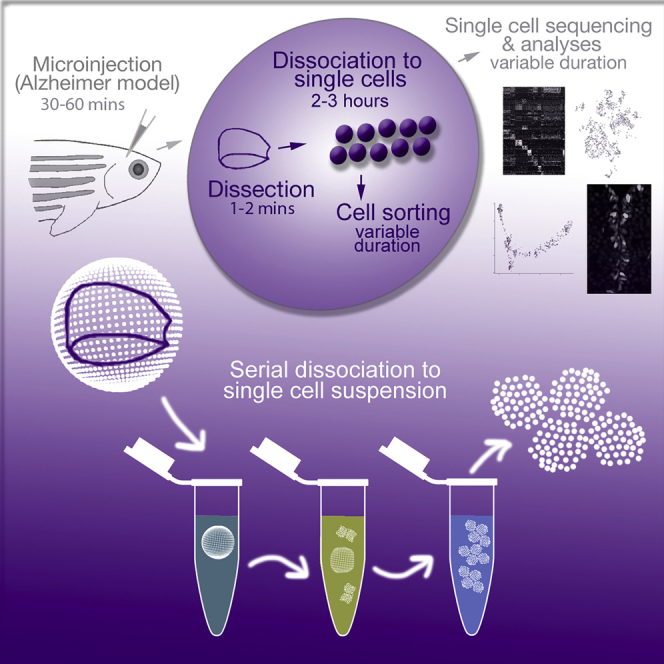
SUMMARY
Single-cell sequencing (sc-Seq) is a powerful tool to investigate the molecular signatures of cell types in a complex mixture of cells. A critical step in sc-Seq is preparing a single-cell suspension with a high number of viable cells. Here, we show how to dissect zebrafish telencephalon and how to dissociate it into a single-cell suspension. This is followed by flow cytometry-based sorting to enrich for neural progenitor stem cells. Our technique typically yields 70,000 live cells from one zebrafish telencephalon.
For complete details on the use and execution of this protocol, please refer to Cosacak et al. (2019).
Graphical Abstract
Highlights
-
•
We describe the dissection of adult zebrafish telencephalon.
-
•
We detail a protocol for dissociation of telencephalon into single-cell suspension.
-
•
We describe a procedure for flow cytometry-based sorting to enrich NSPCs.
-
•
Our technique typically yields 70,000 live cells from one zebrafish telencephalon.
Single-cell sequencing (sc-Seq) is a powerful tool to investigate the molecular signatures of cell types in a complex mixture of cells. A critical step in sc-Seq is preparing a single-cell suspension with a high number of viable cells. Here, we show how to dissect zebrafish telencephalon and how to dissociate it into a single-cell suspension. This is followed by flow cytometry-based sorting to enrich for neural progenitor stem cells. Our technique typically yields 70,000 live cells from one zebrafish telencephalon.
BEFORE YOU BEGIN
Note: Single-cell sequencing can work on complex mixture of cells that are not labeled by any reporter. However, the use of a specific cell population marked by a reporter protein (e.g., GFP) will increase the portion of the resulting data that is useful for the experimental question of interest. For instance, while it is possible to sequence all cells in telencephalon simultaneously, a reporter line can enrich a specific or rare cell type by flow cytometry-based sorting. The advantage of using a reporter line is to enrich a particular cell population among other cell types in the same tissue. While it may be advantageous to sequence all cell types simultaneously, enriching the cell populations can be desirable for financial or technical reasons.
Note: This protocol describes the use of a transgenic zebrafish line expressing green fluorescent protein (GFP) under the her4.1 promoter (Yeo et al., 2007) and marking the astroglia to enrich these cells, which contain neural stem cells (Cosacak et al., 2019). Other reporters can also be used.
Preparations for Dissection
-
1.
Prepare 1X PBS, ice, petri dishes, scalpel, 50 mL tubes, 40-μM cell strainer, glass capillary, rotator and incubator (26.0–28.5°C).
-
2.
Dilute β-mercaptoethanol (0.1% v/v) in 1X PBS.
-
3.
Prepare the glass capillary (Video S1). The orifice of the glass capillary can be adjusted by heating. The smallest orifice must not be too narrow. It should be comparable to the opening of a 200 μL pipette tip.
CRITICAL: The enzymatic dissociation will be too slow or not possible without physical force. As a result, glass capillary is one of the options for dissociation. Trituration by glass capillary must be slow and gentle. The orifice of the glass capillary edge must not be smaller than the diameter of a cell, as it will cause lysis of the cells, which is strictly undesired.
Note: Glass capillary can be reused after washing with 1X-PBS and 70% ethanol.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Transportan signal peptide-coupled Amyloid-beta42 | Bhattarai et al., 2016 | N/A |
| Recombinant human Protein IL4 | Thermo Fischer | Cat# PHC0044 |
| Bovine Serum Albumin (BSA) | Sigma | Cat# A2153 |
| MESAB | Sigma | Cat# A5040 |
| Critical Commercial Assays | ||
| Neural Tissue Dissociation Kit (P) | Miltenyi | Cat# 130-092-628 |
| Deposited Data | ||
| Single-cell transcriptome | Cosacak et al. 2019 | GSE118577 |
| Experimental Models: Organisms/Strains | ||
| Tg(her4.1:GFP) transgenic zebrafish line | (Yeo et al., 2007) | N/A |
| Software and Algorithms | ||
| Cell Ranger 2.01 | Zheng et al., 2016 | RRID:SCR_016957 |
| Transcriptomics dataset website | Cosacak et al. 2019 | https://www.kizillab.org/singlecell |
| Other | ||
| Glass Pasteur Pipets | FisherScientific | Cat# FB50251 |
| Cell strainer | Falcon | Cat# 352340 |
| BD FACSARIA III | BD | RRID:SCR_016695 |
MATERIALS AND EQUIPMENT
Neural Tissue Dissociation Kit solutions (See Video S2)
-
•
Buffer Mix-I: 950 μL Buffer in 1.5 mL tube.
-
-
Add 5.8 μL 1:1,000 diluted β-mercaptoethanol,
-
-
Add 50 μL enzyme P.
-
-
Prepare Buffer Mix-I freshly and warm it up to 28.5°C.
-
•
Buffer Mix-II:
-
-
Add 20 μL enzyme A in 20 μL Buffer-Y
-
-
Keep it on ice until use.
STEP-BY-STEP METHOD DETAILS
Cerebroventricular Microinjection (CVMI)
Timing: 30–60 min
By using CVMI, one can deliver any substance into the zebrafish brain. Here, we describe how to use CVMI to deliver amyloid-β42, IL4, and PBS. This technique has been described in detail previously (Bhattarai et al., 2020, Bhattarai et al., 2016, Bhattarai et al., 2017a, Bhattarai et al., 2017b). This step is not a requirement. Any desired solution can be injected into the adult zebrafish brain using the CVMI method. The experimental protocol and troubleshooting options for CVMI were described in detail previously (Bhattarai et al., 2016, Bhattarai et al., 2017a, Bhattarai et al., 2017b, Cosacak et al., 2019). After the injection, wait for the desired duration to reach the experimental time point.
-
1.
Dissolve Aβ42 and IL4 in PBS and inject into the zebrafish cerebroventricular fluid at 20 μM and 1 μM, respectively, as described before (Bhattarai et al., 2016, Bhattarai et al., 2017b).
-
2.
Keep the fish in water system for 24 h in 14/10 hours light/dark cycle as recommended (Alestrom et al., 2019).
Preparation of Single-Cell Suspension
Timing: 2–3 h
This is the main focus of this protocol. Here, we describe how to dissect the zebrafish telencephalon and dissociate it into single-cell suspension followed by flow cytometry-based sorting (from this point on: FACS). We provide important hints to enhance the dissociation and sorting.
CRITICAL: After starting the dissociation of cells, the protocol has NO STOP until encapsulation of cells and cDNA synthesis. This experiment must be planned for approximately 60 min deviation.
CRITICAL: Before you start, make sure that you will have access to the equipment at the time you need (e.g., the flow cytometer, centrifuges, or the single cell sequencing facility).
CRITICAL: It is always better to first optimize the dissociation and sorting of cells. Determine the optimum number of cells sorted and the percentage of these cells that survive after sorting (should not be less than 95%).
Note: Here, we describe our protocol for dissociation of the telencephalon in the adult zebrafish brain.
CRITICAL: Animals have to be euthanized according to the permission and ethical rules of local authorities.
CRITICAL: Before the step 3.b, animals must be sacrificed, and all further procedures will be performed post-operationally.
-
3.
Dissecting Zebrafish Brain (1–2 min per fish) (Video S3)
-
a.
Euthanize the fish according to the permissions and ethical rules of local authorities.
-
b.
Generate a slit at the caudal of the skull and remove the right flank of the skull carefully.
-
c.
Then remove the left flank carefully.
-
d.
Remove any extra tissues (fat or meninges) carefully.
-
e.
CRITICAL STEP: Cut the telencephalon at the border of the optic tectum and telencephalon.
-
f.
Lift the telencephalon.
-
g.
Olfactory bulb will be automatically released while lifting the telencephalon. If not, remove it carefully.
-
h.
Gently transfer the telencephalon into Buffer Mix-I.
-
i.
Let the telencephalon rotate in a rotator at a temperature between 26.0 and 28.5°C.
-
j.
Continue with the next fish as above.
-
4.
Cell Dissociation (90 min) (Video S4)
-
a.
Rotate the brains for 10 min.
-
b.
Triturate (5 times) with a glass capillary to help physical dissociation (Repeat this step 3 times using the normal orifice size).
CRITICAL: From now on trituration must be slow and gentle.
-
c.
Add 20 μL of Buffer-Y mix (Video S4).
CRITICAL: Glass capillary helps physical dissociation. However, if the orifice size is too small it will induce disruption of the cell. In general, liquid must pass easily through the orifice of the capillary. The tissue chunks should pass easily through the opening. If the orifice is too wide, it can be narrowed down by heating.
-
d.
Incubate for 10 min. Triturate (5 times). Repeat this step 3 times, the orifice can be narrowed down as the chunks get smaller.
-
e.
At the last step, triturate 5 times. If there are still visible chunks, incubate another 10 min.
CRITICAL: Do not use small orifice sizes that might damage the cells (e.g., shearing the cell membrane).
-
5.
Straining and Pelleting the cells (30 min) (Video S5)
-
a.
Put a cell strainer (40 μm) in a 50 mL tube.
-
b.
Pass 5 mL of 4% BSA through the cell strainer for equilibration.
CRITICAL: Always use BSA to prevent cell lysis or adherence of cells to the walls of the tubes. BSA prevents cellular stress and improves the viability significantly.
-
c.
Pass all cell suspension through the strainer using 1 mL pipette tips. Pipetting should be done gently and slowly.
-
d.
Pass 4 mL 1x PBS through the cell strainer to dilute the BSA to 2% BSA.
-
e.
Recycle the extra liquid from the cell strainer to elute all cells.
-
f.
Centrifuge in a benchtop centrifuge at 300 g for 10 min at +4°C.
Note: Another round of centrifugation can decrease the number of particles. This will speed up the FACS, as the number of superfluous events (e.g., cell debris) will be decreased and intact cells will be enriched.
Note: One can remove myelin by using commercial kits.
-
g.
Remove supernatant carefully by inverting the tube.
CRITICAL: Do not keep cells on ice for longer times. Longer time on ice induces cell death.
Note: In order to understand the sensitivity of the cells, one can try keeping cells on ice for 10, 20 and 30 min and then check the percentage of dead cells by using cell viability dyes (e.g., vibrant dye).
-
h.
Add 10 mL 2% BSA. Gently re-suspend the cells.
-
i.
Centrifuge the cells at 300 g for 10 min at +4°C.
-
j.
Remove supernatant carefully by inverting the collection tube.
-
k.
Remove extra liquid and re-suspend the cell pellet in 4% BSA.
-
l.
Put cells on ice until sorting.
-
m.
Proceed as soon as possible to flow cytometry-based sorting.]
-
6.
Flow cytometer-based cell sorting (15 min)
-
a.
Add cell viability dye (Propidium iodide,1:500 dilution)
-
b.
Use 200 μL tubes. Add 2μL of the encapsulation buffer.
CRITICAL: The maximum volume of cell suspension that can be encapsulated in a single 10X Genomics channel is 45 μL. If using a 100 μm nozzle for sorting, this means a maximum of 15,000 cells or particles can be sorted into one tube for a single channel.
-
c.
Perform cell sorting according to the optimized settings of your machine.
CRITICAL: For our publication, we used only one fish. However, if more cells are required, more telencephali must be used.
CRITICAL: First optimize the sorting settings. Here, it is better to test how long the cells survive on ice. If the cells die quickly, try optimizations on every previous step.
CRITICAL: As soon as the cells are sorted, proceed to the cell encapsulation and single-cell sequencing.
EXPECTED OUTCOMES
For the conditions described in the protocol above, we typically get 10% of cells as GFP-positive cells. In general, we can collect more than 70.000 live cells from one fish, this makes approximately 7,000 GFP-positive cells. Depending on the gating properties these numbers can vary.
Figure 1 shows expected outcomes for flow cytometry-based cell sorting.
Figure 1.
Exemplary Histogram of Cell Sorting for Single Cell Sequencing
(A) The cells selected based on side and forward scattering (SSC/FSC).
(B) FSC singlet cells shown.
(C) Propidium iodide dye-negative cells (live cells).
(D) GFP-negative and GFP-positive cells.
(E) Pet-Texas Red (PI-) and GFP-positive cells.
(F) Sorted cells.
(G) Cells sorting tree. Adapted from Cosacak et al. (2019).
LIMITATIONS
Single-cell sequencing is a powerful method for unprecedented analyses of transcriptomics in single-cell resolution. Yet, some limitations do exist, as this technique is quite new and is constantly being improved.
Single-cell sequencing takes into account the intact cells from which RNA reads are obtained. This is fundamentally different than conventional deep sequencing methods that are based on total RNA isolation where compromised cells also contribute to the sequencing. Therefore, in single-cell sequencing, reads from hypothetically more sensitive cell types (e.g., the ones that are compromised during the isolation procedure) could be underrepresented. End users must optimize the cell isolation and readings according to their needs and the cell types. In overall, an optimal sequencing depth and technical quality must be achieved in a particular sequencing experiment. This aspect is not at a universal standard yet and must be determined by individual end users.
TROUBLESHOOTING
Problem
Brain dissociation incomplete.
Potential Solutions
-
1.
The enzymes P and A in Neural Dissociation Kit works better at 37°C, it may require more time to dissociate fish cells at 26–28.5°C.
Note: Longer incubation may alter the effects of the treatment as well as may cause new differentially expressed genes!
-
2.
Sometimes, there are large tissue parts that cannot be dissociated with longer incubation. This tissue may be contamination during dissecting the brain that the enzymes may not affect.
-
3.
Excess of PBS or other liquids may prevent enzymatic reactions. After dissecting the brain, the excess liquid can be removed (e.g., with tissue paper) without touching the brain.
Problem
Insufficient cells after dissociation.
Potential Solutions
-
1.
Always keep cells in BSA after centrifugations, as cells may stick to the plastic tubes with only 1xPBS.
-
2.
Harsh trituration will cause the breaking of cells; as a result trituration must be gentle without air bubbles.
-
3.
Keeping cells on ice for longer time will cause cells to die or get more sensitive to any temperature change. Try to minimize the duration on ice to perform quick sorting.
-
4.
Even healthy cells may be damaged by flow cytometer at high speed and voltage. Minimize the sorting speed and gating voltages. The nozzle size may also have an effect on the cells, increasing the nozzle size will decrease cell death.
Problem
Sequencing depth low.
Potential Solutions
-
1.
In general, protocols are optimized for mouse or human cells (e.g., PMBC cells). For each cell type, the same protocol may not work. Test the protocol on new cell types or cells from a new organism on first use.
-
2.
If the cells are too sensitive they may be disrupted before encapsulation.
-
3.
Incubating cells on ice for longer durations may make cells sensitive to temperature changes or the stress emanating from previous steps during encapsulation. Minimizing incubation during dissociation, sorting will increase effective encapsulation.
Note: Encapsulation is performed at 22–25°C or higher temperatures. Sensitized cells on ice or a sudden temperature difference may cause cell lysis before encapsulation. This will make analysis more complicated; e.g., RNA background in droplets without any cell, RNA contamination from other cells may be amplified with the same barcodes. This may cause background sequencing reads and could influence the downstream analyses. Normally the free-floating RNA has less influence on droplets with cells as it is diluted all over the solution. The bigger problem is the RNA in droplets without a cell – there it generates background, which leads to a lower sequencing depth of the barcodes compared to those droplets that contained a cell.
-
4.
Increase the cycles of paired-end sequencing
-
5.
Always optimize the cell dissociation and cell sorting steps before encapsulation. Improper cell dissociation and sorting may cause stress and cell death.
-
6.
Keeping the cells on ice after sorting will cause cell death or sensitivity. Adding cells to encapsulation will lyse the cells before encapsulation.
-
7.
Suggestion: After sorting the cells, keep cells on ice for 5, 10, 15, and 20 min. Then, pass cells through the flow cytometry and check the viability of the cells. This step has to be optimized for each cell type (e.g., fish cells, mouse cells). For instance, if the cell viability decreases at 15 min incubation on ice, then the time between the sorting and encapsulation must be minimized to maximum 15 min. Otherwise, the cells that are lysed during encapsulation will cause RNA contamination.
ACKNOWLEDGMENTS
This work was supported by German Center for Neurogenerative Disease(DZNE) and the Helmholtz Association Young Investigator Award (VH-NG-1021 to C.K.), Deutsche Forschungsgemeinschaft (DFG) (KI1524/6, KI1524/10, and KI1524/11 to C.K.), Carl-Gustav-Carus Universitätsklinikum Dresden and TU Dresden (FZ-111 and 043_261518 to C.K.). We would like to thank to all facilties within DRESDEN-Concept that helped us to develop this methodology. We also thank all members of the Kizil Lab for critical and helpful discussions.
AUTHOR CONTRIBUTIONS
Conceptualization, M.I.C. and C.K; Investigation, M.I.C., P.B. and C.K.; Writing – Original Draft, M.I.C., P.B., and C.K.; Writing – Review & Editing, M.I.C. and C.K.; Funding Acquisition, C.K.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100042.
Contributor Information
Mehmet Ilyas Cosacak, Email: mehmet.cosacak@dzne.de.
Caghan Kizil, Email: caghan.kizil@dzne.de.
References
- Alestrom P., D'Angelo L., Midtlyng P.J., Schorderet D.F., Schulte-Merker S., Sohm F., Warner S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2019 doi: 10.1177/0023677219869037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai P., Cosacak M.I., Mashkaryan V., Demir S., Popova S., Govindarajan N., Brandt K., Zhang Y., Chang W., Ampatzis K. Neuron-glia interaction through Serotonin-BDNF-NGFR axis enables regenerative neurogenesis in Alzheimer’s model of adult zebrafish brain. PLoS Biol. 2020;18:e3000585. doi: 10.1371/journal.pbio.3000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai P., Thomas A.K., Cosacak M.I., Papadimitriou C., Mashkaryan V., Froc C., Reinhardt S., Kurth T., Dahl A., Zhang Y. IL4/STAT6 signaling activates neural stem cell proliferation and neurogenesis upon amyloid-beta42 aggregation in adult zebrafish brain. Cell Rep. 2016;17:941–948. doi: 10.1016/j.celrep.2016.09.075. [DOI] [PubMed] [Google Scholar]
- Bhattarai P., Thomas A.K., Cosacak M.I., Papadimitriou C., Mashkaryan V., Zhang Y., Kizil C. Modeling amyloid-beta42 toxicity and neurodegeneration in adult zebrafish brain. J. Vis. Exp. 2017 doi: 10.3791/56014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai P., Thomas A.K., Zhang Y., Kizil C. The effects of aging on Amyloid-beta42-induced neurodegeneration and regeneration in adult zebrafish brain. Neurogenesis (Austin) 2017;4:e1322666. doi: 10.1080/23262133.2017.1322666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosacak M.I., Bhattarai P., Reinhardt S., Petzold A., Dahl A., Zhang Y., Kizil C. Single-cell transcriptomics analyses of neural stem cell heterogeneity and contextual plasticity in a zebrafish brain model of amyloid toxicity. Cell Rep. 2019;27:1307–1318.e3. doi: 10.1016/j.celrep.2019.03.090. [DOI] [PubMed] [Google Scholar]
- Yeo S.Y., Kim M., Kim H.S., Huh T.L., Chitnis A.B. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev. Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Zheng G.X., Lau B.T., Schnall-Levin M., Jarosz M., Bell J.M., Hindson C.M., Kyriazopoulou-Panagiotopoulou S., Masquelier D.A., Merrill L., Terry J.M. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



