Highlights
-
•
Due to the rapidly emerging SARS-CoV-2 pandemic and its tremendous public health challenges worldwide, there is a critical demand for rapid and easy to perform diagnostic assays.
-
•
The rapid antigen detection test evaluated here had a high diagnostic sensitivity and specificity in respiratory samples obtained from patients who mainly presented during the first week of COVID-19.
-
•
Rapid antigen detection has the potential to become an important tool for the early diagnosis of SARS-CoV-2, particularly in situations with limited access to molecular methods.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, Diagnosis, Rapid diagnostic test, Antigen
Abstract
Objectives
In the context of the coronavirus disease 2019 (COVID-19) pandemic, the development and validation of rapid and easy-to-perform diagnostic methods are of high priority. This study was performed to evaluate a novel rapid antigen detection test (RDT) for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in respiratory samples.
Methods
The fluorescence immunochromatographic SARS-CoV-2 antigen test (Bioeasy Biotechnology Co., Shenzhen, China) was evaluated using universal transport medium with nasopharyngeal (NP) and oropharyngeal (OP) swabs from suspected COVID-19 cases. Diagnostic accuracy was determined in comparison to SARS-CoV-2 real-time (RT)-PCR.
Results
A total of 127 samples were included; 82 were RT-PCR-positive. The median patient age was 38 years, 53.5% were male, and 93.7% were from the first week after symptom onset. Overall sensitivity and specificity were 93.9% (95% confidence interval 86.5–97.4%) and 100% (95% confidence interval 92.1–100%), respectively, with a diagnostic accuracy of 96.1% and Kappa coefficient of 0.9. Sensitivity was significantly higher in samples with high viral loads.
Conclusions
The RDT evaluated in this study showed a high sensitivity and specificity in samples mainly obtained during the first week of symptoms and with high viral loads, despite the use of a non-validated sample material. The assay has the potential to become an important tool for early diagnosis of SARS-CoV-2, particularly in situations with limited access to molecular methods.
Introduction
Since the first reported cases in December 2019, the rapidly emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has been causing tremendous public health challenges worldwide (WHO, 2020a). Timely detection and isolation of cases and their contacts are considered crucial to help curtail this unprecedented pandemic (Nguyen et al., 2020). This strategy relies on robust, rapid, and easy-to-perform diagnostic tools that can be used to test large numbers of samples in a short period of time. To date, the recommended diagnostic method for SARS-CoV-2 infection (known as coronavirus disease 2019 (COVID-19)) is real-time reverse-transcription PCR (RT-PCR), which was introduced in January 2020 (Corman et al., 2020) and is now applied using World Health Organization (WHO) or US Centers for Disease Control and Prevention (CDC) protocols (WHO, 2020b, CDC, 2020b), as well as various commercial assays (FIND, 2020).
The enormous gap between the large number of patients/contacts and the laboratory capacities to perform RT-PCR in a timely manner is a major limitation of current public health containment strategies (WHO, 2020c). Therefore, there is a critical demand for alternative assays such as antigen detection tests, which, in contrast to antibody tests, can detect the presence of the virus itself in respiratory samples (WHO, 2020c). Tests detecting SARS-CoV-2-specific antigen have recently been developed and many of them are now commercially available (FIND, 2020). However, the real-world performance of these assays is uncertain and their validation is therefore of high priority (ECDC, 2020). Other options include serological tests, but due to their diagnostic limitations in early infections, these tests are currently not recommended for case detection (WHO, 2020c, ECDC, 2020). Among possible test formats, rapid diagnostic tests (RDTs) should be prioritized, since they are timely, easy to perform, and can serve as point-of-care testing (Patel et al., 2020). This study was performed to evaluate a novel antigen-based RDT for the detection of SARS-CoV-2 in respiratory specimens from suspected COVID-19 cases.
Materials and methods
A study of the diagnostic accuracy of a rapid SARS-CoV-2 antigen detection test compared to RT-PCR was conducted. Samples were derived from patients with respiratory symptoms and/or fever and an epidemiological risk factor for SARS-CoV-2 infection (travel or contact with a case), attending Clínica Alemana, a private medical centre in Santiago, Chile (Weitzel et al., 2020), during the first weeks of the outbreak in Chile. Specimens were obtained by trained personnel in a newly created ‘respiratory emergency room’ at the hospital and consisted of a nasopharyngeal (NP) and an oropharyngeal swab (OP), which were placed together in a 3-ml tube of universal transport medium (UTM-RT System; Copan Diagnostics, Murrieta, CA, USA).
The samples were initially examined for SARS-CoV-2 by COVID-19 Genesig Real-Time PCR assay (Primerdesign Ltd, Chandler’s Ford, UK) after RNA extraction with the MagNA Pure Compact System (Roche Molecular Systems Inc., Pleasanton, CA, USA). The Primerdesign RT-PCR was the first European SARS-CoV-2 assay that was commercialized; it has received US Food and Drug Administration (FDA) Emergency Use Authorization (EUA) and is among the WHO Emergency Use Listing (EUL) tests eligible for procurement (https://www.who.int/diagnostics_laboratory/eual/listing/en). The assay includes a positive control template and an RNA internal extraction control. Its target gene is the RNA-dependent RNA polymerase (RdRP); the detection limit reported by the manufacturer is 0.58 copies/μl. Samples showing an exponential growth curve and a cycle threshold (Ct) value ≤40 were considered as positive.
PCR characterized samples (UTM with swabs) were kept at 4 °C and tested within 48 hours with the Bioeasy 2019-Novel Coronavirus (2019-nCoV) Fluorescence Antigen Rapid Test Kit (fluorescence immunochromatographic assay) (Bioeasy Biotechnology Co., Shenzhen, China; catalogue No. YRLF04401025, lot No. 2002N408), which detects SARS-CoV-2 nucleocapsid protein by lateral flow technique. The test uses europium-labelled chicken anti-SARS-CoV-2 IgY antibodies for primary binding and mouse anti-SARS-CoV-2 antibodies for capture; goat anti-chicken IgY antibodies are used for the internal control (Diao et al., 2020).
The manufacturer’s instructions for use recommend direct testing from OP or NP swabs as well as sputum. Our approach using UTM was chosen since it permitted the rapid evaluation of a large number of previously RT-PCR characterized clinical samples. For this procedure, the manufacturer permitted the application of 100 μl of UTM directly into the cassette (Peter Zhong, personal communication).
Positive and negative samples were selected by convenience among the 1453 respiratory specimens processed for SARS-CoV-2 in the clinical laboratory during the study period (March 16–21, 2020). Due to the shortage of available test kits, a 2:1 distribution of positive to negative samples was chosen. The technician performing the RDT was blinded to the RT-PCR results.
The UTM tubes were first vortexed for 20 seconds, then 100 μl of the UTM solution was placed into the sample port of the cassette. This was then incubated at room temperature for 10 minutes and then placed into the fluorescence immunoassay analyser EASY-11 (Bioeasy Biotechnology Co.). The instrument automatically delivers a positive or negative qualitative result with a detection limit of 0.005 ng/ml, according to the manufacturer. All test procedures except the reading of the cassette were performed under a BSL2 cabinet.
Results of the RDT were compared to those of RT-PCR as the reference method. For samples with discordant result, tests were repeated. The demographic and clinical data were obtained from the mandatory national COVID-19 notification forms and were analysed in an anonymized manner. Statistical analysis considered the calculation of sensitivity, specificity, diagnostic accuracy, and Kappa coefficient using standard formulae, and Wilson score confidence interval (CI) at 95% (OpenEpi version 3.01). Test performance was analysed as recommended by current Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2008). Sensitivity was further analysed for certain subgroups such as sex, days of symptoms at sampling, and RT-PCR Ct values.
Test kits used for this evaluation were bought from the local distributor using funds for routine diagnostics of the Clinical Laboratory of Clínica Alemana. The study was approved by the local institutional review board (Comité Etico Científico, Facultad de Medicina Clínica Alemana, Universidad del Desarrollo, Santiago, Chile) and the need for informed consent was waived.
Results
A total of 127 samples were included. Of these, 82 were RT-PCR-positive for SARS-CoV-2 RNA, representing 61% of total positive samples during the study period; 45 samples were RT-PCR-negative. Among the tested cases, 53.5% were male and the median age was 38 years. Most samples were taken during the initial phase of the disease, with a median duration of symptoms of 2 days (interquartile range (IQR) 1–4 days) (Table 1 ). The median Ct value of RT-PCR-positive samples was 17.7 (IQR 14.2–25.1) (Table 1).
Table 1.
Demographic, clinical, and laboratory features of included cases; data represent absolute numbers (%).
| All | PCR-positive | PCR-negative | ||
|---|---|---|---|---|
| Total | 127 | 82 | 45 | |
| Sex | Male | 68 (53.5) | 44 (53.7) | 24 (53.3) |
| Female | 59 (46.5) | 38 (46.3) | 21 (46.7) | |
| Age (years) | Median | 38 | 38 | 38 |
| IQR | 29.5–44 | 31–46.3 | 29–44 | |
| Range | 1–91 | 1–73 | 2–91 | |
| 0–17 | 16 (12.6) | 11 (13.4) | 5 (11.1) | |
| 18–59 | 102 (80.3) | 66 (80.5) | 36 (80.0) | |
| ≥60 | 9 (7.1) | 5 (6.1) | 4 (8.9) | |
| Days post symptom onseta | Median | 2 | 2 | 2 |
| IQR | 1–4 | 1–4 | 1–4 | |
| Range | 0–12 | 0–12 | 0–12 | |
| Day 0–3 | 91 (72.2) | 59 (72.8) | 32 (71.1) | |
| Day 4–7 | 27 (22.4) | 17 (21) | 10 (22.2) | |
| Day ≥8 | 8 (6.3) | 5 (6.2) | 3 (6.7) | |
| Clinical featuresa | Cough | 94 (74.6) | 63 (77.8) | 31 (68.9) |
| Fever | 77 (61.1) | 57 (70.4) | 20 (44.4) | |
| Ct value | Median | 17.7 | ||
| IQR | 14.2–25.1 | |||
| Mean | 20 |
IQR, interquartile range; Ct, cycle threshold of RT-PCR.
At time of sampling.
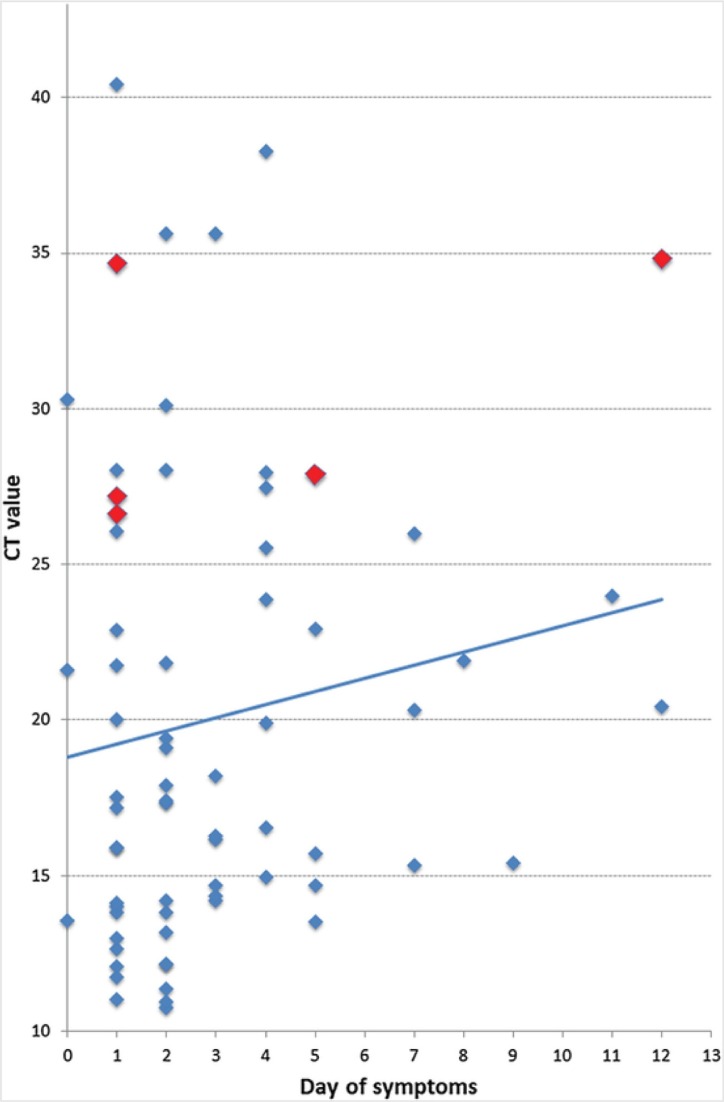
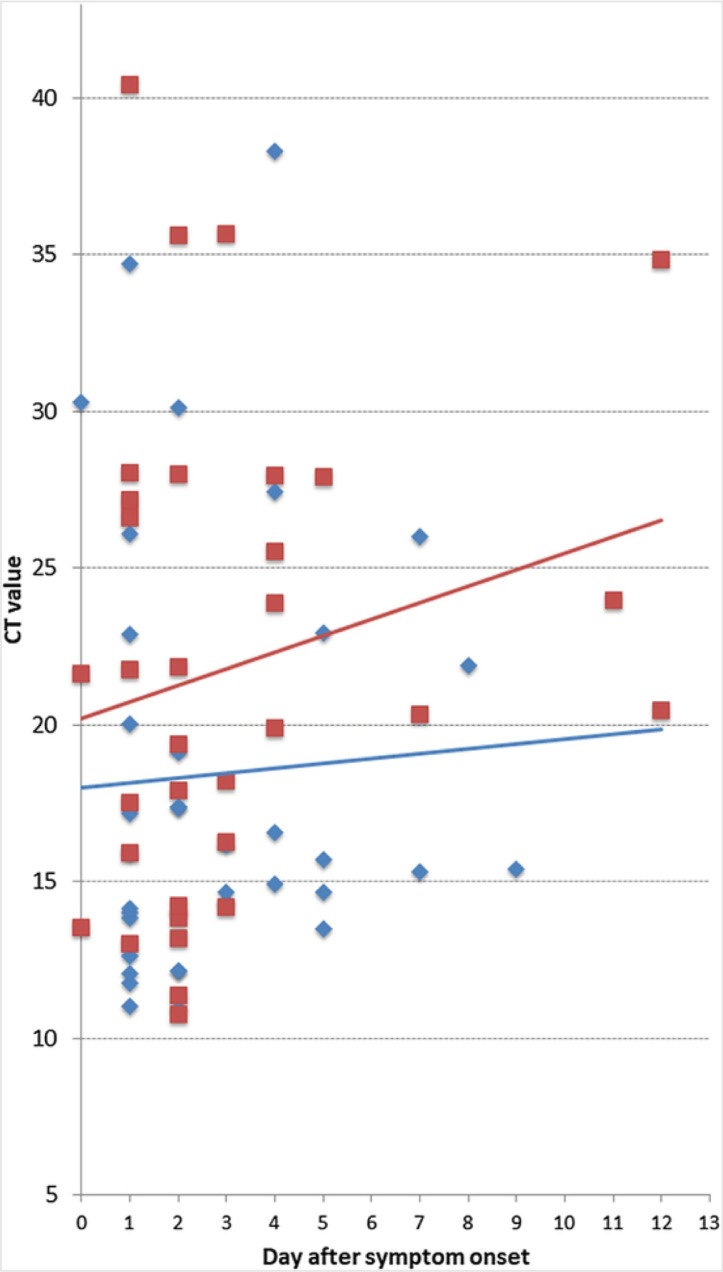
The overall sensitivity and specificity of the evaluated RDT were 93.9% (95% CI 86.5–97.4%) and 100% (95% CI 92.1–100%), respectively (Table 2 ). The diagnostic accuracy was 96.1%, with a Kappa coefficient of 0.9. Sensitivity was significantly reduced in the subgroup of samples with Ct values >25.1, which represented the fourth quartile of Ct values in this population, indicating lower viral loads. No significant difference within other subgroups (Table 2) was identified. All false-negative results (n = 5) corresponded to samples with RT-PCR Ct values >26 (Table 3 ). Ct values of true-positives and false-negatives and their relation to the duration of symptoms are shown in Fig. 1 . A subgroup analysis of Ct values revealed that samples from female patients had higher Ct values and a steeper positive trend line slope over time of infection compared to those from male patients (Fig. 2 ).
Table 2.
Sensitivity and specificity of the antigen detection test in the total sample and in different subgroups of samples
| Antigen detection test | ||||||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Sensitivity | Specificity | |||||
| Samples | RT-PCR | n | n | n | % | 95% CI | % | |
| All | Positive | 82 | 77 | 5 | 93.9 | 86.5–97.4 | 100% | |
| Negative | 45 | 0 | 45 | |||||
| Sex | Male | Positive | 44 | 43 | 1 | 97.7 | 88.2–99.6 | 100% |
| Negative | 24 | 0 | 24 | |||||
| Female | Positive | 38 | 34 | 4 | 89.5 | 75.9–95.8 | 100% | |
| Negative | 21 | 0 | 21 | |||||
| Days post symptom onset | 0–7 | Positive | 76 | 72 | 4 | 94.7 | 87.2–97.9 | 100% |
| Negative | 42 | 0 | 42 | |||||
| 8–12 | Positive | 5 | 4 | 1 | 80.0 | 37.6–96.4 | 100% | |
| Negative | 3 | 0 | 3 | |||||
| Ct values | Quartile 1–3 | Positive | 52 | 52 | 0 | 100 | 89.8–100 | |
| Quartile 4 | Positive | 18 | 13 | 5 | 72.2 | 49.1–87.5 | ||
CI, confidence interval; Ct, cycle threshold of RT-PCR.
Table 3.
Characteristics of RDT false-negative samples
| Number | Sex | Age (years) | Days of symptoms | Fever | Cough | RT-PCR | Ct | RDT |
|---|---|---|---|---|---|---|---|---|
| 4 | Male | 1 | 1 | + | + | Positive | 34.7 | Negative |
| 6 | Female | 51 | 12 | + | + | Positive | 34.8 | Negative |
| 35 | Female | 41 | 1 | + | + | Positive | 26.6 | Negative |
| 79 | Female | 32 | 1 | − | + | Positive | 27.2 | Negative |
| 117 | Female | 73 | 5 | − | + | Positive | 27.9 | Negative |
+, present; −, not present; Ct, cycle threshold; RDT, rapid diagnostic test.
Fig. 1.
Cycle threshold (Ct) values and lineal trend line of 70 RT-PCR-positive samples taken on different days after symptom onset. Dot colours represent false-negative (red) and true-positive (blue) results by antigen detection test.
Fig. 2.
Cycle threshold (Ct) values and lineal trend lines for 33 samples from female patients (red) and 37 from male patients (blue) taken on different days after symptom onset.
Discussion
The novel SARS-CoV-2 antigen test kit from Bioeasy is among the growing number of diagnostic assays available for COVID-19 with CE marking (FIND, 2020), which is based on self-reporting of the manufacturer and can be misused. The challenge of this procedure in light of the rapidly evolving COVID-19 pandemic has recently been addressed by the European Commission (European Commission, 2020).
The test has a cassette format with an external reader and is approved to be used with oropharyngeal swabs, nasopharyngeal swabs, and sputum. In our experience, the system was easy to use and gave a qualitative result for an individual sample in approximately 15 minutes. Depending on the reading mode, the analyser permits a throughput of approximately 5 (standard mode with incubation within the device) to >50 (rapid mode with incubation outside the device) samples per hour. This significant throughput is encouraging given the large number of samples processed in many COVID-19 testing points and the potential use of RDTs as a large-scale decentralized screening tool, e.g. in resource-poor settings. However, the inherent biological hazard requires the handling of specimens in a biosafety cabinet (WHO 2020d), hence slowing down the process and reducing the sample number per hour. This problem could be overcome by the use of extraction buffers or solutions with inactivating capacities.
Within our panel of clinical samples, the novel assay proved to be highly sensitive and specific. Interestingly, a similarly high sensitivity (94%) was reported for the detection of nucleocapsid antigen in early infections with SARS-CoV in a study from 2004 (Che et al., 2004). The sensitivity in the present study (93.9%) was higher than that reported by the manufacturer in the package insert for nasopharyngeal swabs (85.5%) and more than three times higher than the accuracy values reported in the grey literature for a related test with visual read-out. A preprint report from China with participation of the manufacturer found an overall sensitivity of 68% in 208 RT-PCR-positive nasopharyngeal swabs from patients in Hubei Province, China (Diao et al., 2020). However, when analysing the subgroup of samples with Ct values ≤30, the sensitivity of the assay increased to 98%. In the present study, a reduction of the sensitivity to 72% was also observed in samples with higher Ct values.
First information on the dynamics of SARS-CoV-2 demonstrated that viral replication in the pharynx is highest during the first days of clinical disease and declines afterwards (Wölfel et al., 2020, Zou et al., 2020 Zou et al., 2020). This phenomenon was also observed in the analyses of our Ct values (Fig. 1). Interestingly, the decline in viral load seemed more pronounced in female patients (Fig. 2). Accordingly, antigen tests from upper respiratory swabs should be more sensitive in the initial phase of symptomatic infection. Although we could not prove this effect in our study, it is important to highlight that the vast majority of the study samples corresponded to subjects in the early stages of infection (median duration of symptoms 2 days) and patients consulting in the late stage of COVID-19 were largely underrepresented. Furthermore, several samples from the early stage of infection with a low virus concentration were detected. This might be explained by variations associated with the sampling technique or by inaccurate data collection regarding symptom onset. However, the higher overall sensitivity in the present study when compared to the analysis from China is most probably related to the fact that the majority of samples (93.7%) were from patients during their first week of clinical disease. The high sensitivity of SARS-CoV-2 antigen detection in early infection might be a crucial finding for the design of new RDT-based algorithms, which are particularly important in weaker health systems and low-resource settings, where other high burden diseases, like malaria, also need to be considered.
The positive and negative predictive values (PPV and NPV) of the assay were not calculated for the study population. However, a test with a sensitivity of 93.9% and specificity of 100% would have a PPV and NPV of 100% and 99.4%, respectively, if applied for a population with a prevalence of 9%, as observed in our institution during the study period.
The data presented here are critical, not only to support local decision-making, but also for global agencies and governments worldwide in the procurement of simpler, scalable diagnostic tests as an answer to the global call for ‘test, test, test’ (Tedros Adhanom Ghebreyesus, Director General, World Health Organization, March 16, 2020).
This study has some limitations, namely the use of a sample type not specifically permitted in the instructions for use. The advantage of this adapted sample use was that it allowed the comparison of RT-PCR and RDT from the same material, without possible distribution errors from using separate swabs. The UTM volume of 3 ml could have led to a dilution of the antigen and reduction of sensitivity (the assay manufacturer recommends using a single swab and eluting it in 0.5 ml of buffer solution). Another limitation was the retrospective use of clinical data, which were collected under stressful routine work conditions within the ongoing outbreak. Finally, it is important to note that this evaluation was performed during a period of time (late summer in Chile) with a low circulation of other frequent respiratory viruses; therefore the performance of the antigen-based RDT might change in different epidemiological conditions.
In conclusion, the antigen-based immunofluorescence RDT evaluated here showed a high sensitivity and specificity in respiratory samples obtained from patients who mainly presented during the first week of COVID-19. The assay was easy to use and provided results in a timely manner. Hence, it has the potential to become an important tool for the early diagnosis of SARS-CoV-2, particularly in situations with limited access to molecular methods.
Author contributions: LP and TW conceived the study and wrote the first draft. LP, TW, and GP curated the data. LP, PL, XA, and TW analysed the data. LP, PL, and GP performed the investigation. TW and LP administered the project. LP, VV, PL, and TW supervised the study. LP, JMM, RA, XA, PV, MI, SD, and TW validated the data. All authors contributed in reviewing and editing later drafts, and approved the final version.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
All authors declare no competing interests.
References
- CDC (Centers for Disease Control and Prevention), Respiratory Viruses Branch, Division of Viral Diseases. Real-Time RT-PCR Panel for Detection 2019-Novel Coronavirus. Instructions for Use. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/index.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Flab%2Frt-pcr-detection-instructions.html, 2020 (accessed 15 May 2020).
- Che X.Y., Hao W., Wang Y., Di B., Yin K., Xu Y.C. Nucleocapsid protein as early diagnostic marker for SARS. Emerg Infect Dis. 2004;10:1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2008. User Protocol for Evaluation of Qualitative Test Performance; Approved Guideline-Second Edition. CLSI document EP12-A2. [Google Scholar]
- Corman V.M., Landt O., Kaiser M Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C. 2020. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein.https://www.medrxiv.org/content/10.1101/2020.03.07.20032524v2 Preprint at. [Google Scholar]
- ECDC. Coronavirus disease 2019 (COVID-19) pandemic: increased transmission in the EU/EEA and the UK – seventh update, 2020. Stockholm: 2020. https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-coronavirus-disease-2019-covid-19-pandemic (accessed 15 May 2020).
- Commission European. Communication from the Commission: Guidelines on COVID-19 in vitro diagnostic tests and their performance (2020/C 122 I/01) Official Journal of the European Union. 2020 https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:52020XC0415(04)&from=EN (accessed 13 May 2020) [Google Scholar]
- FIND (Foundation for Innovative New Diagnostics). SARS-CoV-2 diagnostic pipeline. 2020 (continuously updated). https://www.finddx.org/covid-19/pipeline (accessed 15 May 2020).
- Nguyen T., Duong Bang D., Wolff A. Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines (Basel) 2020 doi: 10.3390/mi11030306. pii:E306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R., Babady E., Theel E.S., Storch G.A., Pinsky B.A., St George K. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS-CoV-2/COVID-19. mBio. 2020 doi: 10.1128/mBio.00722-20. pii:e00722-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel T., Rodríguez F., Noriega L.M., Marcotti A., Duran L., Palavecino C. Hepatitis B and C virus infection among HIV patients within the public and private healthcare systems in Chile: A cross-sectional serosurvey. PLOS One. 2020;15 doi: 10.1371/journal.pone.0227776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organisation) A coordinated Global Research Roadmap. https://www.who.int/who-documents-detail/a-coordinated-global-research-roadmap, 2020a (accessed 15 May 2020).
- WHO (World Health Organisation) Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. Interim guidance. 2020. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117, 2020b (accessed 15 May 2020).
- WHO (World Health Organisation) Laboratory testing strategy recommendations for COVID-19. Interim guidance. 2020. https://apps.who.int/iris/bitstream/handle/10665/331509/WHO-COVID-19-lab_testing-2020.1-eng.pdf, 2020c (accessed 15 May 2020).
- WHO. Laboratory biosafety guidance related to the novel coronavirus (2019-nCoV). Interim guidance. 2020. https://www.who.int/docs/default-source/coronaviruse/laboratory-biosafety-novel-coronavirus-version-1-1.pdf?sfvrsn=912a9847_2, 2020d (accessed 15 May 2020).
- Wölfel R., Corman V.M., Guggemos, Seilmaier W.M., Zange S., Mueller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 Viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]