Abstract
The black yeast-like fungus Arthrocladium fulminans is known from strains that cause severe and eventually fatal disseminated infections in immunocompromised patients. Given the dramatic outcome of this clinical case, it is essential to understand the virulence potential of this species. The fungus is a member of the family Trichomeriaceae, at some phylogenetic distance from the Herpotrichiellaceae where most infectious fungi in the order Chaetothyriales are located. Main ecological preferences among Trichomeriaceae include colonization of exposed inert surfaces. Currently, black yeasts genomes that are available in public databases cover members of the families Herpotrichiellaceae and Cyphellophoraceae. In the present report, we sequenced the genome of the first member and only clinical representative of the family Trichomeriaceae.
Keywords: black yeast, Chaetothyriales, Trichomeriaceae, whole-genome sequencing
Arthrocladium fulminans is a member of the fungal order Chaetothyriales that covers black yeasts and relatives, known for their potential to cause severe and mutilating infections in immunocompromised as well as in healthy humans. The order comprises five families: Chaetothyriaceae, Cyphellophoraceae, Epibryaceae, Herpotrichiellaceae, and Trichomeriaceae (Réblová et al. 2013). In general, black yeast-like fungi are believed to possess limited competitive abilities toward adjacent microbes (de Hoog 1993), probably having disadvantages for colonializing saprobic environments due to more rapidly growing competitors. Therefore, black yeasts have been commonly isolated from extreme habitats where, where interactions between organisms is limited due to the environmental stress. A remarkably large number of these species have been reported from human infections (Hoog et al. 2019). For example, chromoblastomycosis that affects skin and subcutaneous tissue is caused by clusters of species in the family Herpotrichiellaceae, and cerebral phaeohyphomycosis is mainly caused Cladophialophora bantaina, Fonsecaea monophora, Rhinocladiella mackenziei and Exophiala dermatitidis in the same family (Li and de Hoog 2009; Queiroz-Telles et al. 2016). The few cases of human infection reported outside Herpotrichiellaceae concern superficial skin diseases by members of Cyphellophoraceae (Réblová et al. 2013).
The family Trichomeriaceae comprises mainly rock-inhabiting and epiphytic species (Chomnunti et al. 2012). Phylogenetic analyses have demonstrated that rock-inhabiting fungi often form early diverging groups within the order Chaetothyriales. The non-virulent Trichomeriaceae may be ancestral to the opportunists in Herpotrichiellaceae (Gueidan et al. 2011). In contrast to most genera of Trichomeriaceae having consistent ecology (Selbmann et al. 2017) Arthrocladium includes very rare species with divergent ecological preferences. For example, Arthrocladium tropicale and A. tardum were isolated from ant domatia in Leonardoxa africana (Nascimento et al. 2016) and A. caudatum from leaf litter of Acacia karroo (Papendorf 1969). Arthrocladium fulminans. The single strain known of the latter species caused a fatal disseminated infection in a patient with a GATA-2 disorder, a rare genetic immunodeficiency syndrome (Egenlauf et al. 2019). In addition, A. fulminans was reported causing septic arthritis and osteomyelitis in an immunocompetent patient (Diallo et al. 2017).
Currently, only genomes of two derived families of Chaetothyriales, i.e., Cyphellophoraceae and Herpotrichiellaceae have been sequenced and included in comparative genomic analyses. In order to determine the genomic composition of a basal linage of Chaetothyriales, we sequenced the genome of A. fulminans and functionally annotated their predicted proteins. Comparative analysis was done identifying orthologous clusters shared with other 23 black yeast species. Information about the genome of A. fulminans and members of other families will help to elucidate the origin of opportunism in Herpotrichiellaceae.
Materials and Methods
Strain and sequencing
To extract the genomic DNA, the fungus Arthrocladium fulminans CBS 136243 was cultured in malt extract broth (MEB), with shaking at 150 r.p.m. at 25° for 7 days. DNA was extracted via a cetyltrimethylammonium bromide (CTAB)-based method involving phenol-isoamyl alcoho/isoamyl alcohol (Möller et al. 1992). For genome sequencing, library construction (180 bases-insert library) and genome sequencing (Illumina HiSeq platform) were performed at Eurofins Genomics (Ebersberg, Germany).
Assembly, annotation and comparative analysis
The read quality was assessed by FASTQC v. 0.11.7 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and low-quality sequences were removed by Trimmomatic (Bolger et al. 2014) and adaptors were removed by BBDuk from the BBMap package (https://sourceforge.net/projects/bbmap/) High quality reads were assembled using SPAdes genome assembler v3.10.0 (Bankevich et al. 2012). To find de-novo repeats, the contig swere screened using RepeatModeler v1.0.8. To identify additional copies of de novo repeats across the genome assembly, the library produced by RepeatModeler was used as input for RepeatMasker v4.0.7. Genes were predicted by Augustus (Stanke and Waack 2003) using a training model generated by Genemark-ES v4.30 (Lomsadze 2005). Functional annotations where performed with InterProScan v5.27-66.0 (Quevillon et al. 2005) and BLAST against UniProt SwissProt database. Carbohydrate-active enzymes (CAZymes) were classified using the dbCAN2 meta server (Yin et al. 2012). The mitochondrial genome annotation was performed with MITOS pipeline (Bernt et al. 2013).
Cytochromes P450 genes (CYPs) were annotated by identification of proteins carrying the PFAM domain PF00067 using the InterProScan v5.27-66.0 (Quevillon et al. 2005). Putative CYP450 genes were organized into families and subfamilies as recommended by the International P450 Nomenclature Committee (Nelson 2006). The Mating Type locus (MAT) of A. fulminans was characterized by homology to the MAT1-1 and MAT1-2 reference sequences previously described in related species of Herpotrichiellaceae. (Teixeira et al. 2017). A comparative analyses of melanin-associated genes was done based on the Teixeira et al. (2017) using BLAST with e-value of 1 ☓ 10-5. The results is showed as the supplementary material (Table S1).
Orthologous groups were clustered by comparing the protein sequences of Arthrocladium fulminans to those of 23 previously sequenced black yeasts (Teixeira et al. 2017) using OrthoMCL pipeline (Li 2003) with a Markov inflation index of 1.5 and a maximum e-value of 1 ☓ 10-5. The single-copy genes were extracted of the OrthoMCL output.
Species tree based on orthologous clusters
Single-copy orthologous protein sequences obtained with OrthoMCL were aligned with MUSCLE (Edgar 2004) and poorly aligned regions were automatically removed using TRIMAL (Capella-Gutierrez et al. 2009) under the “-automated1” setting. The sequences were concatenated with FASCONCAT (Kück and Meusemann 2010) v. 1.0 and species trees were inferred by maximum likelihood RAxML (Stamatakis 2006) using PROTGAMMABLOSUM62 and 1000 bootstraps of branch support.
Data availability
Arthrocladium fulminans genome and the Mitochondrial genome have been deposited in the National Centre for Biotechnology Information (NCBI) under the accession numbers GCA_003614865.1 and MN593345. Supplemental material available at figshare: https://doi.org/10.25387/g3.11897388.
Results and Discussion
Assembly, completeness and content of the A. fulminans CBS 136243 genome
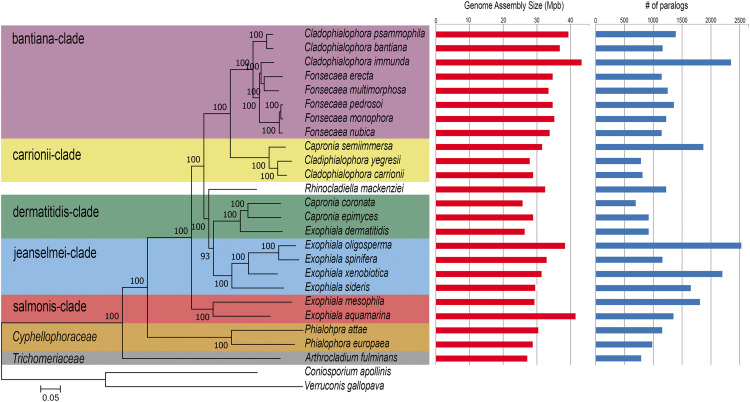
The genome assembly of Arthrocladium fulminans CBS 136243 consists of 27 contigs comprising 27.22 Mb in size, read coverage of 54x and GC content of 51.82%, which is slightly above the average of 51.7% in black yeasts (Table 1). Comparable sizes are found in species belonging to the ‘dermatitidis-clade’ (de Hoog et al. 2011), a group of species with somewhat smaller genomes in Herpotrichiellaceae, where genomes vary between 25.81 Mb to 28.89 Mb in Capronia coronata and in Capronia epimyces, respectively (Figure 1). The repetitive portion of the genome comprises 426,983 bp (1.57%). Low repetitive contents are consistent across chaetothyrialean black yeasts, where repetitive elements are in the range of 0.03–2%.
Table 1. Genome Features.
| Type | Description | Value |
|---|---|---|
| Nuclear | Total sequence length | 27,195,275 |
| Spanned gaps | 2 | |
| Number of scaffolds | 27 | |
| Scaffold N50 | 2,259,535 | |
| Scaffold L50 | 6 | |
| Number of contigs | 34 | |
| Contig N50 | 1,671,613 | |
| Contig L50 | 7 | |
| Mitochondrion | Total sequence length | 24,423 |
| Number of contigs | 1 |
Figure 1.
Genomic landscape of 23 black yeasts belonging to the order Chaetothyriales and the newly sequenced fungus A. fulminans. The species Coniosporium apollinis and Verruconis gallopava were used as outgroup for comparative genomic analyses.
The completeness of the genome assembly of A. fulminans, was accessed with BUSCO (Simão et al. 2015). The genome assembly of A. fulminans contains 97.4%(295 of 303) complete BUSCO genes, being 293 (96.7%) complete and single-copy BUSCOs, 2 complete and duplicated BUSCOs (0.7%), 0,9% (5 of 295) fragmented genes and 3 missing genes.
Genome annotation
The genome of A. fulminans CBS 136243 contains 315 genes coding for putative Carbohydrate-Active Enzymes (CAZymes), families of enzymes playing an essential role in the breakdown, biosynthesis and/or modification of a wide range of carbohydrates. This is the lowest reported number of CAZymes in Chaetothyriales, where they range between 339 genes in the opportunistic species Exophiala dermatitidis to 506 genes in the hydrocarbon-associated species Exophiala xenobiotica. This data suggests that the increased abundance and diversification of CAZymes in black yeasts is a recent evolutionary event in Herpotrichiellaceae. Similar to other species in the order Chaetothyriales, the Glycoside hydrolases (GH) superfamily is the most abundant class of CAZymes in A. fulminans with enlarged families being GH3 (β-glucosidase), GH31 (α-glucosidase), GH16 (xyloglucan: xyloglucosyltransferase), GH13-subfamily 40 (α-amylase) and GH18 (chitinase).
We identified 72 genes likely coding for CYPs in the genome of A. fulminans. This class of enzymes plays an essential role in primary and secondary metabolic pathways as well as in detoxification of xenobiotics. This repertoire of CYPs is comparable to that previously described in species of the ‘dermatitidis-clade’ (de Hoog et al. 2011). Overall, the CYPs were classified into 38 families and 49 subfamilies. Eleven CYPs could not be assigned to any family or subfamily and were considered unique among black yeasts. Among these genes, DZA80_5394 and DZA80_7723 seem to encode an isoform of the CYP51A and CYP51B highly conserved among Eurotiomycetes but absent in the order Chaetothyriales. The gene DZA80_8445 seems to be a P450nor, a unusual P450 reported in Fusarium oxysporum and responsible for reducing NO to N2O rather than catalyzing the monooxygenation reaction (Shiro et al. 1995). Comparative analyses of melanin-associated genes using data set previously reported by Teixeira et al. (2017) supported that yeasts have homologs for the production of melanin by the DHN pathway (Table S1).
The MAT locus of A. fulminans CBS 136243 is composed by a single copy of the MAT1-2 gene (DZA80_6626), which contains the high mobility group box (HMG-box) domain (PF00505). This finding suggests a heterothallic (self-sterility) mating system. In this fungus, the flanking structure of this MAT locus resembles that of usually observed in Eurotiomycetes and includes, in its upstream region, the genes APN2 (DZA80_6625), COX13 (DZA80_6624) and APC5 (DZA80_6623) (Table 2). A similar MAT structure containing the COX13 gene has been only found in unrelated melanized fungi, such as Verruconis gallopava (Venturiales) and Coniosporium apollinis (incertae sedis); possibly COX13 was lost in derived Chaetothyriales species. The gene SLA2, commonly found in the flanking region of the MAT locus in several Eurotiomycetes was found in a distinct genomic region.
Table 2. Genome annotation.
| Features | Value |
|---|---|
| Number of genes | 8,534 |
| Median gene length (bp) | 1,838 |
| Number of exons | 19,553 |
| Coding GC content | 46.9% |
| Median exon length (bp) | 722 |
| Number of introns | 11,022 |
| Median intron length (bp) | 140 |
Comparative analysis
A substantial proportion of the predicted genes (4456) have homologs in other chaetothyrialean black yeast-like species. Two clusters of orthologs were specific to the ‘bantiana-clade’ and A. fulminans. These clusters correspond to a Carboxylesterase type B (IPR019819) and a Pectin lyase fold (IPR011050). Orthologs exclusively shared with the neurotropic species Rhinocladiella mackenziei include the amino acid transporter/polyamine (IPR002293), the Oxoglutarate/iron-dependent dioxygenase (IPR005123), metalloenzymes (IPR029068), Alcohol acetyltransferase/N-acetyltransferase (IPR010828), HNH nuclease (IPR003615), Short-chain dehydrogenase/reductase SDR (IPR002347) and the Aspartic peptidase (IPR021109) with multiple paralogs in A. fulminans and in R.nbsp;mackenziei. Seven orthologous clusters are shared between in A. fulminans and members of the family Cyphellophoraceae, i.e., Phialophora europaea and Phialophora attae, including Carboxylesterase, type B (IPR002018) and Cytochrome P450 (IPR001128). Few orthologous clusters were found specifically among A. fulminans and the ‘salmonis-‘and ‘carrionii-clades’, however most of them without a known biological function. None specific orthologous clusters were found shared by A. fulminans and the ‘jeanselmei-clade’.
Characteristics of the A. fulminans CBS 136243 mitochondrial genome
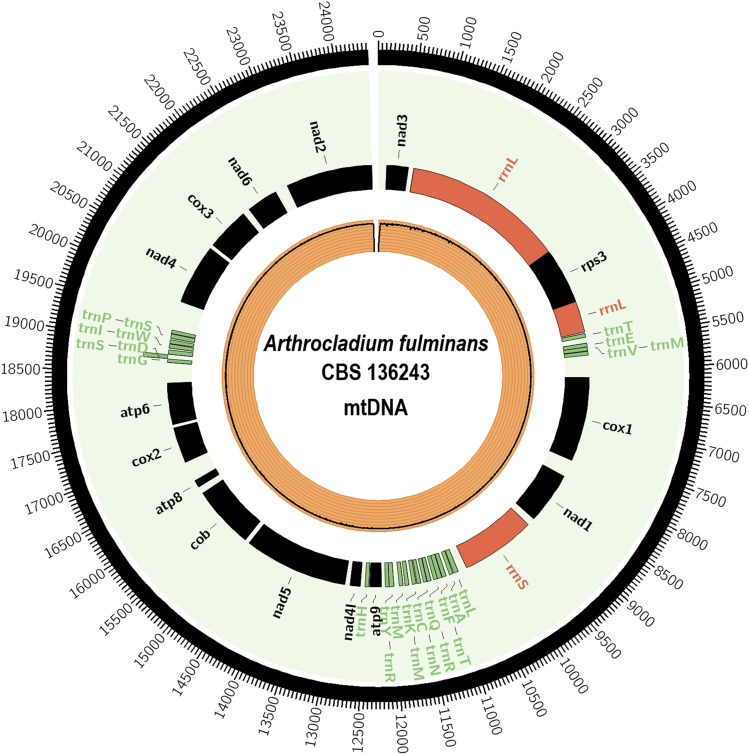
The mitochondrial genome was assembled in a separate 24.5-kb contig with GC content of 27.3%. This sequence contains 25 tRNAs coding for all 20 standard amino acids, 15 polypeptide-encoding genes and two rRNA-encoding genes (Figure 2). The large variable region of the mitochondrial genome, between the genes rrnl and nad2, shows the presence of a nad3 gene and the nuclear ribosomal protein S3 (rps3) which is anchored within the omega intron on the of the rrnl gene. The presence of mitochondrial rps3 is reported in several other fungi and provides insight into its evolution (Korovesi et al. 2018), considered an ancient gene which seems to be evolved within the endosymbiotic model. According to the authors, the mt rps3 proteins are descendants of archaeal and a-proteobacterial homologs, respectively. The nuclear ribosomal protein S3 (rps3) is implicated in the assembly of the ribosomal small subunit. Their C-terminal region is conserved in all Domains of life. Fungi and plants present a gene copy in their mitochondrial (mt) genomes which when rps3 is found in fungal mt genomes, it is either a free-standing gene or an anchored gene within the omega intron.
Figure 2.
Arthocladium Fulminans CBS 136243 mtDNA. Rectangles represent annotated genes: in red rRNAs; in green tRNAs. Orange inner circle shows reads coverage.
Conclusions
The whole genome sequence of the opportunistic, ancestral black yeast-like fungus Arthrocladium fulminans CBS 136243 was determined and compared with closely related species. Gene models were predicted and functionally annotated in order to identify gene families that likely led to the adaptation of extreme habitats by the black yeasts, such as CYPs and CAZymes. Our findings suggested that contrary to what was previously thought, gene family expansion took place later in the evolution of the black yeasts and the repertory of genes associated to resistance and nutrient uptake was reduced, but not absent, in basal lineages of black yeasts.
Acknowledgments
We thank Valter Baura from the Department of Biochemistry, Federal University of Paraná, Curitiba, Brazil for technical assistance. Sequencing of strain Arthrocladium fulminans (CBS 136243) was supported by grant number UTFPR2681-07/2108 from Technological Federal University of Paraná, Ponta Grossa, PR, Brazil. This work was supported by Brazilian Federal Agency for Support and Evaluation of Graduate: Coordination of Improvement of Higher Education Personnel-CAPES (www.capes.gov.br) and National Council for Scientific and Technological Development (http://cnpq.br/), Brazil; the National Institute of Science and Technology of Biological Nitrogen Fixation/CNPq/MCT grant number 573828/2008-3; fellowships from National Council for Scientific and Technological Development (CNPq) grant number 312811/2018-7, Brasilia, Brazil and the Institutional Program of Internationalization - CAPES/PrInt, Brazil.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11897388.
Communicating editor: M. Sachs
Literature Cited
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M. et al. , 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M., Donath A., Jühling F., Externbrink F., Florentz C. et al. , 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69: 313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M., and Gabaldon T., 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomnunti P., Bhat D. J., Jones E. B. G., Chukeatirote E., Bahkali A. H. et al. , 2012. Trichomeriaceae, a new sooty mould family of chaetothyriales. Fungal Divers. 56: 63–76. 10.1007/s13225-012-0197-2 [DOI] [Google Scholar]
- de Hoog G., Vicente V., Najafzadeh M., Harrak M., Badali H., et al. , 2011. Waterborne exophiala species causing disease in cold-blooded animals. Persoonia 27: 46–72. 10.3767/003158511X614258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog G. S., 1993. Evolution of black yeasts: possible adaptation to the human host. Antonie van Leeuwenhoek 63: 105–109. 10.1007/BF00872386 [DOI] [PubMed] [Google Scholar]
- Diallo A., Michaud C., Tabibou S., Raz M., Fernandez C. et al. , 2017. Arthrocladium fulminans arthritis and osteomyelitis. Am. J. Trop. Med. Hyg. 96: 698–700. 10.4269/ajtmh.16-0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egenlauf B., Schuhmann M., Giese T., Junghanss T., Stojkovic M. et al. , 2019. Disseminated mycosis by biarthrocladium/i/b bifulminans/i/b jeopardizing a patient with GATA2 deficiency. Respiration 97: 472–475. 10.1159/000493429 [DOI] [PubMed] [Google Scholar]
- Gueidan C., Ruibal C., de Hoog G., and Schneider H., 2011. Rock-inhabiting fungi originated during periods of dry climate in the late devonian and middle triassic. Fungal Biol. 115: 987–996. 10.1016/j.funbio.2011.04.002 [DOI] [PubMed] [Google Scholar]
- de Hoog G. S., Guarro J., Gené J., Ahmed S., Al-Hatmi A. M. S. et al. , 2019. Atlas of Clinical Fungi, 3rd e-edition. Utrecht / Reus. http://www.clinicalfungi.org/ [Google Scholar]
- Korovesi A. G., Ntertilis M., and Kouvelis V. N., 2018. Mt-rps3 is an ancient gene which provides insight into the evolution of fungal mitochondrial genomes. Mol. Phylogenet. Evol. 127: 74–86. 10.1016/j.ympev.2018.04.037 [DOI] [PubMed] [Google Scholar]
- Kück P., and Meusemann K., 2010. FASconCAT: Convenient handling of data matrices. Mol. Phylogenet. Evol. 56: 1115–1118. 10.1016/j.ympev.2010.04.024 [DOI] [PubMed] [Google Scholar]
- Li D. M., and de Hoog G. S., 2009. Cerebral phaeohyphomycosis—a cure at what lengths? Lancet Infect. Dis. 9: 376–383. 10.1016/S1473-3099(09)70131-8 [DOI] [PubMed] [Google Scholar]
- Li L., 2003. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A., 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33: 6494–6506. 10.1093/nar/gki937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller E., Bahnweg G., Sandermann H., and Geiger H., 1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20: 6115–6116. 10.1093/nar/20.22.6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento M. M., Selbmann L., Sharifynia S., Al-Hatmi A. M., Voglmayr H. et al. , 2016. Arthrocladium, an unexpected human opportunist in trichomeriaceae (chaetothyriales). Fungal Biol. 120: 207–218. 10.1016/j.funbio.2015.08.018 [DOI] [PubMed] [Google Scholar]
- Nelson D. R. Cytochrome P450 nomenclature, 2006. Methods in Molecular Biology. Clifton, N.J. 320: 1–10. 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- Papendorf M., 1969. New south african soil fungi. Trans. Br. Mycol. Soc. 52: 483–489. 10.1016/S0007-1536(69)80132-4 [DOI] [Google Scholar]
- Queiroz-Telles F., de Hoog S., Santos D. W. C. L., Salgado C. G., Vicente V. A. et al. , 2016. Chromoblastomycosis. Clin. Microbiol. Rev. 30: 233–276. 10.1128/CMR.00032-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N. et al. , 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33: W116–W120. 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M., Untereiner W. A., and Réblová K., 2013. Novel evolutionary lineages revealed in the chaetothyriales (fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLoS One 8: e63547 10.1371/journal.pone.0063547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbmann L., Onofri S., Coleine C., Buzzini P., Canini F. et al. , 2017. Effect of environmental parameters on biodiversity of the fungal component in lithic antarctic communities. Extremophiles 21: 1069–1080. 10.1007/s00792-017-0967-6 [DOI] [PubMed] [Google Scholar]
- Shiro Y., Fujii M., Iizuka T., Adachi S., Tsukamoto K. et al. , 1995. Spectroscopic and kinetic studies on reaction of cytochrome p450nor with nitric oxide. J. Biol. Chem. 270: 1617–1623. 10.1074/jbc.270.4.1617 [DOI] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stanke M., and Waack S., 2003. Gene prediction with a hidden markov model and a new intron submodel. Bioinformatics 19: ii215–ii225. 10.1093/bioinformatics/btg1080 [DOI] [PubMed] [Google Scholar]
- Teixeira M., Moreno L., Stielow B., Muszewska A., Hainaut M. et al. , 2017. Exploring the genomic diversity of black yeasts and relatives (chaetothyriales, ascomycota). Stud. Mycol. 86: 1–28. 10.1016/j.simyco.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J., Chen X., Mao F. et al. , 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40: W445–W451. 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Arthrocladium fulminans genome and the Mitochondrial genome have been deposited in the National Centre for Biotechnology Information (NCBI) under the accession numbers GCA_003614865.1 and MN593345. Supplemental material available at figshare: https://doi.org/10.25387/g3.11897388.