Abstract
FUS is a nucleic acid binding protein that, when mutated, cause a subset of familial amyotrophic lateral sclerosis (ALS). Expression of FUS in yeast recapitulates several pathological features of the disease-causing mutant proteins, including nuclear to cytoplasmic translocation, formation of cytoplasmic inclusions, and cytotoxicity. Genetic screens using the yeast model of FUS have identified yeast genes and their corresponding human homologs suppressing FUS induced toxicity in yeast, neurons and animal models. To expand the search for human suppressor genes of FUS induced toxicity, we carried out a genome-scale genetic screen using a newly constructed library containing 13570 human genes cloned in an inducible yeast-expression vector. Through multiple rounds of verification, we found 37 human genes that, when overexpressed, suppress FUS induced toxicity in yeast. Human genes with DNA or RNA binding functions are overrepresented among the identified suppressor genes, supporting that perturbations of RNA metabolism is a key underlying mechanism of FUS toxicity.
Keywords: FUS, ALS, human gene suppressors, yeast, genetic screen
ALS is a neurodegenerative disease characterized by the degeneration of upper and lower motor neurons in the brain and spinal cord leading to progressive paralysis and ultimately death within five years of symptom onset. FUS is a multifunctional RNA-binding protein involved in diverse RNA metabolic processes. Mutations in FUS cause an inherited form of ALS (Kwiatkowski et al. 2009). Several features associated with FUS pathology, including formation of cytoplasmic inclusions and cytotoxicity, have been recapitulated in the budding yeast Saccharomyces cerevisiae, enabling us and others to employ this powerful genetic system to rapidly identify genetic suppressors (Ju et al. 2011; Sun et al. 2011b). A critical resource used in such genetic screens is a library consisting of ∼5,000 sequence verified protein-coding yeast genes (Hu et al. 2007). Screening against this library has identified yeast genes that, when overexpressed, reduce the toxicity of FUS (Ju et al. 2011; Sun et al. 2011b). Consistent with the existence of conserved cellular mechanisms underlying FUS induced toxicity from yeast to human, UPF1, the human homolog of a yeast suppressor gene, shows strong protective effect in yeast, neurons and animal models of ALS (Barmada et al. 2015; Jackson et al. 2015; Xu et al. 2019). UPF1 is an essential component of the nonsense mediated mRNA decay (NMD) pathway, which detects and directs mRNAs with premature stop codons for degradation thus preventing the accumulation of truncated proteins (He and Jacobson 2015). NMD plays a critical role in mRNA surveillance, and is conserved from yeast to human (He and Jacobson 2015). Convergent suppressor mechanisms of FUS induced toxicity in yeast and mammalian systems supports the use of yeast as a model to study FUS toxicity.
We reasoned that direct expression of human genes in yeast may expand the search for genetic modifiers of cytotoxicity induced by disease-associated proteins. The obstacle to set up such a genetic screen is the availability of the full collection of human protein-coding genes cloned in a yeast expression vector. With Gateway cloning technology and a collection of 13570 human genes as entry clones generously provided to us by Dr. Marc Vidal, we generated a library of human genes individually cloned on a yeast expression vector, pAG416GAL-ccdB (Alberti et al. 2007). Using this library, we screened for human suppressor genes of toxicity mediated by ALS-associated protein FUS.
Overexpression genetic screen is typically done by individually transforming a library of plasmid clones into a yeast model of interest (Willingham et al. 2003; Cooper et al. 2006; Ju et al. 2011; Sun et al. 2011b). We developed a more efficient and cost-effective method for the screen that relies on highly efficient yeast mating rather than transformation (Hayden et al. 2018). Using this method, we introduced the arrayed library of human genes into the yeast model of FUS and screened for human suppressor genes. After several round of verifications, we confirmed 37 human genes that, when overexpressed, strongly reduce FUS induced toxicity. Gene ontology (GO) enrichment analysis revealed that FUS suppressors are enriched in genes with nucleic acid or RNA binding functions. Given that our lab and others have previously identified FUS suppressors genes involved in RNA metabolism (Ju et al. 2011; Sun et al. 2011b), expression of the suppressor human RNA-binding proteins in yeast may counter deleterious effects of FUS on RNA processing, transport or stability.
Materials and Mehtods
Human ORF clones
The human ORF clones used to generate the yeast-expression plasmids are Gateway entry clones kindly provided by Dr. Marc Vidal (Center for Cancer Systems Biology at Dana-Farber Cancer Institute and Department of Genetics, Harvard Medical School). The collection of human ORF clones, including the clones from hORFeome version 8.1 (Yang et al. 2011)(http://horfdb.dfci.harvard.edu/) and clones from the ORFeome collaboration (Collaboration 2016)(http://www.orfeomecollaboration.org), have been sequence verified previously. All ORF clones were cloned into the pAG416GAL-ccdB vector (Alberti et al. 2007) using Gateway LR recombination cloning (Hartley et al. 2000; Walhout et al. 2000). The resulting expression clones of human genes were subsequently transformed into the haploid w303 yeast strain (MATα can1-100, leu2-3,112, trp1-1, ura3-1, ade2-1, his3-11,15) using a high-throughput yeast transformation protocol as previously described (Fleming and Gitler 2011; Hayden et al. 2018). Transformed yeast strains bearing the expression plasmids of human genes were arrayed in 96-well plates and stored as glycerol archives at -80°.
Yeast strains and growth media
The 1xFUS integration strain was generated in the haploid W303 yeast strain (MATa can1-100, leu2-3,112, trp1-1, ura3-1, ade2-1, his3-11,15::pRS303Gal1FUS) as previously described (Ju et al. 2011). YPD media (1% yeast extract, 2% peptone and 2% glucose in distilled water) was used for mating. Synthetic dropout media lacking uracil (Ura-) was used for growing the haploid yeast strains containing the expression plasmids of human genes. Synthetic dropout media lacking both uracil and histidine (Ura- His-) was used for growing the diploid yeast strains containing both FUS integration and the human-gene plasmids. Yeast media contained 2% glucose, galactose or raffinose as the carbon source.
Genetic screen and verification
Detailed methods of screening by mating have been previously described (Hayden et al. 2018). Briefly, the haploid 1xFUS integration strain was crossed individually with each of the archived haploid yeast strains containing the expression clones of the human genes. The resulted diploid yeast strains were selected in Ura- His- liquid media, and plated onto Ura- His- agar plates containing glucose and galactose respectively, using the Singer RotoR robotic equipment (Singer Instruments). Following incubation at 30°, pictures of yeast colonies grown on the agar plates were taken daily until day four. Suppressors of FUS were identified by visually inspecting galactose agar plates. Yeast strains that grew clearly better than the background growth of the diploid FUS model on galactose plates were considered as screening hits. The corresponding human gene plasmids of all screening hits were isolated, transformed in the haploid w303 MATα yeast strain. Each transformants was crossed with the haploid 1xFUS integration strain. The resulting diploid strains were then grown to mid-log phase in Ura- His- liquid media containing Raffinose. Next, cell cultures were normalized to OD600 at 1.0, serially diluted by five folds and spotted onto the respective Ura- His- dropout agar plates containing 2% glucose and galactose respectively. Agar plates were incubated at 30° for three days and images of yeast colonies were taken. The identity of each verified human suppressor gene was confirmed by Sanger end-read sequencing and alignment to the expected sequence of the ORF clone.
Go term enrichment analysis
Go term enrichment analysis was conducted using PANTHER overrepresentation test. The reference gene list used to run the analysis included the 13570 Entrez gene IDs corresponding to all the cloned human ORF clones in the library. The annotation datasets used for the analysis was GO molecular function, biological process and cellular component released on 12/09/2019. Statistical analyses include Fisher’s Exact Test followed by FDR correction. All GO terms with a fold enrichment greater than or equal to three and FDR of less than or equal to 0.05 were included in Table 2.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Results and Discussion
Identification of human gene suppressing the toxicity of FUS in yeast
With the ease to carry out genome-wide genetic screens, yeast has served as a model for studying many human disease-associated proteins. One of such genetic screens, involving systematic overexpression of yeast genes, was used to screen yeast suppressor genes of FUS toxicity. Dosage-dependent genetic rescue of FUS toxicity has also been confirmed for the human homolog of a yeast suppressor gene, leading to the identification of conserved mechanisms underlying FUS pathology (Ju et al. 2011; Barmada et al. 2015; Jackson et al. 2015).
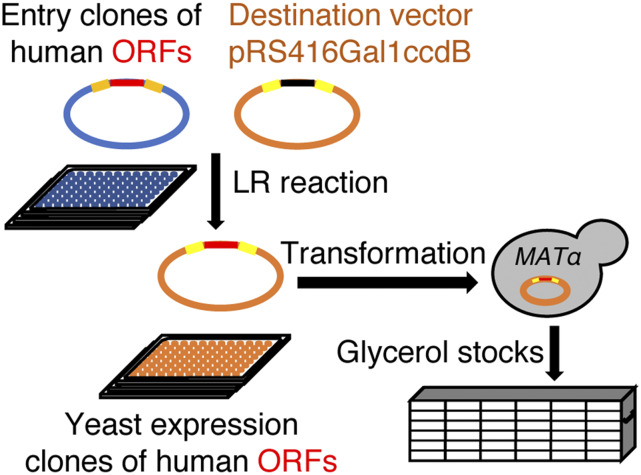
More than 50% of human genes, however, do not have readily detectable homologs in yeast. Human genes without yeast homologs cannot be identified using the current genetic screens in yeast. A lack of sequence similarity does not necessarily exclude the possibility of such non-conserved human genes to function in yeast. Non-conserved human proteins may interact directly with FUS, suppressing its cytotoxicity in yeast. Alternatively, non-conserved human proteins may form inter-species protein-protein interactions with yeast proteins (Zhong et al. 2016), indirectly modulating the toxicity of FUS. To expand the search for new human gene targets related to FUS toxicity in yeast, we developed a new genomic tool that allows systematic overexpression of human genes in yeast (Figure 1). To do so, we first obtained a collection of sequence verified human ORFs as Gateway entry clones (corresponding to 13570 genes) (Yang et al. 2011; Collaboration 2016). While the collection does not contain all human genes, it was developed through unbiased large-scale cloning efforts and represents a key resource for systematic unbiased functional studies of human genes. We individually amplified the plasmid DNA of each entry clone and carried out Gateway LR reaction to transfer each human ORF into a yeast expression vector, pRS416Gal1ccdB (Alberti et al. 2007). The human ORFs cloned in the pRS416Gal1ccdB vector are under the control of the GAL1 promoter, which is induced upon shifting to galactose containing media.
Figure 1.
Generation of yeast-expression human-gene library. The human ORFs were cloned into the yeast expression vector, pRS416Gal1ccdB, by Gateway LR cloning. The yeast-expression plasmids containing the human ORFs (pRS416Gal1hORFs) were isolated and transformed into W303 MATα strain. Yeast strains, each containing an unique human ORF, were arrayed in 96-well microplates, and stored as a glycerol stock at -80 ° for later use.
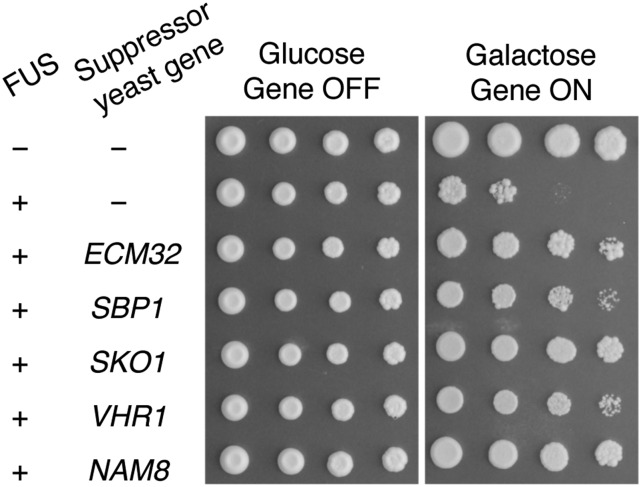
Previously, high-throughput yeast transformation was used to introduce the plasmids bearing the gene to be overexpressed. Preparation of the library of plasmids can be time consuming and costly. Considering that the human-gene library contains many more clones than the yeast-gene libraries, we tested a method that relies on highly efficient yeast mating instead of transformation to introduce the arrayed plasmid library of human ORF clones into the yeast model (Hayden et al. 2018). First, we verified that yeast suppressor genes previously identified in the haploid yeast model of FUS (Ju et al. 2011) also exhibited suppressor effects on FUS toxicity in diploid yeast (Figure 2). This result indicates that screening for suppressors of FUS toxicity may be carried out in a diploid strain background. Next, we introduced the human-gene library into the W303 MATα strain, the isogenic but opposite mating type of the haploid yeast model of FUS (Ju et al. 2011). We arrayed yeast transformants, each with one human-gene expression clone in microplates and archived them as glycerol stock.
Figure 2.
Previously identified yeast suppressor genes rescue FUS toxicity in diploid yeast. Plasmids containing previously identified five yeast genes that suppress FUS toxicity were transformed into a haploid yeast strain w303 MATα. Transformed yeast were crossed with the FUS model generated in the isogenic yeast strain with the opposite mating type, W303 MATa. Diploid yeast were selected, serially diluted, and spotted onto agar plates containing glucose (genes-off condition) and galactose (genes-on condition), respectively. Row 1 shows a control yeast strain containing two empty vectors. Row 2 shows the diploid FUS yeast strain with an empty vector, where expression of FUS is toxic. Rows 3-7 show diploid yeast expressing FUS as well as a suppression gene that can rescue FUS toxicity. The picture, representing three independent experiments, was taken after growth at 30° for three days.
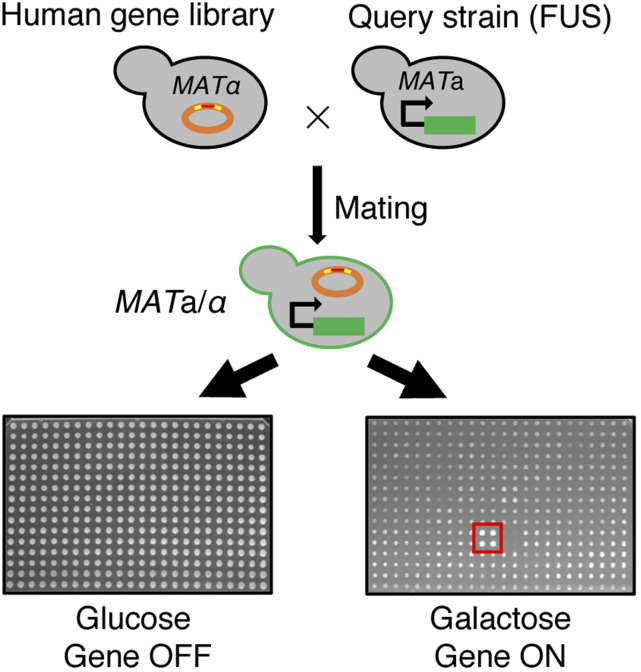
For the screen, we revived the arrayed human-gene library strains and individually crossed to the yeast model of FUS in YEPD. The resulting diploid yeast strains containing both the integrated FUS gene and the human genes from the library were selected. Diploid yeast cells were robotically pinned in quadruplicate onto agar plates containing either glucose or galactose as the carbon source. Since FUS expression is highly toxic to yeast, growth for most diploid strains was reduced on galactose as compare to glucose. In contrast, the growth for a subset of the strains was better, indicating the suppression of FUS toxicity by simultaneously expression of another human gene (Figure 3).
Figure 3.
A mating-based strategy to screen for human suppressor genes rescuing FUS toxicity upon overexpression. W303 MATα yeast containing the expression clones of human ORFs were revived from the glycerol stock and crossed with the FUS model generated in the isogenic yeast strain with the opposite mating type, W303 MATa. Diploid yeast strains were selected and spotted in quadruplicate, using the Singer RotoR robotic equipment, onto agar plates containing glucose (genes-off condition) and galactose (genes-on condition), respectively. On the galactose plates, most yeast had severely reduced growth due to the expression of FUS. The red square labels a screening hit of a human ORF clone that suppresses FUS toxicity allowing yeast to form much larger colonies.
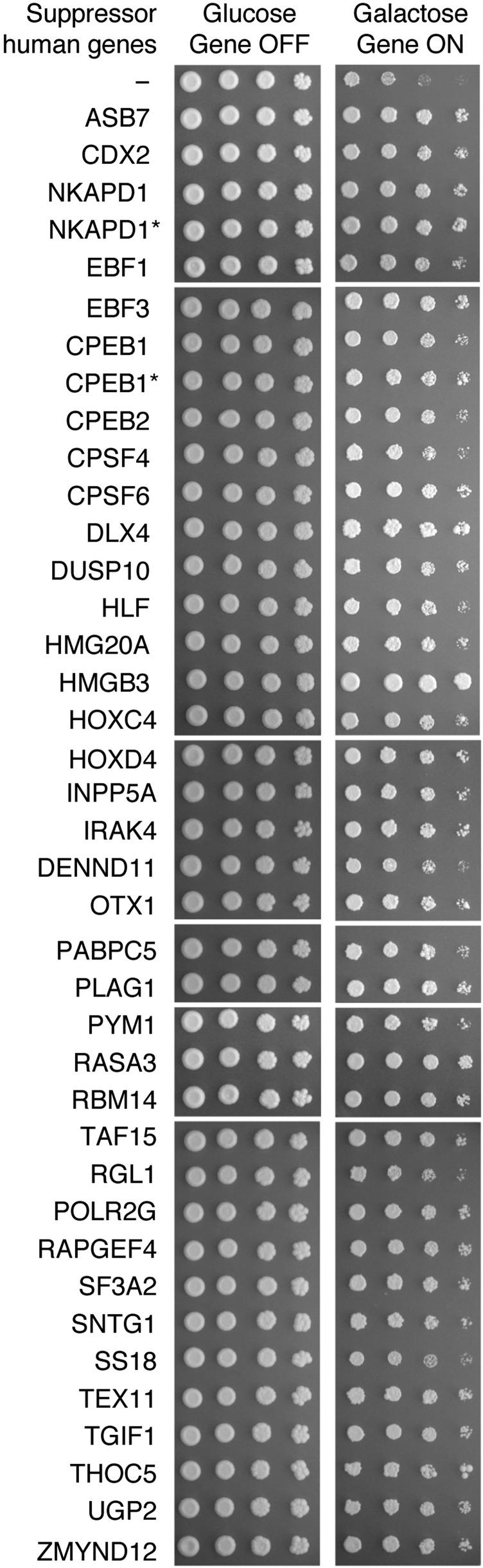
After screening the entire library, we cherrypicked the expression clones of all screening hits and re-transformed them into w303 MATα. We retested the suppressor effect of all new transformants using the mating method. Finally, we confirmed their suppressor phenotype by serial dilution spotting assay (Figure 4). In total, we identified 37 human ORF clones corresponding to 37 human genes (Table 1), that suppress FUS toxicity when overexpressed.
Figure 4.
Human suppressor genes rescue FUS toxicity upon overexpression. W303 MATα containing the empty vector, pRS416Gal1ccdB, or each of the identified human gene that suppress FUS toxicity were crossed with the FUS model generated in the isogenic strain with the opposite mating type, W303 MATa. Diploid yeast strains were selected, serially diluted and spotted onto agar plates containing glucose (genes-off condition) and galactose (genes-on condition), respectively. The picture, representing three independent experiments, was taken after growth at 30° for three days. *indicating different ORF clones of the same gene.
Table 1. List of human genes that suppress FUS toxicity.
| Gene Name | Short Description |
|---|---|
| ASB7 | Belongs to a family of ankyrin repeat proteins that regulate protein turnover by targeting proteins for polyubiquitin and proteasome-mediated degradation |
| CDX2 | Member of caudal-related homeobox transcription factor family |
| CPEB1 | Binding to the polyadenylation element, and regulating mRNA translation and processing |
| CPEB2 | Regulating translation of target mRNAs by binding to the polyadenylation element in the 3′UTR |
| CPSF4 | Component of the cleavage and polyadenylation specificity factor complex |
| CPSF6 | Subunit of a cleavage factor required for 3′ RNA cleavage and polyadenylation processing |
| DENND11 | Probable guanine nucleotide exchange factor (GEF) |
| DLX4 | Member of Distal-less family of homeobox-containing genes |
| DUSP10 | Protein phosphatase involved in the inactivation of MAP kinases |
| EBF1 | Transcription factor expressed in early B-cells, adipocytes, and olfactory neurons. |
| EBF3 | Member of the transcription factors that are involved in B-cell differentiation, bone development and neurogenesis |
| HLF | Homodimers or heterodimers with other PAR family members to activate transcription |
| HMG20A | Chromatin-associated protein playing a role in neuronal differentiation |
| HMGB3 | Associates with chromatin and binds DNA with a preference to non-canonical DNA structures |
| HOXC4 | Member of homeobox family of genes that play important roles in morphogenesis |
| HOXD4 | Member of homeobox family of genes that play important roles in morphogenesis |
| INPP5A | Membrane-associated type I inositol 1,4,5-trisphosphate 5-phosphatase |
| IRAK4 | Serine/threonine-protein kinase that plays a role in initiating innate immune response against foreign pathogens. Involved in Toll-like receptor (TLR) and IL-1R signaling pathways |
| NKAPD1 | Uncharacterized protein C11orf57 |
| OTX1 | Homeodomain-containing transcription factor, playing a role in brain development |
| PABPC5 | Binding to the polyA tail and playing a role in mRNA metabolic processes in the cytoplasm |
| PLAG1 | Proto- oncogene, belongs to the PLAG family of zinc finger transcription factors |
| POLR2G | Subunit of RNA polymerase II, the polymerase responsible for synthesizing mRNA |
| PYM1 | Regulator of the exon junction complex (EJC) as an EJC disassembly factor |
| RAPGEF4 | Guanine nucleotide exchange factor that is activated by binding cAMP |
| RASA3 | Inhibitory regulator of the Ras-c AMP pathway |
| RBM14 | Ribonucleoprotein functioning as a general nuclear coactivator, and an RNA splicing modulator |
| RGL1 | Probable guanine nucleotide exchange factor |
| SF3A2 | A component of the splicing factor SF3A complex involved in pre-mRNA splicing |
| SNTG1 | Member of the syntrophin family, involved in mediating gamma-enolase trafficking to the plasma membrane and in enhancing its neurotrophic activity |
| SS18 | Component of SWI/SNF chromatin remodeling subcomplex |
| TAF15 | Member of the TET family of RNA-binding proteins |
| TEX11 | Regulator of crossing-over during meiosis |
| TGIF1 | Conserved transcription regulator which binds to retinoid X receptor responsive element |
| THOC5 | Component the TREX complex which specifically associates with spliced mRNA, and which is thought to couple mRNA transcription, processing and nuclear export |
| UGP2 | Essential enzyme that catalyzes the conversion of glucose-1- phosphate to UDP-glucose |
| ZMYND12 | Zinc finger MYND domain-containing protein 12 |
Human suppressor genes of FUS toxicity
The identified suppressor genes appear to have a wide range of functions. GO enrichment analysis identified several functional groups of genes are overrepresented (Table 2), particularly those with RNA or DNA binding functions. FUS is a nucleic acid binding protein that has been implicated in transcription regulation, RNA splicing, RNA transport, and RNA stability (Polymenidou et al. 2012; Butti and Patten 2018). The identification of the group of suppressor genes with similar functions supports the idea that expression of FUS perturbs RNA metabolism in yeast, thus inducing cellular toxicity.
Table 2. Enriched functions of the human-gene suppressors of FUS toxicity.
| GO terms | GO ID | GO category | Enrichment folds | P value | FDR |
|---|---|---|---|---|---|
| mRNA 3′-UTR binding | GO:0003730 | Molecular function | 15.8 | 1.43E-04 | 4.92E-02 |
| mRNA binding | GO:0003729 | Molecular function | 10.6 | 1.01E-06 | 2.25E-03 |
| DNA-binding transcription activator activity, RNA polymerase II-specific | GO:0001228 | Molecular function | 6.3 | 4.05E-05 | 3.03E-02 |
| DNA-binding transcription activator activity | GO:0001216 | Molecular function | 6.2 | 4.30E-05 | 2.75E-02 |
| RNA polymerase II regulatory region sequence-specific DNA binding | GO:0000977 | Molecular function | 4.4 | 8.05E-05 | 4.51E-02 |
| RNA polymerase II regulatory region DNA binding | GO:0001012 | Molecular function | 4.4 | 8.16E-05 | 4.06E-02 |
| Transcription regulatory region sequence-specific DNA binding | GO:0000976 | Molecular function | 4.1 | 1.31E-04 | 4.87E-02 |
| Double-stranded DNA binding | GO:0003690 | Molecular function | 3.9 | 9.27E-05 | 4.15E-02 |
| DNA-binding transcription factor activity, RNA polymerase II-specific | GO:0000981 | Molecular function | 3.9 | 1.00E-04 | 4.09E-02 |
| DNA-binding transcription factor activity | GO:0003700 | Molecular function | 3.4 | 1.65E-04 | 4.61E-02 |
| Transcription regulator activity | GO:0140110 | Molecular function | 3.2 | 3.60E-05 | 3.23E-02 |
| RNA binding | GO:0003723 | Molecular function | 3.1 | 5.85E-05 | 3.73E-02 |
| Embryonic organ development | GO:0048568 | Biological process | 6.6 | 2.80E-05 | 2.03E-02 |
| Positive regulation of RNA metabolic process | GO:0051254 | Biological process | 3.7 | 6.31E-07 | 4.81E-03 |
| Positive regulation of transcription, DNA-templated | GO:0045893 | Biological process | 3.7 | 3.73E-06 | 5.68E-03 |
| Positive regulation of macromolecule biosynthetic process | GO:0010557 | Biological process | 3.5 | 6.52E-07 | 3.31E-03 |
| positive regulation of nucleic acid-templated transcription | GO:1903508 | Biological process | 3.4 | 7.93E-06 | 9.30E-03 |
| Positive regulation of RNA biosynthetic process | GO:1902680 | Biological process | 3.4 | 8.01E-06 | 8.72E-03 |
| Positive regulation of nucleobase-containing compound metabolic process | GO:0045935 | Biological process | 3.4 | 2.37E-06 | 4.02E-03 |
| Positive regulation of cellular biosynthetic process | GO:0031328 | Biological process | 3.3 | 1.39E-06 | 4.23E-03 |
| Positive regulation of gene expression | GO:0010628 | Biological process | 3.3 | 1.39E-06 | 3.53E-03 |
| Positive regulation of biosynthetic process | GO:0009891 | Biological process | 3.3 | 1.78E-06 | 3.87E-03 |
| Nuclear chromatin | GO:0000790 | Cellular component | 4.2 | 2.01E-05 | 1.29E-02 |
| Nuclear chromosome | GO:0000228 | Cellular component | 4.0 | 6.54E-06 | 1.26E-02 |
| Chromatin | GO:0000785 | Cellular component | 3.6 | 9.32E-05 | 4.50E-02 |
FDR correction < 0.05; Fold of enrichment > 3.
RNA binding or splicing
Cellular toxicity of FUS has been speculated to involve binding and sequestering of essential proteins and mRNAs into persistent intracellular inclusion bodies (Ciryam et al. 2017; Hayden et al. 2017). Consistent with this idea, previous studies using a yeast model of FUS have shown that FUS aggregation colocalizes with stress granules(Sun et al. 2011b) The identified human-gene suppressors in this group may compete to bind yeast mRNAs and protect them from being sequestered into FUS inclusions.
CPEB1 and CPEB2: Cytoplasmic polyadenylation element-binding protein 1 and 2 are member of the cytoplasmic polyadenylation element binding protein family, which promotes polyadenylation induced translation. CPEB proteins are involved in a wide range of cellular functions, including germ-cell development, cell division, synaptic plasticity, learning and memory (Richter 2007; Ivshina et al. 2014).
CPSF4 and CPSF6: Cleavage and polyadenylation specificity factor subunit 4 and 6 are essential components of the 3′ end processing machinery of pre-mRNAs. The cleavage factor complex plays a key role in pre-mRNAs 3′-cleavage and polyadenylation. Studies have found overexpression of CPSF4 in several types of cancer, such as lung, breast, and colorectal cancer (Zhang et al. 2016; Wu et al. 2019). Mutation studies indicate CPSF6 functions in regulating poly A site selection and preventing premature 3′-UTR cleavage (Sasado et al. 2017). It was reported that the interaction of CPSF6 with HIV-1 capsid plays a critical role for targeting integration of HIV-1 to transcriptionally active chromatin (Sowd et al. 2016). CPSF6 is also implicated in breast cancer as a tumor promoting factor (Binothman et al. 2017).
PABPC5: Poly(A)-binding protein 5 is a protein that binds to the polyA tail of eukaryotic mRNAs. It plays a critical role in the regulation of mRNA transport and mRNA decay in the cytoplasm (Bhattacharjee and Bag 2012).
POLR2G: RNA polymerase II subunit B7 is a conserved protein that is part of the large 12-subunit RNA polymerase II. Together with another subunit B4, POLR2G not only is important for initiation of transcription, but also plays critical roles in diverse cellular processes, such as mRNA export, mRNA decay, DNA repair, protein translation, and stress response (Sharma and Kumari 2013; Kumar et al. 2019).
PYM1: Partner of Y14 and mago (PYM homolog 1) is an exon junction complex-associated factor that play an important role in exon-exon junction complex disassembly. It is also involved in the nonsense-mediated decay pathway and regulation of protein translation (Diem et al. 2007).
RBM14: RNA-binding motif protein 14 is an RNA binding protein that is involved in multiple important cellular processes, such as transcriptional regulation (Li et al. 2017), DNA damage response (Yuan et al. 2014; Simon et al. 2017), genome integrity maintenance (Shiratsuchi et al. 2015; Li et al. 2020), and maintaining the pluripotency of embryonic stem cells (Chen et al. 2018).
SF3A2: Splicing factor 3A subunit 2 is a subunit of the splicing factor 3a protein complex that includes subunits 1, 2 and 3 and is necessary for the assembly of spliceosome that plays a critical role in pre-mRNA splicing (Bennett and Reed 1993). Together with Prp31, it also has a direct role in mitotic chromosome segregation(Pellacani et al. 2018).
TAF15: Mutations in TAF15 were found in familial form of ALS (Couthouis et al. 2011; Ticozzi et al. 2011). TAF15 encodes a TATA-binding protein-associated factor that functions in promoter recognition, transcription initiation complex assembly, and transcription activation (Kapeli et al. 2017).
THOC5: THO complex subunit 5 is a conserved protein in the THO complex that functions in processing and transport of mRNA (Scott et al. 2019). THOC5 is phosphorylated upon extracellular stimuli suggesting its role in DNA damage response (Ramachandran et al. 2011), growth factor/cytokine-mediated differentiation, and cancer development (Wang et al. 2013; Tran et al. 2016). New evidence also implicates this protein in synapse development and dopamine neuron survival (Maeder et al. 2018).
DNA binding or transcription factors
Overexpression of FUS in yeast may affect the expression of some yeast genes and subsequently disrupt their cellular functions and reduce cell fitness. Although the transcription factors identified from this screen may not directly target yeast genes and regulate their expression, it is possible that the suppressor proteins interact with FUS or with other yeast transcriptional factors, thus indirectly reverse the detrimental effect of FUS on yeast gene expression.
CDX2: Homeobox protein CDX-2 is a transcription factor that plays import roles in cell differentiation and proliferation. It is a well-known cancer risk factor. Abnormal CDX2 expression was indicated in metastasis of multiple cancers such as esophagus, stomach, colon, breast, ovarian, prostate cancer, and leukemia (Chawengsaksophak 2019).
DLX4: Homeobox protein DLX-4 is a transcription factor that belongs to the Distal-less (Dlx) family of proteins. DLX4 plays important functions in cell differentiation, proliferation, and migration. Abnormal DLX4 expression is associated with multiple cancers and preeclampsia (Sun et al. 2011a; Zhang et al. 2012). In addition, it was reported that DLX4, functionally replacing c-Myc, promotes generation of human induced pluripotent stem cells (Tamaoki et al. 2014).
EBF1 and EBF3: both genes encode highly conserved Collier/Olf/EBF (COE) family of transcription factors. EBF1 controls the expression of key proteins in B cell differentiation, signal transduction and function (Vilagos et al. 2012; Kong et al. 2016). EBF3 is thought to have functions in the nervous system, as mutations in EBF3 cause hypotonia, ataxia, and delayed development syndrome, a genetic neurodevelopmental syndrome (Chao et al. 2017).
HOXC4 and HOXD4: Homeobox proteins C4 and D4 are a group of related transcription factors that play important roles in specifying regions of the body plan during development, ensuring that the structures form in the correct places (Rastegar et al. 2004; Nolte et al. 2006). Expression of HOXC4 and HOXD4 was elevated in breast cancer, myeloid leukemias and uveal melanoma, suggesting its role as a potential oncogene (Bijl et al. 1997; Wu et al. 2020).
HLF: HLF gene encode a protein called hepatic leukemia factor. A Chimeric protein between HLF and E2A resulting from the leukemogenic translocation is responsible for a rare form of acute lymphoblastic leukemia. This arises from both impairment of normal function of E2A and TCF3 as a tumor suppressor in T lymphocytes, and activation of survival pathway triggered through HLF DNA binding domain (Seidel and Look 2001; Panagopoulos et al. 2012).
HMGB3 and HMG20A: both genes encode members of the HMG-box superfamily of DNA-binding proteins. HMGB 3 encodes the high mobility group protein B3, which plays an important function in maintaining stem cell populations, and increased expression of HMGB3 is a risk factor for several type of cancer, possibly through its regulation on WNT/β-catenin pathway (Gao et al. 2015; Zhang et al. 2017; Lv et al. 2019). HMG20A encodes high mobility group protein 20A, a close homolog HMG20B that plays important roles in neuronal differentiation (Artegiani et al. 2010), and is required for SNAI1-mediated epithelial to mesenchymal transition (Rivero et al. 2015).
HOXC4 and HOXD4: Homeobox proteins C4 and D4 are a group of related transcription factors that play important roles in specifying regions of the body plan during development, ensuring that the structures form in the correct places (Rastegar et al. 2004; Nolte et al. 2006). Expression of HOXC4 and HOXD4 was elevated in breast cancer, myeloid leukemias and uveal melanoma, suggesting its role as a potential oncogene (Bijl et al. 1997; Wu et al. 2020).
OTX1: OTX1 is a member of the bicoid subfamily of homeodomain-containing transcription factors that play critical roles in brain and sensory organ development (Simeone 1998; Zhang et al. 2015). Abnormal expression of OTX1 is also recently reported as a risk factor for various cancers, including neuroblastoma, breast, liver, gastric and colorectal cancer(Li et al. 2016; Qin et al. 2018; Li et al. 2019; Micheloni et al. 2019; Yang et al. 2019).
PLAG1: Pleomorphic adenoma gene 1 (PLAG1) encodes a zinc finger transcription factor, which is developmentally regulated. Study of PLAG1 knockout mice suggests it is an important regulator in postnatal growth and reproduction (Juma et al. 2016). Overexpression and knockdown of PLAG1 also indicate its role in regulating neuronal gene expression and neuronal differentiation of neocortical neural progenitor cells(Sakai et al. 2019).
TGIF1: TGIF1 belongs to the three-amino acid loop extension (TALE) superclass of atypical homeodomain proteins that function as transcription regulators. It plays an important function in normal brain development. Mutations in this gene are associated with holoprosencephaly type 4, which is a structural anomaly of the brain (Wotton and Taniguchi 2018; Zhu et al. 2018). Abnormal expression of TGIF1 has also been implicated in many cancers.
Genes involved in cell signaling
DUSP10: Dual specificity protein phosphatase 10 is an enzyme that inactivates stress-activated kinases, such as p38 and SAPK/JNK, by dephosphorylation. The gene is ubiquitously expressed, and its expression is highly induced upon stress stimuli. Studies indicated a clear role of DUSP10 in inflammation, immunity, and cancer (Jimenez-Martinez et al. 2019).
RAPGEF4: Rap guanine nucleotide exchange factor (GEF) 4 is a protein that is targeted by the second messenger cAMP, and functions as a guanine nucleotide exchange factor for the Ras-like small GTPase Rap upon cAMP stimulation. It is involved in various cellular processes such as integrin-mediated cell adhesion, cell-cell junction formation, cell proliferation and differentiation, cell survival, and neuronal signaling (Bos 2006; Roscioni et al. 2008; Kumar et al. 2018).
RGL1: Ral guanine nucleotide dissociation stimulator-like 1 is a small GTPase functioning in signal transduction (Sood et al. 2000). The Rap1-Rgl-Ral signaling network plays important role in regulating neuroblast cortical polarity and spindle orientation (Carmena et al. 2011).
RASA3: Ras GTPase-activating protein 3, a member of the GAP1 subfamily, is an inositol 1,3,4,5-tetrakisphophate-binding protein (Schurmans et al. 2015). The protein enhances the weak intrinsic GTPase activity of RAS proteins resulting in the inactive GDP-bound form of RAS (Cullen et al. 1995). It plays important roles in erythropoiesis, megakaryopoiesis, megakaryocyte adhesion and migration as well as integrin signaling (Blanc et al. 2012)
INPP5A: The protein encoded by this gene is inositol polyphosphate 5-phosphatase A, a membrane-associated enzyme. It hydrolyzes inositol polyphosphate, which regulates calcium release from intracellular stores and acts as a second messenger regulating cell proliferation and survival. Deletion of INPP5A causes progressive and permanent loss of cerebellar Purkinje cells in mice, suggesting its crucial role in Purkinje cell survival (Ooms et al. 2009; Yang et al. 2015).
IRAK4: Interleukin-1 receptor-associated kinase 4 is one of the four members of the IRAK family that plays an important role in signaling innate immune responses from Toll-like receptors. Accumulated evidence indicates its abnormal expression in inflammatory autoimmune disorders (Su et al. 2020). Interestingly, targeted degradation of IRAK4 was used for the treatment of cancer, neurodegenerative and cardiovascular diseases (Kargbo 2019a; Kargbo 2019b).
Additional genes
ASB7: Ankyrin repeat and SOCS box protein 7 is an E3 ubiquitin ligase. It plays a crucial role in regulating spindle dynamics and genome integrity through targeting DDA3 for proteasomal degradation (Uematsu et al. 2016). A recent study also indicated the expression of ASB7 was elevated upon activation of the unfolded protein response pathway under endoplasmic reticulum stress (Anasa et al. 2018).
DENND11: DENN domain containing 11 protein is highly conserved in a wide range of animal species. Studies indicated its important role in neurogenesis and neuronal recovery in the hippocampus following transient cerebral ischemia (Zhang et al. 2007).
NKAPD1: NKAPD1 encodes an uncharacterized protein C11ORF57, which is also called NKAP Domain Containing 1.
SNTG1: SNTG1 encodes gamma-1 syntrophin, a neuronal cell specific protein. Syntrophins are scaffold cytoplasmic membrane proteins that bind signaling molecules, such as gamma-enolase for its neurotrophic activity (Hogan et al. 2001; Hafner et al. 2010). Gamma-1 synthrophin associates directly with dystrophin, a protein involved in the Duchenne muscular dystrophy (Bashiardes et al. 2004).
SS18: SS18 gene encodes a protein called synovial sarcoma translocated to X chromosome. Disruption of SS18 gene in mouse results in embryonic death due to placental failure. Fusion SS18-SSX1 is believed to underlie the pathogenesis of synovial sarcoma through elevated expression of the key Wnt target AXIN2 (De Bruijn et al. 2006; Cironi et al. 2016).
TEX11: TEX11, a male germ cell specific X-linked gene, encodes a testis expressed sequence 11 protein, which is essential for meiosis and male fertility in animals (Yatsenko et al. 2015).
UGP2: UGP2 gene encodes UTP-glucose-1-phosphate uridylyltransferase, an enzyme conserved from bacteria to human as a key player in glycogenesis and cell wall synthesis. In yeast, its expression contributes to oxidative stress response and long-term cell survival through production of storage carbohydrates. In higher animals, it is highly active in the liver and muscles (Smith and Rutter 2007; Yi and Huh 2015).
ZMYND12: Zinc finger MYND domain-containing protein 12 is conserved among higher eukaryotes. Its function is unknown.
In summary, we screened a previously established yeast model of FUS against a new collection of 13570 human genes and identified 37 suppressor genes of FUS induced toxicity in yeast. Although the identified suppressors have a wide range of cellular functions, genes encoding proteins involved in RNA and DNA binding are significantly overrepresented, suggesting a strong relationship between the toxicity of FUS in yeast and dysregulation in RNA metabolism, which is a widely considered major contributor to ALS pathogenesis (Polymenidou et al. 2012; Butti and Patten 2018).
Acknowledgments
We thank Dr. Marc Vidal and Center for Cancer Systems Biology (CCSB) at Dana-Farber Cancer Institute and Harvard Medical School for sharing the human ORFeome entry clone collections. We thank the members of the Ju and Zhong groups for laboratory assistance and helpful discussions. This work was supported by funding from the Fidelity Bioscience Research Initiative.
Footnotes
Communicating editor: G. Brown
Literature Cited
- Alberti S., Gitler A. D., and Lindquist S., 2007. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24: 913–919. 10.1002/yea.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anasa V. V., Manickam M., Talwar P., and Ravanan P., 2018. Identification of ASB7 as ER stress responsive gene through a genome wide in silico screening for genes with ERSE. PLoS One 13: e0194310 10.1371/journal.pone.0194310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegiani B., Labbaye C., Sferra A., Quaranta M. T., Torreri P. et al. , 2010. The interaction with HMG20a/b proteins suggests a potential role for beta-dystrobrevin in neuronal differentiation. J. Biol. Chem. 285: 24740–24750. 10.1074/jbc.M109.090654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmada S. J., Ju S., Arjun A., Batarse A., Archbold H. C. et al. , 2015. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc. Natl. Acad. Sci. USA 112: 7821–7826. 10.1073/pnas.1509744112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiardes S., Veile R., Allen M., Wise C. A., Dobbs M. et al. , 2004. SNTG1, the gene encoding gamma1-syntrophin: a candidate gene for idiopathic scoliosis. Hum. Genet. 115: 81–89. 10.1007/s00439-004-1121-y [DOI] [PubMed] [Google Scholar]
- Bennett M., and Reed R., 1993. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science 262: 105–108. 10.1126/science.8211113 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee R. B., and Bag J., 2012. Depletion of nuclear poly(A) binding protein PABPN1 produces a compensatory response by cytoplasmic PABP4 and PABP5 in cultured human cells. PLoS One 7: e53036 10.1371/journal.pone.0053036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijl J. J., Rieger E., van Oostveen J. W., Walboomers J. M., Kreike M. et al. , 1997. HOXC4, HOXC5, and HOXC6 expression in primary cutaneous lymphoid lesions. High expression of HOXC5 in anaplastic large-cell lymphomas. Am. J. Pathol. 151: 1067–1074. [PMC free article] [PubMed] [Google Scholar]
- Binothman N., Hachim I. Y., Lebrun J. J., and Ali S., 2017. CPSF6 is a Clinically Relevant Breast Cancer Vulnerability Target: Role of CPSF6 in Breast Cancer. EBioMedicine 21: 65–78. 10.1016/j.ebiom.2017.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc L., Ciciotte S. L., Gwynn B., Hildick-Smith G. J., Pierce E. L. et al. , 2012. Critical function for the Ras-GTPase activating protein RASA3 in vertebrate erythropoiesis and megakaryopoiesis. Proc. Natl. Acad. Sci. USA 109: 12099–12104. 10.1073/pnas.1204948109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., 2006. Epac proteins: multi-purpose cAMP targets. Trends Biochem. Sci. 31: 680–686. 10.1016/j.tibs.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Butti Z., and Patten S. A., 2018. RNA Dysregulation in Amyotrophic Lateral Sclerosis. Front. Genet. 9: 712 10.3389/fgene.2018.00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A., Makarova A., and Speicher S., 2011. The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J. Cell Biol. 195: 553–562. 10.1083/jcb.201108112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao H. T., Davids M., Burke E., Pappas J. G., Rosenfeld J. A. et al. , 2017. A Syndromic Neurodevelopmental Disorder Caused by De Novo Variants in EBF3. Am. J. Hum. Genet. 100: 128–137. 10.1016/j.ajhg.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawengsaksophak K., 2019. Cdx2 Animal Models Reveal Developmental Origins of Cancers. Genes (Basel) 10: 928 10.3390/genes10110928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Zhang D., Zhang L., Feng G., Zhang B. et al. , 2018. RBM14 is indispensable for pluripotency maintenance and mesoderm development of mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 501: 259–265. 10.1016/j.bbrc.2018.04.231 [DOI] [PubMed] [Google Scholar]
- Cironi L., Petricevic T., Fernandes Vieira V., Provero P., Fusco C. et al. , 2016. The fusion protein SS18–SSX1 employs core Wnt pathway transcription factors to induce a partial Wnt signature in synovial sarcoma. Sci. Rep. 6: 22113 10.1038/srep22113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Lambert-Smith I. A., Bean D. M., Freer R., Cid F. et al. , 2017. Spinal motor neuron protein supersaturation patterns are associated with inclusion body formation in ALS. Proc. Natl. Acad. Sci. USA 114: E3935–E3943. 10.1073/pnas.1613854114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaboration O. R., 2016. The ORFeome Collaboration: a genome-scale human ORF-clone resource. Nat. Methods 13: 191–192. 10.1038/nmeth.3776 [DOI] [PubMed] [Google Scholar]
- Cooper A. A., Gitler A. D., Cashikar A., Haynes C. M., Hill K. J. et al. , 2006. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science 313: 324–328. 10.1126/science.1129462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J., Hart M. P., Shorter J., DeJesus-Hernandez M., Erion R. et al. , 2011. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. USA 108: 20881–20890. 10.1073/pnas.1109434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P. J., Hsuan J. J., Truong O., Letcher A. J., Jackson T. R. et al. , 1995. Identification of a specific Ins(1,3,4,5)P4-binding protein as a member of the GAP1 family. Nature 376: 527–530. 10.1038/376527a0 [DOI] [PubMed] [Google Scholar]
- de Bruijn D. R., Peters W. J., Chuva de Sousa Lopes S. M., van Dijk A. H., Willemse M. P. et al. , 2006. Targeted disruption of the synovial sarcoma-associated SS18 gene causes early embryonic lethality and affects PPARBP expression. Hum. Mol. Genet. 15: 2936–2944. 10.1093/hmg/ddl235 [DOI] [PubMed] [Google Scholar]
- Diem M. D., Chan C. C., Younis I., and Dreyfuss G., 2007. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 14: 1173–1179. 10.1038/nsmb1321 [DOI] [PubMed] [Google Scholar]
- Fleming M. S., and Gitler A. D., 2011. High-throughput yeast plasmid overexpression screen. J. Vis. Exp. 53: 2836 10.3791/2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Zou Z., Gao J., Zhang H., Lin Z. et al. , 2015. Increased expression of HMGB3: a novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 8: 345–352. [PMC free article] [PubMed] [Google Scholar]
- Hafner A., Obermajer N., and Kos J., 2010. gamma-1-syntrophin mediates trafficking of gamma-enolase towards the plasma membrane and enhances its neurotrophic activity. Neurosignals 18: 246–258. 10.1159/000324292 [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Temple G. F., and Brasch M. A., 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788–1795. 10.1101/gr.143000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden E., Chen S., Chumley A., Zhong Q., and Ju S., 2018. Mating-based Overexpression Library Screening in Yeast. J. Vis. Exp. 137: 57978 10.3791/57978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden E., Cone A., and Ju S., 2017. Supersaturated proteins in ALS. Proc. Natl. Acad. Sci. USA 114: 5065–5066. 10.1073/pnas.1704885114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., and Jacobson A., 2015. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu. Rev. Genet. 49: 339–366. 10.1146/annurev-genet-112414-054639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan A., Shepherd L., Chabot J., Quenneville S., Prescott S. M. et al. , 2001. Interaction of gamma 1-syntrophin with diacylglycerol kinase-zeta. Regulation of nuclear localization by PDZ interactions. J. Biol. Chem. 276: 26526–26533. 10.1074/jbc.M104156200 [DOI] [PubMed] [Google Scholar]
- Hu Y., Rolfs A., Bhullar B., Murthy T. V., Zhu C. et al. , 2007. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 17: 536–543. 10.1101/gr.6037607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivshina M., Lasko P., and Richter J. D., 2014. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 30: 393–415. 10.1146/annurev-cellbio-101011-155831 [DOI] [PubMed] [Google Scholar]
- Jackson K. L., Dayton R. D., Orchard E. A., Ju S., Ringe D. et al. , 2015. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther. 22: 20–28. 10.1038/gt.2014.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Martinez M., Stamatakis K., and Fresno M., 2019. The Dual-Specificity Phosphatase 10 (DUSP10): Its Role in Cancer, Inflammation, and Immunity. Int. J. Mol. Sci. 20: 1626 10.3390/ijms20071626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S., Tardiff D. F., Han H., Divya K., Zhong Q. et al. , 2011. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 9: e1001052 10.1371/journal.pbio.1001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juma A. R., Damdimopoulou P. E., Grommen S. V., Van de Ven W. J., and De Groef B., 2016. Emerging role of PLAG1 as a regulator of growth and reproduction. J. Endocrinol. 228: R45–R56. 10.1530/JOE-15-0449 [DOI] [PubMed] [Google Scholar]
- Kapeli K., Martinez F. J., and Yeo G. W., 2017. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum. Genet. 136: 1193–1214. 10.1007/s00439-017-1830-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargbo R. B., 2019a PROTAC Degradation of IRAK4 for the Treatment of Cancer. ACS Med. Chem. Lett. 10: 1370–1371. 10.1021/acsmedchemlett.9b00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargbo R. B., 2019b PROTAC Degradation of IRAK4 for the Treatment of Neurodegenerative and Cardiovascular Diseases. ACS Med. Chem. Lett. 10: 1251–1252. 10.1021/acsmedchemlett.9b00385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong N. R., Davis M., Chai L., Winoto A., and Tjian R., 2016. MEF2C and EBF1 Co-regulate B Cell-Specific Transcription. PLoS Genet. 12: e1005845 10.1371/journal.pgen.1005845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Varshney S., Sengupta S., and Sharma N., 2019. A comparative study of the proteome regulated by the Rpb4 and Rpb7 subunits of RNA polymerase II in fission yeast. J. Proteomics 199: 77–88. 10.1016/j.jprot.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Kumar N., Prasad P., Jash E., Saini M., Husain A. et al. , 2018. Insights into exchange factor directly activated by cAMP (EPAC) as potential target for cancer treatment. Mol. Cell. Biochem. 447: 77–92. 10.1007/s11010-018-3294-z [DOI] [PubMed] [Google Scholar]
- Kwiatkowski T. J. Jr, Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R. et al. , 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208. 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- Li H., Miao Q., Xu C. W., Huang J. H., Zhou Y. F. et al. , 2016. OTX1 Contributes to Hepatocellular Carcinoma Progression by Regulation of ERK/MAPK Pathway. J. Korean Med. Sci. 31: 1215–1223. 10.3346/jkms.2016.31.8.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang C., Feng G., Zhang L., Chen G. et al. , 2020. Rbm14 maintains the integrity of genomic DNA during early mouse embryogenesis via mediating alternative splicing. Cell Prolif. 53: e12724 10.1111/cpr.12724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang Y., Rao X., Wang Y., Feng W. et al. , 2017. Roles of alternative splicing in modulating transcriptional regulation. BMC Syst. Biol. 11: 89 10.1186/s12918-017-0465-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhao L. M., Zhang C., Li M., Gao B. et al. , 2020. The lncRNA FEZF1–AS1 promotes the progression of colorectal cancer through regulating OTX1 and targeting miR-30a-5p, Oncol Res; 28: 51–63. 10.3727/096504019X15619783964700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Lv M., Ji X., Xue L., Rui C. et al. , 2019. Down-regulated expressed protein HMGB3 inhibits proliferation and migration, promotes apoptosis in the placentas of fetal growth restriction. Int. J. Biochem. Cell Biol. 107: 69–76. 10.1016/j.biocel.2018.11.007 [DOI] [PubMed] [Google Scholar]
- Maeder, C. I., J. I. Kim, X. Liang, K. Kaganovsky, A. Shen et al., 2018 The THO Complex Coordinates Transcripts for Synapse Development and Dopamine Neuron Survival. Cell 174: 1436–1449 e1420. 10.1016/j.cell.2018.07.046 [DOI] [PMC free article] [PubMed]
- Micheloni, G., G. Millefanti, A. Conti, C. Pirrone, A. Marando et al., 2019 Identification of OTX1 and OTX2 As Two Possible Molecular Markers for Sinonasal Carcinomas and Olfactory Neuroblastomas. J Vis Exp. [DOI] [PubMed]
- Nolte C., Rastegar M., Amores A., Bouchard M., Grote D. et al. , 2006. Stereospecificity and PAX6 function direct Hoxd4 neural enhancer activity along the antero-posterior axis. Dev. Biol. 299: 582–593. 10.1016/j.ydbio.2006.08.061 [DOI] [PubMed] [Google Scholar]
- Ooms L. M., Horan K. A., Rahman P., Seaton G., Gurung R. et al. , 2009. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 419: 29–49. 10.1042/BJ20081673 [DOI] [PubMed] [Google Scholar]
- Panagopoulos I., Micci F., Thorsen J., Haugom L., Tierens A. et al. , 2012. A novel TCF3-HLF fusion transcript in acute lymphoblastic leukemia with a t(17;19)(q22;p13). Cancer Genet. 205: 669–672. 10.1016/j.cancergen.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Pellacani C., Bucciarelli E., Renda F., Hayward D., Palena A. et al. , 2018. Splicing factors Sf3A2 and Prp31 have direct roles in mitotic chromosome segregation. eLife 7: e40325 10.7554/eLife.40325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M., Lagier-Tourenne C., Hutt K. R., Bennett C. F., Cleveland D. W. et al. , 2012. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 1462: 3–15. 10.1016/j.brainres.2012.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S. C., Zhao Z., Sheng J. X., Xu X. H., Yao J. et al. , 2018. Dowregulation of OTX1 attenuates gastric cancer cell proliferation, migration and invasion. Oncol. Rep. 40: 1907–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Tran D. D., Klebba-Faerber S., Kardinal C., Whetton A. D. et al. , 2011. An ataxia-telangiectasia-mutated (ATM) kinase mediated response to DNA damage down-regulates the mRNA-binding potential of THOC5. RNA 17: 1957–1966. 10.1261/rna.2820911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar M., Kobrossy L., Kovacs E. N., Rambaldi I., and Featherstone M., 2004. Sequential histone modifications at Hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol. Cell. Biol. 24: 8090–8103. 10.1128/MCB.24.18.8090-8103.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. D., 2007. CPEB: a life in translation. Trends Biochem. Sci. 32: 279–285. 10.1016/j.tibs.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Rivero S., Ceballos-Chavez M., Bhattacharya S. S., and Reyes J. C., 2015. HMG20A is required for SNAI1-mediated epithelial to mesenchymal transition. Oncogene 34: 5264–5276. 10.1038/onc.2014.446 [DOI] [PubMed] [Google Scholar]
- Roscioni S. S., Elzinga C. R., and Schmidt M., 2008. Epac: effectors and biological functions. Naunyn Schmiedebergs Arch. Pharmacol. 377: 345–357. 10.1007/s00210-007-0246-7 [DOI] [PubMed] [Google Scholar]
- Sakai H., Fujii Y., Kuwayama N., Kawaji K., Gotoh Y. et al. , 2019. Plag1 regulates neuronal gene expression and neuronal differentiation of neocortical neural progenitor cells. Genes Cells 24: 650–666. 10.1111/gtc.12718 [DOI] [PubMed] [Google Scholar]
- Sasado T., Kondoh H., Furutani-Seiki M., and Naruse K., 2017. Mutation in cpsf6/CFIm68 (Cleavage and Polyadenylation Specificity Factor Subunit 6) causes short 3′UTRs and disturbs gene expression in developing embryos, as revealed by an analysis of primordial germ cell migration using the medaka mutant naruto. PLoS One 12: e0172467 10.1371/journal.pone.0172467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmans S., Polizzi S., Scoumanne A., Sayyed S., and Molina-Ortiz P., 2015. The Ras/Rap GTPase activating protein RASA3: from gene structure to in vivo functions. Adv. Biol. Regul. 57: 153–161. 10.1016/j.jbior.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Scott D. D., Aguilar L. C., Kramar M., and Oeffinger M., 2019. It’s Not the Destination, It’s the Journey: Heterogeneity in mRNA Export Mechanisms. Adv. Exp. Med. Biol. 1203: 33–81. 10.1007/978-3-030-31434-7_2 [DOI] [PubMed] [Google Scholar]
- Seidel M. G., and Look A. T., 2001. E2A-HLF usurps control of evolutionarily conserved survival pathways. Oncogene 20: 5718–5725. 10.1038/sj.onc.1204591 [DOI] [PubMed] [Google Scholar]
- Sharma N., and Kumari R., 2013. Rpb4 and Rpb7: multifunctional subunits of RNA polymerase II. Crit. Rev. Microbiol. 39: 362–372. 10.3109/1040841X.2012.711742 [DOI] [PubMed] [Google Scholar]
- Shiratsuchi G., Takaoka K., Ashikawa T., Hamada H., and Kitagawa D., 2015. RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. EMBO J. 34: 97–114. 10.15252/embj.201488979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A., 1998. Otx1 and Otx2 in the development and evolution of the mammalian brain. EMBO J. 17: 6790–6798. 10.1093/emboj/17.23.6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon N. E., Yuan M., and Kai M., 2017. RNA-binding protein RBM14 regulates dissociation and association of non-homologous end joining proteins. Cell Cycle 16: 1175–1180. 10.1080/15384101.2017.1317419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. L., and Rutter J., 2007. Regulation of glucose partitioning by PAS kinase and Ugp1 phosphorylation. Mol. Cell 26: 491–499. 10.1016/j.molcel.2007.03.025 [DOI] [PubMed] [Google Scholar]
- Sood R., Makalowska I., Carpten J. D., Robbins C. M., Stephan D. A. et al. , 2000. The human RGL (RalGDS-like) gene: cloning, expression analysis and genomic organization. Biochim. Biophys. Acta 1491: 285–288. 10.1016/S0167-4781(00)00031-2 [DOI] [PubMed] [Google Scholar]
- Sowd G. A., Serrao E., Wang H., Wang W., Fadel H. J. et al. , 2016. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 113: E1054–E1063. 10.1073/pnas.1524213113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L. C., Xu W. D., and Huang A. F., 2020. IRAK family in inflammatory autoimmune diseases. Autoimmun. Rev. 19: 102461 10.1016/j.autrev.2020.102461 [DOI] [PubMed] [Google Scholar]
- Sun Y. Y., Lu M., Xi X. W., Qiao Q. Q., Chen L. L. et al. , 2011a Regulation of epithelial-mesenchymal transition by homeobox gene DLX4 in JEG-3 trophoblast cells: a role in preeclampsia. Reprod. Sci. 18: 1138–1145. 10.1177/1933719111408112 [DOI] [PubMed] [Google Scholar]
- Sun Z., Diaz Z., Fang X., Hart M. P., Chesi A. et al. , 2011b Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 9: e1000614 10.1371/journal.pbio.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoki N., Takahashi K., Aoki H., Iida K., Kawaguchi T. et al. , 2014. The homeobox gene DLX4 promotes generation of human induced pluripotent stem cells. Sci. Rep. 4: 7283 10.1038/srep07283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N., Vance C., Leclerc A. L., Keagle P., Glass J. D. et al. , 2011. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 156B: 285–290. 10.1002/ajmg.b.31158 [DOI] [PubMed] [Google Scholar]
- Tran D. D., Saran S., Koch A., and Tamura T., 2016. mRNA export protein THOC5 as a tool for identification of target genes for cancer therapy. Cancer Lett. 373: 222–226. 10.1016/j.canlet.2016.01.045 [DOI] [PubMed] [Google Scholar]
- Uematsu K., Okumura F., Tonogai S., Joo-Okumura A., Alemayehu D. H. et al. , 2016. ASB7 regulates spindle dynamics and genome integrity by targeting DDA3 for proteasomal degradation. J. Cell Biol. 215: 95–106. 10.1083/jcb.201603062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilagos B., Hoffmann M., Souabni A., Sun Q., Werner B. et al. , 2012. Essential role of EBF1 in the generation and function of distinct mature B cell types. J. Exp. Med. 209: 775–792. 10.1084/jem.20112422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout A. J., Temple G. F., Brasch M. A., Hartley J. L., Lorson M. A. et al. , 2000. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575–592. 10.1016/S0076-6879(00)28419-X [DOI] [PubMed] [Google Scholar]
- Wang L., Miao Y. L., Zheng X., Lackford B., Zhou B. et al. , 2013. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell 13: 676–690. 10.1016/j.stem.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham S., Outeiro T. F., DeVit M. J., Lindquist S. L., and Muchowski P. J., 2003. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science 302: 1769–1772. 10.1126/science.1090389 [DOI] [PubMed] [Google Scholar]
- Wotton D., and Taniguchi K., 2018. Functions of TGIF homeodomain proteins and their roles in normal brain development and holoprosencephaly. Am. J. Med. Genet. C. Semin. Med. Genet. 178: 128–139. 10.1002/ajmg.c.31612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Miao J., Ding Y., Zhang Y., Huang X. et al. , 2019. MiR-4458 inhibits breast cancer cell growth, migration, and invasiveness by targeting CPSF4. Biochem. Cell Biol. 97: 722–730. 10.1139/bcb-2019-0008 [DOI] [PubMed] [Google Scholar]
- Wu, S., H. Chen, L. Zuo, H. Jiang and H. Yan, 2020 Suppression of Long non-coding RNA MALAT1 inhibits the development of uveal melanoma via microRNA-608-mediated inhibition of HOXC4. Am J Physiol Cell Physiol. 10.1152/ajpcell.00262.2019 [DOI] [PMC free article] [PubMed]
- Xu W., Bao P., Jiang X., Wang H., Qin M. et al. , 2019. Reactivation of nonsense-mediated mRNA decay protects against C9orf72 dipeptide-repeat neurotoxicity. Brain 142: 1349–1364. 10.1093/brain/awz070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A. W., Sachs A. J., and Nystuen A. M., 2015. Deletion of Inpp5a causes ataxia and cerebellar degeneration in mice. Neurogenetics 16: 277–285. 10.1007/s10048-015-0450-4 [DOI] [PubMed] [Google Scholar]
- Yang J., Wu W., Wu M., and Ding J., 2019. Long noncoding RNA ADPGK-AS1 promotes cell proliferation, migration, and EMT process through regulating miR-3196/OTX1 axis in breast cancer. In Vitro Cell. Dev. Biol. Anim. 55: 522–532. 10.1007/s11626-019-00372-1 [DOI] [PubMed] [Google Scholar]
- Yang X., Boehm J. S., Yang X., Salehi-Ashtiani K., Hao T. et al. , 2011. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8: 659–661. 10.1038/nmeth.1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko A. N., Georgiadis A. P., Ropke A., Berman A. J., Jaffe T. et al. , 2015. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 372: 2097–2107. 10.1056/NEJMoa1406192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D. G., and Huh W. K., 2015. UDP-glucose pyrophosphorylase Ugp1 is involved in oxidative stress response and long-term survival during stationary phase in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 467: 657–663. 10.1016/j.bbrc.2015.10.090 [DOI] [PubMed] [Google Scholar]
- Yuan M., Eberhart C. G., and Kai M., 2014. RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget 5: 2820–2826. 10.18632/oncotarget.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Jung B. P., Ho W., Jugloff D. G., Cheung H. H. et al. , 2007. Isolation and characterization of LCHN: a novel factor induced by transient global ischemia in the adult rat hippocampus. J. Neurochem. 101: 263–273. 10.1111/j.1471-4159.2006.04374.x [DOI] [PubMed] [Google Scholar]
- Zhang L., Yang M., Gan L., He T., Xiao X. et al. , 2012. DLX4 upregulates TWIST and enhances tumor migration, invasion and metastasis. Int. J. Biol. Sci. 8: 1178–1187. 10.7150/ijbs.4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Xie Y., Tai Y., Gao Y., Guo W. et al. , 2016. Bufalin Inhibits hTERT Expression and Colorectal Cancer Cell Growth by Targeting CPSF4. Cell. Physiol. Biochem. 40: 1559–1569. 10.1159/000453206 [DOI] [PubMed] [Google Scholar]
- Zhang Y. F., Liu L. X., Cao H. T., Ou L., Qu J. et al. , 2015. Otx1 promotes basal dendritic growth and regulates intrinsic electrophysiological and synaptic properties of layer V pyramidal neurons in mouse motor cortex. Neuroscience 285: 139–154. 10.1016/j.neuroscience.2014.11.019 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Chang Y., Zhang J., Lu Y., Zheng L. et al. , 2017. HMGB3 promotes growth and migration in colorectal cancer by regulating WNT/beta-catenin pathway. PLoS One 12: e0179741 10.1371/journal.pone.0179741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Pevzner S. J., Hao T., Wang Y., Mosca R. et al. , 2016. An inter-species protein-protein interaction network across vast evolutionary distance. Mol. Syst. Biol. 12: 865 10.15252/msb.20156484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Li S., Ramelot T. A., Kennedy M. A., Liu M. et al. , 2018. Structural insights into the impact of two holoprosencephaly-related mutations on human TGIF1 homeodomain. Biochem. Biophys. Res. Commun. 496: 575–581. 10.1016/j.bbrc.2018.01.099 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.