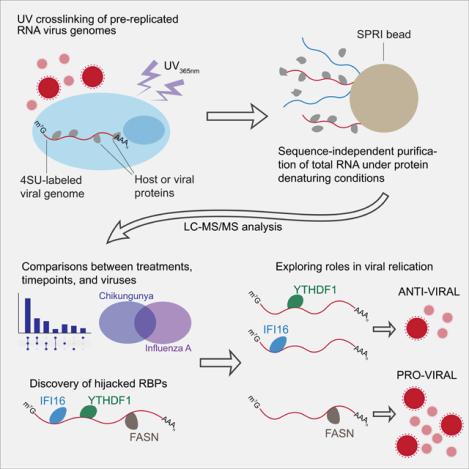
SUMMARY
The primary interactions between incoming viral RNA genomes and host proteins are crucial to infection and immunity. Until now, the ability to study these events was lacking. We developed VIR-CLASP (VIRal Cross-Linking And Solid-phase Purification) to characterize the earliest interactions between viral RNA and cellular proteins. We investigated the infection of human cells using Chikungunya virus (CHIKV) and Influenza A virus and identified hundreds of direct RNA-protein interactions. Here, we explore the biological impact of three protein classes that bind CHIKV RNA within minutes of infection. We find CHIKV RNA binds and hijacks the lipid-modifying enzyme FASN for pro-viral activity. We show that CHIKV genomes are N6-methyladenosine modified and that YTHDF1 binds and suppresses CHIKV replication. Finally, we find that the innate immune DNA sensor IFI16 associates with CHIKV RNA, reducing viral replication and maturation. Our findings have direct applicability to the investigation of potentially all RNA viruses.
Keywords: VIR-CLASP, RNA virus, RNA-binding protein, Innate immunity, Host-pathogen interactions, Interactome capture
Graphical Abstract
eTOC Blurb:
The earliest interactions between incoming RNA viral genomes and host RNA-binding proteins can be crucial to viral replication. Until now, these events couldn’t be distinctly studied. Kim and Arcos et al. develop VIR-CLASP, demonstrate its extensibility across multiple viral families, and report their findings on the CHIKV and IAV interactomes.
INTRODUCTION
Emerging viruses threaten human and livestock populations across the globe. Thus it is imperative to understand differences in viral tropism across species and cell types. The earliest interactions between host protein and RNA viral genomes during an infection can determine viral tropism, yet are unexplored. Before upregulation of interferons, cytokines, and antiviral genes, a cell must rely on mRNAs and proteins already in its cytoplasmic arsenal. These “intrinsic immunity” factors aid host cells in the race between viral replication and cellular production of new antiviral machines (Bieniasz, 2004; Sheehy et al., 2002). The virus also benefits from hijacking host mechanisms before transcription-dependent defenses alter the cellular environment.
Recent technological advances uncovered host RNA-binding proteins (RBPs) that interact with RNA viruses, yet no current method can identify interactions between the infecting, primary viral genome and host proteins (Lenarcic et al., 2013; Phillips et al., 2016). We developed a method to capture interactions between incoming viral RNA genomes and cellular proteins. VIR-CLASP (VIRal Cross-Linking And Solid-phase Purification) differs from RNA-antisense purification masss-pectrometry (RAP-MS) (Phillips et al., 2016) and thiouracil crosslinking mass-spectrometry (TUX-MS) (Lenarcic et al., 2013) in two fundamental ways. First, VIR-CLASP captures interactions with just the pre-replicated viral genome. Second, VIR-CLASP employs sequence-independent purification under protein-denaturing conditions. Here, we use VIR-CLASP to discover the initial host protein-viral RNA interactions between human cells and Chikungunya (CHIKV) or Influenza A (IAV). We selected these human pathogens to represent distinct types of RNA viruses with different strategies for viral replication. CHIKV is a member of the Togaviridae family of positive-sense RNA viruses (Weaver and Lecuit, 2015). CHIKV replication occurs in the cytoplasm, where the genome is a template for translation (Silva and Dermody, 2017). IAV is a member of the Orthomyxoviridae family of negative-sense RNA viruses. Upon infection, IAV translocates to the nucleus before initiating transcription of its segmented genome into the coding positive-sense RNA (Samji, 2009).
VIR-CLASP and mass spectrometry identified of hundreds of proteins that directly bind CHIKV and IAV. We found both shared and distinct host protein interactions with CHIKV and IAV. In-depth study of the CHIKV interactome uncovered condition-dependent interactions, including proteins that bind to CHIKV RNA following pretreatment of cells with interferon or under naïve conditions. We next explored the impact of three different types of CHIKV interacting proteins on viral replication. Fatty Acid Synthase (FASN), an enzyme that generates palmitic acid, was confirmed as a non-canonical viral RBP with an RNA-binding region in the ketoacyl-synthase domain. We found that FASN enhanced viral translation independent of its enzymatic activity and led to elevated levels of CHIKV RNA. VIR-CLASP also revealed that the N6-methyladenosine (m6A)-binding proteins YTH-domain 1–3 (YTHDF1–3) interact with pre-replicated CHIKV RNA. We established that CHIKV genomes contain m6A, an RNA modification found in eukaryotic and viral transcripts, which can regulate mRNA translation, stability, and localization. Overexpression and knockdown studies showed that YTHDF1 suppresses CHIKV replication, while YTHDF2 has the opposite effect. We then found that the DNA-sensor Interferon-inducible protein 16 (IFI16) binds to incoming CHIKV genomes. We showed that IFI16 restricts CHIKV replication independent of interferon-signaling or DNA-binding transcriptional activity (Thompson et al., 2014; Unterholzner et al., 2010). VIR-CLASP uncovered extensive interactions between host proteins and the incoming genomes of CHIKV and IAV, and we found that these RBPs impact CHIKV replication and maturation. VIR-CLASP does not use sequence-specific isolation of RNA; thus, we anticipate that this approach will facilitate study of many pathogenically relevant RNA viruses.
RESULTS
Capturing Viral RNA-Protein Interactions
VIR-CLASP relies on infection of unlabeled host cells with 4-thiouridine (4SU)-labeled viral genomes (Figure 1A, Box 1). Irradiation of infected cells with 365 nm light generates covalent crosslinks between 4SU-labeled viral genomes and interacting host or viral proteins (Figure 1A, Box 2). Solid-phase capture of these complexes with SPRI beads under protein denaturing conditions leads to purification of total RNA but only proteins covalently crosslinked to the 4SU-labeled viral RNA. This purification precedes nuclease (benzonase) digestion and LC-MS/MS identification of the crosslinked proteins (Figure 1A, Box 3). Only incoming viral genomes contain 4SU, so later events of viral replication or interactions between mRNA and protein are not captured. Since protein recovery is sequence-independent and achieved under high stringency, VIR-CLASP facilitates comparisons between time points, viral strains, and cell lines.
Figure 1. VIR-CLASP Isolates Host Proteins Directly interacting with Incoming RNA Viral Genomes.

(A) Box 1, Labeling of RNA viruses. Box 2, Irradiation of infected cells with 365nm light. Box 3, Solid-phase purification of RNP complexes. 4SU: 4-thiouridine; xL: crosslink; ER: endoplasmic reticulum.
(B) SDS-PAGE and silver stain of proteins purified from host cells (U2OS for CHIKV, A549 for EMCV, IAV, VSV and RVFV, DBT-9 for MHV and Huh-7 for ZIKV) infected with unlabeled or 4SU-labeled viruses using VIR-CLASP. Viruses: CHIKV, Chikungunya; EMCV, Encephalomyocarditis; MHV, Mouse Hepatitis; ZIKV, Zika; IAV:, Influenza A; VSV, Vesicular Stomatitis. (*) indicates Benzonase protein band.
(C) SDS-PAGE and silver stain (top) or immunoblot (bottom) of VIR-CLASP on HA-EMC4 CRISPR knock-in (+) or knockout (−) Huh-7 cells infected with unlabeled or 4SU-labeled CHIKV. B: 200 nM bafilomycin for 1 hr; xL: virus was crosslinked with UV365nm before infection.
(D) SDS-PAGE and silver stain of VIR-CLASP performed on lysates from unlabeled or 4SU-labeled, isolated CHIKV virion. E1/E2, E1 and E2 viral glycoproteins. C, viral capsid protein.
(E) SDS-PAGE and silver stain of VIR-CLASP on lysates from U2OS cells infected with unlabeled or 4SU-labeled CHIKV. Cells were pretreated with dinaciclib for 4 hr. Data represent two biologically independent repeats in C–E.
(F) Table showing mapped RNA-sequencing reads from VIR-CLASP using purified CHIKV virions. Reads were mapped to CHIKV strain 181/25 and Mesocricetus auratus genomes.
To demonstrate that SPRI purification of protein requires 4SU-labeled RNA and UV crosslinking, we performed “CLASP” on cellular RNA. We only observed efficient protein purification when RNA was 4SU-labeled and crosslinked (Figure S1A). Also, protein recovery was abolished by benzonase degradation of RNA before purification or inhibition of transcription with dinaciclib to block incorporation of 4SU into cellular RNA (Figures S1B and S1C) (Paruch et al., 2010). These results show that protein purification with CLASP requires UV crosslinking and intact 4SU-labeled RNA. We observed that ribosomal RNA (rRNA) purification with CLASP is unaltered by 4SU incorporation, suggesting unaltered purification of differently structured RNAs (Figure S1B).
We performed VIR-CLASP pilot experiments on viruses spanning seven families and representing plus- and minus-stranded RNA genomes: CHIKV (Togaviridae), EMCV (Encephalomyocarditis, Picornaviridae), MHV (Mouse hepatitis, Coronaviridae), ZIKV (Zika, Flaviviridae), RVFV (Rift Valley Fever, Phenuiviridae), IAV (subtype H3N2, Orthomyxoviridae), and VSV (Vesicular stomatitis, Rhabdoviridae) (Figures 1B and S1D). By silver-stain, we observed diverse bands indicating interactions between cellular proteins and 4SU-labeled viral genomes. We chose to focus on identifying the host interactome with the incoming CHIKV genome using LC-MS/MS. CHIKV is a reemerging alphavirus that causes disease in humans characterized by fever, rash, and arthralgia (Silva and Dermody, 2017; Weaver and Lecuit, 2015). Although CHIKV has caused millions of cases of disease and has significant economic impact, there are no therapies to prevent or treat CHIKV infection.
We first examined whether free 4SU remains in the buffer containing isolated and concentrated CHIKV, despite the stringent purification before infection. Free 4SU could incorporate into RNA of unlabeled and infected cells and contribute to the crosslinked RNA-protein complexes purified with VIR-CLASP. To test this, we treated cells with dinaciclib before infection (Paruch et al., 2010). Dinaciclib inhibits transcription, so if free 4SU incorporates into cellular RNA, this drug should decrease the protein signal observed with VIR-CLASP. We observed no decrease in proteins enriched with VIR-CLASP, indicating that cellular 4SU incorporation does not contribute to the proteins purified in our approach (Figure 1C). We then confirmed that the quantity of protein purified with VIR-CLASP is dependent upon viral multiplicity of infection (MOI) (Figure S1E). Lastly, we performed VIR-CLASP on purified virus without infection into host cells. We observed interactions between viral glycoproteins E1, E2, and viral Capsid protein with the CHIKV genome, and we did not detect mammalian host RBPs in the purified viral particles by silver stain (Figure 1D).
An integral component of the ER membrane complex, EMC4, can facilitate entry of flaviviruses into the host cytoplasm from endosomes (Savidis et al., 2016). CHIKV shares entry mechanisms with flaviviruses, so we hypothesized that CHIKV entry requires EMC4. If so, loss of EMC4 should abolish any protein purified from VIR-CLASP with CHIKV. We performed CHIKV VIR-CLASP in cells containing either a CRISPR knockout for EMC4 or a CRISPR knock-in of HA-EMC4 (Barrows et al., 2019). We also tested two other methods to block viral entry: pretreatment of cells with bafilomycin, a vacuolar ATPase inhibitor (Bowman et al., 1988), or UV-irradiation of 4SU-CHIKV before infection to induce terminal crosslinks within virions. We observed that CHIKV-interacting proteins only enrich with VIR-CLASP when EMC4 is present (Figure 1E). We also observed loss of proteins when cells were pretreated with bafilomycin or pre-crosslinked. These results establish that CHIKV entry into the cytoplasm requires EMC4, that inhibition of viral entry prevents interaction between the viral genome and cytoplasmic proteins, and that isolation of interacting proteins using VIR-CLASP depends upon entry of viral genomes into the host cytoplasm.
We next evaluated the extent of BHK21 (Mesocricetus auratus (golden hamster)) RNA packaged within the viral particle, which may lead to identification of interactions with non-viral RNAs. We performed RNA-seq with RNA from purified CHIKV viral particles and found that of 378.6 million reads across two replicates, over 340 million reads (>90%) mapped to either CHIKV or M. auratus. 98.8% of mapped reads map to the CHIKV genome and about 1% are from M. auratus (Table S1, Figure 1F). We validated by RT-qPCR that the percentage of the 18S and 28S rRNA over the CHIKV genome was less than 0.4% (Table S1). Thus, BHK21 RNA is not a considerable fraction of RNA found within CHIKV virions. We conclude that the potential for identifying interactions with non-viral RNA packaged within virions using VIR-CLASP is likely minimal and stochastic.
To define the period when interactions between incoming viral genomes and host proteins are vital we UV-irradiated 4SU-CHIKV before or during infection. Covalent crosslinks between viral RNA and proteins should be detrimental to production of new virions when the incoming RNA genomes are necessary for critical steps like translation and replication. There should be a time post-infection when formation of 4SU-crosslinks will not affect viral production, marking the period when the incoming viral transcript is no longer required. We crosslinked 4SU-labeled viruses before infection (−1 hour) or after infection (0.2 to 5 hours) in U2OS cells (Ho et al., 2015; J.-W. Lu et al., 2017; Reid et al., 2015; Y.-M. Wang et al., 2016), under naïve or type-I interferon (IFN) stimulated conditions (Figure 2A, top panel). Type-I IFNs are cytokines secreted in response to innate immune activation; IFNs regulate transcription of IFN-stimulated genes (ISGs), including antiviral proteins. As anticipated from Figure 1E, crosslinking before infection decreased viral titer compared to unlabeled (uncrosslinked) control virus in both naïve and IFN-stimulated cells. The initial decrease in titer was from ~109 to 107, so we estimate that ~99% of functional viral particles contain a biologically relevant amount of 4SU. The average amount of 4SU substitution in the purified viral particles was 2.5% (4SU over total uridine) by HPLC (data not shown). Crosslinking of viral particles at 2 hours post-infection (hpi) led to a 10-fold increase in titer compared to crosslinking at 0 or 1 hpi. This indicates that by 2 hpi viral replication has begun but is not independent of the primary genomes. Only when irradiation occurred at 3 hpi did viral titer recover to levels close to control. RT-qPCR analysis showed that CHIKV RNA rapidly increases between 2 and 3 hpi, confirming the start of viral transcription (Figure 2A, bottom panel). We concluded that for CHIKV, the window between viral entry and 3 hpi was the ideal range to capture interactions between pre-replicated viral RNA and host proteins.
Figure 2. VIR-CLASP Profiling of the CHIKV Pre-replicated Interactome.
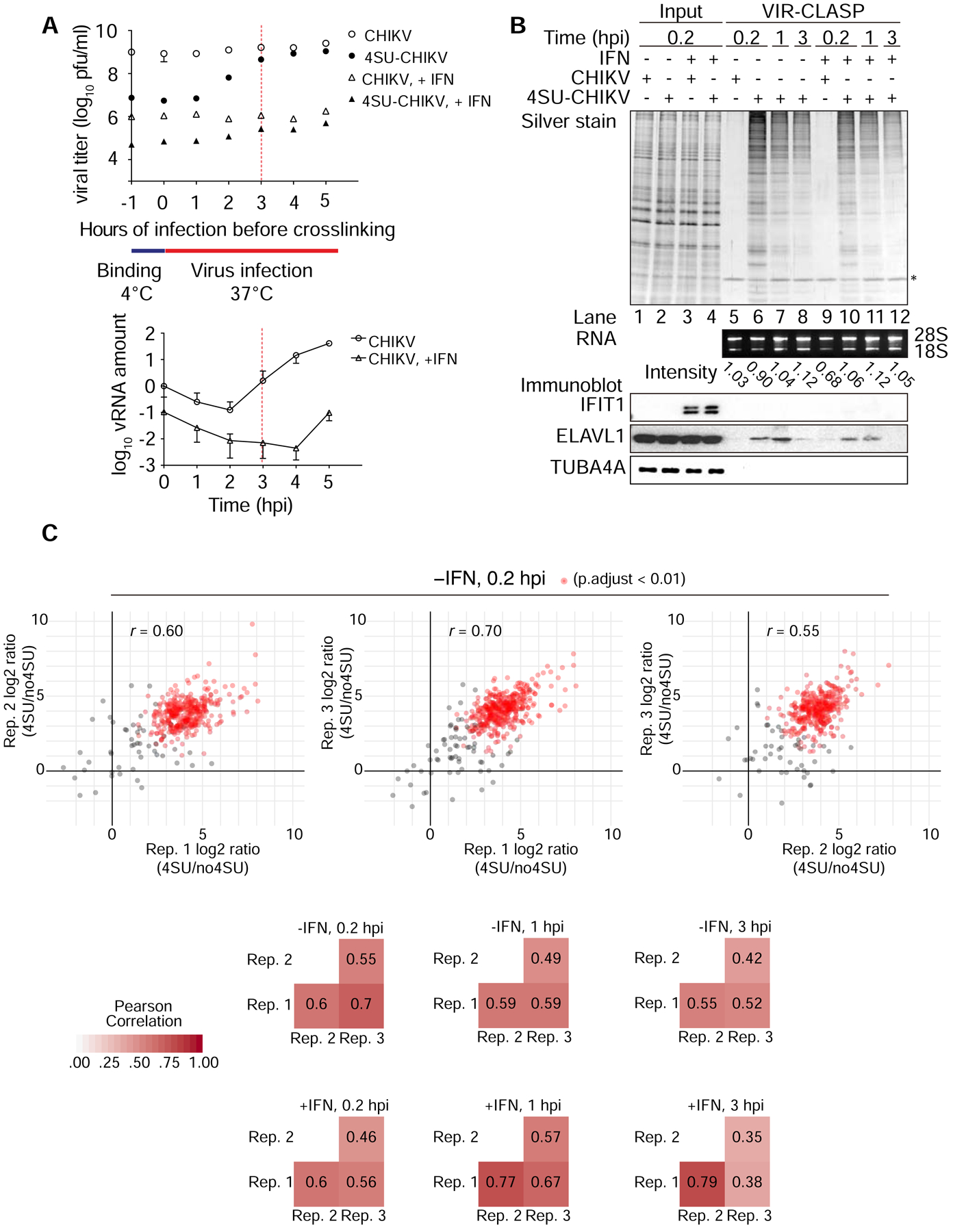
(A) Top, Plaque assay for CHIKV titer from infection with crosslinking (UV365nm) at indicated time point on U2OS cells. Bottom, RT-qPCR analysis of viral RNA in U2OS cells during CHIKV infection. Cells were treated with or without IFN (500U/ml) for 16 hr prior to infection. n = 2, error bars, mean ±SD.
Data represent two biologically independent repeats in A.
(B) SDS-PAGE and silver stain (top), agarose gel (middle), or immunoblot (bottom) of proteins purified from U2OS cells infected with unlabeled or 4SU-labeled CHIKV using VIR-CLASP. IFN: treatment with recombinant interferon-β for 16 hr prior to infection. rRNA band intensity was measured by ImageJ. Data represent three biologically independent repeats in B.
(C) Top, representative scatterplots showing log2-intensity ratios of quantifiable proteins in 4SU versus no4SU samples of three biological replicates of VIR-CLASP for CHIKV (pairwise comparisons). Red dots represent significantly enriched proteins among all 3 replicates. Bottom, summary heatmaps showing the Pearson correlation coefficients for all conditions.
Based on this timeline, we performed VIR-CLASP on 4SU-labeled CHIKV at 0.2, 1, and 3 hpi, with naïve or IFN-treated cells, and performed LC-MS/MS and immunoblot analysis (Figure 2B). Negative controls represent cells infected with unlabeled CHIKV, crosslinked, and processed using VIR-CLASP (Figure 2B, lanes 5 and 9). SDS-PAGE and silver staining of the crosslinked proteins revealed unique protein banding patterns across the 6 conditions. A decrease in interacting protein recovery at 1 and 3 hpi suggests either that host proteins progressively bind less incoming viral RNA over time or that the incoming genomes are decaying. We also validated ELAVL1 (HuR), an RBP that interacts with the CHIKV 3’ UTR (Dickson et al., 2012), by immunoblot (Figures 1E and 2B). We failed to identify antiviral RBP IFIT1 by immunoblot, consistent with previous work showing that IFIT1 binds only to RNA viruses with a 5’ cap-0 structure (Figure 2B) (Daffis et al., 2010).
We performed VIR-CLASP with IAV to test the specificity of VIR-CLASP and to identify differences in host proteins that interact with pre-replicated plus- and minus-strand RNA viruses. We observed that IAV replication is not independent from the pre-replicated genome until ~ 4 hpi (Figure S2A). We chose to perform VIR-CLASP for IAV at 1 hpi in order to capture interactions that occur while viral replication heavily relies on the pre-replicated genome. IAV is known to contain m6A modifications (Courtney et al., 2017; Krug et al., 1976), and we observed an interaction between the m6A-reader YTHDF1 and pre-replicated IAV by western blot (Figure S2B).
Analysis of the CHIKV and IAV Primary Interactomes
To identify proteins in the pre-replicated CHIKV and IAV interactomes we performed proteomics analysis on VIR-CLASP samples from 0.2, 1, and 3 hpi, and with naïve or IFN-treated cells for CHIKV, and at 1 hpi in naïve cells for IAV. To define candidate “VIR-CLASP RBPs” we calculated the peptide intensity ratios between +4SU and −4SU samples, leading to identification of ~400 significantly enriched proteins in each condition (0.01% FDR and fold change > 5) (Figure 2C, S2C, Table S2, and Table S3). Due to the low complexity of the −4SU samples, we could not calculate peptide intensity ratios for all proteins. For the remaining proteins, we used a semiquantitative approach based on the assumption that peptides with no intensity value are below the detection threshold (Garcia-Moreno et al., 2019; Sysoev et al., 2016).
To address the possibility that host proteins package within the virion we performed LC-MS/MS analysis of VIR-CLASP performed on the purified CHIKV virion (Figure 1D). We identified CHIKV Capsid, E1, and E2 proteins with very high intensities (Table S4). We also compared all proteins with non-zero intensity values in one or both replicates of the virion LC-MS/MS to our CHIKV candidate RBPs. 40 proteins had identical peptides present in one or both replicates of the virion LC-MS/MS and the CHIKV candidate list in any condition. Many of the proteins found in both virion and VIR-CLASP are ribosomal proteins. We annotated the 40 proteins as found in both VIR-CLASP and the CHIKV virion in Table S2.
About 50% of proteins identified in VIR-CLASP for IAV were also identified for CHIKV in at least one condition (Figure S2D). The 193 shared proteins include known regulators of viral infection (STAU1 (de Lucas et al., 2010), ZC3HAV1 (Li et al., 2017), and LARP1 (Karlas et al., 2010; Suzuki et al., 2016), and RNA helicases (DHX9, DHX15, DHX29, DHX30, DHX36, and DHX38). We validated unique and overlapping IAV and CHIKV proteins using immunoblot, confirming that FXR1 and DDX21 are specific interactors of pre-replicated CHIKV, WDHD1 and KDMB3 are specific interactors of pre-replicated IAV, and LARP1 interacts with both (Figure S2E).
We identified viral proteins in the pre-replicated interactomes of both CHIKV and IAV. For CHIKV, we identified the Capsid and E1 in all timepoints and conditions; we identified E2 only at 0.2 hpi and 1 hpi (in both +IFN and −IFN). For IAV we identified Nucleoprotein (NP), confirming its interaction with the IAV genome (Area et al., 2004; Das et al., 2010). For the remainder of this report we focus on the CHIKV pre-replicated interactome due to the increased depth of the proteomics data.
The pre-replicated CHIKV interactome contains consistent and dynamic factors among the conditions tested. We found that ~255 of the candidate proteins interact with incoming CHIKV RNA throughout the first 3 hours of infection in both naïve and IFN-treated cells; ~340 proteins were present only in a subset of conditions (Figure 3A) (Lex et al., 2014). We identified 142 proteins unique to the +IFN dataset, while 115 proteins were unique to −IFN (Figure 3A). The +IFN-unique proteins include known regulators of viral infection like SAMD9 (Liu and McFadden, 2015) and PNPT1 (Dhir et al., 2018). 23 proteins were specific to the most “naïve” condition (0 hpi, −IFN), including EIF4G1 and RAC1.
Figure 3. Functional Analysis of the CHIKV Pre-replicated Interactome.

(A) UpSet diagram depicting the overlapping proteins identified in 6 conditions of VIR-CLASP with CHIKV. The bar chart above indicates the count of proteins that are shared in the conditions indicated by the highlighted and connected dots below. The horizontal bar chart on the left indicates the total number of proteins identified in each condition. Shading indicates proteins unique to either (−) or (+) IFN.
(B,C) Dot plots showing top enriched GO biological process (B) and molecular function (C) terms in the pre-replicated CHIKV interactome. Enrichment is based on all human proteins. GO terms were collapsed based on semantic similarity using the clusterProfiler R/Bioconductor package (Yu et al., 2012); the most significant terms across the 6 conditions are shown (p-values adjusted for multiple testing with Benjamini-Hochberg). Color shows p-values; size of dots is Gene Ratio (percentage of VIR-CLASP candidates in the given GO term). Only significant results (p < 0.01) are plotted.
(D) Previously reported RBPs that are Type-1 ISGs in the CHIKV candidate interactome. RBP annotations were compiled from Gene Ontology and previous interactome-capture efforts. ISG annotation was derived from the Interferome (Rusinova et al., 2013).
(E) Dot plot showing enriched PFAM (El-Gebali et al., 2019) domains in either reported or novel RBPs. Enrichment is based on all human proteins. P-values were calculated using Fisher’s exact test and Bonferonni correction. Color shows p-values, size of dots is count of VIR-CLASP candidates containing the given domain.
To understand the functions of the proteins in the pre-replicated CHIKV interactome, we performed GO analysis with candidate VIR-CLASP RBPs (Figures 3B–C, S2F–G and Table S5). Many enriched GO molecular function, biological process, and cellular component terms were related to RNA binding and different aspects of RNA metabolism, including translation initiation (GO: 0006413) (Figures 3B–C and S2F). The CHIKV interactome was also enriched for terms relating to viral infection and immune response (Table S5). Enriched KEGG pathways in the CHIKV interactome include Spliceosome (hsa03040), Ribosome (hsa03010), and other RNA processing pathways (Figure S2G). GO analysis of the IAV pre-replicated interactome also revealed enrichment for molecular function terms related to RNA binding for biological process terms related to RNA localization and viral genome replication, and for KEGG pathways related to RNA processing (Figures S3A–D and Table S6).
Over 50% of the CHIKV interactome proteins were interferon-stimulated genes (ISGs) (Figures S3E–F) (Rusinova et al., 2013). While the proportion of ISGs was consistent among the timepoints and conditions we tested, we were curious whether the ISGs represented potentially novel RBPs. About 60% of the CHIKV interactome is classified as an RBP by GO analysis or by previous interactome identification (Baltz et al., 2012; Castello et al., 2012; Gerstberger et al., 2014; R. Huang et al., 2018; Perez-Perri et al., 2018). The remaining 40% are potentially novel RBPs (Figure S3G). We observed that the proportion of novel RBPs that are also ISGs tended to increase over time or in response to IFN (Figure 3D). Over 50% of the proteins in the IAV interactome were previously classified as RBPs (Figure S3H). Comparison of the PFAM domains (El-Gebali et al., 2019) found in the proteins identified by VIR-CLASP for CHIKV revealed that the previously reported RBPs were enriched in helicase, RNA-recognition motif (RRM) and other nucleotide-binding domains, while the novel RBPs contained Gelsolin repeat, WD40 repeat, and FHA domains (Figure 3E).
We further explored the CHIKV interactome proteins annotated by the GO term “translation initiation” (GO: 0006413) (Figure 4A and Table S5). These included translation initiation factors, RNA helicases, and the m6A-binding proteins YTHDF2 and YTHDF3. While m6A exists in other plus-strand RNA viruses like flaviviruses (Gokhale et al., 2016), whether m6A exists in alphavirus genomes is unknown. The identification by VIR-CLASP of a direct interaction of YTHDF2 and YTHDF3 with pre-replicated CHIKV (Figure 4B) suggests that CHIKV genomes contain m6A.
Figure 4. CHIKV VIR-CLASP Identifies Translation Initiation Factors, Nucleic Acid Sensors, and Metabolic Enzymes.

(A) Table showing pre-replicated CHIKV interactome proteins identified in the GO term “translation initiation” (GO: 0006413).
(B) Immunoblot validation of proteomics results from VIR-CLASP with CHIKV. Data represent two biologically independent samples in B.
(C) Table showing nucleic acid sensors identified in VIR-CLASP for CHIKV. PRR annotations were compiled from recent published reviews (Chow et al., 2018; Ma et al., 2018).
(D) Table showing pre-replicated CHIKV interactome proteins identified in previous screens.
CHIKV VIR-CLASP hits include nucleic acid pattern-recognition receptors (PRRs) (Chow et al., 2018; Ma et al., 2018), such as DHX9, DDX21, OAS3, IFI16, XRCC6, and PRKDC (Figure 4C). Although CHIKV has an RNA genome, we found that pre-replicated CHIKV interacts with IFI16 and the DNA-PK complex (XRCC5, XRCC6, and PRKDC). RNA binding by the DNA-PK complex has been previously reported (Yoo and Dynan, 1998). IFI16 is an antiviral cytosolic-DNA sensor, yet was identified in all timepoints and conditions (Figure 4B). Recent research demonstrated that IFI16 restricts RNA virus infection by regulating transcription of type-I IFNs. Identification of IFI16 as a VIR-CLASP hit and putative RBP suggests a novel mechanism for its restriction of RNA viruses.
We compared our CHIKV VIR-CLASP dataset to previous screens for regulators of CHIKV infection (Figure 4D and Table S7) (Karlas et al., 2016; Ooi et al., 2013; Radoshitzky et al., 2016; Schoggins et al., 2011). VIR-CLASP hits identified in at least one other screen include ADAR, ACLY, and FASN. FASN was also identified as a putative RBP in two other screens to identify novel cellular RBPs (Baltz et al., 2012; Castello et al., 2012). VIR-CLASP expands on these results to demonstrate that FASN binds viral RNA (Figure 4B). The connection between FASN’s role in viral replication and its potential RNA-binding activity is unexplored.
For this report, we follow up on YTHDF1–3, IFI16, and FASN to determine how their interactions with CHIKV affect viral replication.
YTHDFs Modulate CHIKV Viral Replication
m6A is an RNA modification found in mRNA and lncRNA (Jia et al., 2013), and within viral genomes from the Retro-, Orthomyxo-, and Flaviviridae families (Courtney et al., 2017; Gokhale et al., 2016; Krug et al., 1976; Lichinchi et al., 2016; W. Lu et al., 2018). To explore whether CHIKV contains m6A, we performed m6A RNA-immunoprecipitation (MeRIP) on RNA from purified CHIKV virions. CHIKV virion RNA immunoprecipitated with an anti-m6A antibody, but not with an IgG control (Figure 5A). We then performed Me-RIP-RT-qPCR with primers tiled along the CHIKV genome every ~1000 nt. We observed a significant enrichment within 2000 nt at the 5’ end of the genome (Figure 5B).
Figure 5. YTHDF1 Restricts (−) Strand Replication of CHIKV.

(A) Phosphorimage of meRIP prepared from fragmented and 32P-labeled CHIKV virion RNA. RNA was isolated from CHIKV virions purified twice by discontinuous sucrose gradient (30%–60%), and concentrated by sucrose cushion (30%). Data represent two biologically independent replicates in A.
(B) MeRIP-qRT-PCR using primers per 1000 nucleotides (nt) of CHIKV genome showing enrichment of viral RNA in m6A IP over IgG IP. RNA was extracted from purified CHIKV virions as in A. n = 3, error bars, mean ±SD.
(C) Schematic depicting the RNA species present during CHIKV replication.
(D) Strand-specific RT-qPCR showing viral RNA levels in U2OS cells stably expressing Flag-HA-YTHDF1 or parental U2OS. Data were normalized to parental U2OS, MOI 1 within each timepoint. MOI, multiplicity of infection, hpi, hours post infection. n = 4, error bars, mean ±SD.
(E) siRNA knockdown of YTHDF1 or control followed by strand-specific RT-qPCR. CHIKV infection was performed 72 hr after siRNA transfection. MOI, multiplicity of infection, hpi, hours post infection. n = 4, error bars, mean ± SD.
(F) Rescue of viral RNA replication effect by re-expression of YTHDF1. n = 9, error bars, mean ± SD.
(G) Left, CHIKV RNA in supernatants from U2OS cells 24 hpi at MOI=10−4 after siRNA treatment, quantified by RT-qPCR. Right, CHIKV titer measured by plaque assay in Vero cells. MOI, multiplicity of infection, hpi, hours post infection. n = 3, error bars, mean ± SD.
See also Figure S4
We then explored the function of m6A on CHIKV through over- and underexpression of YTHDF1–3. While YTHDF1 was not a CHIKV “candidate RBP”, we decided to study its effect on CHIKV replication given that we identified the other YTHDF proteins by mass-spec, and all were identified by immunoblot (see Figure 4B). Strand-specific RT-qPCR distinguishes plus- and minus-strand CHIKV RNA (Figure 5C). Overexpression of YTHDF1 decreased both strands of CHIKV RNA; knockdown of YTHDF1 increased both strands at 3 and 5 hpi (data normalized within each timepoint) (Figures 5D–E and Figures S4A–D). The increase was highest in the minus-strand. By contrast, YTHDF2 overexpression increased both strands at 5 hpi, while overexpression of YTHDF3 resulted in little change (Figures S4E–F). Knockdown of YTHDF2 had little effect on CHIKV RNA levels at 5 hpi, and knockdown of YTHDF3 slightly increased CHIKV RNA (Figures S4G–H). To examine possible indirect effects of YTHDF1 knockdown we rescued YTHDF1 levels by re-expression. Re-expression of YTHDF1 rescued plus-strand CHIKV to the level of control, while the effect on minus-strand CHIKV was diminished, but not entirely to control levels (Figure 5F). Loss of YTHDF1 led to an increase in translation based on a CHIKV-luciferase reporter indicating that YTHDF1 can also regulate CHIKV protein levels (Figure S4I).
We next tested whether the early effects of YTHDF proteins on CHIKV persist to the release of new virions. Knockdown of YTHDF1 and YTHDF3 increased both extracellular viral RNA levels and mature virions; knockdown of YTHDF2 had the opposite effect (Figure 5G). These data show that YTHDF1 and YTHDF3 restrict CHIKV replication, while YTHDF2 promotes CHIKV replication.
IFI16 is an RNA-Sensor of CHIKV Infection
Interferon-inducible protein 16 (IFI16) is a DNA-binding PRR (Unterholzner et al., 2010). IFI16 also restricts some RNA viruses (Sendai virus, EMCV (Thompson et al., 2014)) through regulating type-I IFN transcription. Our VIR-CLASP data suggest that IFI16 recognizes viral RNA. Therefore, IFI16 may restrict RNA viruses as a PRR. To rule out that VIR-CLASP captures interactions with both RNA and DNA, we treated lysates with DNase, RNase, or Benzonase before purification. Treatment with RNase or Benzonase abolished the recovery of interacting proteins, while DNase treatment only led to a slight decrease in protein recovery compared to the control (Figure 6A). Importantly, IFI16 was present in DNase treated samples by immunoblot.
Figure 6. IFI16 Directly Binds to the Incoming CHIKV Genome and Restricts Replication.

(A) Silver stain (top), agarose gel (middle), or immunoblot (bottom) of CLASP performed on lysates treated with RNase (R), DNase (D) or Benzonase (B). Lysates are from U2OS cells stably expressing Flag-HA-IFI16, cultured in the presence or absence of 100 μM 4SU. Data represent two biologically independent repeats in A.
(B) Left, RIP-RT-qPCR showing enrichment of CHIKV RNA (Top) or TUBA1A mRNA (Bottom) of Flag IP over IgG IP on lysates from U2OS cells expressing Flag-HA-IFI16. Right, Relative enrichment of CHIKV RNA over TUBA1A mRNA from Flag IP (Top) or input (Bottom). IPs were performed with lysates from U2OS cells infected with 4SU-labeled CHIKV and crosslinked with UV365nm. n = 3, error bars, mean ± SD.
(C, D) Strand-specific RT-qPCR showing viral RNA levels in U2OS cells stably expressing IFI16 or parental U2OS at 8 hpi (C), or in cells treated with siRNA for IFI16 or control at 3 hpi or 8 hpi (D). MOI, multiplicity of infection. n = 4, error bars, mean ± SD.
(E) Rescue of viral RNA replication effect by re-expression of IFI16. n = 3, error bars, mean ± SD.
(F) Strand-specific RT-qPCR showing viral RNA levels in U2OS cells stably expressing IFI16 or parental U2OS (8 hpi). Cells were pretreated with 5 nM actinomycin D or 5 μM ruxolitinib for 1 hr. n = 3, error bars, mean ± SD.
(G) Top, CHIKV RNA in supernatants harvested from U2OS cells 24 hpi at MOI=10−4 after 72 hr siRNA treatment, quantified by RT-qPCR. Bottom, CHIKV titer in the supernatants measured by plaque assay in Vero cells. n = 3, error bars, mean ± SD.
See also Figure S5.
We confirmed interaction between IFI16 and CHIKV via RNA-immunoprecipitation (RIP) and RT-qPCR with Flag-IFI16. CHIKV RNA was present in the anti-Flag pull-down but not the IgG control, and was enriched relative to tubulin (Figure 6B). The enrichment of CHIKV RNA was orders of magnitude greater than of tubulin, despite similar levels of CHIKV and tubulin RNA in the input (Figure 6B). Overexpression of IFI16 led to a decrease in both strands of CHIKV RNA (Figures 6C and S5A). Knockdown of IFI16 led to an increase in both strands of CHIKV (Figures 6D and S5B). Rescue of IFI16 levels through combined siRNA knockdown and overexpression restored viral RNA levels (Figure 6E). Interestingly, loss of IFI16 had no effect on translation based on a CHIKV luciferase reporter (Figure S5C). These results expand on previous reports to show that IFI16 can bind viral RNA, and can restrict CHIKV replication (Thompson et al., 2014).
To test whether IFI16 restricts RNA virus replication using a mechanism distinct from regulating IFN transcription (Thompson et al., 2014), we pretreated cells with either ruxolitinib (a JAK/STAT signaling inhibitor (Quintás-Cardama et al., 2010)) or actinomycin D (a transcription inhibitor). Neither treatment abolished the IFI16 knockdown effect, indicating that IFI16 restricts CHIKV replication independent of JAK/STAT-mediated IFN signaling, and that IFI16 retains its activity when transcription is blocked (Figure 6F and S5D). Loss of IFI16 also decreased mature CHIKV virion production (Figure 6G).
FASN Interacts with CHIKV and is Proviral
FASN catalyzes fatty acid formation from acetyl-CoA and malonyl-CoA (Beld et al., 2014). FASN is critical for cellular function and can also regulate viral replication (Heaton et al., 2010; Kulkarni et al., 2017; Sharma-Walia et al., 2010; Yang et al., 2008). A recent siRNA screen found that FASN enzymatic activity promotes CHIKV replication (Karlas et al., 2016). Our VIR-CLASP data establish that FASN interacts with viral RNA. To determine whether FASN binds cellular RNA as well as viral RNA, we performed CLASP and an abbreviated PAR-CLIP (Hafner et al., 2010). The results of both approaches support the model that FASN is a cellular RBP (Figures 7A–B and Figure S6A), corroborating screens that identified FASN as a candidate RBP (Baltz et al., 2012; Castello et al., 2012). A proteome-wide approach to discover RNA-binding domains identified peptides in the FASN N and C termini as putative RNA-binding regions (He et al., 2016). To determine which domains are necessary for RNA binding, we generated constructs from the N or C termini of FASN. CLASP revealed that the ketoacyl synthase (KS) domain and a construct containing the dehydratase (DH) domain were each sufficient for RNA binding (Figures 7C and S6B). Owing to our observation of weak and inconsistent interaction between the C-terminus of FASN and RNA (Figures 7C and S6B), we conclude that the C-terminus is likely not the main RNA-binding region of FASN.
Figure 7. FASN is an RBP that Plays a Proviral Role in CHIKV Replication and Release.
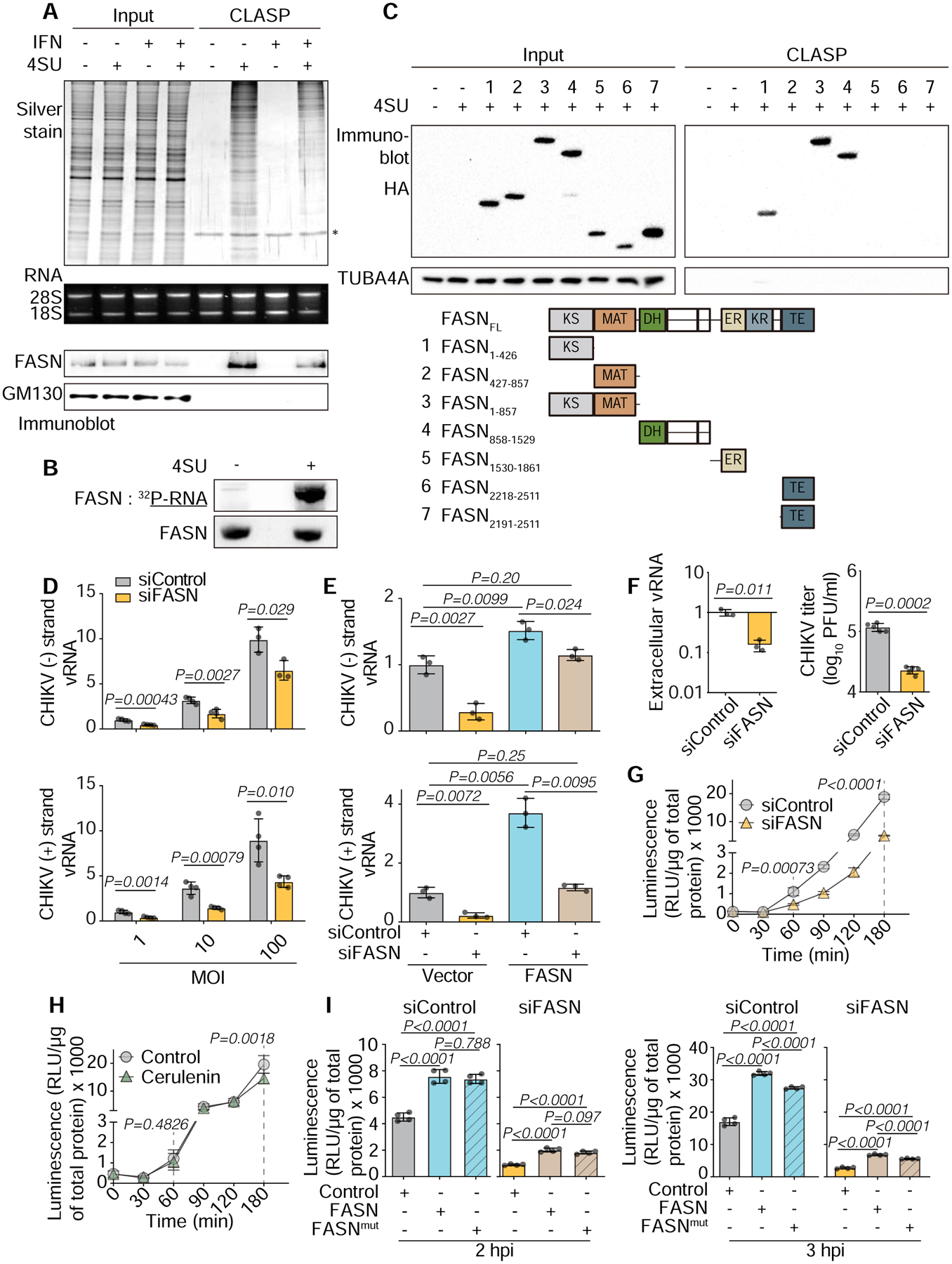
(A) SDS-PAGE and silver stain (top), agarose gel (middle) or immunoblot (bottom) of CLASP performed on lysates from U2OS cells cultured in the presence or absence of 100 μM 4SU, and with or without IFN treatment.
(B) Phosphorimage (top) or immunoblot (bottom) of PAR-CLIP performed on lysates from U2OS cells overexpressing Flag-HA-FASN and cultured in the presence or absence of 100 μM 4SU.
(C) Top, Immunoblot of CLASP performed on lysates from HEK293T cells transiently expressing truncated FASN. Cells were cultured in the presence or absence of 100 μM 4SU. Bottom, Schematic of the protein domains of FASN and its truncated versions. Data represent two biologically independent repeats in A–C.
(D) Strand-specific RT-qPCR of viral RNA levels in U2OS cells treated with siRNA for FASN or control. CHIKV infection was performed 72 hr after siRNA transfection. MOI, multiplicity of infection. n = 4, error bars, mean ± SD.
(E) Rescue of viral RNA replication effect by re-expression of FASN. n = 3, error bars, mean ± SD. *p < 0.05.
(F) Top, CHIKV RNA in supernatants harvested from U2OS cells 24 hpi at an MOI of 10−4 after 72 hr siRNA treatment, quantified by RT-qPCR. Bottom, CHIKV titer in the supernatants as measured by plaque assay in Vero cells. n = 3, error bars, mean ± SD.
(G) Luciferase assay of lysates from cells treated with siRNA for FASN or control. Cells were infected with CHIKV-nLuc at MOI = 10. n = 4, error bars, mean ± SD.
(H) Luciferase assay of cell lysates from cerulenin treated or untreated cells. Cells were infected with CHIKV-nLuc at MOI of 10. Cells were pretreated with 10 μM cerulenin for 2 hr. n = 8, error bars, mean ± SD.
(I) Rescue of viral translation by re-expression of wild-type (FASN) or catalytically-dead mutant (FASNmut) on (G) experimental conditions. n = 4, error bars, mean ± SD.
See also Figure S6
We then examined whether FASN could regulate viral replication. Knockdown of FASN decreased intracellular levels of both CHIKV strands (Figures 7D and S6C), and re-expression of FASN rescued viral RNA levels (Figure 7E). The effect of FASN knockdown persisted through the viral lifecycle, resulting in reduction of mature virions (Figure 7F). We also found that CHIKV replication decreased upon treatment with cerulenin to inhibit FASN enzymatic function(Omura, 1976) (Figure S6D). Knockdown of FASN significantly decreased luciferase activity indicating that FASN also affects translation (Figure 7G and S6E). Importantly, our luciferase results show that the translation effect was not recapitulated by treatment with cerulenin until 3 hpi (Figure 7H). To further explore the non-enzymatic role of FASN in CHIKV translation, we tested whether luciferase activity could be rescued by a catalytically-dead mutant (FASNmut) (Joshi et al., 1998). Overexpression of FASNmut was sufficient to rescue CHIKV translation in both knockdown and cerulenin treated conditions (Figure 7I and S6F). At 2 hpi (before replication of viral RNA (Figure 2A)), wild-type and FASNmut did not show a difference in ability to rescue viral translation (Figure 7I). Strikingly, we found that although FASNmut could partially rescue the knockdown of endogenous FASN at 3 hpi (Figure 7I, right panel), its ability to do so was less than wild-type FASN -- corroborating our results in Fig 7H, which shows FASN catalytic requirements mostly at later stages. Thus FASN can enhance translation levels at an earlier stage and independent of its enzymatic activity.
DISCUSSION
While PRRs and traditional antiviral proteins are important in innate immunity, proteins not considered traditional pro- or antiviral factors also contribute to the outcome of viral infection. “Intrinsic immunity” (Sheehy et al., 2002) factors aid normal cellular function but are co-opted for other processes during infection. An RNA virus can be a substrate for host RBPs, thus providing a mechanism for hijacking host RNA metabolism. RBPs can also aid cellular defense, as they can bind viral RNA before upregulation of traditional antiviral proteins. The earliest timepoints of infection are likely when these roles of RBPs are critical. Thus, VIR-CLASP aims to identify intrinsic and innate immune RBPs that drive initial cellular responses to infection and viral replication.
FASN represents an ideal target for co-option by viruses because it is required for cellular survival. A previous siRNA screen to identify regulators of CHIKV replication examined FASN enzymatic activity (Karlas et al., 2016). VIR-CLASP expands on these observations to show that FASN interacts with CHIKV RNA; whether interaction inhibits or promotes FASN enzymatic function is unknown. Our data indicate that FASN regulates both viral RNA and protein levels. Regulation of viral RNA levels by FASN is dependent on its enzymatic activity, while its effect on viral translation can occur independently. We favor a model in which CHIKV RNA hijacks FASN to localize its enzymatic activity to sites of viral replication, where elevated concentrations of palmitic acid would benefit viral packaging (Heaton et al., 2010) and replication (Zhang et al., 2019). It is unknown if this RNA-binding localization scheme contributes to cellular FASN function, or if other post-translational modification enzymes use RNA-binding to localize to sites of protein synthesis.
m6A RNA modification is necessary for cellular survival (Geula et al., 2015). Whether cells use m6A to mark viruses, or whether RNA viruses adopt m6A to promote their replication appears virus- and cell-type specific (Courtney et al., 2017; Gokhale et al., 2016; Lichinchi et al., 2016). Recent screens identified more m6A-binding proteins, and proteins repelled by m6A (Edupuganti et al., 2017; H. Huang et al., 2018). Thus, the global role of m6A involves combinatorial or competing activities of a growing list of m6A-sensitive RBPs. The reports identifying novel m6A-RBPs investigated steady-state conditions. As many ISGs are also RBPs, there are likely unidentified m6A-sensitive RBPs relevant to innate immunity.
YTHDF1 has a distinct effect on early and late CHIKV replication compared to YTHDF2–3, even though they bind the same RNA modification. YTHDF1 has a more persistent interaction with CHIKV than YTHDF2–3 by immunoblot, which may explain its antiviral potency (see Figure 4C). These observations are consistent with the cellular roles of YTHDF1–3, which also differ: YTHDF1 promotes translation of mRNAs (X. Wang et al., 2015), while YTHDF2 destabilizes mRNA (X. Wang et al., 2014). Whether YTHDF1–3 have similar functions on viral RNAs remains unclear. m6A modifications are present on mRNA and viral RNA, so our observations with CHIKV may be an indirect consequence of YTHDF1–3 acting on mRNA.
Previous efforts to identify novel RBPs used steady-state conditions (Baltz et al., 2012; Castello et al., 2012). VIR-CLASP uses biologically relevant conditions to identify RBPs that may be inactive unless stimulated. One example is that VIR-CLASP identified a novel RNA-binding function of an innate immune PRR. While IFI16 is known to recognize pathogen-associated DNA, our results demonstrate that it can also bind cellular RNA and viral RNA. It is unclear how IFI16 distinguishes between viral and host nucleic acids and whether its functions are distinct when binding foreign DNA versus foreign RNA.
The primary CHIKV interactome contains ~600 distinct proteins across 6 conditions, and the primary IAV interactome at 1 hpi contains ~300 proteins. They include known pro- and antiviral proteins and host proteins involved in cellular homeostasis. Though the high viral inoculum (Kummer et al., 2014; Trinh et al., 2013) used in VIR-CLASP may raise false-positives, all validation experiments used MOI 10 or less. We cannot rule out the possibility that for some viruses, cellular RNA can be packaged stochastically with viral RNA. However, for IAV (Noda et al., 2018) and CHIKV (Kim et al., 2011), little cellular RNA is packaged (IAV: < 3% (Noda et al., 2018), CHIKV: < 1.3% (Kim et al., 2011)) (Table S6). For CHIKV, our data also indicate that few cellular RBPs exist in the viral particle (< 140 proteins with an intensity value > 0 in one or both biological replicates); only 40 RBPs in one or both replicates of the virion mass-spec had identical peptides found by VIR-CLASP post-infection (Figure 1D, Tables S2 and S4). For viruses that are known to package significant amounts of cellular RNA, use of VIR-CLASP may need additional optimization.
Few treatments or vaccines exist for emerging RNA viruses, in part due to their high mutation rates (Steinhauer and Holland, 1987). The identification of RBPs from VIR-CLASP can provide insights into RNA biology, improve our understanding of post-transcriptional gene regulation in host and pathogen, and lead to identification of new targets for therapeutic intervention.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Manuel Ascano (manuel.ascano@vanderbilt.edu)
Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Manuel Ascano (manuel.ascano@vanderbilt.edu). All plasmids and stable cell lines generated in this study are available without restrictions from the Lead Contact and/or through Addgene.
Data and Code Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Deutsch et al., 2017) via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD015863. The RNA-Seq data have been deposited to National Center for Biotechnology Information via Sequence Read Archive (SRA); BioProject ID: PRJNA558784. All code used for proteomic analysis and figure generation is accessible at https://github.com/Ascano-Lab.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and culture
All cell lines except Huh-7 (male) were obtained from ATCC. BHK-21 cells (male), Vero cells (female), A549 cells (male) and HEK-293T cells (female) were maintained in DMEM (Gibco) supplemented to contain 10% fetal bovine serum (FBS from Peak Serum) and 2 mM L-Glutamine (Gibco). U2OS cells (female) were cultured in McCoy’s 5A medium (Gibco) with 10% FBS and 2 mM L-Glutamine. CRISPR knock-in cell line Huh-7 HA-EMC4 (clon: DU12C3) and knockout cell line Huh-7 EMC4 KO (clon: DU13A1) were generated as previously described (Barrows et al., 2019). Cells were treated with the following reagents: Dinaciclib (HY-10492) was from MedChemExpress; Actinomycin D (A9415) and Cerulenin (219557) were from Sigma; Ruxolitinib Phosphate (sc-396768) was from Santa Cruz Biotech.
Viruses
CHIKV strain 181/25 was provided by Terence S. Dermody (University of Pittsburgh School of Medicine). CHIKV was propagated in BHK-21 cells with or without 1mM 4-thiouridine (4SU). Virus stocks were purified by ultracentrifugation of clarified supernatants through a 20% sucrose cushion in TNE buffer (50 mM Tris-HCl [pH 7.2], 0.1 M NaCl, and 1 mM EDTA) at ~125,000 × g for 4 hr in a Beckman SW32Ti rotor. To remove remaining free 4SU, virus pellets were washed three times with TNE buffer, and then re-suspended in virus dilution buffer (DMEM medium containing 10 mM HEPES [Gibco] supplemented to contain 1% FBS), aliquoted, and stored at −70°C. Virus titers were determined by plaque assay using Vero cells.
Encephalomyocarditis virus (EMCV; VR-129B) and zika virus (ZIKV; strain: PRVABC59; VR-1843) were obtained from ATCC, murine hepatitis virus (MHV; strain: MHV-A59) was provided by Mark R. Denison (Vanderbilt University Medical Center), influenza A virus (IAV; strain: California/7/2004 (H3N2)), vesicular stomatitis virus (VSV; strain: Indiana) and rift valley fever virus (RVFV; strain: MP-12) were provided by Thomas G. Voss and James E. Crowe (Vanderbilt Vaccine Center). EMCV, ZIKV, VSV and RVFV were propagated in Vero cells; MHV was propagated in DBT-9 cells; IAV was propagated with TPCK trypsin (2 μg/ml, Pierce) in MDCK cells. 4SU was added during propagation, and the viruses were purified as with CHIKV.
METHOD DETAILS
VIR-CLASP
For VIR-CLASP, cells were infected with 1000 MOI of non-labeled or 4SU-labeled virus for 1 hr at 4°C and uninfected virus was washed away with cold PBS. The infected cells were incubated for the indicated times at 37°C, prior to 365 nm ultraviolet irradiation. For pretreatment with interferon, IFN was added to the media 16 hr before viral infection.
For CLASP, 100 μM 4SU with or without IFN (500 U/mL) was added to the cells and incubated for 16-hr before irradiation. One 15-cm plate was used for each CLASP experiment.
UV365 nm crosslinking
To irradiate with UV365nm, the growth medium was removed and cells were washed with PBS. Cells were irradiated on ice with 365 nm UV light (0.6 J/cm2 × 2 times) in a Stratalinker 2400 (Stratagene). Cells were scraped off in 2.5 ml PBS per plate.
CLASP
Cells were lysed in denaturation buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2.5% SDS, 0.66% NP-40), incubated for 10-min at 95 °C and subsequently slowly cooled to 25°C. Crosslinked RNA-protein complexes were purified by Solid-Phase Reversible Immobilization (SPRI) (Hawkins et al., 1994) beads (GE Healthcare, cat# 65152105050250) under denaturing SPRI buffer (30 mM Tris-HCl, pH 6.8, 6% glycerol, 1.5% SDS, 0.4% NP-40, 1 M NaCl, 8% PEG-8000). To each sample, 0.66× (e.g. 660 μl of beads for 1ml of sample) of SPRI beads (1mg/ml SPRI beads in 10 mM Tris-HCl, pH 8.0, 1 M NaCl, 18% PEG-8000, 1 mM EDTA and 0.055% Tween 20) were added, and samples were incubated at room temperature for 10 minutes. The SPRI beads and complexes were washed 5 times with denaturing SPRI buffer. The crosslinked RNA-protein complexes were eluted for 5min at 37 °C in denaturation buffer (lysis buffer). To reduce non-specific binding on the beads, SPRI purification was repeated.
Benzonase digestion of RNA from crosslinked RNA-protein complexes
An equal volume of 4x Benzonase buffer (80 mM Tris-HCl, pH 7.5, 600 mM NaCl, 20 mM MgCl2, 4 mM DTT, 40% Glycerol) and 2x volume of water were added to eluted samples, followed by the addition of Benzonase (EMD Millipore, cat# 70746–4) to a final concentration of 50 U/ml, and incubation for 2 hr at 37°C. Proteins were precipitated by methanol and chloroform and then re-suspended in 2x NuPAGE LDS Sample Buffer (Thermo Fisher Scientific, cat# NP0007) with 50 mM DTT.
Proteinase K digestion of protein from crosslinked RNA-protein complexes
To measure the efficiency of total RNA purification, with 10% of eluted samples from CLASP, 0.1mg/ml proteinase K (3115879001, Sigma) was treated in proteinase K buffer (50 mM Tris-HCl, pH 7.5, 6.25 mM EDTA-NaOH, pH 8.0, 75 mM NaCl and 1% SDS) for 2 hr at 55°C. RNA was purified using TRIzol™ LS Reagent (Ambion). The RNA was resolved on agarose gel, visualized (ChemiDoc™ MP), and analyzed using ImageJ.
Viral titer with UV365nm crosslinking
Plaque assay with UV365nm crosslinking for CHIKV
Plaque assay with UV365nm crosslinking was performed on U2OS cells. U2OS cells were plated 1 day before. IFN was added to the media 16 hr before viral infection. CHIKV was serially diluted in DMEM medium with 1% FBS and 10mM HEPES and absorbed to U2OS cells for 1 hr at 4 °C. After plates were washed with PBS, infected cells were incubated in cell growth medium at 37 °C until UV365 nm crosslinking. At the indicated time point, the growth medium was removed and cells were washed with PBS. Cells were irradiated on ice with 365 nm UV light (0.6 J/cm2 × 2 times) in a Stratalinker 2400 (Stratagene). Then, cells were overlaid with DMEM with 0.6% SeaPlaque™ Agarose (Lonza, cat# 50100) containing 5% FBS and incubated for 72 hr. Cells were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The resulting plaques were counted.
TCID50 assay of UV365nm cross-linked IAV
A549 cells were infected with 1000 PFU of IAV for 1 hr at 37 °C in DMEM medium with 10 mM HEPES, TPCK trypsin (0.5 μg/ml) and 0.125% BSA (sigma, cat# A8412). After plates were washed with PBS, infected cells were incubated in DMEM medium with 10 mM HEPES, TPCK trypsin (0.5 μg/ml) and 0.125% BSA at 37 °C until UV365 nm crosslinking. At the indicated time point, the growth medium was collected and cells were washed with PBS. Cells were irradiated on ice with 365 nm UV light (0.6 J/cm2 × 2 times) in a Stratalinker 2400 (Stratagene). Then, the collected medium was returned into the culture dish of infected cells. At 8 hr post infection, the growth medium was collected and performed TCID50 assay on MDCK cells.
Mass Spectrometric Analysis
Gel lanes were cut and diced into 1mm3 cubes. Proteins were treated for 30 minutes with 45 mM DTT, and available Cys residues were carbamidomethylated with 100mM iodoacetamide for 45 minutes. Gel pieces were further destained with 50% MeCN in 25mM ammonium bicarbonate, and proteins were digested with trypsin (10ng/uL) in 25mM ammonium bicarbonate overnight at 37°C. Peptides were extracted by gel dehydration with 60% MeCN, 0.1% TFA, the extracts were dried by speed vac centrifugation, and reconstituted in 0.1% formic acid. Peptides were analyzed by LC-coupled tandem mass spectrometry (LC-MS/MS). An analytical column was packed with 22cm of C18 reverse phase material (Jupiter, 3 μm beads, 300Å, Phenomenox) directly into a laser-pulled emitter tip. Peptides were loaded on the capillary reverse phase analytical column (360 μm O.D. × 100 μm I.D.) using a Dionex Ultimate 3000 nanoLC and autosampler. The mobile phase solvents consisted of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). Peptides were gradient-eluted at a flow rate of 350 nL/min, using a 120-minute gradient. The gradient consisted of the following: 1–98 min, 2–40% B; 98–108 min, 40–95% B; 108–110 min, 95% B; 110–111 min, 95–2% B; 111–120 min (column re-equilibration), 2% B. A Q Exactive Plus mass spectrometer (Thermo Scientific), equipped with a nanoelectrospray ionization source, was used to mass analyze the eluting peptides using a data-dependent method. The instrument method consisted of MS1 using an MS AGC target value of 3e6, followed by up to 15 MS/MS scans of the most abundant ions detected in the preceding MS scan. The MS2 AGC target was set to 1e5, dynamic exclusion was set to 20s, HCD collision energy was set to 28 nce, and peptide match and isotope exclusion were enabled.
Bioinformatics analysis of Mass Spectrometry data
Raw data files from the LC-MS/MS instrument were processed and searched using MaxQuant (v1.5.0.35) (Cox and Mann, 2008) to generate peak lists and identify peptide-spectrum matches. Searches were performed using a Uniprot/Swissprot database for Homo sapiens with only reviewed proteins included (downloaded on Feb. 28th, 2018), with added sequences for Benzonase nuclease (Uniprot #P13717), and for CHIKV proteins (Capsid, E1, E2, E3, 6k, nsP1, nsP2, nsP3, nsP4) from strain 181/25 (TSI-GSD-218), and IAV proteins (H9XN78, 79, 80, 81, 83, 84, 85). For the purified CHIKV viral particles, the database used was the Uniprot reference proteome for Mesocricetus auratus (UP000189706- downloaded July 26th, 2019, with one protein sequence per gene) with sequences added for CHIKV proteins (Capsid, E1, E2, E3, 6k, nsP1, nsP2, nsP3, nsP4) from strain 181/25 (TSI-GSD-218). The search parameters for Andromeda were: full tryptic specificity, two missed cleavages allowed, carbamidomethyl (C) fixed modification, and acetylation (N terminal) variable modification. Match between runs was selected, and LFQ normalization was performed in separate parameter groups (+IFN and +4SU, −IFN and +4SU, +IFN and −4SU, and −IFN and −4SU). All other settings used were default, resulting in a protein FDR of < 1% for each dataset. To define the set of pre-replicated CHIKV interacting proteins, we computed peptide intensity ratios between +4SU and −4SU samples for proteins with at least two distinct peptides (Garcia-Moreno et al., 2019; Sysoev et al., 2016). The average log2-intensity ratio for each protein was then tested to be different from 0 using the moderated t-test implemented in the R/bioconductor package limma (Smyth, 2004). P-values were then corrected for multiple testing using the Benjamini-Hochberg method. Proteins with an adjusted p-value < 0.01 and a fold change greater than five were classified as “VIR-CLASP RBPs”.
Due to the low complexity of the −4SU samples, peptide intensity ratios could not be calculated for all proteins. For proteins with intensity values of zero in the −4SU samples, we performed a semiquantitative method that makes the assumption that peptides with no intensity values are below the detection threshold (Garcia-Moreno et al., 2019; Sysoev et al., 2016). This approach determines the number of replicates for +4SU and −4SU samples in which a peptide has an intensity value. For CHIKV, this leads to a matrix of 12 different groups, for peptides that were detected in 0, 1, 2, or 3 +4SU samples and 0, 1, or 2 −4SU samples. For IAV, the matrix contains 9 different groups, representing the 2 replicates performed for +4SU and −4SU each. The FDRs were estimated as described (Sysoev et al., 2016), as the ratios resulting from the division of the transposed matrix. A protein is determined to be a “candidate VIR-CLASP RBP” if it comprises peptides found in cells with an FDR < 0.01.
Correlations between replicates were performed using Pearson correlation and the “pairwise.complete.obs” option in the base R cor function to allow for missing values. Mapping of GO biological process, molecular function, and cellular component, and KEGG pathways was performed using the R/bioconductor packages clusterProfiler (Yu et al., 2012) and DOSE (Yu et al., 2015). The background used was all human genes, and statistical significance was determined using the Fisher’s exact test and Benjamini-Hochberg method to correct for multiple testing. Analysis of enriched PFAM domains was performed using the DAVID Bioinformatics Resource (D. W. Huang et al., 2009a; 2009b) using the Fisher’s exact test and Bonferroni correction for multiple testing. Visualization of overlaps between datasets was performed in R using the UpSetR package (Conway et al., 2017) for UpSet diagrams, and the VennDiagram package for 2-way Venn diagrams. All other data visualizations were made using the R package ggplot2 (Wickham, 2016). All code for the R analysis can be found at https://github.com/Ascano-Lab.
Virion RNA Sequencing and Alignment
CHIKV was purified by ultracentrifugation through a 20% sucrose cushion in TNE buffer as described in method section of viruses. CHIKV was re-suspended in TNE buffer and further purified twice by discontinuous sucrose gradient (30%–60%) in TNE buffer at ~125,000 × g for 3 hr in a Beckman SW32Ti rotor, and concentrated by 30% sucrose cushion. Trizol reagent (Ambion) was added to the virus pellets and viral RNA was extracted. Total RNA was converted into cDNA and sequenced using NEBNext DNA Library Prep Kit for Illumina on the Illumina NovaSeq6000 platform using PE150 at the Vanderbilt Technologies for Advanced Genomics (VUMC VANTAGE). Fastq files were pre-processed with trim-galore with the default settings (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove any adapter contamination and then aligned to the Chikungunya virus strain genome (TSI-GSD-218, GenBank: L27661.3) and Mesocricetrus auratus (GCA_000349665.1) using STAR (“STAR: ultrafast universal RNA-seq aligner.,” 2013) with the default settings. Reference annotation file (GTF) does not exist file for Chikungunya virus strain 181/25 (TSI-GSD-218).
Antibodies and immunoblotting
The antibody to YTHDF1 (anti-YTHDF1; 17479–1-AP) was from Proteintech; anti-FASN(3180), anti-GM130 (12480) and KDM3B (3100) were from Cell Signaling; anti-YTHDH2 (ab170118), anti-YTHDH3 (ab103328), anti-ELAVL1 (ab200342) and anti-TUBA4A (ab7291) were from abcam; anti-IFI16 (sc-8023) and LARP1 (sc-515873 were from Santa Cruz Biotechnology; anti-Flag (F1804) was from Sigma-Aldrich; anti-HA (901502) and WDHD1 (630302) were from BioLegend; anti-FXR1 (MAB2160) was from Millipore.
Samples were separated by SDS-PAGE. After electrophoresis, proteins were semi-dry transferred (Bio-Rad) to nitrocellulose membranes (Hybond-ECL, GE Life Science). Protein membranes were taken through a standard immunoblot protocol followed by enhanced chemiluminescent detection (Luminata Forte ECL, Millipore) using a chemiluminescence imaging system (ChemiDoc MP, Bio-Rad).
Methylated RNA immunoprecipitation (MeRIP)
CHIKV was purified by ultracentrifugation through a 20% sucrose cushion in TNE buffer as described in method section of viruses. CHIKV was re-suspended in TNE buffer and further purified twice by discontinuous sucrose gradient (30%−60%) in TNE buffer at ~125,000 × g for 3 hr in a Beckman SW32Ti rotor, and concentrated by 30% sucrose cushion. Trizol reagent (Ambion) was added to the virus pellets and viral RNA was extracted. The RNA was fragmented by RNaseT1 treatment (0.8 U/μl) for 15 min at 22°C. The fragmented RNA was dephosphorylated with CIP and radiolabeled with T4 PNK and [γ−32P]-ATP. The RNA was incubated with m6A antibody (202 003, Synaptic systems) for 2 hr at 4 °C and pulled down by magnetic protein A beads (Invitrogen). The RNA was eluted in Trizol and separated by 12% UREA gel and analyzed by phosphorimaging.
N6-methyladenosine (m6A) mapping
CHIKV was purified and immunoprecipitated with the m6A antibody described above, with the following changes: the purified CHIKV RNA was treated with RNaseT1 (0.1 U/ μl) for 5 min at 4°C. The fragmented RNA was then extracted with phenol:chloroform and immunoprecipitated with the m6A antibody. The RNA was eluted in Trizol, reverse transcribed using SuperScript III (ThermoFisher) with random hex primers, and quantified by RT-qPCR (see Methods section RT-qPCR analysis) using primers tiled across the CHIKV genome in 1000 nt segments (Supplementary Table 8). Target Ct values were normalized to Input Ct values. Relative enrichment of CHIKV regions in the m6A IP over the IgG IP was then calculated using the ΔΔCt method (2ΔΔCt).
Plasmids and stable cell line
The Flag-HA-tag lentiviral inducible expression vector pLenti CMVtight Blast Flag-HA-DEST was constructed by insertion of Flag-HA-tag from pFRT_TO_DEST Flag-HA (#26361, Addgene) into the plasmid pLenti CMVtight Blast DEST (w762–1) (#26434, Addgene). The genes YTHDF1 (NM_017798.3), YTHDF2 (NM_001172828.1), YTHDF3 (NM_152758.4), FASN (BC_063242.1) and IFI16 (BC_017059.1) were amplified by PCR from the plasmid HsCD00378781 for YTHDF1, HsCD00376823 for YTHDF2 and HsCD00376805 for YTHDF3 (the DNA Resource Core at Harvard Medical School), 6172538 for FASN and 3914632 for IFI16 (Mammalian Gene Collection (MGC) at Dharmacon) with gene-specific primers containing an attB sequence. To generate a catalytic mutatnt of FASN (FASNmut; S581A), two-step overlap-extension PCR was used. Using the Gateway cloning system (Invitrogen), the amplified genes were cloned into pLenti CMVtight Blast Flag-HA-DEST vector.
For producing lentivirus, the plasmids were transfected with packaging vectors psPAX2 (12260, Addgene) and pMD2.G (12259, Addgene) by Lipofectamine 2000 (Invitrogen) in HEK-293T cells. To generate inducible expression cell lines, first a stable tetracycline transactivator (rtTA) expressing U2OS cell line was created by transducing a lentivirus from the plasmid rtTA-N144 (66810, Addgene). Then the cDNA expressing lentiviruses were transduced into rtTA expressing U2OS cells and drug selected.
siRNA knockdown
siRNAs used in this study are listed in Supplementary Table 8. siRNAs were transfected at a final concentration of 20 nM using Lipofectamine® RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Cells were incubated at 37 °C and 5% CO2 for 72-h before infection with indicated MOI. For rescue experiments, FASN expressing plasmid was transfected by Lipofectamine 3000 (Invitrogen) or stable cell lines of Flag-HA tagged YTHDF1 or IFI16 were used.
RT–qPCR analysis
RNA was collected from infected cells using Trizol (Ambion). The RNA concentration was determined using a NanoDrop 2000 (ThermoFisher). Equal amounts of total RNA were reverse-transcribed using SuperScript III (ThermoFisher) with random hex primers or CHIKV strand-specific primers (Plaskon et al., 2009). Real-time PCR reactions were done with FastSYBR Green Plus Master Mix (Applied Biosystems) or TaqMan® Fast Advanced Master Mix (Applied Biosystems) using a StepOnePlus qPCR machine (Applied Biosystems). Oligonucleotides used in this study are listed in Supplementary Table 8. Target Ct values were normalized to TUBA1A Ct values and used to calculate ΔCt. Relative mRNA expression of target genes was then calculated using the ΔΔCt method (2ΔΔCt) except cellular RNA expression in CHIKV virion was calculated using the ΔCt method. To generate a standard curve, a plasmid containing a cDNA copy of the CHIKV strain 181/25 (pSinRep5–181/25ic, #60078, Addgene) was used.
PAR-CLIP
An abbreviated PAR-CLIP protocol was used to determine the extent of crosslinkable RNA that would associate with RBPs of interest; no RNA isolation, cDNA library preparation, and sequencing was performed for this study. Briefly, U2OS cells stably expressing Flag-HA-tagged FASN were labeled with 100 μM 4SU for 16hr, UV365nm irradiated, and lysed with NP-40 lysis buffer. Unlabeled cells were used as control. Immunoprecipitation was carried out with Dyna-protein G beads conjugated to the Flag M2 antibody, and then samples were digested with RNase T1. Bound RNPs were dephosphorylated with CIP and radiolabeled with T4 PNK and [γ−32P]-ATP. The crosslinked RNP complexes were resolved on a NuPAGE 4–12% Bis-Tris Protein Gel (Invitrogen), transferred to nitrocellulose membranes, and analyzed by phosphorimaging and immunoblotting.
Crosslinking RNA immunoprecipitation
For immunoprecipitation of Flag-tagged proteins, U2OS cells overexpressing Flag-HA-tagged IFI16 were infected with 4SU-labeled CHIKV for 1hr at 4°C and uninfected virus was washed away with cold PBS. The infected cells were incubated for 15 minutes at 37 °C, and then crosslinked with UV365 nm. Cells were lysed in RIPA buffer (50 mM Tris-HCl (pH 7.5), 150 mM KCl, 1.0% TritonX-100, 0.5% sodium deoxycholate, 0.1% SDS, 0.5 mM DTT, 200 U/ml SUPERase In and protease inhibitor cocktail). The lysate were diluted 10 fold in binding buffer (50 mM Tris-HCl (pH 7.5), 150 mM KCl, 1.0% TritonX-100, 0.5 mM DTT, 200 U/ml SUPERase In and protease inhibitor cocktail) and then were bound for 1 hr at 4 °C to Dyna-protein G beads with Flag M2 antibody or control IgG. After samples were washed with high salt washing buffer (Binding buffer with 0.05% TritonX-100 and 500mM KCl), the bound Flag-tagged proteins were eluted by incubation of the bound beads for 30 min with 150 ng/μl of 3xFlag peptides in the binding buffer. RNA and protein from the immunoprecipitated samples and from the cell lysates (input) were analyzed by RT-qPCR or immunoblot.
Luciferase reporter assay
The nsP3 fusion nanoLuc reporter CHIKV (CHIKV-nLuc) was created as described previously, except that the plasmid containing a cDNA copy of the CHIKV strain 181/25 (pSinRep5–181/25ic) was used (Sun et al., 2014). U2OS cells were infected with 10 MOI of CHIKV-nLuc. After 1 h of incubation at 4°C, cells were washed with PBS, and appropriate medium was replaced before cells were incubated. At indicated time points post infection, infected cells were washed with PBS, and cell lysates were harvested with 1× passive lysis buffer for luciferase activity assays using a Nano-Glo luciferase assay or Nano-Glo Dual-Luciferase Reporter assay as recommended by the manufacturer (Promega). The luciferase activity was normalized to protein concentration. For dual luciferase assay, firefly luciferase activity was normalized to its mRNA levels, then the ratio of the nano luciferase activity to the firefly luciferase activity (Nluc/Fluc) was calculated.
QUANTIFICATION AND STATISTICAL ANALYSIS
Using the GraphPad PRISM 8 software, a two-tailed Student’s t-test and one-way ANOVA (Tukey) were used for statistical analysis of all data presented except mass-spectrometry and bioinformatics analysis. Numbers of biological replicates of assays (n) are provided and defined within the corresponding figures or figure legends. Error bars shown in the Figures represent means ± SD.
Supplementary Material
Table S1. RNA-seq and RT-qPCR Analysis of the Purified CHIKV Viral Particles, Related to Figure 1
Table S2. Proteomics analysis from all Replicates and Conditions of VIR-CLASP with CHIKV, Related to Figure 2
Table S3. Proteomics analysis from VIR-CLASP with IAV, Related to Figure 2
Table S4. The Proteins Identified using VIR-CLASP with the CHIKV Viral Particle, Related to Figure 1
Table S5. Functional analyses of the CHIKV pre-replicated interactome, Related to Figure 3
Table S6. Functional analyses of the IAV pre-replicated interactome, Related to Figure 3
Table S7. The Candidate CHIKV Proteins that were also Identified in Previous siRNA Screens for CHIKV Pro- and Anti-Viral Factors, Related to Figure 4
Table S8. The Primers and siRNAs Used in this Study, Related to the STAR Methods
Highlights:
VIR-CLASP identifies host protein interactions with incoming RNA viral genomes
VIR-CLASP can reveal the early interactomes of seven different viral families
The CHIKV genome binds to distinct proteins under different conditions and times
(Non)canonical RBPs like FASN, IFI16 and YTHDF1 uniquely regulate CHIKV replication
ACKNOWLEDGMENTS
We would like to thank Matthew Albertolle for technical help in the MS analysis; Neil Sprenkle, James Held for technical help in the molecular studies; Dr. Thomas Voss and Dr. James E. Crowe Jr. (Vanderbilt University Medical Center) for IAV, VSV and RVFV; Dr. Mark R. Denison (Vanderbilt University Medical Center) for MHV; and Dr. Terence S. Dermody (University of Pittsburgh School of Medicine) for CHIKV. Finally, we would like to thank members of the Ascano laboratory for their support, collegiality, and critical review of the manuscript. This work was supported by the National Institutes of Health 1R35GM119569-01 (M.A.), CTSA award No.UL1TR000445 from the National Center for Advancing Translational Sciences (B.K. and M.A.), Vanderbilt University Dept. Biochemistry start-up funds (M.A.), the Chemical Biology of Infectious Disease training grant 5T32AI11254-02 (S.A.), and the Chemistry-Biology Interface training grant 5T32GM065086-14.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors have no competing financial or non-financial interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and eight tables and can be found with this article online at ……
REFERENCES
- Area E, Martín-Benito J, Gastaminza P, Torreira E, Valpuesta JM, Carrascosa JL, Ortín J, 2004. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc Natl Acad Sci USA 101, 308–313. doi: 10.1073/pnas.0307127101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M, 2012. The mRNA-bound proteome and its global occupancy profile on proteincoding transcripts. Molecular Cell 46, 674–690. doi: 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Barrows NJ, Anglero-Rodriguez Y, Kim B, Jamison SF, Le Sommer C, McGee CE, Pearson JL, Dimopoulos G, Ascano M, Bradrick SS, Garcia-Blanco MA, 2019. Dual roles for the ER membrane protein complex in flavivirus infection: viral entry and protein biogenesis. Scientific Reports 9, 9711. doi: 10.1038/s41598-019-45910-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beld J, Lee DJ, Burkart MD, 2014. Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering. Molecular BioSystems 11, 38–59. doi: 10.1039/C4MB00443D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, 2004. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol 5, 1109–1115. doi: 10.1038/ni1125 [DOI] [PubMed] [Google Scholar]
- Bowman EJ, Siebers A, Altendorf K, 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA 85, 7972–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW, 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406. doi: 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Chow KT, Gale M, Loo Y-M, 2018. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu. Rev. Immunol 36, 667–694. doi: 10.1146/annurev-immunol-042617-053309 [DOI] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N, 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. doi: 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR, 2017. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe 22, 377–386.e5. doi: 10.1016/j.chom.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M, 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology 26, 1367–1372. doi: 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T-Y, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi P-Y, Diamond MS, 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature Publishing Group 468, 452–456. doi: 10.1038/nature09489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Aramini JM, Ma L-C, Krug RM, Arnold E, 2010. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol 17, 530–538. doi: 10.1038/nsmb.1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas S, Peredo J, Marión RM, Sánchez C, Ortín J, 2010. Human Staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication. J. Virol 84, 7603–7612. doi: 10.1128/JVI.00504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, Moritz RL, Carver JJ, Wang M, Ishihama Y, Bandeira N, Hermjakob H, Vizcaíno JA, 2017. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Research 45, D1100–D1106. doi: 10.1093/nar/gkw936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rötig A, Crow YJ, Rice GI, Duffy D, Tamby C, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, Proudfoot NJ, 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242. doi: 10.1038/s41586-018-0363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson AM, Anderson JR, Barnhart MD, Sokoloski KJ, Oko L, Opyrchal M, Galanis E, Wilusz CJ, Morrison TE, Wilusz J, 2012. Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J. Biol. Chem 287, 36229–36238. doi: 10.1074/jbc.M112.371203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang S-Y, Baltissen MPA, Jansen PWTC, Rossa M, Müller M, Stunnenberg HG, He C, Carell T, Vermeulen M, 2017. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol 1–15. doi: 10.1038/nsmb.3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD, 2019. The Pfam protein families database in 2019. Nucleic Acids Research 47, D427–D432. doi: 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M, Noerenberg M, Ni S, Järvelin AI, González-Almela E, Lenz CE, Bach-Pages M, Cox V, Avolio R, Davis T, Hester S, Sohier TJM, Li B, Heikel G, Michlewski G, Sanz MA, Carrasco L, Ricci EP, Pelechano V, Davis I, Fischer B, Mohammed S, Castello A, 2019. System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection. Molecular Cell 74, 196–211.e11. doi: 10.1016/j.molcel.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T, 2014. A census of human RNA-binding proteins. Nature Publishing Group 15, 829–845. doi: 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-Haim MS, Eyal E, Yunger S, Pinto Y, Jaitin DA, Viukov S, Rais Y, Krupalnik V, Chomsky E, Zerbib M, Maza I, Rechavi Y, Massarwa R, Hanna S, Amit I, Levanon EY, Amariglio N, Stern-Ginossar N, Novershtern N, Rechavi G, Hanna JH, 2015. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006. doi: 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N, 2004. mTOR signaling to translation. Curr. Top. Microbiol. Immunol 279, 169–197. doi: 10.1007/978-3-642-18930-2_11 [DOI] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, Ilkayeva OR, Law BA, Holley CL, Garcia-Blanco MA, Evans MJ, Suthar MS, Bradrick SS, Mason CE, Horner SM, 2016. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 20, 654–665. doi: 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A-C, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T, 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141. doi: 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins TL, O’Connor-Morin T, Roy A, Santillan C, 1994. DNA purification and isolation using a solid-phase. Nucleic Acids Research 22, 4543–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Sidoli S, Warneford-Thomson R, Tatomer DC, Wilusz JE, Garcia BA, Bonasio R, 2016. High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Molecular Cell 64, 416–430. doi: 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G, 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. U.S.A 107, 17345–17350. doi: 10.1073/pnas.1010811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y-J, Wang Y-M, Lu J-W, Wu T-Y, Lin L-I, Kuo S-C, Lin C-C, 2015. Suramin Inhibits Chikungunya Virus Entry and Transmission. PLoS ONE 10, e0133511. doi: 10.1371/journal.pone.0133511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37, 1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu Y-C, Hüttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan J-L, He C, Yang J, Chen J, 2018. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature Cell Biology 1–18. doi: 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Han M, Meng L, Chen X, 2018. Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl. Acad. Sci. U.S.A 115, E3879–E3887. doi: 10.1073/pnas.1718406115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, He C, 2013. Reversible RNA adenosine methylation in biological regulation. Trends in Genetics 29, 108–115. doi: 10.1016/j.tig.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AK, Witkowski A, Smith S, 1998. The malonyl/acetyltransferase and beta-ketoacyl synthase domains of the animal fatty acid synthase can cooperate with the acyl carrier protein domain of either subunit. Biochemistry 37, 2515–2523. doi: 10.1021/bi971886v [DOI] [PubMed] [Google Scholar]
- Karlas A, Berre S, Couderc TERES, Varjak M, Braun P, Meyer M, Gangneux N, Karo-Astover L, Weege F, Raftery M, nrich GUNSO, Klemm U, Wurzlbauer A, Bracher F, Merits A, Meyer TF, Lecuit M, 2016. A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat Commun 7, 1–14. doi: 10.1038/ncomms11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner K-P, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Mäurer AP, Müller E, Wolff T, Rudel T, Meyer TF, 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822. doi: 10.1038/nature08760 [DOI] [PubMed] [Google Scholar]
- Kim DY, Firth AE, Atasheva S, Frolova EI, Frolov I, 2011. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J. Virol 85, 8022–8036. doi: 10.1128/JVI.00644-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Morgan MA, Shatkin AJ, 1976. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol 20, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Ratcliff AN, Bhat M, Alwarawrah Y, Hughes P, Arcos J, Loiselle D, Torrelles JB, Funderburg NT, Haystead TA, Kwiek JJ, 2017. Cellular fatty acid synthase is required for late stages of HIV-1 replication. Retrovirology 1–12. doi: 10.1186/s12977-017-0368-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer S, Flöttmann M, Schwanhäusser B, Sieben C, Veit M, Selbach M, Klipp E, Herrmann A, 2014. Alteration of protein levels during influenza virus H1N1 infection in host cells: a proteomic survey of host and virus reveals differential dynamics. PLoS ONE 9, e94257. doi: 10.1371/journal.pone.0094257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic EM, Landry DM, Greco TM, Cristea IM, Thompson SR, 2013. Thiouracil cross-linking mass spectrometry: a cell-based method to identify host factors involved in viral amplification. J. Virol 87, 8697–8712. doi: 10.1128/JVI.00950-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H, 2014. UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 20, 1983–1992. doi: 10.1109/TVCG.2014.2346248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MMH, Lau Z, Cheung P, Aguilar EG, Schneider WM, Bozzacco L, Molina H, Buehler E, Takaoka A, Rice CM, Felsenfeld DP, MacDonald MR, 2017. TRIM25 Enhances the Antiviral Action of Zinc-Finger Antiviral Protein (ZAP). PLoS Pathog 13, e1006145. doi: 10.1371/journal.ppat.1006145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM, 2016. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nature Microbiology 1, 16011. doi: 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, McFadden G, 2015. SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J. Virol 89, 1925–1931. doi: 10.1128/JVI.02262-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J-W, Hsieh P-S, Lin C-C, Hu M-K, Huang S-M, Wang Y-M, Liang C-Y, Gong Z, Ho Y-J, 2017. Synergistic effects of combination treatment using EGCG and suramin against the chikungunya virus. Biochem. Biophys. Res. Commun 491, 595–602. doi: 10.1016/j.bbrc.2017.07.157 [DOI] [PubMed] [Google Scholar]
- Lu W, Tirumuru N, St Gelais C, Koneru PC, Liu C, Kvaratskhelia M, He C, Wu L, 2018. N6-methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J. Biol. Chem doi: 10.1074/jbc.RA118.004215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Ni G, Damania B, 2018. Innate Sensing of DNA Virus Genomes. Annu Rev Virol 5, 341–362. doi: 10.1146/annurev-virology-092917-043244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Murakami S, Nakatsu S, Imai H, Muramoto Y, Shindo K, Sagara H, Kawaoka Y, 2018. Importance of the 1+7 configuration of ribonucleoprotein complexes for influenza A virus genome packaging. Nat Commun 9, 54. doi: 10.1038/s41467-017-02517-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S, 1976. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev 40, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M, 2013. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog 9, e1003835. doi: 10.1371/journal.ppat.1003835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paruch K, Dwyer MP, Alvarez C, Brown C, Chan T-Y, Doll RJ, Keertikar K, Knutson C, McKittrick B, Rivera J, Rossman R, Tucker G, Fischmann T, Hruza A, Madison V, Nomeir AA, Wang Y, Kirschmeier P, Lees E, Parry D, Sgambellone N, Seghezzi W, Schultz L, Shanahan F, Wiswell D, Xu X, Zhou Q, James RA, Paradkar VM, Park H, Rokosz LR, Stauffer TM, Guzi TJ, 2010. Discovery of Dinaciclib (SCH 727965): A Potent and Selective Inhibitor of Cyclin-Dependent Kinases. ACS Med Chem Lett 1, 204–208. doi: 10.1021/ml100051d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perri JI, Rogell B, Schwarzl T, Stein F, Zhou Y, Rettel M, Brosig A, Hentze MW, 2018. Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat Commun 9, 4408–13. doi: 10.1038/s41467-018-06557-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA, 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 47, D442–D450. doi: 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SL, Soderblom EJ, Bradrick SS, Garcia-Blanco MA, 2016. Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication. MBio 7, e01865–15. doi: 10.1128/mBio.01865-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaskon NE, Adelman ZN, Myles KM, 2009. Accurate strand-specific quantification of viral RNA. PLoS ONE 4, e7468. doi: 10.1371/journal.pone.0007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintás-Cardama A, Vaddi K, Liu P, Manshouri T, Li J, Scherle PA, Caulder E, Wen X, Li Y, Waeltz P, Rupar M, Burn T, Lo Y, Kelley J, Covington M, Shepard S, Rodgers JD, Haley P, Kantarjian H, Fridman JS, Verstovsek S, 2010. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood 115, 3109–3117. doi: 10.1182/blood-2009-04-214957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky SR, Pegoraro G, Chī X, Dǒng L, Chiang C-Y, Jozwick L, Clester JC, Cooper CL, Courier D, Langan DP, Underwood K, Kuehl KA, Sun MG, Caì Y, Yú S, Burk R, Zamani R, Kota K, Kuhn JH, Bavari S, 2016. siRNA Screen Identifies Trafficking Host Factors that Modulate Alphavirus Infection. PLoS Pathog 12, e1005466–30. doi: 10.1371/journal.ppat.1005466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Tritsch SR, Kota K, Chiang C-Y, Dǒng L, Kenny T, Brueggemann EE, Ward MD, Cazares LH, Bavari S, 2015. Sphingosine kinase 2 is a chikungunya virus host factor colocalized with the viral replication complex. Emerg Microbes Infect 4, e61. doi: 10.1038/emi.2015.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ, 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Research 41, D1040–6. doi: 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samji T, 2009. Influenza A: understanding the viral life cycle. Yale J Biol Med 82, 153–159. [PMC free article] [PubMed] [Google Scholar]
- Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, Guo Z, Green S, Kowalik TF, Brass AL, 2016. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep 16, 232–246. doi: 10.1016/j.celrep.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM, 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485. doi: 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma-Walia N, Paul AG, Bottero V, Sadagopan S, Veettil MV, Kerur N, Chandran B, 2010. Kaposi’s sarcoma associated herpes virus (KSHV) induced COX-2: a key factor in latency, inflammation, angiogenesis, cell survival and invasion. PLoS Pathog 6, e1000777. doi: 10.1371/journal.ppat.1000777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH, 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650. doi: 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Silva LA, Dermody TS, 2017. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. Journal of Clinical Investigation 127, 737–749. doi: 10.1172/JCI84417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article3. doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Steinhauer DA, Holland JJ, 1987. Rapid evolution of RNA viruses. Annu. Rev. Microbiol 41, 409–433. doi: 10.1146/annurev.mi.41.100187.002205 [DOI] [PubMed] [Google Scholar]
- Sun C, Gardner CL, Watson AM, Ryman KD, Klimstra WB, 2014. Stable, high-level expression of reporter proteins from improved alphavirus expression vectors to track replication and dissemination during encephalitic and arthritogenic disease. J. Virol 88, 2035–2046. doi: 10.1128/JVI.02990-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Chin W-X, Han Q, Ichiyama K, Lee CH, Eyo ZW, Ebina H, Takahashi H, Takahashi C, Tan BH, Hishiki T, Ohba K, Matsuyama T, Koyanagi Y, Tan Y-J, Sawasaki T, Chu JJH, Vasudevan SG, Sano K, Yamamoto N, 2016. Characterization of RyDEN (C19orf66) as an Interferon-Stimulated Cellular Inhibitor against Dengue Virus Replication. PLoS Pathog 12, e1005357. doi: 10.1371/journal.ppat.1005357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysoev VO, Fischer B, Frese CK, Gupta I, Krijgsveld J, Hentze MW, Castello A, Ephrussi A, 2016. Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat Commun 7, 12128–11. doi: 10.1038/ncomms12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, Fitzgerald KA, Kurt-Jones EA, 2014. Interferon γ-inducible protein (IFI) 16 transcriptionally regulates type i interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J. Biol. Chem 289, 23568–23581. doi: 10.1074/jbc.M114.554147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh HV, Grossmann J, Gehrig P, Roschitzki B, Schlapbach R, Greber UF, Hemmi S, 2013. iTRAQ-Based and Label-Free Proteomics Approaches for Studies of Human Adenovirus Infections. Int J Proteomics 2013, 581862–16. doi: 10.1155/2013/581862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG, 2010. IFI16 is an innate immune sensor for intracellular DNA. Nature Publishing Group 11, 997–1004. doi: 10.1038/ni.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C, 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C, 2015. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-M, Lu J-W, Lin C-C, Chin Y-F, Wu T-Y, Lin L-I, Lai Z-Z, Kuo S-C, Ho Y-J, 2016. Antiviral activities of niclosamide and nitazoxanide against chikungunya virus entry and transmission. Antiviral Research 135, 81–90. doi: 10.1016/j.antiviral.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Lecuit M, 2015. Chikungunya virus and the global spread of a mosquito-borne disease. N. Engl. J. Med 372, 1231–1239. doi: 10.1056/NEJMra1406035 [DOI] [PubMed] [Google Scholar]
- Wickham H, 2016. ggplot2. Springer, Cham.
- Yang W, Hood BL, Chadwick SL, Liu S, Watkins SC, Luo G, Conrads TP, Wang T, 2008. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology 48, 1396–1403. doi: 10.1002/hep.22508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Dynan WS, 1998. Characterization of the RNA binding properties of Ku protein. Biochemistry 37, 1336–1343. doi: 10.1021/bi972100w [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y, 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Yan G-R, He Q-Y, 2015. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609. doi: 10.1093/bioinformatics/btu684 [DOI] [PubMed] [Google Scholar]
- Zhang N, Zhao H, Zhang L, 2019. Fatty Acid Synthase Promotes the Palmitoylation of Chikungunya Virus nsP1. J. Virol 93, 662. doi: 10.1128/JVI.01747-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. RNA-seq and RT-qPCR Analysis of the Purified CHIKV Viral Particles, Related to Figure 1
Table S2. Proteomics analysis from all Replicates and Conditions of VIR-CLASP with CHIKV, Related to Figure 2
Table S3. Proteomics analysis from VIR-CLASP with IAV, Related to Figure 2
Table S4. The Proteins Identified using VIR-CLASP with the CHIKV Viral Particle, Related to Figure 1
Table S5. Functional analyses of the CHIKV pre-replicated interactome, Related to Figure 3
Table S6. Functional analyses of the IAV pre-replicated interactome, Related to Figure 3
Table S7. The Candidate CHIKV Proteins that were also Identified in Previous siRNA Screens for CHIKV Pro- and Anti-Viral Factors, Related to Figure 4
Table S8. The Primers and siRNAs Used in this Study, Related to the STAR Methods
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Deutsch et al., 2017) via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD015863. The RNA-Seq data have been deposited to National Center for Biotechnology Information via Sequence Read Archive (SRA); BioProject ID: PRJNA558784. All code used for proteomic analysis and figure generation is accessible at https://github.com/Ascano-Lab.


