by surveying the fitness effects of all point mutations across all characterized genotypes in FLfepB, FLarti, and FLdmsC. Point mutations in 9-nt genotypes can be categorized into 108 types in terms of nine nucleotide positions (Fig. 6A) and 12 nucleotide substitutions. For each point mutation type, we inspected the relationship between the fitness of each genotype and its fitness change upon gaining the mutation. We selected general trends and presented them as overlaid graphs in Figure 6 (for a complete analysis, see Supplemental Figs. S16–S21). From low-fitness to high-fitness genetic backgrounds, we observed diminishing returns in the fitness effect of C→G (marked as “Experiment” in Fig. 6B), C→A, U→G, U→A, and A→G mutations, which were overall beneficial according to Figure 5A. Was such background dependence caused by higher-order epistasis? We addressed this by examining the mutational effects predicted by the additive model (Supplemental Figs. S22–S27). This approach distinguished the effect of higher-order epistasis as the model considered just the fitness contribution of RBS contexts, single nucleotides, and pairwise epistasis (Fig. 5E). Contrastingly, the additive model predicted the beneficial effect of C→G mutations to increase with background fitness (Fig. 6B), suggesting the involvement of higher-order epistasis in experimental data. We sought the cause of higher-order epistasis by inspecting the impact of C→G mutations on SD:aSD base-pairing energy (Fig. 6C; Supplemental Figs. S28, S29). The RNA folding algorithm overall predicted the diminishing effect of C→G mutations in genetic backgrounds with strong base-pairing energy (Lorenz et al. 2011). Yet the effect of C→G mutations on SD:aSD base pairing did not turn deleterious, unlike the experimental data, which showed a negative fitness effect in high-fitness backgrounds (Fig. 6B). This difference might result from the increasing guanine content in high-fitness genotypes (Fig. 3A), which promoted base-pairing interaction between SD and its upstream U-rich or C-rich regions (Fig. 1A). Although this hypothesis was consistent with RNA folding prediction (Fig. 6D; Supplemental Fig. S30), further investigation would be needed for verification.
Figure 6.
Global spectrum of mutational effects. (A) Markers of nucleotide positions. (B,C) Relationship between the fitness (Log[GFP]) (B) and SD:aSD base-pairing energy (ΔG) (C) of genotypes and the effects of C→G mutations. (D) Relationship between the guanine content of SD and the RNA folding energy (ΔG) of 30-nt RBS. Genotypes are ranked by the guanine content and grouped into 10 bins. Lines and bars indicate group means and standard deviations, respectively. (E–G) Relationship between the fitness (E), SD:aSD base-pairing energy (F), and SD:aSD duplex length (G) of genotypes and the effects of G→C mutations. (B,C,E–G) Genotypes are ranked by the considered phenotypes and grouped into 20, 20, 20, 20, and 10 equal-sized bins, respectively. For each phenotype-ranked group, the mean phenotypic effects of mutations at each nucleotide position in SD are computed and shown by lines styled according to A. (C,D,F) The predictions of ΔG are ≤0 and shown as absolute values for simplicity. (B,E) The additive model predicts mutational effects based on the fitness contribution of RBS contexts (N0), single nucleotides (N1), and pairwise epistasis (N2). (H) Chain-breaking model. Mismatch mutations occurring at the internal (red) and the edge (blue) of a RNA duplex are highlighted: (ΔG1) energy penalty caused by base-pairing mismatches; (ΔG2) energy penalty caused by breaking the base-pairing chain. (I) Probability of chain-breaking mutations as a function of the nucleotide position and the duplex length formed by two 9-nt position-aligned RNA strands made of any sequences. The chain-breaking probability at each nucleotide position is shown by lines styled according to A.
Conversely, from low-fitness to high-fitness backgrounds, experimental data showed the aggravating trends of G→C (Fig. 6E), G→U, G→A, A→C, and A→U mutations, whose effects were overall deleterious. Part of the tendency could be explained by the additive model, but the strongly negative correlation between the mutational effect and background fitness suggested the influence of higher-order epistasis. In addition, the negative correlation seemed more pronounced at the internal than the edge positions in SD. We propose a mechanistic model termed “chain-breaking” to explain these two related phenomena (Fig. 6H): The fitness of genotypes increases generally with the SD:aSD duplex length (Supplemental Fig. S4B). A deleterious (i.e., mismatch) mutation thus has a greater chance to interrupt the SD:aSD base-pairing chain when it occurs in high-fitness genetic backgrounds or at the center of SD. Consequently, this chain-breaking mutation destabilizes the SD:aSD duplex in addition to the base mismatch, causing a significant drop in fitness. To verify this model, we applied the same analysis to SD:aSD base-pairing predictions. Indeed, the in silico trends of G→C mutations bore striking resemblance to those found in vivo, with regard to either SD:aSD binding energy (Fig. 6F; Supplemental Figs. S28, S29) or the duplex length (Fig. 6G; Supplemental Figs. S31, S32). To investigate the generality of chain breakage in RNA base pairing, we mathematically described the probability of chain-breaking mutations (i.e., mismatch mutations interrupting RNA duplexes) as a function of the mutation position and the duplex length formed by two single-stranded RNA made of any sequences (Eqs. 8–10 in Supplemental Text). As expected, the probability function predicted chain-breaking mutations more prevalent at the center of RNA strands and in genetic backgrounds with longer duplex length (Fig. 6I).
Discussion
Through exploring the entire genotypic space of SD under three RBS contexts, we generated empirical data sets valuable for studying prokaryotic translation initiation, guiding bioinformatic analysis of synthetic and endogenous RBS, and illuminating the fundamentals of fitness landscapes and molecular evolution. Despite the distinct fitness distribution of FLfepB, FLarti, and FLdmsC, we showed that the underlying trends of mutational effects, biochemical principles, and epistatic interaction were similar. In addition to canonical AG-rich motifs frequently found in endogenous SD (e.g., “GGAGG” frequency = 327/4566), we also observed strong and weak translation initiation by SD bearing poly(G) and poly(U) tracts, respectively, both of which were uncommon in E. coli (e.g., “GGGGG” frequency = 73/4566 and “UUUUU” frequency = 0/4566) (Hayashi et al. 2006). This suggests that endogenous SD represents the evolutionary product, and its sequence features do not necessarily reflect the full recognition capacity of E. coli ribosomes. Therefore, future bioinformatic studies of SD or other genetic elements should consider approaches alternative to scanning canonical sequence motifs.
The positive effect of poly(G) tracts on mRNA-ribosome binding and translation initiation likely results from G·U wobble base pairing between SD and aSD (Varani and McClain 2000). Unlike AG-rich motifs that establish tighter but strict Watson-Crick base pairing with the aSD, poly(G) tracts form weaker wobble pairing with aSD but permit multiple base-pairing configurations. The correlation between the guanine content and the translation efficiency of SD seems straightforward given the CU-rich nature of aSD, but it has not been reported previously. The capability to experimentally synthesize and characterize massive genotypic libraries likely overcomes the limitation of prior work, which mainly studies endogenous SD with narrower sequence and function variation. Uncovering this “G-more” rule will simplify the task of designing RBS for building synthetic gene circuits or genomes (Salis et al. 2009; Bonde et al. 2016). Conversely, in vitro and in vivo assays showed that E. coli ribosomes were able to use poly(U) SD for translation initiation. Synthetic RBS examined here resembles weakly structured RBS in bacteroides and cyanobacteria (Nakagawa et al. 2010; Hockenberry et al. 2018) but differs from those in prior studies concerning the interaction between poly(U) tracts and the ribosomal protein S1 (Boni et al. 1991; Zhang and Deutscher 1992; Duval et al. 2013). In these studies, RBS forms strong secondary structure, the poly(U) tract sits upstream of the SD region, and the SD region contains AG-rich motifs. Future characterization of our synthetic RBS will provide mechanistic insights into the translation initiation of non-SD-led RBS.
From the G-P mapping between or within FLfepB, FLarti, and FLdmsC, we discerned pervasive epistasis and its profound influence (Figs. 1C, 5C). Epistasis convolutes G-P mapping, intensifies the ruggedness of fitness landscapes, and has been shown to decelerate or constrain adaptive evolution (Weinreich et al. 2006; Chou et al. 2011; Tokuriki et al. 2012; Harms and Thornton 2014). Epistasis is frequently noted in large-scale G-P mapping, but elucidating its molecular basis remains challenging (Li et al. 2016; Sarkisyan et al. 2016; Aguilar-Rodríguez et al. 2017). Through global analysis of mutational effects, we revealed the exacerbation of deleterious mutations in high-fitness genotypes (Fig. 6). Supported by RNA base-pairing prediction, our chain-breaking model suggested the negative tendency partly attributed to mismatch mutations disrupting the SD:aSD duplex irrespective of the exact sequence composition. This chain-breaking epistasis is unique in that it stems from the stability constraint of macromolecules, analogous to the effect of stabilizing/destabilizing mutations on protein folding (Tokuriki and Tawfik 2009). Chain-breaking epistasis may profoundly impact the evolution and function of RNA splicing, microRNA/lncRNA regulation, and CRISPR-Cas immunity, as these RNA-mediated processes all involve base pairing.
Extending the implication beyond protein translation and RNA function, our SD G-P fitness landscapes provide an exciting paradigm to investigate the principles of evolutionary genetics. In Maynard Smith's essay concerning the protein sequence space, he conjectured that similar genotypes more likely showed similar phenotypes, and such G-P correlation would facilitate adaptive evolution of fit genotypes by reducing their deleterious mutation load (Maynard Smith 1970). More recently, Wagner and others showed that G-P correlations promoted evolution of promiscuous function by creating neutral networks for DNA, RNA, and proteins to accumulate cryptic genetic variation (Gupta and Tawfik 2008; Hayden et al. 2011; Payne and Wagner 2014). Besides reaffirming the aforementioned attributes, our results further suggest that G-P correlations may support adaptive evolution by the stable supply of beneficial mutations over broad altitudes of fitness landscapes. This feature may be particularly important for biological systems in which adaptation is not supported because of the disproportional scarcity of high-fitness genotypes, like FLarti and FLdmsC (Fig. 1B). As more comprehensive G-P mapping data are becoming available, soon we will be able to tell if this trend of beneficial mutations is SD-specific or intrinsic to biology.
Methods
Primers, plasmids, and strains
All primers were synthesized by Integrated DNA Technologies (Supplemental Table S1). The procedures for constructing plasmids (Supplemental Table S2) were described in Supplemental Text. Four strains of E. coli, 10G (Lucigen), MG1655 (Chou et al. 2015), EK222, and MRE600 (Kurylo et al. 2016), were used in this study. E. coli 10G was applied to plasmid construction and generation of the SD sequence variant libraries. Plasmids and SD variant libraries were then transformed into E. coli MG1655 via electroporation to examine cellular expression of GFP or YFP (Cormack et al. 1996; Nagai et al. 2002). E. coli EK222 served as the negative control for quantification of cellular GFP production. EK222 was constructed via the λ-Red-mediated gene replacement (Datsenko and Wanner 2000). A 1.4-kb fragment, containing the ykgC::kan allele of E. coli JW5040 (Baba et al. 2006), was amplified by PCR using primer pair HCEp165/HCEp166 and transformed into E. coli MG1655 via electroporation. Successful allelic replacement of the chromosomal ykgC locus was screened in terms of kanamycin resistance and confirmed by PCR amplification using primer pairs K1/HCEp168 and K2/HCEp167. E. coli MRE600 was used for purification of ribosomal subunits.
Construction of SD variant libraries
All enzymes were purchased from Thermo Fisher Scientific unless specified otherwise. Three replicates of the FLfepB, FLarti, and FLdmsC SD variant libraries and one replicate of the FLarti-Y library were constructed by the following procedures. Construction of these four libraries began with inverse PCR of template plasmids pYC09, pYC08, pYC20, and pHC199v by Q5 DNA polymerase (NEB) using primer pairs YC09SDRf/YC09SDRr, YC08SDRf/YC08SDRr, YC12SDRf/YC12SDRr, and 199vSDRf/YC08SDRr, respectively. For each of these primer pairs, the 5′ ends of the forward and reverse primers consisted of five and four randomized nucleotides, respectively, which jointly formed the 9-nt SD sequence upstream of GFP or YFP genes later on. The 5′ ends of these primers were phosphorylated to facilitate the ligation step. Following inverse PCR, the reaction product was purified by the Zymo DNA Clean & Concentrator and went through DpnI (NEB) digestion. The remaining 4.4-kb linear DNA was self-ligated by T4 DNA ligase to form circular plasmids. Circular plasmids were transformed into E. coli 10G via electroporation. Cells were revived in the manufacturer's recovery medium for 1 h and then were grown in 12.5 mL of LBK medium at 37°C and 225 rpm overnight. The amount of viable transformants, estimated by serial dilution, ranged from 6.3 × 107 to 3.2 × 108 cells per library. Total plasmids in E. coli 10G were extracted by the Qiagen Plasmid Miniprep Kit and retransformed into E. coli MG1655 via electroporation. The amount of viable transformants at this step was 1.0–5.5 × 108 cells per library. Cells were revived in SOC medium for 1 h and then were grown in 12.5 mL of LBK medium at 37°C and 225 rpm. Next day E. coli MG1655 cells in the overnight culture were collected by centrifugation and resuspended in 1 mL of PBS containing 25% glycerol (v/v). This cell suspension was stored at −80°C as 20 equal aliquots. Individual genotypes isolated directly from these libraries were assigned a unique serial number beginning with “pLK” (Supplemental Tables S2, S3).
FACS experiments
SD variant libraries hosted by E. coli MG1655 were revived by inoculation of one aliquot of frozen cell suspension into 20 mL of LBKG medium. Meanwhile, spike-in variants (Supplemental Table S3), individual genotypes with known sequences and translation efficiencies, were revived by inoculation of 1 μL of frozen stocks into 1 mL LBKG medium. Both SD libraries and spike-in variants were grown overnight at 37°C and 225 rpm. Subsequently, 50 μL and 2.5 μL of the library and spike-in precultures were transferred to 20 mL and 1 mL LBKG medium, respectively. Both cultures were grown at 37°C and 225 rpm until their OD reached 0.55–0.65, during which cell fluorescence was relatively steady (Supplemental Fig. S2B). All spike-in variants were evenly pooled together and added into the library culture in a 1:2500 ratio (v/v). This mixture was diluted 10-fold with PBS and was stored on ice.
FACS was performed by a BD FACSJazz cell sorter, and cells were maintained at 4°C throughout. The sensitivity of the photomultiplier tube (PMT) was calibrated based on two negative control strains (E. coli MG1655 bearing pTK03 or pTK06) and two positive control strains (E. coli MG1655 bearing pYC08 or pHC199v) for GFP and YFP, respectively. The PMT was adjusted such that the GFP and YFP signals of strains bearing pYC08 and pHC199v centered upon 2.86 and 2.81, respectively, in the logarithmic scale. The fluorescence distribution of each library was divided into eight ranks except that the third FLfepB replicate library was divided into 15 ranks for technical comparison (Supplemental File S1). Ranks were numbered in the ascending order according to the fluorescence intensity. For GFP-based libraries with eight ranks, the fluorescence peak positions of pTK03 and pYC08 set the lower and upper bounds of a range in which Ranks 2–7 were evenly spaced; Ranks 1 and 8 were defined as the zones below and above this range, respectively. The eight ranks of the FLarti-Y library were defined similarly except using pTK06 and pHC199v, respectively, as the lower and upper bounds of Ranks 2–7. For the third FLfepB replicate library, the fluorescence peaks of pTK03 and pYC08, respectively, set the lower and upper bounds of a range in which Ranks 2–13 were evenly spaced. The zone below this range was defined as Rank 1, and the zone above this range was further divided into Ranks 14 and 15 in terms of the peak position of spike-in variant pLK170110v71. A total of 10,000,000 cells were collected per sort-seq experiment, and the amount of cells collected for each rank was proportional to its relative abundance in the library (Supplemental File S1). After FACS, cells of each rank were immediately grown in 10 mL of LBKG medium at 37°C and 225 rpm overnight. Subsequently plasmids of each rank were extracted by the Qiagen Plasmid Miniprep Kit. To inspect the FACS purity of each rank, 5 μL of overnight cultures were inoculated to 2 mL of LBKG medium incubated at 37°C and 225 rpm. The fluorescence distribution of each culture was examined by the cell sorter when its OD reached 0.55–0.65. The FACS purity of a rank was defined as the percentage of 50,000 cells falling in that rank and its bordering ranks because the latter likely represented phenotypic variation of individual genotypes rather than sorting errors (Supplemental Text).
Deep sequencing
The SD genotypes on plasmids were sequenced by the Illumina HiSeq 2500 system. Preparation of sequencing libraries followed the Illumina two-step PCR procedure. A 0.3-kb fragment including the 24-bp 5′ reporter gene, RBS, and the further upstream plasmid region of the FLfepB, FLarti, and FLdmsC libraries, was amplified by amplicon PCR using KAPA HiFi DNA polymerase (Roche), one forward primer ILp2, and four reverse primers ILp6, ILp6a, ILp6b, and ILp6c. Amplicon PCR of the FLarti-Y library was performed in a similar manner except for four reverse primers ILp1, ILp1a, ILp1b, and ILp1c. For each rank, amplicon PCR products were purified by AMPure XP Beads (Beckman) and then were subjected to index PCR using KAPA HiFi DNA polymerase and a unique pair of index primers from the Nextera XT Index Kit (Illumina). Index PCR products of each rank were purified by AMPure XP Beads, and their DNA concentrations were measured by a Qubit fluorometer (Thermo Fisher Scientific). Index PCR products of each rank in a SD library were pooled together in terms of its relative abundance in that library. This multiplexed sample was subjected to 50-nt single-read sequencing. Deep sequencing generated about 50,000,000 reads per sort-seq experiment. All of the raw data passed the FastQC quality control score (Q = 25). The amount of reads in each rank should be proportional to its library abundance measured by the cell sorter (Supplemental File S1). In cases in which the library percentage of the former was lower than that of the latter by 10%, a rank was subjected to resequencing, and the original and resequencing reads of that rank were merged for data processing and analysis.
Fitness calculation using sort-seq data
Sequence hard-matching by the Linux grep command removed irrelevant or indel-containing reads from raw sequencing data. In-house scripts (Supplemental File S6) were then executed to extract the 9-nt SD sequence from each read and count the occurrence of each genotype in each rank (Sanner 1999; R Core Team 2019). One read count table summarizing the results was generated for each sort-seq experiment. The read distribution of each genotype across ranks was used to estimate its fitness. The rank mean, defined as the averaged rank value of a genotype weighted by its read counts in each rank, was computed (Eqs. 1 and 2 in Supplemental Text). Following identical procedures, the rank means of spike-in SD variants were computed. Linear regression of the rank means of spike-in variants and their fitness measured separately by the cell sorter was performed to generate a standard curve (Supplemental Table S4). Based on this standard curve, a linear equation converted the rank mean of each genotype to fitness (Eq. 3 in Supplemental Text). We combined fitness quantified by three sort-seq experiments with 25 or more reads into a union table for FLfepB, FLarti, and FLdmsC libraries (Supplemental Files S2–S4). The fitness of a genotype was reported as the average of one to three replicate measurements. The fitness of genotypes in FLarti-Y is available in Supplemental File S5.
Prediction of SD:aSD base pairing and local mRNA folding
SD:aSD base pairing and mRNA folding were predicted by the Vienna RNA package (Lorenz et al. 2011). Base pairing between each of the 262,144 9-nt SD genotypes and aSD (5′-ACCUCCUUA-3′) was predicted by the RNAsubopt program. SD:aSD base pairing energy (ΔG) was predicted at 37°C with a contribution from dangling ends. For each SD genotype, we accepted the prediction with the lowest ΔG. Based on the predicted base-pairing pattern, we calculated the length of the SD:aSD duplex. We operationally defined the duplex length as the longest base-pairing region uninterrupted by mismatches for two reasons. First, the predicted minimal ΔG of SD:aSD base pairing was −13.0 kcal/mol. Empirically, the RNAsubopt program assigned a large energy penalty (3∼4 kcal/mol) per internal mismatch such that the predicted ΔG of a mismatch-containing duplex was quantitatively similar to that of the longest uninterrupted base-pairing region of the same duplex. Second, it was technically difficult to convert internal mismatches into the measure of the duplex length. The influence of SD genotypes on mRNA folding was predicted by the RNAfold program. Unless indicated otherwise, we used the 60-nt sequence, containing both the 30-nt regions upstream of and downstream from the gfp start codon, as the input and accepted the predicted structure with the lowest ΔG.
Data access
The sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA516114.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Y.-C. Lin, D.-H. Kuo, and Y.-S. Cheng for experimental assistance; National Taiwan University (NTU) College of Life Science (CLS) Technology Commons for instrument support; Taiwan National Core Facility for Biopharmaceuticals for sequencing; and J.-Y. Leu, D. Agashe, B.-Y. Liao, and C.-C. Wu for valuable comments. We are thankful for support of a Marie Curie fellowship to initiate this project at Cambridge (to H.D.C.) and for European Research Council Advanced Grant ERC AdG 695669 (to F.H.). This work was funded by Ministry of Science and Technology, Taiwan (MOST) grants 105-2311-B-002-004-MY2, 106-2628-B-002-002-MY4, and 106-2311-B-002-012-MY2, and NTU grants 107L7738, 108L7721, and 108L7867.
Author contributions: Conceptualization, H.D.C.; methodology, J.W., F.H., and H.D.C.; investigation, Y.-J.C., S.K., Y.-L.C., and Y.L.; formal analysis, R.J., S.K., J.W., and H.D.C.; software, R.J. and S.K.; data curation, R.J. and S.K.; visualization, R.J., S.K., and H.D.C.; writing—original draft, J.W. and H.D.C.; writing—review and editing, J.W., F.H., and H.D.C.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.260182.119.
References
- Aguilar-Rodríguez J, Payne JL, Wagner A. 2017. A thousand empirical adaptive landscapes and their navigability. Nat Ecol Evol 1: 45 10.1038/s41559-016-0045 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco C, Janzen E, Pressman A, Saha R, Chen IA. 2019. Molecular fitness landscapes from high-coverage sequence profiling. Annu Rev Biophys 48: 1–18. 10.1146/annurev-biophys-052118-115333 [DOI] [PubMed] [Google Scholar]
- Bonde MT, Pedersen M, Klausen MS, Jensen SI, Wulff T, Harrison S, Nielsen AT, Herrgård MJ, Sommer MOA. 2016. Predictable tuning of protein expression in bacteria. Nat Methods 13: 233–236. 10.1038/nmeth.3727 [DOI] [PubMed] [Google Scholar]
- Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. 1991. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res 19: 155–162. 10.1093/nar/19.1.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Halgamuge S, Tang SL. 2006. Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene 373: 90–99. 10.1016/j.gene.2006.01.033 [DOI] [PubMed] [Google Scholar]
- Chen H, Bjerknes M, Kumar R, Jay E. 1994. Determination of the optimal aligned spacing between the Shine–Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res 22: 4953–4957. 10.1093/nar/22.23.4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Chiu HC, Delaney NF, Segre D, Marx CJ. 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332: 1190–1192. 10.1126/science.1203799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Marx CJ, Sauer U. 2015. Transhydrogenase promotes the robustness and evolvability of E. coli deficient in NADPH production. PLoS Genet 11: e1005007 10.1371/journal.pgen.1005007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173: 33–38. 10.1016/0378-1119(95)00685-0 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Rubin AJ, Sauer RT. 2011. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res 39: 1131–1141. 10.1093/nar/gkq810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit MH, van Duin J. 1990. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci 87: 7668–7672. 10.1073/pnas.87.19.7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo J, Diss G, Lehner B. 2018. Pairwise and higher-order genetic interactions during the evolution of a tRNA. Nature 558: 117–121. 10.1038/s41586-018-0170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo J, Baeza-Centurion P, Lehner B. 2019. The causes and consequences of genetic interactions (epistasis). Annu Rev Genomics Hum Genet 20: 433–460. 10.1146/annurev-genom-083118-014857 [DOI] [PubMed] [Google Scholar]
- Duval M, Korepanov A, Fuchsbauer O, Fechter P, Haller A, Fabbretti A, Choulier L, Micura R, Klaholz BP, Romby P, et al. 2013. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol 11: e1001731 10.1371/journal.pbio.1001731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espah Borujeni A, Channarasappa AS, Salis HM. 2014. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res 42: 2646–2659. 10.1093/nar/gkt1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat Rev Genet 8: 610–618. 10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Gao R, Yu K, Nie JK, Lian TF, Jin JS, Liljas A, Su XD. 2016. Deep sequencing reveals global patterns of mRNA recruitment during translation initiation. Sci Rep 6: 30170 10.1038/srep30170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorochowski TE, Chelysheva I, Eriksen M, Nair P, Pedersen S, Ignatova Z. 2019. Absolute quantification of translational regulation and burden using combined sequencing approaches. Mol Syst Biol 15: e8719 10.15252/msb.20188719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RD, Tawfik DS. 2008. Directed enzyme evolution via small and effective neutral drift libraries. Nat Methods 5: 939–942. 10.1038/nmeth.1262 [DOI] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW. 2014. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature 512: 203–207. 10.1038/nature13410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, et al. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2: 2006.0007 10.1038/msb4100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EJ, Ferrada E, Wagner A. 2011. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474: 92–95. 10.1038/nature10083 [DOI] [PubMed] [Google Scholar]
- Hockenberry AJ, Stern AJ, Amaral LAN, Jewett MC. 2018. Diversity of translation initiation mechanisms across bacterial species is driven by environmental conditions and growth demands. Mol Biol Evol 35: 582–592. 10.1093/molbev/msx310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM. 2013. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci 110: 14024–14029. 10.1073/pnas.1301301110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurylo CM, Alexander N, Dass RA, Parks MM, Altman RA, Vincent CT, Mason CE, Blanchard SC. 2016. Genome sequence and analysis of Escherichia coli MRE600, a colicinogenic, nonmotile strain that lacks RNase I and the Type I methyltransferase, EcoKI. Genome Biol Evol 8: 742–752. 10.1093/gbe/evw008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. 2005. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123. 10.1128/MMBR.69.1.101-123.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qian WF, Maclean CJ, Zhang JZ. 2016. The fitness landscape of a tRNA gene. Science 352: 837–840. 10.1126/science.aae0568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. 1970. Natural selection and the concept of a protein space. Nature 225: 563–564. 10.1038/225563a0 [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90. 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Niimura Y, Miura K, Gojobori T. 2010. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc Natl Acad Sci 107: 6382–6387. 10.1073/pnas.1002036107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotajo D, Tate T, Cho H, Choudhary M. 2015. Distribution and diversity of ribosome binding sites in prokaryotic genomes. BMC Genomics 16: 604 10.1186/s12864-015-1808-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. 2005. The genetic theory of adaptation: a brief history. Nat Rev Genet 6: 119–127. 10.1038/nrg1523 [DOI] [PubMed] [Google Scholar]
- Osterman IA, Evfratov SA, Sergiev PV, Dontsova OA. 2013. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res 41: 474–486. 10.1093/nar/gks989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne JL, Wagner A. 2014. The robustness and evolvability of transcription factor binding sites. Science 343: 875–877. 10.1126/science.1249046 [DOI] [PubMed] [Google Scholar]
- Podgornaia AI, Laub MT. 2015. Pervasive degeneracy and epistasis in a protein-protein interface. Science 347: 673–677. 10.1126/science.1257360 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: http://www.R-project.org/. [Google Scholar]
- Rodnina MV. 2016. The ribosome in action: tuning of translational efficiency and protein folding. Protein Sci 25: 1390–1406. 10.1002/pro.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. 2009. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27: 946–950. 10.1038/nbt.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner MF. 1999. Python: a programming language for software integration and development. J Mol Graph Model 17: 57–61. [PubMed] [Google Scholar]
- Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, Ivankov DN, Bozhanova NG, Baranov MS, Soylemez O, et al. 2016. Local fitness landscape of the green fluorescent protein. Nature 533: 397–401. 10.1038/nature17995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Childs L, Walther D, Bock R. 2011. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet 7: e1002155 10.1371/journal.pgen.1002155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254: 34–38. 10.1038/254034a0 [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. 2009. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19: 596–604. 10.1016/j.sbi.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, Tawfik DS. 2012. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun 3: 1257 10.1038/ncomms2246 [DOI] [PubMed] [Google Scholar]
- Varani G, McClain WH. 2000. The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep 1: 18–23. 10.1093/embo-reports/kvd001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. 2008. Robustness and evolvability: a paradox resolved. Proc Biol Sci 275: 91–100. 10.1098/rspb.2007.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312: 111–114. 10.1126/science.1123539 [DOI] [PubMed] [Google Scholar]
- Zhang J, Deutscher MP. 1992. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc Natl Acad Sci 89: 2605–2609. 10.1073/pnas.89.7.2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.