Abstract
LRRK2 mutations cause Parkinson’s, but the molecular link from increased kinase activity to pathological neurodegeneration remains undetermined. Previous in vitro assays indicate that LRRK2 substrates include at least 8 Rab GTPases. We have now examined this hypothesis in vivo in a functional, electroretinogram screen, expressing each Rab with/without LRRK2-G2019S in selected Drosophila dopaminergic neurons. Our screen discriminated Rab10 from Rab3. The strongest Rab/LRRK2-G2019S interaction is with Rab10; the weakest with Rab3. Rab10 is expressed in a different set of dopaminergic neurons from Rab3. Thus, anatomical and physiological patterns of Rab10 are related. We conclude that Rab10 is a valid substrate of LRRK2 in dopaminergic neurons in vivo. We propose that variations in Rab expression contribute to differences in the rate of neurodegeneration recorded in different dopaminergic nuclei in Parkinson’s.
Keywords: LRRK2, G2019S, Rab10, Rab3, Drosophila melanogaster, Parkinson’s disease, Parkinson’s UK K-1704 G-1804
Inherited mutations in LRRK2 (Leucine-rich-repeat kinase 2) are a common cause of Parkinson’s. A single amino-acid change, G2019S, increases LRRK2 kinase activity (Greggio and Cookson 2009). This mutation results in a toxic cascade that kills substantia nigra dopaminergic neurons. However, the main steps in this pathological signaling pathway remain to be determined. Partly this is because LRRK2 is potentially a multi-functional protein, with kinase, GTPase and protein-binding domains. A diverse range of >30 proteins that might be phosphorylated by LRRK2 have been reported, suggesting it is a generalized kinase (Tomkins et al. 2018). However, several research teams have recently reported that LRRK2 is a more specific kinase, phosphorylating a range of Rab GTPases (Thirstrup et al. 2017; Steger et al. 2017; Fan et al. 2018; Liu et al. 2018; Jeong et al. 2018; Kelly et al. 2018).
Rabs are a plausible LRRK2 substrate leading to neurodegeneration, as they act as molecular switches interacting with a range of proteins (GEFs, GAPs and GDIs) regulating supply and delivery of cargo to membranes. Indeed many of the 66 Rabs in humans have been linked to neurodegenerative disorders (Kiral et al. 2018). Mutations in Rabs 29 and 39 cause Parkinson’s (MacLeod et al. 2013; Beilina et al. 2014; Wilson et al. 2014). Biochemically, at least 8 seem to be directly phosphorylated by LRRK2 [Rabs 3, 5, 8, 10, 12, 29, 35 and 43] (Steger et al. 2017). However, it is not clear which of the more than 60 Rabs are actually phosphorylated in vivo. In mammals, analysis of the role of the Rabs is complex because individual Rabs may have similar, or even compensatory functions, which may differ by tissue (Chen et al. 2012; Kelly et al. 2018). The situation is simpler in the fly, because there are fewer Rabs - only 23 mammalian orthologs. Here, we use a Drosophila screen to assess the link from LRRK2 to Rabs in vivo using the Tyrosine Hydroxylase (TH) GAL4 to achieve dopamine specific expression. UAS-LRRK2-G2019S (Liu et al. 2008) was driven with and without each Rab gene (Zhang et al. 2006).
We measured a visual phenotype using the SSVEP (Steady State Visual Evoked Potential). Although the outer structure of the eye differs markedly between flies and mammals, the retinal circuitry is highly similar (Cajal and Sanchez 1915; Sanes and Zipursky 2010) – importantly both contain dopaminergic neurons. In the human, the retinal dopaminergic neurons die in Parkinson’s (Harnois and Di Paolo 1990), while in the TH > G2019S model of Parkinson’s, the retina has visual deficits, including neurodegeneration (Hindle et al. 2013; Afsari et al. 2014; West et al. 2015a). We can now use the ability of the SSVEP assay to separate and quantify the response of the photoreceptors and lamina neurons to go beyond measuring neurodegeneration, but to test for a synergistic interaction of a Parkinson’s related gene with potential substrates. Notably, we can do this in vivo in young flies before degeneration has set in.
We determined that, in vivo, Rab10 has the strongest synergy with LRRK2-G2019S, Rab3 the weakest. We validated the physiological results by showing differences in the expression of Rab10 and Rab3 in visual dopaminergic interneurons.
Materials and methods
Flies (Drosophila melanogaster) were raised and manipulated according to standard fly techniques. Fly stocks are listed in Table 1. Crosses were raised at 25° on a 12:12 light-dark cycle. On the day of emergence, female flies were placed in the dark at 29°.
Table 1. List of Fly stocks.
| Genotype | kind gift of |
|---|---|
| LRRK2-G2019S | Wanli Smith |
| TH-GAL4 | Serge Birman |
| w1118 (w¯) | Sean Sweeney |
| eIf-4A3-GFP | Andreas Prokop |
| Genotype | Bloomington Stock |
| RedStinger4 nRFP | 8546 |
| empty vector | 36303 |
| Rab10 GAL4 | 51588 |
| Rab3 GAL4 | 51582 |
| Rab line Bloomington Stock number | |
| Rab1 24104 | Rab18 9796 |
| Rab2 23246 | Rab19 24150 |
| Rab3 9762 | Rab21 23242 |
| Rab4 9767 | Rab23 9802 |
| Rab5 50788 | Rab26 23245 |
| Rab6 23251 | Rab27 9810 |
| Rab7 23270 | Rab30 9812 |
| Rab8 9782 | Rab32 9815 |
| Rab9 9784 | Rab35 9821 |
| Rab10 24097 | Rab39 9825 |
| Rab11 50782 | Rab40 9830 |
| Rab14 9794 | |
Screen design
Virgins from the TH-GAL4, or from a TH-GAL4::UAS-LRRK2-G2019S (THG2) recombinant were crossed with males carrying UAS-Rab, for each of the Rabs that are homologous to those of mammals.
The principle of the SSVEP screen is shown in Figure 1. The visual response of flies stimulated with a flickering blue light was recorded. Young, 4-12 hr old, PD-mimic flies show visual hyperexcitability, particularly in the lamina neurons (Afsari et al. 2014; Himmelberg et al. 2017). This includes the THG2 flies. As they age, the visual response gets weaker and vanishes by 28 days. We therefore chose to test flies aged for 24-36 hr (1 day) or 1 week - between the time at which G2019S expression results in hyperexcitability and the time at which degeneration is first noted. At these time points, the mean visual response of dark-reared THG2 flies was similar to the TH/+ controls.
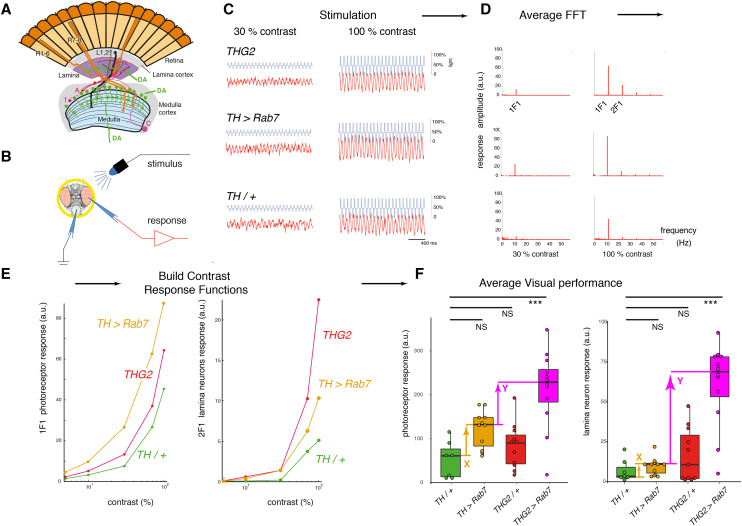
Figure 1.
SSVEP (Steady State Visual Evoked Potential Analysis) measures the contrast response function of the insect eye. A. The fly eye consists of ∼800 ommatidia, each containing 8 photoreceptors. Their axons project to the lamina and medulla, where they synapse with the second- and third order neurons (lamina and medulla neurons). The medulla contains intrinsic dopaminergic neurons (MC, also called Mi15 neurons (Davis et al. 2020)), while some dopaminergic neurons from the CNS project to the lamina. B. Recording the fly visual response: A fly, restrained in a pipette tip, is illuminated with blue light from a LED, and the voltage across the eye is amplified and recorded. C. Repetitive stimuli given to the fly about a fixed mean light level evoke a contrast response increasing with the peak-peak excursion of the stimulus waveform. D. The response to a series of identical stimuli is analyzed by the Fast Fourier Transform, and averaged. This shows a response at the stimulus frequency (1F1) and additional components at multiples of the input, notably twice the input frequency (2F1). Genetic dissection shows that the 1F1 component is mostly generated by the photoreceptors and the 2F1 by the lamina neurons (Afsari et al. 2014; Nippe et al. 2017). E. Plotting the amplitude of the 1F1 and 2F1 components against the stimulus contrast generates a CRF (Contrast Response Function), which differs from fly to fly. F. The averaged maximum CRF is dependent on genotype, with THG2 (flies expressing LRRK2-G2019S in their dopaminergic neurons under the Tyrosine Hydroxylase-GAL4, TH) and TH > Rab7 both showing a similar mean response to control flies (TH/+). However, flies expressing both G2019S and Rab7 in their dopaminergic neurons (THG2 > Rab7) have a larger mean response than any other genotype, indicating synergy. The differences marked X (between the mean TH/+ and TH > Rab7) and Y (between the mean TH > Rab7 and THG2 > Rab7) are used as the X and Y axes of Fig. 2A. Box-plot representing median and interquartile range. Exact genotypes and sample sizes: TH/+, TH/w1118, N= 7; THG2, TH::G2019S/ w1118, N = 11; TH > Rab7, N = 11; THG2 > Rab7, N = 12.
Sample test for synergy
We test for an interaction between Rab7 and G2019S in dopaminergic neurons as follows: we compare flies expressing both Rab7 and G2019S transgenes (THG2 > Rab7) with flies expressing just one transgene (TH > Rab7 or THG2) and control flies with no transgene expression (TH/+) (Figure 1F). The average visual response of TH > Rab7 and THG2 flies is very similar to the control flies – there is no mean difference for either the photoreceptors or lamina neurons. We do note that the THG2 flies have a larger variability than the TH/+ flies, particularly in the lamina neurons (Figure 1F). However, in flies with dopaminergic expression of both G2019S and Rab7, the photoreceptor and lamina neuron responses were much increased (4.1x and 8.8x, both P < 0.001). This demonstrates that dopaminergic neurons with both Rab7 and G2019S have a synergistic hyperexcitable visual phenotype.
SSVEP preparation
At the required age, flies were prepared for SSVEP measurements using a pooter and nail polish to secure them in the cut-off tip of a pipette tip, without anesthesia (Figure 1B). Each fly was presented 5 times with a set of 9 flickering stimuli. In each stimulus, the average light intensity was the same, but the amplitude of the flicker was adjusted from 10 to 100%, giving a range of contrasts. Sample stimuli are shown in Figure 1C. Offline, the Fast-Fourier Transform was applied to the responses, to separate the first harmonic (1F1), due to the photoreceptors from the second harmonic (2F1), due to the lamina neurons (Figure 1D). Other harmonics present in the data were not analyzed. For these first two harmonics, we plotted the contrast response function for each fly (Figure 1E) and determined the best response of that fly. This allowed us to determine the average visual performance for each cross (Figure 1F). This data pipeline is the same as that devised by Afsari et al. (2014), but using an Arduino Due to generate the stimuli and record the responses instead of a PC. Data were analyzed in Matlab, Excel and R. Full code at https://github.com/wadelab/flyCode.
Immunocytochemistry:
This was performed as described recently (Cording et al. 2017). Tyrosine hydroxylase was detected with Mouse anti TH Immunostar (22941, 1:1000). Fluorescent reporters (nRFP, eIf-GFP) were expressed in dopaminergic neurons using the TH-GAL4. Images were prepared for publication using ImageJ; original images are available on request.
Western blots:
Blots for EYFP, encoded in each Rab transgene were made from non-boiled fly head lysates, run on Novex pre-cast mini gels (NuPAGE 4–12% Bis-Tris Gels, NP0322BOX, Thermo Scientific) in 1 x MOPS buffer and transferred onto PVDF membranes using a Hoefer wet transfer tank (TE22) at 100V for 1 hr. Membranes were probed with Guinea pig anti-GFP (Synaptic Systems, 1:1000). For detection of LRRK2 protein, boiled lysates were run on 4–20% Mini-PROTEAN TGX Precast gradient gels and transferred using the same method. Membranes were probed with anti-LRRK2 (Neuromab, clone N241A/34, 1:1000). α-drosophila synaptotagmin was used as a loading control (West et al. 2015b). Densitometric analysis was carried out using ImageJ.
Statistics:
These were calculated in R, with the mean ± SE reported by error bars or median ± interquartile range in box plots. Post-hoc tests were calculated for ANOVA using the Dunnett test.
Data availability
Data tables (Excel sheets) and R code are open access on GitHub: https://github.com/wadelab/flyCode/tree/master/analyzeData/fly_arduino/G3. Raw images and SSVEP traces are available on request. No new reagents are described.
Results
A visual expression screen identifies that Rab10 has the strongest genetic interaction With LRRK2-G2019S; Rab3 the weakest
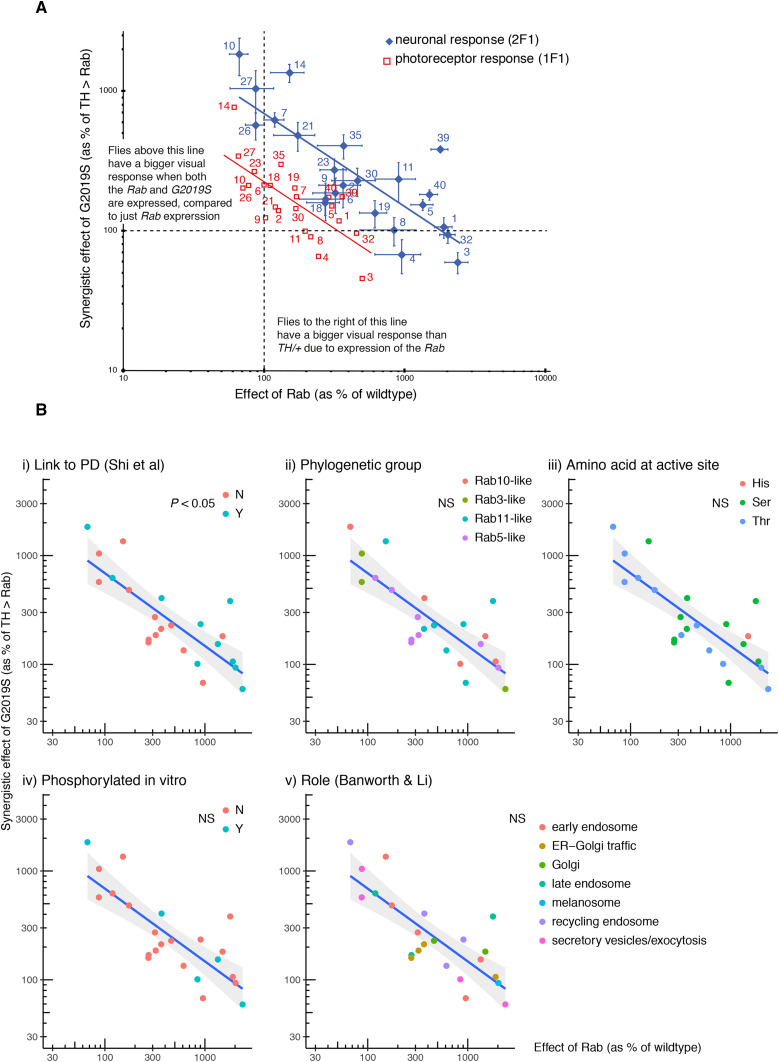
In order to identify the Rabs which show a strong synergy with LRRK2-G2019S we compared the increase in visual response due to expression of the Rab by itself (X in Figure 1F, X-axis in Figure 2A) against the further increase in visual response when both Rab and G2019S are expressed (Y in Figure 1F, Y-axis in Figure 2A). This plot places the Rabs along a spectrum, from those that interact synergistically with G2019S (top left) to those with a little or no interaction (bottom right). Thus, for some Rabs (10, 14, 27, 26) expression of both G2019S and the Rab in dopaminergic neurons leads to a big increase in the lamina neuron response. Interestingly, these Rabs have little effect when expressed alone. The converse is also true: for the Rabs with the biggest effect (3, 32, 1), adding G2019S has no further effect. This is true for both components of the SSVEP signal – that from the lamina neurons is higher, but parallel to the photoreceptor signal.
Figure 2.
Expression screen highlights Rab10 with the strongest synergy with LRRK2-G2019S, and Rab3 as the weakest. Each Rab was expressed in dopaminergic neurons (using TH-GAL4) by itself (TH > Rab), or along with G2019S (THG2 > Rab) and the visual response measured after 24-36 hr (labeled 1 day) or 7 days in the dark. A. Rab10 has the strongest synergy with G2019S. Relationship of Rab and G2019S showing their inverse relationship. Rabs (3, 32, 1) which have a big effect on vision when expressed on their own have little further consequence when G2019S is also expressed; but other Rabs (10, 27, 14, 26) which have little visual impact on their own have a strong synergy with G2019S. B. An established role in Parkinson’s is the only factor that influences the inverse relationship between TH > Rab and THG2 > Rab. The LRRK2 ↔ Rab 1 day data in Fig. 2A are replotted here to test if it is affected by factors that have been proposed to influence LRRK2 ↔ Rab interactions. (i). Rabs previously linked to Parkinson’s (Shi et al. 2017) have a stronger Rab ↔ G2019S response than those which do not influence Parkinson’s, since a higher proportion of the magenta points lie above the line (Fisher’s exact test, P = 0.036). B(ii). Sensitivity is not linked to the phylogenetic grouping of the fly Rabs (Zhang et al. 2006). B(iii). Rabs usually have a serine (Ser) or threonine (Thr) where they could be phosphorylated by LRRK2, though Rab40 has a histidine (His) (Zhang et al. 2006). Although a preference for LRRK2 to phosphorylate Rabs with a threonine was suggested by in vitro assays (Steger et al. 2016), in vivo this is not detected. B(iv). Some Rabs are phosphorylated by LRRK2 in vitro (Steger et al. 2017), but these Rabs are not more sensitive to G2019S in vivo. B(v). The proposed main functional role of the Rab (Banworth and Li 2018) does not affect the regression. Total flies: 1119, at least 9 for each data point. Bars represent SE.
We wanted to examine which factors controlled this synergy. A number of hypotheses have been put forward in the LRRK2/Rab literature. First, Rabs previously linked to Parkinson’s (Shi et al. 2017), either through population studies or through potential actions with Parkinson’s-related genes, generally have a stronger response to G2019S than others (Figure 2Bi). Indeed, the Rab furthest above the regression line is one that causes Parkinson’s, Rab39 (Wilson et al. 2014). Next, we tested if Rabs with a high degree of phylogenetic similarity clustered systematically, but did not find any difference (Figure 2Bii). Then we examined where, on our spectrum, the Rabs phosphorylated in vitro [3, 5, 8, 10, 12, 29, 35 and 43] might lie. There is no close homolog for Rabs 12, 29 or 43. Rabs 3, 5 and 8 are on the right of the spectrum, Rab35 in the middle and Rab10 on the top-left, so no clear pattern emerges. The in vitro data suggest that LRRK2-G2019S preferentially phosphorylates Rabs with Thr rather than Ser at the active site (Steger et al. 2016), but this is not evident from the spectrum. Neither in vitro evidence for phosphorylation of the Rab by LRRK2, nor the amino-acid at the active site affects the regression (Figure 2Biii,iv). We also noted that, in cell based assays, LRRK/Rab interactions have been noted at mitochondria (Wauters et al. 2019), lysosomes (Eguchi et al. 2018) or Golgi (Liu et al. 2018). Thus, we tested if the ‘main’ organelle associated with the Rab (Banworth and Li 2018) affected the position of a Rab on the spectrum, but found no sign that this affected the regression (Figure 2Bv). However, the Rabs placed in the middle of the spectrum [2, 6, 9, 18] are linked to traffic in the Golgi or ER-Golgi.
Thus, the relationship between visual impact of Rab and impact of Rab + G2019S identifies 10, 14, 27 as having the strongest synergy. This holds for the responses of both photoreceptors and lamina neurons, with the same Rabs found clustered at each end of the spectrum.
The Rab10/G2019S interaction enhances neuronal signaling
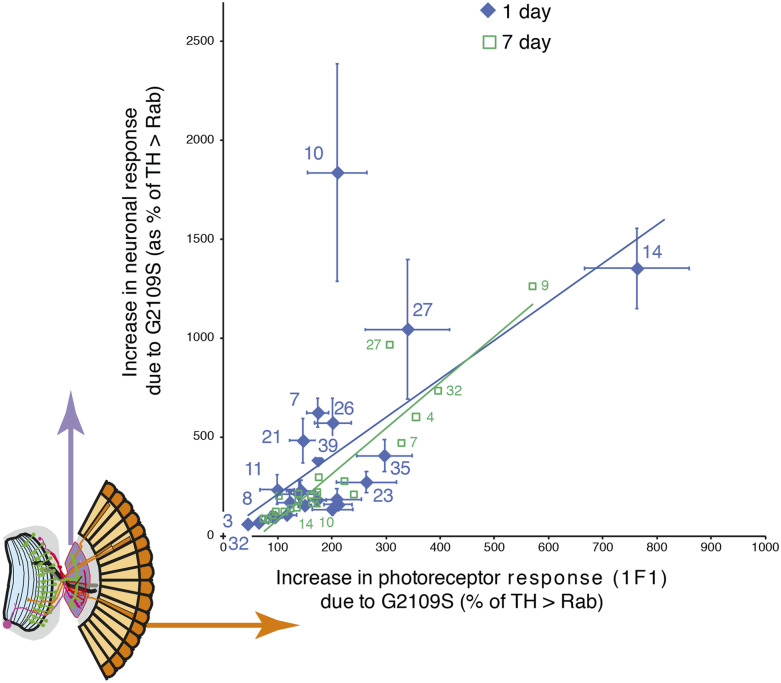
Normally, flies with more excitable photoreceptors activate the lamina neurons more strongly, though there is some adaptation. The SSVEP response can be decomposed into two components – 1F1 and 2F1, corresponding to activity in the photoreceptors and lamina neurons respectively. This allows us to test the physiological relationship between the photoreceptors and lamina neurons, and to see if any Rab disrupts the retinal neuronal circuitry. Generally, as the photoreceptor response increases, so does the lamina neuron response (Figure 3). This relationship is remarkably similar in young (day 1) and older (day 7) flies. However, there is a one marked outlier, Rab10, where the lamina neuron response at day 1 is ∼5 times the value expected from the regression, and at day 7 is substantially below the line. Thus, in young THG2 > Rab10 flies there is much greater neuronal activity than expected, but in 1-week old THG2 > Rab10 flies we observe reduced activity, suggesting neurodegeneration has begun. Young and old Rab3 flies lie on the regression, close to the origin, very different from Rab10.
Figure 3.
Standout role of Rab10 with G2019S in neuronal signaling. The SSVEP response is split into two components, representing the photoreceptors and lamina neurons (inset orange and purple). For each Rab, the increase in lamina neuronal signaling due to G2019S is plotted as a function of the photoreceptor signal. The increase in lamina neuron response is highly correlated with the response of the photoreceptors, with one outlying exception, Rab10 at 1 day. Total flies: 1119, at least 9 for each data point. Bars represent SE.
Thus our screen highlights a major difference between two of the Rabs that are phosphorylated in vitro: Rab3 expression in dopaminergic neurons has a big increase in visual sensitivity, but no further effect when G2019S is added, whereas Rab10 expression has little effect by itself, but a massive effect in young flies with G2019S.
Why might G2019S interact so strongly With Rab10 but have no effect on Rab3?
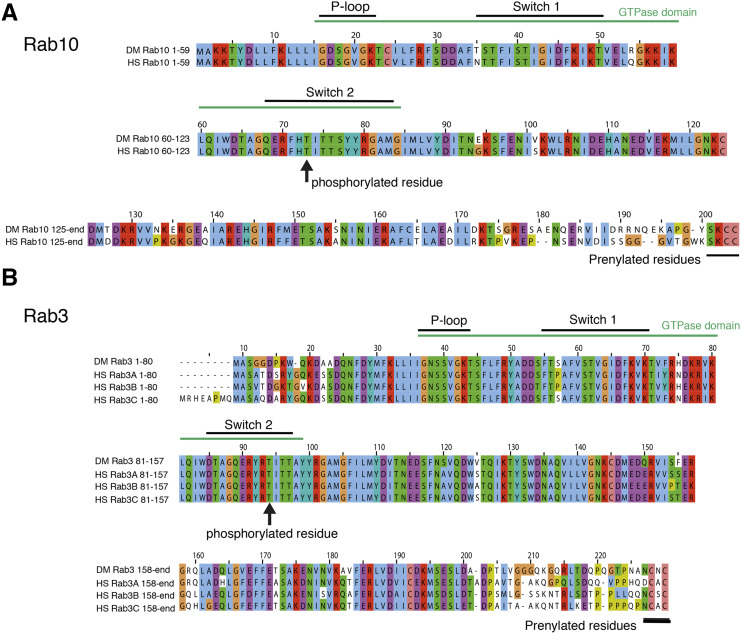
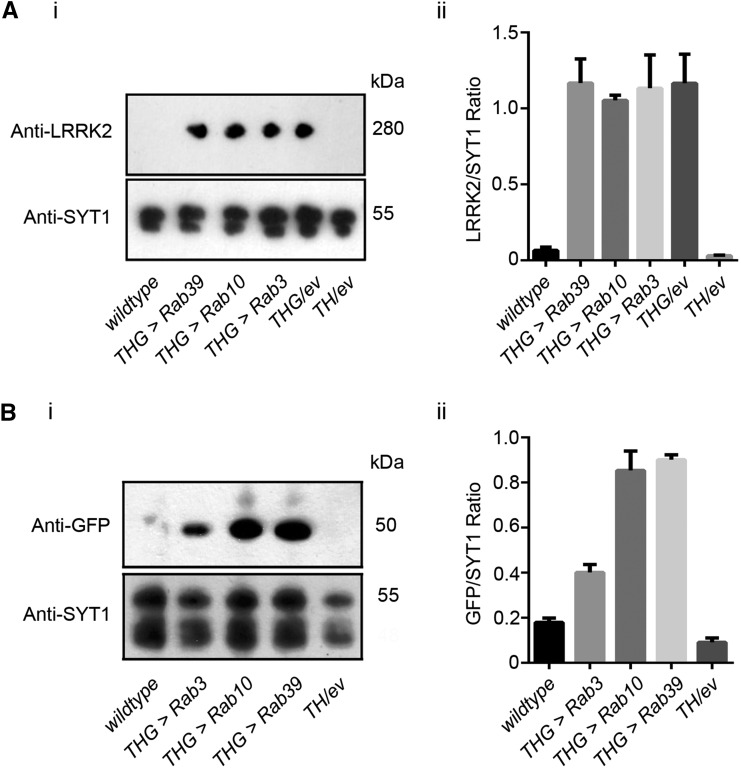
As LRRK2 is a human protein, and the Rabs we expressed were native Drosophila proteins, one possibility is that the fly and human Rabs are sufficiently different that LRRK2-G2019S can phosphorylate fly Rab10 but not fly Rab3. This seems very unlikely as the hRab3 / dRab3 and dRab10 / hRab10 sequences are very similar, indeed they are identical over the GTP binding domain and LRRK2 phosphorylation sites (Figure 4). A second explanation for the difference between Rab10 and Rab3 is that the Rab and/or G2019S is not expressed to the same extent. Western Blots of THG2 > Rabs 3, 39 or 10 were probed for LRRK2, and compared with THG2. Essentially the same level of protein was measured (Figure 5A). This is not unexpected, as each cross contains the same GAL4 and UAS-LRRK2-G2019S constructs. A second set of blots were probed for EYFP, as each of the UAS-Rab lines carries an EYFP fusion. This showed that the levels of Rab10 and Rab39 were similar, though Rab3 was less at about 50% (Figure 5B). The reduced level of Rab3 may arise from the different insertion site, or from a more rapid breakdown during synaptic signaling. The differences in level of Rab proteins are not sufficient to explain the physiological differences.
Figure 4.
High sequence homology between Drosophila and human Rabs. Comparison of fly and human Rab10 (A) and Rab3 (B) showing conservation in the GTPase domain and prenylated region. Also shown is the region which is phosphorylated in vitro by LRRK2, again highly conserved.
Figure 5.
Similar Expression of LRRK2-G2019S and Rab-GFP in dopaminergic neurons. A. Co-expression of a Rab-GFP transgene does not affect the levels of LRRK2-G2019S. (i) Sample blot, (ii) Quantification of 3 replicates. B. Similar levels of Rab10 and Rab39, and less Rab3 when driven with LRRK2-G2019S. (i) Sample blot, (ii) Quantification of 3 replicates. wild-type is CS/w¯, TH/+ is TH/empty vector.
We therefore wondered if the stronger synergy between G2019S and Rab10, compared with Rab3, might result from a difference in the anatomical distribution of the Rabs (along with their GEFS, GAPs and effectors) among fly dopaminergic neurons.
Rab10 and Rab3 are found in different dopaminergic neurons
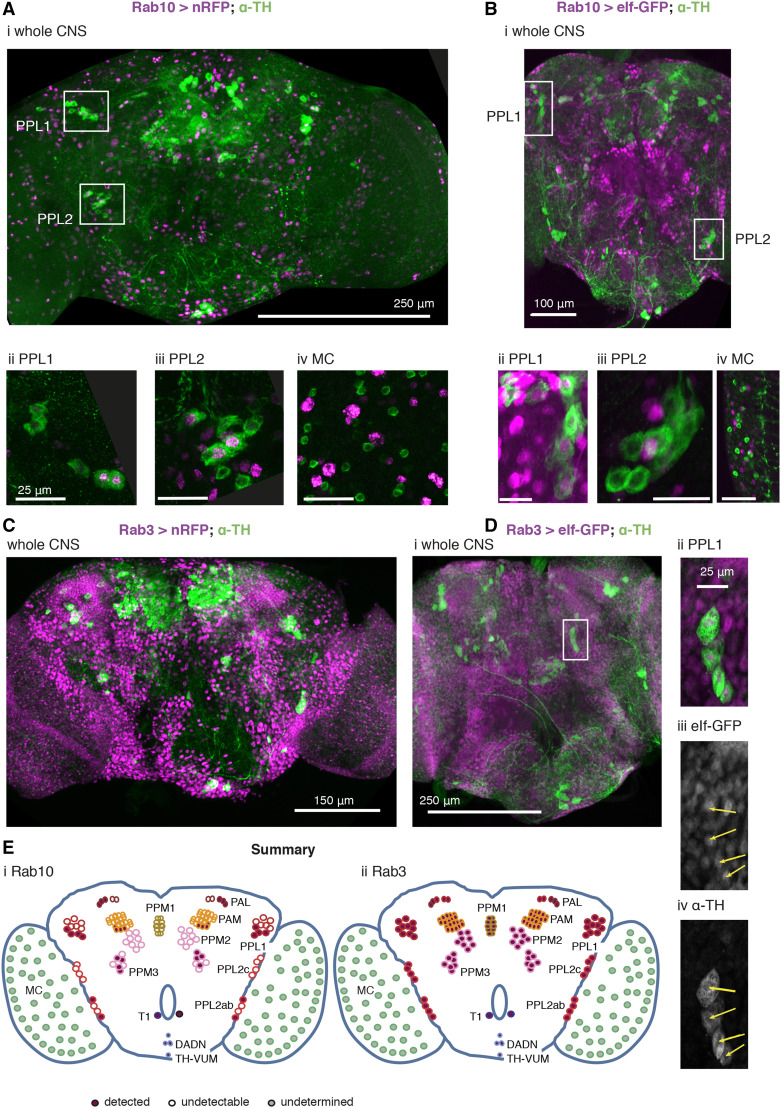
The fly visual system is innervated by three kinds of dopaminergic neurons (Hindle et al. 2013), the MC neurons in the medulla, and two type of PPL neurons, which innervate either lobula or lamina respectively. These, and the other clusters of dopaminergic neurons, are reliably marked by α-TH antibody, which binds in the cytoplasm.
To examine the overall distribution of Rab10, we used Rab10-GAL4 (Chan et al. 2011) to express either a RFP which strongly localises to the nucleus, or a GFP with mainly nuclear localization. These fluorescent constructs have two advantages: (i) they provide a reduced background compared with membrane localized reporters, and (ii) the nuclear fluorescence is contained within the cytoplasmic signal from α-TH, reducing the problems of determining co-localization.
Only a small proportion of CNS neurons are Rab10 positive (Figure 6). We find that some (by no means all) dopaminergic neurons are Rab10 positive (Figure 6A,B). Even within a cluster, we only detect Rab10 in some neurons; in other neurons in the same cluster Rab10 is undetectable (e.g., PAL, PPL2ab, PPM3 and PAM). The individually identifiable neurons (TH-VUM, TH_AUM, the DADN pair, and T1 pair) were consistently clearly marked. However, in two clusters we saw no evidence for Rab10 driven fluorescence (PPL2c and PPM1/2).
Figure 6.
Rab10 and Rab3 are located in different subsets of the dopaminergic neurons. A, B. Rab10 is detected in some of the dopaminergic neurons that control vision (PPL1, Aii, Bii; PPL2 Aiii, Biii). Not all dopaminergic neurons, identified by a cytosolic α-Tyrosine Hydroxylase antibody (α-TH, green), are indicated by Rab10-GAL4 expression of a strong nuclear RFP or the mainly nuclear eIf-GFP (magenta). The dopaminergic MC neurons in the visual lobes do not stain well with fluorescent reporters (Nassel and Elekes 1992; Hindle et al. 2013) and we could not detect Rab10-driven fluorescence (MC, Aiv, Biv, marked with gray in E). C, D. Rab 3 is present in all dopaminergic neurons. Rab3-GAL4 driven nuclear RFP or eIf-GFP (magenta) marks most neurons, including nearly all that are dopaminergic (green). The PPL neurons not marked by Rab10 expression are included (Dii-iv). E. Summary of the expression pattern of (i) Rab10 and (ii) Rab3. The MC neurons in the optic lobe (Nassel et al. 1988) are also called Mi15 neurons (Davis et al. 2020). Ai, Bi, Ci and Di: projection of confocal stacks through the whole CNS; Aii, Aiii, Bii, Biii, Dii-iv projections of confocal stacks through the cell groups, approximately marked in the whole CNS image; Aiv and Biv sections from a separate preparation to Ai and Bi. Data representative of at least nine brains (from at least 3 crosses), 3-7 days old. The Rab3 > nRFP flies were raised at 18 °C to improve viability. Exact genotypes: +; RedStinger4 nRFP/+; Rab10 Gal4/+; or +; RedStinger4 nRFP/+; Rab3 Gal4/+; or +; eIf-4A3-GFP/+; Rab10 Gal4/+; or +; eIf-4A3-GFP/+; Rab3 Gal4/+;
When we used Rab3-GAL4 to drive the same RFP/GFP almost all the neurons were marked (Figure 6 C, Di). This includes the majority of the dopaminergic neurons, including all the PPL1 (Figure 6 Dii-iv) and PPL2 neurons.
The MC neurons in the optic lobes were not marked in either the Rab10 or Rab3 experiments (Figure 6 Aiv, Biv), though other Rab10 / Rab3 positive neurons are present nearby. Since the MC neurons do not generally stain well with GFP (Nassel and Elekes 1992; Hindle et al. 2013), we tested if the MC neurons were detected with TH-GAL4 > nRFP. This marked all the neurons highlighted by α-TH, except the MC neurons. The MC neurons do express TH, along with other genes linked to dopamine - Ddc (dopa decarboxylase), Vmat (vesicular monoamine transporter) and DAT (dopamine transporter) (Davis et al. 2020) so are genuinely dopaminergic. The MC neurons are one of three kinds of Medulla intrinsic neurons that express Rab10 at high levels, while all the optic lobe neurons (including MC) have high expression of Rab3 (Davis et al., 2020, extended data at http://www.opticlobe.com/).
Thus, we conclude that some of the dopaminergic neurons in the visual system are Rab10 positive. These are some of the PPL cluster that innervate the lobula or project to the lamina, and the MC neurons in the medulla. All dopaminergic neurons are Rab3 positive.
Differences in the loss of dopaminergic neurons between neuronal clusters
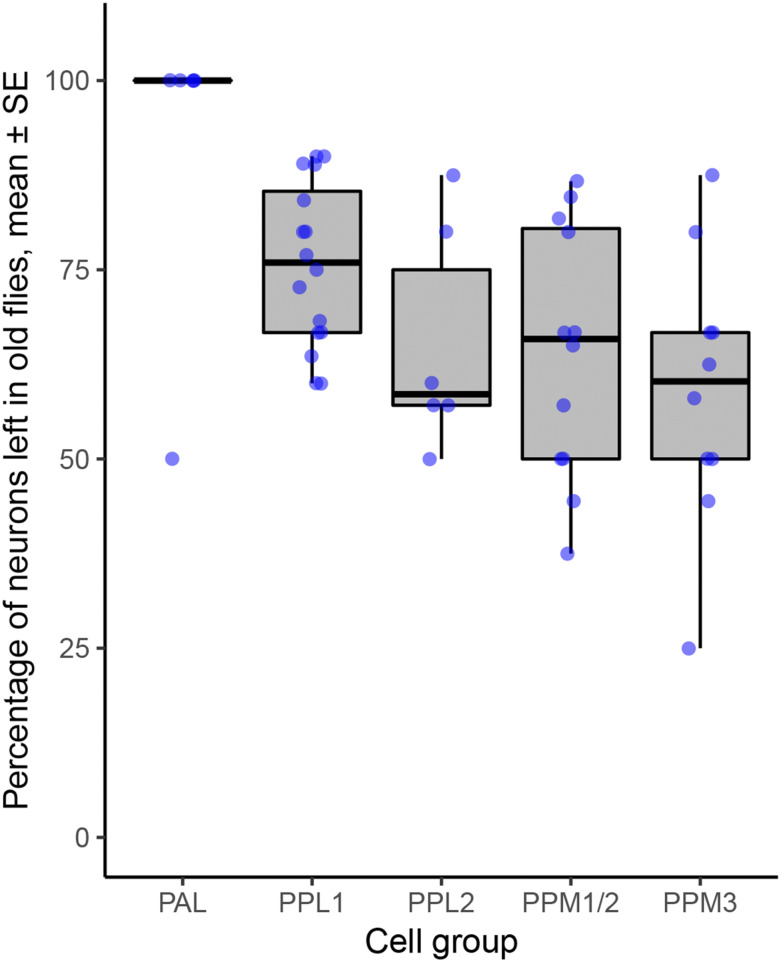
Drosophila models of Parkinson’s have consistently shown loss of dopaminergic neurons with age when LRRK2, α-synuclein or parkin were manipulated. For LRRK2, most of the published information is for the Parkinson’s-causative mutations G2019S or I2020T, driven by Ddc-GAL4. This expresses in the dopaminergic and some serotonergic neurons. By 6-7 weeks (about two-thirds of the fly lifespan), about 25–50% of the dopaminergic neurons have been lost. For each cluster, there is quite a spread of the data (Figure 7), which is most likely due to differences in the food used to feed the flies or the genetic background (Lavoy et al. 2018; Chittoor-Vinod et al. 2020). However, overall, the PAL cluster is much less susceptible to cell loss than the PPL1, PPL2, PPM1/2 or PPM3 clusters.
Figure 7.
Differences in neuron survival in dopaminergic clusters when an increased kinase mutation (G2019S or I2020T) is expressed with Ddc-GAL4 (which expresses in dopaminergic and serotonergic neurons) (ANOVA, 4,45 df, P < 0.002). Data collected from (Liu et al. 2008; Ng et al. 2009; Xiong et al. 2012; Angeles et al. 2014, 2016; Martin et al. 2014; Nucifora et al. 2016; Lin et al. 2016; Sun et al. 2016; Basil et al. 2017; Marcogliese et al. 2017; Yang et al. 2018; Lavoy et al. 2018; Sim et al. 2019; Maksoud et al. 2019; Chittoor-Vinod et al. 2020). Differences in the extent of degeneration within a neuronal cluster may be partially explained by differences in the composition of the fly food (Chittoor-Vinod et al. 2020).
Discussion
Rab10 shows a strong synergy with LRRK2-G2019S
The key observation from the screen was that two of the Rabs suggested to be substrates of LRRK2 in vitro behave quite differently in vivo, in a physiological response to expression in dopaminergic neurons. Rab10 shows a strong synergy with G2019S; Rab3 none. The existence of (Drosophila) Rab10 in the tyrosine hydroxylase positive neurons controlling vision (MC and PPL2ab neurons) argues that LRRK2 might indeed phosphorylate dRab10 directly. Thus our in vivo results both support the in vitro (biochemical and cell culture) data in which LRRK2 directly phosphorylates hRab10 (Thirstrup et al. 2017; Steger et al. 2017; Fan et al. 2018; Liu et al. 2018; Jeong et al. 2018; Kelly et al. 2018). It also implies that the MC / PPL2ab cells contain Rab10 effectors which interact with phospho-dRab10. The results of this will include changes to the cellular homeostasis and physiological responsiveness of dopaminergic neurons. One possible physiological outcome is that Rab10 phosphorylation reduces retinal dopamine release onto the photoreceptors. This will increase the amplitude and speed of the photoreceptor response (Chyb et al. 1999). Dopamine may also affect the lamina neurons, and third order medulla neurons, but it remains to be determined if they have dopamine receptors. It is also possible that p-Rab10 modulates the release of co-transmitters or growth factors from dopaminergic neurons.
A unique feature of the screen is that when G2019S and Rab10 are expressed together the lamina neuron response is much bigger than that predicted from the photoreceptor response. This might arise from the unusual double role of Rab10 – in both exo- and in endo-cytosis (Larance et al. 2005; Glodowski et al. 2007; Chua and Tang 2018). The best defined role of Rab10 in exocytosis is in adipocytes, as part of the insulin-stimulated release of GLUT4 vesicles, linked to AS/160 (see for review (Jaldin-Fincati et al. 2017)). In endocytosis, the effects of Rab10 are mediated through a different pathway, including the EHBP1-EHD2 complex. In the follicle cells of Drosophila, ehbp1 expression and knockdown phenocopy Rab10 manipulations (Isabella and Horne-Badovinac 2016), while EHBP1 was also identified by a systematic proteomic analysis as indirectly phosphorylated by LRRK2 in HEK293 cells (Steger et al. 2017) and a lysosomal assay (Eguchi et al. 2018). The phosphorylation of Rab10 by LRRK2 may switch its effector, and so activate a different pathway.
A spectrum of Rab ↔ G2019S interactions in vision
Our screen placed the Rabs along a spectrum, ranging from those with a strong synergy with G2019S to those which had a strong effect when expressed by themselves.
Among the Rabs which show little synergy with G2019S but have strong visual effect are 1, 3, 5, 6 and 11. Two of these Rabs [3,5] are phosphorylated by LRRK2 in vitro (Steger et al. 2017), but neither synergise with LRRK2-G2019S in the visual assay. Our data suggest Rab3 is not a major substrate of LRRK2-G2019S in these dopaminergic neurons, possibly because Rab3 is located synaptically. This may be far from LRRK2 at the trans-Golgi network (Liu et al. 2018). The difference between Rab3 and 10 (at opposite ends of our spectrum) is notable because in vitro mammalian cell assays have highlighted similar roles of Rabs 3 and 10 in lysosome exocytosis, (Encarnação et al. 2016; Vieira 2018).
Rabs 10, 14 and 27 have the strongest synergy with G2019S, though by themselves they have little effect on visual sensitivity. Like Rab10, Rabs 14 and 27 have defined roles in exocytosis (Larance et al. 2005; Ostrowski et al. 2010).
Some Rabs are in the middle of the spectrum [2, 6, 9, 18], with a 2-3 fold increase in visual response when the Rab is expressed alone, and a further 2-3 fold increase when both Rab and G2019S are expressed. These Rabs have been linked to the Golgi, or to Golgi-ER traffic (Banworth and Li 2018). Thus a cellular phenotype parallels the physiological response.
Our observation that every Rab seems to have some effect on dopaminergic signaling in the visual system goes some way to explain why many studies of individual Rabs have demonstrated effects with LRRK2; Rab3a (Islam et al. 2016); Rab5 (Shin et al. 2008); Rab7 (Dodson et al. 2012); Rab29 (Beilina et al. 2014). Although cellular studies support binding of Rab29 to LRRK2 (Purlyte et al. 2018), the closest fly homolog (Rab32) shows little synergy with G2019S in our screen.
The availability of Rab transgenic flies facilitates screening in Drosophila. Screens have identified key roles for Rab2 in muscle T-tubule development (Fujita et al. 2017); Rabs 2, 7, 19 in loss of huntingtin (White et al. 2015), 1, 5, 7, 11 and 35 in the Drosophila renal system (Fu et al. 2017), Rab32 in lipid storage (Wang et al. 2012) and Rab39 in tracheal formation (Caviglia et al. 2016). The varied outcomes of these screens indicate the validity of the LRRK2-G2019S ↔ Rab10 relationship reported here.
Each dopaminergic neuron has its own palette of Rab expression
Finally, we note that not all dopaminergic neurons are equally susceptible in Parkinson’s. A long-standing observation is that the dopaminergic neurons in the VTA (ventral tegmental area) do not degenerate in the same way as those in the substantia nigra. More particularly, even within the substantia nigra there is a range of outcomes, with dopaminergic neurons in the pars compacta dying more than those in the dorsal and lateral zones (Damier et al. 1999). The same is true for the fly brain: the neurons in the PPM clusters degenerate more than the PAL (though no data are available for the visual MC neurons). If anything, our data suggest the clusters with less Rab10 have more neurodegeneration. Previously, faster neurodegeneration has been ascribed to increased cytosolic dopamine levels (Burbulla et al. 2017), to intracellular effects of glutamate (Steinkellner et al. 2018), to increased calcium influx (Guzman et al. 2010), to more action potentials (Subramaniam et al. 2014), or to longer axons with more synapses (Pacelli et al. 2015). It has not escaped our notice that faster degeneration in some neurons may be the result of their different palettes of Rab proteins and their effectors.
Acknowledgments
We are grateful for the gifts of flies from Wanli Smith and the Bloomington Drosophila Supply Center. We also thank Olivia Compton and Martin France who helped with pilot studies, the University of York Biology Technology Facility and Flybase. Ian Martin kindly provided unpublished details of dopaminergic cell loss. We are particularly grateful to Parkinson’s UK and to their volunteers for support (K-1704, G-1804).
Footnotes
Communicating editor: R. Anholt
Literature Cited
- Afsari F., Christensen K. V., Smith G. P., Hentzer M., Nippe O. M. et al. , 2014. Abnormal visual gain control in a Parkinson’s disease model. Hum. Mol. Genet. 23: 4465–4478. 10.1093/hmg/ddu159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles D. C., Ho P., Chua L. L., Wang C., Yap Y. W. et al. , 2014. Thiol-peroxidases ameliorate LRRK2 mutant-induced mitochondrial and dopaminergic neuronal degeneration in Drosophila. Hum. Mol. Genet. 23: 3157–3165. 10.1093/hmg/ddu026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles, D. C., P. Ho, B. W. Dymock, K. Lim, Z. Zhou et al., 2016 Antioxidants inhibit neuronal toxicity in Parkinson’s disease-linked LRRK2. Ann. Clin. Transl. Neurol. 3: 288–294. 10.1002/acn3.282. [DOI] [PMC free article] [PubMed]
- Banworth M. J., and Li G., 2018. Consequences of Rab GTPase dysfunction in genetic or acquired human diseases. Small GTPases 9: 158–181. 10.1080/21541248.2017.1397833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basil A. H., Sim J. P. L., Lim G. G. Y., Lin S., Chan H. Y. et al. , 2017. AF-6 Protects Against Dopaminergic Dysfunction and Mitochondrial Abnormalities in Drosophila Models of Parkinson’s Disease. Front. Cell. Neurosci. 11: 241 10.3389/fncel.2017.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A., Rudenko I. N., Kaganovich A., Civiero L., Chau H. et al. , 2014. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. USA 111: 2626–2631. 10.1073/pnas.1318306111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulla L. F., Song P., Mazzulli J. R., Zampese E., Wong Y. C. et al. , 2017. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 357: 1255–1261. 10.1126/science.aam9080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal S. R., and Sanchez D., 1915. Contribucion al conocimiento de los centros nerviosos de los insectos. Parte 1. Retina y centros opticos. Trab. Lab Invest. Bio. Univ. Madrid 13: 1–168. [Google Scholar]
- Caviglia S., Brankatschk M., Fischer E. J., Eaton S., and Luschnig S., 2016. Staccato/Unc-13–4 controls secretory lysosome-mediated lumen fusion during epithelial tube anastomosis. Nat. Cell Biol. 18: 727–739. 10.1038/ncb3374 [DOI] [PubMed] [Google Scholar]
- Chan C.-C., Scoggin S., Wang D., Cherry S., Dembo T. et al. , 2011. Systematic discovery of Rab GTPases with synaptic functions in Drosophila. Curr. Biol. 21: 1704–1715. 10.1016/j.cub.2011.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang Y., Zhang J., Deng Y., Jiang L. et al. , 2012. Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. J. Cell Biol. 198: 545–560. 10.1083/jcb.201111091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittoor-Vinod V. G., Villalobos-Cantor S., Roshak H., Shea K., and Martin I., 2020. Dietary Amino Acids Impact LRRK2-induced Neurodegeneration in Parkinson’s Disease Models. bioRxiv 10.1101/2020.01.13.905471 [DOI] [PMC free article] [PubMed]
- Chua C. E. L., and Tang B. L., 2018. Rab 10—a traffic controller in multiple cellular pathways and locations. J. Cell. Physiol. 233: 6483–6494. 10.1002/jcp.26503 [DOI] [PubMed] [Google Scholar]
- Chyb S., Hevers W., Forte M., Wolfgang W. J., Selinger Z. et al. , 1999. Modulation of the light response by cAMP in Drosophila photoreceptors. J. Neurosci. 19: 8799–8807. 10.1523/JNEUROSCI.19-20-08799.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cording A. C., Shiaelis N., Petridi S., Middleton C. A., Wilson L. G. et al. , 2017. Targeted kinase inhibition relieves slowness and tremor in a Drosophila model of LRRK2 Parkinson’s. npj Park. Dis. 3: 34 10.1038/s41531-017-0036-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damier P., Hirsch E. C., Agid Y., and Graybiel A. M., 1999. The substantia nigra of the human brain: II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain 122: 1437–1448. 10.1093/brain/122.8.1437 [DOI] [PubMed] [Google Scholar]
- Davis F. P., Nern A., Picard S., Reiser M. B., Rubin G. M. et al. , 2020. A genetic, genomic, and computational resource for exploring neural circuit function. eLife 9: e50901 10.7554/eLife.50901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M. W., Zhang T., Jiang C., Chen S., and Guo M., 2012. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum. Mol. Genet. 21: 1350–1363. 10.1093/hmg/ddr573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi T., Kuwahara T., Sakurai M., Komori T., Fujimoto T. et al. , 2018. LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc. Natl. Acad. Sci. USA 115: E9115–E9124. 10.1073/pnas.1812196115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encarnação M., Espada L., Escrevente C., Mateus D., Ramalho J. et al. , 2016. A Rab3a-dependent complex essential for lysosome positioning and plasma membrane repair. J. Cell Biol. 213: 631–640. 10.1083/jcb.201511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Howden A. J. M., Sarhan A. R., Lis P., Ito G. et al. , 2018. Interrogating Parkinson’s disease LRRK2 kinase pathway activity by assessing Rab10 phosphorylation in human neutrophils. Biochem. J. 475: 23–44. 10.1042/BCJ20170803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Zhu J., Zhang F., Richman A., Zhao Z. et al. , 2017. Comprehensive functional analysis of Rab GTPases in Drosophila nephrocytes. Cell Tissue Res. 368: 615–627. 10.1007/s00441-017-2575-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Huang W., Lin T.-H., Groulx J.-F., Jean S. et al. , 2017. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife 6: e23367 10.7554/eLife.23367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glodowski D. R., Chen C. C.-H., Schaefer H., Grant B. D., and Rongo C., 2007. RAB-10 regulates glutamate receptor recycling in a cholesterol-dependent endocytosis pathway. Mol. Biol. Cell 18: 4387–4396. 10.1091/mbc.e07-05-0486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E., and Cookson M. R., 2009. Leucine-Rich Repeat Kinase 2 Mutations and Parkinson’s Disease: Three Questions. ASN Neuro 1: AN20090007 10.1042/AN20090007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman J. N., Sanchez-Padilla J., Wokosin D., Kondapalli J., Ilijic E. et al. , 2010. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 468: 696–700. 10.1038/nature09536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnois C., and Di Paolo T., 1990. Decreased dopamine in the retinas of patients with Parkinson’s disease. Invest. opthal. vis. sci. 31: 2473–2475. [PubMed] [Google Scholar]
- Himmelberg, M. M., R. J. H. West, C. J. H. Elliott, and A. R. Wade, 2017 Abnormal visual gain control and excitotoxicity in early-onset Parkinson’s disease Drosophila models. J. Neurophysiol. 119: 957–970. 10.1152/jn.00681.2017 [DOI] [PMC free article] [PubMed]
- Hindle S. J., Afsari F., Stark M., Middleton C. A., Evans G. J. O. et al. , 2013. Dopaminergic expression of the Parkinsonian gene LRRK2–G2019S leads to non-autonomous visual neurodegeneration, accelerated by increased neural demands for energy. Hum. Mol. Genet. 22: 2129–2140. 10.1093/hmg/ddt061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella A. J., and Horne-Badovinac S., 2016. Rab10-Mediated Secretion Synergizes with Tissue Movement to Build a Polarized Basement Membrane Architecture for Organ Morphogenesis. Dev. Cell 38: 47–60. 10.1016/j.devcel.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. S., Nolte H., Jacob W., Ziegler A. B., Pütz S. et al. , 2016. Human R1441C LRRK2 regulates the synaptic vesicle proteome and phosphoproteome in a Drosophila model of Parkinson’s disease. Hum. Mol. Genet. 500: 5365–5382. 10.1093/hmg/ddw352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaldin-Fincati J. R., Pavarotti M., Frendo-Cumbo S., Bilan P. J., and Klip A., 2017. Update on GLUT4 Vesicle Traffic: A Cornerstone of Insulin Action. Trends Endocrinol. Metab. 28: 597–611. 10.1016/j.tem.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Jeong G. R., Jang E.-H., Bae J. R., Jun S., Kang H. C. et al. , 2018. Dysregulated phosphorylation of Rab GTPases by LRRK2 induces neurodegeneration. Mol. Neurodegener. 13: 8 10.1186/s13024-018-0240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K., Wang S., Boddu R., Liu Z., Moukha-Chafiq O. et al. , 2018. The G2019S mutation in LRRK2 imparts resiliency to kinase inhibition. Exp. Neurol. 309: 1–13. 10.1016/j.expneurol.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiral F. R., Kohrs F. E., Jin E. J., and Hiesinger P. R., 2018. Rab GTPases and Membrane Trafficking in Neurodegeneration. Curr. Biol. 28: R471–R486. 10.1016/j.cub.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larance M., Ramm G., Stöckli J., van Dam E. M., Winata S. et al. , 2005. Characterization of the Role of the Rab GTPase-activating Protein AS160 in Insulin-regulated GLUT4 Trafficking. J. Biol. Chem. 280: 37803–37813. 10.1074/jbc.M503897200 [DOI] [PubMed] [Google Scholar]
- Lavoy S., Chittoor-Vinod V. G., Chow C. Y., and Martin I., 2018. Genetic Modifiers of Neurodegeneration in a Drosophila Model of Parkinson’s Disease. Genetics 209: 1345–1356. 10.1534/genetics.118.301119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-H., Lin H.-I., Chen M.-L., Lai T.-T., Cao L.-P. et al. , 2016. Lovastatin protects neurite degeneration in LRRK2–G2019S parkinsonism through activating the Akt/Nrf pathway and inhibiting GSK3β activity. Hum. Mol. Genet. 25: 1965–1978. 10.1093/hmg/ddw068 [DOI] [PubMed] [Google Scholar]
- Liu Z., Bryant N., Kumaran R., Beilina A., Abeliovich A. et al. , 2018. LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum. Mol. Genet. 27: 385–395. 10.1093/hmg/ddx410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang X., Yu Y., Li X., Wang T. et al. , 2008. A Drosophila model for LRRK2-linked parkinsonism. Proc. Natl. Acad. Sci. USA 105: 2693–2698. 10.1073/pnas.0708452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D. A., Rhinn H., Kuwahara T., Zolin A., Di Paolo G. et al. , 2013. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron 77: 425–439. 10.1016/j.neuron.2012.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksoud E., Liao E. H., and Haghighi A. P., 2019. A Neuron-Glial Trans-Signaling Cascade Mediates LRRK2-Induced Neurodegeneration. Cell Rep. 26: 1774–1786.e4. 10.1016/j.celrep.2019.01.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcogliese P. C., Abuaish S., Kabbach G., Abdel-Messih E., Seang S. et al. , 2017. LRRK2(I2020T) functional genetic interactors that modify eye degeneration and dopaminergic cell loss in Drosophila. Hum. Mol. Genet. 26: 1247–1257. 10.1093/hmg/ddx030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I., Kim J. W., Lee B. D., Kang H. C., Xu J.-C. et al. , 2014. Ribosomal Protein s15 Phosphorylation Mediates LRRK2 Neurodegeneration in Parkinson’s Disease. Cell 157: 472–485. 10.1016/j.cell.2014.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassel D. R., and Elekes K., 1992. Aminergic neurons in the brain of blowflies and Drosophila: dopamine- and tyrosine hydroxylase-immunoreactive neurons and their relationship with putative histaminergic neurons. Cell Tissue Res. 267: 147–167. 10.1007/BF00318701 [DOI] [PubMed] [Google Scholar]
- Nassel D. R., Elekes K., Johansson K. U., and Nässel D. R., 1988. Dopamine-immunoreactive neurons in the blowfly visual system: light and electron microscopic immunocytochemistry. J. Chem. Neuroanat. 1: 311–325. [PubMed] [Google Scholar]
- Ng C.-H. H., Mok S. Z. S., Koh C., Ouyang X., Fivaz M. L. et al. , 2009. Parkin Protects against LRRK2 G2019S Mutant-Induced Dopaminergic Neurodegeneration in Drosophila. J. Neurosci. 29: 11257–11262. 10.1523/JNEUROSCI.2375-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nippe O. M., Wade A. R., Elliott C. J. H., and Chawla S., 2017. Circadian Rhythms in Visual Responsiveness in the Behaviorally Arrhythmic Drosophila Clock Mutant Clk Jrk. J. Biol. Rhythms 32: 583–592. 10.1177/0748730417735397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora F. C., Nucifora L. G., Ng C. H., Arbez N., Guo Y. et al. , 2016. Ubiqutination via K27 and K29 chains signals aggregation and neuronal protection of LRRK2 by WSB1. Nat. Commun. 7: 11792 10.1038/ncomms11792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G. et al. , 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12: 19–30. 10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- Pacelli C., Giguère N., Bourque M.-J., Lévesque M., Slack R. S. et al. , 2015. Elevated Mitochondrial Bioenergetics and Axonal Arborization Size Are Key Contributors to the Vulnerability of Dopamine Neurons. Curr. Biol. 25: 2349–2360. 10.1016/j.cub.2015.07.050 [DOI] [PubMed] [Google Scholar]
- Purlyte E., Dhekne H. S., Sarhan A. R., Gomez R., Lis P. et al. , 2018. Rab29 activation of the Parkinson’s disease-associated LRRK2 kinase. EMBO J. 37: 1–18. 10.15252/embj.201798099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J. R., and Zipursky S. L., 2010. Design principles of insect and vertebrate visual systems. Neuron 66: 15–36. 10.1016/j.neuron.2010.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Shi C.-H., and Xu Y., 2017. Rab GTPases: The Key Players in the Molecular Pathway of Parkinson’s Disease. Front. Cell. Neurosci. 11: 81 10.3389/fncel.2017.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N., Jeong H., Kwon J., Heo H. Y., Kwon J. J. et al. , 2008. LRRK2 regulates synaptic vesicle endocytosis. Exp. Cell Res. 314: 2055–2065. 10.1016/j.yexcr.2008.02.015 [DOI] [PubMed] [Google Scholar]
- Sim J. P. L., Ziyin W., Basil A. H., Lin S., Chen Z. et al. , 2019. Identification of PP2A and S6 Kinase as Modifiers of Leucine-Rich Repeat Kinase-Induced Neurotoxicity. Neuromolecular Med. 10.1007/s12017-019-08577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M., Diez F., Dhekne H. S., Lis P., Nirujogi R. S. et al. , 2017. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 6: e31012 10.7554/eLife.31012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M., Tonelli F., Ito G., Davies P., Trost M. et al. , 2016. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5: e12813 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkellner T., Zell V., Farino Z. J., Sonders M. S., Villeneuve M. et al. , 2018. Role for VGLUT2 in selective vulnerability of midbrain dopamine neurons. J. Clin. Invest. 128: 774–788. 10.1172/JCI95795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam M., Althof D., Gispert S., Schwenk J., Auburger G. et al. , 2014. Mutant α-Synuclein Enhances Firing Frequencies in Dopamine Substantia Nigra Neurons by Oxidative Impairment of A-Type Potassium Channels. J. Neurosci. 34: 13586–13599. 10.1523/JNEUROSCI.5069-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ran D., Zhao X., Huang Y., Long S. et al. , 2016. Melatonin attenuates hLRRK2-induced sleep disturbances and synaptic dysfunction in a Drosophila model of Parkinson’s disease. Mol. Med. Rep. 13: 3936–3944. 10.3892/mmr.2016.4991 [DOI] [PubMed] [Google Scholar]
- Thirstrup K., Dächsel J. C., Oppermann F. S., Williamson D. S., Smith G. P. et al. , 2017. Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells. Sci. Rep. 7: 10300 10.1038/s41598-017-10501-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins J. E., Dihanich S., Beilina A., Ferrari R., Ilacqua N. et al. , 2018. Comparative Protein Interaction Network Analysis Identifies Shared and Distinct Functions for the Human ROCO Proteins. Proteomics 18: e1700444 10.1002/pmic.201700444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira O. V., 2018. Rab3a and Rab10 are regulators of lysosome exocytosis and plasma membrane repair. Small GTPases 9: 349–351. 10.1080/21541248.2016.1235004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., and Huang X., 2012. Rab32 is important for autophagy and lipid storage in Drosophila. PLoS One 7: e32086 10.1371/journal.pone.0032086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters F., Cornelissen T., Imberechts D., Martin S., Koentjoro B. et al. , 2019. LRRK2 mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy 16: 203–222. 10.1080/15548627.2019.1603548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. J. H., Furmston R., Williams C. A. C., and Elliott C. J. H., 2015a Neurophysiology of Drosophila models of Parkinson’s disease. Parkinsons Dis. 2015: 381281 10.1155/2015/381281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. J. H., Lu Y., Marie B., Gao F.-B., and Sweeney S. T., 2015b Rab8, POSH, and TAK1 regulate synaptic growth in a Drosophila model of frontotemporal dementia. J. Cell Biol. 208: 931–947. 10.1083/jcb.201404066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. A., Anderson E., Zimmerman K., Zheng K. H., Rouhani R. et al. , 2015. Huntingtin differentially regulates the axonal transport of a sub-set of Rab-containing vesicles in vivo. Hum. Mol. Genet. 24: 7182–7195. 10.1093/hmg/ddv415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. R., Sim J. C. H., McLean C., Giannandrea M., Galea C. A. et al. , 2014. Mutations in RAB39B cause X-linked intellectual disability and early-onset Parkinson disease with α-synuclein pathology. Am. J. Hum. Genet. 95: 729–735. 10.1016/j.ajhg.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Yuan C., Chen R., Dawson T. M., and Dawson V. L., 2012. ArfGAP1 is a GTPase activating protein for LRRK2: reciprocal regulation of ArfGAP1 by LRRK2. J. Neurosci. 32: 3877–3886. 10.1523/JNEUROSCI.4566-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Thomas J. M., Li T., Lee Y., Liu Z. et al. , 2018. The Drosophila hep pathway mediates Lrrk2-induced neurodegeneration. Biochem. Cell Biol. 96: 441–449. 10.1139/bcb-2017-0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Schulze K. L., Hiesinger P. R., Suyama K., Wang S. et al. , 2006. Thirty-One Flavors of Drosophila Rab Proteins. Genetics 176: 1307–1322. 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data tables (Excel sheets) and R code are open access on GitHub: https://github.com/wadelab/flyCode/tree/master/analyzeData/fly_arduino/G3. Raw images and SSVEP traces are available on request. No new reagents are described.