Abstract
High-throughput genetic screens are powerful methods to interrogate gene function on a genome-wide scale and identify genes responsible to certain stresses. Here, we developed a piggyBac strategy to deliver pooled sgRNA libraries stably into cell lines. We used this strategy to conduct a screen based on genome-wide clustered regularly interspaced short palindromic repeat technology (CRISPR)-Cas9 in Bombyx mori cells. We first constructed a single guide RNA (sgRNA) library containing 94,000 sgRNAs, which targeted 16,571 protein-coding genes. We then generated knockout collections in BmE cells using the piggyBac transposon. We identified 1006 genes that are essential for cell viability under normal growth conditions. Of the identified genes, 82.4% (829 genes) were homologous to essential genes in seven animal species. We also identified 838 genes whose loss facilitated cell growth. Next, we performed context-specific positive screens for resistance to biotic or nonbiotic stresses using temperature and baculovirus separately, which identified several key genes and pathways from each screen. Collectively, our results provide a novel and versatile platform for functional annotations of B. mori genomes and deciphering key genes responsible for various conditions. This study also shows the effectiveness, practicality, and convenience of genome-wide CRISPR screens in nonmodel organisms.
A major task in biology is to functionally annotate genomes and to identify genetic elements underlying normal cellular processes or diseases. In the past decade, genome-wide loss-of-function screens, which is a powerful hypothesis-free approach to discover genes that underlie certain biological processes, have been used successfully to address many fundamental biological questions (Boutros et al. 2004; Carette et al. 2009). In diploid eukaryotic cells, high-throughput loss-of-function screens have primarily been achieved by RNA interference (RNAi), either in well-by-well arrays or in barcoded pooled libraries (Mohr et al. 2014). Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated protein 9 (Cas9) (CRISPR) system has been explored for pooled genome-scale functional screening (Shalem et al. 2014; Wang et al. 2014). The CRISPR system largely overcomes major drawbacks of RNAi such as incomplete loss-of-function and off-target effects. More fitness genes can be identified using the CRISPR system compared to using RNAi (Hart et al. 2014; Shalem et al. 2014; Wang et al. 2014). Furthermore, the CRISPR system has been confirmed to be efficient in identifying genes for drug resistance (Koike-Yusa et al. 2014; Hou et al. 2017), tumorigenesis (Chen et al. 2015; Song et al. 2017; Xu et al. 2017), immune response (Parnas et al. 2015), host-pathogen interactions (Ma et al. 2015; Kim et al. 2018), and cancer immunotherapy (Patel et al. 2017).
Currently, CRISPR library screenings are only available in mammalian cells, bacteria (Liu et al. 2017; Wang et al. 2018), and Drosophila cells (Bassett et al. 2015; Viswanatha et al. 2018). To gain a comprehensive understanding of the genomic basis of universal biological processes or particular phenomena, genome-wide loss-of-function screens in more organisms is indispensable and urgently needed. Bombyx mori is a lepidopteran insect with both research and agricultural importance. Genetics and genomics studies in B. mori have provided insight into many fundamental biological questions, such as domestication history, sex determination, metamorphosis, and silk production (Goldsmith et al. 2005; Xia et al. 2009; Omenetto and Kaplan 2010; Kiuchi et al. 2014; Xia et al. 2014). However, approximately >80% of the genes in B. mori remain functionally uncharacterized.
In the current study, we developed a novel method to conduct genome-wide CRISPR screens in B. mori cells. A plasmid library containing 94,000 single guide RNAs (sgRNAs) was generated and stably delivered into BmE cells using the piggyBac transposon. Moreover, we showed that this system can be used to identify genes essential for cell viability in normal conditions and to screen resistance genes for both biotic and nonbiotic stresses. This system provides a powerful platform to rapidly elucidate the functional genome of B. mori and to identify key genes and mechanisms associated with particular biological processes. Furthermore, because piggyBac is a universal tool that can drive gene delivery in a wide range of organisms, including insects, plants and animals, this approach should be amenable to adapting CRISPR screens in other cell lines.
Results
Development of a piggyBac library delivery method for B. mori cells
In mammalian cells, pooled DNA libraries are often delivered by lentiviral vectors. However, in B. mori, lentiviral vectors are extremely inefficient. As an alternative strategy to deliver the sgRNA library, we chose piggyBac, which has been shown to have high transformation activity in both B. mori germline and cultured cells. Previously, we established a binary transient CRISPR vector for gene knockout in the B. mori embryonic cell line BmE (Ma et al. 2017). To allow for the integration of both Cas9 and sgRNA expression cassettes into the genome of a single cell, we combined two cassettes into an all-in-one vector, pB-CRISPR (Supplemental Materials). A zeocin expression cassette in which a zeocin antibiotic resistance gene is driven by the IE2 promoter, was also included in pB-CRISPR to facilitate the establishment of stable transgenic lines (Fig. 1A).
Figure 1.
The piggyBac library delivery method for gene knockout in B. mori cells. (A) Schematic of the piggyBac library delivery vector (pB-CRISPR): (ITR) inverted terminal repeats of piggyBac; (IE2) IE2 promoter; (Zeocin) zeocin selection marker gene; (Ser1) Sericin 1 poly(A); (gRNA) sgRNA and Scaffolds; (U6) polymerase III U6- promoter; (Hsp70) hsp70 promoter; (SpCas9) B. mori codon-optimized SpCas9 coding sequence; (SV40) SV40 poly(A). (B) Flow cytometry analysis (upper) and statistical analysis (lower) of BmE-Mi-EGFP cells transduced with pB-CRISPRs: (mock) no vector; (NS-1) pB-CRISPR-NS-1; (NS-2) pB-CRISPR-NS-2; (EGFP-1) pB-CRISPR- EGFP-1; (EGFP-2) pB-CRISPR-EGFP-2; (EGFP-3) pB-CRISPR-EGFP-3. (C) Fluorescent images of BmE-Mi-EGFP knockout experiments. (D) Sanger sequence analysis of the ratio of indels for EGFP-1 (left), EGFP-2 (middle), and EGFP-3 (right) target regions of BmE-Mi-EGFP cells.
To test the knockout efficiency of pB-CRISPR, we first established a cell line that stably expressed enhanced green fluorescent protein (EGFP; BmE-EGFP) using a second general transposon, Minos (Supplemental Fig. S1A,C; Supplemental Materials). Three sgRNAs targeting EGFP were designed and constructed into pB-CRISPR, and two nonspecific sgRNAs were used as controls (Supplemental Fig. S2; Supplemental Table S1). Each vector was cotransfected with a piggyBac transposase expression vector (A3-Helper) into BmE-EGFP. After 2 mo of selection with zeocin, EGFP fluorescence was totally abolished in cells transfected with all three EGFP targeting sgRNAs, while cells transfected with either control sgRNA remained unchanged (Fig. 1C; Supplemental Fig. S1D). Flow cytometry (FCM) analysis also showed a significant decrease in EGFP signals in the targeting sgRNA transfected cells, whereas the signals were not decreased in control cells (Fig. 1B).
The targeting regions of EGFP were amplified by the polymerase chain reaction (PCR) and cloned for sequencing. Sequencing results from a total of 44 clones showed that nearly 100% of the EGFP alleles had genetic variations with >70% causing frame shift mutations (Fig. 1D; Supplemental Fig. S1B). The editing efficiency was much higher than that achieved by transient transfection with binary vectors (Ma et al. 2017). Similar to our previous observations, small deletions dominated the types of mutations (71.4%–92.3%) (Ma et al. 2014). From these results, we concluded that piggyBac was suitable to stably deliver CRISPR constructs into the B. mori genome and that our established pB-CRISPR vector was suitable for efficient gene knockout in BmE cells.
Design and construction of the CRISPR sgRNA library
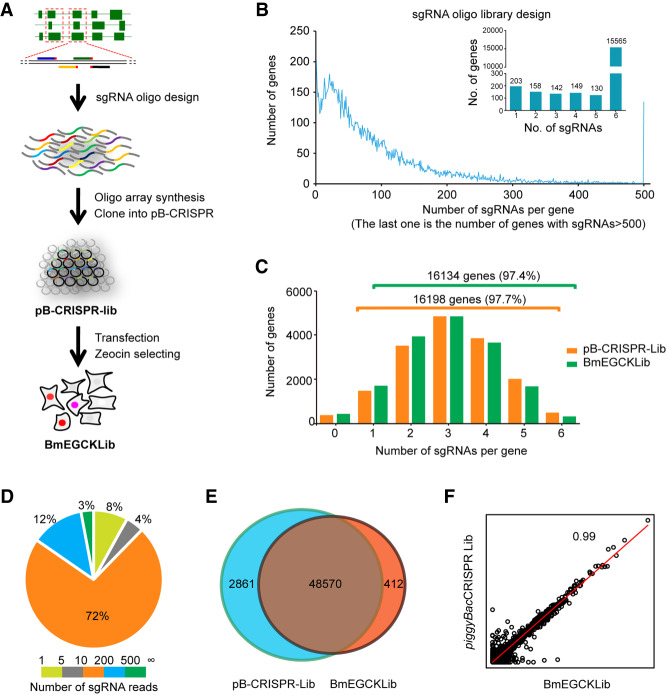
To construct the genome-wide sgRNA library, we designed a strategy as illustrated in Figure 2A. The library was constructed according to the following procedure. (1) The whole genome of B. mori was searched for all exons which were used to design a total of 1,534,227 sgRNAs (Fig. 2B). The sgRNAs were then ranked using the following two criteria. First, location of an sgRNA within the first half of the coding region to ensure any frame shift at the target site could disrupt the protein function. Second, to avoid potential off-target effects, sgRNA sequences were unique at the seed region located 12 bp downstream from the protospacer adjacent motif (PAM) and were as unique as possible in the nonseed region. Finally, 94,000 sgRNAs were chosen (approximately six sgRNAs per gene) and synthesized on a microarray chip (Supplemental Table S2). (2) The sgRNA oligonucleotide pool was subsequently amplified by PCR and cloned into pB-CRISPR to form the CRISPR knockout plasmid library (pB-CRISPR-lib; Fig. 2A). More than 108 clones on 240 petri dishes were selected with ampicillin, and >1000 clones per sgRNA were carried to maintain the sgRNA library diversity. Then we evaluated the quality of library by deep sequencing. A total of 51,431 sgRNAs were detected, which covered 96.3% of all B. mori genes. More than 87.4% of the genes had more than two sgRNAs (Fig. 2C), and 72% of sgRNAs had 11–200 reads (Fig. 2D). These results suggested that the coverage, accuracy, and diversity of the pB-CRISPR-lib were sufficient for the subsequent experiments. (3) Then the pB-CRISPR-lib and the piggyBac transposase expression vector (A3-Helper) were cotransfected into BmE cells. To maintain the high diversity of the original library, ∼2000 cells per sgRNA were transfected and ∼2000 cells per passage carried. To enrich sgRNA-harboring cells, the transfected cells were selected with zeocin for 2 mo followed by culturing with complete medium without antibiotic. Cells surviving these conditions composed the BmE genome-scale CRISPR-Cas9 knockout library (BmEGCKLib).
Figure 2.
Design and construction of the CRISPR sgRNA library for B. mori cells. (A) Flowchart for constructing the CRISPR sgRNA library for B. mori cells. (B) Design for all sgRNAs in all B. mori gene exons (lower), and the sgRNAs selected for the subsequent experiment (upper); see Supplemental Table S2. (C) Second-generation sequence analysis of the sgRNA distribution for the pB-CRISPR library (orange) and BmEGCKLib (green). (D) Pie chart showing the percent distribution of the sgRNAs. (E) Overlap in sgRNA distribution between the pB-CRISPR library and BmEGCKLib. (F) Correlation analysis between the pB-CRISPR library and BmEGCKLib.
To evaluate the coverage and diversity of the BmEGCKLib, genomic DNA was extracted from 4 × 107 cells, and the sgRNA regions were amplified by PCR for deep sequencing. A total of 48,982 sgRNAs, which accounted for 95.2% of the pB-CRISPR-lib, were detected with at least one read (Fig. 2E). The compositions of the BmEGCKLib and pB-CRISPR-lib libraries were highly correlated (correlation coefficient = 0.99) (Fig. 2F). The number of genes containing 1–6 sgRNAs in the BmEGCKLib was basically the same as the corresponding number in the pB-CRISPR-lib vector library (Fig. 2C). These results indicated that the genome-wide knockout cell library of BmE was successfully generated with sufficient coverage to perform further genetic screens.
Screening of essential and growth-restricting genes under normal conditions
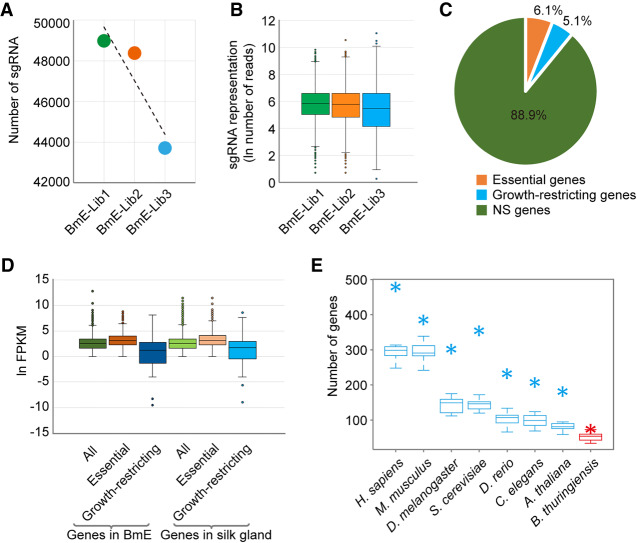
After generating the BmEGCKLib, we next aimed to identify essential genes that affected the survival on normal growth of BmE based on depletion of their sgRNAs in the library population, as well as growth-restricting genes that provided growth advantages based on sgRNA enrichment over time. BmEGCKLib cells were grown under normal conditions, and ∼4 × 107 cells were harvested at each of three time points: immediately (BmE-Lib1), 1 mo (BmE-Lib2), and 2 mo (BmE-Lib3) after completion of zeocin selection (Supplemental Fig. S3A). Deep sequencing of the libraries at different time points revealed a reduction in the correlation coefficients over time (Supplemental Fig. S3B); the number of sgRNAs also decreased in the BmE-Lib2 and BmE-Lib3 groups (Fig. 3A). Although the overall abundance of sgRNAs showed a gradual depletion, some sgRNAs were significantly enriched (Fig. 3B). These observations suggested that BmEGCKLib could be used for screening essential and growth-restricting genes.
Figure 3.
Screening of essential and growth-restricting genes in BmE cells. (A,B) Changes in sgRNAs contained in BmEGCKLib over time. Approximately 4 × 107 cells of BmEGCKLib were harvested at each of three time points: immediately (BmE-Lib1), 1 mo (BmE-Lib2), and 2 mo (BmE-Lib3) after completion of zeocin selection. Overall the sgRNAs were gradually depleted but some of them were enriched over time. (C) Percentages of essential genes and growth-restricting genes in BmE cells. (D) Expression levels among all genes, essential genes, and growth-restricting genes in BmE cells and silk gland cells. (E) The overlap of essential genes among B. mori and diverse eukaryotic species (Homo sapiens, Mus musculus, Drosophila melanogaster, Saccharomyces cerevisiae, Danio rerio, Caenorhabditis elegans, and Arabidopsis thaliana), and a prokaryotic species (Bacillus thuringiensis). (Asterisks) The intersection of 1006 essential genes with B. mori orthologs of essential genes in the eight species; (box plots) the intersection of 1000 random genes with B. mori orthologs of essential genes in the eight species.
We used the MAGeCK program (Li et al. 2014) to compute the negative (sgRNA depleted) or positive (sgRNA enriched) scores for each gene. Both the log10 fold-changes of individual sgRNAs and log10(negative scores) of genes showed good correlations between two biological replicates (r = 0.87 and r = 0.76, respectively) (Supplemental Fig. S4; Supplemental Table S3), indicating a considerable high reproducibility of our fitness screens. All of the B. mori genes were ranked by negative or positive scores and a P-value less than 0.05 was used as a threshold value. We identified 1006 genes as essential for normal growth (Supplemental Fig. S3C; Supplemental Table S4) and 838 genes as growth-restricting genes (Supplemental Fig. S3D; Supplemental Table S4). The relative ratios of essential or growth-restricting genes to the total number of coding genes (∼6.1% and ∼5.1%, respectively) were equal to those reported in other species (Fig. 3C). Furthermore, we compared the 1006 essential genes to those identified in Homo sapiens, Mus musculus, Drosophila melanogaster, Saccharomyces cerevisiae, Danio rerio, Caenorhabditis elegans, Arabidopsis thaliana, and Bacillus thuringiensis. Considerable overlap was found between B. mori essential genes and available eukaryotic species; however, essential genes of BmE only partially overlapped with the essential genes reported for the prokaryote B. thuringiensis (Fig. 3E). Because true essential genes must be transcriptionally active, we measured the expression level of essential genes using RNA-seq data to validate the CRISPR results (Fig. 3D; Supplemental Table S5). As expected, the essential genes showed higher transcriptional levels compared to the average expression level of all genes, whereas the transcriptional levels for the growth-restricting genes were lower. The same tendency was also found among three gene groups in the silk gland, suggesting that the essential genes defined in BmE cells may also be essential in specialized tissues, or even in whole organisms (Fig. 3D).
Analysis of essential genes for B. mori cells
We next investigated the essential and growth-restricting genes of B. mori cells from different aspects, including genome-wide distribution, functional categorization, and cellular localization. Comparison of all genes density and selected genes revealed that overall both essential and growth-restricting genes were evenly distributed across all chromosomes, with the exception of a few specific chromosomal regions (Supplemental Fig. S5). Random distribution of essential and growth-restricting genes is also observed in mammalian cells (Yilmaz et al. 2018).
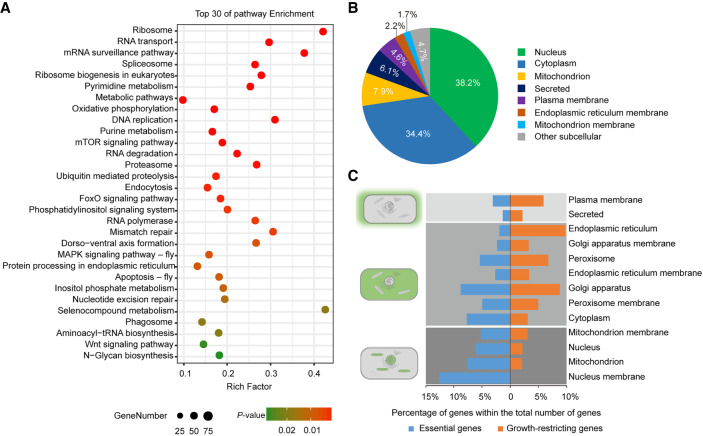
Functional categorization by Gene Ontology (GO) analysis revealed that the essential genes were primarily related to core cell components (cell part, binding, and catalytic activity) and metabolism (Supplemental Fig. S6A). A majority of basic Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were significantly enriched in essential genes (Fig. 4A), including Ribosome (42.1%), RNA transport (29.5%), mRNA surveillance (37.5%), Spliceosome (26.2%), Ribosome biogenesis in eukaryotes (27.5%), and Pyrimidine metabolism (25.0%). These pathways are mainly involved in the fundamental biological processes that maintain cell growth, proliferation, and survival. For example, three of the most enriched categories pertained to DNA processing (11 genes, “DNA replication”; six genes, “mismatch repair”), RNA processing (36 genes, “RNA transport”; 24 genes, “mRNA surveillance”; 32 genes, “spliceosome”; 14 genes, “RNA degradation”), and protein processing (53 genes, “Ribosome”; 10 genes, “proteasome”). We also found that some signaling pathways, such as mTOR and FoxO, were significantly enriched. Inactivation of TOR in both Drosophila and mammalian cells has shown that TOR controls both cell size and proliferation in early embryos (Murakami et al. 2004). FOXO1 has also been found to be an essential regulator of pluripotency in human embryonic stem cells and developing embryos (Yu et al. 2018). Therefore, we suspect that mTOR and FoxO signaling pathways were enriched because BmE is an embryonic cell line of B. mori. Unlike the essential genes, the growth-restricting genes were primarily enriched in lysosome and diverse metabolic pathways which are dispensable for cell survival but critical for growth control and secondary metabolism (Supplemental Fig. S6B).
Figure 4.
Analysis of essential genes for BmE cells. (A) Top 30 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of essential genes. (B) Distribution of essential genes assigned to subcellular localization. (C) Percent distribution of essential and growth-restricting genes assigned to subcellular localization within the total number of genes.
Subcellular localization analysis showed that the majority of the essential genes encoded proteins that localized to nucleus (38.2%), cytoplasm (34.4%), and mitochondrion (7.9%), and the rest encoded secreted proteins or proteins that localized to other cellular compartments, such as the plasma membrane, endoplasmic reticulum membrane, and mitochondrial membrane (Fig. 4B). To further investigate the importance of subcellular localization of selected genes, we analyzed the percentage of essential genes among all genes assigned to different categories of subcellular localization. We found that significantly more essential genes than growth-restricting genes were localized in the nucleus and mitochondrion, and a higher proportion of growth-restricting genes than essential genes were assigned to cellular compartments related to the extracellular space such as plasma membrane and secreted proteins (Fig. 4C). The observed biased cellular localizations of essential and growth-restricting genes have also been observed in mammalian cells (Yilmaz et al. 2018).
Use of B. mori CRISPR screens to identify genes responsible for temperature challenge
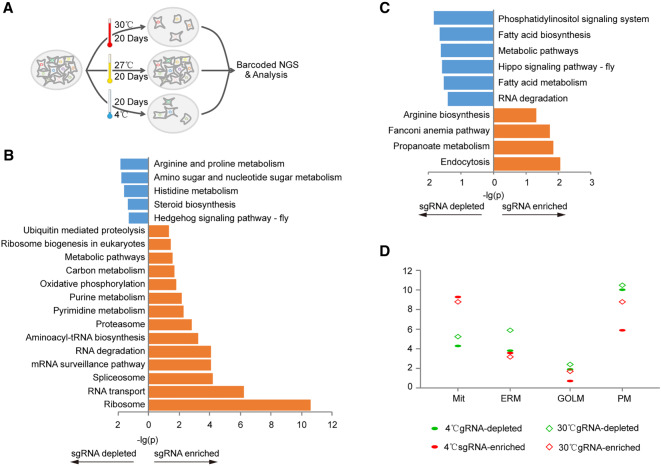
We next asked whether our BmEGCKLib library could be used as a CRISPR screening platform to identify genes that respond to environmental stimuli. As a proof-of-principle investigation, we chose temperature challenge. Each group of about 4 × 107 BmEGCKLib cells was exposed to 4°C and 30°C for 20 d; cells grown at 27°C were used as control (Fig. 5A). Only a few cells survived in the 4°C and 30°C groups, whereas cells in the control group grew well. The sgRNAs of surviving cells were analyzed for enrichment and depletion using similar methods to those used to identify the essential and growth-restricting genes. Apparently, several KEGG pathways were significantly enriched in cells challenged at 4°C and overlapped highly with some pathways for essential genes. The overlapped pathways were categorized to RNA protein processing. However, none of the pathways pertaining to DNA processing that were enriched in essential genes were found in the 4°C challenged group (Fig. 5B; Supplemental Table S6). We also found that genes with depleted sgRNAs in the 4°C group were significantly enriched in the steroid biosynthesis pathway, and genes with depleted sgRNAs in the 30°C group were highly enriched in fatty acid biosynthesis and fatty acid metabolism pathways (Fig. 5B,C; Supplemental Table S6). Both steroids and fatty acids are known to be key regulators of cell membrane fluidity (Singer and Nicolson 1972; Ipsen et al. 1987). These results are consistent with their reported role in membrane fluidity (Ipsen et al. 1987) as well as their protective roles at low temperatures (Xu and Siegenthaler 1997). Comparing the proportions of enriched genes on cytomembrane, subcellular organ membranes, and other nonmembrane subcellular organs, we found genes that anchored on biomembranes, especially on the cytomembrane, were likely to be associated with depleted sgRNAs in both the 4°C and 30°C groups (Fig. 5D). This suggested that biomembrane systems were more likely to be destroyed by extreme temperatures. We also identified more genes with enriched sgRNAs compared to those with depleted sgRNAs in the mitochondria of cells from both temperature challenge groups (Fig. 5D; Supplemental Table S6). This indicated that decreased energy metabolism may help cells survive against extreme ambient temperatures.
Figure 5.
Screening of genes responsible for temperature challenge. (A) Flowchart for the screening of genes involved in a response to temperature challenges. (B,C) KEGG pathway enrichment for the genes in cells cultured at 4°C (B) and 30°C (C). (D) Percentage of major types of subcellular localization genes within the total number of genes at 4°C and 30°C: (Mit) mitochondrion; (ERM) endoplasmic reticulum membrane; (GOLM) Golgi apparatus membrane; (PM) plasma membrane.
Use of B. mori CRISPR screens to identify host-pathogen interactions
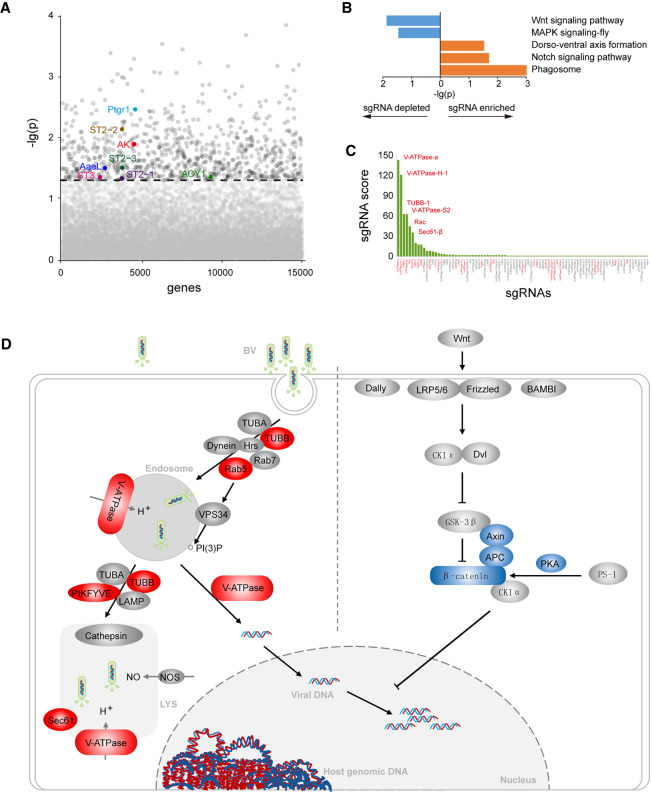
Last, we aimed to take advantage of the hypothesis-free application of the BmEGCKLib screen platform to identify genes involved in host-pathogen interactions between BmNPV and B. mori. BmNPV, a typical species of Baculoviridae, is a natural B. mori pathogen that causes enormous economic losses in the sericulture industry every year. BmEGCKLib cells were infected with BmNPV four times at a high level of infection once every 2 d. The few cells surviving among approximately 4 × 107 cells infected were harvested for analysis of host-pathogen interactions. Using a P-value <0.05 as a threshold, we identified a positive selection of 811 genes and negative selection of 809 genes, in which most highly ranked genes were represented by multiple independent sgRNAs. Many genes previously reported to play anti-BmNPV roles were found in the group of positively selected genes (Fig. 6A; Supplemental Table S7), suggesting that the BmEGCKLib can be used to identify genes involved in B. mori-BmNPV interactions and that the 1614 selected genes may be truly responsible for BmNPV infection.
Figure 6.
Screening of genes involved in the interaction between BmE cells and B. mori nucleopolyhedrovirus (BmNPV). (A) Plot of genes ranked by negative score. The portion of genes that associated with virus resistance are highlighted. (B) Genes for BmE-BmNPV interactions uncovered by the KEGG pathway analysis. (C) sgRNA score of genes in the phagosome pathway. (D) Schematic of B. mori cell interactions with BmNPV.
We then mapped the selected genes to KEGG pathways (Fig. 6B). We observed enrichment for genes involved in phagosome and notch signaling pathways, both of which are represented in genome-wide screens for influenza virus and have been shown by several previous studies to be involved in the host response to influenza virus, vesicular stomatitis virus, and Autographa californica multiple nucleopolyhedrovirus (AcMNPV) (Volkman and Goldsmith 1985; Maxfield and Yamashiro 1987; Sun et al. 2005). To investigate further the possible mechanism of BmNPV entry into B. mori cells, we analyzed in detail the genes in the most enriched pathway, the phagosome pathway. We found five of 12 genes were the vacuolar-type H+-ATPase (V-ATPase) subunits (Fig. 6C; Supplemental Table S7). As a key enzyme of endosomal acidification, V-ATPase has been shown to play an important role during influenza virus entry into mammalian cells (Rossmann and Rao 2012). Previously, V-ATPase was found to show a higher expression level in NPV-resistant B. mori strains (Lu et al. 2013). A more recent study showed that inhibition of endosome acidification by ammonium chloride treatment in B. mori cells results in significant inhibition of virus reproduction (Feng et al. 2018). In addition to V-ATPase, five other components of the endocytosis pathways, including TUBB, Sec61, Rab5, Rac, and PIKFYVE, were significantly enriched (Fig. 6D). These results suggest that endocytosis, especially V-ATPase-mediated endosomal acidification, plays a critical role during the entry of BmNPV into B. mori cells.
To reveal further the genes and pathways involved in host cell defense against virus infection, we also analyzed negatively selected genes. These genes were primarily classified in the Wnt signaling and MAPK signaling pathways (Fig. 6B). Wnt is a cellular pathway related to cell cycle signaling that has also been shown to be affected by diverse human viruses, such as Epstein–Barr virus (Birdwell et al. 2018), human herpesvirus (Liu et al. 2016), human papillomavirus (Bello et al. 2015), and hepatitis B virus (Yin et al. 2017). Thus, the manipulation of Wnt signaling is recognized as a critical generalized process for viral pathogenesis (Hao et al. 2015). The Wnt pathway has also been identified in several genome-wide genetics screens in human cells or Drosophila against Rift Valley fever virus and Sendai virus (Baril et al. 2013; Harmon et al. 2016). However, whether the Wnt pathway has antiviral activity against BmNPV or whether BmNPV modulates Wnt signaling to evade this antiviral host response remains unknown. Our results showed that a core component of the Wnt pathway, β-catenin, was highly ranked among the negatively selected genes (Fig. 6D). This supports the supposition that the Wnt pathway may play an important role in B. mori for host response against BmNPV.
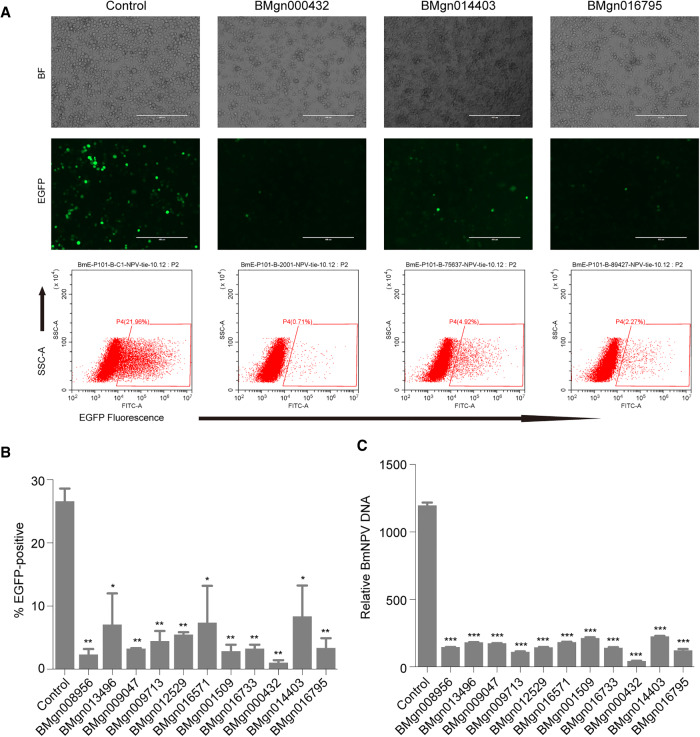
To validate whether the screened genes play important roles in the BmNPV infection, we randomly chose nine genes from the top 30 positively screened genes and two genes encoding V-ATPase subunits, and we constructed 11 knockout cell lines. BmNPV infections were performed with MOI = 1 in 11 cell lines; BmE cells were used as control. The fluorescent microscopy observation and flow cytometry analysis at 72 hpi both revealed that BmNPV positive cells were significantly fewer in all 11 knockout cell lines than that in control BmE cells (Fig. 7A,B; Supplemental Fig. S7). Quantification of viral DNA in infected cells using qPCR showed that the DNA content of BmNPV were significantly lower in all knockout cell lines (Fig. 7C). These results indicated the crucial roles for 11 genes under BmNPV infection and the accuracies of our screens results.
Figure 7.
Functional analysis of genes involved in B. mori-BmNPV interactions. (A) Fluorescent images analysis (upper) and flow cytometry analysis (lower) of the BmE knockout cells infected with BmNPV at an MOI of 1. (B,C) Statistical flow cytometry analysis (B) and relative BmNPV DNA level (C) in 11 BmE knockout cells infected with BmNPV at an MOI of 1. Statistically significant differences were determined by Student's t-test: (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Discussion
Genome-wide CRISPR-based screening technologies have greatly accelerated the functional annotation of genomes and provided a hypothesis-free, cost-effective pipeline to uncover target genes in certain contexts. Delivery of genome-wide CRISPR libraries is achieved mostly by lentiviral vectors in mammalian cells and recently by site-specific recombination in Drosophila cells (Shalem et al. 2014; Viswanatha et al. 2018). However, these approaches are currently inapplicable for B. mori and many other nonmodel organisms, possibly as a result of their low efficiency. In the current study, we showed that the piggyBac transposon could be used as an alternative strategy to deliver multiplexed DNA libraries. The coverage, accuracy, and sgRNA distribution of the newly constructed BmEGCKLib were sufficient to perform the screening of essential and growth-restricting genes under normal conditions, as well as context-specific screening under abiotic (such as temperature challenge) and biotic (such as host-pathogen interactions) stresses. The piggyBac transposon, a mobile genetic element present in a diverse range of species, can efficiently transpose between vectors and chromosomes in nearly all organisms tested. Because piggyBac transposes via a “cut and paste” mechanism, the inserted copy number per cell can be easily controlled by quantifying the transfected donor plasmid. In the present study, we observed that >95% cells in the BmEGCKLib harbored only a single sgRNA insertion as revealed by single-cell sequencing (Supplemental Fig. S8). Furthermore, the piggyBac transposon has the largest reported cargo capacity, up to 207 kb (Li et al. 2013), for these kinds of vectors, allowing codelivery of other functional elements with a CRISPR library in complex screens. Thus, we believe that the piggyBac strategy developed in our study possesses several advantages over lentiviral or recombinase-based strategies and will be versatile for diverse cell types and organisms.
B. mori is among the most important economically beneficial insects and a powerful model system for Lepidoptera because of its finely decoded genomic sequence (Xia et al. 2008), rich genetic resources (Goldsmith et al. 2005), and multiple available tools for genetic manipulation (Tamura et al. 2000; Takasu et al. 2010; Ma et al. 2012; Wang et al. 2013). After the completion of B. mori genome sequencing, the systematical functional investigation of all B. mori genes has been a major challenge. Transgenic and genome editing technologies have been established in an effort to achieve this goal. However, the function of >80% of the B. mori genes remains unclear, partially because of the low efficiency of hypothesis-driven research strategies, which heavily rely on previous knowledge and well-grounded hypotheses. In the current study, we presented the first hypothesis-free genetic screen approach to rapidly investigate the function of B. mori genes on a genome-wide scale. A total of 1006 genes essential for cell viability, 838 genes that restricted cell growth, 3013 genes responsible for ambient temperature change, and 1614 genes involved in BmNPV infection were identified using our genetic screen approach.
The identification and analysis of genes essential for cell viability in B. mori also implies some interesting aspects of cell biology in general. First, we found that the essential genes in B. mori shared considerable overlap with all seven eukaryotic species compared, whereas very limited overlap existed with the prokaryote B. thuringiensis, indicating an evolutionary conservation of basic biological processes for eukaryotic cell survival. Second, essential genes primarily encoded proteins involved in fundamental biological processes such as DNA, RNA, and protein processing, and overall were evenly distributed across all chromosomes, suggesting that the random chromosomal distribution of essential genes may have a protective role in the core biological processes. Third, these results indicated that essential genes leading to cell survival under normal conditions were located primarily in the nucleus, cytoplasm and mitochondrion, whereas genes located on the cell membrane and extracellular matrix were much lower in the numbers detected by our screen. That these have also been observed in mammalian cells (Yilmaz et al. 2018) suggests that (1) essential genes may have different roles in regulating cell growth; (2) essential genes may have different levels of functionally redundant genes in the different cellular compartments; and (3) most proteins secreted outside cells are not crucial to cell viability and their loss may even promote cell growth.
Because the ambient temperature has a great impact on the ectotherm-like insect, we performed the CRISPR screening to identify genes that respond to environmental stimuli in BmE cells. Our screening showed DNA maintenance and replication were the core events of the cells and their disruption probably threatens cell survival; however, two other fundamental biological processes crucial to the maintenance of cell growth in normal conditions, RNA processing and protein processing, were not. In addition, gene function clustering revealed steroids and fatty acids played important roles in membrane fluidity and protected cells from extreme ambient temperatures. Additionally, subcellular localization analyzing suggested that biomembrane systems were very fragile at extreme temperatures, and the decreased energy metabolism may help cells survive against extreme ambient temperatures. Because global climate change has become the most serious environmental issue, the extreme temperatures appear more frequently. Therefore, the study of the genes responsible for the temperature challenge in BmE cells is helpful to understand the effects of climate change on insects. Because insects are the main agricultural pest, our research also provided enlightenment for pest control under global climate changes.
BmNPV is the most important pathogen of B. mori. The research of the interaction mechanism between BmNPV and host B. mori was important not only for sericulture but also for pest control. Taken together, our BmEGCKLib screening confirmed that previously described biological processes, such as phagosome and notch signaling pathways, are involved in the interaction between BmNPV and host B. mori cells and uncovered some novel processes, such as dorsal-ventral axis formation and the Wnt pathways associated with the infection of BmNPV. In addition, we identified more than 493 significantly selected genes whose functions are currently unknown owing to their limited homology with identified genes of model organisms. We propose that some of the selected genes may also be involved in the interaction between BmNPV and host B. mori cells and may provide a reliable resource for defining new targets for antiviral drugs and breeding silkworm strains genetically resistant to this widespread pathogen. It is the first time host-pathogen interactions were uncovered in an insect through a genome-scale knockout library approach. Our strategy can provide new ideas for insect antiviral research.
We developed a functional screening platform in B. mori using piggyBac-delivered CRISPR libraries. We showed that CRISPR screening is a powerful tool for the rapid investigation of gene functions on a large scale and may serve as a powerful resource for investigating long-standing questions in B. mori and entomology. The value of our platform was shown by the discovery and highlighting of a large set of new genes and pathways that participate in cell variability, cell growth, ambient temperature stimuli, and BmNPV infection. We believe that further validation and investigation of the candidate target genes revealed by our screening will shed more light on the molecular mechanisms of host response to ambient temperature stimuli and BmNPV infection. More importantly, the present study lays the ground for further genetic screens against diverse biological processes.
Methods
Design and construction of the vectors
The piggyBac transposase expression vector, A3-helper was from stocks stored in our laboratory. The CRISPR library delivery vector, pB-CRISPR, was constructed with the process described in the Supplemental Materials. The target vectors used to test the knockout efficiency of pB-CRISPR was constructed using library construction methods. The EGFP expression cassette was delivered by Minos transposons, the vector named PUC57-Mi-puro-EGFP. The Minos transposase gene expression cassette was synthesized and inserted into pUC57-T-simple and the plasmid named Mi-helper. The details were available in the Supplemental Materials. The whole sequences are available in Supplemental Materials.
sgRNA library design, synthesis, and construction
The sgRNAs were designed using the CasFinder method (Aach et al. 2014). All sgRNAs selected had a 5′ G added to improve the U6 transcription efficiency. The library of sgRNAs were encoded within 70-nt oligonucleotides and synthesized on the 94,000 arrays using the services of BGI. U6 promoter, multiple sgRNAs, and sgRNA scaffolds were linked together using overlap PCR, then cloned into the AscI/NheI site of pB-CRISPR vector to construct the CRISPR knockout plasmid library (pB-CRISPR library). The details are available in the Supplemental Materials.
Genome-scale screening in B. mori
For essential or growth-restricting genes screening, three parts of 4 × 107 BmEGCKLib cells were harvested at three time points. For the screening of genes responsible for temperature challenging, three sets of 4 × 107 cells of the BmEGCKLib in complete medium were exposed for 20 d to different temperatures. For the screening to identify genes involved in host-pathogen interactions, 4 × 107 cells of the BmEGCKLib were supplemented with BmNPV (with an EGFP tag, stored in our laboratory) for four rounds of infection. The genomic DNA was extracted from each group, and the sgRNA distribution analyzed by second-generation sequencing (Mega Genomic). The details are available in the Supplemental Materials.
Pooled sgRNA sequences and data analysis
All of the genomic DNA extracted from the cell libraries was PCR amplified. The PCR products were gel-purified and sequenced by Illumina. The raw data were uploaded to a server in our laboratory and filtered. The paired sequences were then spliced using flash software. Then the number of reads for each sgRNA was calculated using Bowtie 2 (Langmead and Salzberg 2012) and performed the next analysis using MAGeCK. The details are available in the Supplemental Materials.
The analysis of EGFP knockout efficiency of pB-CRISPR
To test the knockout efficiency of pB-CRISPR, three sgRNAs targeting EGFP were constructed into pB-CRISPR, and two nonspecific (NS) sgRNAs were used as controls (Supplemental Fig. S2; Supplemental Table S1). Each vector was cotransfected with A3-Helper into the BmE-EGFP cell line. After 2 mo of selection with zeocin, all the samples underwent flow cytometry analysis, fluorescence imaging, and Sanger sequencing. The details are available in the Methods section of the Supplemental Materials.
Data access
The raw data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA602216.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Prof. Marian R. Goldsmith for her valuable advice and help in the preparation of this manuscript. This work was supported by grants from the National Natural Science Foundation of China (Nos. 31802011, 31530071), and Chongqing Research program of basic Research and Frontier Technology (Nos. cstc2017jcyjAX0349, cstc2018jcyjAX0471). None of the funding entities played roles in the design of the study; collection, analysis, and interpretation of data; or in writing the manuscript.
Author contributions: J.C. and S.M. designed the experiments. J.C. and K.Y. performed most of the experiments with help from R.W., Y.L., R.S., and X.W. R.W. performed the functional validation experiments during the revision process. T.Z. and X.C. performed the bioinformatics analysis. J.C. and S.M. wrote the manuscript with help from Q.X. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.249045.119.
References
- Aach J, Mali PR, Church GM. 2014. CasFinder: flexible algorithm for identifying specific Cas9 targets in genomes. bioRxiv 10.1101/005074 [DOI]
- Baril M, Es-Saad S, Chatel-Chaix L, Fink K, Pham T, Raymond VA, Audette K, Guenier AS, Duchaine J, Servant M, et al. 2013. Genome-wide RNAi screen reveals a new role of a WNT/CTNNB1 signaling pathway as negative regulator of virus-induced innate immune responses. PLoS Pathog 9: e1003416 10.1371/journal.ppat.1003416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Kong LS, Liu JL. 2015. A genome-wide CRISPR library for high-throughput genetic screening in Drosophila cells. J Genet Genomics 42: 301–309. 10.1016/j.jgg.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello JOM, Nieva LO, Paredes AC, Gonzalez AMF, Zavaleta LR, Lizano M. 2015. Regulation of the Wnt/β-catenin signaling pathway by human papillomavirus E6 and E7 oncoproteins. Viruses 7: 4734–4755. 10.3390/v7082842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdwell CE, Prasai K, Dykes S, Jia Y, Munroe TGC, Bienkowska-Haba M, Scott RS. 2018. Epstein-Barr virus stably confers an invasive phenotype to epithelial cells through reprogramming of the WNT pathway. Oncotarget 9: 10417–10435. 10.18632/oncotarget.23824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Heidelberg Fly Array Consortium, Paro R, Perrimon N. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835. 10.1126/science.1091266 [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. 2009. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326: 1231–1235. 10.1126/science.1178955 [DOI] [PubMed] [Google Scholar]
- Chen SD, Sanjana NE, Zheng KJ, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. 2015. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160: 1246–1260. 10.1016/j.cell.2015.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M, Zhang JJ, Xu WF, Wang HP, Kong XS, Wu XF. 2018. Bombyx mori nucleopolyhedrovirus utilizes a clathrin and dynamin dependent endocytosis entry pathway into BmN cells. Virus Res 253: 12–19. 10.1016/j.virusres.2018.05.020 [DOI] [PubMed] [Google Scholar]
- Goldsmith MR, Shimada T, Abe H. 2005. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol 50: 71–100. 10.1146/annurev.ento.50.071803.130456 [DOI] [PubMed] [Google Scholar]
- Hao HP, Wen LB, Li JR, Wang Y, Ni B, Wang R, Wang X, Sun MX, Fan HJ, Mao X. 2015. LiCl inhibits PRRSV infection by enhancing Wnt/β-catenin pathway and suppressing inflammatory responses. Antivir Res 117: 99–109. 10.1016/j.antiviral.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Harmon B, Bird SW, Schudel BR, Hatch AV, Rasley A, Negrete OA. 2016. A genome-wide RNA interference screen identifies a role for Wnt/β-catenin signaling during Rift Valley fever virus infection. J Virol 90: 7084–7097. 10.1128/JVI.00543-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. 2014. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol 10: 733 10.15252/msb.20145216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou PP, Wu C, Wang YC, Qi R, Bhavanasi D, Zuo ZX, Dos Santos C, Chen SL, Chen Y, Zheng H, et al. 2017. A genome-wide CRISPR screen identifies genes critical for resistance to FLT3 inhibitor AC220. Cancer Res 77: 4402–4413. 10.1158/0008-5472.CAN-16-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsen JH, Karlstrom G, Mouritsen OG, Wennerstrom H, Zuckermann MJ. 1987. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim Biophys Acta 905: 162–172. 10.1016/0005-2736(87)90020-4 [DOI] [PubMed] [Google Scholar]
- Kim HS, Lee K, Kim SJ, Cho S, Shin HJ, Kim C, Kim JS. 2018. Arrayed CRISPR screen with image-based assay reliably uncovers host genes required for coxsackievirus infection. Genome Res 28: 859–868. 10.1101/gr.230250.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T, et al. 2014. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509: 633–636. 10.1038/nature13315 [DOI] [PubMed] [Google Scholar]
- Koike-Yusa H, Li YL, Tan EP, Velasco-Herrera MD, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32: 267–273. 10.1038/nbt.2800 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RB, Zhuang Y, Han M, Xu T, Wu XH. 2013. piggyBac as a high-capacity transgenesis and gene-therapy vector in human cells and mice. Dis Model Mech 6: 828–833. 10.1242/dmm.010827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao TF, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hancock M, Workman A, Doster A, Jones C. 2016. β-Catenin, a transcription factor activated by canonical Wnt signaling, is expressed in sensory neurons of calves latently infected with bovine herpesvirus 1. J Virol 90: 3148–3159. 10.1128/JVI.02971-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gallay C, Kjos M, Domenech A, Slager J, van Kessel SP, Knoops K, Sorg RA, Zhang JR, Veening JW. 2017. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 13: 931 10.15252/msb.20167449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Xia HC, Gao L, Pan Y, Wang Y, Cheng X, Lu HG, Lin F, Chen L, Yao Q, et al. 2013. V-ATPase is involved in silkworm defense response against Bombyx mori nucleopolyhedrovirus. PLoS One 8: e64962 10.1371/journal.pone.0064962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SY, Zhang SL, Wang F, Liu Y, Liu YY, Xu HF, Liu C, Lin Y, Zhao P, Xia QY. 2012. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One 7: e45035 10.1371/journal.pone.0045035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SY, Chang JS, Wang XG, Liu YY, Zhang JD, Lu W, Gao J, Shi R, Zhao P, Xia QY. 2014. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci Rep 4: 4489 10.1038/srep04489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HM, Dang Y, Wu YG, Jia GX, Anaya E, Zhang JL, Abraham S, Choi JG, Shi GJ, Qi L, et al. 2015. A CRISPR-based screen identifies genes essential for West-Nile-virus-induced cell death. Cell Rep 12: 673–683. 10.1016/j.celrep.2015.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SY, Liu Y, Liu YY, Chang JS, Zhang T, Wang XG, Shi R, Lu W, Xia XJ, Zhao P, et al. 2017. An integrated CRISPR Bombyx mori genome editing system with improved efficiency and expanded target sites. Insect Biochem Molec 83: 13–20. 10.1016/j.ibmb.2017.02.003 [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Yamashiro DJ. 1987. Endosome acidification and the pathways of receptor-mediated endocytosis. Adv Exp Med Biol 225: 189–198. 10.1007/978-1-4684-5442-0_16 [DOI] [PubMed] [Google Scholar]
- Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. 2014. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol 15: 591–600. 10.1038/nrm3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol 24: 6710–6718. 10.1128/MCB.24.15.6710-6718.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omenetto FG, Kaplan DL. 2010. New opportunities for an ancient material. Science 329: 528–531. 10.1126/science.1188936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. 2015. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162: 675–686. 10.1016/j.cell.2015.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, et al. 2017. Identification of essential genes for cancer immunotherapy. Nature 548: 537–542. 10.1038/nature23477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Rao VB. 2012. Viruses: sophisticated biological machines. Viral Mol Mach 726: 1–3. 10.1007/978-1-4614-0980-9_1 [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SJ, Nicolson GL. 1972. The fluid mosaic model of the structure of cell membranes. Science 175: 720–731. 10.1126/science.175.4023.720 [DOI] [PubMed] [Google Scholar]
- Song CQ, Li YX, Mou HW, Moore J, Park A, Pomyen Y, Hough S, Kennedy Z, Fischer A, Yin H, et al. 2017. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology 152: 1161–1173.e1. 10.1053/j.gastro.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Yau VK, Briggs BJ, Whittaker GR. 2005. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338: 53–60. 10.1016/j.virol.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Takasu Y, Kobayashi I, Beumer K, Uchino K, Sezutsu H, Sajwan S, Carroll D, Tamura T, Zurovec M. 2010. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem Molec 40: 759–765. 10.1016/j.ibmb.2010.07.012 [DOI] [PubMed] [Google Scholar]
- Tamura T, Thilbert C, Royer C, Kanda T, Abraham E, Kamba M, Komoto N, Thomas JL, Mauchamp B, Chavancy G, et al. 2000. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol 18: 81–84. 10.1038/71978 [DOI] [PubMed] [Google Scholar]
- Viswanatha R, Li ZC, Hu YH, Perrimon N. 2018. Pooled genome-wide CRISPR screening for basal and context-specific fitness gene essentiality in Drosophila cells. eLife 7: e36333 10.7554/eLife.36333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman LE, Goldsmith PA. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143: 185–195. 10.1016/0042-6822(85)90107-2 [DOI] [PubMed] [Google Scholar]
- Wang YQ, Li ZQ, Xu J, Zeng BS, Ling L, You L, Chen YZ, Huang YP, Tan AJ. 2013. The CRISPR/Cas System mediates efficient genome engineering in Bombyx mori. Cell Res 23: 1414–1416. 10.1038/cr.2013.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TM, Guan CG, Guo JH, Liu B, Wu YA, Xie Z, Zhang C, Xing XH. 2018. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat Commun 9: 2475 10.1038/s41467-018-04899-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia QY, Wang J, Zhou ZY, Li RQ, Fan W, Cheng DJ, Cheng TC, Qin JJ, Duan J, Xu HF, et al. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Molec 38: 1036–1045. 10.1016/j.ibmb.2008.11.004 [DOI] [PubMed] [Google Scholar]
- Xia QY, Guo YR, Zhang Z, Li D, Xuan ZL, Li Z, Dai FY, Li YR, Cheng DJ, Li RQ, et al. 2009. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326: 433–436. 10.1126/science.1176620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia QY, Li S, Feng QL. 2014. Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu Rev Entomol 59: 513–536. 10.1146/annurev-ento-011613-161940 [DOI] [PubMed] [Google Scholar]
- Xu YN, Siegenthaler PA. 1997. Low temperature treatments induce an increase in the relative content of both linolenic and Δ3-trans-hexadecenoic acids in thylakoid membrane phosphatidylglycerol of squash cotyledons. Plant Cell Physiol 38: 611–618. 10.1093/oxfordjournals.pcp.a029211 [DOI] [Google Scholar]
- Xu CL, Qi XL, Du XG, Zou HY, Gao F, Feng T, Lu HX, Li SL, An XM, Zhang LJ, et al. 2017. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc Natl Acad Sci 114: 722–727. 10.1073/pnas.1615735114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Peretz M, Aharony A, Sagi I, Benvenisty N. 2018. Defining essential genes for human pluripotent stem cells by CRISPR–Cas9 screening in haploid cells. Nat Cell Biol 20: 610–619. 10.1038/s41556-018-0088-1 [DOI] [PubMed] [Google Scholar]
- Yin YQ, Li F, Li SL, Cai JJ, Shi J, Jiang YH. 2017. TLR4 influences hepatitis B virus related hepatocellular carcinoma by regulating the Wnt/β-catenin pathway. Cell Physiol Biochem 42: 469–479. 10.1159/000477594 [DOI] [PubMed] [Google Scholar]
- Yu F, Wei R, Yang J, Liu JL, Yang K, Wang HN, Mu YM, Hong TP. 2018. FoxO1 inhibition promotes differentiation of human embryonic stem cells into insulin producing cells. Exp Cell Res 362: 227–234. 10.1016/j.yexcr.2017.11.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.