Abstract
Mouse sex chromosomes are enriched for co-amplified gene families, present in tens to hundreds of copies. Co-amplification of Slx/Slxl1 on the X chromosome and Sly on the Y chromosome are involved in dose-dependent meiotic drive, however the role of other co-amplified genes remains poorly understood. Here we demonstrate that the co-amplified gene family on the X chromosome, Srsx, along with two additional partial gene annotations, is actually part of a larger transcription unit, which we name Laidx. Laidx is harbored in a 229 kb amplicon that represents the ancestral state as compared to a 525 kb Y-amplicon containing the rearranged Laidy. Laidx contains a 25,011 nucleotide open reading frame, predominantly expressed in round spermatids, predicted to encode an 871 kD protein. Laidx has orthologous copies with the rat and also the 825-MY diverged parasitic Chinese liver fluke, Clonorchis sinensis, the likely result of a horizontal gene transfer of rodent Laidx to an ancestor of the liver fluke. To assess the male reproductive functions of Laidx, we generated mice carrying a multi-megabase deletion of the Laidx-ampliconic region. Laidx-deficient male mice do not show detectable reproductive defects in fertility, fecundity, testis histology, and offspring sex ratio. We speculate that Laidx and Laidy represent a now inactive X vs. Y chromosome conflict that occurred in an ancestor of present day mice.
Keywords: amplicon, X chromosome, horizontal gene transfer, male fertility, testicular germ cells, mouse, Genetics of Sex
The mouse has highly co-amplified gene families on the X and Y chromosomes (Soh et al. 2014). These co-amplified X- (Slx, Slxl1, Sstx, Srsx, Astx) and Y-linked (Sly, Ssty1, Ssty2, Srsy, Asty) gene families are predominantly expressed in post-meiotic testicular germ cells, suggesting an important function in reproduction and testicular germ cell development (Mueller et al. 2008; Soh et al. 2014; Szot et al. 2003; Reynard et al. 2009; Comptour et al. 2014; Touré et al. 2005). The co-amplification of gene families on the X and Y chromosomes is thought to have arisen because of meiotic drive, the unequal transmission of an allele to the next generation (Soh et al. 2014; Kruger et al. 2019; Rathje et al. 2019). Slx, Slxl1, and Sly are meiotic drivers, where increases in gene expression generates a competitive advantage of X- or Y-bearing sperm (Kruger et al. 2019; Cocquet et al. 2012). The Slx/Slxl1 gene family is also required for male fertility, highlighting how a co-amplified gene family can become essential for male fertility (Kruger et al. 2019; Cocquet et al. 2010). However, little is known about the biological functions or evolutionary origins of other X- and Y-linked co-amplified gene families.
We chose to explore the evolutionary origins and reproductive function of Srsx for multiple reasons. First, Srsx and Srsy share the highest level of nucleotide identity (∼95%) of the X- and Y-linked co-amplified gene families in mice (Soh et al. 2014). Second, it is unclear if Srsx encodes a protein. Third, while we know Srsx is present in ∼14 copies within a ∼2 Mb amplicon array on the X chromosome (Mueller et al. 2008; Bennett-Baker and Mueller 2017), the evolutionary origins of Srsx and the amplicon containing it are not well-defined. Finally, similar to Slx/Slxl1/Sly, Srsx is predominantly expressed in round spermatids (Mueller et al. 2008; Soh et al. 2014), suggesting a potential role in meiotic drive and male fertility.
Materials and Methods
BAC sequencing and assembly
BAC sequencing and assembly was performed as previously described (Vollger et al. 2019). Briefly, DNA from BAC RP23-106J7 was isolated using a High Pure Plasmid Isolation Kit (Roche Applied Science) following the manufacturer’s instructions. Approximately 1 μg of BAC DNA was sheared using a Covaris g-TUBE. Libraries were processed using the PacBio SMRTbell Template Prep kit following the protocol “Procedure and Checklist—20 kb Template Preparation Using BluePippin Size-Selection System” with the addition of barcoded SMRTbell adaptors. Library size was measured using a FEMTO Pulse. The pooled library was size-selected on a Sage PippinHT with a start value of 12,000 and an end value of 50,000. BACs were sequenced on a PacBio Sequel with version 3.0 chemistry on one SMRT cell and then demultiplexed using LIMA in SMRTlink6.0. Demultiplexed reads were run through the CCS algorithm in SMRTlink6.0. CCS reads were filtered for contaminating E. coli reads, and the resulting filtered fasta file was used as input for assembly using Canu v1.8.
Dot plots
Dot plots showing sequence identity within one sequence and between two sequences were generated using fastdotplot, a custom Perl script that can be found at https://www.pagelab.wi.mit.edu/materials-request. Nucleic acid sequence alignments between Mus musculus, Rattus norvegicus, and Clonorchis sinensis was performed using the blastn algorithm for somewhat similar sequences (Agarwala et al. 2018). Amino acid alignments between Mus musculus, Rattus norvegicus, and Clonorchis sinensis was performed using the blastp algorithm (Agarwala et al. 2018).
mRNA-seq
Total RNA quality was assessed using the 2100 Bioanalyzer (Agilent). 400ng of total RNA (RIN >6) from testis was used to generate polyA-selected libraries with Kapa mRNA HyperPrep kits (Roche) with indexed adaptors. Libraries were assessed on the Tapestation 2200 (Agilent) and quantitated by Kapa qPCR. Pooled libraries were subjected to 50 bp paired-end sequencing according to the manufacturer’s protocol (Illumina NovaSeq6000). Bcl2fastq2 Conversion Software (Illumina) was used to generate de-multiplexed Fastq files.
RNA-seq and ChIP-seq mapping
RNA-seq and ChIP-seq analyses were conducted on previously published datasets. Specifically, mouse tissue panel data were analyzed from SRP016501 (Merkin et al. 2012), oocyte data from SRP061454 (Yu et al. 2016a), germinal vesicle data from SRP065256 (Yu et al. 2016b), and sorted round spermatid data from SRP111389 (Wichman et al. 2017). Rat testis data were analyzed from ERR3417900. Alignments were performed with Tophat, using genomic sequence from the representative X- and Y-amplicons as the reference genome. Due to the ampliconic nature of these sequences–max-multihits were set to 1 and–read-mismatches set to 0; otherwise, standard default parameters were used. We used Cufflinks, with the refFlat RefSeq gene annotation file, to estimate expression levels as fragments per kilobase per millions of mapped fragments (FPKM).
RNA and RT-PCR
Total testis RNA was extracted from C57BL/6J males (wild-type) and Laidx-/Y using Trizol (Life Technologies) according to manufacturer’s instructions. Ten μg of total RNA was DNase treated using Turbo DNAse (Life Technologies) and reverse transcribed using Superscript II (Invitrogen) using Laidx-specific primers following manufacturer’s instructions. Intron-spanning primers were used to perform RT-PCR on adult testis cDNA preparations for Laidx and a round spermatid-specific gene Trim42 (Supplemental Tables 1 and 2).
Transgenic lines
To generate mice with multi-megabase deletions of the Laidx ampliconic region, loxP sites were sequentially integrated upstream and downstream of the Laidx ampliconic region via CRISPR/Cas9. LoxP sites were introduced via cytoplasmic injection of Cas9 mRNA, an sgRNA targeting unique sequence flanking the ampliconic region, and a single stranded oligo donor carrying the loxP sequence (Supplemental Table 3). Cytoplasmic injections were performed on zygotes derived from a F1 (DBA/2JxC57BL/6J) male and a C57BL/6J female to ensure all targeted X chromosomes were of C57BL/6J origin. Floxed mice were mated against C57BL/6J EIIa-Cre mice (Jackson Laboratory: stock #003724, backcrossed more than 10 generations to C57BL/6J) resulting in mice carrying either a Laidx region deletion or duplication. Two independent deletion and duplication lines were generated. No differences were observed between independent lines and data were therefore compiled. All deletion or duplication carrying males were derived from heterozygous female mice that had been backcrossed to C57BL/6J males for at least four generations. Mice were genotyped by extracting DNA from a tail biopsy using Viagen DirectPCR lysis reagent using primers that flank loxP sites (Supplemental Table 3).
Histology
Testes were collected from 2-6 month old mice. The tunica albuginea was nicked and fixed with Bouins Fixative overnight at 4°. Testes were then washed through a series of ethanol washes (25%, 50%, 75% EtOH) before being stored in 75% EtOH at 25°. Testes were paraffin embedded and sectioned to 5μm. Sections were stained with Periodic Acid Schiff (PAS) and hematoxylin, visualized using a light microscope, and staged (Ahmed and de Rooij 2009). Specific germ cell populations were identified based upon their location, nuclear size, and nuclear staining pattern (Ahmed and de Rooij 2009).
Fertility, fecundity, and sex ratio distortion assessments
The fecundity of males was assessed by mating at least three deletion and duplication males 2-6 months of age to 2-6 month old CD1 females and monitoring females for copulatory plugs. The fertility of all lines was compared to that of C57BL/6J littermate control males (wild-type). When littermate controls were not available we used age-matched C57BL/6J male controls. Offspring sex ratio data were compiled by sex genotyping offspring (postnatal day 1 or 2) with PCR using primers specific to Uba1x/y (Supplemental Table 3), from the aforementioned fecundity assays of males bred with CD1 females. Sperm counts were conducted on sperm isolated from the cauda epididymis. Briefly, the cauda epididymis was isolated and nicked three times to allow sperm to swim out. The nicked epididymis was then rotated for 1hr at 37° in Toyoda Yokoyama Hosi media (TYH). Sperm were fixed in 4% PFA and counted using a hemocytometer. For each genotype, at least three male mice were counted with three technical replicates performed for each mouse and averaged. Testes were collected from 2-6 month old males for all experiments and weighed, along with total body, in order to determine relative testis weight.
Sperm swim-up assay
Mouse cauda epididymis were dissected from Laidx-/Y mice and wild-type littermate controls. The cauda epididymis was nicked three times to allow sperm to swim out and placed in a 2ml round bottom Eppendorf tube filled with 1.1ml of 37° Human Tubal Fluid media (Millipore). Sperm were placed in a 37° incubation chamber and allowed to swim out for 10 min before the cauda epididymis was removed. A 30µl aliquot was removed as a pre-swim-up reference. Samples were centrifuged for 5 min at 2000 rpm, placed at a 45° angle in a 37° incubation chamber, and sperm allowed to swim out of the pellet for one hour. The pre- and post-swim-up sperm were counted using a hemocytometer and percent motility calculated. Three technical replicates were performed per mouse.
Data availability
The complete BAC sequence of RP23-106J7 generated in this study with Laidx annotation is available from NCBI GenBank (Accession MN842289). All mRNA-seq libraries generated in this study are available at NCBI SRA (PRJNA595938). Laidx-/Y and LaidxDup/Y mice are available upon request. Supplemental material available at figshare: https://doi.org/10.25387/g3.11962158.
Results
The Srsx-amplicon is rearranged on the Y chromosome
To define the genomic structure of a single amplicon containing Srsx, we generated a high-quality assembly of BAC RP23-106J7 using PacBio sequencing. Comparison of the 209 kb BAC sequence to the mouse reference genome (mm10) revealed the amplicon size is 20 kb larger than the BAC (Figure 1a). We used this 229 kb representative amplicon (chrX:123,326,277-123,555,768; mm10) for all subsequent analyses (Figure 1a, Supplemental Figure 1). The representative Srsx-amplicon sequence shares 99.1% sequence identity with another Srsx-amplicon (chrX:123,104,838-123,326,276; mm10), differing primarily by L1 and ERVK transposable element content (Supplemental Figure 2). There are truncated forms of the 229 kb amplicon ranging from 120-130 kb in length, which are arranged in tandem in the opposite (palindromic) orientation of the full-length amplicons (Figure 1a, Supplemental Figure 1). Within both the full-length and truncated Srsx-amplicons are two internal tandem repeats, one that is 1149 bps, repeated ∼12.5 times and shares 93% sequence identity between repeats, while the other is 381 bps, repeated five times, and shares 96% sequence identity between repeats (Figure 1b). We expect the entire Srsx ampliconic region is comprised primarily of these full-length and truncated amplicons, but because of multiple gaps across the region the genomic architecture remains unresolved (Figure 1a, Supplemental Figure 1).
Figure 1.
Amplicons containing Srsx and Srsy are present in multiple copies within ampliconic regions of the mouse X and Y chromosomes. (A) Dot plot comparing the representative Srsx-containing amplicon to the entire ampliconic region (chrX:123,050,000-126,250,000; mm10). Each dot represents 100% sequence identity in a 100 bp window. Blue arrows indicate position and orientation of Srsx-containing amplicons. The representative sequence used for subsequent analyses is indicated by a blue bar. Vertical dotted gray lines mark the boundaries of each amplicon. Dark gray bars mark gaps in the mm10 reference genome sequence. (B) Self-symmetry triangular dot plots of X- and Y-amplicons are shown with each amplicon compared to itself. Each dot represents a perfect match of 50 nucleotides. Horizontal lines indicate tandemly-arrayed amplicons. The chromosomal regions shown are chrX:123,326,277-123,555,768 and chrY:49,567,447-50,092,166 (mm10). (C) Dot plots of DNA sequence identity between the X- and Y-amplicons from (B), on the Y and X axes, respectively. Each dot represents 100% sequence identity in a 25 bp window. Blue highlights indicate regions of sequence identity between the two sequences. The red, yellow, and blue Y-amplicon subunits are shown at top. Testis RNA-seq reads for each region are illustrated along the respective axes. The positions of Sly and Ssty1/2 have been previously mapped (Soh et al. 2014) and are excluded from the Y-amplicon for simplicity.
Defining the Srsx-amplicon allows us to perform a more accurate comparison with Srsy-amplicons. Srsy is within a 525 kb amplicon on the Y chromosome also containing Sly, Ssty1 and Ssty2 (Soh et al. 2014). The 525 kb representative Srsy-amplicon consists of three subunits, labeled red, yellow, and blue, with the yellow subunit duplicated within the amplicon (Figure 1b) (Soh et al. 2014). Pairwise sequence comparisons between Srsx- and Srsy-amplicons (Figure 1c), reveal previously observed regions with high levels of nucleotide identity (Soh et al. 2014). We additionally find the Srsx-amplicon is not contained in its entirety, nor contiguously, within the larger Srsy-amplicon. For example, a 34.2 kb region of the Srsx-amplicon is represented once in each yellow repeat, as well as twice in degenerated form in the red repeat, while a different part of the Srsx-amplicon is duplicated in the blue repeat sequence (Figure 1c). Based on these observations, we speculate the Srsx-amplicon represents the ancestral state of a common sequence shared on the X and Y chromosomes.
Laidx is a large gene in the Srsx-amplicon
We examined how differences between Srsx- and Srsy-amplicon affect transcription in the testis. Mapping of previously published total RNA-seq sequences (Wichman et al. 2017) from round spermatids to the Srsx- and Srsy-amplicons reveals a single, long transcription unit in the Srsx-amplicon. Sequences homologous to the long Srsx-amplicon transcription unit are rearranged within the Srsy-amplicon (Figure 1c), suggesting the Srsy-amplicon lacks the ability to generate a contiguous transcript homologous to a transcript from the Srsx-amplicon. Instead, the rearranged Srsy-amplicon sequences produce several, separate transcripts with homology to small segments of the X-amplicon transcript, including Srsy, Asty, and Gm28689. These Y-specific transcripts are detected at low levels (FPKM = 0.03 – 2.13; Supplemental Figure 3). The presence of a single, long, transcription unit within the Srsx-amplicon, as compared to fragmented transcripts on the Srsy-amplicon further supports the Srsx-amplicon as the ancestral state.
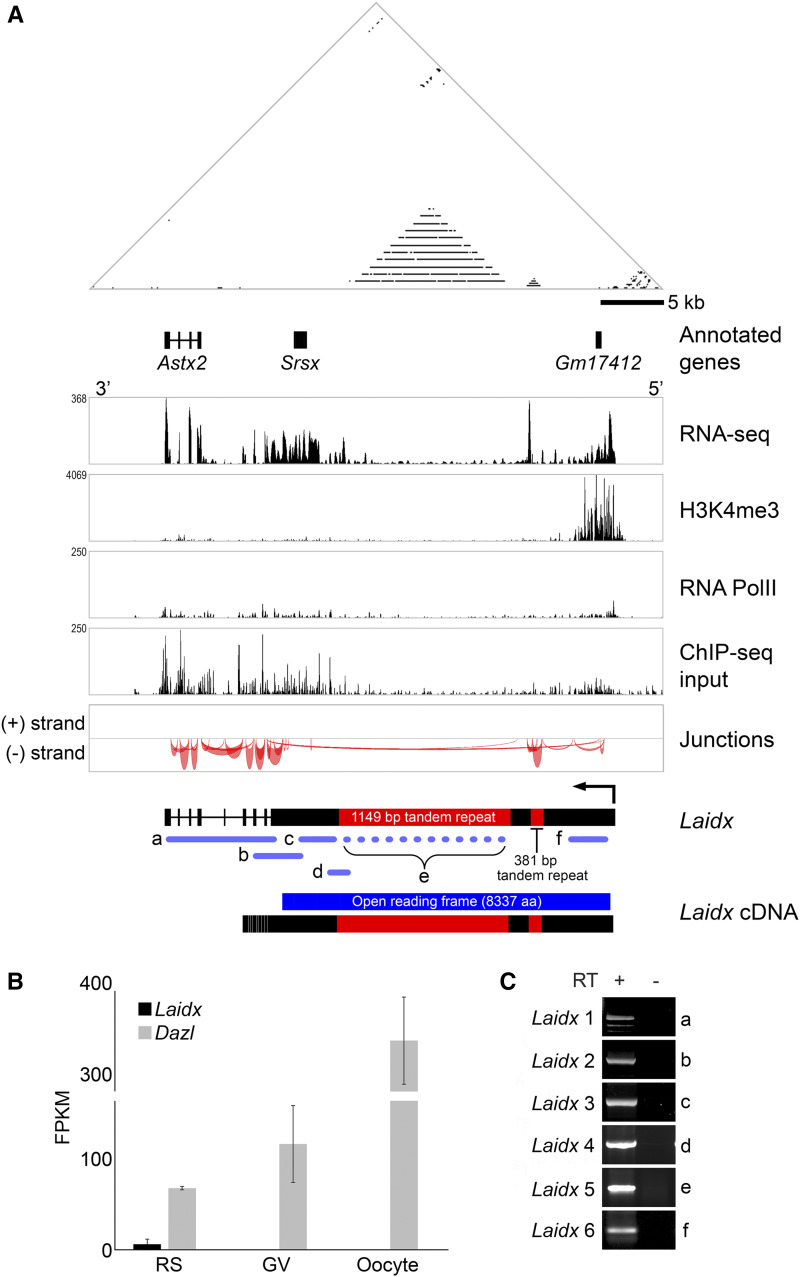
We further characterized the large transcription unit within the Srsx-amplicon to determine whether it encodes a protein. The large transcription unit spans 35.6 kb and encodes a 28.2 kb mature transcript comprising nine exons (Figure 2a). Consistent with a single transcription start site, reanalysis of ChIP-seq data from round spermatids (Hammoud et al. 2014) reveals a small enrichment of RNA polymerase II and a broad enrichment of H3K4me3 overlapping the transcription start site (Figure 2a). This long transcription unit encompasses Srsx and two other partial gene annotations also co-amplified on the X and Y chromosomes, Astx2 and Gm17412 (Touré et al. 2005). There is no enrichment of H3K4me3 at the annotated start sites of Srsx, Astx2, and Gm17412, suggesting they are not independently transcribed in round spermatids. This long transcription unit contains nine exons with exon 1 comprising >93% of the predicted transcript and the entirety of the predicted open reading frame. This long transcription unit is detectable in round spermatids of the testis (Figure 2b, Supplemental Figure 4) and undetectable across several somatic tissues (Supplemental Figure 4), germinal vesicles or oocytes (Figure 2b), consistent with previous studies (Touré et al. 2005). Altogether, we find three partially-annotated genes (Srsx, Astx, and Gm17412) are contained within a single transcriptional unit expressed in round spermatids.
Figure 2.
A large transcription unit, Laidx, encompasses three partially annotated genes, including Srsx. (A) Self-symmetry triangular dot plot of a 45.5 kb region encompassing the large transcription unit. The coordinates of the chromosomal regions shown are chrX:123,443,060-123,488,539 (mm10). Positions of three partially annotated genes (Astx2, Srsx, and Gm17412) are indicated. Below the dotplot and gene annotations are aligned reads from RNA-seq performed on round spermatids, showing transcription extending from upstream of Gm17412 to the end of Astx2. In addition, ChIP-seq on round spermatids revealed modest enrichment of RNA polymerase II along the transcription unit and a small amount of enrichment at the transcription start site, along with broad enrichment of H3K4me3, a modification associated with active promoters. RNA-seq and ChIP-seq alignments were performed on repeat masked sequence (Smit et al. 2015). “Junctions” illustrates predicted splice sites based on RNA-seq. The height and thickness of the arcs are proportional to read depth spanning the junction, up to 50 reads. The predicted genomic organization of the Laidx gene is illustrated below. Purple bars represent select RT-PCR assays used to verify expression and are lettered to correspond with labels in (C). (B) Quantitation of RNA-seq data from round spermatids (RS), germinal vesicles (GV), and oocytes demonstrating transcription in the male but not female germline. Dazl, a gene expressed in both male and female germlines, is used as a control. FPKM, Fragments Per Kilobase per Million reads. (C) RT-PCR on RT (+) and no RT controls (-) performed on RNA isolated from adult testis.
To validate this novel long gene, we performed RT-PCR with primers specific to different regions of the putative transcription unit (Figures 2a and c, Supplemental Figure 5). We used sequences spanning the intron-exon junctions predicted by Cufflinks (Trapnell et al. 2010) to design primers that amplify products spanning multiple exons. These RT-PCR products confirm the expression of a single large transcription unit in testis. While it is not clear if this transcript produces a protein, there is an open reading frame of 25,011 base pairs encoding a large predicted protein of 8337 amino acids (871 kD). This protein has no known functional motifs and is predicted to be an intrinsically disordered protein (Supplemental Figure 6). We name this new gene, Laidx (Large amplified intrinsically disordered protein-coding gene on the X). Based on genomic rearrangements and RNA-seq data (Figure 1c), we consider Laidy to be pseudogenized in present day mice. The remainder of this study will focus on Laidx.
Laidx migrated from murine rodents to fluke via a single horizontal gene transfer event
Laidx is detectable and potentially amplified on the rat X chromosome, but not detectable in the genomes of guinea pig or deer mouse. We detect 76% nucleotide sequence identity between a high-quality rat BAC assembly (CH230-1D6; GenBank Accession AC130042) containing Laidx sequence and mouse Laidx (Supplemental Figure 7a). The 1149 bp mouse repeat has 12.5 copies, while the rat repeat is truncated (∼650bp), single copy, and shares 70% nucleotide sequence identity with the mouse repeat. Similarly, the 381 bp mouse repeat (five copies) is single copy and truncated (228bp) in rat with approximately 79% sequence identity with the mouse repeat. While the rat Laidx sequence is interrupted by multiple LINE elements, there is a large open reading frame encoding a predicted protein of 1806 amino acids with 43% identity with mouse LAIDX (Supplemental Figure 7b). Thus, the Laidx gene is intact in rats, with the single copy, truncated, and diverged internal repeats maintaining the rat open reading frame. A comparison of the rat BAC containing Laidx BAC to a rat Y BAC (RNAEX-9O8; GenBank Accession AC279156) reveals rat Laidy is rearranged, with most of the large open reading frame deleted from the rat Y (Supplemental Figure 8). In both rat and mice, rearrangement of Laidy disrupts the coding potential seen in Laidx. The conservation of a large open reading frame in mouse and rat suggests Laidx is translated.
Mouse LAIDX protein shares ∼43% sequence identity with a 5280 amino acid hypothetical protein CSKR_14446s (GenBank: RJW68620.1) in the ∼825 MY diverged Chinese liver fluke, Clonorchis sinensis (Figure 3a). Consistent with the protein similarities, the mouse and Clonorchis gene sequences share 69% nucleotide identity across 60% of Laidx exon 1 (Figure 3b). The 1149 bp and 381 bp mouse repeats are found in a single copy and maintain the open reading frame in Clonorchis. Comparisons of rat and Clonorchis genes and proteins reveal 67% nucleotide identity and 40% amino acid sequence identity, respectively (Supplemental Figure 9).
Figure 3.
High Laidx sequence identity between mouse and Chinese liver fluke suggests a horizontal gene transfer event. (A) Blastp alignment of the 5280 amino acid Clonorchis protein CSKR_14446s and the predicted 8337 amino acid LAIDX protein. (B) TBLASTN alignment of Laidx with Clonorchis genomic sequence encoding CSKR_14446s. (C) Evolutionary history of Laidx. Species that carry a Laidx ortholog are marked with a “+”. An orange arrow and orange branches mark a proposed horizontal gene transfer event that occurred between a mouse/rat ancestor and the liver fluke. Mya, Million years ago.
The conservation of Laidx between Clonorchis, Rattus, and Mus, but not in other lineages, including other mammals, insects, and nematodes, suggests Laidx moved between rodents and Clonorchis via a single horizontal gene transfer event in the last 82 million years (Figure 3c). To determine the directionality of the horizontal gene transfer we examined transposable element content in each genomic region. Several murine rodent-specific ERVK transposable elements are present in the Clonorchis sequence encoding CSKR_14446s and non-mammalian transposable elements were not detected in the mouse Laidx-amplicon. The presence of murine rodent lineage-specific transposable elements in the Clonorchis genome, near this gene, suggests the single horizontal gene transfer event occurred from a murine rodent ancestor to an ancestor of the Chinese liver fluke.
Laidx deletion and duplication mice do not exhibit overt reproductive defects
To explore the function of Laidx in the mouse germline, we generated precise and complete multi-megabase Laidx deletions (Laidx-/Y) and duplications (LaidxDup/Y) using CRISPR and Cre/loxP (Figure 4a). Deletions were confirmed by RT-PCR assays specific to three different regions of Laidx (Figure 4b). While one assay demonstrated loss of the transcript in the Laidx-/Y mice, the other two assays yielded RT-PCR products. Presence of these products could indicate either incomplete deletion or expressed sequences with high sequence identity on the Y chromosome, such as Srsy or Asty. Sanger sequencing of PCR products from wild-type mice reveals sequence variants between Laidx and Srsy/Asty on the Y chromosome. However, RT-PCR products from the deletion mice contained only Y-specific variants (Figure 4c), supporting that all RT-PCR products are derived from sequences on the Y chromosome, consistent with a complete deletion of the Laidx-ampliconic region.
Figure 4.
Generation of Laidx-/Y and LaidxDup/Y transgenic mice. (A) Schematic representation of the mouse X chromosome. Ampliconic regions are shown in blue, centromere in gray, and pseudoautosomal region (PAR) in green. The region of the X chromosome carrying the Laidx-containing amplicons is expanded to show a representation of the repeat structure (blue arrows). Red arrows denote loxP sites. Mice carrying loxP sites flanking the ampliconic region were mated to Ella-Cre mice to generate Laidx-/Y mice. (B) RT-PCR on RT (+) and no RT controls (-) performed on RNA isolated from adult testis from WT and Laidx-/Y mice. Trim42 is a testis-specific gene used as a positive control. Primer pairs for each assay are indicated (see Supplemental Tables 1 and 2). (C) Sanger sequencing chromatograms from Laidx RT-PCR product in WT (top) and Laidx-/Y (bottom) mice. The WT product contains multiple sequence variants that are specific to both the X and Y chromosome. The Laidx-/Y product contains only variant sequences specific to the Y chromosome. (D) mRNA-seq was performed on testes from WT, Laidx-/Y, and LaidxDup/Y mice.
To further confirm successful deletion and duplication of Laidx, mRNA-seq was performed on testes from wild-type, Laidx-/Y, and LaidxDup/Y mice. Due to the high sequence identity between these regions on the X and Y chromosomes, mRNA-seq reads were mapped to a Laidx cDNA sequence that is masked across regions with 100% sequence identity between the X and Y chromosome. FPKM of Laidx-/Y testes was reduced compared to wild-type mice (FPKM = 0.042 vs. 0.354, respectively) supporting deletion of the region (Figure 4d). LaidxDup/Y mice displayed approximately double the level of gene expression (FPKM = 1.81), consistent with a duplication (Figure 4d).
Laidx-/Y mice do not display notable reproductive deficits on a C57BL/6J genetic background. Testicular morphology, sperm development, and timing of spermatogenic events are not different compared to wild type controls (Supplemental Figure 10). To test the effects of Laidx deletion on fertility and fecundity, we bred three Laidx-/Y males and two wild-type littermates to wild-type CD1 female mice. Laidx-/Y and LaidxDup/Y male mice have normal fertility and fecundity compared to wild-type (Figure 5a). Wild-type males sired 180 pups in 15 litters (mean = 12.0), Laidx-/Y males sired 286 pups in 23 litters (mean = 12.4) (P = 0.55), and LaidxDup/Y males sired 220 pups in 23 litters (mean = 9.6) (P = 0.06). In addition, litters show no differences in the ratio of male to female pups (Figure 5b). Litters sired by wild-type, Laidx-/Y, and LaidxDup/Y males are 57% (66/115), 49% (79/160; P = 0.22), and 52% (127/243; P = 0.49) male, respectively. Compared to wild-type, Laidx-/Y males do not exhibit statistically significant differences in sperm count or motility, as assessed by the sperm swim-up assay (Figure 5c-d). Laidx-/Y males do exhibit a 24% reduction in sperm count and 15% reduction in motile sperm, as compared to wild-type males. Laidx−/− females are able to produce offspring, consistent with lack of expression in oocytes and germinal vesicles (Figure 2b) but a comprehensive characterization of female fecundity has yet to be performed.
Figure 5.
Deletion or duplication of Laidx has no detectable effect on male fertility, fecundity, sperm count or sperm motility. (A) Male fecundity as a function of litter size. Each data point represents the number of pups from a single litter. The horizontal line indicates the mean, with error bars representing standard deviation. P-value was determined by Student’s t-test. (B) The proportion of male offspring is shown as a percentage along with the number of pups screened in parentheses. P-values were calculated using Fisher’s Exact Test. (C) Sperm counts. Each data point represents the average sperm count from an individual mouse. The horizontal line indicates the mean while error bars represent standard deviation. P-value determined by Student’s t-test. (D) Sperm motility. Each data point is the percentage of motile sperm from an individual mouse. The horizontal line indicates the mean while error bars represent standard deviation. P-value determined by Two Proportion Z Test.
Discussion
We have identified Laidx, a novel, large, and amplified gene encompassing three previously-annotated genes co-amplified on the mouse X and Y chromosomes. Laidx consists of nine exons and encodes an 8337 amino acid (871 kD) putative protein, which shares sequence similarity with a protein in the Chinese liver fluke, Clonorchis sinensis. Laidx is predominantly expressed in post-meiotic testicular germ cells, suggesting a role in spermatogenesis and reproduction. However, male mice with Laidx deletion and duplication exhibit fertility, fecundity, testis histology, and offspring sex ratio similar to wild-type, indicating the role of Laidx may be uncovered under other conditions (e.g., stress, old age). The reduced sperm counts and sperm motility, though not statistically significant, suggests loss or lower copy number of Laidx could impact the fitness of males in wild populations. Our resolving the entire gene structure of the large and complex Laidx gene sequence combined with the generation of mutant mouse models provides the foundation for future studies exploring the role of Laidx in post-meiotic germ cell development.
Laidx is present in mouse, rat, and the Chinese liver fluke, Clonorchis sinensis and is not detectable in other mammals, suggesting a single horizontal gene transfer event. Rat is a definitive host of Clonorchis (Chai et al. 2005), thus providing an opportunity for such a horizontal gene transfer event, which can occur from host to parasite (Wijayawardena et al. 2013). The presence of host transposable element sequences in parasite genomes at sites of horizontal gene transfer is evidence of this directionality. We found murine rodent-specific ERVK sequences near the Clonorchis orthologous gene, supporting that Laidx was transferred from murine rodent to the fluke. However, as there is no complete Clonorchis genome sequence assembly, we cannot determine the boundaries of the horizontal gene transfer event, which could give insight into the mechanism of transfer. It is difficult to validate a host-parasite horizontal gene transfer event, in part because contamination of a parasite sample with host DNA can create false positive results. However, the liver flukes used to generate the Clonorchis reference genome sequence were isolated from cat (Wang et al. 2011), making contamination of the reference sequence with rat genomic sequence unlikely. Consistent with this, the sequence and structural divergence between mouse Laidx and Clonorchis CSKR_14446s was higher than would be expected if it was derived from contaminating rodent sequence. The high sequence identity between Clonorchis with both rat and mouse make it difficult to predict whether the horizontal gene transfer event occurred from an animal on the rat or mouse lineage, or a common ancestor. Conservation of a Laidx open reading frame (ORF) in mouse, rat, and liver fluke along with confirmation of expression in the mouse and rat testis provides additional support for the ancestral state of Laidx gene structure and suggests it is important for reproduction. Considering the germ cell expression of Laidx in mouse and rat, it will be interesting to examine whether the Clonorchis ortholog also functions in germ cells.
The co-amplification of genes on the mouse X and Y chromosomes is thought to have arisen through meiotic drive, whereby gene duplication confers a competitive advantage in X- or Y-bearing sperm (Cocquet et al. 2012). An example of this phenomenon can be seen in Sly, Slx, and Slxl1 (Kruger et al. 2019; Cocquet et al. 2012). Characterization of Laidx reveals that, while there is considerable sequence identity between the Laidx/y-amplicons, these chromosomes produce dramatically different transcripts. Given the high similarity of LAIDX to Clonorchis CSKR_14446s protein, we propose that Laidx represents the rat/mouse ancestral gene. It is unclear if the massive amplification of Laidy is due to functional selection for one of the smaller transcripts, or if it is a passenger resulting from amplification of Sly, Ssty1, and Ssty2, which share the same Y-amplicon (Soh et al. 2014). Comparative genomic studies of Laidx/y in mammals that predate mouse-rat divergence may provide insights into their role in meiotic drive and the origin of this large predicted protein-coding gene.
Acknowledgments
We would like to acknowledge Dirk de Rooij at Utrecht University for his assistance with histology evaluation, the University of Michigan Histology Core, the University of Michigan Advanced Genomics Core for Sanger and Illumina sequencing and mRNA-seq library preparations, Melanie Sorensen at University of Washington for BAC sequencing, Katherine Stansifer at Ohana Biosciences for the sperm swim-up assay protocol, and the Transgenic Animal and Genome Editing Core at Cincinnati Children’s Hospital Medical Center. We thank David Page and colleagues at the Whitehead Institute and the Human Genome Sequencing Center at Baylor College of Medicine for the public database deposits of the finished rat Y BAC sequences used in our analyses. We thank David Ginsburg for sharing EIIa-Cre mice. We thank Emma Gerlinger and Melody Gorishek for assistance with genotyping. We thank Alyssa Kruger, Callie Swanepoel, and Eden Dulka for helpful comments. This work was supported by National Institutes of Health grant R01HD094736.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11962158.
Communicating editor: F. Pardo-Manuel de Villena
Literature Cited
- Agarwala R., Barrett T., Beck J., Benson D. A., Bollin C. et al. , 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46: D8–D13. 10.1093/nar/gkx1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed E. A., and de Rooij D. G., 2009. Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 558: 263–277. 10.1007/978-1-60761-103-5_16 [DOI] [PubMed] [Google Scholar]
- Bennett-Baker P. E., and Mueller J. L., 2017. CRISPR-mediated isolation of specific megabase segments of genomic DNA. Nucleic Acids Res. 45: e165 10.1093/nar/gkx749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J. Y., Darwin Murrell K., and Lymbery A. J., 2005. Fish-borne parasitic zoonoses: status and issues. Int. J. Parasitol. 35: 1233–1254. 10.1016/j.ijpara.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J., Mahadevaiah S. K., Affara N. A., Vaiman D. et al. , 2012. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 8: e1002900 10.1371/journal.pgen.1002900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J., Yamauchi Y., Riel J. M., Karacs T. P. et al. , 2010. Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol. Biol. Cell 21: 3497–3505. 10.1091/mbc.e10-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comptour A., Moretti C., Serrentino M. E., Auer J., Ialy-Radio C. et al. , 2014. SSTY proteins co-localize with the post-meiotic sex chromatin and interact with regulators of its expression. FEBS J. 281: 1571–1584. 10.1111/febs.12724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S. S., Low D. H., Yi C., Carrell D. T., Guccione E. et al. , 2014. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15: 239–253. 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Kruger A.N., Brogley M. A., Huizinga J. L., Kidd J. M., de Rooij D. G. et al. , 2019. A Neofunctionalized X–Linked Ampliconic Gene Family Is Essential for Male Fertility and Equal Sex Ratio in Mice. Curr Biol 29: 3699–3706.e5. 10.1016/j.cub.2019.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J., Russell C., Chen P., and Burge C. B., 2012. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338: 1593–1599. 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J. L., Mahadevaiah S. K., Park P. J., Warburton P. E., Page D. C. et al. , 2008. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 40: 794–799. 10.1038/ng.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathje C.C., Johnson E. E. P., Drage D., Patinioti C., Silvestri G. et al. , 2019. Differential Sperm Motility Mediates the Sex Ratio Drive Shaping Mouse Sex Chromosome Evolution. Curr Biol 29: 3692–3698.e4. 10.1016/j.cub.2019.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard L. N., Cocquet J., and Burgoyne P. S., 2009. The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol. Reprod. 81: 250–257. 10.1095/biolreprod.108.075382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A. F. A., Hubley R., & Green P., 2015. RepeatMasker Open-4.0. 2013–2015. http://www.repeatmasker.org
- Soh Y. Q., Alfoldi J., Pyntikova T., Brown L. G., Graves T. et al. , 2014. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159: 800–813. 10.1016/j.cell.2014.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szot M., Grigoriev V., Mahadevaiah S. K., Ojarikre O. A., Toure A. et al. , 2003. Does Rbmy have a role in sperm development in mice? Cytogenet. Genome Res. 103: 330–336. 10.1159/000076821 [DOI] [PubMed] [Google Scholar]
- Touré A., Clemente E. J., Ellis P., Mahadevaiah S. K., Ojarikre O. A. et al. , 2005. Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol. 6: R102 10.1186/gb-2005-6-12-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G. et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollger M. R., Dishuck P. C., Sorensen M., Welch A. E., Dang V. et al. , 2019. Long-read sequence and assembly of segmental duplications. Nat. Methods 16: 88–94. 10.1038/s41592-018-0236-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen W., Huang Y., Sun J., Men J. et al. , 2011. The draft genome of the carcinogenic human liver fluke Clonorchis sinensis. Genome Biol. 12: R107 10.1186/gb-2011-12-10-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman L., Somasundaram S., Breindel C., Valerio D. M., McCarrey J. R. et al. , 2017. Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility. Biol. Reprod. 97: 313–323. 10.1093/biolre/iox084 [DOI] [PubMed] [Google Scholar]
- Wijayawardena B. K., Minchella D. J., and DeWoody J. A., 2013. Hosts, parasites, and horizontal gene transfer. Trends Parasitol. 29: 329–338. 10.1016/j.pt.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Yu C., Ji S. Y., Dang Y. J., Sha Q. Q., Yuan Y. F. et al. , 2016a Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. 26: 275–287. 10.1038/cr.2016.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Ji S. Y., Sha Q. Q., Dang Y., Zhou J. J. et al. , 2016b BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 23: 387–394. 10.1038/nsmb.3204 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete BAC sequence of RP23-106J7 generated in this study with Laidx annotation is available from NCBI GenBank (Accession MN842289). All mRNA-seq libraries generated in this study are available at NCBI SRA (PRJNA595938). Laidx-/Y and LaidxDup/Y mice are available upon request. Supplemental material available at figshare: https://doi.org/10.25387/g3.11962158.