Graphical abstract
Keywords: Severe acute respiratory coronavirus 2, Infection, Stem cells, Immunomodulation, Repair
Highlights
-
•
ACE2+ stem cells of the respiratory tracts are likely involved in SARS-CoV-2 infection.
-
•
Dysregulatiton of immune response is underlying the physiopathology of fata ARDS and ALI.
-
•
MSCs appear to be a promising cell therapy because they favourably modulate the immune response to reduce lung injury.
-
•
Due to viral infection targeting pulmonary stem cells, lung might loss endogenous repair capability.
-
•
Transplantation of exogenous stem cells may be needed to facilitate the regeneration of lung tissue.
-
•
Vaccines, NK cells and CTLs may offer urgent therapeutics and prevention measures of CoV-SARS-2 reemergence.
Abstract
The emergence of the novel severe acute respiratory coronavirus 2 (SARS-CoV-2) in China and its rapid national and international spread have created a global health emergency. The resemblance with SARS-CoV in spike protein suggests that SARS-CoV-2 employs spike–driven entry into angiotensin-converting enzyme 2 (ACE2)-expressing cells. From a stem cell perspective, this review focuses on the possible involvement of ACE2+ stem/progenitor cells from both the upper and lower respiratory tracts in coronavirus infection. Viral infection-associated acute respiratory distress syndrome and acute lung injury occur because of dysregulation of the immune response. Mesenchymal stem cells appear to be a promising cell therapy given that they favorably modulate the immune response to reduce lung injury. The use of exogenous stem cells may lead to lung repair. Therefore, intervention by transplantation of exogenous stem cells may be required to replace, repair, remodel, and regenerate lung tissue in survivors infected with coronavirus. Ultimately, vaccines, natural killer cells and induced-pluripotent stem cell-derived virus-specific cytotoxic T lymphocytes may offer off-the-shelf therapeutics for preventing coronavirus reemergence.
1. Introduction
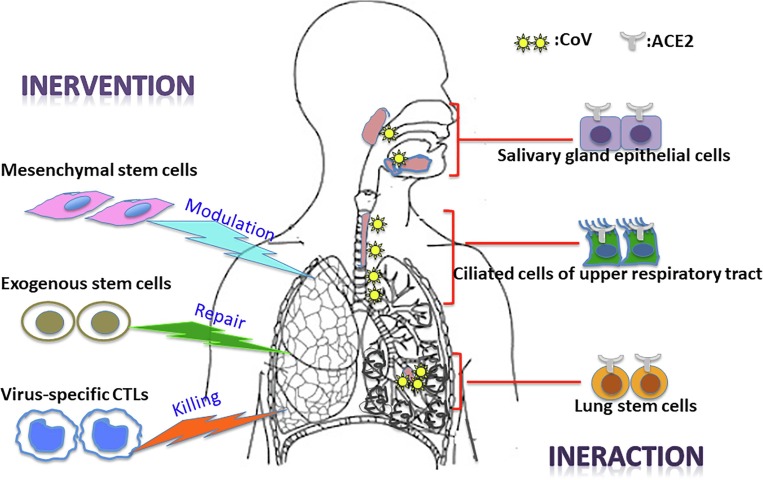
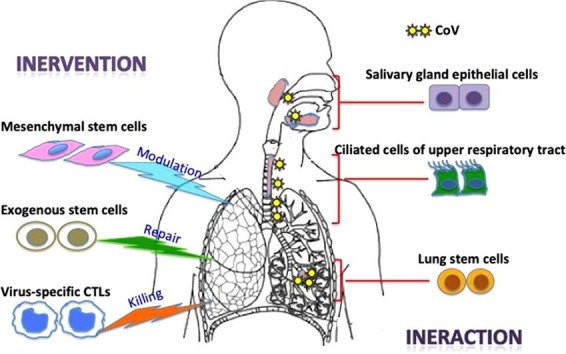
The ongoing outbreak of viral pneumonia in the city of Wuhan, China, which began in December 2019, has now spread to various countries worldwide. The etiologic agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a novel form of coronavirus that causes coronavirus disease 2019 (COVID-19), which is closely related to SARS-CoV. Our understanding of the novel virus is either based on the established knowledge of SARS-CoV or just beginning to emerge. Studies have shown that SARS-CoV-2 shares a receptor with SARS-CoV, providing clues regarding its cell entry, viral infection route, and pathogenic mechanisms at the cellular level. Except for asymptomatic carriers, infection with this virus causes a large fraction of patients to develop moderate clinical illness in a short period, with a small but a substantial number of patients experiencing severe disease characterized by fatal acute lung injury (ALI) and acute respiratory distress syndrome (ARDS). Stem cells in the respiratory tract /lung parenchyma appear to play a major role by interacting with the virus to cause entry infection, lung damage, and repair. Exogenous mesenchymal stem cells (MSCs) represent a promising cell therapy for modulating the immune dysregulation underlying ALI/ARDS to restore lung function. Transplantation of exogenous stem cells may lead to lung repair and regeneration. To be prepared for the worst-case scenario of coronavirus reemergence, it is important to develop vaccines and therapeutics. Stem cell-derived natural killer cells (NK cells) and off-the-shelf virus-specific cytotoxic T lymphocytes (CTLs) may be useful for meeting this urgent medical need. In this review, we discuss recent advances in our understanding of coronavirus infection and pathogenesis, with a special emphasis on the involvement of stem cells (Fig. 1 ).
Fig. 1.
Schematic diagram illustrating the involvement of stem cell interaction and intervention in SARS-CoV infection.
2. Epidemiology
In December 2019, a series of pneumonia cases of unknown cause emerged in Wuhan, Hubei Province, China. Patients presented with fever, dry cough, dyspnea, and bilateral ground-glass opacities on chest computed tomography (CT) scans. Organ dysfunction, for example, shock, ARDS, acute cardiac injury, acute kidney injury, and even death occurred in severe cases (Huang et al., 2020).
On January 3, 2020, a novel coronavirus subsequently named SARS-CoV-2 was detected by deep sequencing of the bronchoalveolar lavage fluid from patients, and was believed to be the etiologic agent of the new viral pneumonia (Lu et al., 2020). Phylogenic analysis showed that SARS-CoV-2 forms a distinct clade from SARS-CoV but shares 79.5% homology with SARS-CoV. On February 11, 2020, the World Health Organization (WHO) and the International Committee on Taxonomy of Viruses (ICTV) have officially announced the disease as COVID-19 and designated the virus itself as SARS-CoV-2.
Some initially infected patients had a history of exposure to the Huanan Seafood Wholesale Wet Market, potentially involving animal contact. The virus is thus thought to have a zoonotic origin, after which human-to-human transmission occurred. Its genetic sequences have shown high similarities to bat coronaviruses (96%), indicating the likelihood of bats being the ultimate origin (Zhou et al., 2020). Although an intermediate reservoir such as a pangolin was proposed (Tao Zhang and Zhang, 2020), the exact source of the virus and how it crosses the species barrier and jumps to humans remains largely unknown (Yu et al., 2020).
The contagious epidemic outbreak spread rapidly from Wuhan to other areas in China and the world. By February 12, 2020, a total of 59,805 cases were reported in China. Internationally, 499 exported cases were confirmed in 26 countries across 5 continents.
2.1. Spike protein and ACE2 receptor
Thus far, very little is known regarding the mode of SARS-CoV-2 interactions with host cells at the onset of infection. Fortunately, the quick release of the genomic sequences by Chinese scientists revealed that both SARS-CoV-2 and SARS-CoV belong to the β-genus of coronaviruses (Lu et al., 2020, Zhou et al., 2020). Coronaviruses are a large family of single-stranded enveloped RNA viruses. The positive-sense RNA genome encodes for four structural proteins including spike (S) glycoprotein, matrix (M) protein, small envelope (E) protein, and nucleocapsid (N) protein (Li et al., 2003). It is well-known that the spike glycoprotein mediates receptor binding and membrane fusion and is crucial for determining host tropism and transmission capacity (Marra et al., 2003, Turner et al., 2004).
A longer spike protein is encoded by SARS-CoV-2 genome compared to that of SARS-CoV (1273 versus 1255 amino acids) because of 3 short insertions at the N terminus (Lu et al., 2020, Zhou et al., 2020). The overall sequence similarities between the SARS-CoV-2 spike and SARS-CoV spike (isolated from human, civet, or bat) are approximately 76–78% for the whole protein, 73–76% for the receptor-binding domain, and 50–53% for the receptor-binding motif(Wan et al.,2020). SARS-CoV spike protein engages angiotensin-converting enzyme 2 (ACE2) as an entry receptor. Importantly, among the 14 key ACE2-contacting residues in the receptor-binding domain previously identified in SARS-CoV, 9 are fully conserved and 4 are partially conserved among SARS-CoV-2 and SARS-CoV from human, civet, and bat (Wan et al., 2020). Based on a previous study of SARS-CoV and sequence similarity, structural modeling strongly predicted that SARS-CoV-2 employs the ACE2 receptor for cell entry as well. In agreement with these findings, directed expression of human ACE2 allowed S protein pseudotyped-particle entry into the otherwise non-susceptible BHK cells(Hoffmann et al., 2020), indicating that SARS-CoV-2 like SARS-S uses ACE2 for cellular entry. In addition, a publication in Nature described that when directly combined SARS-CoV-2 with HeLa cells, SARS-CoV-2 could use ACE2 receptors from human, bat, civet, and swine, but not mouse, as an entry receptor in ACE2-expressing HeLa cells, except in cells without ACE2 expression (Zhou et al., 2020). Together, the structural modeling, pseudotyping, and infection data provide strong evidence that human ACE2 is the receptor for SARS-CoV-2.
2.2. ACE2 expression in pulmonary stem cells
As a functional receptor for coronavirus, studying the tissue distribution of ACE2 has major implications for understanding the targeted cell types and infection routes. ACE2 is a homolog of ACE, which plays an important role in the renin–angiotensin system for blood pressure homeostasis (Turner et al., 2004); however, its physiological role in the airways is currently unknown. A rapid report indicated no significant disparities in ACE2 gene expression between racial groups (Asian vs Caucasian), age groups (>60 vs < 60 years) or gender groups (male vs female). However, significantly higher ACE2 gene expression was observed in smoker samples compared to in non-smoker samples, suggesting that this group is more vulnerable to coronavirus infection (Cai, 2020). ACE2 mRNA is known to be present in virtually all organs, particularly in the bronchus and lung parenchyma as well as in the heart, kidney, and gastrointestinal tract (Harmer et al., 2002). The most remarkable finding was the surface expression of ACE2 protein on lung alveolar epithelial cells and enterocytes of the small intestine, correlating with symptoms seen in patients such as ARDS and diarrhea. The protein expression of ACE2 in the upper respiratory tract remains controversial. In one study, ACE2 protein expression was not detected on the surface of epithelial cells lining the oral cavity, nasal mucosa, and nasopharynx (Hamming et al., 2004). Another study showed that ACE2 expression is positively correlated with the differentiation state of the epithelia. ACE2 protein was more abundantly expressed on the apical than on the basolateral surface of polarized airway epithelia (Jia et al., 2005). The study demonstrated SARS-CoV preferentially infects well-differentiated ciliated epithelial cells expressing ACE2 in an in vitro model.
The outbreak of COVID-19 triggered a new wave of revisiting studies of ACE2 expression, and many are based on single-cell RNA-sequencing (scRNA-seq). Immunohistochemistry on human tissues confirmed that ACE2 is expressed within type II pneumocytes of human lung [15,16]. Notably, the not previously investigated nasal ciliated epithelial cells were shown to have one of the highest ACE2 mRNA expression levels among investigated cell types in the respiratory tract, which was validated independently at protein level by another group using immunostaining on human tissues (Ivan T Lee, 2020, Sungnak et al., 2020).
The pulmonary system contains functionally distinct candidate stem/progenitor cells which reside in distinct anatomical locations. The basal cells (Hong et al., 2004), Club cells (Rawlins et al., 2009), bronchoalveolar stem cells (BASCs) (Kim et al., 2005) and type II pneumocytes (Fehrenbach, 2001) are the candidate stem/progenitor cells which can repair the injured lungs and contribute to local needs in times of tissue damage. In addition to Type II pneumocytes, several studies revealed that a subset of murine and human Oct4+ pulmonary stem cells expressing ACE2 is the prime target of SARS-CoV infection (Chen et al., 2007, Ling et al., 2006, Mallick et al., 2009). The infected cells support active virus replication, which leads to their own destruction (Ling et al., 2006). This might explain the long course of illness, in the context of continued deterioration of lung tissues and apparent loss of capacity of lung repair observed in SARS patients. This could be also true for CoV-SARS-2 since they share same ACE2 receptor for cell entry.
2.3. Involvement of upper airway in coronavirus infection
Our current understanding of the target cells of SARS-CoV in the lower airway/lung parenchyma is largely based on analysis of autopsy specimens from patients who died of SARS. The early involvement of the upper airway has been less widely studied and is poorly understood. Although SARS-CoV mainly targets the lung, not all patients develop pulmonary destruction symptoms. In one report, approximately one-third of SARS-infected patients had self-limited symptoms with no clinical or radiological evidence of progression to pneumonitis (Tsui et al., 2003), but upper respiratory tract symptoms occurred in a minority of patients with SARS (Peiris et al., 2003). In another study, larger amount of SARS-CoV RNA was found in the saliva from all specimens of 17 patients with SARS, including 4 patients before the development of lung lesions and in some nasopharyngeal aspirates (Wang et al., 2004). Convincing evidence of airborne spread of the SARS-CoV droplets was presented in affected apartment in Hong Kong (Yu et al., 2004). Using a non-human primate model, Liu et al found that ACE2-positive cells were widely distributed in the upper respiratory tract in Chinese macaques, and ACE2 epithelial cells lining the salivary gland ducts were early target cells that were productively infected (Liu et al., 2011). Further studies are warranted to examine whether those cells are stem cells or well-differentiated cells. Another in vitro study showed that SARS-CoV efficiently infected ciliated cells when applied to the apical surface. The virus then replicated in polarized epithelia and preferentially exited via the apical surface (Jia et al., 2005). It also would be of interest to test if the mucociliary clearance function was impaired in order to facilitate the releasing of virus into the lumen of the human lung. Collectively, both clinical and experimental data suggest a critical role for viral production in upper respiratory sites, including salivary gland ducts, in the early phase of infection. Similarly, SARS-CoV-2 has a lot of resemblance to SARS-CoV, for instance, respiratory droplet transmission (Lai et al., 2020). While revising the manuscript, newly emerging data demonstrated that ciliated cells in nasal epithelia have the highest gene expression of ACE2 across airway (Sungnak et al., 2020). And in both asymptomatic and symptomatic patients, nasal swabs have yielded higher CoV-SARS-2 viral loads than throat swabs (To et al., 2020, Zhou et al., 2020). These data strongly suggest nasal epithelium as a more important portal within upper airway for initial virus infection, production and transmission.
2.4. Stem cells for modeling
However, in vivo confirmation of the identity of virus-targeted cells in the early phase is difficult because of the lack of availability of samples from patients who died within 3 days after illness onset or even before onset. Most data from human respiratory tissue of patients with SARS were obtained at two or more weeks following disease onset (Franks et al., 2003, To et al., 2004). Although SARS-CoV-2 and SARS-CoV showed numerous similarities, direct evidence regarding how SARS-CoV-2 infects cells in the respiratory tract is lacking. Airway stem cells may offer a quick in vitro solution for these studies. We and others have shown that basal cells from the nasal and nasopharyngeal mucosa exhibit cardinal features of stem cells (Hong et al., 2004, Yu et al., 2017, Yu et al., 2012, Zhao et al., 2012). When cultured at the air-liquid interface on Transwell (Yu et al., 2017, Zhao et al., 2012), they can different into pseudostratified mucociliary epithelium that recapitulates the morphological and physiological features of the human conducting airway in vivo. SARS-CoV-2 then can be applied to the pseudostratified epithelium at the apical or basolateral side to examine the potential for infection. While some animal models are under development, most of the virus research is carried out in immortalized simian Vero E6 cell line. Although Vero E6 cell model is convenient and useful, it has limitations and does not allow for the understanding of virus biology and evolution. This ex vivo Transwell model will constitute a convenient tool to study the viral infection and test antiviral drugs. In the same way, Oct4+ BASCs at the bronchioalveolar duct junction can readily form expandable colonies in vivo (Kim et al., 2005), which can be used to test infection in the lower airway. Although transgenic mice and non-human primate models are the next step for conducting these studies, they are might be taking long time and costly. Additionally, there are some differences between species. For example, ACE2 is widely distributed in the entire upper airway in Chinese macaques, which contradicts most data from humans (Liu et al., 2011).
2.5. Stem cells for therapy
Severe pneumonia caused by pathogenic coronaviruses is often associated with rapid virus replication, massive inflammatory cell infiltration, and elevated pro-inflammatory cytokine/chemokine responses (cytokine storm) resulting in ALI and ARDS (Channappanavar and Perlman, 2017). The pathological features of COVID-19 strongly resemble those of ARDS associated with SARS and Middle Eastern respiratory syndrome coronavirus infections (Xu et al., 2020), such as massive alveolar damage and progressive respiratory failure.
The current treatments for ARDS associated with coronavirus infection include early combination use of antiviral drugs (e.g., remdesvir lopinavir plus ritonavir) plus steroids (hydrocortisone, interferon) (Tsui et al., 2003), and supportive measures, such as lung-protective ventilation and conservative fluid management (Acute Respiratory Distress Acute Respiratory Distress Syndrome et al., 2000, National Heart et al., 2006). No pharmacological therapies appear to effectively halt the disease, as it progresses very rapidly. Studies of humans who died from SARS and COVID-19 suggested that a dysregulated immune response occurred, resulting in excessive inflammation and lethal ARDS (Channappanavar and Perlman, 2017, Nicholls et al., 2003, Xu et al., 2020, Zumla et al., 2020). Thus, immunomodulation should be an effective therapy, such as by using stem cells. MSCs represent one of the most promising candidates since their safety and efficacy have been shown in pre-clinical models of ARDS (Hayes et al., 2012, Horie et al., 2018). During revision of this manuscript, a study using MSCs to treat patients with COVID-19 was published. Seven critically ill patients were intravenously administered MSCs at 1 × 106 cells per kilogram body weight, followed by 14-day observation. The treatment was safe and had no adverse effects. Inflammatory and immune functions were corrected based on measurement of cytokine levels (i.e., TNF-α, IL-10) and a subset of immune cells (D4, CD8, NK, DC cells). Importantly, the pulmonary function and symptoms of these seven patients were significantly improved at 2 days after MSC transplantation (Leng et al., 2020). Additionally, the MSCs were ACE2- and TMPRSS2-negative at the gene level, suggesting the unlikelihood of virus infection in the transplanted cells. Together with local hospitals in Wuhan, we treated 24 critically ill patients with COVID-19 via intravenous administration of clinical-grade human MSCs derived from umbilical cord. The treatment was safe and resulted in improved functional outcomes in patients (manuscript under preparation). In short, MSCs appear show the greatest immediate potential for clinical translation for treating COVID-19, given that they may favorably modulate the immune response to reduce lung injury while maintaining host immune-competence.
2.6. Stem cells for repair and regeneration
SARS targets endogenous lung stem cells, which likely also occurs for SARS-CoV-2. This could explain the apparent loss in the capacity for lung repair after coronavirus infection and later phases of lung failure in severely diseased patients (Peiris et al., 2003). Long-term survivors exhibit aberrant or excess remodeling involving tissue fibrosis. Regeneration of the injured distal lung epithelium and restoration of alveolar barrier function are prerequisite for re-establishing proper gas exchange. Transplantation of exogenously derived stem cells may be an option for this situation. The molecular mechanisms underlying stem/progenitor cell-mediated regenerative responses have not been well-characterized. Particularly, the impact of coronavirus infection on endogenous lung stem cells and their regenerative responses remain unclear. This type of data is not yet available for patients with SARS or COVID-19 (For general lung injury or ARDS and stem cell repair, please see reviews elsewhere (Hayes et al., 2012, Neuringer and Randell, 2004)). However, many studies have been conducted in animal models to evaluate cell-remodeled airways and populate the damaged alveolar parenchyma upon widespread destruction of airway and alveolar epithelial cells following severe influenza infection (Kumar et al., 2011, Ray et al., 2016, Vaughan et al., 2015, Zuo et al., 2015). In a murine model, a highly pathogenic influenza virus displays a strong tropism to and infected a subset of lung stem/progenitor cells phenotyped as EpCamhighCD24lowintegrin(α6)high. Of significance, influenza virus-infected stem cells exhibited severely impaired renewal capacity because of virus infection-induced blockade of β-catenin-dependent Fgfr2b signaling, as evidenced by in vivo loss of alveolar tissue repair capacity after intrapulmonary stem cell transplantation (Quantius et al., 2016). In contrast, transplantation of non-infected stem cells restored alveolar barrier function and increased survival following influenza viral pneumonia in response to the application of exogenous Fgf10 (Quantius et al., 2016). These results provide insight into the feasibility of using exogenous stem cell therapy for treating coronavirus infected patients.
We and others have demonstrated that many types of stem/progenitor cells are present along the proximodistal axis of the mammalian respiratory tract (Fehrenbach, 2001, Hong et al., 2004, Kim et al., 2005, Rawlins et al., 2009, Yu et al., 2017, Yu et al., 2012, Zhao et al., 2012). Additionally, various other cell types such as MSCs, endothelial progenitor cells, embryonic stem cells, and induced pluripotent stem cells (iPSCs) are present. Each of these candidates show potential for lung regeneration following coronavirus infection with advantages and disadvantages (Hayes et al., 2012). However, significant deficits remain in our knowledge regarding the mechanisms of action of stem cells, their efficacy in relevant pre-clinical models, and their safety. These gaps must be addressed before the enormous therapeutic potential of stem cells for ALI/ARDS can be realized.
2.7. Stem cells for prevention
In patients who have recovered from SARS, data strongly suggest that neutralizing antibodies and CTLs plays a central role in the clearance of SARS-CoV (Wang and Chen, 2004). In COVID-19, neutralizing antibodies were detected in recovered patients and showed therapeutic potential for treating acute infections. The counts of peripheral CD4 and CD8 T cells were substantially reduced, whereas their status was hyperactivated (Huang et al., 2020). This indicates that both humoral immunity and cellular immunity are involved in SARS-CoV-2 infections.
Responding to the outbreak of COVID-19, world scientists have been moving at record speed to develop vaccines for SARS-CoV-2. The vaccine production platforms and technologies include RNA vaccines, DNA vaccines, recombinant protein vaccines, viral vector-based vaccines, live attenuated vaccines and inactivated vaccines. An mRNA-based vaccine developed by Moderna (ClinicalTrials.gov: NCT04283461) and an inactivated vaccine developed by Chinese Academy of Medical Sciences are currently the furthest along (Gao et al., 2020). For details of the current status and timeline please refer to the report in (Amanat and Krammer, 2020).
A new study showed COVID-19 patients experience functional exhaustion of antiviral lymphocytes (Zheng et al., 2020). Therefore adoptive immune therapy is a promising alternative. On January 29, 2020, Sorrento Therapeutics and Celularity Inc initiated emergency allogeneic natural killer (NK) cell therapy development for coronavirus infection with Chinese leading experts and scientists. The objective of the collaboration is to expand the therapeutic use of Celularity's CYNK-001, an allogeneic, off-the-shelf, placental-derived NK cell therapy, for the treatment and prevention of coronavirus infections. Technical details, such as dosing levels and timing, are not yet available. The Yingfeng Biological Group houses one of seven largest banks of cord blood, cord tissue, and placenta in China. We are currently actively exploring the development of stem cell therapeutics to fight the virus by taking advantage of our abundant resources.
Adoptive transfer of virus-specific CTLs has already shown efficacy for treating viral pathogens. The cells often are from seropositive third-party donors (Ando and Nakauchi, 2017, Leen et al., 2013, Vickers et al., 2014). However, broader implementation of this therapeutic approach is limited by time, cost, complexity, and lack of seropositive donors for individualized product manufacture, which may preclude the immediate availability of CTLs for urgent medical needs. Coronavirus poses a serious threat of reemergence, and thus an immediately available off-the-shelf product is needed. Such feasibility was only recently demonstrated (Minagawa et al., 2018, Nishimura et al., 2013, Tzannou et al., 2017). The innovative idea is that antigen-specific CTL clones are first established from antigen-specific CTLs obtained from the peripheral blood. These clones are reprogrammed into iPSCs (T-iPSCs), which can re-differentiate into CTLs that retain their ancestral antigen specificity and cytotoxicity, but with 100~1000-fold greater expansion capability. Thus, these cells can be banked as unlimited off-the-shelf therapeutics to meet urgent needs (Minagawa et al., 2018, Nishimura et al., 2013, Tzannou et al., 2017). Although there are safety concerns that must be addressed, such as undesirable rearrangement of the TCRA gene during reprogramming and redifferentiation, anti-virus activity was demonstrated in models of killing human immunodeficiency virus and Epstein-Barr virus in vitro and in vivo (Ando and Nakauchi, 2017, Doubrovina et al., 2012, Nishimura et al., 2013, Vickers et al., 2014). However, SARS-COV-2-specific cytotoxic T lymphocyte infusion as a possible treatment option for COVID-19 patients with severe disease, has not received enough attention till today.
3. Concluding remarks
Pulmonary stem cells are among a variety of cell types along respiratory tract to lung parenchyma targeted by coronavirus infection. The consequences of infection include ARDS/ALI, and eventually, loss of the ability to endogenously repair the lungs. Mesenchymal stem cells constitute a promising therapeutic strategy for patients suffering from coronaviruses infection associated ALI/ARDS by immune modulation. Transplantation of exogenous stem cells may directly replace injured lung tissues. Ultimately, while vaccines are under development, NK cells and virus-specific CTLs engineered using iPSC technology may offer an off-the-shelf cell therapeutic option for preventing the reemergence of coronaviruses.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The manuscript was prepared in early February. During the period of submission and revision, new data on SARS-CoV-2 continued to emerge. The authors regret that they cannot cite those valuable work done afterwards. We thank Professor Aaron Drake for his critical reading of our manuscript.
Author contributions
FY conceived the review. FY, RJ, YT, JL and BW wrote the manuscript.
References
- Acute Respiratory Distress Syndrome N., Brower R.G., Matthay M.A., Morris A., Schoenfeld D., Thompson B.T., Wheeler A. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N. Engl. J. Med. 2000;342(18):1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando M., Nakauchi H. 'Off-the-shelf' immunotherapy with iPSC-derived rejuvenated cytotoxic T lymphocytes. Exp. Hematol. 2017;47:2–12. doi: 10.1016/j.exphem.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Cai, G. (2020). Tobacco-Use Disparity in Gene Expression of ACE2, the Receptor of 2019-nCov. Preprints, Available from: https://www.preprints.org/manuscript/202002.200051/v202001.
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chan V.S., Zheng B., Chan K.Y., Xu X., To L.Y., Lin C.L. A novel subset of putative stem/progenitor CD34+Oct-4+ cells is the major target for SARS coronavirus in human lung. J. Exp. Med. 2007;204(11):2529–2536. doi: 10.1084/jem.20070462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doubrovina E., Oflaz-Sozmen B., Prockop S.E., Kernan N.A., Abramson S., Teruya-Feldstein J., O'Reilly R.J. Adoptive immunotherapy with unselected or EBV-specific T cells for biopsy-proven EBV+ lymphomas after allogeneic hematopoietic cell transplantation. Blood. 2012;119(11):2644–2656. doi: 10.1182/blood-2011-08-371971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbach H. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir. Res. 2001;2(1):33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.J., Chong P.Y., Chui P., Galvin J.R., Lourens R.M., Reid A.H., Travis W.D. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 2003;34(8):743–748. doi: 10.1016/s0046-8177(03)00367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q., Bao, L., Mao, H., Wang, L., Xu, K., Yang, M., . . . Qin, C. (2020). Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Science, eabc1932. doi:10.1126/science.abc1932. [DOI] [PMC free article] [PubMed]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532(1–2):107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Hayes M., Curley G., Ansari B., Laffey J.G. Clinical review: Stem cell therapies for acute lung injury/acute respiratory distress syndrome - hope or hype? Crit. Care. 2012;16(2):205. doi: 10.1186/cc10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Krüger, N., Müller, M., Drosten, C., & Pöhlmann, S. (2020). The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv[Preprint], Available from:https://doi.org/10.1101/2020.1101.1131.929042. doi:10.1101/2020.01.31.929042.
- Hong K.U., Reynolds S.D., Watkins S., Fuchs E., Stripp B.R. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am. J. Pathol. 2004;164(2):577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie S., Gonzalez H.E., Laffey J.G., Masterson C.H. Cell therapy in acute respiratory distress syndrome. J Thorac Dis. 2018;10(9):5607–5620. doi: 10.21037/jtd.2018.08.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan T Lee, T. N., Chien-Ting Wu, Yury Goltsev, Sizun Jiang, Phillip A Gall, Chun-Kang Liao, Liang-Chun Shih, Christian M Schurch, David R McIlwain, Pauline Chu, Nicole A Borchard, David Zarabanda, Sachi S Dholakia, Angela Yang, Dayoung Kim, Tomoharu Kanie, Chia-Der Lin, Ming-Hsui Tsai, Katie M Phillips, Raymond Kim, Jonathan B Overdevest, Matthew A Tyler, Carol H Yan, Chih-Feng Lin, Yi-Tsen Lin, Da-Tian Bau, Gregory J Tsay, Zara M Patel, Yung-An Tsou, Chih-Jaan Tai, Te-Huei Yeh, Peter H Hwang, Garry P Nolan, Jayakar V Nayak, Peter K Jackson. (2020). Robust ACE2 protein expression localizes to the motile cilia of the respiratory tract epithelia and is not increased by ACE inhibitors or angiotensin receptor blockers. MedRxiv[Preprint], Available from: https://doi.org/10.1101/2020.1105.1108.20092866.
- Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79(23):14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.F., Jackson E.L., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Kumar P.A., Hu Y., Yamamoto Y., Hoe N.B., Wei T.S., Mu D., McKeon F. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147(3):525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leen A.M., Bollard C.M., Mendizabal A.M., Shpall E.J., Szabolcs P., Antin J.H., Heslop H.E. Multicenter study of banked third-party virus-specific T cells to treat severe viral infections after hematopoietic stem cell transplantation. Blood. 2013;121(26):5113–5123. doi: 10.1182/blood-2013-02-486324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Z., Zhu R., Hou W., Feng Y., Yang Y., Han Q., Zhao R.C. Transplantation of ACE2(-) Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020;11(2):216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling T.Y., Kuo M.D., Li C.L., Yu A.L., Huang Y.H., Wu T.J., Yu J. Identification of pulmonary Oct-4+ stem/progenitor cells and demonstration of their susceptibility to SARS coronavirus (SARS-CoV) infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 2006;103(25):9530–9535. doi: 10.1073/pnas.0510232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Alvarez X., Wang H., Du Y., Zhu H., Chen Z. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J. Virol. 2011;85(8):4025–4030. doi: 10.1128/JVI.02292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick B., Ghosh Z., Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE. 2009;4(11) doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Minagawa, A., Yoshikawa, T., Yasukawa, M., Hotta, A., Kunitomo, M., Iriguchi, S., . . . Kaneko, S. (2018). Enhancing T Cell Receptor Stability in Rejuvenated iPSC-Derived T Cells Improves Their Use in Cancer Immunotherapy. Cell Stem Cell, 23(6), 850-858 e854. doi:10.1016/j.stem.2018.10.005. [DOI] [PubMed]
- National Heart, L., Blood Institute Acute Respiratory Distress Syndrome Clinical Trials, N., Wiedemann, H. P., Wheeler, A. P., Bernard, G. R., Thompson, B. T., . . . Harabin, A. L. (2006). Comparison of two fluid-management strategies in acute lung injury. N Engl J Med, 354(24), 2564-2575. doi:10.1056/NEJMoa062200. [DOI] [PubMed]
- Neuringer I.P., Randell S.H. Stem cells and repair of lung injuries. Respir. Res. 2004;5:6. doi: 10.1186/1465-9921-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Peiris J.S. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–1778. doi: 10.1016/s0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Kaneko S., Kawana-Tachikawa A., Tajima Y., Goto H., Zhu D., Nakauchi H. Generation of rejuvenated antigen-specific T cells by reprogramming to pluripotency and redifferentiation. Cell Stem Cell. 2013;12(1):114–126. doi: 10.1016/j.stem.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Peiris, J. S., Chu, C. M., Cheng, V. C., Chan, K. S., Hung, I. F., Poon, L. L., . . . Group, H. U. S. S. (2003). Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet, 361(9371), 1767-1772. doi:10.1016/s0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed]
- Quantius J., Schmoldt C., Vazquez-Armendariz A.I., Becker C., El Agha E., Wilhelm J., Herold S. Influenza Virus Infects Epithelial Stem/Progenitor Cells of the Distal Lung: Impact on Fgfr2b-Driven Epithelial Repair. PLoS Pathog. 2016;12(6) doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins E.L., Okubo T., Xue Y., Brass D.M., Auten R.L., Hasegawa H., Hogan B.L. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4(6):525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Chiba N., Yao C., Guan X., McConnell A.M., Brockway B., Stripp B.R. Rare SOX2(+) Airway Progenitor Cells Generate KRT5(+) Cells that Repopulate Damaged Alveolar Parenchyma following Influenza Virus Infection. Stem Cell Rep. 2016;7(5):817–825. doi: 10.1016/j.stemcr.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., Network H.C.A.L.B. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Zhang, Q. W., Zhigang Zhang. (2020). Pangolin homology associated with 2019-nCoV. bioRxiv[Preprint], Available from: https://doi.org/10.1101/2020.1102.1119.950253.
- To K.F., Tong J.H., Chan P.K., Au F.W., Chim S.S., Chan K.C., Ng H.K. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 2004;202(2):157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C., Yuen K.Y. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;ciaa149 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui P.T., Kwok M.L., Yuen H., Lai S.T. Severe acute respiratory syndrome: clinical outcome and prognostic correlates. Emerg. Infect. Dis. 2003;9(9):1064–1069. doi: 10.3201/eid0909.030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A.J., Hiscox J.A., Hooper N.M. ACE2: from vasopeptidase to SARS virus receptor. Trends Pharmacol. Sci. 2004;25(6):291–294. doi: 10.1016/j.tips.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzannou I., Papadopoulou A., Naik S., Leung K., Martinez C.A., Ramos C.A., Omer B. Off-the-Shelf Virus-Specific T Cells to Treat BK Virus, Human Herpesvirus 6, Cytomegalovirus, Epstein-Barr Virus, and Adenovirus Infections After Allogeneic Hematopoietic Stem-Cell Transplantation. J. Clin. Oncol. 2017;35(31):3547–3557. doi: 10.1200/JCO.2017.73.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A.E., Brumwell A.N., Xi Y., Gotts J.E., Brownfield D.G., Treutlein B., Chapman H.A. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517(7536):621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers M.A., Wilkie G.M., Robinson N., Rivera N., Haque T., Crawford D.H., Turner M.L. Establishment and operation of a Good Manufacturing Practice-compliant allogeneic Epstein-Barr virus (EBV)-specific cytotoxic cell bank for the treatment of EBV-associated lymphoproliferative disease. Br. J. Haematol. 2014;167(3):402–410. doi: 10.1111/bjh.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94(7):e00127–00120. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.K., Chen S.Y., Liu I.J., Chen Y.C., Chen H.L., Yang C.F., Hospital S.R.G., o. t. N. T. U. N. T. U. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg. Infect. Dis. 2004;10(7):1213–1219. doi: 10.3201/eid1007.031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.D., Chen W.F. Detecting specific cytotoxic T lymphocytes against SARS-coronavirus with DimerX HLA-A2: Ig fusion protein. Clin Immunol. 2004;113(2):151–154. doi: 10.1016/j.clim.2004.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Lu Y., Tao L., Jiang Y.Y., Lin D.C., Wang L., Loh K.S. Non-malignant epithelial cells preferentially proliferate from nasopharyngeal carcinoma biopsy cultured under conditionally reprogrammed conditions. Sci. Rep. 2017;7(1):17359. doi: 10.1038/s41598-017-17628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Zhao X., Li C., Li Y., Yan Y., Shi L., Wang D.Y. Airway stem cells: review of potential impact on understanding of upper airway diseases. Laryngoscope. 2012;122(7):1463–1469. doi: 10.1002/lary.23320. [DOI] [PubMed] [Google Scholar]
- Yu I.T., Li Y., Wong T.W., Tam W., Chan A.T., Lee J.H., Ho T. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N. Engl. J. Med. 2004;350(17):1731–1739. doi: 10.1056/NEJMoa032867. [DOI] [PubMed] [Google Scholar]
- Yu W.B., Tang G.D., Zhang L., Corlett R.T. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2 / HCoV-19) using whole genomic data. Zoological Research. 2020;41(3):247–257. doi: 10.24272/j.issn.2095-8137.2020.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yu F., Li C., Li Y., Chao S.S., Loh W.S., Wang D.Y. The use of nasal epithelial stem/progenitor cells to produce functioning ciliated cells in vitro. Am J Rhinol Allergy. 2012;26(5):345–350. doi: 10.2500/ajra.2012.26.3794. [DOI] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Azhar E.I., Memish Z.A., Maeurer M. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet. 2020;395(10224):e35–e36. doi: 10.1016/S0140-6736(20)30305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W., Zhang T., Wu D.Z., Guan S.P., Liew A.A., Yamamoto Y., McKeon F. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517(7536):616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]