Abstract
Aging varies among individuals due to both genetics and environment, but the underlying molecular mechanisms remain largely unknown. Using a highly recombined Saccharomyces cerevisiae population, we found 30 distinct quantitative trait loci (QTLs) that control chronological life span (CLS) in calorie-rich and calorie-restricted environments and under rapamycin exposure. Calorie restriction and rapamycin extended life span in virtually all genotypes but through different genetic variants. We tracked the two major QTLs to the cell wall glycoprotein genes FLO11 and HPF1. We found that massive expansion of intragenic tandem repeats within the N-terminal domain of HPF1 was sufficient to cause pronounced life span shortening. Life span impairment by HPF1 was buffered by rapamycin but not by calorie restriction. The HPF1 repeat expansion shifted yeast cells from a sedentary to a buoyant state, thereby increasing their exposure to surrounding oxygen. The higher oxygenation altered methionine, lipid, and purine metabolism, and inhibited quiescence, which explains the life span shortening. We conclude that fast-evolving intragenic repeat expansions can fundamentally change the relationship between cells and their environment with profound effects on cellular lifestyle and longevity.
Aging is a progressive decline in biological functions occurring in almost all living organisms that ultimately leads to death (Finch 1990; Jones et al. 2014). The first life span regulating genes were identified in the beginning of the 1990s (Johnson 1990; Kenyon et al. 1993; Sun et al. 1994). Today, hundreds have been uncovered (Kenyon 2010), although most are of small effect and few explain aging variation between individuals. Besides genetics, environmental factors, such as calorie restriction (CR) (Klass 1977; Weindruch et al. 1986; Jiang 2000; Pletcher et al. 2002; Colman et al. 2009), reduced oxygen exposure (Rascon and Harrison 2010; Leiser et al. 2013), and low temperature (Sestini et al. 1991; Conti et al. 2006; Leiser et al. 2011), extend longevity. How genetics and environment interact to control variation in life span and by which mechanisms remains poorly understood. The beneficial effect of calorie restriction on longevity in organisms ranging from yeast (Lin et al. 2000) to primates (Mattison et al. 2017) has been known for >80 yr (McCay et al. 1935) and is still the most successful intervention to delay aging, although its impact on life span has sometimes been disputed (Liao et al. 2010; Schleit et al. 2013). Cellular mediation of CR is at least in part occurring through nutrient-sensitive signaling networks, including the insulin/IGF1, MTOR (target of rapamycin), cAMP-PKA, and AMPK pathways. These regulate life span by controlling stress responses, mitochondrial respiration, redox homeostasis, genome stability, autophagy, energy, and fat metabolism (Schulz et al. 2007; Weinberger et al. 2007; Hansen et al. 2008; Madia et al. 2008; Wei et al. 2008; Alvers et al. 2009; Molin et al. 2011; Yuan et al. 2012). Pharmaceutical control of some of these pathways can extend longevity in model organisms. Rapamycin, a clinically approved TOR inhibitor (Eisenberg et al. 2009; Harrison et al. 2009; Martin-Montalvo et al. 2013; De Haes et al. 2014), extends life span by mimicking CR (Blagosklonny 2010), but undesirable side effects in humans restrict its usage (Kaeberlein 2014).
The budding yeast Saccharomyces cerevisiae has been pivotal in elucidating mechanisms regulating aging. Yeast aging can be studied through two approaches: replicative life span (RLS) and chronological life span (CLS). Replicative lifespan is the number of mitotic divisions before senescence and is used as a paradigm to study aging of proliferative tissues, such as stem cells (Mortimer and Johnston 1959; Steinkraus et al. 2008). Chronological life span is the time yeasts survive in nonproliferative conditions and models the aging of post-mitotic cells, such as neurons (Longo et al. 1996, 2012). Hundreds of genes whose disruption affects the CLS of lab-domesticated yeast in calorie-rich (Powers et al. 2006; Fabrizio et al. 2010; Garay et al. 2014) and calorie-restricted (Matecic et al. 2010; Campos et al. 2018) environments have been identified. However, most studies relied on artificial gene deletions and were performed in lab-domesticated strains, which are highly atypical (Warringer et al. 2011), maintained as haploids rather than diploids (Peter et al. 2018), carry auxotrophies that alter life span (Gomes et al. 2007; Boer et al. 2008), and have never been exposed to natural selection (Kaya et al. 2015). Thus, genetic variants that control natural life span variation are still largely unknown. Crosses between natural yeast strains have the potential to uncover these variants (Brem 2002; Steinmetz et al. 2002) but remain poorly explored. Previous work linked natural polymorphisms in the ribosomal DNA and in the sirtuin SIR2 (Stumpferl et al. 2012; Kwan et al. 2013), as well as telomere maintenance (Kwan et al. 2011) and serine biosynthesis (Jung et al. 2018) to life span variation. Lack of genetic diversity, mapping resolution, and power has prevented more exhaustive exploration. Here, we leveraged a highly recombined S. cerevisiae population derived from natural isolates with the aim to map genetic variants controlling yeast chronological life span. We generated a thousand unique diploid individuals and monitored their longevity with and without treatments promoting life span to provide a detailed portrait of naturally occurring yeast life span variants.
Results
Calorie restriction and rapamycin extend life span through different genetic variants
We crossed a long-lived North American (NA) oak tree bark strain (YPS128) with a short-lived West African (WA) palm wine strain (DBVPG6044) which differ at 0.53% of nucleotide sites (Liti et al. 2009; Parts et al. 2011). A pool of F12 segregants of opposite mating types were then mated to generate 1056 diploids with hybrid, phased genomes, termed Phased Outbred Lines (POLs) (Hallin et al. 2016). POLs were individually cultivated in calorie-rich (synthetic dextrose complete, SDC), calorie restricted, or rapamycin-supplemented (RM) environments for the whole experiment, and viability was measured by high throughput flow cytometry at 7, 21, and 35 d after media exhaustion. A total of 52,466 genetic markers were called and used to run a genome-wide linkage analysis.
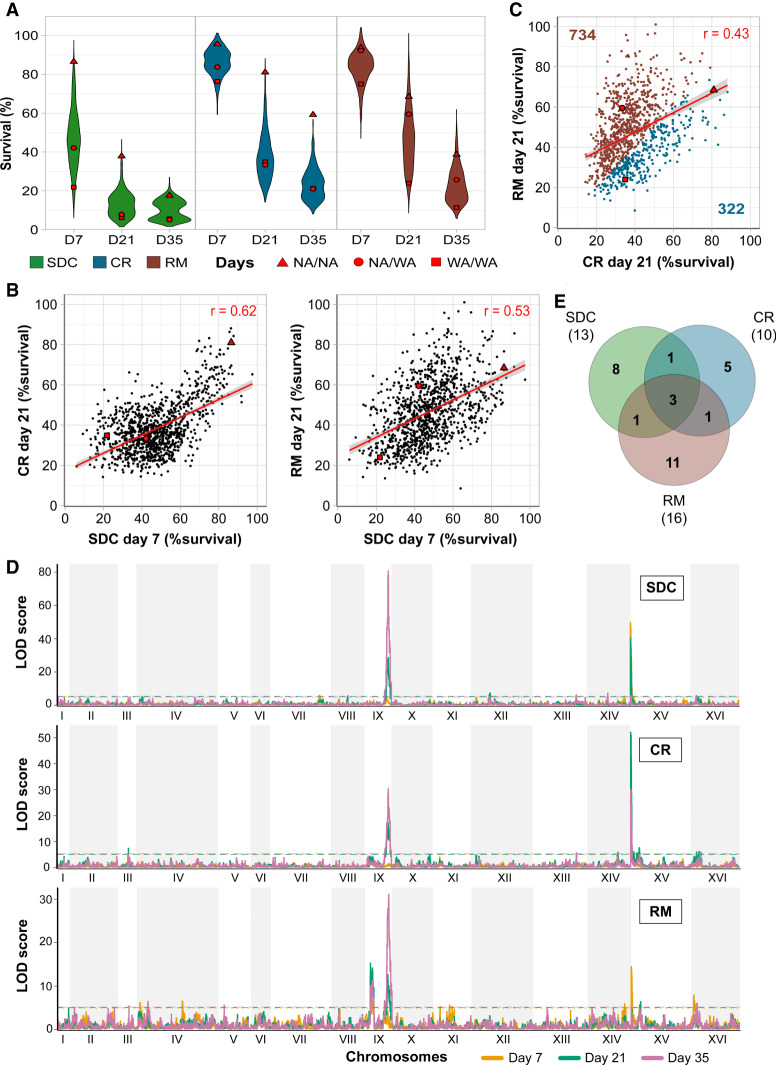
We found a broad lifespan diversity (Fig. 1A; Supplemental Table S3), with survival rates ranging from 6% to 97% already after 7 d in SDC (47% mean viability). Calorie restriction (86% mean viability at day 7) and RM (83% mean viability at day 7) extended life spans of all genotypes (with a single exception in RM) (Fig. 1A; Supplemental Fig. S1A). Life span was fairly correlated across environments (Pearson's r = 0.62 for SDC vs. CR, 0.53 for SDC vs. RM) (Fig. 1B), implying that CLS is mainly regulated by shared genetic effects across environments. Nevertheless, the more modest correlation between CR and RM (Pearson's r = 0.43) (Fig. 1C) suggested that they extend life span through partially distinct mechanisms.
Figure 1.
Calorie restriction and rapamycin extend life span through different genetic variants. Chronological life span of 1056 diploid segregant lineages from an F12 NA/WA advanced intercross. CLS was measured by counting viable cells (%) 7, 21, and 35 d after entry into quiescence, following growth in calorie-sufficient (SDC), -restricted (CR), and rapamycin (RM) media. Red: Founder homozygote parents (NA/NA, WA/WA) and their F1 hybrid (NA/WA). (A) CLS distributions across time points and conditions. (B) Comparing CLS across environments and time points. Red line: linear regression, with 95% confidence interval. (C) CLS comparison in RM and CR. Numbers: lineages living longer in CR (blue) or RM (brown). (D) Linkage analysis of CLS. Panels: calorie-rich (top), -restricted (middle), and rapamycin (bottom) media. Line color: 7 (yellow), 21 (green), and 35 (purple) days after entry into quiescence. y-axis: LOD score, x-axis: genome position. Dashed lines: Significance QTL (α = 0.05). (E) QTLs private to and shared between environments. Numbers in parentheses: total QTLs per environment.
We found a total of 30 unique QTLs associated with chronological aging (Fig. 1D; Supplemental Fig. S1B) that explained up to 40% of life span variation. QTLs were mostly private to one environment, and only three were detected in all (Fig. 1E). Two of these, located on Chr IX and Chr XV, were stronger than others and explained up to ∼30% and ∼20% of life span variation, respectively (Supplemental Table S4). Although both major QTLs were ubiquitous, the Chr XV QTL was partially masked by RM treatment, whereas the Chr IX QTL became significant only at advanced age (days 21 and 35). Most of the remaining QTLs were time- and environment-dependent (Fig. 1E; Supplemental Fig. S1C) and explained much less (mean: ∼3%) of the life span variation (Supplemental Table S4). Thus, CLS was largely determined by a few, very strong QTLs that were shared across calorie-rich and calorie-restricted environments. Chronological life span was then fine-tuned by mechanisms private to each environment, although the conservative threshold for calling QTLs may lead us to somewhat underestimate the shared QTLs.
Natural variations in the cell wall glycoproteins Hpf1p and Flo11p control chronological life span
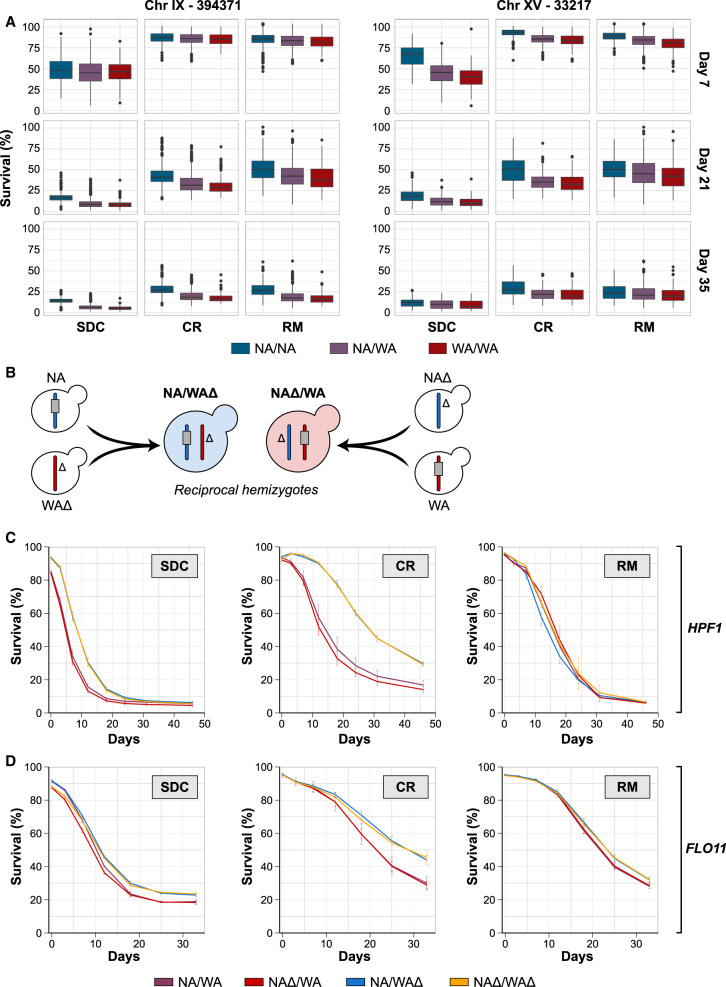
The two major QTLs peaked within FLO11 (Chr IX) and HPF1 (Chr XV). Both encode secreted cell wall glycoproteins with no known connections to life span. Hpf1p is functionally uncharacterized, while Flo11p regulates cell adhesion, pseudohyphae, and biofilm formation (Guo et al. 2000; Douglas et al. 2007; Váchová et al. 2011). We found a shorter life span for WA-FLO11 and HPF1 compared to NA homozygotes, with the WA short life span alleles being completely dominant (Fig. 2A). We validated these effects in a reciprocal hemizygosity assay (Fig. 2B; Methods; Steinmetz et al. 2002); a NA/WA hybrid deleted for the WA-HPF1 allele lived 70% longer, regardless of the presence of NA-HPF1 (Fig. 2C). Life span extension by rapamycin, but not by calorie restriction, rescued the WA-HPF1 induced life span shortening, consistent with the QTL being weaker in rapamycin (Fig. 2C,D). The WA-FLO11 also shortened life span but less than expected from the QTL strength, likely due to linkage or epistasis with other variants. As for HPF1, removing the WA-FLO11 or both the WA and NA-FLO11 alleles extended life span, while removing the NA-FLO11 had no effect (Fig. 2D). The negative effect of WA-FLO11 increased with age and was not rescued by rapamycin, again as expected from linkage analysis (Figs. 2D, 1D). Removing both alleles of either HPF1 or FLO11 extended the life span of WA/WA but not NA/NA homozygotes and had no effect in the domesticated reference strain, S288C (Supplemental Fig. S2A–C). We found one additional major RM-specific QTL located on the left arm of Chr IX. We tested four candidate genes (FKH1, ASG1, RPL16A, RPI1) located close to the QTL peak by reciprocal hemizygosity; however, allelic variation in these genes had no effect on CLS (Supplemental Fig. S3). The width of this QTL (40 kb, >30 genes) makes identifying the causative variants challenging.
Figure 2.
Natural allelic variations in the HPF1 and FLO11 control chronological life span. (A) Chronological life span of the 1056 POLs separated according to genotype at the markers with highest LOD score in each of the two major QTLs: 394,381 kb in Chromosome IX (in FLO11) and 33,217 kb in Chromosome XV (in HPF1). (B) Schematic representation of the NA/WA reciprocal hemizygosity design used to validate the CLS effect of the HPF1 and FLO11 WA alleles. Color: NA (blue) and WA (red) chromosomes. Gray rectangle: candidate gene (HPF1, FLO11), Δ: gene deletion. (C) Reciprocal hemizygosity. CLS of NA/WA hemizygotes for NA (blue; WAΔ) and WA (red; NAΔ) HPF1, heterozygote for HPF1 (purple; NA/WA), and lacking HPF1 (yellow; NAΔ/WAΔ). (D) As in C, but for FLO11.
Massive intragenic tandem repeat expansions within HPF1 shorten life span
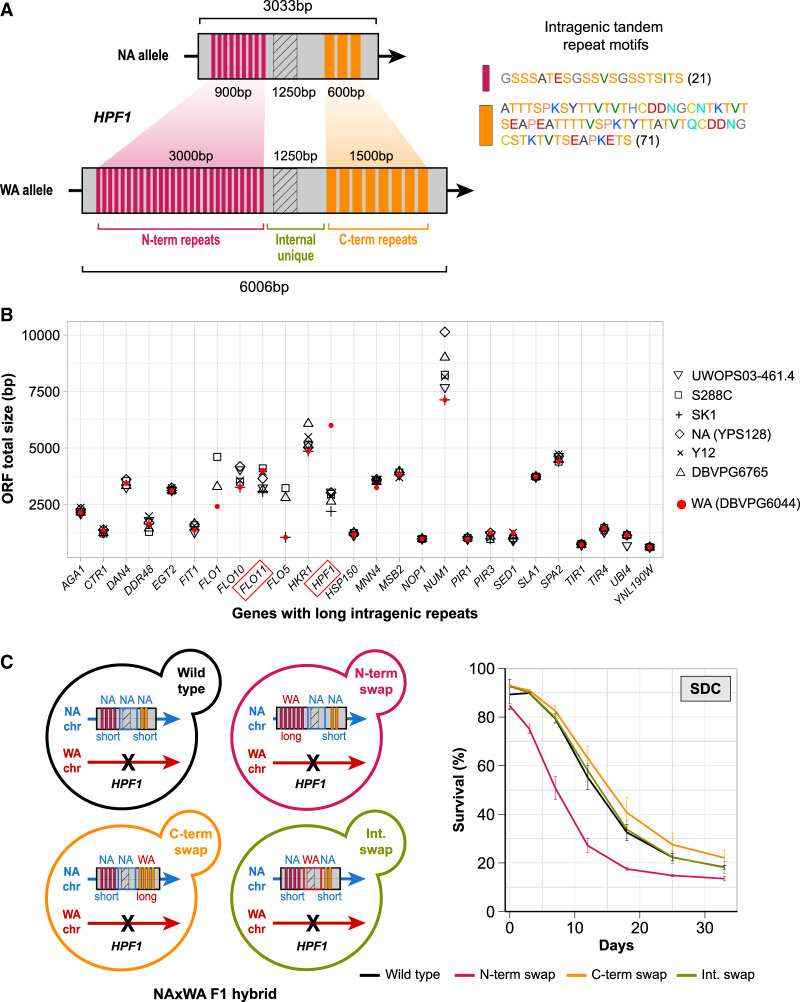
Inspecting complete genome assemblies, we found that FLO11 and HPF1 both carry intragenic tandem repeats that are expanded in the WA allele. WA-HPF1 is twice as long (6006 vs. 3033 bp) and WA-FLO11 is 10% longer (4014 vs. 3654 bp) than their NA counterparts (Fig. 3A; Supplemental Fig. S4A,B; Supplemental Table S6). Repeat motifs were between 21 and 71 amino acids and mainly composed of threonine and serine, as reported for repeat motifs in other cell wall proteins (Fig. 3A; Verstrepen and Klis 2006). The partial degeneration of WA-HPF1 and WA-FLO11 repeat motifs prevented pinpointing their exact patterns and boundaries. Such large repeat expansions in the WA isolate is specific to HPF1; of 26 genes containing very long tandem repeats (Verstrepen et al. 2005), only HPF1 was massively expanded in WA relative to six other strains for which complete genome assemblies exist (Fig. 3B; Yue et al. 2017).
Figure 3.
Massive intragenic tandem repeat expansions within HPF1 shorten life span. (A) Schematic representation of the intragenic repeats (colored rectangles) in HPF1 for NA and WA alleles. Hatched rectangle: a HPF1 internal unique region with high sequence variation between NA and WA alleles. Right: repeat motif units. Amino acids are colored according to the RasMol nomenclature. Numbers = motif size (amino acids). (B) Size variation of genes containing long intragenic repeats in seven diverged S. cerevisiae strains (Verstrepen et al. 2005; Yue et al. 2017). Diamonds: North American, red circles: West African. (C) Left panel: Design of allele swaps of HPF1 segments in the NA/WA F1 hybrid. The WA-HPF1 allele was deleted (black cross), while the NA-HPF1 was kept unchanged (wild type), or a segment was replaced by the corresponding WA-HPF1 segment. N-term: N-terminal repeats, C-term: C-terminal repeats, Int: internal unique region. Right panel: CLS for allele swapped constructs in SDC media.
We hypothesized that the WA-HPF1 massive repeat expansions explained the life span shortening and tested this by swapping HPF1 alleles in the F1 NA/WA hybrid. We removed WA-HPF1, while we engineered the remaining NA allele to contain specific segments of the WA allele (see Methods). Swapped segments corresponded to N- and C-terminal blocks of tandem repeats and to the highly polymorphic internal unique domain (Fig. 3A,C). Substituting the C-terminal repeats or the internal domain of NA-HPF1 with its WA counterpart did not shorten CLS, but replacing the N-terminal repeats shortened CLS as much as the native WA allele (Fig. 3C). Likewise, inserting the WA N-terminal repeats into a NA homozygote diploid shortened life span, while inserting the NA N-terminal repeats into a WA homozygote diploid extended its life span to a comparable extent (Supplemental Fig. S4C). We performed the same assay for FLO11, shifting the complete array of WA-FLO11 tandem repeats into the NA allele in the NA/WA hybrid background while removing WA-HPF1. In contrast to what we observed in HPF1, insertion of WA-FLO11 repeats had no effect on CLS, meaning that the life span shortening caused by WA-FLO11 is driven by variants other than its repeat expansions (Supplemental Fig. S4D).
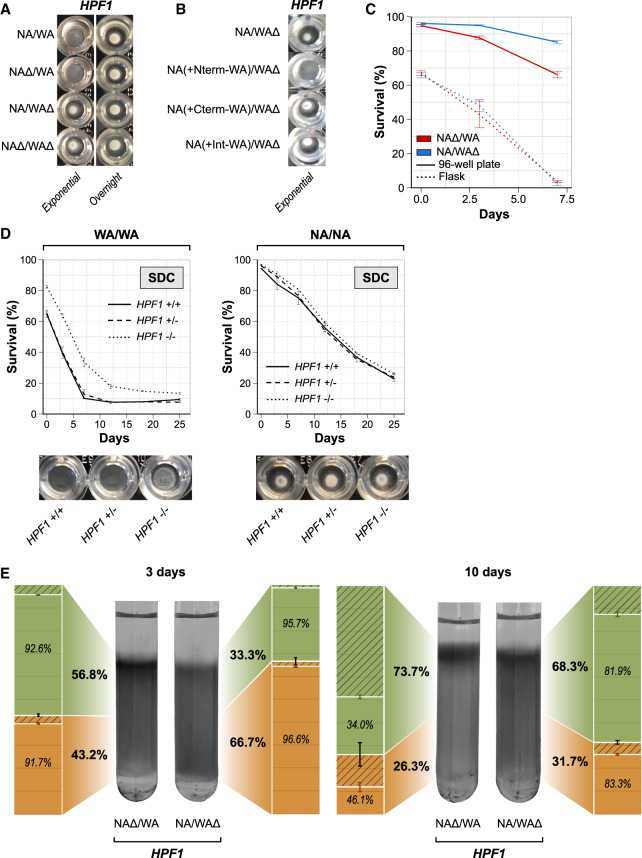
Buoyancy triggered by HPF1 N-terminal repeat expansion impairs quiescence
We observed that NA/WA hybrids carrying the WA-HPF1 became buoyant during exponential growth, i.e., they shifted to a free-floating lifestyle. Following entry into stationary phase, cells sedimented again, returning to a sedentary lifestyle (Fig. 4A). FLO11 alleles had no effect on buoyancy (Supplemental Fig. S5A). We showed that the shift to a buoyant lifestyle was caused by the WA-HPF1 N-terminal repeat expansion (Fig. 4B). To probe whether buoyancy per se shortens lifespan, we repeated the CLS assay under conditions of high aeration in intensely shaken flasks rather than in static 96-well plates. Because intense shaking forces all yeast cells to remain in suspension, we postulated that it would eliminate the HPF1 allelic effect only if it was due to buoyancy. In line with this assumption, we found that buoyancy enforced by shaking reduced life span and completely negated the effect of HPF1 allelic variation on life span (Fig. 4C). A shorter life span in shaking cultures has previously been explained as a result of higher exposure to oxygen (Longo et al. 1999; Fabrizio et al. 2003), although a too low access to oxygen also impairs the life span by reducing mitochondrial respiration (Ocampo et al. 2012). We therefore probed whether increased oxygen exposure as a consequence of a buoyant lifestyle could explain the shorter life span of WA-HPF1 cells. We cultivated cells with WA and NA HPF1 alleles, respectively, in static, sealed tubes and varied the volume of air in each tube. Cultivation with lower air volume, and therefore with lesser oxygen exposure, completely suppressed the shorter life span of cells with the WA-HPF1 allele (Supplemental Fig. S5B). Thus, by promoting cellular buoyancy in static cultures, WA-HPF1 exposes cells to higher oxygen levels and shortens life span. Neither secretion of Hpf1p to the medium nor variations in medium acidity had any life span effect (Supplemental Fig. S5C,D). Complete HPF1 removal in the parental WA homozygote nullified the shift to a buoyant lifestyle and increased CLS, while HPF1 removal affected neither buoyancy nor life span in the parental NA homozygote background (Fig. 4D).
Figure 4.
Buoyancy triggered by HPF1 N-terminal repeat expansions shortens life span. (A,B) Buoyancy of cells cultivated for 7 h (exponential phase) or overnight in calorie-rich medium in a 96-well plate. (A) HPF1 hemizygotes. (B) HPF1 allele swaps (as described in Fig. 3C). (C) Comparing CLS of HPF1 hemizygote cells cultivated in shake flasks and 96-well plates. Shake flasks had a 1:5 medium/volume ratio and were shaken at 220 rpm. Ninety-six-well plates were filled with 200 µL medium, with no shaking. (D) CLS and buoyancy (96-well plates; exponential phase) of WA and NA homozygotes parents with no (full line), 1 (dashed lines), or both copies (dotted lines) of HPF1 deleted in calorie-rich medium. (E) Percoll density gradients with HPF1 hemizygotes incubated in SDC media in a 96-well plate for either 3 d (left panel) or 10 d (right panel). The upper (nonquiescent cells) and lower (quiescent cells) phases were isolated by pipetting. The fraction (bold) and viability (italics) of cells in each phase were measured by flow cytometry (bar plots). Green: Upper/nonquiescent fraction, orange: lower/quiescent fraction, hatched area: dead cell fraction.
Higher external oxygen increases intracellular superoxide anion levels and thereby prevents cells from arresting growth and entering into the aging-resistant quiescent state upon nutrient exhaustion (Weinberger et al. 2010). We therefore tested whether WA-HPF1-driven buoyancy impairs quiescence. We measured both the fraction and viability of quiescent cells in HPF1 reciprocal hemizygotes, using density gradients (Allen et al. 2006). We found a higher fraction of quiescent cells in the sedentary NA-HPF1 hemizygote than in the buoyant WA-HPF1 3 d post-inoculation (Fig. 4E). After 10 d, the number of quiescent cells decreased for both hemizygotes, as previously observed (Madia et al. 2009). At this stage, we found only a minor viability difference between upper and lower phases (Fig. 4E), possibly because the enrichment for quiescent cells in the lower phase is less pronounced in natural than in lab strains or because there may be multiple quiescence states with distinct cell properties (Klosinska et al. 2011; Miles et al. 2019).
Many cell wall proteins are involved in cell-cell cohesion, cell-surface adhesion, often through pseudohyphal growth, which could affect buoyancy (Lo and Dranginis 1998; Verstrepen and Klis 2006). However, we found no role of HPF1 in flocculation or pseudohyphal growth (Supplemental Figs. S5E, S6). Complete loss of HPF1 decreased surface adhesion in the NA homozygote parent (Supplemental Fig. S7). Because neither the WA homozygote nor the NA/WA hybrid adhered to surfaces, we could not test if HPF1 reduced surface adhesion in these backgrounds (Supplemental Fig. S7). Overall, the lack of these traits in the NA/WA hybrid background rules out their contribution to buoyancy and life span.
HPF1-induced buoyancy reprograms methionine, lipid, and purine metabolism
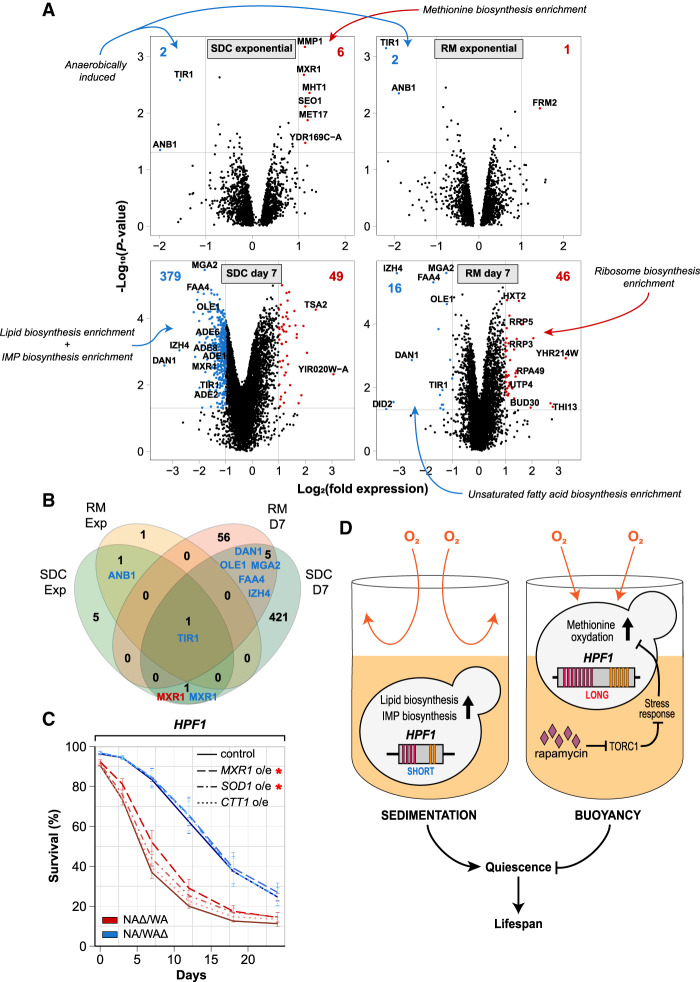
We hypothesized that buoyant WA-HPF1 cells exposed to higher oxygen experience altered cellular redox homeostasis and that this required reprogramming redox metabolism and gene expression. We therefore compared the transcriptomes of the two HPF1 reciprocal hemizygous hybrids before (exponential growth) and after (7 d) the onset of aging, with or without rapamycin exposure (Supplemental Table S5). During exponential growth, buoyancy and higher oxygen exposure induced by the WA-HPF1 only weakly affected relative transcript abundances (eight and three genes changing greater than twofold in calorie-rich and rapamycin media, respectively) (Fig. 5A,B). The known low oxygen responders TIR1 and ANB1 (Lowry and Lieber 1986; Cohen et al. 2001) were repressed by oxygen (i.e., in WA-HPF1). Five of the six transcripts induced by oxygenation encode proteins with a role in methionine metabolism (Fig. 5A), notably MXR1, which encodes a methionine-S-sulfoxide reductase known to control life span (Koc et al. 2004). In contrast, during chronological aging, the WA-HPF1 cells expressed less MXR1 (Fig. 5A,B). We overexpressed (over 10-fold on a transcript level) MXR1 and found this overexpression to partially rescue the short life span of WA-HPF1 cells (Fig. 5C). This suggests that the lower MXR1 expression in WA-HPF1 cells during chronological aging limited their life span. Overexpression of SOD1, encoding the mostly cytosolic superoxide dismutase, also improved the CLS of WA-HPF1 cells somewhat, while overexpression of the cytosolic catalase CTT1 did not (Fig. 5C). This further supports that the buoyant WA-HPF1 cells experience an increased oxidative burden, although this is not enough to cause oxidative distress and reduce growth (Supplemantal Fig. S8A). We found that methionine restriction extended and methionine supplementation shortened the life span of cells (Supplemental Fig. S8B), as shown before (Johnson and Johnson 2014; Ruckenstuhl et al. 2014). Neither of these regimens suppressed the effect of WA-HPF1-induced buoyancy, however, suggesting that their premature aging was neither due to excessive nor deficient intracellular methionine pools. Rapamycin supplementation nullified the induction of methionine metabolism and partly restored MXR1 expression, despite cells being buoyant, potentially explaining why rapamycin prevents the WA-HPF1 from shortening life span (Figs. 5A,B,D, 2C).
Figure 5.
HPF1-induced buoyancy reprograms methionine, lipid, and purine metabolism. (A) Transcriptome changes induced by WA-HPF1-dependent buoyancy. NA/WA hybrids hemizygotes for WA or NA-HPF1 were cultivated in calorie-rich (left panels) or rapamycin (right panels) medium, and RNA was extracted and sequenced from exponential phase (top panels) or aging (bottom panels; 7 d after entry into quiescence) cells. y-axis: −log10 (P-value), x-axis: log2 (HPF1 NAΔ/WA) – log2 (HPF1 NA/WAΔ). Blue: transcripts more (>2×, P < 0.05) abundant in HPF1 NA/WAΔ, red: transcripts more (>2×, P < 0.05) abundant in HPF1 NAΔ/WA. The total number of transcripts passing each criterion are reported (top corners, blue and red text). Gene Ontology classifications enriched among transcripts passing each criterion are indicated (blue and red arrows) (Supplemental Table S5). (B) Comparing the number of transcripts more (>2×, P < 0.05) abundant in HPF1 NA/WAΔ (blue) or in HPF1 NAΔ/WA (red) across environments (SDC and RM, in exponential phase and after 7 d of aging [D7]). (C) CLS of HPF1 reciprocal hemizygotes overexpressing (o/e) either MXR1 (dashed), SOD1 (dot-dashed), or CTT1 (dotted) in SDC media in 96-well plate. Red asterisk indicates significance (P < 0.05 unpaired Student's t-test) at 7 d (MXR1) and 12 d (MXR1 and SOD1). (D) Model for how the intragenic tandem repeat expansions in WA-HPF1 shortens the life span by shifting cells from a sedentary (left) to a buoyant lifestyle (right), exposing them to more oxygen and causing mild oxidation of, in particular, methionine, which impairs entry into quiescence. Rapamycin prevents the accumulation of oxidized methionine, possibly through inhibiting TORC1 and through the activation of stress response genes.
In sharp contrast to before the onset of aging, we found the transcriptome to be fundamentally reprogrammed in WA-HPF1 cells during aging (428 and 62 genes changing greater than twofold, in calorie-rich and rapamycin media) (Fig. 5A,B). The high oxygen exposure broadly repressed lipid and purine biosynthesis transcripts (Fig. 5A,D), whose expression is known to promote a long life span (Matecic et al. 2010; Garay et al. 2014; Handee et al. 2016; Arlia-Ciommo et al. 2018). OLE1, a known hypoxia responder encoding the fatty acid desaturase (Kwast et al. 1999), was much less expressed in the buoyant WA-HPF1 hemizygote. Adding one of its main products, oleic acid, improved the CLS of both hemizygotes but could not rescue the short life span of WA-HPF1-induced buoyancy (Supplemental Fig. S8C).
Discussion
We found calorie restriction to delay chronological aging for each of the 1056 yeast genotypes studied. This effect of calorie restriction is fully in line with reports on single gene knockouts, where calorie restriction extends chronological life span regardless of what gene is missing (Matecic et al. 2010). In contrast, calorie restriction only extends the replicative life span of approximately half of single gene knockouts, with many being negatively affected (Schleit et al. 2013). The diverging effects of calorie restriction on chronological and replicative life span may reflect the natural yeast life cycle; wild yeasts spend a significant part of their life span as starved, quiescent cells (Liti 2015; De Chiara et al. 2020) and survival in this state may be very strongly selected. In contrast, the mother-aged, extensively replicated state is extremely rare in natural yeast populations and unlikely to be under selection.
Except for the universally positive effects of calorie restriction, we found chronological life span to be extremely genotype-dependent, with a total of 30 distinct QTLs explaining up to 41% of life span variation. Among those, two major QTLs drove most of the life span variation (∼30% and ∼20%), while the remaining QTLs were more time- and environment-specific and contributed less (∼3% each). We linked the two major QTLs to the cell wall encoding genes FLO11 (Chr IX) and HPF1 (Chr XV), with West African alleles having a pronounced and dominant life span shortening effect. The cell wall integrity pathway reacts to cell wall perturbations, induced by, e.g., heat shock or starvation (Krause and Gray 2002), and regulates both chronological and replicative life spans (Kaeberlein and Guarente 2002; Ray et al. 2003; Stewart et al. 2007; Matecic et al. 2010). We found that the life span shortening induced by the WA-HPF1 was not due to cell wall damage but to a shift in lifestyle. Cells carrying a North American HPF1 had long, sedentary lives in sedimented yeast populations, while cells carrying a WA-HPF1 lived shorter lives as buoyant, free-floating yeasts. The buoyancy directly caused the shorter life span, since forcing cells carrying the NA-HPF1 to a free-floating state through vigorous shaking shortened the life span to the same levels as that of cells carrying the WA-HPF1. In contrast, we observed no buoyancy effect of FLO11 variation, although FLO11-induced buoyancy has been previously reported (Fidalgo et al. 2006).
Buoyancy likely shortens life span by shifting cells from a semi-anaerobic state within yeast sediments to a highly oxygenated state while floating. Higher oxygen exposure implies increased levels of oxidized molecules and a reprogramming of the cellular redox homeostasis. We found hypoxic genes to be down-regulated in buoyant yeasts, while systems dealing with methionine oxidation and metabolism, in contrast, were broadly induced during the exponential phase. Nonoxidized methionine is essential for proper folding and function of proteins, synthesis of the central signaling molecule S-adenosine methionine, and the maintenance of glutathione pools, a key redox buffer (Brown-Borg and Buffenstein 2017). However, neither methionine supplementation nor restriction helped cells overcome the impact of buoyancy on life span. Because WA-HPF1 cells both grew and expressed canonical oxidative stress responders normally, buoyancy impaired life span without causing detectable oxidative distress. One explanation is that cells experience very mild elevations in oxidation levels: enough to trigger a ROS signal but not enough to impose oxidative distress. Evidence does point to a role of oxidation in regulating cellular signaling that is partially disconnected from the handling of oxidative distress (Hanzén et al. 2016). For instance, upon exposure to a superoxide-generating agent, cells express 100-fold more Sod1p than is required for handling the associated distress, and this serves to promote Sod1p signaling through the yeast casein kinase Yck1p and through Sod1p translocating to the nucleus, where it serves as a transcription factor (Reddi and Culotta 2013; Tsang et al. 2014). At the end of the growth phase, cells sediment and become sedentary regardless of which HPF1 allele they carry. Many yeast cells at this stage of their life cycle enter a quiescent state in which life span often is longer (Allen et al. 2006; Aragon et al. 2008; Davidson et al. 2011). WA-HPF1 cells exhibit a reduced tendency to do so. Higher levels of superoxide anions impede entry into quiescence (Weinberger et al. 2010), and a mild elevation of superoxide anions could explain why WA-HPF1 cells are less prone to enter quiescence. Increased methionine oxidation in WA-HPF1 cells may be a downstream consequence of such a mild superoxide elevation, and an attractive speculation is that the methionine oxidation serves as the quiescence-inhibiting signal. This would explain why increasing the expression of the methionine reductase gene MXR1 partially rescued the life span defect in WA-HPF1 cells. Rapamycin supplementation, which restored MXR1 expression upon chronological aging and rescued the short life span of WA-HPF1 cells, in spite of these cells still being buoyant, could impinge directly on this superoxide/methionine oxidation signaling. TORC1 inhibits the expression of stress response genes, and its repression by rapamycin may prevent the increased methionine oxidation and allow WA-HPF1 cells to enter quiescence normally (Fig. 5D). Furthermore, metabolomic analyses support extensive remodeling of methionine metabolism in aged flies (Avanesov et al. 2014), and the enzyme methionine sulfoxide reductase (Mxr) is a conserved modulator of the rate of aging in yeasts, flies, and mice (Moskovitz et al. 2001; Ruan et al. 2002; Kaya et al. 2010), suggesting that buoyancy-induced methionine oxidation in natural yeast strains impinges on a metabolic trait regulating aging in organisms from yeasts to mice.
HPF1 and FLO11, like the vast majority of genes encoding cell wall proteins, contain intragenic tandem repeats (Verstrepen et al. 2005). Intragenic tandem repeats are dynamic in size, both due to strand-slippage during replication and ectopic recombination (Pâques et al. 1998; Fan and Chu 2007), and were found to fuel rapid yeast evolution (Verstrepen et al. 2005; Gemayel et al. 2012). Here, we showed that expansion of the N-terminal intragenic repeats within WA-HPF1 was sufficient to shift yeasts toward a buoyant lifestyle, which reduced their life span. The majority (14/21) of amino acids in the expanded N-terminal repeat motif are serines or threonines, a huge overrepresentation compared to the 12% expected by their general prevalence in proteins (Kozlowski 2017). Serine and threonine are unique among amino acids in containing hydroxyl groups that directly facilitate hydrogen bonding with surrounding water molecules, an effect known to enhance the solubility of organic particles. An enticing possibility is therefore that the serine/threonine richness of the Hpf1 intragenic repeat expansion directly induces buoyancy. Furthermore, serine and threonine residues in cell wall and secreted proteins are highly O-glycosylated. Repeat expansion could thus increase cell wall glycosylation, which is predicted to improve solubility. The dynamic repeat expansions and contractions of HPF1 may serve as a lifestyle switch, allowing rapid evolutionary shifts between buoyant and sedentary lifestyles in evolution, as dictated by fluctuating, opposing selection pressures. We note that the hydrophobic/hydrophilic properties of the yeast cell wall have been linked to buoyancy before (Palmieri et al. 1996; DeSousa et al. 2003; Fidalgo et al. 2006).
The WA-HPF1 induced shift to a buoyant lifestyle in exponential phase, with a concomitant life span shortening, has no immediate parallels in higher multicellular organisms, such as humans. Nevertheless, stem cell fate and proliferation are determined by physical constraints and other environmental cues imposed by the surrounding extracellular matrix (Campisi 2001; Rando 2006; Guilak et al. 2009). For instance, fibroblast senescence can be reversed by culturing old cells in a young extracellular matrix (Choi et al. 2011). In addition, high oxygen exposure shortens the replicative life span of stem cells (Campisi 2001), while physiological levels of oxygen promote stem cell self-renewal (Topchiy et al. 2013; Sheshadri et al. 2015). Such cell-matrix interactions have been proposed to control human aging: the crosslinking theory of aging postulating that aging is a consequence of the progressive crosslinking of the extracellular matrix that impairs tissue homeostasis (Bjorksten 1968). It is quite likely that tandem repeats have a role in mediating these and other cell-environment interactions of relevance to human health and development. Intragenic tandem repeats occur in 17% of the genes in the human genome (Legendre et al. 2007) and are enriched in genes encoding extracellular proteins (Legendre et al. 2007; Gemayel et al. 2010). Repeat expansions in the transmembrane glycoprotein Muc1 improve tumor cell adhesion to lung tissue and thereby contributes to metastasis (Ciborowski and Finn 2002). Tandem repeat expansions have also been associated with human diseases, such as Huntington's chorea, Fragile X syndrome (Orr and Zoghbi 2007; Gemayel et al. 2010), and progeroid syndromes, such as myotonic dystrophy (Meinke et al. 2018). Besides controlling cell-extracellular matrix interactions, these tandem repeat variations tune human gene expression (Gymrek et al. 2016; Quilez et al. 2016), control nucleosome positioning (Volle and Delaney 2012), regulate circadian clocks (Sawyer et al. 1997), affect organismal morphology (Fondon and Garner 2004), and drive the evolution of pathogenic bacteria (Stern et al. 1986). It is now recognized that intragenic tandem repeat polymorphisms explain parts of the missing heritability that have evaded detection in genome-wide association studies (Hannan 2018; Gardiner et al. 2019). We expect that the ongoing development of long-read sequencing, which allows the rapid detection of intragenic repeat polymorphisms, will help illuminate their roles in many other classes of phenotypic variation, as illustrated here.
Methods
Strains
Phased Outbred Lines were derived from a cross between a North American oak tree strain (YPS128) and a West African palm wine strain (DBVPG6044) (Liti et al. 2009). Heterothallic (ho::HygMX) ancestral parents carrying LYS2 or URA3 at the LYS2 locus (lys2::URA3) (Cubillos et al. 2009) were first mated to generate the NA/WA F1 hybrid (Supplemental Table S1). This F1 hybrid was used to generate a very large pool of progeny (106–107 cells) that was cycled through 12 rounds of alternating random mating, diploid selection, meiosis, sporulation, and haploid selection, resulting in a final pool of F12 outbred haploids (Parts et al. 2011). Eighty-six F12 haploid segregants of each mating type were randomly isolated from the outcrossed pool and sequenced, and their genotype was inferred using a set of 52,466 markers (Illingworth et al. 2013). The selected haploid F12 segregants genotypes allow systematic crossing (MATa, ura3::KanMX, ho::HygMX paired with MATα; ura3::KanMX; ho::HygMX; lys2::URA3) to generate the prototroph POLs as described (Hallin et al. 2016) with minor modifications. F12 segregants were randomly paired and mated (in liquid YPD) in 96-well plates, and 1056 unique diploids with known, phased genomes were then selected during three consecutive diploid selective cultivations on liquid minimal media. Diploids were arrayed, stored, and analyzed in 96-well plates, each plate containing eight (three each of NA/NA and WA/WA, and two of NA/WA) internal controls used for life span normalization (Supplemental Table S1).
Reciprocal hemizygotes at the HPF1 and FLO11 loci were constructed in a NA/WA diploid hybrid, using genetically tractable NA and WA haploids, as described (Cubillos et al. 2009). Native HPF1 and FLO11 genes were deleted in haploids by homologous recombination with a NatMX4 cassette, using the lithium acetate/PEG transformation protocol (Cubillos et al. 2009), before being mated to the appropriate counterpart to generate diploids hemi- or homozygote for HPF1 and FLO11. The same procedure was followed to build FKH1, ASG1, RPL16A, and RPI1 reciprocal hemizygotes.
HPF1 allele swapping was performed in two steps. First, part of HPF1 (N-terminal repeats, C-terminal repeats, or internal part) was deleted in NA and WA haploids using homologous recombination with a URA3 cassette. Then, the HPF1 segments to be swapped were PCR-amplified from the desired alleles with Platinum SuperFi (Thermo Fisher Scientific) DNA polymerase and swapped into the orthologous position of the recipient strain using homologous recombination (targeting identical nonrepeated sequences for both alleles) and selected on 5-FOA. Strains obtained were then mated to hpf1::NatMX4 haploids to generate the indicated diploids. The same procedure was followed to perform FLO11 allele swapping. A summary of strains and primers used in this study can be found in Supplemental Tables S1 and S2, respectively.
Media
YPD (1% yeast extract, 2% peptone, 2% dextrose, 2% agar [MP Biomedicals]) was used for all matings. Mated cells were streaked on synthetic minimal medium (2% dextrose [Sigma-Aldrich], 0.675% yeast nitrogen bases [Formedium], pH set to 6.0 with 2.5 M NaOH) to select for diploids. Counter selection of URA3 cells for allele swapping was made on 5-FOA plates (2% dextrose [Sigma-Aldrich], 0.675% yeast nitrogen base [Formedium], 0.088% uracil drop-out [Formedium], 0.005% uracil [Sigma-Aldrich], 2% agar, 0.1% 5-FOA [Sigma-Aldrich]). Pseudohyphal growth was induced on SLAHD plates (2% dextrose [Sigma-Aldrich], 1.7 g/L yeast nitrogen base without ammonium sulfate [Formedium], 6.6 mg/L ammonium sulfate [Formedium], 42 mg/L histidine [Sigma-Aldrich], 2% agar) (Hope and Dunham 2014).
Life span was estimated in: (1) calorie-rich synthetic dextrose complete (SDC) media (2% dextrose, 0.675% yeast nitrogen base [Formedium], 0.088% complete amino acid supplement [Formedium], pH set to 6.0 with 2.5 M NaOH); (2) calorie-restricted media (SDC as above, but with 0.5% dextrose instead of 2%) (Jiang 2000; Lin et al. 2000; Smith et al. 2007); and (3) rapamycin supplemented media (SDC supplemented with 0.025 µg/mL rapamycin [Sigma-Aldrich]) (Vázquez-García et al. 2017; Li et al. 2019). Methionine supplementation was performed in SDC supplemented with 500 mg/L methionine (15× Met), while methionine restriction was performed in SDC completely deprived of methionine (0× Met, complete amino acid supplement substituted with methionine drop-out). Oleic acid supplementation was performed in SDC supplemented with 0.1% oleic acid.
Chronological life span assay
Cells cultivated overnight in calorie-rich (SDC) media were diluted (1:100) in 200 µL of either fresh SDC, or CR, or RM media in a 96-well plate. Cultivation plates were sealed with adhesive aluminum foil to prevent evaporation and incubated at 30°C. Aging was considered to start at saturation of the culture, 72 h post-inoculation (Fabrizio and Longo 2007), and cells were kept in saturated media for the whole duration of the experiment unless otherwise specified. To generate hypoxia, overnight SDC cultures were diluted (1:100) and aged in different volumes (3, 15, 30, or 50 mL) of fresh SDC in static sealed FALCON tubes. When CLS was performed in water, 72-h stationary cultures were centrifuged, and cells were washed 3× before being resuspended and kept in the same volume of distilled water. For oleic acid and methionine supplementation or restriction, overnight SDC cultures were diluted (1:100) and aged in 200 µL of the appropriate SDC-based media (see Media subsection).
Aging was measured as viable cells (%) by flow cytometry based on the uptake of the fluorescent molecules propidium iodide (PI) and YO-PRO-1 iodide (YP). Propidium iodide and YO-PRO-1 are membrane-impermeable nucleic acid binding molecules that enter into necrotic but not into alive cells. Therefore, nonfluorescent cells are alive, while fluorescent cells are not. YO-PRO-1 penetrates also into apoptotic cells (Herker et al. 2004; Wlodkowic et al. 2009). At each aging time point (7, 21, 35 d after entry into quiescence unless otherwise stated), 5 µL of cells were transferred into 100 µL of staining solution (phosphate buffer saline + 3 µM propidium iodide [Sigma-Aldrich] + 200 nM YP [Thermo Fisher Scientific]) in a 96-well plate and incubated for 5 min in the dark at 30°C. The samples were analyzed on a FACS-Calibur flow cytometer (Becton Dickinson) using a High Throughput Sampler (Becton Dickinson) device to process 96-well plates and detect fluorescence with FL-1 (YP) and FL-3 (PI) channels. POLs experiments, where each allele is present and therefore replicated in many lineages, were run in single replicate, and viability estimates were normalized to those of eight internal controls run on the same plate. All other experiments were run at least in triplicate.
Linkage analysis
Linkage analysis was performed as described (Hallin et al. 2016). Briefly, mapping of life span QTLs was done using the normalized POL life spans, the R/qtl package in R (Broman et al. 2003), and the marker regression method in the scanone function. Significance thresholds were calculated with 1000 permutations to call QTLs with a significance level of 0.05. Confidence intervals for the peaks were calculated using a 1.8-LOD drop using the lodint function in R/qtl. We corrected for population structure by using the deviation of the life span of each POL from the parent mean.
Fraction and viability of quiescent cells
Isolation of quiescent cells was performed as described (Allen et al. 2006) with minor modifications. Cells were grown overnight in SDC and 1:100 diluted into 200 µL of fresh SDC in a 96-well plate. Each strain was distributed 96 times in the same 96-well plate and incubated for either 3 d or 10 d (1 strain per 96-well plate per time point). At the indicated time point, the 96 replicates of each strain were collected in a single FALCON tube, washed once with Tris buffer (50 mM, pH 8), and resuspended in 1 mL Tris buffer. Cells were then gently overlaid on the top of the preformed Percoll gradient (8 mL) and centrifuged at 400g at 20°C for 1 h. Upper and lower phases were equally split using a ruler and isolated by pipetting in distinct FALCON tubes. Ten milliliters of PBS were added into each tube prior to vortexing to homogeneously resuspend the cells. Two microliters of each tube were sampled and mixed with 100 µL of PI/YP staining solution, and viability was determined by cytometry as described in the chronological life span assay above. Cell concentration was given by the cytometer and used to calculate the ratio of quiescent/nonquiescent cells. The whole experiment was run with two independent replicates for each strain.
RNA-seq
We extracted RNA from SDC- and RM-cultivated cells in exponential phase and after 7 d of aging using the KAPA Biosystems Hyperplus kit with RiboErase, as per the manufacturer's instructions at 500 ng input. RNA integrity was assessed on an Agilent Bioanalyzer using the RNA Pico kit to determine RIN scores. Library quality was assessed using an Agilent Bioanalyzer using the DNA 1000 kit. Libraries were quantified using a KAPA Biosystems library quantification kit. The libraries were normalized, pooled, and loaded onto a NextSeq 500/550 High Output v2 kit (300 cycles) flow cell and sequenced on a NextSeq 500 instrument.
We performed transcript-level abundance quantification by pseudo-aligning RNA-seq reads to the coding sequences of a Saccharomyces Genome Database (SGD) yeast reference gene set, using kallisto (v0.44.0) (Bray et al. 2016). In this way, we obtained the transcripts per kilobase million (TPM) value for each gene in each sample as its normalized expression level, which is directly comparable, both among different genes and among different samples. Sleuth (0.30.0) (Pimentel et al. 2017) was further used to assess the statistical difference of the same gene between different samples by decoupling biological differences from experimental noise. False discovery rate (FDR; α = 0.05) adjustment (Benjamini and Hochberg 1995) was further applied for multiple test correction.
Total RNA was extracted from two biological replicates either during exponential growth or after 7 d of aging in SDC or in RM. Global expression level was analyzed by pairwise comparison of the NA/WA hybrids hemizygotes for WA and NA-HPF1 within each environment and time point to identify differentially expressed transcripts. Standard GO term analysis was performed on greater than twofold differentially expressed genes with the GO Term Finder tool available at SGD, with an FDR corrected α threshold of 0.01.
CRISPR-Cas9-mediated genomic insertion
The MXR1, SOD1, and CTT1 open reading frames were first cloned in frame with the TEF1 promoter and CYC1 terminator using BamHI and EcoRI restriction. The overexpression cassettes were then amplified with the primers 1492 and 1461 (Supplemental Table S2), containing 50-bp homology to target the HO locus, and 30-bp and 23-bp homology with TEF1p and CYC1t, respectively. The PCR product was provided as donor DNA. Genomic integration was mediated by CRISPR-Cas9 based on the pUDP004 single gRNA and Cas9 system (Addgene no. 101165) (Gorter de Vries et al. 2017). The amidase selection marker was swapped for nourseothricin (Nat) for antibiotic resistance by Gibson assembly using NEBuilder HiFi DNA Assembly of the amplified pUDP004 backbone, yielding the pL59-Nat plasmid. Two highly conserved gRNA sites in the HO gene were identified—gRNA1: TATTTTTATAAAGATTGGAG and gRNA2: TAAAGACATCGCAAACGTCA, 100% identical in all S. cerevisiae and S. paradoxus strains in the yeast population reference panel (Yue et al. 2017). These gRNAs were inserted into the BsaI-cut pL59-Nat plasmid by Gibson assembly of two synthesized fragments containing gRNA1 and gRNA2, respectively, yielding the pL90-Nat-2×HO plasmid. The reverse complement of the first six bases of the gRNA were included for splicing with the hammerhead ribozyme. For CRISPR-Cas9 editing, 500 ng of pL90-Nat-2×HO and 50 µL of donor DNA PCR reactions were transformed following the lithium acetate/PEG protocol (Cubillos et al. 2009). pL59-Nat and the pL90-Nat-2×HO were submitted to Addgene (139069 and 140465, respectively).
Pseudohyphal growth and surface adhesion
For pseudohyphal growth, yeasts were grown overnight in YPD, and single cells were dropped at precise locations on a SLAHD plate (see Media subsection) by micromanipulation. Cells were incubated at 30°C for 24 h before acquiring pictures. For surface adhesion, 10 µL of YPD overnight culture were dropped on a YPD plate and incubated at 30°C. After 6 d, yeast patches were washed with sterile water and smoothly rubbed with a finger.
Growth rate
Cells were grown overnight in SDC before being diluted 1:100 into 200 µL of fresh SDC in a 96-well plate and incubated at 30°C without shaking. Every 2 h, cells were resuspended by pipetting, and the optical density (600 nm) was measured using a Tecan Infinite M200 plate reader.
Software
Data analysis, plots, and statistical tests were performed with R (R Core Team 2019) and RStudio (https://www.rstudio.com). Figures were prepared using Adobe Illustrator CC and Adobe Photoshop CC. Genetic engineering, molecular cloning, and self-alignment of HPF1 and FLO11 were designed using Geneious R9.1.8 (https://www.geneious.com).
Data access
Raw RNA-seq data from this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA544860.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by Agence Nationale de la Recherche (ANR-11-LABX-0028-01, ANR-13-BSV6-0006-01, ANR-15-IDEX-01, ANR-16-CE12-0019, ANR-18-CE12-0004), Fondation de France (00087525), the Swedish Research Council (2014-6547, 2014-4605, and 2018-03638), and the Swedish Cancer Fund (2017-778). The PhD fellowship for B.P.B. was supported by La Ligue Contre le Cancer (GB-MA-CD-11287) and Fondation pour la Recherche Médicale (FDT20170437209). We also acknowledge the IRCAN Flow Cytometry Facility CytoMed.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.253351.119.
Freely available online through the Genome Research Open Access option.
References
- Allen C, Büttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, et al. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol 174: 89–100. 10.1083/jcb.200604072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA Jr, Aris JP. 2009. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 8: 353–369. 10.1111/j.1474-9726.2009.00469.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon AD, Rodriguez AL, Meirelles O, Roy S, Davidson GS, Tapia PH, Allen C, Joe R, Benn D, Werner-Washburne M. 2008. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol Biol Cell 19: 1271–1280. 10.1091/mbc.e07-07-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlia-Ciommo A, Leonov A, Beach A, Richard VR, Bourque SD, Burstein MT, Kyryakov P, Gomez-Perez A, Koupaki O, Feldman R, et al. 2018. Caloric restriction delays yeast chronological aging by remodeling carbohydrate and lipid metabolism, altering peroxisomal and mitochondrial functionalities, and postponing the onsets of apoptotic and liponecrotic modes of regulated cell death. Oncotarget 9: 16163–16184. 10.18632/oncotarget.24604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanesov AS, Ma S, Pierce KA, Yim SH, Lee BC, Clish CB, Gladyshev VN. 2014. Age- and diet-associated metabolome remodeling characterizes the aging process driven by damage accumulation. eLife 3: e02077 10.7554/eLife.02077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bjorksten J. 1968. The crosslinkage theory of aging. J Am Geriatr Soc 16: 408–427. 10.1111/j.1532-5415.1968.tb02821.x [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. 2010. Calorie restriction: decelerating mTOR-driven aging from cells to organisms (including humans). Cell Cycle 9: 683–688. 10.4161/cc.9.4.10766 [DOI] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. 2008. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci 105: 6930–6935. 10.1073/pnas.0802601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brem RB. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science (80-) 296: 752–755. 10.1126/science.1069516 [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Buffenstein R. 2017. Cutting back on the essentials: Can manipulating intake of specific amino acids modulate health and lifespan? Ageing Res Rev 39: 87–95. 10.1016/j.arr.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. 2001. From cells to organisms: Can we learn about aging from cells in culture? Exp Gerontol 36: 607–618. 10.1016/S0531-5565(00)00230-8 [DOI] [PubMed] [Google Scholar]
- Campos SE, Avelar-Rivas JA, Garay E, Juárez-Reyes A, DeLuna A. 2018. Genomewide mechanisms of chronological longevity by dietary restriction in budding yeast. Aging Cell 17: e12749 10.1111/acel.12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HR, Cho KA, Kang HT, Bin LJ, Kaeberlein M, Suh Y, Chung IK, Park SC. 2011. Restoration of senescent human diploid fibroblasts by modulation of the extracellular matrix. Aging Cell 10: 148–157. 10.1111/j.1474-9726.2010.00654.x [DOI] [PubMed] [Google Scholar]
- Ciborowski P, Finn OJ. 2002. Non-glycosylated tandem repeats of MUC1 facilitate attachment of breast tumor cells to normal human lung tissue and immobilized extracellular matrix proteins (ECM) in vitro: potential role in metastasis. Clin Exp Metastasis 19: 339–345. 10.1023/A:1015590515957 [DOI] [PubMed] [Google Scholar]
- Cohen BD, Sertil O, Abramova NE, Davies KJA, Lowry CV. 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res 29: 799–808. 10.1093/nar/29.3.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons A, Kemnitz JW, et al. 2009. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science (80-) 325: 201–204. 10.1126/science.1173635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti B, Sanchez-Alavez M, Winsky-Sommerer R, Morale MC, Lucero J, Brownell S, Fabre V, Huitron-Resendiz S, Henriksen S, Zorrilla EP, et al. 2006. Transgenic mice with a reduced core body temperature have an increased life span. Science 314: 825–828. 10.1126/science.1132191 [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Louis EJ, Liti G. 2009. Generation of a large set of genetically tractable haploid and diploid Saccharomyces strains. FEMS Yeast Res 9: 1217–1225. 10.1111/j.1567-1364.2009.00583.x [DOI] [PubMed] [Google Scholar]
- Davidson GS, Joe RM, Roy S, Meirelles O, Allen CP, Wilson MR, Tapia PH, Manzanilla EE, Dodson AE, Chakraborty S, et al. 2011. The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures. Mol Biol Cell 22: 988–998. 10.1091/mbc.E10-06-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chiara M, Barré B, Persson K, Chioma AO, Irizar A, Warringer J, Liti G. 2020. Domestication reprogrammed the budding yeast life cycle. bioRxiv 10.1101/939314 [DOI] [PubMed]
- De Haes W, Frooninckx L, Van Assche R, Smolders A, Depuydt G, Billen J, Braeckman BP, Schoofs L, Temmerman L. 2014. Metformin promotes lifespan through mitohormesis via the peroxiredoxin PRDX-2. Proc Natl Acad Sci 111: E2501–E2509. 10.1073/pnas.1321776111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSousa SR, Oliveira KF, Souza CS, Kilikian BV, Laluce C. 2003. Yeast flotation viewed as the result of the interplay of supernatant composition and cell-wall hydrophobicity. Colloids Surfaces B Biointerfaces 29: 309–319. 10.1016/S0927-7765(03)00019-5 [DOI] [Google Scholar]
- Douglas LM, Li L, Yang Y, Dranginis AM. 2007. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot Cell 6: 2214–2221. 10.1128/EC.00284-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. 2009. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11: 1305–1314. 10.1038/ncb1975 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. 2007. The chronological life span of Saccharomyces cerevisiae. Methods Mol Biol 371: 89–95. 10.1007/978-1-59745-361-5_8 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Liou LL, Moy VN, Diaspro A, Valentine JS, Gralla EB, Longo VD. 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, Giaever G, Nislow C, Longo VD. 2010. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet 6: e1001024 10.1371/journal.pgen.1001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Chu JY. 2007. A brief review of short tandem repeat mutation. Genomics, Proteomics Bioinforma 5: 7–14. 10.1016/S1672-0229(07)60009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Barrales RR, Ibeas JI, Jimenez J. 2006. Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci 103: 11228–11233. 10.1073/pnas.0601713103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. 1990. Longevity, senescence and the genome. Univ. of Chicago Press, Chicago. [Google Scholar]
- Fondon JW, Garner HR. 2004. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci 101: 18058–18063. 10.1073/pnas.0408118101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay E, Campos SE, González de la Cruz J, Gaspar AP, Jinich A, DeLuna A. 2014. High-resolution profiling of stationary-phase survival reveals yeast longevity factors and their genetic interactions. PLoS Genet 10: e1004168 10.1371/journal.pgen.1004168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner SL, Trompet S, Sabayan B, Boogaard MW, Jukema JW, Slagboom PE, Roos RAC, van der Grond J, Aziz NA. 2019. Repeat variations in polyglutamine disease–associated genes and cognitive function in old age. Neurobiol Aging 84: 236.e17–236.e28. 10.1016/j.neurobiolaging.2019.08.002 [DOI] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. 2010. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet 44: 445–477. 10.1146/annurev-genet-072610-155046 [DOI] [PubMed] [Google Scholar]
- Gemayel R, Cho J, Boeynaems S, Verstrepen KJ. 2012. Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes (Basel) 3: 461–480. 10.3390/genes3030461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes P, Sampaio-Marques B, Ludovico P, Rodrigues F, Leão C. 2007. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mech Ageing Dev 128: 383–391. 10.1016/j.mad.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Gorter de Vries AR, de Groot PA, van den Broek M, Daran JG. 2017. CRISPR-Cas9 mediated gene deletions in lager yeast Saccharomyces pastorianus. Microb Cell Fact 16: 222 10.1186/s12934-017-0835-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. 2009. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 5: 17–26. 10.1016/j.stem.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Styles CA, Feng Q, Fink GR. 2000. A Saccharomyces gene family involved in invasive growth, cell–cell adhesion, and mating. Proc Natl Acad Sci 97: 12158–12163. 10.1073/pnas.220420397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, Daly MJ, Price AL, Pritchard JK, Sharp AJ, et al. 2016. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet 48: 22–29. 10.1038/ng.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin J, Märtens K, Young AI, Zackrisson M, Salinas F, Parts L, Warringer J, Liti G, Visscher PM, Brown MA, et al. 2016. Powerful decomposition of complex traits in a diploid model. Nat Commun 7: 13311 10.1038/ncomms13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handee W, Li X, Hall KW, Deng X, Li P, Benning C, Williams BL, Kuo M-H. 2016. An energy-independent pro-longevity function of triacylglycerol in yeast. PLoS Genet 12: e1005878 10.1371/journal.pgen.1005878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan AJ. 2018. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet 19: 286–298. 10.1038/nrg.2017.115 [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. 2008. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4: e24 10.1371/journal.pgen.0040024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzén S, Vielfort K, Yang J, Roger F, Andersson V, Zamarbide-Forés S, Andersson R, Malm L, Palais G, Biteau B, et al. 2016. Lifespan control by redox-dependent recruitment of chaperones to misfolded proteins. Cell 166: 140–151. 10.1016/j.cell.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson EJ, Frenkel K, Carter CS, et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460: 392–395. 10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Jungwirth H, Lehmann KA, Maldener C, Fröhlich KU, Wissing S, Büttner S, Fehr M, Sigrist S, Madeo F. 2004. Chronological aging leads to apoptosis in yeast. J Cell Biol 164: 501–507. 10.1083/jcb.200310014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope EA, Dunham MJ. 2014. Ploidy-regulated variation in biofilm-related phenotypes in natural isolates of Saccharomyces cerevisiae. G3 (Bethesda) 4: 1773–1786 10.1534/g3.114.013250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth CJR, Parts L, Bergström A, Liti G, Mustonen V. 2013. Inferring genome-wide recombination landscapes from advanced intercross lines: application to yeast crosses. PLoS One 8: e62266 10.1371/journal.pone.0062266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JC. 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J 14: 2135–2137. 10.1096/fj.00-0242fje [DOI] [PubMed] [Google Scholar]
- Johnson TE. 1990. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science (80-) 249: 908–912. 10.1126/science.2392681 [DOI] [PubMed] [Google Scholar]
- Johnson JE, Johnson FB. 2014. Methionine restriction activates the retrograde response and confers both stress tolerance and lifespan extension to yeast, mouse and human cells. PLoS One 9: e97729 10.1371/journal.pone.0097729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones OR, Scheuerlein A, Salguero-Gómez R, Camarda CG, Schaible R, Casper BB, Dahlgren JP, Ehrlén J, García MB, Menges ES, et al. 2014. Diversity of ageing across the tree of life. Nature 505: 169–173. 10.1038/nature12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung PP, Zhang Z, Paczia N, Jaeger C, Ignac T, May P, Linster CL. 2018. Natural variation of chronological aging in the Saccharomyces cerevisiae species reveals diet-dependent mechanisms of life span control. Aging Mech Dis 4: 3 10.1038/s41514-018-0022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. 2014. Rapamycin and ageing: when, for how long, and how much? J Genet Genomics 41: 459–463. 10.1016/j.jgg.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Guarente L. 2002. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 160: 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya A, Koc A, Lee BC, Fomenko DE, Rederstorff M, Krol A, Lescure A, Gladyshev VN. 2010. Compartmentalization and regulation of mitochondrial function by methionine sulfoxide reductases in yeast. Biochemistry 49: 8618–8625. 10.1021/bi100908v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya A, Ma S, Wasko B, Lee M, Kaeberlein M, Gladyshev VN. 2015. Defining molecular basis for longevity traits in natural yeast isolates. npj Aging Mech Dis 1: 15001 10.1038/npjamd.2015.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. 2010. The genetics of ageing. Nature 464: 504–512. 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtlang R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- Klass MR. 1977. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev 6: 413–429. 10.1016/0047-6374(77)90043-4 [DOI] [PubMed] [Google Scholar]
- Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR. 2011. Yeast cells can access distinct quiescent states. Genes Dev 25: 336–349. 10.1101/gad.2011311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A, Gasch AP, Rutherford JC, Kim H-Y, Gladyshev VN. 2004. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc Natl Acad Sci 101: 7999–8004. 10.1073/pnas.0307929101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski LP. 2017. Proteome-pI: proteome isoelectric point database. Nucleic Acids Res 45: D1112–D1116. 10.1093/nar/gkw978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause SA, Gray JV. 2002. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr Biol 12: 588–593. 10.1016/S0960-9822(02)00760-1 [DOI] [PubMed] [Google Scholar]
- Kwan EX, Foss E, Kruglyak L, Bedalov A. 2011. Natural polymorphism in BUL2 links cellular amino acid availability with chronological aging and telomere maintenance in yeast. PLoS Genet 7: e1002250 10.1371/journal.pgen.1002250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, Raghuraman MK, Brewer BJ, Kennedy BK, Bedalov A. 2013. A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet 9: e1003329 10.1371/journal.pgen.1003329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Staahl BT, Poyton RO. 1999. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc Natl Acad Sci 96: 5446–5451. 10.1073/pnas.96.10.5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Pochet N, Pak T, Verstrepen KJ. 2007. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res 17: 1787–1796. 10.1101/gr.6554007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser SF, Begun A, Kaeberlein M. 2011. HIF-1 modulates longevity and healthspan in a temperature-dependent manner. Aging Cell 10: 318–326. 10.1111/j.1474-9726.2011.00672.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser SF, Fletcher M, Begun A, Kaeberlein M. 2013. Life-span extension from hypoxia in Caenorhabditis elegans requires both HIF-1 and DAF-16 and is antagonized by SKN-1. Journals Gerontol - Ser A Biol Sci Med Sci 68: 1135–1144. 10.1093/gerona/glt016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Vázquez-García I, Persson K, González A, Yue J-X, Barré B, Hall MN, Long A, Warringer J, Mustonen V, et al. 2019. Shared molecular targets confer resistance over short and long evolutionary timescales. Mol Biol Evol 36: 691–708. 10.1093/molbev/msz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. 2010. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9: 92–95. 10.1111/j.1474-9726.2009.00533.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science (80-) 289: 2126–2128. 10.1126/science.289.5487.2126 [DOI] [PubMed] [Google Scholar]
- Liti G. 2015. The fascinating and secret wild life of the budding yeast S. cerevisiae. eLife 4: e05835 10.7554/eLife.05835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W-S, Dranginis AM. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell 9: 161–171. 10.1091/mbc.9.1.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Gralla EB, Valentine JS. 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: mitochondrial production of toxic oxygen species in vivo. J Biol Chem 271: 12275–12280. 10.1074/jbc.271.21.12275 [DOI] [PubMed] [Google Scholar]
- Longo VD, Liou LL, Valentine JS, Gralla EB. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys 365: 131–142. 10.1006/abbi.1999.1158 [DOI] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy BK. 2012. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16: 18–31. 10.1016/j.cmet.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry CV, Lieber RH. 1986. Negative regulation of the Saccharomyces cerevisiae ANB1 gene by heme, as mediated by the ROX1 gene product. Mol Cell Biol 6: 4145–4148. 10.1128/MCB.6.12.4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Gattazzo C, Wei M, Fabrizio P, Burhans WC, Weinberger M, Galbani A, Smith JR, Nguyen C, Huey S, et al. 2008. Longevity mutation in SCH9 prevents recombination errors and premature genomic instability in a Werner/Bloom model system. J Cell Biol 180: 67–81. 10.1083/jcb.200707154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Wei M, Yuan V, Hu J, Gattazzo C, Pham P, Goodman M, Longo VD. 2009. Oncogene homologue Sch9 promotes age-dependent mutations by a superoxide and Rev1/Polζ-dependent mechanism. J Cell Biol 186: 509–523. 10.1083/jcb.200906011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, et al. 2013. Metformin improves healthspan and lifespan in mice. Nat Commun 4: 2192 10.1038/ncomms3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. 2010. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet 6: e1000921 10.1371/journal.pgen.1000921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, Ingram DK, Weindruch R, De Cabo R, Anderson RM. 2017. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun 8: 14063 10.1038/ncomms14063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. 1935. The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr 10: 63–79. 10.1093/jn/10.1.63 [DOI] [PubMed] [Google Scholar]
- Meinke P, Hintze S, Limmer S, Schoser B. 2018. Myotonic dystrophy—a progeroid disease? Front Neurol 9: 601 10.3389/fneur.2018.00601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles S, Li LH, Melville Z, Breeden LL. 2019. Ssd1 and the cell wall integrity pathway promote entry, maintenance, and recovery from quiescence in budding yeast. Mol Biol Cell 30: 2205–2217. 10.1091/mbc.E19-04-0190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin M, Yang J, Hanzén S, Toledano MB, Labarre J, Nyström T. 2011. Life span extension and H2O2 resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol Cell 43: 823–833. 10.1016/j.molcel.2011.07.027 [DOI] [PubMed] [Google Scholar]
- Mortimer R, Johnston J. 1959. Life span of individual yeast cells. Nature 183: 1751–1752. 10.1038/1831751a0 [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. 2001. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci 98: 12920–12925. 10.1073/pnas.231472998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. 2012. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab 16: 55–67. 10.1016/j.cmet.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. 2007. Trinucleotide repeat disorders. Annu Rev Neurosci 30: 575–621. 10.1146/annurev.neuro.29.051605.113042 [DOI] [PubMed] [Google Scholar]
- Palmieri MC, Greenhalf W, Laluce C. 1996. Efficient flotation of yeast cells grown in batch culture. Biotechnol Bioeng 50: 248–256. [DOI] [PubMed] [Google Scholar]
- Pâques F, Leung W-Y, Haber JE. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol 18: 2045–2054. 10.1128/MCB.18.4.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parts L, Cubillos FA, Warringer J, Jain K, Salinas F, Bumpstead SJ, Molin M, Zia A, Simpson JT, Quail MA, et al. 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res 21: 1131–1138. 10.1101/gr.116731.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, De Chiara M, Friedrich A, Yue J-X, Pflieger D, Bergström A, Sigwalt A, Barré B, Freel K, Llored A, et al. 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol 12: 712–723. 10.1016/S0960-9822(02)00808-4 [DOI] [PubMed] [Google Scholar]
- Powers RW III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev 20: 174–184. 10.1101/gad.1381406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilez J, Guilmatre A, Garg P, Highnam G, Gymrek M, Erlich Y, Joshi RS, Mittelman D, Sharp AJ. 2016. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res 44: 3750–3762. 10.1093/nar/gkw219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA. 2006. Stem cells, ageing and the quest for immortality. Nature 441: 1080–1086. 10.1038/nature04958 [DOI] [PubMed] [Google Scholar]
- Rascon B, Harrison JF. 2010. Lifespan and oxidative stress show a non-linear response to atmospheric oxygen in Drosophila. J Exp Biol 213: 3441–3448. 10.1242/jeb.044867 [DOI] [PubMed] [Google Scholar]
- Ray A, Hector RE, Roy N, Song JH, Berkner KL, Runge KW. 2003. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat Genet 33: 522–526. 10.1038/ng1132 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Reddi AR, Culotta VC. 2013. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 152: 224–235. 10.1016/j.cell.2012.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Tang XD, Chen ML, Joiner MLA, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, et al. 2002. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci 99: 2748–2753. 10.1073/pnas.032671199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckenstuhl C, Netzberger C, Entfellner I, Carmona-Gutierrez D, Kickenweiz T, Stekovic S, Gleixner C, Schmid C, Klug L, Sorgo AG, et al. 2014. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet 10: e1004347 10.1371/journal.pgen.1004347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP. 1997. Natural variation in a Drosophila clock gene and temperature compensation. Science (80-) 278: 2117–2120. 10.1126/science.278.5346.2117 [DOI] [PubMed] [Google Scholar]
- Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, Hsieh EJ, Moller RM, Wasko BM, Delaney JR, et al. 2013. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell 12: 1050–1061. 10.1111/acel.12130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. 2007. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab 6: 280–293. 10.1016/j.cmet.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Sestini EA, Carlson JC, Allsopp R. 1991. The effects of ambient temperature on life span, lipid peroxidation, superoxide dismutase, and phospholipase A2 activity in Drosophila melanogaster. Exp Gerontol 26: 385–395. 10.1016/0531-5565(91)90050-V [DOI] [PubMed] [Google Scholar]
- Sheshadri P, Ashwini A, Jahnavi S, Bhonde R, Prasanna J, Kumar A. 2015. Novel role of mitochondrial manganese superoxide dismutase in STAT3 dependent pluripotency of mouse embryonic stem cells. Sci Rep 5: 9516 10.1038/srep09516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL Jr, McClure JM, Matecic M, Smith JS. 2007. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell 6: 649–662. 10.1111/j.1474-9726.2007.00326.x [DOI] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. 2008. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol 24: 29–54. 10.1146/annurev.cellbio.23.090506.123509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW. 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. 10.1038/416326a [DOI] [PubMed] [Google Scholar]
- Stern A, Brown M, Nickel P, Meyer TF. 1986. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell 47: 61–71. 10.1016/0092-8674(86)90366-1 [DOI] [PubMed] [Google Scholar]
- Stewart MS, Krause SA, McGhie J, Gray JV. 2007. Mpt5p, a stress tolerance- and lifespan-promoting PUF protein in Saccharomyces cerevisiae, acts upstream of the cell wall integrity pathway. Eukaryot Cell 6: 262–270. 10.1128/EC.00188-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpferl SW, Brand SE, Jiang JC, Korona B, Tiwari A, Dai J, Seo JG, Jazwinski SM. 2012. Natural genetic variation in yeast longevity. Genome Res 22: 1963–1973. 10.1101/gr.136549.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kale SP, Childress AM, Pinswasdi C, Jazwinski SM. 1994. Divergent roles of RAS1 and RAS2 in yeast longevity. J Biol Chem 269: 18638–18645. [PubMed] [Google Scholar]
- Topchiy E, Panzhinskiy E, Griffin WST, Barger SW, Das M, Zawada WM. 2013. Nox4-generated superoxide drives angiotensin II-induced neural stem cell proliferation. Dev Neurosci 35: 293–305. 10.1159/000350502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XFS. 2014. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun 5: 3446 10.1038/ncomms4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váchová L, Šťovíček V, Hlaváček O, Chernyavskiy O, Štěpánek L, Kubínová L, Palková Z. 2011. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J Cell Biol 194: 679–687. 10.1083/jcb.201103129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-García I, Salinas F, Li J, Warringer J, Mustonen V, Liti G, Fischer A, Barré B, Hallin J, Bergström A, et al. 2017. Clonal heterogeneity influences the fate of new adaptive mutations. Cell Rep 21: 732–744. 10.1016/j.celrep.2017.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Klis FM. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol 60: 5–15. 10.1111/j.1365-2958.2006.05072.x [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. 2005. Intragenic tandem repeats generate functional variability. Nat Genet 37: 986–990. 10.1038/ng1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volle CB, Delaney S. 2012. CAG/CTG repeats alter the affinity for the histone core and the positioning of DNA in the nucleosome. Biochemistry 51: 9814–9825. 10.1021/bi301416v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J, Zörgö E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, Forsmark A, Durbin R, Omholt SW, Louis EJ, et al. 2011. Trait variation in yeast is defined by population history. PLoS Genet 7: e1002111 10.1371/journal.pgen.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li LM, Longo VD. 2008. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet 4: e13 10.1371/journal.pgen.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Feng L, Paul A, Smith DL, Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman JA, Burhans WC. 2007. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS One 2: e748 10.1371/journal.pone.0000748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Mesquita A, Carroll T, Marks L, Yang H, Zhaojie Z, Ludovico P, Burhans WC. 2010. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany NY) 2: 709–726. 10.18632/aging.100215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie A. 1986. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. Am Inst Nutr 116: 641–654. 10.1093/jn/116.4.641 [DOI] [PubMed] [Google Scholar]
- Wlodkowic D, Skommer J, Darzynkiewicz Z. 2009. Flow cytometry-based apoptosis detection. Methods Mol Biol 559: 19–32. 10.1007/978-1-60327-017-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Kadiyala CS, Ching TT, Hakimi P, Saha S, Xu H, Yuan C, Mullangi V, Wang L, Fivenson E, et al. 2012. Enhanced energy metabolism contributes to the extended life span of calorie-restricted Caenorhabditis elegans. J Biol Chem 287: 31414–31426. 10.1074/jbc.M112.377275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Li J, Aigrain L, Hallin J, Persson K, Oliver K, Bergström A, Coupland P, Warringer J, Lagomarsino MC, et al. 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet 49: 913–924. 10.1038/ng.3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.