Abstract
Despite the interest in characterizing genomic variation, the presence of large repeats at the breakpoints hinders the analysis of many structural variants. This is especially problematic for inversions, since there is typically no gain or loss of DNA. Here, we tested novel linkage-based droplet digital PCR (ddPCR) assays to study 20 inversions ranging from 3.1 to 742 kb flanked by inverted repeats (IRs) up to 134 kb long. Of those, we validated 13 inversions predicted by different genome-wide techniques. In addition, we obtained new experimental human population information across 95 African, European, and East Asian individuals for 16 inversions, including four already validated variants without high-throughput genotyping methods. Through comparison with previous data, independent replicates and both inversion breakpoints, we demonstrate that the technique is highly accurate and reproducible. Most studied inversions are widespread across continents, and their frequency is negatively correlated with genetic length. Moreover, all except two show clear signs of being recurrent, and we could better define the factors affecting recurrence levels and estimate the inversion rate across the genome. Finally, the generated genotypes have allowed us to check inversion functional effects, validating gene expression differences reported before for two inversions and finding new candidate associations. Therefore, the developed methodology makes it possible to screen these and other complex genomic variants quickly in a large number of samples for the first time, highlighting the importance of direct genotyping to assess their potential consequences and clinical implications.
Over the last 15 years, a substantial amount of information has accumulated about different types of genomic changes, ranging from single nucleotide polymorphisms (SNPs) to more complex structural variants (SVs) (Sudmant et al. 2015; The 1000 Genomes Project Consortium 2015; Audano et al. 2019; Chaisson et al. 2019; Levy-Sakin et al. 2019). However, inversions remain as one of the most difficult classes of variation to identify and characterize. Polymorphic inversions have been studied for a long time and are known to have adaptive value and to be associated with phenotypic effects in many organisms (Hoffmann and Rieseberg 2008; Kirkpatrick 2010; Wellenreuther and Bernatchez 2018). In humans, although hundreds of inversions have been predicted (Martínez-Fundichely et al. 2014; Puig et al. 2015a), it is not possible to detect reliably both orientations for many of them due to their balanced nature and the complexity of their breakpoints, and only a few have been analyzed in detail (Stefansson et al. 2005; Salm et al. 2012; González et al. 2014; Puig et al. 2015b; Giner-Delgado et al. 2019).
Approximately half of human inversions have inverted repeats (IRs) at their breakpoints (Martínez-Fundichely et al. 2014; Puig et al. 2015a), which can be hundreds of kilobases long. Therefore, short second-generation sequencing reads (∼100–150 bp) (Sudmant et al. 2015; Hehir-Kwa et al. 2016; Collins et al. 2019), and even longer reads from single-molecule sequencing technologies (∼10–20 kb) (Huddleston et al. 2017; Shao et al. 2018; Audano et al. 2019) or paired-end mapping (PEM) data from large fragments (∼40-kb fosmid clones) (Kidd et al. 2008), are often not able to jump across the breakpoint IRs and determine the inversion orientation. New methods like single-cell sequencing of one DNA strand (Strand-seq) (Sanders et al. 2016; Chaisson et al. 2019) or Bionano genome optical maps based on linearized DNA molecules labeled at particular sequences (Levy-Sakin et al. 2019) have demonstrated their ability to detect inversions despite the presence of long IRs. Nevertheless, these techniques are not suitable for the analysis of large numbers of individuals.
The recent targeted genotyping of 45 inversions in 551 individuals by a combination of inverse PCR (iPCR) and multiplex ligation-dependent probe amplification (MLPA) (Giner-Delgado et al. 2019) has increased considerably the available human inversion data, although only IRs up to ∼25–30 kb could be spanned. At the other end, fluorescence in situ hybridization (FISH) has been used to study inversions with large IRs, but FISH is very time-consuming and can be applied only to inverted segments in the Mb scale where probes are separated enough (Zody et al. 2008; Antonacci et al. 2009; Salm et al. 2012). This leaves a set of potential inversions with IRs too large for iPCR-based techniques and inverted regions too small for FISH analysis that cannot be validated or genotyped in multiple samples.
Moreover, the great majority of IR-mediated inversions have been shown to occur recurrently several times both within the human lineage and in nonhuman primates (Cáceres et al. 2007; Zody et al. 2008; Antonacci et al. 2009; Aguado et al. 2014; Vicente-Salvador et al. 2017; Giner-Delgado et al. 2019). These recurrent inversions tend to have low linkage disequilibrium (LD) to nearby nucleotide variants (Giner-Delgado et al. 2019) and, as a consequence, they have probably been missed in existing genome-wide association studies (GWAS) of different phenotypes. In fact, inversions have potential important effects on gene expression, phenotypic traits, and disease susceptibility (de Jong et al. 2012; Salm et al. 2012; González et al. 2014; Puig et al. 2015a,b; Giner-Delgado et al. 2019). Therefore, it is necessary to expand the set of analyzed inversions by developing new tools able to assess directly the order of the sequences around the breakpoints, rather than relying on linkage to other variants easier to genotype or computational methods based on SNP combinations (Ma and Amos 2012; Cáceres and González 2015; Ruiz-Arenas et al. 2019).
In this context, droplet digital PCR (ddPCR) provides the opportunity to fill the void in inversion characterization by detecting linkage between amplicons located at both sides of the breakpoint repeats and thereby jump long genomic distances. This technology has already been used to detect copy number variation (Boettger et al. 2012), viral load (Strain et al. 2013), low-frequency transcripts (Hindson et al. 2013), rare mutations, or cell-free DNA (Olmedillas-López et al. 2017; Camunas-Soler et al. 2018), among other applications. Recently, ddPCR-based linkage calculations have also been used to phase variants separated by up to 200 kb (Regan et al. 2015), including deletions (Boettger et al. 2016), or for fusion transcript detection (Hoff et al. 2016). Here, we have developed new ddPCR assays to genotype quickly and reliably human polymorphic inversions flanked by large IRs that could not be studied before, showing that most of them are recurrent and that inversion alleles are associated with gene-expression changes.
Results
Inversion genotyping
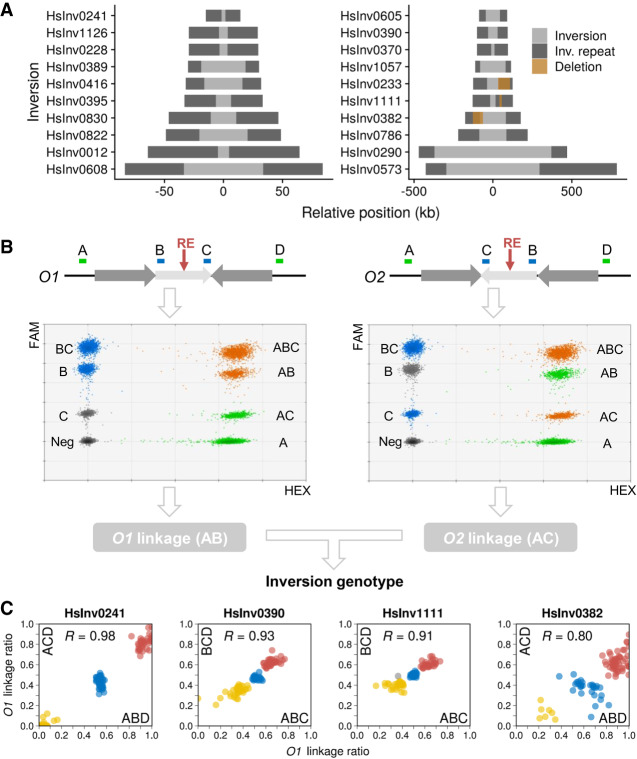
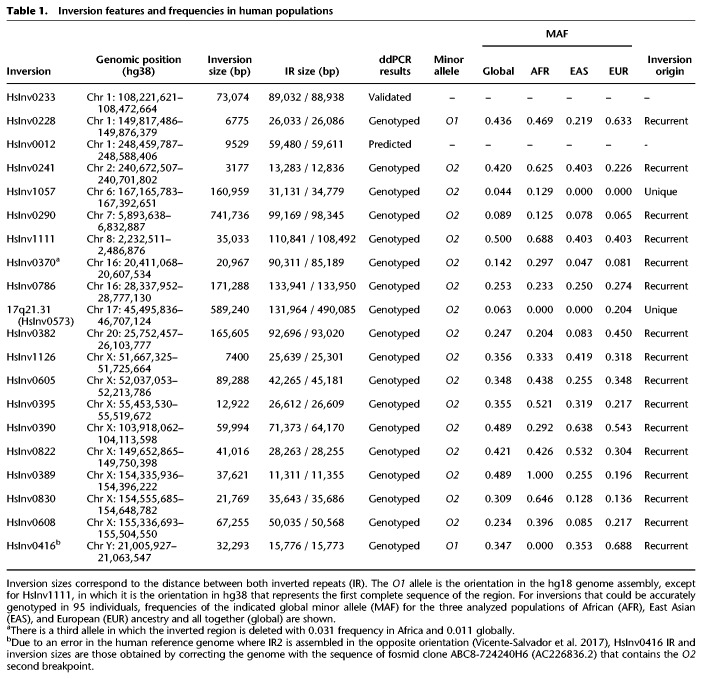
To test the ddPCR application for inversion genotyping, we analyzed a representative sample of 20 well-supported inversions mediated by IRs of different sizes from the InvFEST database (Martínez-Fundichely et al. 2014). We excluded predictions on very complex regions full of segmental duplications (SDs) or with assembly gaps, in which the genomic organization is not totally clear and could differ frequently between individuals. Fourteen inversions were initially predicted from fosmid PEM data (Kidd et al. 2008), although five had additional supporting evidence (Supplemental Table S1). The rest were validated inversions for which simple experimental genotyping methods did not exist (the well-known 17q21 inversion [or HsInv0573], HsInv0390, HsInv0290, and HsInv0786) and two control inversions already genotyped by iPCR-based assays (HsInv0241 and HsInv0389) (Supplemental Table S1). Minimal inversion sizes range from 3.1 to 741.7 kb and IRs at breakpoints between 11.3 and 134 kb (Fig. 1A; Table 1).
Figure 1.
Inversion characterization by ddPCR genotyping. (A) Main features of the studied polymorphic inversions, with the inverted region shown in gray and the IRs as dark boxes. Brown boxes denote deletions affecting inversion breakpoints. (B) Strategy for ddPCR genotyping of an inversion (light gray arrow) mediated by IRs at inversion breakpoints (dark gray arrows) using as an example the simultaneous triplex amplification of amplicons A, B, and C (shown on top), although the same procedure was used for other amplicon combinations (BCD, ABD, or ACD). O1 linkage (AB, left) and O2 linkage (AC, right) were calculated separately from the triplex ddPCR results, ignoring, respectively, amplicons C (left graph) or B (right graph). Colors of droplet clusters indicate if they are used to estimate the molecules containing only amplicon A (green), only the B or C internal amplicon (blue), two amplicons at the same time, either A and B or A and C (orange), or if they are considered as negative (gray). (C) Plots of O1 linkage ratios obtained from two different triplex ddPCR reactions (indicated in each axis) interrogating both breakpoints of four inversions in 95 analyzed individuals and the correlation between them (R), which show clearly differentiated genotype groups (O1/O1, red; O1/O2, blue; O2/O2, yellow). HsInv0241, where DNA was digested to separate both breakpoints with a restriction enzyme (RE, red arrow in B), shows the expected linkage ratios, but homozygote and heterozygote clusters are closer in HsInv0390, HsInv1111, and HsInv0382 analyzed using undigested DNA. The effect of a large deletion within breakpoint AB in HsInv0382 can be seen as higher O1 linkage ratio values in the horizontal axis. The gray dot is a sample with altered amplicon ratios in HsInv1111 ABC breakpoint.
Table 1.
Inversion features and frequencies in human populations
ddPCR technology allows us to quantify how close two sequences are within a DNA molecule from their simultaneous amplification in a higher number of droplet partitions than expected by chance (Regan et al. 2015). Thus, for each inversion, we designed three or four amplicons located in the unique sequence outside the IRs (A and/or D) and at the ends of the inverted segment (B and/or C) (Fig. 1B). Then, one combination of three amplicons (ABC, BCD, ABD, or ACD) (Supplemental Table S2) was used to determine the percentage of DNA molecules containing amplicons A and B (or C and D), which support orientation 1 (O1 linkage), and A and C (or B and D), which support orientation 2 (O2 linkage). Inversions were finally genotyped by the ratio between O1 linkage and the total linkage for both orientations (O1+O2 linkage), with ideal expected values of 1 (O1/O1), 0.5 (O1/O2), and 0 (O2/O2). A different strategy was used for inversion 17q21, where all breakpoints contain variable repeat blocks of >200 kb except for AB (132-kb repeat) (Zody et al. 2008; Boettger et al. 2012; Steinberg et al. 2012). Therefore, we measured only the O1 linkage (AB) and compared it to a reference linkage (Ref) between two products located at the same relative distance in both orientations, resulting in O1/Ref linkage ratios of 1 (O1/O1), 0.5 (O1/O2), and 0 (O2/O2). In addition, to simplify the process, each individual genotype was obtained by amplifying the three amplicons from a given inversion in a single ddPCR reaction using different amounts of two FAM-labeled probes (Fig. 1B; Supplemental Table S2). These assays were tested and optimized in a small sample of 7–15 individuals, including those in which the inversions were predicted.
Three main problems for ddPCR genotyping were found. First, in small inversions, linkage can be detected between A and C in O1 chromosomes, or A and B in O2 chromosomes, for example, creating linkage ratios for homozygotes that are closer to heterozygotes. In most cases, the three genotypes can still be distinguished reliably (Fig. 1C; Supplemental Fig. S2), but whenever possible we separated both breakpoints by DNA digestion with a restriction enzyme that cuts within the inverted sequence but not the IRs (Fig. 1B; Supplemental Table S2). The exception was the 9.5 kb-inversion HsInv0012 (Fig. 1A; Table 1), in which there were no suitable restriction sites and amplicons B and C were equally linked to D in all samples, so it was not analyzed further. Second, increasing distance between amplicons results in less molecules long enough to contain both of them. Thus, in the inversions with the largest IRs, true linkage values become closer to background values and DNA integrity limits the ability to resolve the genotypes. Third, IR size variation caused by large indels can change the distance between amplicons in the population. The selected regions were relatively stable, and by checking SV information in dbVar (Lappalainen et al. 2013a) and analyzing alternative sequences of these regions, we found that many of the indels are too small to be detected (<25% of the distance between amplicons), they include the amplicons (and they would have been detected in our samples), or were predicted only in a few individuals. Taking this into account, there are five inversions with polymorphic deletions within one of the IRs that could alter linkage values (Supplemental Table S2). However, linkage was only affected significantly in HsInv0382 (62-kb AB deletion within a 92.7-kb IR), with three separated genotype groups anyway (Fig. 1C), and HsInv0233 (74.5-kb deletion within an 89-kb IR), in which the large deletion combined with the relatively small inversion size prevents assigning genotypes with confidence (Supplemental Fig. S1).
Next, we genotyped 19 inversions (excluding HsInv0012) in 95 individuals included in different genomic projects (Lappalainen et al. 2013b; The 1000 Genomes Project Consortium 2015) with African (32 YRI), East Asian (EAS) (16 CHB and 16 JPT), and European (31 CEU) ancestry (Supplemental Table S3). All experiments were performed in duplicate, except for HsInv0389 that had been previously genotyped (Giner-Delgado et al. 2019), and for five inversions the two breakpoints were analyzed independently (HsInv0241, HsInv0233, HsInv0382, HsInv0390, and HsInv1111) (Fig. 1C). In HsInv0233, we identified a few samples showing distinctive O1/O1 and O2/O2 genotypes with no signs of the large deletion, but we could not genotype reliably all individuals (Supplemental Fig. S1). Final genotypes for the rest of the inversions were called using a statistical clustering method that takes into account the linkage ratios of every replicate, which was especially important for inversions analyzed using undigested DNA (Fig. 1C; Supplemental Fig. S2). In general, ddPCR results were very robust, as shown by the high correlation of O1 linkage ratios between replicates of the same or different breakpoints (median R = 0.91) (Fig 1C; Supplemental Fig. S2) and the grouping of the samples into well-defined clusters with good genotype score values (Supplemental Fig. S3; Supplemental Table S3). Excluding HsInv0233, only 29 genotypes (1.7%) were not determined because of low linkage (13), inconclusive clustering results (12), or altered amplicon ratios due to copy number variants (CNVs) affecting linkage calculation (4) (Supplemental Table S4).
ddPCR genotyping accuracy was assessed by comparison with the published genotypes for HsInv0241 and HsInv0389 (Giner-Delgado et al. 2019), plus a few genotypes from two other inversions obtained by FISH or Southern blot (Supplemental Table S5). In addition, we genotyped by haplotype fusion PCR (HF-PCR) (Turner et al. 2006) inversion HsInv0395 (90 CEU individuals) and HsInv0605 (eight individuals) with medium-sized IRs (26.6–45.2 kb) (Supplemental Fig. S4). HF-PCR creates fusion products from amplicons at both sides of a breakpoint, but it is difficult to set up and not very robust. Of the 242 genotypes compared, only those of two African individuals for HsInv0241 (0.8%) differed between iPCR (O2/O2) and ddPCR (O1/O2) (Supplemental Table S5). HsInv0241 genotyping of 76 additional African individuals from the YRI and LWK populations by ddPCR (Supplemental Table S3) identified four extra samples with the same discrepancies (Supplemental Table S5). Given that ddPCR results of both breakpoints are very clear (Fig. 1C), this suggests that some unknown variants affect HsInv0241 O1 chromosome detection by iPCR, which was already shown to have problems (Aguado et al. 2014; Giner-Delgado et al. 2019). Similarly, 17q21 inversion ddPCR genotypes match perfectly those inferred from the commonly used tag SNPs (Steinberg et al. 2012). In contrast, in HsInv0786, nine of the 81 genotypes (11.1%) computationally predicted from SNP data (González et al. 2014) are incorrect (Supplemental Table S5), with four caused by inversion recurrence (see below) and the rest belonging to one particular O2 haplotype group, which illustrates the limitations of indirect methods.
When ddPCR data were compared to recent inversion predictions in multiple human genomes, different results were obtained depending on the technique used (Supplemental Fig. S5). Despite most of the inversions being relatively common, only HsInv0241, with some of the smallest IRs, was detected using short reads and two more (HsInv0370 and HsInv1126) using long reads (Supplemental Table S1; Supplemental Fig. S5). On the other hand, Bionano optical maps (Levy-Sakin et al. 2019) or a multiplatform study relying mainly on Strand-seq (Chaisson et al. 2019) identified 14 inversions each, with nine detected by both (Supplemental Table S1). Although in most cases genotypes were not provided, based on the presence or not of the inverted allele of the identified inversions in common individuals, short and long reads showed again the lowest performance, whereas the error rate for the Bionano and the multiplatform strategy was, respectively, 23% and 0% (Supplemental Fig. S5). This emphasizes the amount of information still missing from the studied genomes and the need of specific methods for the analysis of IR-mediated inversions.
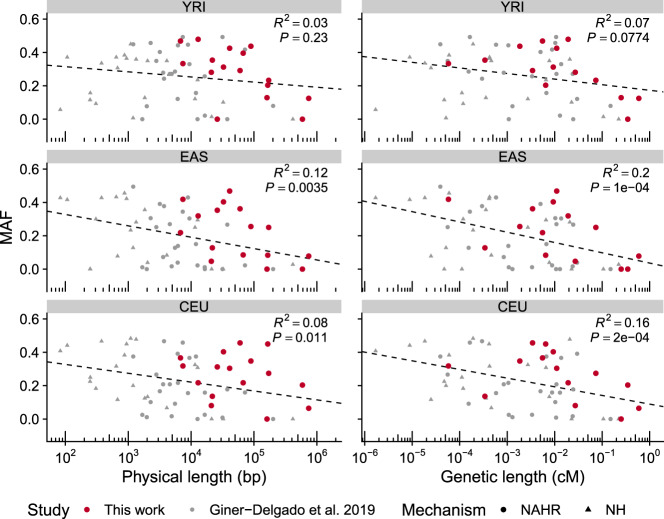
Of the genotyped inversions, all are in Hardy-Weinberg equilibrium in the analyzed populations and just three are not widespread with global minor allele frequencies (MAF) >0.1: HsInv1057, found exclusively in Africans; the 17q21 inversion, detected only in Europeans where a higher O2 frequency has been described (Stefansson et al. 2005; Steinberg et al. 2012; Alves et al. 2015); and HsInv0290 with low frequency everywhere (Table 1). Several inversions also have variable frequencies between continents, although only FST values of HsInv0389 are within the top 5% of the expected distribution (Supplemental Table S6; Giner-Delgado et al. 2019). Finally, the inclusion of longer inversions strengthened the negative correlation between inversion size and MAF observed before (Giner-Delgado et al. 2019), considering either the inversions analyzed here alone or all inversions together, and these correlations tend to be higher with the inversion genetic length than the physical length (Fig. 2).
Figure 2.
Correlation between inversion size and frequency. Graphs show a negative correlation between the logarithm of the minimal physical or genetic length and minor allele frequency (MAF) of inversions in three populations with African (YRI), East Asian (EAS), and European (CEU) ancestry. Values are those from 61 inversions, including the 16 ddPCR-analyzed inversions (in red) and 45 previously described inversions (in gray), which comprise inversions generated by nonhomologous mechanisms (NH) or nonallelic homologous recombination (NAHR), although similar results were obtained with each separate data set (see Supplemental Methods for details).
Nucleotide variation analysis
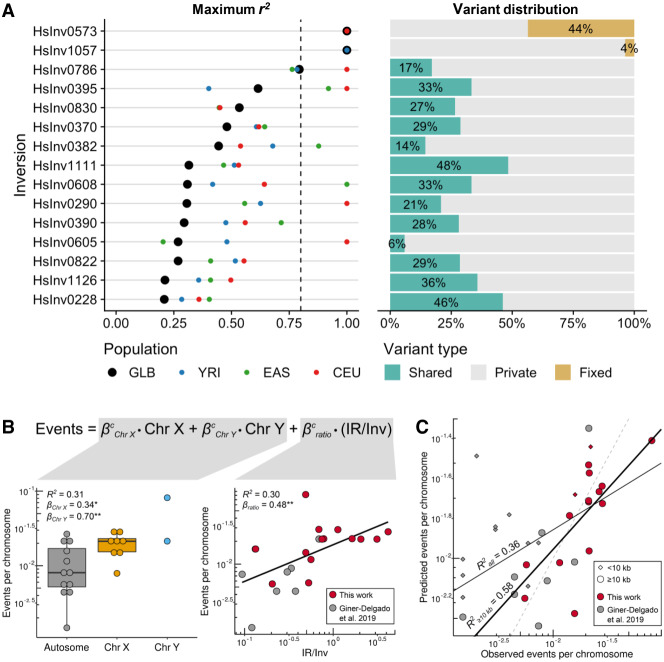
LD with 1000 Genome Project (1000GP) nucleotide variants (92 individuals in common) was calculated for 15 newly genotyped inversions (excluding HsInv0416 in Chr Y without 1000GP data). Only population-specific inversions 17q21 and HsInv1057 have tag SNPs in perfect LD (r2 = 1), while for the rest of the inversions, the maximum r2 values range between 0.21 and 0.79, and just six of them have r2 > 0.8 in at least one of the populations (CEU, EAS, or YRI) (Fig. 3A; Supplemental Table S7). We also classified the variants within the inverted region as shared between orientations, private to one of them, or fixed (i.e., in perfect LD) (Fig. 3A; Supplemental Table S7). The 17q21 and HsInv1057 inversions show no reliable shared SNPs, consistent with the inhibition of recombination throughout the inverted region. In contrast, shared SNPs represent an important fraction (6%–48%) of the variation within the entire length of the remaining inversions (Supplemental Fig. S6A), a pattern consistent with several inversion events on different haplotypes. Actually, longer inversions tend to have higher LD with other variants and a lower proportion of shared SNPs, although these trends are stronger for the ratio between IR and inversion size (IR/Inv ratio) (Supplemental Fig. S6B).
Figure 3.
Nucleotide variation and recurrence analysis of genotyped inversions in human populations. (A) For the 15 newly genotyped autosomal and Chr X inversions, maximum LD (r2) with 1000GP variants located 1 Mb at each side of the inversion in all individuals together (black dots) and different populations (colored dots) is shown at the left, and the proportion of SNPs within each inversion classified as fixed (yellow), shared between orientations (green), or private of one orientation (gray) at the right. (B) Effect size (β) and variance explained (R2) by the chromosome type (left) and the IR/inverted region (Inv) size ratio (right) of the logarithm-transformed number of independent inversion events per chromosome was estimated for 22 inversions >10 kb from this work and Giner-Delgado et al. (2019). P values: (*) P < 0.05, (**) P < 0.01. (C) Adjustment of the observed inversion events and the expected number calculated by applying the developed model to all inversions in both studies. The number of observed events is underestimated in small inversions (<10 kb; diamonds), which results in a lower R2 value for all inversions (all, thin black line) than for those >10 kb (≥10 kb, thick black line). Dashed line represents the 1:1 equivalence.
Next, we estimated the independent inversion events by phasing inversion genotypes into 1000GP haplotypes of the inverted region (Supplemental Fig. S7). While in the two inversions with perfect tag SNPs, all O2 haplotypes cluster together, in the other inversions different clusters of similar haplotypes containing both orientations can be identified, which are typical of recurrent inversion and re-inversion events (Giner-Delgado et al. 2019). We identified between two and five inversion events in the 14 new recurrent inversions, and 27 of the 33 additional inversions/re-inversions detected were shared by more than one individual (Supplemental Table S8). This suggests that they are real evolutionary events rather than mutations generated in the lymphoblastoid cell lines (LCLs) used as a DNA source. Possible phasing errors in inversion heterozygotes were removed by checking if switching the orientation of both haplotypes still supported recurrence or not. Also, haplotypes formed by blocks from other haplotypes (either due to recombination or SNP phasing errors) were not classified as recurrent. The only inversion not affected by these complications is HsInv0416, in which the O1 and O2 distribution in the known phylogeny of Chr Y haplogroups (Poznik et al. 2016) clearly supports four independent inversion events (Supplemental Table S8) and results in an inversion mutation rate of 1.29 × 10−4 inversions per generation, very similar to that described for another Chr Y inversion (0.53 × 10−4 inversions per generation) (Giner-Delgado et al. 2019).
We also investigated the effect of different parameters on recurrence levels by combining all the inversions mediated by NAHR analyzed so far. Focusing on the 22 inversions >10 kb, in which the higher number of SNPs increases the ability to distinguish haplotype clusters and detect recurrence, the only significant variables were the IR/Inv ratio and chromosome type (autosome, Chr X, or Chr Y), with IR/Inv ratio and sex chromosomes being positively correlated with the number of inversion events (Fig. 3B). The resulting model fits very well the real data (R2 = 0.58) and evidences the underestimation of recurrence events in smaller inversions (<10 kb) (Fig. 3C).
Functional effects of inversions
Inversion functional consequences were first investigated through the association of the generated genotypes in 59 CEU and YRI samples and available LCL gene expression data (Lappalainen et al. 2013b). We identified two inversions associated with gene-expression changes in this small sample (Fig. 4A; Supplemental Table S9). The main effects were found for inversion 17q21, which is the lead variant (r2 ≥ 0.8 with top eQTLs) for an antisense RNA and three pseudogenes at the gene level, as well as for specific transcripts from protein-coding genes KANSL1 and LRRC37A2, plus other pseudogenes and noncoding RNAs (Supplemental Table S9). To increase the power to detect associations, we extended the analysis to a larger sample of 387 Europeans for five inversions whose genotypes could be inferred through perfect tag SNPs (r2 = 1) in the CEU population (Fig. 3A). These inversions were significantly associated with expression changes in 42 genes and 103 transcripts (Fig. 4A; Supplemental Table S9), including multiple other genes for inversion 17q21. In addition, HsInv0786 appeared as potential lead eQTL for gene NFATC2IP and APOBR, IL27, and EIF3C transcripts.
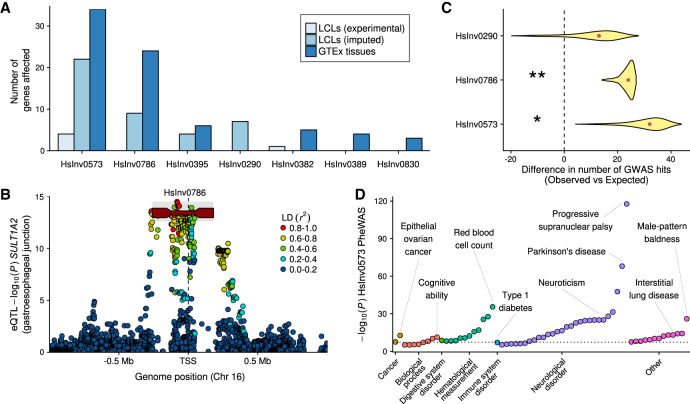
Figure 4.
Inversion effects on gene expression and phenotypic traits. (A) Number of significant associations at the gene level in each of the differential expression analyses (excluding the associations with specific transcripts). (B) Manhattan plot for cis-eQTL GTEx associations of gene SULT1A2 in gastroesophageal junction, showing inversion HsInv0786 (dark red line with rectangles representing the IRs) as potential lead variant. HsInv0786 eQTL P values and LD with neighboring variants (r2) were calculated by permuting samples with the same ancestry proportions as in GTEx samples (see Methods), and the P-value imputation range is shown in gray. (C) Enrichment of GWAS Catalog signals within individual inversions measured by the deviation in the observed minus expected value. Density distribution represents the 95% one-sided confidence interval with the median indicated by a red dot; one-tailed permutation test, (*) P < 0.05, (**) P < 0.01. (D) Phenome-wide association study (PheWAS) for inversion 17q21 (HsInv0573). Significant reported traits (P < 10−5) were grouped by GWAS Catalog ontology categories. Dotted line indicates genome-wide significance threshold (P = 5 × 10−8).
Since more than 60% of genes close to our inversions (±1 Mb) are not expressed in LCLs, we imputed inversion association P values in other tissues by estimating LD patterns between inversion alleles and SNPs identified as eQTLs in GTEx (The GTEx Consortium 2017) (see Methods). We found 73 genes associated with six inversions in different tissues, although the majority involve inversions 17q21 and HsInv0786, whose effects are easier to infer due to the high LD with neighboring eQTLs (including 37 genes for which the inversions were potential lead variants) (Fig. 4A,B; Supplemental Table S10; Supplemental Fig. S8). However, the high recurrence levels of most analyzed inversions prevent inferring their contribution to gene expression variation.
We also checked whether inversions account for phenotypic variation by using the GWAS Catalog information (MacArthur et al. 2017). We found a 2.8-fold enrichment (P = 0.026) of GWAS Catalog signals within inversion regions, with 17q21 (P = 0.028), HsInv0786 (P = 0.009), and HsInv0290 (P = 0.157) apparently driving this enrichment (Fig. 4C). Moreover, several of them showed an enrichment of GWAS-reported genes for certain traits within 150 kb of the analyzed inversions (Supplemental Table S11), such as brain-related traits for 17q21 and immunological disorders and body mass characteristics for HsInv0786. For seven inversions in high LD (r² ≥ 0.8) with SNPs in at least one population, we looked for GWAS hits associated with those SNPs in the corresponding population or continent (Supplemental Table S12). The 17q21 inversion has already been linked to many traits (Puig et al. 2015a), and a total of 64 potential associations with P < 10−6 from 35 studies were found in this analysis (Fig. 4D), including lung function, neurological traits or disorders, ovarian cancer, blood profiles, and diabetes, which illustrates the pleiotropic consequences of this inversion. HsInv0786, previously associated with an asthma and obesity phenotype (González et al. 2014), was associated with several pediatric autoimmune diseases (Li et al. 2015b) and the presence of type 1 diabetes autoantibodies (Plagnol et al. 2011), among other traits, which, together with the enrichment of immunological GWAS hits in the inversion region, suggests a role in the immune system that may be related to the expression changes in genes like IL27 and NFATC2IP (Supplemental Tables S9, S10).
Discussion
To fill the void in the study of polymorphic inversions, especially of those flanked by large IRs, we have taken advantage of the ability of ddPCR technology to measure linkage between sequences at long distances to genotype inversions with IRs up to ∼150 kb efficiently and reliably. Using this method, we have validated 13 inversion predictions and generated new experimentally resolved genotypes for 16 inversions, most of which are analyzed in detail for the first time. In comparison with the recent analysis of 24 inversions flanked by IRs (Giner-Delgado et al. 2019), on average our inversions are longer (138.9 kb vs. 20.7 kb) and have much bigger IRs (74.2 kb vs. 6.9 kb). Almost all these inversions are missed by massively parallel sequencing with short or long reads (Supplemental Fig. S5). In addition, although the multiplatform and Bionano approaches together (Chaisson et al. 2019; Levy-Sakin et al. 2019) identified 95% of them (Supplemental Table S1), an acceptable genotyping accuracy (>90%) is only achieved by combining different techniques, which complicates large-scale analyses. Therefore, the novel ddPCR application represents a valuable resource for the targeted characterization of inversions and complex SVs, providing also accurate information on other associated variants (like indels or CNVs) (Supplemental Table S4).
In comparison with iPCR-based techniques (Aguado et al. 2014; Giner-Delgado et al. 2019), the ddPCR design has less requirements and is much more flexible, allowing us to test inversions for which no assays were available before and correct possible errors, as in HsInv0241. In fact, half of the inversions mediated by inverted SDs in InvFEST (Martínez-Fundichely et al. 2014) have IRs 25–150 kb long that could be analyzed using ddPCR. The main limitations are restricted to extremely long IRs (>100 kb), small inversions (<5–10 kb) where breakpoints cannot be separated by restriction-enzyme digestion, and CNVs altering significantly the distance between amplicons. We have overcome these problems by using good-quality high-molecular-weight DNA and a clustering method to distinguish genotype groups independently of the magnitude of the linkage ratio differences. In the near future, the possibility to interrogate several inversions simultaneously using amplicons labeled with different fluorochromes and even longer DNA molecules will expand the range of studied inversions and reduce costs, making it easier to undertake more ambitious ddPCR genotyping projects.
Consistent with previous results (Giner-Delgado et al. 2019), we have shown that the vast majority of inversions mediated by IRs are generated multiple times on different haplotypes by NAHR and cannot be easily imputed from SNP data (as exemplified by half of the HsInv0786 imputation errors). Inversion recurrence or some other mechanism able to exchange variants between the two IRs had been already suggested for HsInv0390 (Beck et al. 2015), HsInv0830 (Aradhya et al. 2001), and HsInv0290 (Hayward et al. 2007). The only exceptions are the two inversions with more restricted geographical distributions: HsInv1057 and 17q21 (Stefansson et al. 2005; Steinberg et al. 2012; Alves et al. 2015). This indicates that, although a few inversions can be indirectly imputed by tag SNPs, the only way to genotype accurately recurrent inversions is to interrogate experimentally the sequences flanking the breakpoints with techniques like the one developed here.
Actually, the higher resolution provided by the longer inversions analyzed in this work has allowed us to estimate more precisely recurrence levels and determine that chromosome type and IR/inversion size ratio together explain a very significant part (up to 58%) of recurrence variance between inversions. This fits well with the expectations, since repeat length and distance have been found to affect the generation of recurrent pathological rearrangements (Liu et al. 2011), suggesting that the closer and longer the IRs are, the more likely they are to pair and recombine. In addition, increased NAHR within the unpaired Chr X is probably the cause of the higher frequency of the hemophilia A inversion in the male germ line (Antonarakis et al. 1995). According to the estimated values for two Chr Y inversions, the model results in predicted NAHR mutation rates for the analyzed autosomal and Chr X inversions of 0.9–4.4 × 10−5 and 1.9–7.4 × 10−5 inversions/generation, respectively, which illustrate the relevance of this phenomenon. However, other factors, like DNA 3D conformation or recombination motifs, might affect recurrence as well.
Inversion genotypes have also allowed us to carry out a complete analysis of the potential functional consequences of these variants and associate eight inversions with gene-expression changes across different tissues. In particular, the inversions with the largest effects were 17q21 and HsInv0786, which were the top eQTLs for many genes. In this case, our analyses confirmed most expression changes already reported for 17q21 in blood, cerebellum, and cortex (de Jong et al. 2012), and some of those for HsInv0786 in blood and LCLs (González et al. 2014), and identified expression differences in additional genes and tissues never examined before (Supplemental Table S10; Supplemental Fig. S8). Moreover, the same two inversions are enriched in GWAS signals, and they are in LD with multiple variants associated with different phenotypes. Nevertheless, the inversion functional analysis is limited by the small number of genotyped individuals with expression data (59), which results in low statistical power and in only large differences being detectable, especially for low-frequency inversions. In addition, for many inversions, the lack of LD with neighboring variants makes it difficult to infer reliably their association with gene expression in other tissues or with phenotypic traits from nongenotyped individuals. Thus, although our results show that inversions could have an important role in gene expression and clinically relevant disorders, we are probably missing a significant fraction of their effects, emphasizing the need for inversion genotyping in a larger set of individuals.
Another important effect of inversions is the generation of aberrant chromosomes by recombination within the inverted region in heterozygotes (Hoffmann and Rieseberg 2008; Kirkpatrick 2010). By extending the analysis to a broader set of inversions, we have reinforced the idea that negative selection related to their influence on fertility could be an important factor in determining inversion frequency (Giner-Delgado et al. 2019). In that sense, it is noteworthy that some of the longer inversions, such as 17q21 (589 kb) and HsInv0786 (171 kb), are the best examples of inversions with functional consequences at different levels. This suggests that their functional effects compensate in part the potential negative costs associated with inversion length, and it has already been proposed that the 17q21 inversion has been positively selected in European populations through increased fertility in carrier females (Stefansson et al. 2005).
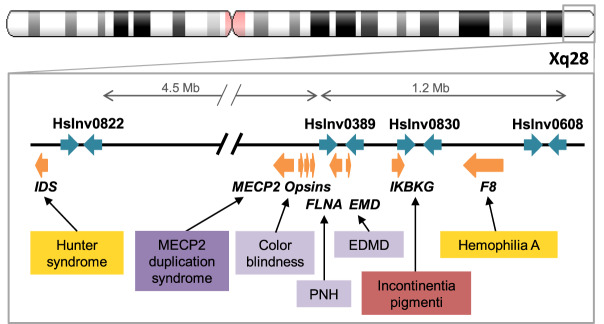
Finally, inversions could also predispose to other pathological SVs in the region, due either to recombination problems in heterozygotes or changes in the orientation of repeats (Puig et al. 2015a). Recently, a complete catalog of inversions in nine individuals has shown that a high proportion of them overlap critical regions of microdeletion and microduplication syndromes (Chaisson et al. 2019). However, these inversions tend to be mediated by large and complex repeats and are difficult to characterize with simple methods. In our case, seven of the eight Chr X inversions analyzed are located in regions where additional disease-associated SVs have been described (Supplemental Table S13). These involve recurrent mutations mediated by repeats within the polymorphic inversions, like the hemophilia A inversion (Antonarakis et al. 1995) or the deletion causing incontinentia pigmenti (Aradhya et al. 2001), different duplication-inverted triplication-duplication (DUP-TRP/INV-DUP) rearrangements due to DNA polymerase stalling during IR replication that affect dose-sensitive genes such as MECP2 and PLP1 (Carvalho et al. 2011; Beck et al. 2015), or deletions with one breakpoint mapping within or nearby the inversion IRs involved in X-linked intellectual disability (Grau et al. 2017). Specifically, within the 6-Mb Xq28 telomeric region, there are seven genomic disorders caused by rearrangements overlapping or in close proximity to four polymorphic inversions (Deeb et al. 1992; Bondeson et al. 1995; Small et al. 1997; Aradhya et al. 2001; Clapham et al. 2012; Fusco et al. 2012; Li et al. 2015a), making this region a possible hotspot for genome reorganization (Fig. 5). The novel ddPCR application offers now the opportunity to easily study these inversions in parents of patients and determine their role in the generation of pathological variants, contributing to a more complete picture of the genomic impact of inversions.
Figure 5.
Polymorphic inversions in chromosome region Xq28 and genomic disorders caused by other rearrangements. Polymorphic inversion IRs are represented as blue arrows and genes as orange arrows. Boxes indicate disorders associated with different types of recurrent pathological rearrangements (inversions in yellow, deletion in red), and complex events resulting in deletion (light purple) or in a duplication-inverted triplication-duplication structure (DUP-TRP/INV-DUP) (dark purple). (EDMD) Emery-Dreifuss muscular dystrophy, (PNH) periventricular nodular heterotopia.
Methods
Human samples and DNA isolation
High-molecular-weight genomic DNA of 95 unrelated human samples (Supplemental Table S3) was isolated from ∼20-mL culture of an Epstein-Barr virus-transformed B-lymphoblastoid cell line of each individual (Coriell Cell Repository) by standard phenol:chloroform extraction (see Supplemental Methods). To preserve DNA integrity, all steps that involved handling of the DNA were done pipetting gently with wide-bore tips, and DNA was stored at 4°C. Identity of the isolated DNAs was confirmed by microsatellite analysis.
Droplet digital PCR genotyping
Inversion genotyping by ddPCR assays was carried out by quantitative amplification of three different products simultaneously with six primers and three fluorescent probes labeled with HEX or FAM (Supplemental Table S14) in aqueous droplets within an oil phase (emulsion PCR) using the QX200 ddPCR system (Bio-Rad). Since only two colors can be detected, we used different concentrations of two FAM probes to separate the clusters of droplets containing the different amplicon combinations (Fig. 1B). All primers and probes were tested in duplex experiments before optimizing the triplex reactions. Final ddPCR reactions were prepared in 96-well plates in a total volume of 22 μL with 450 nM–2.5 μM of each primer, 75–275 nM of each probe, 1× ddPCR Supermix for Probes (No dUTP) (Bio-Rad), and 50 ng of genomic DNA. The ddPCR mix was mixed by gently pipetting up and down 5–10 times with wide-bore tips to avoid breaking the DNA molecules. Droplet generation, thermal cycling, and fluorescence reading were performed as explained before (McDermott et al. 2013). Linkage between each pair of targeted amplicons (percentage of DNA molecules containing both amplicons) (Fig. 1B) was obtained as the excess of double-positive droplets over what is statistically expected (Regan et al. 2015) using the QuantaSoft version 1.7.4 software (Bio-Rad). For inversions with restriction enzyme targets inside the inverted region but not within the breakpoint IRs, DNA was digested prior to ddPCR quantification in 10-μL reactions including 2.5 U of restriction enzyme, 1× of the digestion buffer, and 250 ng of genomic DNA at 37°C for 3 h, and then 2 μL (50 ng) were directly used as ddPCR template. Since DNA molecule size decreases over time, inversion genotyping was performed in inverse order of IR size, starting with those with higher DNA quality requirements.
ddPCR results were not considered valid when (1) total linkage was <7.5% (or <15% if just one measurement was available), which was only an issue in those inversions with the largest IRs; (2) droplet count was below 10,000 due to the presence of extremely long undigested DNA molecules; and (3) small deletions or duplications of one or more of the amplicons result in ratios between them different than 1 in certain individuals, which, in the case of copy number increases, makes it very difficult to interpret linkage values (Supplemental Table S4). These ddPCR reactions and others with intermediate linkage ratios between the expected values for homozygotes and heterozygotes (e.g., ∼0.25 or ∼0.75) were repeated using a fresh DNA dilution from the stock or storing the diluted DNA at 4°C for a few days, and, except for the altered amplicon ratios, usually the problems were solved. Inversion genotypes were called by hierarchical clustering of the Euclidean distance between every pair of individuals calculated from all valid O1 linkage ratios of the different replicates (see Supplemental Fig. S2; Supplemental Methods). To assess the uncertainty of sample classification, we clustered two-thirds of our samples selected at random 10,000 times, and their percentage of inclusion in the most supported cluster was assigned as the genotype score (Supplemental Fig. S3; Supplemental Table S3). Individuals that were included >5% of times in a different cluster were not genotyped.
Haplotype fusion PCR
HF-PCR was carried out in an oil and water emulsion to generate the fused amplification product, followed by a regular PCR with nested primers (Supplemental Table S15). Emulsion PCR reactions were performed in 96-well plates with 25 ng of digested DNA in 25 μL for 40 cycles as previously described (Turner et al. 2006; Turner and Hurles 2009), and the aqueous phase was recovered by centrifugation of the emulsion (see Supplemental Methods for details).
Linkage disequilibrium analysis
Pairwise LD between genotypes of inversions and neighboring biallelic SNPs and small indels from 1000GP Phase 3 (inversion region ±1 Mb) was calculated using the r2 statistic with PLINK v1.9 (Purcell et al. 2007) separately for each population group and the 92 samples common to both data sets. SNPs were further classified as shared (unambiguously polymorphic in the two orientations), private (polymorphic only in one orientation), or fixed (in perfect LD with the inversion) (Giner-Delgado et al. 2019). To minimize possible errors, only reliable variants, defined as those located in accessible regions according to the 1000GP strict accessibility mask and that do not overlap known SDs, were used (Handsaker et al. 2015; Sudmant et al. 2015; The 1000 Genomes Project Consortium 2015; Audano et al. 2019).
Recurrence analysis
To generate haplotypes of the inverted regions, available 1000GP Phase 3 haplotypes were used as scaffolds by selecting reliable variants present in at least two genotyped chromosomes (except if there were less than 50 variants, when those accessible according to the 1000GP pilot criteria were also included to maximize information). The phase of the inversion genotypes was inferred with MVNcall (Menelaou and Marchini 2013) by positioning inversions in the middle as another variant. Haplotypes were clustered by similarity, and their relationships were visualized using the iHPlots strategy (Giner-Delgado et al. 2019). The putative ancestral orientation and the original inversion event were defined based on the O1 and O2 haplotype diversity and the frequency and geographical distribution of haplotypes (normally taking as ancestral those found in African samples) (Supplemental Table S8). Additional independent inversion or re-inversion events were identified conservatively as clusters including haplotypes with an unexpected orientation compared to the rest (Supplemental Fig. S7B,C), after eliminating possible errors in inversion phase in heterozygotes. To avoid artifacts caused by mixed haplotype blocks formed by recombination or SNP phasing errors, possible recurrent haplotypes were revised manually and only those clearly differentiated from other O1 or O2 haplotype clusters by at least three positions (although usually there are many more) extending over most of the inverted region (and spanning >3 kb for the smallest inversions) were considered. For HsInv0416, we estimated the recurrence rate based on the known Chr Y haplogroup information (Poznik et al. 2016) as in Giner-Delgado et al. (2019) (Supplemental Methods). To determine the effect on the number of recurrent events per chromosome, we tested different variables: chromosome type (autosome, Chr X, or Chr Y), inversion and IR length, IR/Inv size ratio, IR identity, and PRDM9 motifs/kb within IRs (Myers et al. 2008). The model was built by stepwise regression with forward selection using the robustbase::lmrob R function (http://robustbase.r-forge.r-project.org/) and logarithm-transformed values to remove outliers.
Functional analysis
Inversion effects on LCL gene expression were first analyzed in 30 CEU and 29 YRI experimentally genotyped individuals from the Geuvadis project (Lappalainen et al. 2013b), excluding HsInv0416 in Chr Y due to the low statistical power to detect differences only in males. Inversions 17q21, HsInv0290, HsInv0395, HsInv0605, and HsInv0786 were also imputed in 328 additional Geuvadis Europeans (59 CEU, 91 TSI, 86 GBR, and 92 FIN) and HsInv1057 in 58 YRI individuals using a representative tag SNP in each population (which, except for HsInv0290, belongs to a larger set of SNPs in LD in the expanded population). Expression quantification and cis-eQTL association for genes and transcripts located up to 1 Mb from the studied inversions were done following a similar approach as in previous studies (The GTEx Consortium 2017; Giner-Delgado et al. 2019; see Supplemental Methods for details). Significant eQTLs correspond to a Q value false-discovery rate (FDR) <0.05 (Storey and Tibshirani 2003). Inversion gene-expression effects in other tissues were estimated through imputation of the inversion association P values from the LD between inversion alleles and eQTLs in GTEx V7 release (The GTEx Consortium 2017) using FAPI v0.1 (Kwan et al. 2016). LD between inversions and GTEx eQTLs was calculated by permutations of samples of experimentally genotyped individuals simulating the ethnic composition in GTEx (see Supplemental Methods for details). For both Geuvadis and GTEx, the most significant associated variants for each gene/transcript were designated as lead eQTLs. Moreover, inversions in high LD with the top marker (r2 ≥ 0.8) were indicated as potential lead eQTLs.
The impact of inversions in relevant phenotypic traits was assessed with the GWAS Catalog curated collection of the most significant SNPs associated with a particular phenotype (P < 10−5) (http://www.ebi.ac.uk/gwas/; release 2018-06-25, v1.0) (MacArthur et al. 2017). First, we explored the enrichment of GWAS SNPs within inversions as described in the Supplemental Methods in detail. Also, we compared the proportion of genes related to particular clinically relevant traits or diseases as reported in the GWAS Catalog inside or around (±150 kb) each inversion with respect to the rest of the genome (Supplemental Table S11). Finally, we crossed GWAS Catalog variants with those in high LD with our inversions (r2 ≥ 0.8) in the corresponding population or the closest one available in our data, while the global LD was used for GWAS with populations from different continents (Supplemental Table S12).
Data access
All inversion information from this study is available at InvFEST (http://invfestdb.uab.cat), and genotypes from this study have been submitted to the NCBI database of human genomic structural variation (dbVar; https://www.ncbi.nlm.nih.gov/dbvar) under accession number nstd185.
Competing interest statement
The authors declare the following competing financial interests: Bio-Rad Laboratories, Inc. markets and sells the QX200 Droplet Digital PCR System. J.F.R. and G.K.-N. are or were employees of Bio-Rad Laboratories, Inc. at the time the study was performed, and G.K.-N. owns Bio-Rad stock.
Supplementary Material
Acknowledgments
We thank Salvador Bartolomé, Xavier Alba, and James Thomas for technical support and advice; Alba Vilella, Marina Laplana, and Sergi Villatoro for help with DNA isolations and microsatellite analysis; and Xavier Estivill and Marta Morell for the European and African lymphoblastoid cell lines. This work was supported by research grants BFU2013-42649-P and BFU2016-77244-R funded by the Agencia Estatal de Investigación (AEI, Spain) and the European Regional Development Fund (FEDER, EU), ERC Starting Grant 243212 (INVFEST) from the European Research Council under the European Union Seventh Research Framework Programme (FP7), and 2017-SGR-1379 from the Generalitat de Catalunya (Spain) to M.C., and a La Caixa Doctoral fellowship to J.L.-J. M.G.-V. was supported by POCI-01-0145-FEDER-006821 funded through the Operational Programme for Competitiveness Factors (COMPETE, EU) and UID/BIA/50027/2013 from the Foundation for Science and Technology (FCT, Portugal). ddPCR reagents used in this study were provided by Bio-Rad Laboratories, Inc.
Author contributions: M.P. and M.C. designed the genotyping assays and oversaw all steps; M.P., S.P., D.I., and A.D. performed experiments; C.G.-D., M.G.-V., M.P., and M.C. analyzed evolutionary data; J.L.-J. and M.P. analyzed functional effects; M.P., S.P., J.F.R., G.K.-N., and M.C. contributed to ddPCR assay development and optimization; M.P., J.L.-J., and M.C. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.255273.119.
Freely available online through the Genome Research Open Access option.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado C, Gayà-Vidal M, Villatoro S, Oliva M, Izquierdo D, Giner-Delgado C, Montalvo V, García-González J, Martínez-Fundichely A, Capilla L, et al. 2014. Validation and genotyping of multiple human polymorphic inversions mediated by inverted repeats reveals a high degree of recurrence. PLoS Genet 10: e1004208 10.1371/journal.pgen.1004208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves JM, Lima AC, Pais IA, Amir N, Celestino R, Piras G, Monne M, Comas D, Heutink P, Chikhi L, et al. 2015. Reassessing the evolutionary history of the 17q21 inversion polymorphism. Genome Biol Evol 7: 3239–3248. 10.1093/gbe/evv214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacci F, Kidd JM, Marques-Bonet T, Ventura M, Siswara P, Jiang Z, Eichler EE. 2009. Characterization of six human disease-associated inversion polymorphisms. Hum Mol Genet 18: 2555–2566. 10.1093/hmg/ddp187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Rossiter JP, Young M, Horst J, de Moerloose P, Sommer SS, Ketterling RP, Kazazian HH Jr, Négrier C, Vinciguerra C, et al. 1995. Factor VIII gene inversions in severe hemophilia A: results of an international consortium study. Blood 86: 2206–2212. 10.1182/blood.V86.6.2206.bloodjournal8662206 [DOI] [PubMed] [Google Scholar]
- Aradhya S, Bardaro T, Galgóczy P, Yamagata T, Esposito T, Patlan H, Ciccodicola A, Munnich A, Kenwrick S, Platzer M, et al. 2001. Multiple pathogenic and benign genomic rearrangements occur at a 35 kb duplication involving the NEMO and LAGE2 genes. Hum Mol Genet 10: 2557–2567. 10.1093/hmg/10.22.2557 [DOI] [PubMed] [Google Scholar]
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Carvalho CMB, Banser L, Gambin T, Stubbolo D, Yuan B, Sperle K, McCahan SM, Henneke M, Seeman P, et al. 2015. Complex genomic rearrangements at the PLP1 locus include triplication and quadruplication. PLoS Genet 11: e1005050 10.1371/journal.pgen.1005050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger LM, Handsaker RE, Zody MC, McCarroll SA. 2012. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet 44: 881–885. 10.1038/ng.2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger LM, Salem RM, Handsaker RE, Peloso GM, Kathiresan S, Hirschhorn JN, McCarroll SA. 2016. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat Genet 48: 359–366. 10.1038/ng.3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson ML, Dahl N, Malmgren H, Kleijer WJ, Tönnesen T, Carlberg BM, Pettersson U. 1995. Inversion of the IDS gene resulting from recombination with IDS-related sequences in a common cause of the Hunter syndrome. Hum Mol Genet 4: 615–621. 10.1093/hmg/4.4.615 [DOI] [PubMed] [Google Scholar]
- Cáceres A, González JR. 2015. Following the footprints of polymorphic inversions on SNP data: from detection to association tests. Nucleic Acids Res 43: e53 10.1093/nar/gkv073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program, Sullivan RT, Thomas JW. 2007. A recurrent inversion on the eutherian X chromosome. Proc Natl Acad Sci 104: 18571–18576. 10.1073/pnas.0706604104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camunas-Soler J, Lee H, Hudgins L, Hintz SR, Blumenfeld YJ, El-Sayed YY, Quake SR. 2018. Noninvasive prenatal diagnosis of single-gene disorders by use of droplet digital PCR. Clin Chem 64: 336–345. 10.1373/clinchem.2017.278101 [DOI] [PubMed] [Google Scholar]
- Carvalho CMB, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, et al. 2011. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet 43: 1074–1081. 10.1038/ng.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, Gardner EJ, Rodriguez OL, Guo L, Collins RL, et al. 2019. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 10: 1784 10.1038/s41467-018-08148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham KR, Yu TW, Ganesh VS, Barry B, Chan Y, Mei D, Parrini E, Funalot B, Dupuis L, Nezarati MM, et al. 2012. FLNA genomic rearrangements cause periventricular nodular heterotopia. Neurology 78: 269–278. 10.1212/WNL.0b013e31824365e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Khera AV, Francioli LC, Gauthier LD, Wang H, Watts NA, et al. 2019. An open resource of structural variation for medical and population genetics. bioRxiv 10.1101/578674 [DOI]
- Deeb SS, Lindsey DT, Hibiya Y, Sanocki E, Winderickx J, Teller DY, Motulsky AG. 1992. Genotype-phenotype relationships in human red/green color-vision defects: molecular and psychophysical studies. Am J Hum Genet 51: 687–700. [PMC free article] [PubMed] [Google Scholar]
- de Jong S, Chepelev I, Janson E, Strengman E, van den Berg LH, Veldink JH, Ophoff RA. 2012. Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics 13: 458 10.1186/1471-2164-13-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco F, Paciolla M, Napolitano F, Pescatore A, D'Addario I, Bal E, Lioi MB, Smahi A, Miano MG, Ursini MV. 2012. Genomic architecture at the Incontinentia Pigmenti locus favours de novo pathological alleles through different mechanisms. Hum Mol Genet 21: 1260–1271. 10.1093/hmg/ddr556 [DOI] [PubMed] [Google Scholar]
- Giner-Delgado C, Villatoro S, Lerga-Jaso J, Gayà-Vidal M, Oliva M, Castellano D, Pantano L, Bitarello B, Izquierdo D, Noguera I, et al. 2019. Evolutionary and functional impact of common polymorphic inversions in the human genome. Nat Commun 10: 4222 10.1038/s41467-019-12173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JR, Cáceres A, Esko T, Cuscó I, Puig M, Esnaola M, Reina J, Siroux V, Bouzigon E, Nadif R, et al. 2014. A common 16p11.2 inversion underlies the joint susceptibility to asthma and obesity. Am J Hum Genet 94: 361–372. 10.1016/j.ajhg.2014.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau C, Starkovich M, Azamian MS, Xia F, Cheung SW, Evans P, Henderson A, Lalani SR, Scott DA. 2017. Xp11.22 deletions encompassing CENPVL1, CENPVL2, MAGED1 and GSPT2 as a cause of syndromic X-linked intellectual disability. PLoS One 12: e0175962 10.1371/journal.pone.0175962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, McCarroll SA. 2015. Large multiallelic copy number variations in humans. Nat Genet 47: 296–303. 10.1038/ng.3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward BE, De Vos M, Valleley EMA, Charlton RS, Taylor GR, Sheridan E, Bonthron DT. 2007. Extensive gene conversion at the PMS2 DNA mismatch repair locus. Hum Mutat 28: 424–430. 10.1002/humu.20457 [DOI] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Marschall T, Kloosterman WP, Francioli LC, Baaijens JA, Dijkstra LJ, Abdellaoui A, Koval V, Thung DT, Wardenaar R, et al. 2016. A high-quality human reference panel reveals the complexity and distribution of genomic structural variants. Nat Commun 7: 12989 10.1038/ncomms12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. 2013. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10: 1003–1005. 10.1038/nmeth.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff AM, Alagaratnam S, Zhao S, Bruun J, Andrews PW, Lothe RA, Skotheim RI. 2016. Identification of novel fusion genes in testicular germ cell tumors. Cancer Res 76: 108–116. 10.1158/0008-5472.CAN-15-1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst 39: 21–42. 10.1146/annurev.ecolsys.39.110707.173532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J, Chaisson MJP, Steinberg KM, Warren W, Hoekzema K, Gordon D, Graves-Lindsay TA, Munson KM, Kronenberg ZN, Vives L, et al. 2017. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res 27: 677–685. 10.1101/gr.214007.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64. 10.1038/nature06862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol 8: e1000501 10.1371/journal.pbio.1000501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JS, Li M-X, Deng J-E, Sham PC. 2016. FAPI: fast and accurate P-value imputation for genome-wide association study. Eur J Hum Genet 24: 761–766. 10.1038/ejhg.2015.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen I, Lopez J, Skipper L, Hefferon T, Spalding JD, Garner J, Chen C, Maguire M, Corbett M, Zhou G, et al. 2013a. dbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res 41: D936–D941. 10.1093/nar/gks1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedländer MR, ‘t Hoen PAC, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. 2013b. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Sakin M, Pastor S, Mostovoy Y, Li L, Leung AKY, McCaffrey J, Young E, Lam ET, Hastie AR, Wong KHY, et al. 2019. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun 10: 1025 10.1038/s41467-019-08992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JN, Carrero IG, Dong JF, Yu FL. 2015a. Complexity and diversity of F8 genetic variations in the 1000 genomes. J Trombos Haemost 13: 2031–2040. 10.1111/jth.13144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, Li J, Zhao SD, Bradfield JP, Mentch FD, Maggadottir SM, Hou C, Abrams DJ, Chang D, Gao F, et al. 2015b. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat Med 21: 1018–1027. 10.1038/nm.3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR. 2011. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am J Hum Genet 89: 580–588. 10.1016/j.ajhg.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Amos CI. 2012. Investigation of inversion polymorphisms in the human genome using principal components analysis. PLoS One 7: e40224 10.1371/journal.pone.0040224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45: D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Fundichely A, Casillas S, Egea R, Ràmia M, Barbadilla A, Pantano L, Puig M, Cáceres M. 2014. InvFEST, a database integrating information of polymorphic inversions in the human genome. Nucleic Acids Res 42: D1027–D1032. 10.1093/nar/gkt1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott GP, Do D, Litterst CM, Maar D, Hindson CM, Steenblock ER, Legler TC, Jouvenot Y, Marrs SH, Bemis A, et al. 2013. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal Chem 85: 11619–11627. 10.1021/ac403061n [DOI] [PubMed] [Google Scholar]
- Menelaou A, Marchini J. 2013. Genotype calling and phasing using next-generation sequencing reads and a haplotype scaffold. Bioinformatics 29: 84–91. 10.1093/bioinformatics/bts632 [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. 2008. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet 40: 1124–1129. 10.1038/ng.213 [DOI] [PubMed] [Google Scholar]
- Olmedillas-López S, García-Arranz M, García-Olmo D. 2017. Current and emerging applications of droplet digital PCR in oncology. Mol Diagn Ther 21: 493–510. 10.1007/s40291-017-0278-8 [DOI] [PubMed] [Google Scholar]
- Plagnol V, Howson JMM, Smyth DJ, Walker N, Hafler JP, Wallace C, Stevens H, Jackson L, Simmonds MJ, Type 1 Diabetes Genetics Consortium, et al. 2011. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 7: e1002216 10.1371/journal.pgen.1002216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznik GD, Xue Y, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, Ayub Q, McCarthy SA, Narechania A, Kashin S, et al. 2016. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat Genet 48: 593–599. 10.1038/ng.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Casillas S, Villatoro S, Cáceres M. 2015a. Human inversions and their functional consequences. Brief Funct Genomics 14: 369–379. 10.1093/bfgp/elv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Castellano D, Pantano L, Giner-Delgado C, Izquierdo D, Gayà-Vidal M, Lucas-Lledó JI, Esko T, Terao C, Matsuda F, et al. 2015b. Functional impact and evolution of a novel human polymorphic inversion that disrupts a gene and creates a fusion transcript. PLoS Genet 11: e1005495 10.1371/journal.pgen.1005495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JF, Kamitaki N, Legler T, Cooper S, Klitgord N, Karlin-Neumann G, Wong C, Hodges S, Koehler R, Tzonev S, et al. 2015. A rapid molecular approach for chromosomal phasing. PLoS One 10: e0118270 10.1371/journal.pone.0118270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Arenas C, Cáceres A, López-Sánchez M, Tolosana I, Pérez-Jurado L, González JR. 2019. scoreInvHap: inversion genotyping for genome-wide association studies. PLoS Genet 15: e1008203 10.1371/journal.pgen.1008203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salm MPA, Horswell SD, Hutchison CE, Speedy HE, Yang X, Liang L, Schadt EE, Cookson WO, Wierzbicki AS, Naoumova RP, et al. 2012. The origin, global distribution, and functional impact of the human 8p23 inversion polymorphism. Genome Res 22: 1144–1153. 10.1101/gr.126037.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AD, Hills M, Porubský D, Guryev V, Falconer E, Lansdorp PM. 2016. Characterizing polymorphic inversions in human genomes by single cell sequencing. Genome Res 26: 1575–1587. 10.1101/gr.201160.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Ganesamoorthy D, Duarte T, Cao MD, Hoggart CJ, Coin LJM. 2018. npInv: accurate detection and genotyping of inversions using long read sub-alignment. BMC Bioinformatics 19: 261 10.1186/s12859-018-2252-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K, Iber J, Warren ST. 1997. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet 16: 96–99. 10.1038/ng0597-96 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, et al. 2005. A common inversion under selection in Europeans. Nat Genet 37: 129–137. 10.1038/ng1508 [DOI] [PubMed] [Google Scholar]
- Steinberg KM, Antonacci F, Sudmant PH, Kidd JM, Campbell CD, Vives L, Malig M, Scheinfeldt L, Beggs W, Ibrahim M, et al. 2012. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet 44: 872–880. 10.1038/ng.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8: e55943 10.1371/journal.pone.0055943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Hurles ME. 2009. High-throughput haplotype determination over long distances by Haplotype Fusion PCR and Ligation Haplotyping. Nat Protoc 4: 1771–1783. 10.1038/nprot.2009.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Shendure J, Porreca G, Church G, Green P, Tyler-Smith C, Hurles ME. 2006. Assaying chromosomal inversions by single-molecule haplotyping. Nat Methods 3: 439–445. 10.1038/nmeth881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Salvador D, Puig M, Gayà-Vidal M, Pacheco S, Giner-Delgado C, Noguera I, Izquierdo D, Martínez-Fundichely A, Ruiz-Herrera A, Estivill X, et al. 2017. Detailed analysis of inversions predicted between two human genomes: errors, real polymorphisms, and their origin and population distribution. Hum Mol Genet 26: 567–581. 10.1093/hmg/ddw415 [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol 33: 427–440. 10.1016/j.tree.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Zody MC, Jiang Z, Fung H-C, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, et al. 2008. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet 40: 1076–1083. 10.1038/ng.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.