Abstract
Identification of gene expression traits unique to the human brain sheds light on the molecular mechanisms underlying human evolution. Here, we searched for uniquely human gene expression traits by analyzing 422 brain samples from humans, chimpanzees, bonobos, and macaques representing 33 anatomical regions, as well as 88,047 cell nuclei composing three of these regions. Among 33 regions, cerebral cortex areas, hypothalamus, and cerebellar gray and white matter evolved rapidly in humans. At the cellular level, astrocytes and oligodendrocyte progenitors displayed more differences in the human evolutionary lineage than the neurons. Comparison of the bulk tissue and single-nuclei sequencing revealed that conventional RNA sequencing did not detect up to two-thirds of cell-type-specific evolutionary differences.
The human brain is an enormously complex organ that has expanded greatly in comparison with the brains of our closest relatives: chimpanzees, bonobos, and other apes. Increased size alone, however, fails to explain cognitive abilities unique to humans (Semendeferi and Damasio 2000; Elston et al. 2006; Teyssandier 2008; Semendeferi et al. 2011; Barger et al. 2012). Functional changes acquired on the human lineage are likely to be mediated by alterations in gene expression and cell-type composition in particular brain structures (O'Bleness et al. 2012; Sousa et al. 2017b; McKenzie et al. 2018). Yet we currently lack a comprehensive understanding of these uniquely human evolutionary differences.
Gene expression within the human brain differs substantially among regions and anatomical structures, both within neocortex and among subcortical areas (Kang et al. 2011; Hawrylycz et al. 2012). Initial studies comparing gene expression in humans to non-human primates (NHPs) examined one or several brain regions with the main focus on the prefrontal area of the neocortex (Enard et al. 2002; Caceres et al. 2003; Marvanová et al. 2003; Khaitovich et al. 2004b). These studies identified multiple expression differences specific to humans and revealed an acceleration of expression evolution on the human lineage. Although the expression differences shared among brain regions often represent molecular and functional changes not specific to the brain (Khaitovich et al. 2004a, 2005), differences particular to individual brain regions tend to be associated with specific brain functions (Khaitovich et al. 2004a). Recent studies examining eight and 16 brain regions in humans and closely related NHPs expanded these results further by revealing the rapid expression evolution of several subcortical regions in addition to the neocortical areas (Sousa et al. 2017a; Xu et al. 2018).
Although the brain is composed of functionally diverse anatomical regions, each brain region is composed of multiple cell types (Lein et al. 2007). Single-cell RNA sequencing provides an opportunity to decompose gene expression within brain regions and compare homologous cell types across species (La Manno et al. 2016; Saunders et al. 2018; Tosches et al. 2018; Zeisel et al. 2018). A single-cell level comparison between human and macaque transcriptomes in prenatal and adult dorsolateral prefrontal cortices indeed showed that all detected human cell types had a close homolog in macaques, and vice versa (Zhu et al. 2018). Furthermore, this study identified genes differentially expressed between humans and macaques in individual cell types. Similar results were obtained in studies of single-cell expression in human and chimpanzee cerebral organoids, indicating that cell-type composition can be accurately matched between closely related primate species and expression differences within each type identified (Mora-Bermúdez et al. 2016; Kanton et al. 2019; Pollen et al. 2019).
Here, we report transcriptome maps of the human, chimpanzee, bonobo, and macaque brain constructed using conventional RNA sequencing (bulk RNA-seq) and single-nuclei sequencing (snRNA-seq).
Results
Global gene expression variation analysis
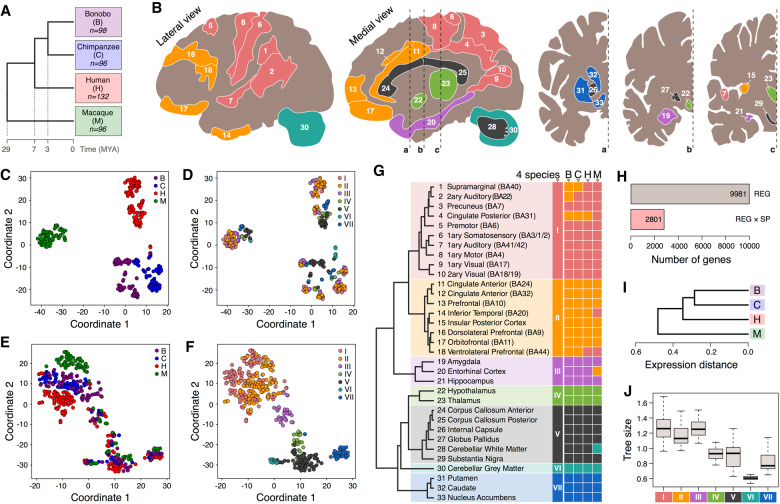
We used bulk RNA-seq to examine RNA expression in 33 brain regions from four humans, three chimpanzees, three bonobos, and three rhesus monkeys (Fig. 1A,B; Supplemental Table S1). The visualization of expression variation among samples revealed separation of species, consistent with their phylogenetic relationship (Fig. 1C,D; Supplemental Figs. S1, S2). Removal of inter-individual variation further revealed a common pattern of differences among the 33 brain regions within each brain (Fig. 1E,F; Supplemental Fig. S1). Accordingly, the 33 brain regions further separated into seven clusters shared across individuals and species (Fig. 1G; Supplemental Fig. S3). The clusters largely corresponded to anatomical areas, with three clusters representing cortical regions: primary and secondary cortices (cluster I), limbic and association cortices (cluster II), and archicortex (cluster III); whereas the remaining four clusters contained subcortical structures: thalamus and hypothalamus (cluster IV), white matter structures (cluster V), cerebellar gray matter (cluster VI), and striatum (cluster VII). This clustering was consistent with the one reported in humans based on gene expression analysis of 120 brain regions (Supplemental Fig. S4; Hawrylycz et al. 2012).
Figure 1.
Gene expression variation analysis in 33 brain regions. (A) Phylogenetic relationship among analyzed species. Numbers indicate the number of analyzed brain samples. (B) Anatomical localization of 33 analyzed brain regions within the human brain. Colors represent expression-based regional clusters, defined in G. (C–F) t-SNE plots based on expression variation among all 422 analyzed samples: (C,D) the total variation; (E,F) the residual variation after removal of the average species’ and individual differences. Each circle represents a sample. Circle colors represent species (C,E) or expression-based regional clusters (D,F). (G) Unsupervised hierarchical clustering of brain regions based on the average gene expression values of all 11,176 detected genes in four species. Regions within each species are assigned to the nearest cluster. The clustering based on each individual sample is shown in Supplemental Figure S3. (H) Numbers of genes differentially expressed among brain regions (REG), or among species with a significant dependence on the region (REG × SP) in ANOVA. (I) Average phylogenetic tree reconstructed based on the expression differences identified using ANOVA with species and regions used as factors. The trees reconstructed for each brain region are shown in Supplemental Figure S5. (J) Total branch length calculated for the reconstructed phylogenetic trees for each of the 33 brain regions grouped by expression-based regional clusters I–VII, defined in G.
Of the 11,176 detected orthologous protein-coding genes, 2801 showed brain-region-dependent species differences (ANOVA, BH-adjusted P < 0.00002) (Fig. 1H). Assigning differences among species to the evolutionary lineages recovered the phylogenetic relationship among the four species (Fig. 1I; Supplemental Fig. S5). Region-dependent expression differences accumulated faster in the three cortical clusters compared to the subcortical regions across the phylogenetic tree (Fig. 1J; Supplemental Fig. S6), and this result was robust to intra-human individual variation (Supplemental Fig. S7).
Regional analysis of the human-specific gene expression differences
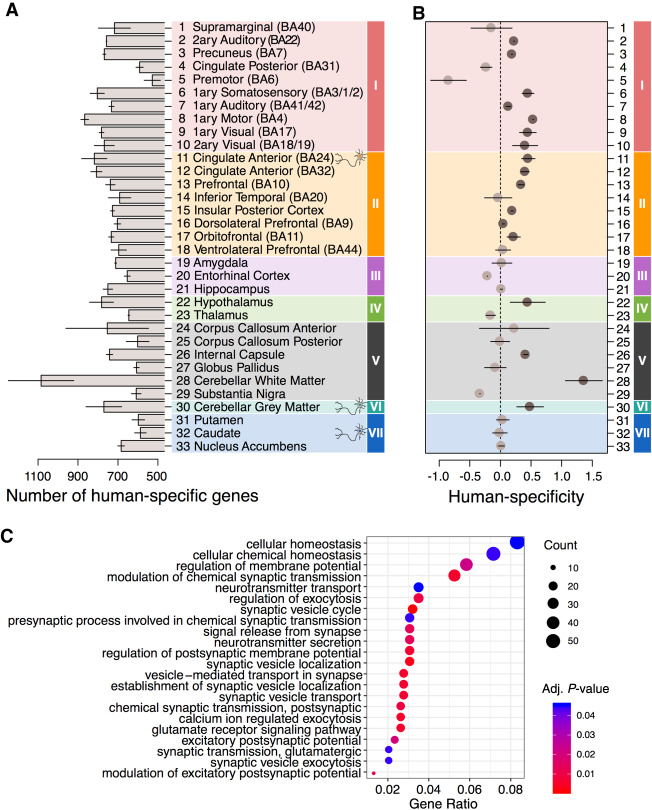
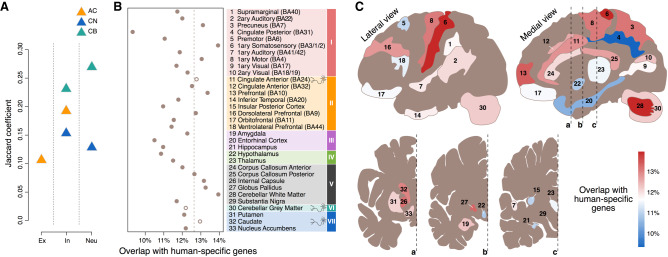
Among 2801 expression differences, we assigned the ones distinguishing humans from non-human primates (NHPs) to the human evolutionary lineage (Supplemental Fig. S8; Supplemental Table S2). The distribution of these differences was not uniform across brain regions. Neocortical regions represented in clusters I and II showed higher than the average proportion of human-specific expression differences, whereas the highest number of differences (1079 genes) was found in cerebellar white matter (Fig. 2A; Supplemental Table S2). Although >95% of these differences represented quantitative expression alterations, some appeared to reflect a complete expression loss either in humans or in non-human primates (Supplemental Fig. S9). The number of human-specific expression differences in a region did not depend on the number of expressed genes and was robust to the definition of human-specific expression differences (Supplemental Figs. S10, S11).
Figure 2.
Region-dependent analysis of human-specific gene expression differences. (A) Numbers of genes showing human-specific expression differences in each brain region. The differences were defined as those showing twofold greater human-macaque expression difference relative to the chimpanzee-macaque or bonobo-macaque difference. The bars show the mean of the chimpanzee-based and bonobo-based comparisons. The error bars span the difference between chimpanzee-based and bonobo-based estimates. Colors represent expression-based clusters of brain regions defined in Figure 1G. (B) The human-specificity ratio of gene expression estimated in each brain region as the ratio of human-specific expression differences and chimpanzee-specific or bonobo-specific expression differences. Circles show the mean of chimpanzee-based and bonobo-based comparisons, and lines span the difference between the two estimates. Darker circles mark brain regions showing an excess of human-specific expression differences compared to both ape species. (C) Top Gene Ontology (GO) functional terms enriched in the human-specific expression differences present in more than 10 of the 33 brain regions. The size of circles reflects the proportion of genes within the GO term among genes detected in the brain region using snRNA-seq data (Gene Ratio) (Yu et al. 2012). The color of circles indicates the BH-adjusted enrichment P-value.
Normalizing the number of human-specific differences by the number of chimpanzee-specific and bonobo-specific ones (human-specificity ratio) revealed a similar nonuniform distribution of human-specific evolutionary differences. Although, in agreement with previous works (Sousa et al. 2017a; Xu et al. 2018), more differences mapped to the human lineage on the average (n = 712) than to the chimpanzee (n = 641) and bonobo (n = 640) lineages, not all brain regions showed human-specific evolutionary acceleration. Specifically, we found no excess of human-specific expression differences in all regions representing archicortex and striatum, in the thalamus, and four of the six white matter structures (Fig. 2B). In contrast, the majority of neocortical regions in clusters I and II (72%), as well as the hypothalamus, internal capsule, and cerebellar white and gray matter, showed a substantial excess of the human-specific differences (Fig. 2B). This result was robust to the definition of species-specific expression differences, intra-species individual variation, and covariate adjustment procedure (Supplemental Figs. S10–S17).
Cognitive functions particularly pronounced in humans involve multiple brain regions (Genon et al. 2018). Accordingly, we tested whether genes showing human-specific expression differences in multiple brain regions were enriched in functional processes associated with cognition. Indeed, uniquely human differences detected in more than 10 brain regions displayed significant enrichment in Gene Ontology (GO) terms linked to the neuronal function: synaptic transmission, regulation of exocytosis, neurotransmitter secretion, and others (hypergeometric test, BH-adjusted P < 0.01) (Fig. 2C; Supplemental Table S3). This enrichment remained stable using more stringent gene selection cutoff (Supplemental Fig. S18).
We further compared the relative expression human specificity of brain regions determined in our study with previous reports. Despite the experimental and statistical differences of analyses, our results correlated positively and significantly with the published data (Spearman's correlation, P < 0.01) (Supplemental Fig. S19; Sousa et al. 2017a).
Single-nuclei transcriptome analysis
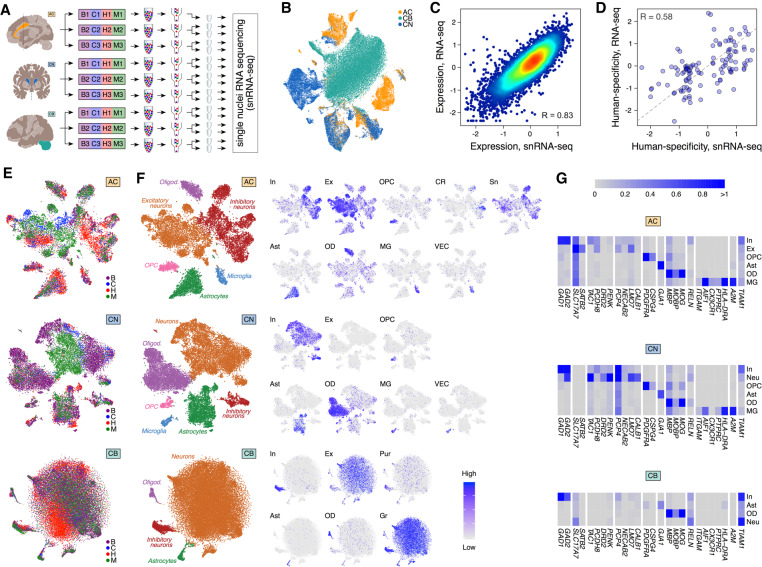
To investigate evolutionary differences accumulating within cell types, we sequenced RNA from individual nuclei (snRNA-seq) in three of the 33 brain regions of four species, anterior cingulate cortex (AC), caudate nucleus (CN), and cerebellar gray matter (CB), in three individuals per species (Supplemental Table S1). To reduce experimental variation among species, tissue samples from one individual of each species were pooled and processed in parallel in each brain region (Fig. 3A). The nuclei species’ identity was then recovered computationally, based on the sequence differences between species. Each pool, except one (AC1), yielded two independent snRNA-seq libraries, resulting in a total of 88,047 nuclei with at least 500 unique detected molecules: 7337 ± 5548 nuclei per species per region. Of them, 21% were derived from AC, 29% from CN and 50% from CB. Within each brain region, the humans were on average represented by 37% of the nuclei, chimpanzees by 10%, bonobos by 32%, and macaques by 21% (Supplemental Table S4). Visualization of the total expression variation across these nuclei revealed a separation of the three brain regions (Fig. 3B; Supplemental Fig. S20). The average expression levels of the nuclei within each tissue of each species correlated well with the corresponding bulk RNA-seq data and published single-cell RNA-seq data (Fig. 3C; Supplemental Figs. S21, S22; Pollen et al. 2019). Similarly, the extent of human-specific expression divergence relative to the chimpanzee-specific or bonobo-specific divergence agreed well between the averaged snRNA-seq and the bulk RNA-seq data (Fig. 3D; Supplemental Fig. S23). Within each region of each species, the nuclei formed six main clusters in AC and CN and four in CB, each enriched in known cell-type markers (Fig. 3E–G; Supplemental Fig. S24; Supplemental Table S5). For purposes of our analysis, we focused on these broad cell classifications, but we are aware that subtypes could be more finely resolved and characterized (Supplemental Fig. S25). Furthermore, comparisons to published finely defined subtype clusters revealed a correct assignment of multiple subtypes to clusters defined in our study (Supplemental Figs. S26–S28; Hodge et al. 2019). Most of our analysis, however, could not be confidently performed at such a finer subtype definition because of insufficient numbers of nuclei per subtype (Supplemental Figs. S29–S31; Supplemental Table S4).
Figure 3.
Single-nuclei transcriptomics in three brain regions. (A) Design of the snRNA-seq experiment. (B) t-SNE plot of 88,047 single nuclei colored by brain regions after integration with Seurat 3.0 (Stuart et al. 2019). (C) Correlation of gene expression levels between bulk RNA-seq and averaged snRNA-seq data sets in human AC. Dots represent genes, and colors show the density of the dots. (D) Correlation of human-specificity ratios between bulk RNA-seq and averaged snRNA-seq data sets in AC for genes passing the human-chimpanzee difference cutoff in either data set. Each dot represents a gene. The dashed line indicates the linear relationship with a slope of 1 and an intersect of 0. (E) t-SNE plot of nuclei colored by species in each of the three brain regions after integration with Seurat 3.0 (Stuart et al. 2019). (F) The cumulative cell-type annotation of t-SNE clusters (left) and projection of expression levels averaged across cell-type marker genes onto the t-SNE plots. Abbreviations next to t-SNE plots mark cell types: (In) inhibitory neurons; (Ex) excitatory neurons; (Sn) spindle neurons; (Pur) Purkinje cells; (OPC) oligodendrocyte progenitor cells; (Ast) astrocytes; (OD) oligodendrocytes; (CR) Cajal-Retzius cells; (MG) microglia; (VEC) vascular endothelial cells. (G) Average expression levels of cell-type marker genes in t-SNE clusters. The same marker genes were used in F.
Cell-type-based analysis of human expression evolution in three brain regions
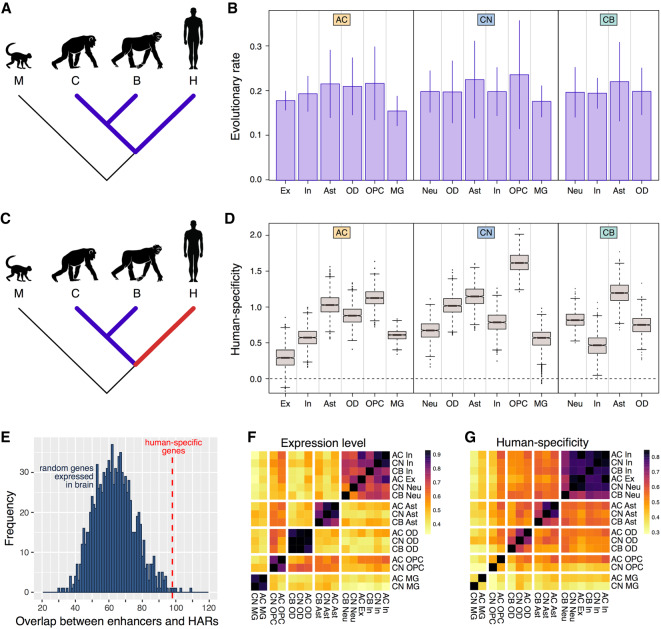
The calculation of the expression evolution rate within each cell type across the human, chimpanzee, and bonobo lineages revealed no statistically significant increase or decrease in any of the analyzed cell types (Fig. 4A,B). Of note, broadly defined neuronal-cell-type clusters showed greater heterogeneity but did not display an increase in the evolutionary rate (Supplemental Fig. S32; Zeisel et al. 2018).
Figure 4.
Cell-type-based analysis of the expression evolution in three brain regions. (A) Phylogenetic tree highlighting the branches used in the evolutionary rate analysis. (B) The evolutionary rate of cell types within each brain region. Error bars mark the standard deviation of the average estimates. (C) Phylogenetic tree highlighting the branches used in the human-specificity ratio analysis. (D) Human-specificity ratio calculated within each t-SNE cluster in each of the three brain regions. The ratio represents the number of genes with human-specific expression divided by the number of genes with chimpanzee-specific and bonobo-specific expression. Boxes mark the median and the first and the third quartiles of the distribution, and whiskers extend to the 1.5 interquartile ranges. The cell types are abbreviated as in Figure 3F. (E) Overlap between enhancers linked to 1271 genes showing human-specific expression in snRNA-seq data and brain-active cis-regulatory elements located in HARs (Vermunt et al. 2016). The histogram represents the distribution of the overlap values calculated by random subsampling of 1271 genes from the 9138 genes expressed in the brain 1000 times. The red dashed line marks an actual overlap (n = 98). (F) The expression level similarity among t-SNE clusters based on the average gene expression levels within clusters in humans. The colors in F and G indicate Pearson correlation coefficients. (G) The similarity of human-specificity ratio estimates among t-SNE clusters calculated based on the comparison to chimpanzee and bonobo in 1000 bootstraps of cells.
Although the evolutionary rate did not differ substantially among the cell types, the comparison of the rates between the lineages revealed significant differences (Fig. 4C,D). Specifically, neuronal clusters tended to show smaller excess of human-specific expression differences over the chimpanzee- or bonobo-specific ones, compared to the other cell types (Fig. 4D). In contrast, astrocytes and oligodendrocyte progenitor cells consistently showed the largest excess of human-specific expression differences in all examined brain regions (Fig. 4D; Supplemental Fig. S33). The human-to-NHPs evolutionary ratio estimates calculated independently using chimpanzee or bonobo data were highly consistent (Supplemental Fig. S34).
Despite differences, all cell types displayed greater evolutionary rate on the human evolutionary lineage compared to both ape species. Thus, we tested whether human-specific expression acceleration could be linked to the genomic human-accelerated regions (HARs) (Vermunt et al. 2016). Genes showing human-specific expression in the cell types indeed situated in the vicinity of the enhancers overlapping with brain-active cis-regulatory elements located in HARs more often than expected by chance (98 out of 248 HARs, permutation P < 0.005) (Fig. 4E; Supplemental Table S6). Furthermore, 35 out of 42 transcription factors (TFs) showing human-specific expression in our data have binding sites in these 98 HAR-enhancer overlaps. Moreover, 13 of these 35 TFs bind to the HAR-enhancer overlaps more frequently than expected by chance (permutation P < 0.05; BH correction) (Supplemental Table S6). In contrast to snRNA-seq data, there was no association between HARs and genes showing human-specific expression based on bulk tissue RNA-seq data set (permutation P > 0.05).
Gene expression within each cell type correlated well across brain regions (Fig. 4F; Supplemental Fig. S35). In contrast, human-specific expression differences correlated well among neuronal subtypes excluding cerebellar granule cells, but not among astrocytes, oligodendrocytes, oligodendrocyte precursors, or microglia originating from different brain regions (Fig. 4G; Supplemental Fig. S35). This result indicates that most neuronal subtypes might share a characteristic signature of human-specific expression differences.
Deconvolution of bulk human-specific expression differences using neuronal evolutionary signature
The existence of the neuronal evolutionary signature shared among brain regions could be used to deconvolute bulk RNA-seq data, analogous to the cell type marker-based deconvolution procedure (Wang et al. 2019). Supporting this notion, human-specificity ratios of genes preferentially expressed in neuronal subtypes correlated positively and significantly between single nuclei and bulk RNA-seq data sets, and this result was robust to the definition of human-specific expression differences (Fig. 5A; Supplemental Figs. S36–S39).
Figure 5.
Deconvolution of bulk human-specific expression differences using neuronal evolutionary signature. (A) Overlap of human-specific genes, defined as those showing twofold greater human-macaque expression difference relative to the chimpanzee-macaque and bonobo-macaque difference, between bulk RNA-seq and snRNA-seq data sets for genes preferentially expressed in a specific neuronal subtype (Supplemental Table S5). Colors indicate brain regions. The x-axis labels indicate neuronal subtypes. (B) Percentages of genes showing human-specific expression in each brain region in bulk RNA-seq data set overlapping with genes showing human-specific expression in neuronal subtypes in snRNA-seq data. Empty circles indicate the three brain regions used in the snRNA-seq experiment. The dashed line represents an average overlap for three brain regions used in the snRNA-seq experiment. (C) Anatomical localization of 33 regions in the human brain colored according to the overlap between human-specific expression differences in bulk RNA-seq and in neuronal subtypes.
To estimate the extent of human-specific neuronal differences in each of the 33 brain regions using bulk RNA-seq data, we calculated the overlap between genes showing human-specific expression differences in bulk RNA-seq data and in neuronal subtypes. Because this overlap should be positively biased toward the three brain regions used in both bulk and single-nuclei analysis, most of the brain regions contained fewer neuronal human-specific differences compared to them (Fig. 5B). Although the overall variation of the overlap with human-specific neuronal differences was small, seven brain regions stood out by displaying more differences than the three regions used in the snRNA-seq experiment (Fig. 5B,C). This result was robust to the nuclei subsampling (Supplemental Fig. S40). Thus, although indirect, this observation suggests that these seven regions, which include primary somatosensory cortex, internal capsule, and cerebellar white matter, might display substantial human-specific alterations of neuronal expression.
Gene expression differences detected by snRNA-seq and bulk RNA-seq
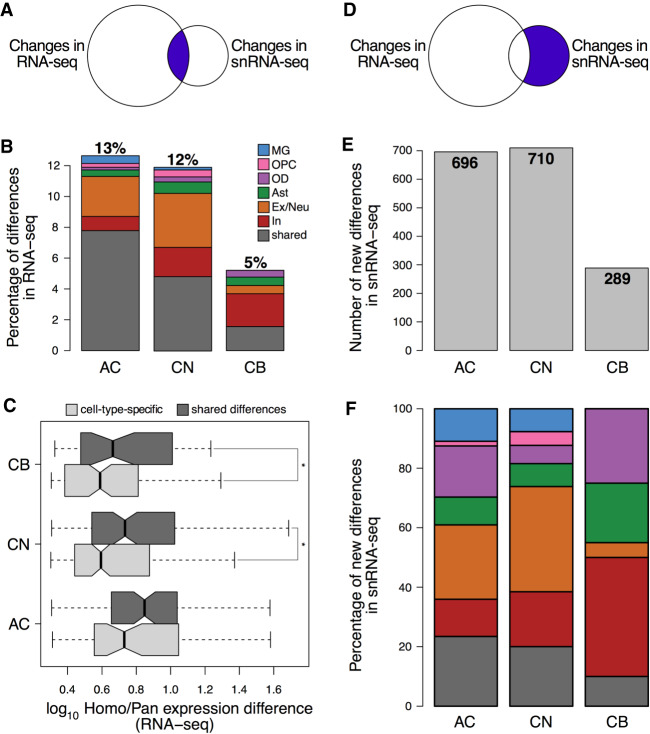
Although expression differences separating humans from apes correlated well between bulk RNA-seq and snRNA-seq data sets, including individual cell types (Fig. 3D; Supplemental Fig. S41), only a fraction of them was significant in both (absolute greater than twofold difference in Homo/Pan comparison, BH-adjusted P < 0.05, two-sided t-test for RNA-seq, Wilcoxon test implemented in Seurat 3.0 for snRNA-seq) (Fig. 6). Specifically, up to 13% of significant differences identified in bulk tissue were also significant in the snRNA-seq data set (Fig. 6A,B). Nearly half of these differences were observed in multiple cell types and showed substantially higher amplitudes in bulk tissue (Fig. 6B,C). This is expected because >90% of genes detected in the brain are expressed in multiple cell types. Hence, the ubiquitous differences are reflected better in bulk tissue expression.
Figure 6.
Gene expression differences detected by snRNA-seq and bulk RNA-seq. (A) Schematic representation and (B) percentage and the cell type specificity of the expression differences present in bulk RNA-seq and snRNA-seq, defined as absolute greater than twofold difference in human samples compared to a pool of chimpanzee and bonobo samples (Methods). (C) The amplitude of expression differences detected in one (specific) or multiple (shared) cell types in bulk RNA-seq. (*) P < 0.05, one-sided t-test. (D) Schematic representation, (E) numbers, and (F) cell type specificity of the expression differences solely detected by snRNA-seq. Color as in A.
On the other hand, expression differences present in a particular cell type might be attenuated or lost in the bulk tissue samples. Indeed, using snRNA-seq, we identified 696 genes showing the expression differences separating humans from apes in AC, not detectable in bulk RNA-seq data (Fig. 6D,E). Similarly, there were 710 such differences in CN and 289 in CB. On average, these differences constitute more than two-thirds of the differences found using snRNA-seq.
Among the 696 expression differences found in AC, 25% localized in excitatory neurons, and 12.5% localized in inhibitory neurons (Fig. 6F). The differences particular to excitatory neurons were enriched in Gene Ontology (GO) terms involved in regulation of neuron projection development, whereas the differences detected in microglia (10.9%) were enriched in immune response and lipid localization functions (hypergeometric test, BH-adjusted P < 0.01) (Supplemental Fig. S42).
Gene expression differences detected by immunohistochemistry
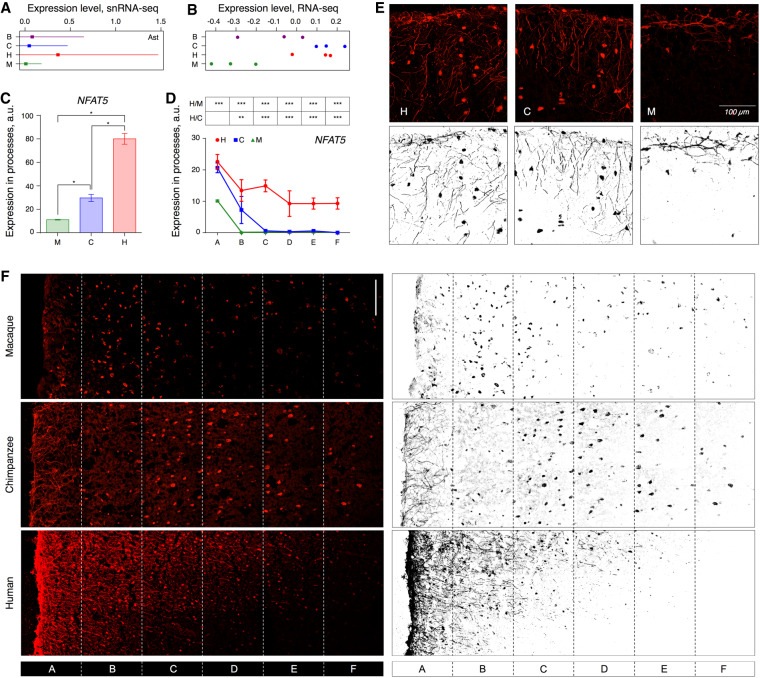
We visualized the spatial distribution of expression differences revealed by snRNA-seq in AC using immunohistochemistry (IHC). As neurons consistently showed the smallest excess of human-specific expression differences in snRNA-seq data, we focused on the other cell types for IHC analysis. Specifically, we selected two genes, MSI2 and NFAT5, which displayed higher expression in human astrocytes according to snRNA-seq data (two-sided t-test, P-value < 10−10) (Fig. 7A; Supplemental Figs. S43, S44). Both genes generated clear and specific staining in the frozen AC tissue sections. Of note, although MSI2 similarly showed an increased expression in human AC in conventional RNA-seq data (two-sided t-test, P = 0.0001) (Supplemental Fig. S44), there was no such expression increase for NFAT5 (two-sided t-test, P = 0.5) (Fig. 7B). Immunohistochemical staining assisted by the common astrocytic and neuronal marker proteins, GFAP and RBFOX3 (also known as NeuN), placed MSI2 and NFAT5 proteins within astrocytic processes and neuronal cell bodies in human, chimpanzee, and macaque tissue sections (Supplemental Figs. S45–S48). Both proteins showed significantly higher fluorescent intensity in human astrocytic processes compared to those in NHPs (two-sided t-test, P < 0.05) (Fig. 7C). Furthermore, whereas the processes localized in the uppermost cortical layer in chimpanzees and macaques, they spread to deeper laminar structures, including layers two and three, in humans (two-sided t-test, P < 0.05) (Fig. 7D,E; Supplemental Fig. S49). This observation adds to reports of functional heterogeneity and rapid evolution of astrocytic cell types in primates (Oberheim et al. 2009; Haim and Rowitch 2017).
Figure 7.
Gene expression differences detected by IHC. (A) The mean log10-transformed expression level of NFAT5 mRNA in AC astrocytes (squares), and the standard deviation of the mean (horizontal lines). (B) The log10-transformed read counts normalized for the median of NFAT5 mRNA in bulk AC data. Circles indicate individual samples. Average fluorescent intensities of NFAT5 IHC signal in the astrocytic processes of macaques, chimpanzees, and humans across cortical layers (C) and at different cortical depth (D). Error bars show the standard deviation of the mean. (***) P < 0.0005; (**) P < 0.005; (*) P < 0.05, two-sided t-test, Holm-Sidak correction; (H/C) human-chimpanzee comparison; (H/M) human-macaque comparison. Symbols indicate cortical sections located at increasing depth, depicted in panel F. (E) IHC (upper) and its binarized representation (lower) of NFAT5 protein in the uppermost layer of AC sections. (F) Immunostaining (left) and its binarized representation (right) of NFAT5 protein in the three upper layers of AC sections in macaques, chimpanzees, and humans (for GFAP and DAPI staining of these sections, see also Supplemental Figs. S45–S49). Sections A–F indicate segmentation areas used in the analysis presented in panel D. (Scale bar) 100 µm.
Discussion
We show that the distribution of human-specific expression differences separating us from our closest living relatives, chimpanzees and bonobos, was not uniform across 33 examined brain regions. Instead, our results suggest an intricate pattern of the expression evolution of the human brain involving both neocortical and subcortical regions, analogous to functional networks defined by magnetic resonance imaging studies (Supplemental Fig. S50; Greicius et al. 2003; Shukla et al. 2010; Washington et al. 2014; Patel et al. 2015; Maximo and Kana 2019).
Our analysis of human-specific expression features conducted at the single nuclei level further provided the following insights. First, we detected multiple expression differences between species within each cell type. Although approximately one-third of these differences, especially those present in multiple cell types, could be detected using conventional bulk RNA-seq data, the remaining differences could only be revealed using cell-type-specific methodologies. Conversely, some of the expression differences detected in bulk tissue but not in snRNA-seq data could be caused by cell-type composition alterations between humans and NHPs. Indeed, such differences in cell-type composition, including a decrease in neuron-to-glia ratio and increase in the proportion of von Economo neurons, were described (Allman et al. 2011; Evrard et al. 2012; Herculano-Houzel 2014). The number of technical and biological replicates included in our study, however, was not sufficient to assess subtle quantitative differences in cell-type composition between humans and NHPs.
Second, non-neuronal cell types showed substantially greater excess of human-specific expression differences in all three examined brain regions compared to neurons. Among them, astrocytes and oligodendrocyte progenitors displayed the largest excess of human-specific expression differences. These human-specific differences were particular to each brain region, precluding deconvolution of bulk RNA-seq data for non-neuronal cell types. It has to be noted that the number of nuclei covered by our study was not sufficient to accurately examine expression evolution in specific neuronal subtypes, such as von Economo neurons, known to be overrepresented in human AC and the frontoinsular cortex (Yang et al. 2019), or the rosehip neurons present in human but not rodent cortical layer I (Boldog et al. 2018). Thus, our results do not exclude the existence of specialized neuronal populations showing rapid expression evolution in humans. Furthermore, application of neuronal human-specific expression signature detected in snRNA-seq data to 33 brain regions revealed seven, including primary somatosensory cortex, internal capsule, and cerebellar white matter, showing the greater extent of human-specific expression differences characteristic of neuronal cells.
Although all reported cell-type-specific evolutionary differences are novel, they concur with previous observations. The regional specificity of the astrocytic human-specific expression differences matches reports of molecular and functional heterogeneity of astrocytes in adult brain regions (Haim and Rowitch 2017). In turn, excess of astrocytic human-specific expression differences matches histological differences reported between humans and the other primate species for interlaminar astrocytes, polarized astrocytes, and varicose projection astrocytes (Oberheim et al. 2009; Falcone et al. 2018). Similarly, oligodendrocyte progenitor cells showing rapid expression evolution in human caudate nucleus were reported to dysfunction in the caudate nucleus of schizophrenia patients (Georgieva et al. 2006; Uranova et al. 2007; Cassoli et al. 2015; Mauney et al. 2015), a disorder suggested to affect aspects of cognition particular to humans (Dean 2009; Konopka and Geschwind 2010).
Taken together, our results show that systematic investigation of gene expression evolution across a large number of brain regions and cell types has the potential to reveal evolutionary patterns reflecting the emergence of the human brain functionality. An important component that was missing from our study, an analysis of temporal patterns of expression evolution in the developing brain, analogous to the one presented in Zhu et al. (2018), would further increase the power to associate expression differences with cognitive functions.
Methods
Samples
Human samples were obtained from the Chinese Brain Bank Center. Informed consent for the use of human tissues for research was obtained in writing from all donors or their next of kin. All subjects were defined as healthy with respect to the sampled brain tissue by forensic pathologists at the tissue bank. All subjects suffered sudden death with no prolonged agony state from causes not related to brain function. All non-human primates used in this study have suffered sudden deaths for reasons other than their participation in this study and without any relation to the tissue used. Chimpanzee samples were obtained from the Southwest National Primate Research Center in San Antonio, Texas. Bonobo samples were obtained from the Lola Ya Bonobo Sanctuary, Democratic Republic Congo, where they were maintained in natural habitat and suffered death from natural causes not related to brain tissue function. Rhesus monkey samples were obtained from the Suzhou Experimental Animal Center, China. All monkeys represented a control animal group, which was not subjected to any pharmaceutical or immunologically related treatment anytime during their lifetime. The use and care of the animals in this research was reviewed and approved by the Institutional Animal Care and Use Ethics Committee at the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (CAS).
Sample dissection
A total of 422 brain samples were dissected from the brains of 14 individuals with at least three individuals per species (Supplemental Table S1). For each individual, samples were dissected from 33 brain regions covering all major anatomical and functional brain structures (Supplemental Table S1). See Supplemental Methods for the dissection procedure details.
RNA sequencing (RNA-seq)
Total RNA was isolated using Direct-zol-96 RNA (Zymo Research) from 10 mg of the frozen tissue per sample. Sequencing libraries were prepared with NEBNext Ultra II RNA Library Prep Kit (New England Biolabs) according to the manufacturer's instructions. Libraries were then sequenced on the Illumina HiSeq 4000 system using the 150-bp paired-end sequencing protocol. See Supplemental Methods for the library preparation details.
Single-nuclei sequencing (snRNA-seq)
Frozen tissue samples of cingulate anterior cortex (BA24), cerebellar gray matter, and caudate nucleus from brains of three individuals per species were used for the intact nuclei isolation. For each brain region, three pooled sample sets were prepared. Each sample set contained pooled equal tissue samples of 5 mg for one human, one chimpanzee, one bonobo, and one macaque individual (Fig. 3A). See Supplemental Methods for the library preparation details.
Single-nuclei experiments were performed using a 10x Chromium single cell 3′ v2 reagent kit by precisely following the detailed protocol of the manufacturer to construct 10x Genomics single-cell 3′ libraries. Single nucleus libraries were pooled at equal ratios and run using paired-end sequencing on the NovaSeq 6000 platform (Illumina) according to the manufacturer's instructions.
Immunohistochemistry
For multiple immunofluorescent histochemistry, 20 µm thick cryosections were prepared from samples of the anterior cingulate cortex (BA24) from three humans, three chimpanzees, and three rhesus monkeys (Supplemental Table S1). See Supplemental Methods for the immunohistochemistry procedure details (Supplemental Fig. S51; Supplemental Table S7). Images were obtained by Zeiss LSM 800 AiryScan system with C Plan-Apochromat 40x/1,3 Oil DIC UV-VIS-IR objective.
RNA-seq data processing
In total, we obtained 7,483,498,084 RNA-seq reads, with an average sample coverage of approximately 17.7 million reads (Supplemental Table S8). See Supplemental Methods for the read processing details (Supplemental Table S9).
t-SNE was applied to visualize the samples (Fig. 1C,D). To reduce individual-to-individual variability, gene expression values were additionally normalized by the median expression level among regions in each individual brain (Fig. 1E,F; Supplemental Figs. S1, S2).
We calculated variance explained by each known covariate (sex, age, RIN, hemisphere, individual) using manova function in R for all expressed genes using the following model: lm ∼ Species + Regions + Sex + Age + RIN + Hemisphere + Individual (R Core Team 2017). For this analysis, expressed genes were defined as those showing non-zero expression in at least one species, and the age of non-human primates was adjusted to the human scale as described in Somel et al. (2009). Sex, age, RIN, and hemisphere covariates accounted only for <2.5% of variance each (Supplemental Fig. S14), and individual covariate explained 5.9% of the variance, suggesting that there might be an additional hidden source of individual-to-individual variability. As the order of covariates in the model is important for the calculation of the explained variance, we transformed our model into the following: lm ∼ Species + Regions + Individual + Sex + Age + RIN + Hemisphere. Under this model, individual covariate explained 12.8% of the variance, and sex, age, RIN, and hemisphere covariates accounted for 0% of the variance (Supplemental Fig. S14). Therefore, normalization to the median of the individual was necessary and sufficient.
To further validate this approach, we adjusted gene expression values for technical covariates with a linear model as described in Berto et al. (2019) using the individual from which the sample was collected as a covariate. We used the residuals to calculate the human specificity and compared the results to our original approach based on the additional normalization by the median expression level among regions in each individual brain (Supplemental Fig. S15).
Complete linkage method of unsupervised hierarchical clustering with one minus Pearson correlation coefficient as a distance metric was used to cluster brain regions based on average gene expression values among four species (Fig. 1G). Further, we calculated the average gene expression values within each cluster. Based on them, we assigned each brain region of each species to the nearest cluster using one minus Pearson correlation coefficient as a distance metric to assess the conservation of clustering among four species (Fig. 1G; Supplemental Fig. S3).
Expression differences between species
To identify expression differences among species that varied significantly depending on the brain region, ANOVA was applied with both species and regions variables as factors in the following model: lm ∼ Regions + Species + Regions : Species (Fig. 1H). To reconstruct the phylogenetic tree based on the identified expression differences, UPGMA method was used with one minus Pearson correlation coefficient as a distance metric (Fig. 1I). Similarly, phylogenetic trees were reconstructed for each brain region separately (Supplemental Fig. S5), and, for each tree, the total branch length was calculated (Supplemental Figs. S6, S7).
To assign the expression differences to the evolutionary lineages, we classified human-specific expression differences as those showing twofold greater human/macaque difference relative to chimpanzee/macaque or bonobo/macaque differences (Fig. 2A). Chimpanzee-specific expression differences were defined as those showing twofold greater chimpanzee/macaque difference relative to human/macaque difference. Bonobo-specific expression differences were defined similarly. The human-specificity ratio of each brain region was estimated as the number of human-specific genes divided by the number of chimpanzee-specific or bonobo-specific genes (Fig. 2B).
We next explored whether the number of human-specific expression differences in a given region depended on the number of genes expressed in this region (Supplemental Fig. S10). We defined genes expressed in a given region as genes showing average log10 TPM values in this region >10% of average log10 TPM values per region calculated among all 2801 genes identified as species-specific in ANOVA analysis.
To verify the validity of our definition of human-specific gene expression changes, we applied an alternative procedure and compared the obtained results. In this alternative definition, we excluded genes that changed expression in either chimpanzee or bonobo alone and changed our criterion of human specificity to |H-M|/|C-M| > 2 AND |H-M|/|B-M| > 2. Additionally, to eliminate possible false positives caused by taking the ratio of small nonsignificant differences, we excluded 10% of the smallest gene expression differences, requiring (|H-M| > Q or |C-M| > Q) AND (|H-M| > Q or |B-M| > Q), where Q was calculated as 10% among all |H-M|, |C-M|, and |B-M| differences. Of note, the amount of possible false positives would be approximately the same for the nominator and denominator of the ratio; therefore, given a large number of observations (i.e., genes), the overall ratio is not expected to be affected. Indeed, human-specificity ratios calculated using the first procedure described above and the alternative, more strict definition, correlated extremely well (Pearson's R = 0.99) (Supplemental Fig. S12). Moreover, numbers of human-specific genes per region correlated highly and significantly between the two procedures (Pearson's R = 0.93) (Supplemental Fig. S11), although the absolute numbers of human-specific genes dropped in the second procedure, as expected, because of the use of a more stringent criterion: AND instead of OR.
To test the dependence of results on the choice of the fold-change cutoff, we applied different cutoffs (2, 1.5, and 1) and compared the obtained results. Correlation between numbers of human-specific genes calculated with different cutoffs was high (Pearson's R = 0.99 for cutoffs 2 and 1.5, Pearson's R = 0.94 for cutoffs 2 and 1) (Supplemental Fig. S13). Correlation between human-specificity ratios calculated with different cutoffs was even higher (Pearson's R = 0.99 for cutoffs 2 and 1.5, Pearson's R = 0.98 for cutoffs 2 and 1) (Supplemental Fig. S13). Thus, we can conclude that our results do not depend on the choice of the fold-change cutoff.
To cluster genes with expression differences among species that varied significantly depending on the brain region, UPGMA method was used with one minus Spearman's correlation coefficient as a distance metric (Supplemental Fig. S52). A minimal number of modules (n = 3) were selected for further functional network analysis (Supplemental Fig. S50): M1 (1389 genes), M2 (1267 genes), and M3 (145 genes).
Functional enrichment analysis for genes sharing human-specific expression differences in more than 10 of the 33 brain regions was performed using clusterProfiler (Yu et al. 2012). Method enrichGO was used with BP ontology, BH correction for multiple testing, Q value cutoff = 0.1, and 2801 genes significant in ANOVA as the background. Parameter showCategory = 30 was used to visualize the result. To assess the dependence of the result on the cutoff used to define the number of brain regions sharing human-specific expression, we repeated the same analysis using genes showing human-specific expression differences in >8, >9, >10, >11, and >12 brain regions (Supplemental Fig. S18).
Single-nuclei data processing
A total of 3,081,653,593 paired-end sequencing reads of snRNA-seq were processed using publicly available 10x Genomics software, Cell Ranger v2.2.0 (Zheng et al. 2017). See Supplemental Methods for the read processing details (Supplemental Figs. S53, S54; Supplemental Table S10).
The sparse expression matrix generated by cellranger analysis pipeline with the list of 88,047 nuclei assigned to species was used as input to Seurat 3.0 (Stuart et al. 2019). To account for technical variation, we performed cross-species integration. At the first step, for each species separately, we performed normalization using “LogNormalize” with the scale factor of 10,000 and identified 2000 variable features. Next, we performed cross-species integration by finding corresponding anchors between the species using 30 dimensions. We then computed 50 principal components and tested their significances by jackstraw (Chung et al. 2015). We selected the first 30 principal components for subsequent t-SNE and clustering analyses.
To compare snRNA-seq gene expression levels with the RNA-seq data set, we calculated average gene expression values across all nuclei per region per species. In each species and region, Pearson correlation coefficients were calculated between snRNA-seq and bulk RNA-seq values for all genes expressed in both data sets (Fig. 3C; Supplemental Fig. S21). To calculate bulk RNA-seq values for this analysis, we normalized log-transformed read counts per gene by the sample median but did not normalize by the median expression level among regions in each individual brain, for consistency with the snRNA-seq data set. Additionally, we calculated Pearson correlation coefficients between snRNA-seq and RNA-seq human-specificity ratios for genes with high (greater than 100 times) human/chimpanzee gene expression differences in either snRNA-seq or bulk RNA-seq (Fig. 3D; Supplemental Fig. S23). Using a more stringent definition of human-specific expression differences (|H-M|/|C-M| > 2 AND |H-M|/|B-M| > 2), we calculated Jaccard coefficients estimating an overlap of human-specific and panspecific genes between bulk RNA-seq and snRNA-seq data sets (Supplemental Fig. S55).
We further compared the average snRNA-seq expression values with the published single-cell RNA-seq data set (Supplemental Fig. S22; Pollen et al. 2019). For this analysis, we obtained average gene expression values for the primary brain samples and the organoid models from GEO accession GSE124299, log-transformed them, and normalized for the sample median.
Seurat 3.0 package (Stuart et al. 2019) was used to visualize nuclei using t-SNE (Fig. 3B,E), to perform nuclei clustering (Fig. 3F; Supplemental Fig. S25), and to infer and plot marker genes of nuclei clusters (Supplemental Fig. S24). For nuclei clustering, resolution parameters 0.095 (CN), 0.04 (CB), and 0.15 (AC) were used. To assign cell type identity to clusters, we chose cell-type marker genes based on literature: GAD1, GAD2 (inhibitory neurons) (Lake et al. 2016); SLC17A7, SATB2 (excitatory neurons) (Lake et al. 2016); TAC1, PCDH8, DRD2, ADORA2A, PENK (spindle neurons) (Gokce et al. 2016; McCullough et al. 2018); PCP4, NECAB2, LMO7, CALB1 (Purkinje cells) (Uhlen et al. 2015); PDGFRA, CSPG4 (oligodendrocyte precursor cells) (McKenzie et al. 2018; Zeisel et al. 2018), GJA1 (astrocytes) (Zeng et al. 2012; McKenzie et al. 2018); MBP, MOBP, MOG (oligodendrocytes) (Zeng et al. 2012); RELN (Cajal-Retzius) (D'Arcangelo et al. 1997); AIF1, CX3CR1, PTPRC, HLA−DRA (microglia) (McKenzie et al. 2018); A2M (endothelial vascular cells) (Zeng et al. 2012); TIAM1 (granular cells) (Uhlen et al. 2015). We further plotted average expression levels of selected marker genes in each nucleus (Fig. 3F) and in each t-SNE cluster (Fig. 3G). We discarded cells in finely resolved clusters (Cluster 6 in AC; Cluster 5 in CN; Cluster 2 in CB) (Supplemental Fig. S25) as they showed higher percentage of mitochondrial genes per cell.
To show the robustness of our batch integration, we have used two alternative, independent batch integration procedures: harmony (Korsunsky et al. 2019) and liger (Welch et al. 2019) and compared their results with Seurat 3.0. First, we performed independent batch integration using either harmony (Korsunsky et al. 2019) or liger (Welch et al. 2019). Then, we selected the number of clusters in harmony or liger integration in such a way that each Seurat 3.0 cluster would match at least one of the harmony or liger clusters. We observed that both harmony and liger approaches yielded similar results with Seurat 3.0 (Supplemental Figs. S56, S57).
Human-specific expression differences in snRNA-seq data set
Similar to RNA-seq data analysis, we classified human-specific expression differences in each cell type as those showing twofold greater human/macaque difference relative to chimpanzee/macaque and bonobo/macaque differences. Pan-specific expression differences were defined as those showing twofold greater chimpanzee/macaque and bonobo/macaque difference relative to human/macaque difference. To identify significant expression differences among species, we used ANOVA with both species and cell type variables included as factors in the following model: lm ∼ Species + Cell Type + Species : Cell Type. If either the species factor or the interaction term showed a significant impact on gene expression (BH-adjusted P < 0.05, aov function in R followed by drop1 function with test = “F” parameter), the gene was selected for further analysis. To balance the number of nuclei between cell types, we bootstrapped the nuclei 1000 times by randomly sampling 25 nuclei per cell type per region per species without replacement in all downstream analyses. To assess the evolutionary rate of a particular cell type, we calculated the average number of human-specific, chimpanzee-specific, and bonobo-specific genes among 1000 bootstraps for each cell type in three brain regions (Fig. 4A,B).
Further, the human-specificity ratio of each cell type was estimated as the number of human-specific genes divided by the number of chimpanzee-specific or bonobo-specific genes (Fig. 4C,D). To estimate the number of nuclei required for robust human-specificity calculations, we also bootstrapped the nuclei 1000 times by randomly sampling 15 and 5 nuclei per cell type per region per species without replacement, and we obtained highly similar results (Supplemental Fig. S33). To validate this result using a less stringent definition of human specificity, we calculated the human-specificity ratio as the number of genes with human-specific expression divided by the number of genes with chimpanzee-specific or bonobo-specific expression, and we obtained highly consistent results (Supplemental Fig. S34). To assess heterogeneity of nuclei within clusters, we reclassified human-specific genes while bootstrapping the nuclei 1000 times to one nucleus per cell type per region. Then, we calculated the pairwise overlap/union of human-specific genes among 1000 bootstraps within each cell type and each region (Supplemental Fig. S32).
Analysis of snRNA-seq data resulted in a list of 1271 unique genes showing human-specific expression differences in at least one cell type (FindMarkers function in Seurat 3.0; human nuclei compared to chimpanzee, bonobo, and macaque nuclei combined in each t-SNE cluster separately; fold-change threshold = 2; Wilcoxon rank-sum test; Bonferroni adjusted P-value < 0.05). Using the GeneHancer database (Fishilevich et al. 2017), we formed a list of enhancers associated with these genes. We then counted the enhancers corresponding to the brain-active cis-regulatory elements located in HARs (284 predicted HARs from Vermunt et al. (2016) (Fig. 4E; Supplemental Table S6). To estimate the significance of this overlap, we performed random subsampling of 1271 genes from the 9138 genes expressed in the brain 1000 times (Fig. 4E). To analyze TFs binding sites within these 98 enhancers, we used the distribution of TFs binding sites from the GeneHancer database based on ChIP-seq analysis (Supplemental Table S6).
We further averaged gene expression values per cell type per region in snRNA-seq data. Based on these values, we calculated the average human specificity per cell type per region as the mean of log-transformed ratios of human/macaque difference to chimpanzee/macaque or bonobo/macaque difference in snRNA-seq data. The number of nuclei per cell type per region per species was balanced to 25 nuclei for this analysis. Pearson correlation coefficients were calculated between average gene expression values in human (Fig. 4F) and between average human specificity per cell type per region (Fig. 4G).
To deconvolute bulk human-specific expression differences using a neuronal evolutionary signature, we first compared human-specific genes preferentially expressed in neuronal subtypes (Supplemental Table S5) in the snRNA-seq data set with human-specific genes in the bulk RNA-seq data set. We first calculated correlations of human-specificity ratios defined as the human-macaque difference relative to chimpanzee-macaque or bonobo-macaque difference between bulk RNA-seq and averaged snRNA-seq data sets for genes passing human-chimpanzee (Supplemental Fig. S36) or human-bonobo (Supplemental Fig. S37) 10-fold difference cutoffs in either data set (Supplemental Fig. S38). An overlap between the two gene lists was estimated using the Jaccard coefficient (Fig. 5A; Supplemental Fig. S39). For each of 33 brain regions, we further calculated the percentage of genes showing human-specific expression in the bulk RNA-seq data set, which overlapped with genes showing human-specific expression in the neuronal subtypes in snRNA-seq data (Fig. 5B,C). An average overlap was calculated by random subsampling of 25 nuclei per species per cell type among all neuronal subtypes and all brain regions analyzed in the snRNA-seq experiment 1000 times (Supplemental Fig. S40). The result was robust to the definition of human-specific expression differences (Supplemental Fig. S40).
Gene expression differences detected by snRNA-seq and bulk RNA-seq
Expression differences separating humans from chimpanzees and bonobos in bulk RNA-seq data set were defined as absolute greater than twofold difference in human samples compared to a pool of chimpanzee and bonobo samples (BH-adjusted P < 0.05, two-sided t-test). For this analysis, we normalized log-transformed read counts per gene by the sample median but did not normalize by the median expression level among regions in each individual brain, for consistency with snRNA-seq data set.
In the snRNA-seq data set, expression differences separating humans from chimpanzees and bonobos were defined in each cell type as absolute greater than twofold difference in human nuclei compared to a pool of chimpanzee and bonobo nuclei (BH-adjusted P < 0.05, Wilcoxon test implemented in Seurat 3.0 function FindMarkers) (Stuart et al. 2019). Only genes that were detected in a minimum fraction of 0.1 nuclei in humans or in a pool of chimpanzees and bonobos were tested.
To find an overlap between gene expression differences detected by snRNA-seq and bulk RNA-seq, we calculated the percentage of differences detected by both snRNA-seq and bulk RNA-seq relative to the total number of differences detected by bulk RNA-seq (Fig. 6A,B). Cell-type-specific differences were solely detected in one cell type, whereas shared differences were detected in more than one cell type by snRNA-seq (Fig. 6B). We further calculated an absolute log10-transformed amplitude of these cell-type-specific and shared expression differences measured using bulk RNA-seq between humans and the average of two ape species (Fig. 6C).
Next, we focused on gene expression differences detected by snRNA-seq but not by RNA-seq (Fig. 6D,E). Functional enrichment analysis was performed for each group of cell-type-specific and shared differences using clusterProfiler (enrichGO function, BP ontology, genes expressed in AC snRNA-seq data set as a background, BH-adjusted P < 0.01) (Supplemental Fig. S42; Yu et al. 2012). We calculated the percentage of cell-type-specific and shared differences relative to the total number of differences solely detected by snRNA-seq (Fig. 6F). Because there was an unequal number of nuclei per cell type, we observed more significant expression differences separating humans from chimpanzees and bonobos for cell types containing more nuclei because of the higher statistical power owing to the larger sample sizes. Thus, for the unbiased percentage calculation, we balanced the number of nuclei per cell type to 110 human nuclei and a pool of 110 chimpanzee and bonobo nuclei by subsampling the nuclei 100 times and counting expression differences separating humans from chimpanzees and bonobos in ≥0.9 subsampling iterations.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE127898 and GSE127774. Custom scripts used in this study are available as Supplemental Code and at https://cb.skoltech.ru/~khrameeva/brainmap/code/. We provide an interactive website at https://nucseq.cobrain.io/, reproducing key analyses of RNA-seq and snRNA-seq data with variable parameters. The website can be browsed by gene, with information on its expression level in cell types of four species.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the Chinese Brain Bank Center for providing the human samples. We also thank the Southwest National Primate Research Center, Lola Ya Bonobo Sanctuary, and the Suzhou Experimental Animal Center for providing the chimpanzee, bonobo, and macaque samples. We are grateful to CoBrain IT team and personally to Dmitry Vinogradov for the technical support. We thank Grace Cuddihy for her comments on the manuscript. This work was supported by the Russian Foundation for Basic Research (Grant 20-34-70077), Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB13010200); the National Natural Science Foundation of China (Grant 31420103920); the National One Thousand Foreign Experts Plan (Grant WQ20123100078); the National Key R&D Program of China (Grant 2017YFA0505700); and the Russian Science Foundation (Grant 16-14-00220).
Author contributions: B.T., J.G.C., S.P., C.C.S., and P.K. designed the research. D.H., P.G., S.K., M.San., M.B., O.E., and Z.Q. performed research. E.K., I.K., S.R., P.M., M.Sab., and A.T. analyzed the data. All authors contributed to writing the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.256958.119.
Freely available online through the Genome Research Open Access option.
References
- Allman JM, Tetreault NA, Hakeem AY, Park S. 2011. The von Economo neurons in apes and humans. Am J Hum Biol 23: 5–21. 10.1002/ajhb.21136 [DOI] [PubMed] [Google Scholar]
- Barger N, Stefanacci L, Schumann CM, Sherwood CC, Annese J, Allman JM, Buckwalter JA, Hof PR, Semendeferi K. 2012. Neuronal populations in the basolateral nuclei of the amygdala are differentially increased in humans compared with apes: a stereological study. J Comp Neurol 520: 3035–3054. 10.1002/cne.23118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berto S, Mendizabal I, Usui N, Toriumi K, Chatterjee P, Douglas C, Tamminga CA, Preuss TM, Yi SV, Konopka G. 2019. Accelerated evolution of oligodendrocytes in the human brain. Proc Natl Acad Sci 116: 24334–24342. 10.1073/pnas.1907982116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog E, Bakken TE, Hodge RD, Novotny M, Aevermann BD, Baka J, Bordé S, Close JL, Diez-Fuertes F, Ding S-L, et al. 2018. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat Neurosci 21: 1185–1195. 10.1038/s41593-018-0205-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. 2003. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci 100: 13030–13035. 10.1073/pnas.2135499100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassoli JS, Guest PC, Malchow B, Schmitt A, Falkai P, Martins-de-Souza D. 2015. Disturbed macro-connectivity in schizophrenia linked to oligodendrocyte dysfunction: from structural findings to molecules. NPJ Schizophr 1: 15034 10.1038/npjschz.2015.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung NC, Storey JD. 2015. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31: 545–554. 10.1093/bioinformatics/btu674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcangelo G, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Curran T. 1997. Reelin is a secreted glycoprotein recognized by the CR-50 monoclonal antibody. J Neurosci 17: 23–31. 10.1523/JNEUROSCI.17-01-00023.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean B. 2009. Is schizophrenia the price of human central nervous system complexity? Aust N Z J Psychiatry 43: 13–24. 10.1080/00048670802534416 [DOI] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, Elston A, Zietsch B, Defelipe J, Manger P, Casagrande V, Kaas JH. 2006. Specializations of the granular prefrontal cortex of primates: implications for cognitive processing. Anat Rec A Discov Mol Cell Evol Biol 288A: 26–35. 10.1002/ar.a.20278 [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zöllner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340–343. 10.1126/science.1068996 [DOI] [PubMed] [Google Scholar]
- Evrard HC, Forro T, Logothetis NK. 2012. Von Economo neurons in the anterior insula of the macaque monkey. Neuron 74: 482–489. 10.1016/j.neuron.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Falcone C, Wolf-Ochoa M, Amina S, Hong T, Vakilzadeh G, Hopkins WD, Hof PR, Sherwood CC, Manger PR, Noctor SC, et al. 2018. Cortical interlaminar astrocytes across the therian mammal radiation. J Comp Neurol 527: 1654–1674. 10.1002/cne.24605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M, et al. 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017: bax028 10.1093/database/bax028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genon S, Reid A, Langner R, Amunts K, Eickhoff SB. 2018. How to characterize the function of a brain region. Trends Cogn Sci 22: 350–364. 10.1016/j.tics.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva L, Moskvina V, Peirce T, Norton N, Bray NJ, Jones L, Holmans P, MacGregor S, Zammit S, Wilkinson J, et al. 2006. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc Natl Acad Sci 103: 12469–12474. 10.1073/pnas.0603029103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhof TC, Quake SR, 2016. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep 16: 1126–1137. 10.1016/j.celrep.2016.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. 2003. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci 100: 253–258. 10.1073/pnas.0135058100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim LB, Rowitch DH. 2017. Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci 18: 31–41. 10.1038/nrn.2016.159 [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489: 391–399. 10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. 2014. The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 62: 1377–1391. 10.1002/glia.22683 [DOI] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. 2019. Conserved cell types with divergent features in human versus mouse cortex. Nature 573: 61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature 478: 483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchís-Calleja F, Guijarro P, Sidow L, Fleck JS, Han D, et al. 2019. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574: 418–422. 10.1038/s41586-019-1654-9 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. 2004a. Regional patterns of gene expression in human and chimpanzee brains. Genome Res 14: 1462–1473. 10.1101/gr.2538704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Pääbo S. 2004b. A neutral model of transcriptome evolution. PLoS Biol 2: e132 10.1371/journal.pbio.0020132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pääbo S. 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854. 10.1126/science.1108296 [DOI] [PubMed] [Google Scholar]
- Konopka G, Geschwind DH. 2010. Human brain evolution: harnessing the genomics (r)evolution to link genes, cognition, and behavior. Neuron 68: 231–244. 10.1016/j.neuron.2010.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P, Raychaudhuri S. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16: 1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S, et al. 2016. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352: 1586–1590. 10.1126/science.aaf1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. 2016. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167: 566–580.e19. 10.1016/j.cell.2016.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Marvanová M, Ménager J, Bezard E, Bontrop RE, Pradier L, Wong G. 2003. Microarray analysis of nonhuman primates: validation of experimental models in neurological disorders. FASEB J 17: 929–931. 10.1096/fj.02-0681fje [DOI] [PubMed] [Google Scholar]
- Mauney SA, Pietersen CY, Sonntag KC, Woo TW. 2015. Differentiation of oligodendrocyte precursors is impaired in the prefrontal cortex in schizophrenia. Schizophr Res 169: 374–380. 10.1016/j.schres.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximo JO, Kana RK. 2019. Aberrant “deep connectivity” in autism: a cortico-subcortical functional connectivity magnetic resonance imaging study. Autism Res 12: 384–400. 10.1002/aur.2058 [DOI] [PubMed] [Google Scholar]
- McCullough KM, Daskalakis NP, Gafford G, Morrison FG, Ressler KJ. 2018. Cell-type-specific interrogation of CeA Drd2 neurons to identify targets for pharmacological modulation of fear extinction. Transl Psychiatry 8: 164 10.1038/s41398-018-0190-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, Hurd YL, Dracheva S, Casaccia P, Roussos P, et al. 2018. Brain cell type specific gene expression and co-expression network architectures. Sci Rep 8: 8868 10.1038/s41598-018-27293-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Bermúdez F, Badsha F, Kanton S, Camp JG, Vernot B, Köhler K, Voigt B, Okita K, Maricic T, He Z, et al. 2016. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife 5: e18683 10.7554/eLife.18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JHC, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, et al. 2009. Uniquely hominid features of adult human astrocytes. J Neurosci 29: 3276–3287. 10.1523/JNEUROSCI.4707-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. 2012. Evolution of genetic and genomic features unique to the human lineage. Nat Rev Genet 13: 853–866. 10.1038/nrg3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel GH, Yang D, Jamerson EC, Snyder LH, Corbetta M, Ferrera VP. 2015. Functional evolution of new and expanded attention networks in humans. Proc Natl Acad Sci 112: 9454–9459. 10.1073/pnas.1420395112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, et al. 2019. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176: 743–756.e17. 10.1016/j.cell.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. 2018. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174: 1015–1030.e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semendeferi K, Damasio H. 2000. The brain and its main anatomical subdivisions in living hominoids using magnetic resonance imaging. J Hum Evol 38: 317–332. 10.1006/jhev.1999.0381 [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, Travis K, Buckwalter J. 2011. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb Cortex 21: 1485–1497. 10.1093/cercor/bhq191 [DOI] [PubMed] [Google Scholar]
- Shukla DK, Keehn B, Lincoln AJ, Müller R-A. 2010. White matter compromise of callosal and subcortical fiber tracts in children with autism spectrum disorder: a diffusion tensor imaging study. J Am Acad Child Adolesc Psychiatry 49: 1269–1278.e2. 10.1016/j.jaac.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. 2009. Transcriptional neoteny in the human brain. Proc Natl Acad Sci 106: 5743–5748. 10.1073/pnas.0900544106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, Stutz B, Meyer KA, Li M, Kawasawa YI, et al. 2017a. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358: 1027–1032. 10.1126/science.aan3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Meyer KA, Santpere G, Gulden FO, Sestan N. 2017b. Evolution of the human nervous system function, structure, and development. Cell 170: 226–247. 10.1016/j.cell.2017.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssandier N. 2008. Revolution or evolution: the emergence of the Upper Paleolithic in Europe. World Archaeol 40: 493–519. 10.1080/00438240802452676 [DOI] [Google Scholar]
- Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G. 2018. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360: 881–888. 10.1126/science.aar4237 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347: 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Uranova NA, Vostrikov VM, Vikhreva OV, Zimina IS, Kolomeets NS, Orlovskaya DD. 2007. The role of oligodendrocyte pathology in schizophrenia. Int J Neuropsychopharmacol 10: 537 10.1017/S1461145707007626 [DOI] [PubMed] [Google Scholar]
- Vermunt MW, Tan SC, Castelijns B, Geeven G, Reinink P, de Bruijn E, Kondova I, Persengiev S, Bontrop R, Cuppen E, et al. 2016. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat Neurosci 19: 494–503. 10.1038/nn.4229 [DOI] [PubMed] [Google Scholar]
- Wang X, Park J, Susztak K, Zhang NR, Li M. 2019. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun 10: 380 10.1038/s41467-018-08023-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington SD, Gordon EM, Brar J, Warburton S, Sawyer AT, Wolfe A, Mease-Ference ER, Girton L, Hailu A, Mbwana J, et al. 2014. Dysmaturation of the default mode network in autism. Hum Brain Mapp 35: 1284–1296. 10.1002/hbm.22252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. 2019. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177: 1873–1887.e17. 10.1016/j.cell.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Li Q, Efimova O, He L, Tatsumoto S, Stepanova V, Oishi T, Udono T, Yamaguchi K, Shigenobu S, et al. 2018. Human-specific features of spatial gene expression and regulation in eight brain regions. Genome Res 28: 1097–1110. 10.1101/gr.231357.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Yang Y, Yuan J, Sun Y, Dai J, Su B. 2019. Transcriptomic landscape of von Economo neurons in human anterior cingulate cortex revealed by microdissected-cell RNA sequencing. Cerebral Cortex 29: 838–851. 10.1093/cercor/bhy286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. 2018. Molecular architecture of the mouse nervous system. Cell 174: 999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, et al. 2012. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 149: 483–496. 10.1016/j.cell.2012.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, Esteller-Cucala P, Juan D, Ferrández-Peral L, Gulden FO, et al. 2018. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362: eaat8077 10.1126/science.aat8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.