Abstract
The phenotypic variation of living organisms is shaped by genetics, environment, and their interaction. Understanding phenotypic plasticity under natural conditions is hindered by the apparently complex environment and the interacting genes and pathways. Herein, we report findings from the dissection of rice flowering-time plasticity in a genetic mapping population grown in natural long-day field environments. Genetic loci harboring four genes originally discovered for their photoperiodic effects (Hd1, Hd2, Hd5, and Hd6) were found to differentially respond to temperature at the early growth stage to jointly determine flowering time. The effects of these plasticity genes were revealed with multiple reaction norms along the temperature gradient. By coupling genomic selection and the environmental index, accurate performance predictions were obtained. Next, we examined the allelic variation in the four flowering-time genes across the diverse accessions from the 3000 Rice Genomes Project and constructed haplotypes at both individual-gene and multigene levels. The geographic distribution of haplotypes revealed their preferential adaptation to different temperature zones. Regions with lower temperatures were dominated by haplotypes sensitive to temperature changes, whereas the equatorial region had a majority of haplotypes that are less responsive to temperature. By integrating knowledge from genomics, gene cloning and functional characterization, and environment quantification, we propose a conceptual model with multiple levels of reaction norms to help bridge the gaps among individual gene discovery, field-level phenotypic plasticity, and genomic diversity and adaptation.
Understanding the genetic and environmental mechanisms of phenotypic plasticity has been a long-term challenge (Marais et al. 2013; Josephs 2018). Advances in genomics and molecular biology allowed further exploration of the genetic architecture of complex traits under natural field conditions (Marais et al. 2013; Blackman 2017; Li et al. 2018; Millet et al. 2019). Rice (Oryza sativa L.), with a large number of genes cloned for agronomic traits, is well positioned to bridge the gap among genomic diversity, individual genes, and field-level phenotypic plasticity dissection (Nicotra et al. 2010; Chen et al. 2019). Rice accounts for about one-fifth of the world's caloric intake. Population growth drives up the global demand for food, which is expected to increase for the next several decades (Godfray et al. 2010). Producing more rice from less arable land under fluctuating climatic conditions requires a concerted effort from both public and private sectors. Although advances in genomics, breeding, and precision agriculture have been recognized as solution components for global food security (McCouch et al. 2013; Huang et al. 2015; Zeng et al. 2017; Wang et al. 2018), enriched knowledge of the varied performance of genotypes across environments, or phenotypic plasticity (Nicotra et al. 2010), is required to design and implement the best genetic deployment and agronomic management practices.
Flowering time plays a critical role in rice adaptation and production, and modification of flowering time is determined by genetic pathways that integrate endogenous and exogenous signals (Hori et al. 2016). The worldwide distribution of rice is the manifestation of the large genetic diversity adapted to varying environmental conditions (Wang et al. 2018). Because flowering time also affects other agronomic traits, genetic improvement in rice often involves the selection of flowering time for yield optimization.
Molecular mechanisms underlying the timing of transition from vegetative to reproductive growth have been extensively studied in rice (Xue et al. 2008; Huang et al. 2012b; Yano et al. 2016), a model crop species. Heading date 1 (Hd1), a homolog of CONSTANS from Arabidopsis, is a central regulator of flowering time in rice, a facultative short day–length (SD) plant (Yano et al. 2000). Hd1 promotes Heading date 3a (Hd3a; a florigen-coding gene) expression in SD and suppresses the expression of Hd3a in long day–length (LD) conditions (Hayama et al. 2003; Tamaki et al. 2007; Nuñez and Yamada 2017). OsGIGANTEA (OsGI) acts as an activator of Hd1 under SD. In addition to the OsGI-Hd1-Hd3a photoperiodic pathway branch, a second branch, Ghd7-Ehd1-RFT1, also affects rice flowering time (Hayama et al. 2003; Shrestha et al. 2014; Zhang et al. 2017). Ehd1 promotes flowering by activating the expression of Hd3a and RFT1 (another florigen-coding gene). Ghd7 acts as a LD-preferential repressor by blocking the expression of Ehd1 (Xue et al. 2008). Hd2 and Hd5 are key regulators in the Ehd1 pathway, whereas Hd6 plays a critical role in regulating Hd1 activity (Takahashi et al. 2001; Wei et al. 2010; Koo et al. 2013). In addition, as in Arabidopsis, flowering in rice is induced by the coincidence of circadian and solar rhythms for Hd1 and Ehd1, consistent with the external coincidence model (Yeang 2013).
Although natural variants of these genes were associated with flowering time in diverse rice accessions (Zhao et al. 2011; Huang et al. 2012b; Yano et al. 2016), integration of these findings to explain and predict complex phenotypic plasticity observed in natural fields has received limited attention. A recent review highlighted the research need to examine gene–environmental interaction and gene–gene interaction (Chen et al. 2019). An earlier study examined the flowering-time plasticity in a sorghum biparental population (Li et al. 2018), but how findings from a narrow genetic background can be connected with diverse germplasm remains unclear. Herein, we show that the key flowering-time genes (Hd1, Hd2, Hd5, and Hd6), originally discovered for their photoperiodic responses, are the major loci responding to the varied temperature at nine different environments for a genetic mapping population. Their dynamic effects and interactions shape the complex phenotypic plasticity landscape. Reaction norms of gene effects can be obtained along the temperature gradient differentiating these environments. Moving into a broader genetic context, we constructed the multigene haplotypes for the diverse accessions from the 3000 Rice Genomes Project. By leveraging the known functional polymorphisms of these well-studied genes, we assigned the slope parameter values obtained from the biparental population to these haplotypes. A clear preferential distribution pattern emerged for haplotypes with different sensitivity to temperature zones. Finally, we propose a conceptual model to illustrate phenotypic complexity using the multiple levels of reaction norms along an environmental gradient.
Results
Rice flowering-time plasticity in natural long-day environments
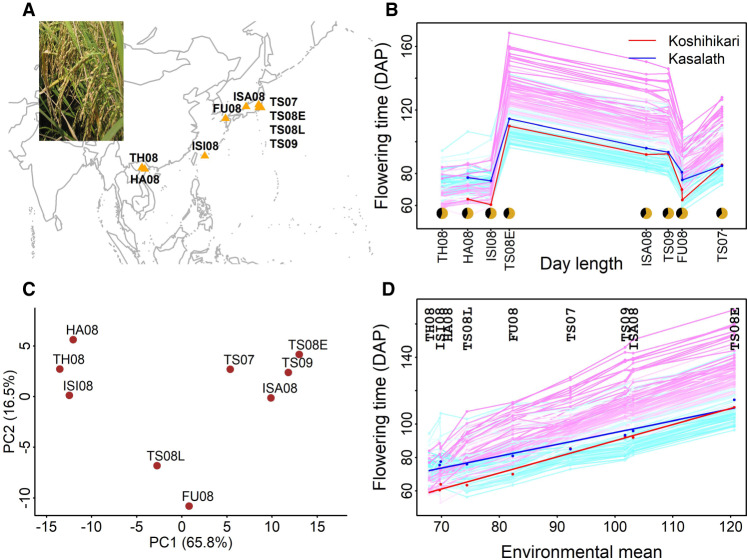
Complex flowering-time (heading date) variation was observed in a rice genetic mapping population grown in nine natural environments (Fig. 1A,B; Supplemental Table S1). This population is a group of 174 backcross inbred lines (BILs) derived from crossing two parents. One parent is a japonica cultivar, Koshihikari, and the other parent is an indica cultivar, Kasalath. The latitudes of field sites range from 21°01′N (Vietnam) to 36°01′N (Japan), and planting time ranged from March to June. The environmental mean, the average flowering time of the whole population within each environment, varied from 68 to 121 d after planting (DAP). Trait correlation and prediction of flowering time for each pair of environments ranged from 0.20 to 0.99 and from 0.17 to 0.91, respectively (Supplemental Fig. S1). Flowering time expressed as DAP showed a pattern that can be readily modeled, unlike using growing degree days (GDD) as the flowering-time unit (Supplemental Fig. S2).
Figure 1.
Flowering-time plasticity in rice. (A) Nine natural field environments. (B) Reaction norm for flowering time based on average day length (from planting to 50 days after planting [DAP]). (C) Principal component analysis of G × E from the additive main effect and multiplicative interaction (AMMI) model. (D) Reaction norm based on a numerical order of environmental mean. Regression fitted lines are indicated for two parents in panel D. Dots are the observed flowering-time values.
We first conducted several analyses to understand the overall phenotypic variation and genotype by environment interaction (G × E) (Malosetti et al. 2013). Variance component analysis partitioned the phenotypic variance into environment (73.3%), genotype (17.6%), and G × E (8.9%) (Supplemental Tables S2, S3). We then partitioned the G × E variance into lack of genetic correlation (79.0%) and heterogeneity of genotypic variance (21.0%) (Supplemental Table S4). By following the additive main effects and multiplicative interaction (AMMI) model, we found that the first two principal components accounted for 82.3% of the G × E (Fig. 1C). With the joint regression analysis approach (Finlay and Wilkinson 1963; Eberhart and Russell 1966), G × E was partitioned into heterogeneity in slopes (73.3%) and error (26.7%) (Supplemental Table S5).
We focused our attention on joint regression analysis, which models the overall phenotypic variation with a numerical index (environmental mean) to connect all environments (Fig. 1D). By regressing the observations of individual genotypes on environmental mean, one can obtain the expected reaction norm of this genetic population. Despite two parents showing relatively small differences, their progenies showed transgressive segregation in flowering time (Fig. 1D). Although grouping patterns emerged in both genotypes and environments, this traditional joint regression analysis relies on environmental mean and could not discern the underlying biological mechanisms or allow performance prediction of other environments.
Temperature defines the environmental index capturing flowering-time variation
A better understanding of the general environmental pattern is needed to facilitate both systematic gene effect comparison and performance prediction. As an aggregate measure, environmental mean represents the outcome of the interplay between genetics and environment. If we regard the whole population as a single genotype, environmental mean informs us about differences among environments. However, environmental mean can only be obtained after the actual experiment and is specific to the tested set of environments, lacking the capability of inference.
We examined ways to replace environmental mean by an explicit, performance-derived environmental index (Supplemental Table S6). After testing temperature (using GDD), photoperiod, and photothermal time (PTT) from different growth windows (Supplemental Fig. S3), we found that average temperature from 9–50 DAP (denoted as GDD9–50) can serve as an environmental index to best characterize the environments and replace environmental mean (Fig. 2A,B). Besides having a strong correlation (r = −0.990, P = 3 × 10−7) (Fig. 2C), GDD9–50 involves a biologically relevant window covering the early growth stage when plants process the environmental cues to determine the timing of transition to reproductive development. All nine environments were regarded as LD environments because the average daily photoperiod before flowering ranged from 13.9 to 15.5 h (Supplemental Fig. S4), >13.5-h threshold for the general SD and LD classification (Itoh et al. 2010). Correlation between photoperiod and environmental mean was only 0.38 in the same 9–50 DAP window (Supplemental Fig. S3). Because of the negligible increase in correlation strength when considering photoperiod as an additional environmental factor to pair with temperature, PTT (r = −0.991) was not chosen (Supplemental Fig. S3). In addition, subsampling analyses verified the general consistency of this GDD9–50 across both subsets of environments and subsets of genotypes (Supplemental Fig. S5).
Figure 2.
Identifying an environmental index from the performance data. (A) Rice development and temperature (in GDD) profiles across different environments. (B) Search for the most indicative growth window within which average temperature is highly correlated to environmental mean of flowering time. Temperature within the window of 9–50 DAP was chosen as the environmental index and denoted as GDD9–50. (C) Significant correlation between GDD9–50 and environmental mean. (D) Regression-fitted reaction norm using the environmental index (GDD9–50) as the explanatory variable.
With the identified environmental index, the fitted reaction norm of this genetic population was obtained by regressing flowering-time observations from each genotype on the values of GDD9–50 (Fig. 2D). The varied responses among individual genotypes can be described by two reaction-norm parameters: (1) intercept, quantifying the overall expected flowering time, and (2) slope, quantifying the sensitivity to environmental changes. A one-unit increase in temperature promoted the whole population to flower 1.94 d earlier, whereas this value varied between 0.6 and 3.6 d for different genotypes (i.e., different slopes).
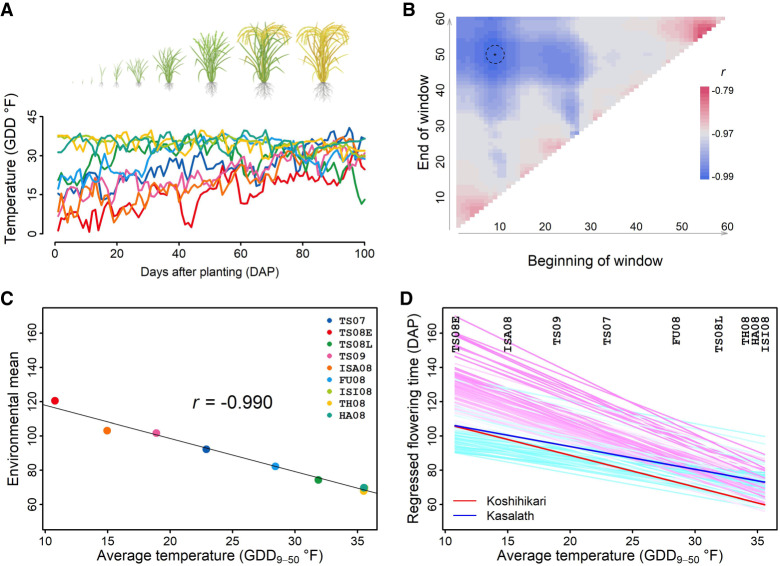
Performance prediction through JGRA
To first focus on the whole-plant performance level, we implemented joint genomic regression analysis (JGRA) to model and predict flowering time for this rice population by integrating the identified environmental index, joint regression analysis, and genomic selection. With the whole population being genotyped, we tested two approaches: (1) genomic predicted reaction-norm parameters for individual genotypes (Fig. 3A–C) and (2) genome-wide marker effect continua for individual markers (Fig. 3D–F). All 162 markers across the genome were used to construct the genomic relationship matrix for rection norm parameters or to derive the genome-wide marker effects. Although this marker number appeared small, it is known that the requirement for the number of markers in biparental population is generally low (Bernardo and Yu 2007). We examined three scenarios by splitting environments and genotypes into either tested or untested.
Figure 3.
Performance prediction of flowering time with joint genomic regression analysis (JGRA) to leverage environmental index and genomic prediction. (A–C) JGRA using reaction-norm parameters. (D–F) JGRA using genome-wide marker effect continua. The three scenarios are predicting performance for tested genotypes in untested environments (A,D), predicting performance for untested genotypes in tested environments (B,E), and predicting performance for untested genotypes in untested environments (C,F). Prediction accuracy within each individual environment (in parentheses) and across all environments (r) are indicated; the diagonal line indicates the exact match between observed and predicted values.
High prediction accuracy (i.e., correlation between predicted and observed values) was obtained from both JGRA approaches for predicting performance of tested genotypes in untested environments (leave-one-environment-out cross-validation): 0.97 and 0.94 (Fig. 3A,D). Prediction accuracy within individual environment varied from 0.82 to 0.99 for the reaction-norm parameter approach and from 0.71 to 0.90 for the marker effect continuum approach. The predicted values were very close to the observed values, with a ratio close to one (Supplemental Fig. S6). For the second scenario, predicting the performance of untested genotypes in tested environments (leave-one-half-genotypes-out cross-validation), prediction accuracy was 0.91 and 0.89 (Fig. 3B,E). This scenario describes when only a proportion of possible genotypes can be tested in different environments (either because of limited resources or by design) and predictions of the remaining untested genotypes are made. Prediction accuracy within individual environment ranged from 0.45–0.66 and 0.46–0.67 for the two approaches, lower than the first scenario. The ratio of predicted values to observed values was close to one on average in the second scenario (Supplemental Fig. S6).
For the most challenging scenario, predictions for untested genotypes in untested environments, connecting tested and untested genotypes with genome-wide markers and connecting tested and untested environments with environmental index were needed (Fig. 3C,F). The leave-one-environment-and-one-half-genotypes-out cross-validation resulted in an accuracy of 0.88 for the reaction-norm parameter approach and 0.90 for the marker effect continuum approach and in an individual-environment accuracy of 0.44–0.67. We further showed that in all three scenarios, the series of environment-specific predictions generated from JGRA were superior to the fixed predictions using average performance across tested environments (best linear unbiased estimator [BLUE]) (Supplemental Fig. S6).
Prediction accuracy across all environments was higher than those for individual environments. This was expected given the wider context for the across-all-environments prediction (i.e., range change for x-y correlation of predicted-observed) and given that the across-all-environments correlation contains the environment effect that was well captured by the environmental index. Biologically, unique local environmental conditions may alter flowering time in different directions from the main environmental factor across environments and thus reduce prediction accuracy.
By following the framework of joint regression analysis (Finlay and Wilkinson 1963; Eberhart and Russell 1966), we partitioned the overall variance in the predicted values into components (Supplemental Table S7). As expected, contribution of environment to phenotypic variance was the highest, followed by genotype and G × E.
Genetic dissection of flowering-time plasticity
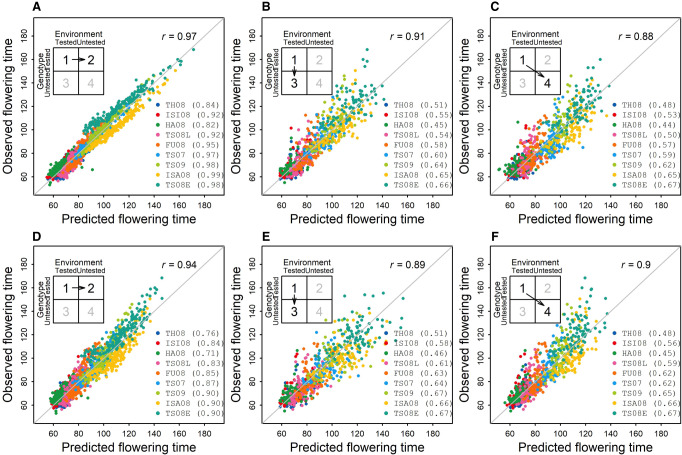
To reveal the underlying genetic loci and their effect dynamics, we conducted QTL linkage mapping using flowering-time observations within individual environments, mapping using flowering-time observations across environments, and mapping using two reaction-norm parameters (intercept and slope). Although the environment effect accounted for a large part of the overall phenotypic variation in the multienvironment trial (73.3%), genotypic variance was twice of G × E variance and entry-mean-based heritability of flowering time was 0.946 (Supplemental Tables S2–S4). The detection of QTLs was not affected because the heritability for single environment was moderately high (0.662). Consistently, four loci corresponding to Hd1, Hd2, Hd5, and Hd6 were detected by all mapping approaches (Fig. 4A,B; Supplemental Figs. S7, S8).
Figure 4.
Varied effects at four loci along the temperature gradient underlying rice flowering-time plasticity. (A) Reaction norms of genetic effects at the single-locus level along the environmental index by temperature (GDD9–50). Dots show the effects detected from individual environment analysis. Dashed vertical line shows the positions of intercepts at the average temperature value. (B) Loci detected by mapping using the parameters (intercept and slope) derived from the reaction norms of genotypes. Horizontal lines in LOD plots show the significance threshold. Additive effect in descending absolute-value order for intercept (Hd5, Hd1, Hd6, and Hd2) and for slope (Hd1, Hd2, Hd5, and Hd6). (C) Known or potential functional polymorphisms between two alleles in four flowering-time genes from sequence analysis. (D) Positions of four genes highlighted in the rice flowering control pathway under natural long day-length (LD) conditions. These genes originally discovered for photoperiodic response are found to be involved in sensing temperature differences in LD. Pathway diagram is modified following the Figure 1 in the work by Matsubara et al. (2014).
Given the well-studied photoperiod response in rice (Matsubara et al. 2014) and available genome sequence information of two parental inbreds (Wang et al. 2018), we verified the functional polymorphisms in these four genes (Hd1, Hd2, Hd5, and Hd6) (Fig. 4C; Supplemental Fig. S9; Supplemental Table S8). In addition, all four genes were originally identified by map-based cloning involving Kasalath, one of the mapping parents, and Nipponbare, the reference genome. All this information allowed us to connect these four cloned genes with the mapped QTLs. We adapted the known pathway (Matsubara et al. 2014) to show their additional roles for temperature sensing to control flowering time (Fig. 4D). The effect of temperature on flowering time and the effect through genes identified from the photoperiodic response pathway were documented in the earlier studies (Vergara and Chang 1985; Li et al. 2015b). In the current study, the Koshihikari allele of Hd1 delayed flowering in most of the environments, which agrees with the function of Hd1 as a repressor of Hd3a in LD condition, and the effect was dependent on the temperature. In comparison with the Hd2 allele from Kasalath, the Hd2 allele from Koshihikari promoted flowering in environments with GDD9–50 > 20 while repressing flowering in environments with GDD9–50 < 20. The negative genetic effects of Hd5 and Hd6 indicated that Koshihikari alleles promoted flowering across the temperature range captured in these tested environments.
Having the same set of loci detected from mapping using flowering-time observations (Fig. 4A) and mapping using reaction-norm parameters (Fig. 4B) indicated these plasticity genes were responding to the temperature gradient that differentiated the environments (Nicotra et al. 2010). In addition, different orders of QTL effects were identified: Hd5, Hd1, Hd6, and Hd2 for intercept but Hd1, Hd2, Hd5, and Hd6 for slope (Fig. 4B), which agreed with reaction norms of genetic effects at single-locus level along the environmental index defined by temperature (Fig. 4A). By using effect estimates (Fig. 4B) from mapping with reaction-norm parameters (Fig. 2D), reaction norms of genetic effects for four loci can also be plotted, which is comparable to the plot from mapping first and regression next (Fig. 4A). Reaction norms at different resolution levels showed consistent patterns (Supplemental Methods; Supplemental Figs. S10–S12).
To examine contributions from additive effect and QTL by environment interaction (QEI) effect, we conducted QEI mapping (Supplemental Fig. S8; Li et al. 2015a). Agreeance was found between the results from QTL mapping with intercept and slope. Hd1, Hd5, and Hd6 were detected mainly by additive effect, whereas Hd1 and Hd2 were detected mainly by QEI effect. Hd2 was the most significant gene controlling QEI as the effect of Hd2 is bidirectional, promoting or repressing flowering depending on the environment it was exposed to. To discover the interaction between genes, we performed QEI mapping on epistasis and detected strong interaction signals between Hd2 and Hd6 and between Hd1 and Hd5 (Supplemental Fig. S8). Additionally, other signals with intermediate strength across the genome were detected, indicating that many small-effect interactions also contributed to the flowering-time variation. It appears that even though complex pathways and networks might have been involved, strong marginal effects and two-way interactions can still be detected.
Alternative to genomic prediction using genome-wide markers, flowering time can also be predicted by constructing a model with only markers tagging these four genes (Supplemental Fig. S13). Comparable results were obtained, which is expected given the common framework of environmental index search and joint regression analysis.
Haplotype networks of four flowering-time plasticity genes in global germplasm
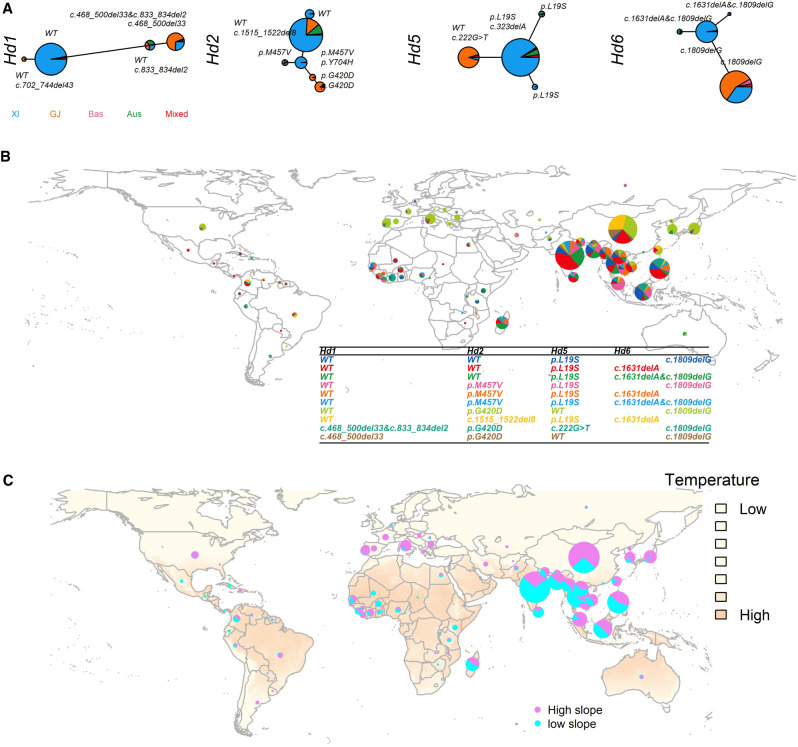
It is interesting to see how different alleles and allele combinations are present in diverse rice accessions. This is because we observed the differential temperature responses of these four genes originally discovered from their photoperiod effects, and a recent genome scan with whole-genome sequencing identified Hd1, Hd2, and Hd6 underlying flowering time across a diverse rice accessions (Yano et al. 2016). We constructed haplotype networks of these four genes for the 3010 diverse cultivated rice genomes from the 3000 Rice Genomes Project (Supplemental Fig. S14; Supplemental Table S9; Wang et al. 2018). From 29 million single-nucleotide polymorphisms (SNPs), there were more than 100 SNPs for each gene (Hd1, Hd2, Hd5, and Hd6). These four genes were found to have multiple haplotypes but were primarily separated into xian/indica (XI) and geng/japonica (GJ) accessions.
Haplotype analysis identified four major haplotypes for Hd1, each with more than 100 accessions. We aligned these SNP-defined haplotypes with characterized functional polymorphisms (Fig. 5A; Supplemental Table S10). One major haplotype with the largest number of accessions was dominated by XI accessions with a wild-type allele, whereas another major haplotype with the second largest number of accessions was dominated by GJ accessions carrying either a c.468_500del33 or c.833_834del2 allele (Fig. 5A). We identified six major haplotypes for Hd2. Three haplotypes mainly consisted of XI accessions and two mainly GJ accessions. Wild-type haplotype was only detected in XI accessions. Three functional sites, c.1515_1522del8, M457V, and Y704H, were also uncovered in XI accessions, whereas the majority of GJ accessions carried the G420D allele. We found Hd5 had four major haplotypes, with two of them containing more than 800 accessions. One was found to contain mostly XI accessions with either a L19S or c.323delA allele, whereas the second haplotype contained mostly GJ accessions with either a wild-type or c.222G > T allele. Additionally, Hd6 had four major haplotypes, with 1398 and 945 accessions for the first two haplotypes. Out of 1398 accessions sharing the same haplotype carrying c.1809delG, 57% were XI accessions and 35% were GJ accessions. The second haplotype was mainly shared by XI accessions with the allele of c.1631delA&c.1809delG. The number of accessions for Hd6 wild-type allele was 48, and this small group of accessions was not considered as a major haplotype.
Figure 5.
Natural allelic variations in the Hd1, Hd2, Hd5, and Hd6 genes among 3000 rice genome accessions. (A) Haplotype network of four genes from 3010 rice accessions. Haplotype frequencies are proportional to the sizes of the circles. Each major haplotype is annotated with the functional sites. The total number of accessions used for haplotype analysis varies among four genes. The different colors represent the classification of rice accessions. (B) Geographic distribution of the four-gene combinations in rice accessions. The different colors represent different combinations of Hd1, Hd2, Hd5, and Hd6. The relative size of each pie indicates the ratio of accessions within each combination in a given country. (C) Geographic distribution of the multigene haplotypes with different slope values indicating the sensitivity to temperature. High slope, absolute value ≥ 2; low slope, absolute value < 2.
Geographic origins of the 3010 accessions were primarily composed of accessions from East Asia, Southeast Asia, and India (Fig. 5B; Supplemental Fig. S15). We identified a total of 158 haplotype combinations (Hd1: Hd2: Hd5: Hd6) by aggregating functional alleles from four genes (Supplemental Table S11). Ten combinations with more than 50 accessions were regarded as major haplotypes. The combination of WT: p.G420D: WT: c.1809delG dominated in the northern region, whereas the combination of WT: WT: p.L19S: X (X is designated to any haplotype of Hd6) accounted for South Asia. China and India contributed the largest numbers of accessions with different haplotype combinations: WT: p.G420D: WT: c.1809delG and WT: c.1515_1522del8: p.L19S: c.1631delA in China and WT: WT: p.L19S: c.1631delA and WT: WT: L19S: c1809delG&c.1631delA in India. To test the significance of genetic differentiation on the country-level, we assessed population genetic differentiation based on SNPs within the gene loci (Supplemental Table S12). The Gst value (Nei and Chesser 1983) was 0.55 from genetic differentiation between indica and japonica (Huang et al. 2012a). We obtained the mean Gst values of all pairwise comparisons among 89 countries, which were 0.43 (Hd1), 0.37 (Hd2), 0.45 (Hd5), 0.41 (Hd6), and 0.42 (overall across four genes). These numbers suggested a strong population differentiation and suggested that these genes were selected for local adaptation.
To connect the findings from two rice populations (the biparental population and the diverse accessions), we further classified haplotypes at these genes into two functional classes: WT and non-WT. With this designation, we were able to leverage the slope value of reaction norm obtained from the biparental population. In the biparental population, parent Koshihikari has the wild-type alleles for Hd1, Hd2, and Hd5 but the mutant allele for Hd6 (Supplemental Table S13). Accordingly, the slope value for WT was from the wild-type allele; for non-WT, from the mutant allele. As a result, the identified 158 major haplotype combinations in diverse rice genomes were categorized into 16 groups (24 = 16), each with a unique slope value obtained from the corresponding haplotype combination in the biparental population (Supplemental Fig. S11). From the geographic distribution of haplotypes, we observed that regions with lower mean annual temperature were dominated by haplotypes sensitive to temperature changes (absolute slope value ≥ 2), whereas regions with higher temperature had a majority of haplotypes less responsive to temperature changes (absolute slope value < 2) (Fig. 5C). This global distribution suggested that combinations of flowering-time plasticity genes have contributed to the rice expansion and adaptation to around 18 regions and 89 countries.
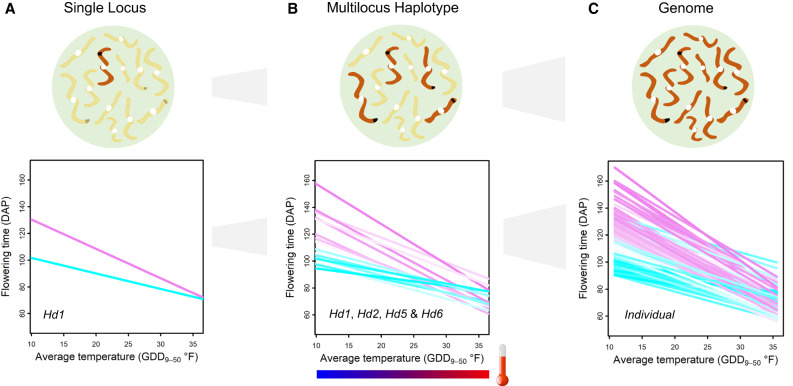
Multiple levels of reaction norms underlying phenotypic complexity
To encapsulate our understanding of phenotypic complexity, we diagramed multiple reaction norms along the temperature gradient using findings from the current study as an example. At the single-locus level, reaction norms of two homozygous genotypes are represented by two alleles of a major-effect gene (Fig. 6A). Within the range of environmental input, they may show a noncrossover gene-by-environment interaction like Hd1, a crossover interaction like Hd2, or not much interaction like Hd5 and Hd6. These patterns emerged out of sorting by individual locus agree with the reaction norms of genetic effects. Reaction norms at the multilocus haplotype level can be revealed by plotting the reaction norms of allelic combinations of major loci (Fig. 6B). Compared with a single locus, multilocus haplotypes account for both additive effects and interactions among genes, reflecting the combined effect from these loci in relevant pathways and networks. The haplotype combination from alleles with the higher slopes was most plastic along the environmental index; conversely, most stable (least responsive) was the haplotype combination from alleles with the lower slopes. Reaction norms of genotypes observed as individual organisms involved both major genes and many other background genes on the genome (Fig. 6C).
Figure 6.
A conceptual model to explain phenotypic complexity using reaction norms at multiple levels with varied environmental inputs. (A) Reaction norms at the single-locus level to temperature changes. Two homozygous genotype classes are represented by two alleles for the gene Hd1. (B) Reaction norms at the multilocus haplotype level to temperature changes. The 24 = 16 haplotype (homozygous genotype) classes are shown for four genes (Hd1, Hd2, Hd5, and Hd6). (C) Reaction norms at the genome level observed as individual organisms to temperature changes.
Reaction norm at the genome-wide marker effect level has a similar general pattern, representing the outcome of a genome-wide approach to partition the observed phenotypic variation, different from other reaction norms. In our study, this series of reaction norms constitute the systematic view of the interplay between genetics and environment in generating the observed phenotypic variation at different resolution levels (Figs. 1, 2, 4, 6; Supplemental Figs. S10–S12).
Discussion
Uncovering genetic architecture and molecular mechanisms of complex traits is important research in biology, evolution, agriculture, and medical science (Mackay et al. 2009; Marais et al. 2013). This current study showcased the benefit of pattern discovery in natural environments to explore the interdependent relationship of genetics and environments behind phenotypic plasticity. Specifically, temperature at the early growth stage was found to differentiate the nine environments in which this rice population was grown, and an explicit environmental index was identified to explain, model, and predict rice flowering time. More importantly, changes in size and direction of genetic effects were systematically revealed with reaction norms along the temperature gradient. Genes known to respond to day length changes were found to respond to temperature in these long-day environments. This can be explained by the interconnected pathways in plants in processing different environmental cues (Blackman 2017; Scheres and van der Putten 2017).
Unlike the earlier study in sorghum in which a single biparental population was examined (Li et al. 2018), in the current study, we uncovered the patterned geographic distributions of multigene haplotypes using the whole-genome sequencing data from a set of diverse rice accessions. By projecting the plasticity parameter value (the slope) obtained from the biparental population to the diverse accessions, we revealed that these multigene haplotypes with either a higher slope value (sensitive to temperature change) or a lower slope value (less responsive to temperature change) are preferentially distributed in different regions. Further research into the complex haplotype networks of flowering-time genes under broad agroecological conditions may eventually explain the patterns of global rice germplasm adaptation at a finer resolution.
Transition to flowering, a well-recognized trait of plasticity, is one of the most critical stages in the life cycle of a plant. In this study, the same group of genes were found to be underlying both reaction norm parameters, namely, intercept and slope, even though their effect sizes varied, similar to the earlier study in sorghum (Li et al. 2018). In both cases, the identified environmental index explained a large proportion of variation in environmental mean. Therefore, it appears that a set of genes from overlapping pathways and networks are underlying phenotypic plasticity, but the exact dynamics may differ depending on different input values of the primary environmental factor(s) differentiating these natural field conditions with respect to flowering time. Distinct genetic architectures for intercept and slope detected in a recent maize study (Kusmec et al. 2017), in which environmental mean was used, may be owing to joint effects from multiple environmental indices or the high genetic diversity involved.
Building on earlier works about dynamic gene effects (Marais et al. 2013; Li et al. 2018) and the omnigenic model (Boyle et al. 2017), we propose a conceptual model to reveal the layers of complexity of a quantitative trait using a set of reaction norms along an environmental index: reaction norms of genetic effects at the single-locus level, reaction norms of genotypes at the single-locus level, reaction norms of genotypes at the multilocus combination level, reaction norms of genome-wide marker effect continua, and reaction norms of genotypes (observed as individuals). Our findings highlighted the need, as well as the gain in clarity, of quantifying the relevant environmental context when we define and estimate the effects of genes underlying complex traits (Marais et al. 2013). Besides these revealed patterns of genetics and environment interplay, we showed that accurate performance prediction can be achieved through the integrated approaches with genome-wide SNPs and the identified environmental index. Finally, we expect to see further integration of knowledge and approaches in studying the genotype–phenotype relationship from the detailed molecular mechanisms perspective (e.g., gene mapping and cloning, molecular and developmental biology, and genome editing) and the broad genomic and environmental diversity perspective (e.g., sequencing, genome-wide association studies, genomic prediction, high-throughput phenotyping, and crop model and physiology).
Assessing genetic effects along the environmental factors provides a way to optimize the utilization of genetic resources in plant breeding process. Abundant allelic variations have been observed at many loci in rice flowering time. Complementary to genome-wide prediction, quantifying the allelic effects and haplotype effects at multiple loci across different environmental gradients can help breeders fine-tune flowering time in rice cultivars with designed introgression or targeted editing (Zeng et al. 2017; Chen et al. 2019). In addition, resource allocation of testing environments can be optimized to better capture major environmental gradients. Besides flowering time, other agriculturally and economically important traits are also strongly affected by environmental conditions and can be examined using the same approaches. Besides temperature and day length, other quantifiable factors, such as soil properties, including soil water retention and plant available water, can also be examined and potentially factored into constructing the environmental index. Understanding the genetic and environmental mechanisms underlying complex traits helps improve crops to satisfy the demand for food supply for still increasing world population under climate change.
Methods
Genetic population and phenotyping
The genetic mapping population from Koshihikari and Kasalath was developed by the National Institute of Agrobiological Sciences Rice Genome Resource Center, Japan. Phenotype and genotype data were detailed in previous studies (Ma et al. 2002; Onogi et al. 2016) and are available at GitHub (https://github.com/Onogi/HeadingDatePrediction). Genotypes and a linkage map of 162 restriction fragment length polymorphism markers are available at https://www.rgrc.dna.affrc.go.jp/ineKKBIL182.html. All these markers are biallelic. This population of 174 BILs was evaluated in six experimental fields across 3 yr. The nine environments were: Tsukuba 2007 (TS07), Tsukuba 2008 early planting (TS08E), Tsukuba 2008 late planting (TS08L), Tsukuba 2009 (TS09), Ishikawa 2008 (ISA08), Fukuoka 2008 (FU08), Ishigaki 2008 (ISI08), Thai Nguyen 2008 (TH08), and Ha Noi 2008 (HA08) (Supplemental Table S1). Planting dates for these trials varied from March 31 to June 30. At each site, seeds were pregerminated in water and planted to seedling trays filled with soil. Seedlings were transplanted at the three-to-four-leaf stage. Heading dates were recorded for five plants from the middle of each row, the averaged value was recorded, and we used term flowering time to represent heading date. A randomized complete block design (RCBD) was applied in the field, each environment had two replications, and the average value was used for each line. Environmental mean of flowering time was calculated as the average heading date for the whole population at each environment.
Identifying the environmental index
Environment data for temperature and day length (based on civil twilight) were retrieved from the National Oceanic and Atmospheric Administration (NOAA: https://www.noaa.gov/weather) and the Astronomical Applications Department of the U.S. Naval Observatory (https://www.usno.navy.mil/USNO/astronomical-applications). Daily temperature (°F) was converted to GDD for rice with the formula: GDD = [(maximum temperature + minimum temperature) / 2] – 50.
We tested three environmental parameters: temperature (expressed as GDD), day length, and PTT (GDD × day length) (Supplemental Table S6). From each window during development, the mean value of each parameter was calculated first, and then its correlation with environmental mean of flowering time was obtained. Each window was from a starting day (i) to an end day (j), and both were expressed as DAP. The search was conducted with consecutive starting and ending days, and only windows before the trait expression were considered. The parameter–window combination with the highest correlation and reasonable biological interpretation was chosen as the final environmental index.
Genetic mapping and flowering-time genes
We conducted inclusive composite interval mapping with the software ICIM (Meng et al. 2015). A total of 162 markers were used. QTL mapping with additive and epistasis were conducted for flowering time in individual environments and for QEIs using multienvironment trials (Supplemental Methods; Li et al. 2006, 2015a).
Intercept and slope were calculated by regressing phenotypes of each individual onto the environmental index using R (R Core Team 2019). For mapping of reaction-norm parameters, intercept and slope were treated as phenotypes to detect QTL with ICIM. The significant thresholds were determined with 1000-time permutations.
Functional polymorphisms of four flowering-time genes (Hd1, Hd2, Hd5, and Hd6) underlying the detected QTLs for the two parental inbreds (Koshihikari and Kasalath) (Supplemental Table S13) were tabulated from literatures (Yano et al. 2000; Takahashi et al. 2001; Wei et al. 2010; Koo et al. 2013). From the 3000 Rice Genomes Project (Wang et al. 2018), we obtained the known or potential functional polymorphisms between two parents in four flowering-time genes (Supplemental Methods).
Additional information about reaction norms at multiple levels is presented in the Supplemental Methods.
Geographical distribution of haplotypes of flowering-time genes
Across 3010 rice accessions, polymorphisms (SNPs and small indels) for four genes (Hd1, Hd2, Hd5, and Hd6) were extracted from 3000 Rice Genomes Project (Wang et al. 2018; http://iric.irri.org/resources/3000-genomes-project). There were 143, 475, 282, and 184 SNPs within the gene regions for Hd1, Hd2, Hd5, and Hd6, respectively. After filtering with missing rate and minor allele frequency less than 0.05, the number of high-quality SNPs were 11, 74, 14, and 21 for Hd1, Hd2, Hd5, and Hd6, respectively. These high-quality SNPs were used to conduct haplotype determination and analyses by using R package “pegas” (Paradis 2010). The minimum number of shared accessions was set as 100 to select major haplotypes.
Documented functional sites (small insertions, deletions, and large structural variations) were compiled from literatures charactering Hd1, Hd2, Hd5, and Hd6 (Yano et al. 2000; Takahashi et al. 2001; Wei et al. 2010; Koo et al. 2013) to annotate the haplotypes constructed by SNPs in “pegas.”
The global temperature visualized in the geographic map was the mean annual temperature for the years 1970–2000 with R package “raster” and function “getData.” The temperature data were from the database “worldclim,” which collects global interpolated climate data.
Population genetic differentiation analysis
To test whether accessions with different haplotype combinations were selected for local adaptation, we grouped the 3010 accessions into 89 countries. The R package “mmod” was used to analyze population genetic differentiation (Winter 2012). We applied the function “diff_stats” to calculate three different statistics of differentiation using SNPs within the genes extracted from sequence data. The three statistics are heterozygosity of subpopulations (Hs), heterozygosity of the total population (Ht), and Nei's Gst (Nei and Chesser 1983), all of which were estimated for individual genes and four genes together (Supplemental Table S12).
JGRA for performance prediction
Performance prediction with JGRA was conducted for three scenarios: predicting the performance of tested genotypes in untested environments, predicting the performance of untested genotypes in tested environments, and predicting the performance of untested genotypes in untested environments (Supplemental Methods). Unlike the traditional joint regression analysis, JGRA involved performance prediction of individuals without performance data through genomics and performance prediction for untested environments through environmental index (Li et al. 2018).
JGRA through reaction-norm parameter approach obtained the intercept and slope by regressing an individual's performance on environmental index, and the connection of individuals with and without performance data was established with the genome-wide relationship (Supplemental Methods). JGRA through a genome-wide marker effect continuum approach obtained genome-wide marker effects at each environment, and these effects were then regressed on environmental index to obtain fitted values for prediction. JGRA is a generic framework that can be applied for input data sets with sizes ranging from small to large. Both genomic prediction with the genome-wide relationship and genome-wide marker effect estimation use rrBLUP as a default setting, which can be customized to accommodate other methods.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work is supported by ISU Plant Sciences Institute and ISU Raymond F. Baker Institute for Plant Breeding.
Author contributions: J.Y. and X.L. designed research; T.G., X.L., Q.M., J.W., A.V., A.O., and H.I. performed research; and T.G., X.L., and J.Y. wrote the paper with input from all other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.255703.119.
References
- Bernardo R, Yu J. 2007. Prospects for genomewide selection for quantitative traits in maize. Crop Sci 47: 1082–1090. 10.2135/cropsci2006.11.0690 [DOI] [Google Scholar]
- Blackman BK. 2017. Changing responses to changing seasons: natural variation in the plasticity of flowering time. Plant Physiol 173: 16–26. 10.1104/pp.16.01683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169: 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Huang X, Tian Z, Wing RA, Han B. 2019. The genomics of Oryza species provides insights into rice domestication and heterosis. Annu Rev Plant Biol 70: 639–665. 10.1146/annurev-arplant-050718-100320 [DOI] [PubMed] [Google Scholar]
- Eberhart ST, Russell W. 1966. Stability parameters for comparing varieties. Crop Sci 6: 36–40. 10.2135/cropsci1966.0011183X000600010011x [DOI] [Google Scholar]
- Finlay K, Wilkinson G. 1963. The analysis of adaptation in a plant-breeding programme. Aust J Agric Res 14: 742–754. 10.1071/AR9630742 [DOI] [Google Scholar]
- Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. 2010. Food security: the challenge of feeding 9 billion people. Science 327: 812–818. 10.1126/science.1185383 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. 2003. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. 10.1038/nature01549 [DOI] [PubMed] [Google Scholar]
- Hori K, Matsubara K, Yano M. 2016. Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor Appl Genet 129: 2241–2252. 10.1007/s00122-016-2773-4 [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wang Z-X, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, et al. 2012a. A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XH, Zhao Y, Wei XH, Li CY, Wang A, Zhao Q, Li WJ, Guo YL, Deng LW, Zhu CR, et al. 2012b. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44: 32–39. 10.1038/ng.1018 [DOI] [PubMed] [Google Scholar]
- Huang X, Yang S, Gong J, Zhao Y, Feng Q, Gong H, Li W, Zhan Q, Cheng B, Xia J, et al. 2015. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun 6: 6258 10.1038/ncomms7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. 2010. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638. 10.1038/ng.606 [DOI] [PubMed] [Google Scholar]
- Josephs EB. 2018. Determining the evolutionary forces shaping G × E. New Phytol 219: 31–36. 10.1111/nph.15103 [DOI] [PubMed] [Google Scholar]
- Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang ZY, Li JJ, Li ZC, Paek NC. 2013. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6: 1877–1888. 10.1093/mp/sst088 [DOI] [PubMed] [Google Scholar]
- Kusmec A, Srinivasan S, Nettleton D, Schnable PS. 2017. Distinct genetic architectures for phenotype means and plasticities in Zea mays. Nat Plants 3: 715–723. 10.1038/s41477-017-0007-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ye G, Wang J. 2006. A modified algorithm for the improvement of composite interval mapping. Genetics 175: 361–374. 10.1534/genetics.106.066811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang J, Zhang L. 2015a. Inclusive composite interval mapping of QTL by environment interactions in biparental populations. PLoS One 10: e0132414 10.1371/journal.pone.0132414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu H, Wang M, Liu H, Tian X, Zhou W, Lu T, Wang Z, Chu C, Fang J, et al. 2015b. Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China. J Integr Plant Biol 57: 698–707. 10.1111/jipb.12326 [DOI] [PubMed] [Google Scholar]
- Li X, Guo TT, Mu Q, Li XR, Yu JM. 2018. Genomic and environmental determinants and their interplay underlying phenotypic plasticity. Proc Natl Acad Sci 115: 6679–6684. 10.1073/pnas.1718326115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Shen R, Zhao Z, Wissuwa M, Takeuchi Y, Ebitani T, Yano M. 2002. Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant and Cell Physiology 43: 652–659. 10.1093/pcp/pcf081 [DOI] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. 2009. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10: 565–577. 10.1038/nrg2612 [DOI] [PubMed] [Google Scholar]
- Malosetti M, Ribaut JM, van Eeuwijk FA. 2013. The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front Physiol 4: 44 10.3389/fphys.2013.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais DLD, Hernandez KM, Juenger TE. 2013. Genotype-by-environment interaction and plasticity: exploring genomic responses of plants to the abiotic environment. Annu Rev Ecol Evol Syst 44: 5–29. 10.1146/annurev-ecolsys-110512-135806 [DOI] [Google Scholar]
- Matsubara K, Hori K, Ogiso-Tanaka E, Yano M. 2014. Cloning of quantitative trait genes from rice reveals conservation and divergence of photoperiod flowering pathways in Arabidopsis and rice. Front Plant Sci 5: 193 10.3389/fpls.2014.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch S, Baute GJ, Bradeen J, Bramel P, Bretting PK, Buckler E, Burke JM, Charest D, Cloutier S, Cole G, et al. 2013. Agriculture: feeding the future. Nature 499: 23–24. 10.1038/499023a [DOI] [PubMed] [Google Scholar]
- Meng L, Li H, Zhang L, Wang J. 2015. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. The Crop Journal 3: 269–283. 10.1016/j.cj.2015.01.001 [DOI] [Google Scholar]
- Millet EJ, Kruijer W, Coupel-Ledru A, Alvarez Prado S, Cabrera-Bosquet L, Lacube S, Charcosset A, Welcker C, van Eeuwijk F, Tardieu F. 2019. Genomic prediction of maize yield across European environmental conditions. Nat Genet 51: 952–956. 10.1038/s41588-019-0414-y [DOI] [PubMed] [Google Scholar]
- Nei M, Chesser RK. 1983. Estimation of fixation indices and gene diversities. Ann Hum Genet 47: 253–259. 10.1111/j.1469-1809.1983.tb00993.x [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, et al. 2010. Plant phenotypic plasticity in a changing climate. Trends Plant Sci 15: 684–692. 10.1016/j.tplants.2010.09.008 [DOI] [PubMed] [Google Scholar]
- Nuñez FD, Yamada T. 2017. Molecular regulation of flowering time in grasses. Agronomy 7: 17 10.3390/agronomy7010017 [DOI] [Google Scholar]
- Onogi A, Watanabe M, Mochizuki T, Hayashi T, Nakagawa H, Hasegawa T, Iwata H. 2016. Toward integration of genomic selection with crop modelling: the development of an integrated approach to predicting rice heading dates. Theor Appl Genet 129: 805–817. 10.1007/s00122-016-2667-5 [DOI] [PubMed] [Google Scholar]
- Paradis E. 2010. pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26: 419–420. 10.1093/bioinformatics/btp696 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Scheres B, van der Putten WH. 2017. The plant perceptron connects environment to development. Nature 543: 337–345. 10.1038/nature22010 [DOI] [PubMed] [Google Scholar]
- Shrestha R, Gómez-Ariza J, Brambilla V, Fornara F. 2014. Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann Bot-London 114: 1445–1458. 10.1093/aob/mcu032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M. 2001. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc Natl Acad Sci 98: 7922–7927. 10.1073/pnas.111136798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. 2007. Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. 10.1126/science.1141753 [DOI] [PubMed] [Google Scholar]
- Vergara BS, Chang TT. 1985. The flowering response of the rice plant to photoperiod. IRRI, Los Banos. [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. 2018. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557: 43–49. 10.1038/s41586-018-0063-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei XJ, Xu JF, Guo HN, Jiang L, Chen SH, Yu CY, Zhou ZL, Hu PS, Zhai HQ, Wan JM. 2010. DTH8 Suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153: 1747–1758. 10.1104/pp.110.156943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DJ. 2012. MMOD: an R library for the calculation of population differentiation statistics. Mol Ecol Resour 12: 1158–1160. 10.1111/j.1755-0998.2012.03174.x [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767. 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2483. 10.1105/tpc.12.12.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yamamoto E, Aya K, Takeuchi H, Lo PC, Hu L, Yamasaki M, Yoshida S, Kitano H, Hirano K, et al. 2016. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet 48: 927–934. 10.1038/ng.3596 [DOI] [PubMed] [Google Scholar]
- Yeang HY. 2013. Solar rhythm in the regulation of photoperiodic flowering of long-day and short-day plants. J Exp Bot 64: 2643–2652. 10.1093/jxb/ert130 [DOI] [PubMed] [Google Scholar]
- Zeng D, Tian Z, Rao Y, Dong G, Yang Y, Huang L, Leng Y, Xu J, Sun C, Zhang G, et al. 2017. Rational design of high-yield and superior-quality rice. Nat Plants 3: 17031 10.1038/nplants.2017.31 [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Hu W, Shen GJ, Liu HY, Hu Y, Zhou XC, Liu TM, Xing YZ. 2017. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep-Uk 7: 5388 10.1038/s41598-017-05873-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, et al. 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2: 467 10.1038/ncomms1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.