Abstract
Background
The 4-component (4C) model is a criterion method for human body composition that separates the body into fat, water, mineral, and protein, but requires 4 measurements with significant cost and time requirements that preclude wide clinical use. A simplified model integrating only 2 measurements—dual-energy X-ray absorptiometry (DXA) and bioelectrical impedance analysis (BIA)—and 10 min of patient time has been proposed.
Objective
We aimed to validate a rapid, simplified 4C DXA + BIA body composition model in a clinical population.
Design
This was a cross-sectional observational study of 31 healthy adults. Participants underwent whole-body DXA, segmental BIA, air displacement plethysmography (ADP), and total body water (TBW) measurement by deuterium (D2O) dilution. 4C composition was calculated through the use of the Lohman model [DXA mineral mass, D2O TBW, ADP body volume (BV), scale weight] and the simplified model (DXA mineral mass and BV, BIA TBW, scale weight). Accuracy of percentage of fat (%Fat) and protein measurements was assessed via linear regression. Test-retest precision was calculated with the use of duplicate DXA and BIA measurements.
Results
Of 31 participants, 23 were included in the analysis. TBWBIA showed good test-retest precision (%CV = 5.2 raw; 1.1 after outlier removal) and high accuracy to TBWD2O [TBWD2O = 0.956*TBWBIA, R2= 0.92, root mean squared error (RMSE) = 2.2 kg]. %Fat estimates from DXA, ADP, D2O, and BIA all showed high correlation with the Lohman model. However, only the 4C simplified model provides high accuracy for both %Fat (R2 = 0.96, RMSE = 2.33) and protein mass (R2= 0.76, RMSE = 1.8 kg). %Fat precision from 4C DXA + BIA was comparable with DXA (root mean square-SD = 0.8 and 0.6 percentage units, respectively).
Conclusions
This work validates a simplified 4C method that measures fat, water, mineral, and protein in a 10-min clinic visit. This model has broad clinical application to monitor many conditions including over/dehydration, malnutrition, obesity, sarcopenia, and cachexia.
Keywords: body composition, DXA, bioelectrical impedance, obesity, lean mass
INTRODUCTION
Four-component (4C) models of body composition—those that divide the body into fat, water, protein, and mineral masses—are considered the reference within the research community. Importantly, 4C models do not assume a fixed hydration as is the case in simpler body composition models (1). This is important for assessment of undernutrition in children, which is often associated with dehydration (2), as well as assessment of lean mass in older adults, which has been shown to have significantly different hydration than that found in younger adults (3). Altered lean mass hydration in older adults may explain why dual-energy X-ray absorptiometry (DXA)-measured lean mass (which assumes fixed hydration) is a poor predictor of mortality and functional strength compared with simple handgrip strength (4). Direct measurement of lean mass water and protein content is therefore particularly useful in the presence of wasting conditions associated with aging such as sarcopenia and cachexia (5).
Despite its advantages, 4C body composition is seldom used in the clinic because it requires several different measurements that are time consuming and costly. The Lohman 4C model, for example, includes bone mineral measurements from DXA, body mass from a scale, total body volume (BV), and total body water (TBW) from labeled water dilution (6). Furthermore, precision of the conventional 4C model is difficult to quantify owing to the multiple measures and time constant of dilution for deuterium. Wilson et al. (7) proposed a simplified model for clinically viable 4C body composition that uses DXA-calculated BV in place of air displacement plethysmography (ADP) and bioelectrical impedance analysis (BIA)-calculated TBW in place of labeled water dilution. We sought to validate the precision and accuracy of this rapid, simplified 4C method against the reference Lohman method as well as against 2-component (BIA, ADP, and TBW) and 3-component models (DXA) of percentage of fat (%Fat).
METHODS
A cross-sectional convenience sample of healthy adults underwent whole-body DXA, multifrequency BIA, ADP, height and weight, and TBW deuterium dilution measurements. 4C body composition was calculated with the use of the Lohman method (8) and the simplified DXA + BIA approach of Wilson et al. (7). Linear regression analysis was performed to determine the agreement between body composition methods. We describe the details of each part of the study below.
Participants
Thirty-one healthy adults >18 y of age were enrolled in a prospective open recruitment during the time period of November 2016 to April 2017. Each participant received duplicate measures with repositioning for whole-body DXA and segmented multifrequency BIA scans, and singleton measures of deuterium dilution and ADP (owing to time considerations for these techniques). Exclusion criteria included a history of body-altering surgery, significant nonremovable metallic implants, height >73 inches (in) (185 cm), and weight >250 pounds (lb) (113 kg) (to ensure whole-body fit within the dimensions of the DXA scan table). Recruitment was performed with the use of flyers posted around the University of California, San Francisco (UCSF) Parnassus campus. All participants provided informed consent. The study protocol was approved by the UCSF Committee on Human Research (Institutional Review Board #16–19,342).
DXA
Whole-body DXA scans were acquired on a Hologic Discovery/W system (Hologic Inc., Marlborough, MA). All scans were analyzed at Hologic, Inc. by a single International Society for Clinical Densitometry–certified technologist using Hologic APEX software (version 4.6.0.4) with NHANES body composition correction disabled. Participants were clothed in form-fitting undergarments, without shoes, and positioned on the scanner table with arms out to the side, hands flat on the table, and feet in planarflex position, in accordance with the manufacturer's standard protocols. The scanner was kept in regular calibration through the use of daily and weekly quality control protocols scanning spine and soft tissue phantoms according to International Society for Clinical Densitometry guidelines.
BIA
Whole-body segmented multifrequency BIA measurements were acquired on an InBody S10 system (InBody Inc., Cerritos, CA). Measurements were performed with the participant in supine position immediately after DXA scans. Contact sites on the fingers and ankles were cleaned before measurement with a sterile antimicrobial tissue provided by the manufacturer. Touch type electrodes were used in accordance with standard protocols. Participants were scanned a total of 3 times to allow for assessment of measurement precision with and without repositioning. TBW and %Fat measurements were recorded directly from the device. The average of 2 TBW measurements (with repositioning) was used for 4C analysis.
ADP
Whole-body volume measurements were taken through the use of ADP in a BodPod (v5.4.1, COSMED USA, Inc., Concord, CA). Measurements were taken via the manufacturer's standard protocol. Participants were clothed in form-fitting clothing and a swim cap. Lung volume was measured directly with the use of the built-in breathing tube system. The BodPod was regularly calibrated with the use of a known-volume cylinder and known-mass weights in accordance with the manufacturer's guidelines. The BodPod provided BV measurements for the 4C models as well as its own estimate of %Fat.
Deuterium dilution
TBW was assessed through the use of a 4-h deuterium (D2O) dilution protocol as defined in the International Atomic Energy Agency standards (9). In summary, participants were provided with a measured dose of D2O in local drinking water (100 mL total volume) to achieve 0.05 g of excess2H per kilogram of body weight. Three 2.5-mL saliva samples were collected: 1 at baseline (before dose consumption), one 3 h postdosing, and one 4 h postdosing. Participants were allowed to void and/or drink small amounts (<500 mL) of water during the 4-h protocol; all fluid changes were measured and recorded as change in body weight with the use of a high-precision scale. TBW was calculated by measuring D2O enrichment in the saliva samples against baseline dose and drinking water samples (9) which included the correction factor of 1.041 for nonaqueous exchange of deuterium. Fat mass was estimated from TBW with the use of a fixed hydration constant of 0.732 for lean mass: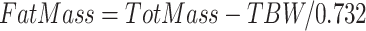 . All samples were analyzed at the University of Wisconsin Biotechnology Center.
. All samples were analyzed at the University of Wisconsin Biotechnology Center.
4C models
4C models divide the body into fat, water, protein, and mineral masses. We calculated 4C composition via the model of Lohman et al. (6), also described in Heymsfield et al. (10), and reproduced here.
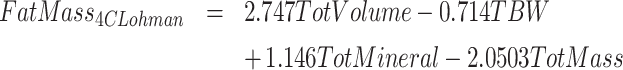 |
(1) |
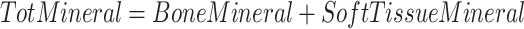 |
(2) |
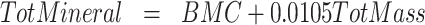 |
(3) |
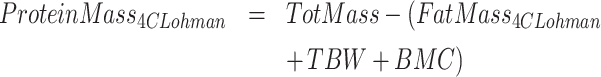 |
(4) |
ADP, D2O dilution, DXA, and scale weight measurements were used for TotVolume, TBW, BMC (bone mineral content), and TotMass, respectively. The Lohman model served as criterion method for fat and protein measurements. It may be noted that this model is often misrepresented to include only bone mineral mass instead of total mineral mass, which includes mineral in both bone and soft tissues. Bartok-Olson et al. (11) described the discrepancy and its implications elsewhere. Note also that the residual mass (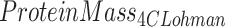 ) contains a small amount of carbohydrate in addition to protein.
) contains a small amount of carbohydrate in addition to protein.
The simplified DXA + BIA 4C model described by Wilson et al. (7) uses the same form as the Lohman 4C model, but BV calculated from DXA instead of ADP, and water mass measured by BIA instead of D2O. Specifically, Wilson et al. (12) showed that BV could be accurately calculated through the use of calibrated fat, lean, and bone densities along with the measured masses from whole-body DXA scans:
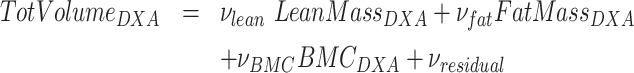 |
(5) |
where inverse density coefficients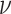 are calibrated for each make of DXA scanner. These coefficients were published in earlier cross-sectional studies with matched DXA and ADP measurements that used multiple linear regression with the 3-component DXA masses as input and ADP volume as output. Wilson et al. derived separate volume coefficients for Hologic (12) and GE DXA systems (7), and reported test-retest precision of root mean square (RMS)-%CV = 1.1 in total body DXA-volume. In the present study, Hologic calibration values were used (
are calibrated for each make of DXA scanner. These coefficients were published in earlier cross-sectional studies with matched DXA and ADP measurements that used multiple linear regression with the 3-component DXA masses as input and ADP volume as output. Wilson et al. derived separate volume coefficients for Hologic (12) and GE DXA systems (7), and reported test-retest precision of root mean square (RMS)-%CV = 1.1 in total body DXA-volume. In the present study, Hologic calibration values were used (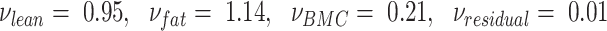 ). We sought to validate Wilson et al.'s DXA + BIA 4C model and demonstrate hardware independence by using an independent recruitment and different devices (Hologic DXA and InBody BIA, compared with GE DXA and Impedimed BIA).
). We sought to validate Wilson et al.'s DXA + BIA 4C model and demonstrate hardware independence by using an independent recruitment and different devices (Hologic DXA and InBody BIA, compared with GE DXA and Impedimed BIA).
Statistical analysis
Linear regressions were performed to assess the agreement between different modalities. TBW from BIA was compared against deuterium dilution criterion measurement. Percentage body fat was compared between DXA, ADP, BIA, D2O, and the proposed 4C DXA + BIA model against the 4C Lohman criterion method. Constant intercepts were included in linear models only if significant atP< 0.05. Test-retest precision was quantified with the use of RMS-%CV for mass and volume measurements, and RMS-SD for %Fat measurements as described elsewhere (13). Outlier detection thresholds for test-retest measurements were conservatively defined at 6 SDs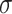 away from 0, where
away from 0, where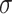 was estimated by the sample median absolute difference (MAD) between repeat measurements (14). Statistical analyses were performed with the use of pandas 0.20.1 (Python Software Foundation, Wilmington, DE), SAS version 9.4 (SAS Institute, Cary, NC), and MATLAB R2017a (The MathWorks Inc., Natick, MA).
was estimated by the sample median absolute difference (MAD) between repeat measurements (14). Statistical analyses were performed with the use of pandas 0.20.1 (Python Software Foundation, Wilmington, DE), SAS version 9.4 (SAS Institute, Cary, NC), and MATLAB R2017a (The MathWorks Inc., Natick, MA).
RESULTS
Of the 31 participants that completed the study protocol, 23 had complete valid measurements. There was an error in the dose preparation for the first batch of 8 participants that invalidated measurements; these participants’ data were used for precision analysis only. Summary demographics of the participants included in the analysis are shown inTable 1.
TABLE 1.
Summary demographic statistics of the 23 participants with complete data included in the present analysis (11 male)1
| Variable | Mean ± SD | Min | Max |
|---|---|---|---|
| Age, y | 33.8 ± 12.3 | 22 | 63 |
| Height, cm | 168.4 ± 11.0 | 148.7 | 188.6 |
| Mass, kg | 73.2 ± 12.6 | 54.1 | 104.1 |
| BMI, kg/m2 | 25.6 ± 4.1 | 20.2 | 36.9 |
| Total bone mineral content (DXA), kg | 2.40 ± 0.43 | 1.57 | 3.29 |
| Intracellular water (BIA), kg | 25.1 ± 5.7 | 15.5 | 39.0 |
| Extracellular water (BIA), kg | 14.9 ± 3.1 | 9.7 | 22.5 |
| Total body volume, L | |||
| ADP | 70.4 ± 12.0 | 56.9 | 104.9 |
| DXA | 70.9 ± 12.1 | 57.4 | 104.7 |
| Total body water, kg | |||
| D2O | 38.4 ± 7.9 | 26.1 | 53.9 |
| BIA | 40.0 ± 8.8 | 25.1 | 61.5 |
| Fat percentage | |||
| DXA | 26.9 ± 10.7 | 9.9 | 45.1 |
| ADP | 26.0 ± 11.2 | 13.0 | 50.0 |
| BIA | 25.1 ± 11.8 | 8.9 | 52.3 |
| D2O | 27.9 ± 10.0 | 11.2 | 47.0 |
ADP, air displacement plethysmography; BIA, bioelectrical impedance analysis; DXA, dual-energy X-ray absorptiometry; D2O, deuterium; Max, maximum; Min, minimum.
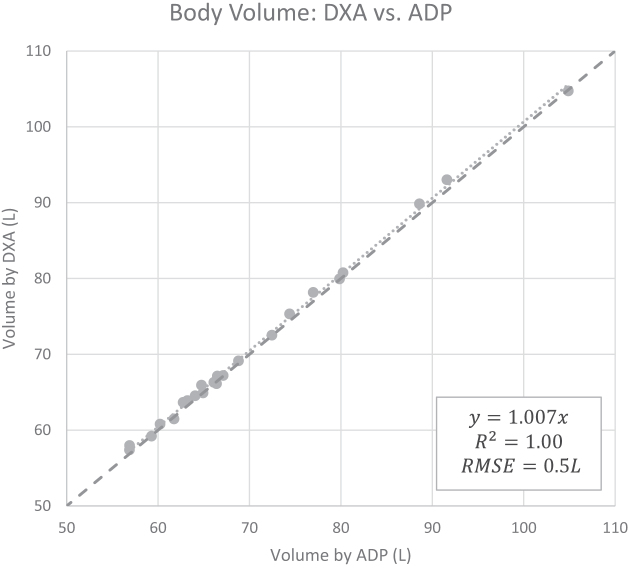
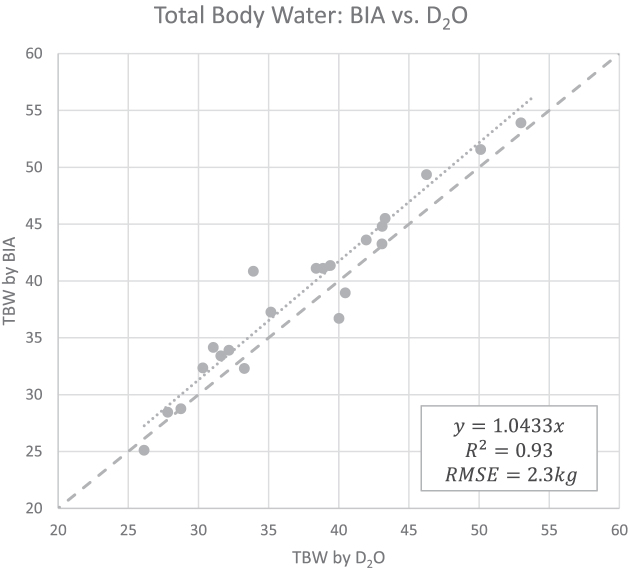
BV measurements calculated from DXA showed excellent agreement with ADP (Figure 1), BVADP = 0.993 BVDXA (95% CI: 0.990, 0.996). TBW measurements from BIA also showed strong agreement with criterion measurements from D2O (Figure 2), TBWD2O = 0.956 TBWBIA (95% CI: 0.932, 0.979).
FIGURE 1.
Linear regression between BV measurements from DXA and ADP (n = 23). High correlation was observed, although a slope significantly different from 1 was detected. These data were used to determine a linear correction equation for BV from DXA: BVADP = 0.993 BVDXA (95% CI: 0.990, 0.996). The dashed line is the line of identity. ADP, air displacement plethysmography; BV, body volume; DXA, dual-energy X-ray absorptiometry; RMSE, root mean squared error.
FIGURE 2.
Linear regression between TBW measurements from BIA and D2O (n = 23). High correlation was observed, although a slope significantly different from 1 was detected. These data were used to determine a linear correction equation for TBW from BIA: TBWD2O = 0.956 TBWBIA (95% CI: 0.932, 0.979). The dashed line is the line of identity. BIA, bioelectrical impedance analysis; D2O, deuterium; RMSE, root mean squared error; TBW, total body water.
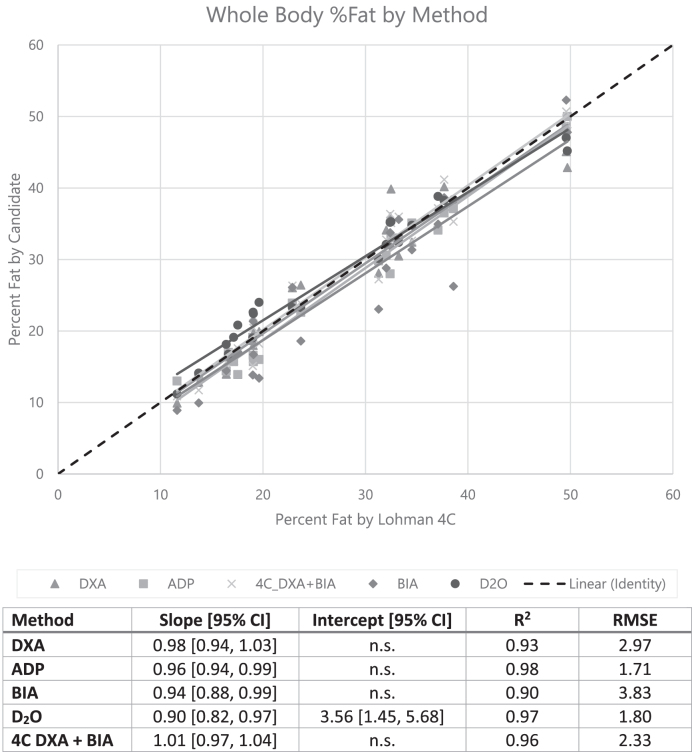
Whole-body %Fat was calculated with the use of the 4C Lohman model as presented earlier after correcting for the small biases in the DXA volume and BIA water measures through the use of the equations inFigures 1 and 2. Regression results for the 2- and 3-component and 4C methods are shown inFigure 3. Each of the DXA, BIA, ADP, and D2O estimates of %Fat showed strong agreement with the Lohman model, with R2 ≥ 0.90. D2O dilution was the only modality to exhibit a significant bias. BIA exhibited the highest root mean squared error (RMSE) at 3.83 percentage units, whereas ADP had the lowest at 1.71; however, this is likely due to the fact that the 4C Lohman %Fat equation is dominated by BV (here measured by ADP). Note that the Hologic DXA %Fat results were calculated with NHANES correction (15) disabled. Enabling the NHANES correction resulted in overestimated DXA fat values compared with the 4C Lohman model (see Supplemental Text andSupplemental Figure 1).
FIGURE 3.
Linear regression between whole-body %Fat from the 2- and 3-component and 4C body composition assessment methods in this study and the reference 4C Lohman model (n = 23). “n.s.” indicates that the regression intercept was nonsignificant (P> 0.05) and set to 0. ADP, air displacement plethysmography; BIA, bioelectrical impedance analysis; DXA, dual-energy X-ray absorptiometry; D2O, deuterium; RMSE, root mean squared error; 4C, 4-component; %Fat, percentage of fat.
The simplified 4C DXA + BIA model closely agreed with the Lohman 4C reference. The 4C DXA + BIA model can be generalized through the use of the following equation.
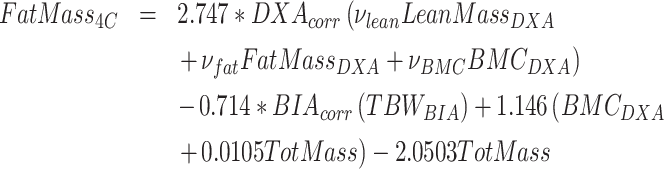 |
(6) |
where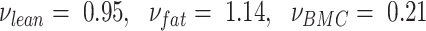 , and
, and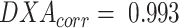 for the present Hologic DXA system, and
for the present Hologic DXA system, and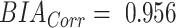 for the present InBody BIA system.
for the present InBody BIA system.
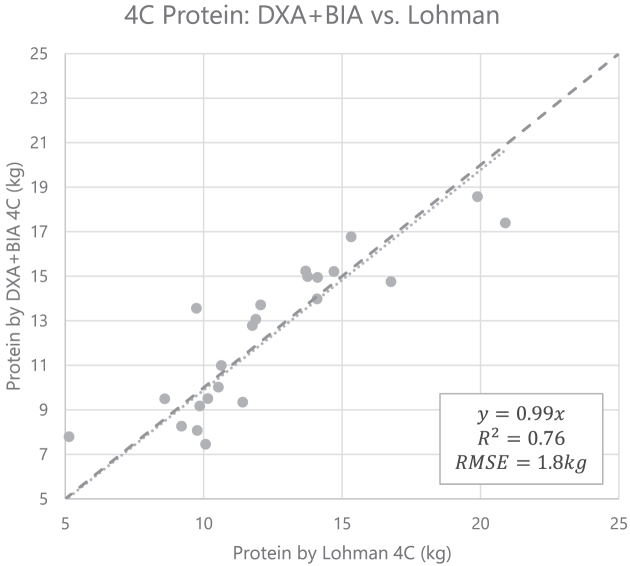
Residual protein measurements from the 4C DXA + BIA model compared with the Lohman model are shown inFigure 4. Whole-body 4C protein measured by DXA + BIA closely approximates the Lohman reference method.
FIGURE 4.
Linear regression of whole-body 4C residual protein measured by the DXA + BIA method compared with the Lohman reference method (n = 23). The equation for the line of best fit is ProtDXA+BIA 4C = 0.99 ProtLohman 4C (95% CI: 0.93, 1.05). The dashed line is the line of identity. BIA, bioelectrical impedance analysis; DXA, dual-energy X-ray absorptiometry; RMSE, root mean squared error; 4C, 4-component.
Duplicate DXA and BIA measurements were available for all 31 participants. Test-retest precision results for BIA TBW and fat mass, DXA fat mass and volume, and 4C DXA + BIA fat mass and protein mass are shown inTable 2. Repeat TBWBIA measurements (with immediate repositioning) showed a %CV = 5.2. The MAD in the test-retest data was 0.3 kg. Outliers were defined conservatively as the test-retest difference exceeding 6 times the MAD, or 1.8 kg. Four outlier pairs were identified in the set of 31 test-retest measurements. Excluding these outliers results in %CV = 1.1. As shown inTable 2, precision in TBWBIA significantly affects the precision of 4C DXA + BIA fat and protein measurements. Observed test-retest RMS-SDs for BIA, DXA, and 4C DXA + BIA %Fat (after BIA TBW outlier removal) were 0.9, 0.6, and 0.8 percentage units, respectively. Least significant changes implied by these intramethod errors [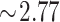 times the test-retest precision, per the International Society for Clinical Densitometry (16)] are smaller than the corresponding intermethod errors compared with Lohman 4C %Fat of 3.83, 2.97, and 2.33 percentage units, respectively (Figure 3).
times the test-retest precision, per the International Society for Clinical Densitometry (16)] are smaller than the corresponding intermethod errors compared with Lohman 4C %Fat of 3.83, 2.97, and 2.33 percentage units, respectively (Figure 3).
TABLE 2.
Test-retest precision for the BIA and DXA measurements utilized in the study1
| Variable | Before BIA outlier removal (n = 31) | After BIA outlier removal (n = 27) |
|---|---|---|
| BIA total body water, %CV | 5.2 | 1.1 |
| DXA total body volume, %CV | 0.2 | 0.3 |
| BIA %Fat, RMS-SD | 3.8 | 0.9 |
| DXA %Fat, RMS-SD | 0.6 | 0.6 |
| 4C DXA + BIA %Fat, RMS-SD | 1.9 | 0.8 |
| 4C DXA + BIA protein mass, %CV | 6.1 | 4.4 |
Duplicate measurements were collected via each method, with repositioning. High variability in BIA TBW measurements leads to imprecision in BIA fat mass and 4C DXA + BIA fat and protein masses. Removal of outlier BIA TBW measurements as described in the text results in significantly improved precision for each of those measurements. BIA, bioelectrical impedance analysis; DXA, dual-energy X-ray absorptiometry; RMS, root mean square; TBW, total body water; 4C, 4-component; %Fat, percentage of fat.
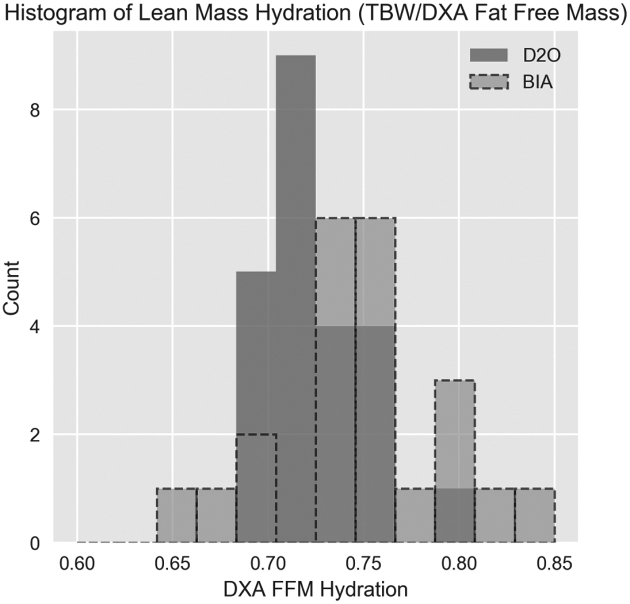
Lean mass hydration was calculated as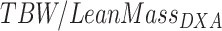 where TBW was measured with the use of either D2O or BIA. Histograms of participants’ lean mass hydration levels are shown inFigure 5. Greater mean lean mass hydration was observed for TBWBIA (mean = 0.755) than for TBWD2O (mean = 0.726). Greater spread in lean mass hydration levels was observed in TBWBIA (SD = 0.047) than in TBWD2O (SD = 0.027). Hydration results generally agree well with a review of cadaver studies by Wang et al. (1) in which the hydration mean ± SD was found to be 0.737 ± 0.036. The range of D2O hydration values is relatively narrow, with only a single data point >1 SD away from the 0.737 mean reported by Wang et al. However, the range of BIA hydration values was much larger, with 5 data points >1 SD away from 0.737.
where TBW was measured with the use of either D2O or BIA. Histograms of participants’ lean mass hydration levels are shown inFigure 5. Greater mean lean mass hydration was observed for TBWBIA (mean = 0.755) than for TBWD2O (mean = 0.726). Greater spread in lean mass hydration levels was observed in TBWBIA (SD = 0.047) than in TBWD2O (SD = 0.027). Hydration results generally agree well with a review of cadaver studies by Wang et al. (1) in which the hydration mean ± SD was found to be 0.737 ± 0.036. The range of D2O hydration values is relatively narrow, with only a single data point >1 SD away from the 0.737 mean reported by Wang et al. However, the range of BIA hydration values was much larger, with 5 data points >1 SD away from 0.737.
FIGURE 5.
Histograms of FFM hydration (defined as TBW divided by FFM from DXA) (n = 23). More variance was observed with the use of BIA for TBW than with the use of D2O for TBW. The observed range of hydration values extends beyond physiologic bounds for healthy adults in the sample; definition of thresholds on plausible hydration levels may provide criteria to validate BIA TBW measurements. BIA, bioelectrical impedance analysis; DXA, dual-energy X-ray absorptiometry; D2O, deuterium; FFM, fat-free mass; TBW, total body water.
DISCUSSION
4C body composition is a well-established method for assessment of metabolic status and health. In this study we found that %Fat measurements from several different technologies that used both 2-component (ADP, BIA, and D2O) and 3-component (DXA) models agreed well with the criterion 4C Lohman model. Each of these devices should provide accurate, reliable measurements of adiposity in the clinical setting when normal hydration is expected. The best agreement with the Lohman model for both %Fat and protein measures was found to be with the rapid 4C DXA + BIA method.
Agreement between the two 4C methods relies on the agreement of BVDXA with BVADP, and TBWBIA with TBWD2O. Although we found high agreement between the 2 BV measurements, a small but significant difference was observed (TBWD2O/TBWBIA = 0.993). BV is highly weighted in the Lohman 4C model, so it is important that BV measures are accurately calibrated. The 2 methods have very different underlying assumptions and it is unclear which method more accurately measures volume in an individual. The BodPod is calibrated through the use of a reference object periodically so that accuracy on solid volumes is ensured. However, in vivo error sources include uncertainty in the lung and gastric volumes. DXA systems only measure solid volume and are unaffected by lung and gastric voids. Potential errors in the DXA volume include the extrapolation of the lean and soft tissue masses over regions containing bone and the lack of existing quality assurance methods to validate soft tissue mass accuracy (standard phantoms and protocols exist only for calibration of bone mineral mass). Clearly, a quality control method for DXA that ensures mass accuracies to better than 0.5% is warranted to ensure agreement between DXA systems in the field.
In a similar but larger study, Smith-Ryan et al. (17) rederived coefficients for BVDXA calibrated to BVADP using the same make and model of DXA system. Smith-Ryan et al. also did not use the NHANES correction, and found coefficients (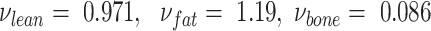 ) similar to those originally published by Wilson et al. (12):
) similar to those originally published by Wilson et al. (12): . In this study, we found that BVDXA derived through the use of Wilson et al.'s coefficients agreed with BVADP to within 0.3%. The differences seen by Smith-Ryan et al. were likely due to slight calibration differences in the DXA systems and could have been corrected with the use of a reference quality control phantom for soft tissue masses.
. In this study, we found that BVDXA derived through the use of Wilson et al.'s coefficients agreed with BVADP to within 0.3%. The differences seen by Smith-Ryan et al. were likely due to slight calibration differences in the DXA systems and could have been corrected with the use of a reference quality control phantom for soft tissue masses.
We found high agreement between the 2 TBW measurements with a small but significant difference (TBWD2O/TBWBIA = 0.956). Although we used a trained laboratory for D2O spectroscopy and clinical staff to measure and administer the doses, there was still 1 measurement that appeared outside realistic biological bounds. Potential errors in D2O TBW measurements include subject noncompliance with the fasting and resting requirements before and during the protocol. Strenuous activity or significant food and drink consumption, particularly in the hours immediately before dose consumption and sample collection, can significantly affect the accuracy of D2O TBW measurements (9). Potential sources of BIA error include electrode placement inaccuracy, poor electrode contact, and significant variability in body shape (18). Nonetheless, BIA is an appealing method for clinical TBW measurement owing to its low cost, rapid results, and amenability to field calibration with the use of stable phantoms. Significant outliers in TBWBIA can be detected by applying thresholds of agreement between duplicate TBWBIA measurements, or comparison to reference physiologic hydration ranges for singleton measurements. Using these outlier exclusion methods, the TBWBIA test-retest precision was 1.1%. If the difference between the 2 measurements exceeds 1.8 kg, we recommend collecting a third measurement and averaging of the 2 closest measurements. Further validation of TBWBIA precision in different models and with the use of different electrodes (adhesive gel pads compared with touch type) might be useful to expand the utility of this method. Without these outlier detection methods, the observed TBWBIA test-retest precision was 5.2%. Vaché et al. (19) reported precision of 4.1%CV for test-retest measurements collected 8 h apart. Further precision studies would be needed to assess the long-term precision of BIA for TBW.
Precision of %Fat measurements from the 4C DXA + BIA method (RMS-SD = 0.8% units after BIA outlier removal) was found to be comparable to that of DXA (RMS-SD = 0.6% units), suggesting that the method may be suitable for monitoring individual %Fat in longitudinal studies. Precision of 4C DXA + BIA protein measurements was somewhat lower (%CV = 4.4) because protein is a smaller fraction of total body mass. This suggests that 4C DXA + BIA protein may be more suitable for population analysis and individual classification than monitoring of individual protein mass changes.
Our results on the accuracy of 4C DXA + BIA protein measurements compared with a reference Lohman model (R2 = 0.76, RMSE = 1.8 kg) are nearly identical to the results found by Wilson et al. (7) who compared 4C DXA + BIA protein with neutron activation analysis (R2 = 0.77, RMSE = 1.8 kg).
This study has several strengths. To our knowledge, our study is the first to validate the use of BIA for TBW measurements in a 4C model, and the first to quantify test-retest precision of 4C DXA + BIA measurements. Notably, the present study validates the use of specific DXA and BIA systems (from Hologic and InBody) whereas Wilson et al.'s (7) seminal study used systems from different manufacturers (GE and Impedimed). Smith-Ryan et al. (17) used a Hologic DXA system and an Impedimed BIA system. Although the equations presented herein are specifically calibrated to the particular devices (Hologic DXA and InBody BIA) in this study, the success of this validation demonstrates the hardware agnosticism of the 4C DXA + BIA approach. Second, we showed how BIA and DXA can be combined to measure hydration. Third, we showed how total body protein can be estimated independently of water status.
However, there are also limitations. Namely, the limited sample of participants included only healthy and normally hydrated individuals. Recruitment was not stratified to target a wide range of body sizes, ethnicities, and ages. We also did not include strength metrics that could have shown if protein is a superior estimate of function to DXA lean mass. Future studies may investigate the robustness of the 4C DXA + BIA model in populations with impaired metabolic and functional profiles, older ages, and abnormal hydration status.
In summary, this work validates the accuracy and precision of a clinically viable technique incorporating DXA and BIA technology for 4C body composition. Translation to clinical practice would enable fast, accessible 4C assessment including fat, protein, and hydration status—measures important for monitoring a wide variety of conditions including dieting, sarcopenia, cachexia, and performance training. Validation in such special populations is warranted.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Nisa Kelly (UCSF) for subject recruitment and implementation of the study protocol, as well as Timothy Shriver (University of Wisconsin) for processing and analyzing all deuterium samples.
The authors’ contributions were as follows—BKN and JAS: drafted the manuscript and had primary responsibility for final content; and all authors: reviewed and approved the final manuscript. WW, TLK, and KEW are employees and shareholders of Hologic, Inc. SBH serves on the Medical Advisory Board for Tanita Corporation. JAS receives research funding from Hologic, Inc. and GE Healthcare. The remaining authors report no conflicts of interest.
Notes
Supported in part by Hologic Investigator-Initiated Study Grant no. P0515173.
Supplemental Figure 1 is available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents athttps://academic.oup.com/ajcn/.
Abbreviations used:
- ADP
air displacement plethysmography
- BIA
bioelectrical impedance analysis
- BMC
bone mineral content
- BV
body volume
- DXA
dual-energy X-ray absorptiometry
- D2O
deuterium
- MAD
median absolute difference
- RMS
root mean square
- RMSE
root mean squared error
- TBW
total body water
- 4C
4-component body composition model (herein: water, bone, fat, and protein)
- %Fat
percentage of fat
REFERENCES
- 1. Wang Z,Deurenberg P,Wang W,Pietrobelli A,Baumgartner RN,Heymsfield SB. Hydration of fat-free body mass: review and critique of a classic body-composition constant.Am J Clin Nutr.1999;69:833–41. [DOI] [PubMed] [Google Scholar]
- 2. Mwangome MK,Fegan G,Prentice AM,Berkley JA. Are diagnostic criteria for acute malnutrition affected by hydration status in hospitalized children? A repeated measures study.Nutr J.2011;10:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bossingham MJ,Carnell NS,Campbell WW. Water balance, hydration status, and fat-free mass hydration in younger and older adults.Am J Clin Nutr.2005;81:1342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Newman AB,Kupelian V,Visser M,Simonsick EM,Goodpaster BH,Kritchevsky SB,Tylavsky FA,Rubin SM,Harris TB. Strength, but not muscle mass, is associated with mortality in the Health, Aging and Body Composition Study cohort.J Gerontol A Biol Sci Med Sci.2006;61:72–7. [DOI] [PubMed] [Google Scholar]
- 5. von Haehling S,Morley JE,Anker SD. An overview of sarcopenia: facts and numbers on prevalence and clinical impact.J Cachexia Sarcopenia Muscle.2010;1:129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lohman TG.Applicability of body composition techniques and constants for children and youths.Exerc Sport Sci Rev.1986;14:325–57. [PubMed] [Google Scholar]
- 7. Wilson JP,Strauss BJ,Fan B,Duewer FW,Shepherd JA. Improved 4-compartment body-composition model for a clinically accessible measure of total body protein.Am J Clin Nutr.2013;97:497–504. [DOI] [PubMed] [Google Scholar]
- 8. Lohman TG,Going SB. Multicomponent models in body composition research: opportunities and pitfalls.Basic Life Sci.1993;60:53–8. [DOI] [PubMed] [Google Scholar]
- 9. IAEA.Introduction to body composition assessment using the deuterium dilution technique with analysis of saliva samples by Fourier transform infrared spectrometry,Vienna, Austria:IAEA;2011;[cited 2017 May 25]. Available from:http://www-pub.iaea.org/books/IAEABooks/8369/Introduction-to-Body-Composition-Assessment-Using-the-Deuterium-Dilution-Technique-with-Analysis-of-Saliva-Samples-by-Fourier-Transform-Infrared-Spectrometry. [Google Scholar]
- 10. Heymsfield SB,Ebbeling CB,Zheng J,Pietrobelli A,Strauss BJ,Silva AM,Ludwig DS. Multi-component molecular-level body composition reference methods: evolving concepts and future directions.Obes Rev.2015;16:282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartok-Olson CJ,Schoeller DA,Sullivan JC,Clark RR. The “B” in the Selinger four-compartment body composition formula should be body mineral instead of bone mineral.Ann N Y Acad Sci.2000;904:342–4. [DOI] [PubMed] [Google Scholar]
- 12. Wilson JP,Fan B,Shepherd JA. Total and regional body volumes derived from dual-energy X-ray absorptiometry output.J Clin Densitom.2013;16:368–73. [DOI] [PubMed] [Google Scholar]
- 13. Glüer CC,Blake G,Lu Y,Blunt BA,Jergas M,Genant HK. Accurate assessment of precision errors: how to measure the reproducibility of bone densitometry techniques.Osteoporos Int J Establ Result Coop Eur Found Osteoporos Natl Osteoporos Found USA.1995;5:262–70. [DOI] [PubMed] [Google Scholar]
- 14. Hampel FR.The influence curve and its role in robust estimation.J Am Stat Assoc.1974;69:383–93. [Google Scholar]
- 15. Schoeller DA,Tylavsky FA,Baer DJ,Chumlea WC,Earthman CP,Fuerst T,Harris TB,Heymsfield SB,Horlick M,Lohman TG,et al.. QDR 4500A dual-energy X-ray absorptiometer underestimates fat mass in comparison with criterion methods in adults.Am J Clin Nutr.2005;81:1018–25. [DOI] [PubMed] [Google Scholar]
- 16. FAQ—International Society for Clinical Densitometry (ISCD)[Internet].[cited 2016 Apr 18]. Available from:http://www.iscd.org/resources/calculators/precision-calculator/faq/. [Google Scholar]
- 17. Smith-Ryan AE,Mock MG,Ryan ED,Gerstner GR,Trexler ET,Hirsch KR. Validity and reliability of a 4-compartment body composition model using dual energy X-ray absorptiometry-derived body volume.Clin Nutr.2017;36:825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chumela WC, Guo SS. Bioelectrical impedance: a history, research issues, and recent consensus. In:Carlson-Newberry SJ,Costello RB, editors.Emerging technologies for nutrition reserarch: Potential for assessing military performance capability.Washington (DC):National Academies Press (US);1997:169-92. [PubMed] [Google Scholar]
- 19. Vaché C,Rousset P,Gachon P,Gachon AM,Morio B,Boulier A,Coudert J,Beaufrère B,Ritz P. Bioelectrical impedance analysis measurements of total body water and extracellular water in healthy elderly subjects.Int J Obes Relat Metab Disord.1998;22:537–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.