Abstract
Understanding the genomic basis of adaptative intraspecific phenotypic variation is a central goal in conservation genetics and evolutionary biology. Lake trout (Salvelinus namaycush) are an excellent species for addressing the genetic basis for adaptive variation because they express a striking degree of ecophenotypic variation across their range; however, necessary genomic resources are lacking. Here we utilize recently-developed analytical methods and sequencing technologies to (1) construct a high-density linkage and centromere map for lake trout, (2) identify loci underlying variation in traits that differentiate lake trout ecophenotypes and populations, (3) determine the location of the lake trout sex determination locus, and (4) identify chromosomal homologies between lake trout and other salmonids of varying divergence. The resulting linkage map contains 15,740 single nucleotide polymorphisms (SNPs) mapped to 42 linkage groups, likely representing the 42 lake trout chromosomes. Female and male linkage group lengths ranged from 43.07 to 134.64 centimorgans, and 1.97 to 92.87 centimorgans, respectively. We improved the map by determining coordinates for 41 of 42 centromeres, resulting in a map with 8 metacentric chromosomes and 34 acrocentric or telocentric chromosomes. We use the map to localize the sex determination locus and multiple quantitative trait loci (QTL) associated with intraspecific phenotypic divergence including traits related to growth and body condition, patterns of skin pigmentation, and two composite geomorphometric variables quantifying body shape. Two QTL for the presence of vermiculations and spots mapped with high certainty to an arm of linkage group Sna3, growth related traits mapped to two QTL on linkage groups Sna1 and Sna12, and putative body shape QTL were detected on six separate linkage groups. The sex determination locus was mapped to Sna4 with high confidence. Synteny analysis revealed that lake trout and congener Arctic char (Salvelinus alpinus) are likely differentiated by three or four chromosomal fissions, possibly one chromosomal fusion, and 6 or more large inversions. Combining centromere mapping information with putative inversion coordinates revealed that the majority of detected inversions differentiating lake trout from other salmonids are pericentric and located on acrocentric and telocentric linkage groups. Our results suggest that speciation and adaptive divergence within the genus Salvelinus may have been associated with multiple pericentric inversions occurring primarily on acrocentric and telocentric chromosomes. The linkage map presented here will be a critical resource for advancing conservation oriented genomic research on lake trout and exploring chromosomal evolution within and between salmonid species.
Keywords: Linkage map, Salvelinus, QTL, RAD, genomics, lake trout
Maintaining adaptive phenotypic diversity is a central tenet of conservation biology. In many taxa, diversity is produced through selective pressures that favor reduced intraspecific competition and trophic specialization (Skulason and Smith 1995; Robinson and Schluter 2000; Whiteley 2007). The evolution of trophically specialized morphotypes has been observed in multiple fish species including Arctic char (Snorrason et al. 1994), lake trout (Eshenroder 2008; Muir et al. 2016), multiple coregonid species (Lu and Bernatchez 1999; Thomas et al. 2019), and African cichlids (Rüber et al. 1999), and represents an important pathway by which phenotypic diversity is generated and maintained in nature (Pfennig and Pfennig 2012). Intraspecific diversity can promote community and ecosystem stability (Schindler et al. 2010); however, the genomic basis for this variation is often poorly understood for non-model species. Advancement of our understanding is largely limited by a lack of genomic resources.
Lake trout (Salvelinus namaycush) are a salmonid fish species endemic to North America with substantial cultural, ecological, and economic importance. Across their range, lake trout are often the keystone predator of lentic ecosystems (Ryder et al. 1981) and historically supported valuable commercial and subsistence fisheries (Waters 1987; Hansen 1999; Brenden et al. 2013). Lake trout express a large degree of sympatric phenotypic variation (Muir et al. 2016) making them a useful species for exploring the genomic basis for phenotypic diversity. Multiple morphotypes exist across the species range (Muir et al. 2016; Marin et al. 2017), with diversification largely associated with the ability to exploit resources and habitats at varying depths in large post-glacial lakes (Zimmerman et al. 2006; Stafford et al. 2013; Muir et al. 2014; Marin et al. 2017). In the Great Lakes, trophic specialization has resulted in the evolution of three widely recognized morphotypes — leans, siscowets, and humpers — that are differentiated by patterns of skin pigmentation, size-at-age, body shape, tissue lipid content, habitat use, and diet (Thurston 1962; Eschmeyer and Phillips 1965; Burnham-Curtis 1994; Harvey et al. 2003; Alfonso 2004; Zimmerman et al. 2007; Zimmerman et al. 2009; Goetz et al. 2013). Similar patterns of divergence exist in other lake trout populations (Blackie et al. 2003; Zimmerman et al. 2006; Hansen et al. 2012; Marin et al. 2016; Chavarie et al. 2015), with some degree of morphological and phenological variation existing among individuals of the same morphotype (Bronte 1993; Bronte and Moore 2007).
Previous studies have evaluated differences in gene expression and signals of adaptive divergence between lake trout morphotypes (Goetz et al. 2010; Bernatchez et al. 2016; Perreault-Payette et al. 2017). However, no study has explicitly evaluated which loci control variation in specific traits that underly morphotype divergence. Additionally, these studies have relied on de novo assembled markers distributed anonymously across the genome. Although these approaches can be powerful (Davey et al. 2013), fully interpreting results requires some knowledge of how loci are ordered along chromosomes. All scans for adaptively significant loci and genotype-phenotype associations inherently take advantage of linkage disequilibrium between genotyped markers and causal loci. Without knowing the relative locations of loci, it can be difficult to determine if genotype-phenotype associations or signals of selection are associated with a single genomic region or multiple regions distributed widely across the genome. Information on the order of loci along chromosomes can be readily attained via linkage mapping or assembly of a reference genome; however, linkage maps are often needed a priori to produce chromosome-scale genome assemblies.
Linkage maps have been used to map loci associated with disease resistance (Houston et al. 2008; Moen et al. 2009), life history and physiological trait variation (Rogers and Bernatchez 2007; Miller et al. 2012; Gagnaire et al. 2013a; Sutherland et al. 2017; Pearse et al. 2019), and commercially valuable traits (Gonzalez-Pena et al. 2016) in salmonids and have been instrumental in the assembly of salmonid reference genomes (Lien et al. 2016, Christensen et al. 2018a, 2018b; Pearse et al. 2019; Sävilammi et al. 2019). A linkage map for lake trout would enable the application of cutting-edge genomic tools to questions in lake trout management and evolution and would aid in the identification of loci underlying phenotypic variation and local adaptation. Specifically, a linkage map would increase the strength of inference from genome-wide association studies and scans for selection (Bradbury et al. 2013; Gagnaire et al. 2013b; McKinney et al. 2016) and allow for the localization of quantitative trait loci (Peichel et al. 2001; Qiu et al. 2018) and tracts of admixture and homozygosity, and the estimation of historical effective population sizes and admixture dynamics (Hollenbeck et al. 2016; Leitwein et al. 2018). This information would be valuable for selecting stocks for reintroduction and translocation and for estimating the adaptive potential of intact populations under changing climate and abiotic conditions (Leitwein et al. 2016; Bay et al. 2017).
Comparative analysis of linkage maps and genome assemblies from related species can also shed light on chromosomal evolution and speciation (Rastas et al. 2015; Sutherland et al. 2016; Hale et al. 2017). Chromosomal inversions appear to have played an important role in speciation and adaptive divergence within the salmonid lineage (Miller et al. 2012; Sutherland et al. 2016, Pearse et al. 2019) and within other taxa (Lowry and Willis 2010; Küpper et al. 2016; for review see Wellenreuther and Bernatchez 2018). Instances of reduced hybrid fitness and hybrid inviability are widespread within the family Salmonidae (Leary et al., 1993; Fugjiwara et al. 1997; Muhlfeld et al. 2009). Information on the locations of inversions differentiating species and phenotypically divergent populations could shed light on the genetic basis for these phenomena. Inversions can contribute to isolation between species and populations because they can suppress recombination over large chromosomal regions, allowing for adaptive differences to accumulate between inverted and non-inverted haplotypes even in the presence of gene flow (Berg et al. 2017; Wellenreuther and Bernatchez 2018). Inversions can also produce post-zygotic isolation between incipient species if crossing over within heterozygous individuals results in formation of abnormal or inviable gametes (Wellenreuther and Bernatchez 2018). An improved understanding of the extent to which pericentric (including the centromere) and paracentric (outside the centromere) inversions can accumulate between salmonid species over varied evolutionary time scales, could provide clues about pre- and post-zygotic isolation mechanisms that contributed to adaptive divergence and incipient speciation within salmonids.
Linkage maps have been constructed for multiple salmonid species including rainbow trout (Miller et al. 2012; Palti et al. 2012; Gonzalez-Pena et al. 2016), chinook salmon (Brieuc et al. 2014; McKinney et al. 2016; McKinney et al. 2019), coho salmon (Kodama et al. 2014), sockeye salmon (Everett, Miller and Seeb 2012; Larson et al. 2015; Limborg et al. 2015), chum salmon (Waples et al. 2016); pink salmon (Spruell et al. 1999; Lindner et al. 2000), Atlantic salmon (Moen et al. 2008; Lien et al. 2011; Brenna-Hansen et al. 2012; Gonen et al. 2014), Arctic char (Nugent et al. 2017; Christensen et al. 2018a), brook trout (Sauvage et al. 2012; Sutherland et al. 2016; Hale et al. 2017), brown trout (Leitwein et al. 2017), European grayling (Sävilammi et al. 2019), lake whitefish (Rogers and Bernatchez 2007; Gagnaire et al. 2013a), and European whitefish (De-Kayne and Feulner 2018). No linkage map has been constructed for lake trout (but see May et al. 1979, Johnson et al. 1987, for work on segregation patterns in lake trout x brook trout hybrids), although the lake trout karyotype has been characterized in multiple previous studies (Phillips and Zajicek 1982; Reed and Phillips 1995) providing a reference for the number of expected chromosomes.
Here we present a high-density linkage map for lake trout generated using restriction site associated DNA (RAD) capture (Rapture; Ali et al. 2016), a modified RAD sequencing protocol that allows variable loci to be preferentially genotyped. The map was used to characterize the lake trout karyotype, estimate recombination rates, determine centromere locations, map the sex determination locus, and identify chromosomal inversions and translocations differentiating lake trout from other salmonids. We demonstrate the utility of the linkage map by using available phenotype data to map quantitative trait loci (QTL) associated with pigmentation patterns, growth and condition related traits, and variation in body shape — all traits hypothesized to be adaptive in lake trout and other salmonids.
Materials And Methods
Linkage mapping families
Two F1 full-sibling families were created by crossing Seneca Lake hatchery strain females with Parry Sound strain males (Table 1, Figure 1). The Seneca Lake strain was founded using individuals from Seneca Lake, New York and this strain has contributed disproportionately to restoring lake trout populations in the Great Lakes (Scribner et al. 2018). The Parry Sound strain was founded by wild individuals collected from Georgian Bay in Lake Huron. The Seneca and Parry Sound strains are genetically divergent (FST = 0.089) based on a previous study using microsatellites (Scribner et al. 2018). Crosses were produced in 2017 using adult lake trout and housed at Pendills Creek National Fish Hatchery (U.S. Fish and Wildlife Service, Figure 1). Eggs were fertilized, incubated in Heath trays at ambient temperature, and raised until swim-up phase. Offspring were then killed using a lethal dose of MS-222 and preserved in 95% ethanol. Genetic sex was determined for offspring using a sdY presence-absence quantitative PCR (qPCR) assay designed using the approach of Anglès d’Auriac et al. (2014; see Trait Mapping methods below). These families were ultimately used for constructing the linkage map and localizing the lake trout sex determination locus.
Table 1. Family IDs, cross type (diploid or gynogenetic diploid), number of genotyped offspring per family, and maternal and paternal origins for the five families used for linkage and QTL mapping.
| Family | Type | No. Offspring | Mother Origin | Father Origin |
|---|---|---|---|---|
| S1 | Diploid | 88 | Seneca Lake | Parry Sound |
| S2 | Diploid | 91 | Seneca Lake | Parry Sound |
| P1 | Diploid | 91 | Killala X Kingscote F1 | Killala X Kingscote F1 |
| P3 | Diploid | 88 | Killala X Kingscote F1 | Killala X Kingscote F1 |
| G1 | Gynogenetic Diploid | 45 | Killala X Kingscote F1 | None |
Figure 1.
Map displaying the locations of hatchery facilities (dots) and locations of wild progenitor populations (diamonds) used for mapping. Locations of hatchery facilities used for conducting crosses are marked with black circles. The locations of the progenitor populations are identified with black diamonds. Longitude is displayed on the Y-axis and latitude is displayed on the X-axis.
An additional F2 half-sibling family was created using adult lake trout from the Killala Lake hatchery strain and wild individuals from Kingscote Lake, Ontario (Table 1, Figure 1). The Killala Lake strain was founded by individuals from Killala Lake, Ontario, which is within the Lake Superior drainage. This hatchery strain is most similar to lean form hatchery strains derived from Lake Superior based on a previous allozyme genotyping study (Marsden et al. 1993). Individuals from the Kingscote Lake strain also resemble lean lake trout; however, they are small bodied and lack spots and vermiculations (Wilson and Evans 2010). Examination of F2 offspring at age 3 revealed substantial variation in pigmentation, weight and length at age, and body shape among individuals. These traits are commonly recognized as being adaptively differentiated between lake trout populations and ecophenotypes (Eshenroder 2008; Muir et al. 2016). Body shape and early growth rate in particular have been recognized as important traits for differentiating lean, siscowet, and humper ecophenotypes (Moore and Bronte 2001; Hansen et al. 2016). The observation that skin pigmentation patterns vary between ecophenotypes and across depth strata in some lake trout populations also suggests that pigmentation traits might be an important axis of ecophenotypic divergence within lake trout (Zimmerman et al. 2006). The F2 Kingscote x Killala family was used for linkage map construction, localization of the sex determination locus, and QTL mapping. Crosses, culture conditions, and phenotyping procedures are described below.
Initial Kingscote x Killala F1 crosses were produced using adult lake trout using a 2x2 factorial mating design. In 2012, mature adults from initial crosses were mated to produce F2 families. Eggs from each family were incubated in Heath trays at ambient temperature (2-5°). Prior to swim-up, hatched sac fry from families were transferred to 36L laundry tubs (200 fry per tub) where they remained until age 1+. Families were manually fed 1% of tank biomass twice-daily and family sizes were periodically reduced by culling to avoid overcrowding. At age 1+, families were transferred to 700L circular tanks with ambient lighting and fed to satiation on an EWOS pellet diet. At age 3, fork length and weight were determined and lateral photographs were collected using the protocol from Bernatchez et al. (2016). Fish were photographed using a Nikon Coolpix P7700 digital camera with a focal length of 50mm mounted on a tripod in fixed position. Fish were photographed with the head facing to the left and were cradled in a stretched mesh net as in Zimmerman et al. (2006) in order to avoid distorting body shape. Fin clips were collected and preserved in 95% ethanol. Photographs were later used for morphometric analysis and scoring individuals for presence-absence of spots and vermiculations (see Trait Mapping methods section below).
An additional gynogenetic diploid family was created using a female F1 resulting from initial Kingscote x Killala crosses using a protocol similar to that of Thorgaard et al. (1983). This family was used for mapping centromeres using half-tetrad analysis (Thorgaard et al. 1983; Limborg et al. 2016). Sperm from a male lake trout was diluted 10:1 using sperm extender (9.2 g Tris buffer, 1.05 g citric acid, 4.81 g glycine, 2.98 g KCl, 100g PVP-40, and 1 liter of distilled water), mixed thoroughly in a 9x13x2 inch glass pyrex dish, placed on ice, and irradiated for 2 min using a 25-watt germicidal UV lamp placed 20 centimeters from the dish. Eggs and sperm were then mixed and sperm was activated by adding water. Ten minutes after fertilization, eggs were heat shocked at 26° for 10 min, water hardened, transferred to Heath trays for incubation, and raised using the same conditions described for diploid families. All Kingscote X Killala families were produced at the Codrington Fisheries Research Facility (Ontario Ministry of Natural Resources and Forestry; Figure 1; Codrington, Ontario). This facility has a surface water supply which undergoes seasonal and diel temperature variation ranging between 2-5° in winter and 9-16° in summer.
Sample preparation
For all Kingscote x Killala families, DNA from offspring and parents (Table 1) was extracted using the high-throughput SPRI bead-based extraction protocol described in Ali et al. (2016) with Serapure beads (described in Rohland and Reich 2012) substituted for Ampure XP beads. For the Seneca x Parry Sound crosses, DNA was extracted using Qiagen DNeasy Blood and Tissue extraction kits (69506, Qiagen, Hilden, Germany) using manufacturer recommendations. DNA quality was initially assessed using a Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts) by evaluating 260/230 and 260/280 absorbance ratios. Samples were diluted to less than 100ng/ul based on Nanodrop readings, then diluted 10-fold before determining double-stranded DNA (dsDNA) concentrations using Quantit Picogreen assays (Thermo Fisher Scientific, Waltham, Massachusetts).
Sequencing library preparation
DsDNA concentrations were normalized to 10ng/ul using an Eppendorf epMotion 2750 TMX liquid handling robot (Eppendorf, Hamburg, Germany) before proceeding with the bestRAD protocol and RAD-capture using 100ng of total input DNA (Ali et al. 2016). Modifications to the protocol are noted below with a detailed description of methods provided in supplementary material (Document S1). First, the enzyme PstI was substituted for SbfI and PstI was heat-killed at 80° rather than 65°. After ligating bestRAD adapters and pooling samples, shearing was carried out using a Covaris E220 Ultrasonicator (Covaris Inc., Woburn, Massachusetts) using the recommended settings for a 300bp mean fragment length. Finished libraries were amplified for 10 cycles, pooled equally in sets of two, and bead cleaned twice using a 0.9:1 bead-to-DNA ratio Ampure XP cleanup (A63881, Beckman Coulter, Brea, California). The two resulting pools were then enriched for 58,889 RAD loci that were previously found to be variable in lake trout populations in the Great Lakes, Seneca Lake, Ontario, Montana, and Alaska using the RAD-capture protocol (Ali et al. 2016). Target enrichment reactions were carried out using a MyBaits Custom Target Enrichment kit using manufacturer recommendations (MycroArray, Ann Arbor, Michigan; Protocol Version 3; for more information on capture and bait selection see Document S1). Finished capture reactions were amplified for an additional 9 cycles, pooled, and sequenced in three lanes of an Illumina HiSeq X instrument (2 X 150 bp paired end reads; Illumina, San Diego, California) by the Novogene Corporation (Novogene, Sacremento California).
Bioinformatics and genotyping
Read quality was initially assessed using FastQC v0.11.5 (Andrews 2014), and a custom script was used to re-orient paired end reads such that individual specific barcodes and restriction enzyme overhang sequences were always located at the beginning of the first read. Reads were demultiplexed using process_radtags v2.2, duplicate reads were removed using clone_filter v2.2 (Catchen et al. 2013; Rochette et al. 2019), and adapter sequences were clipped from reads using Trimmomatic v0.36 (Bolger et al. 2014). At this point, we produced two sets of fastq files: one conservatively filtered dataset used for de novo assembly of RAD loci and a slightly less conservatively filtered dataset used for calculating genotype likelihoods that would ultimately be used for linkage mapping and other analyses. For the de novo assembly dataset, reads were trimmed whenever the mean base quality across a sliding window of 4bp dropped below Q20, read pairs were removed if one or both reads in a pair were less than 140bp in length after trimming, and reads were cropped to a length of 140bp such that all reads were of identical length. For the dataset used to calculate genotype likelihoods, reads were trimmed whenever the mean base quality across a sliding window of 4bp dropped below Q15 and excluded if one or both reads in the pair were less than 50bp after trimming.
The stringently filtered dataset (read length =140bp, trimming threshold of Q20) was used to assemble RAD loci de novo using modules available in Stacks v2.2 (Rochette et al. 2019). RAD loci were identified for individuals using ustacks v2.2, which was run with a minimum depth of coverage of 3 (-m 3), a maximum distance between stacks of 3 (-M 3), a maximum distance to align secondary reads to primary stacks of 2 (-N 2), a minimum of 2 stacks at each de novo locus (–max_locus_stacks 2), and disabling calling haplotypes from secondary reads (-H). We then created a catalog of RAD loci for the parents of crosses using cstacks v2.2, allowing for up to two mismatches between sample loci when building the locus catalog (-n 2). Putative RAD loci alleles for all individuals were matched to this catalog using sstacks v2.2, converted to bam format using tsv2bam v2.2, and then assembled using gstacks v2.2. Consensus sequences for RAD loci were obtained by passing the “–fasta-loci” flag to the populations v2.2 module. The fasta file containing RAD locus consensus sequences was normalized using Picard NormalizeFasta v2.8 (http://broadinstitute.github.io/picard/), indexed using bwa index v7.15 (Li 2013) and samtools faidx v1.3 (Li et al. 2009), and used as a de novo reference for subsequent analysis.
Next, the larger set of variable length paired end reads that were trimmed using a Q-threshold of 15 were mapped to the de novo assembly using bwa mem v7.15 (Li 2013) with default setting. Genotype likelihoods were calculated for single nucleotide polymorphisms (SNPs) within RAD loci using Lepmap3 v0.2 and associated modules (Rastas et al. 2015). SAM files produced by bwa-mem were converted to bam format and sorted using samtools v1.3, then converted to mpileup format using a minimum mapping quality of 30 and a minimum base quality of 20. The resulting file was filtered using the script pileupParser2.awk using a minimum read depth of 3 and a missingness threshold of 0.3. Genotype likelihoods were calculated using the pileup2posterior.awk script distributed with LepMap3 v0.2 (Rastas et al. 2015, Rastas 2017). We opted to use pileup2posterior.awk to calculate genotype likelihoods because the LepMap3 pipeline was originally validated using likelihoods calculated using this program (Rastas 2017).
Linkage map construction
Linkage mapping and additional data filtering were carried out using various programs distributed with LepMap3 v0.2 (Rastas 2017). First, any missing parental SNP genotypes were imputed using ParentCall2. Second, SNPs showing evidence of segregation distortion were removed using Filtering2 with a p-value (–dataTolerance) threshold of 0.01. We required that SNPs be informative for linkage mapping in at least 1 family and removed SNPs with minor allele frequencies less than 0.05.
SNPs were assigned to linkage groups (LGs) using SeparateChromosomes2 run with logarithm of odds ratios (LOD) thresholds ranging from 8 to 60 and a minimum LG size of 50 SNPs. No single LOD threshold produced the expected number of LGs (n = 42; Phillips and Zajicek 1982; Reed and Phillips 1995). Beginning with the map produced using a universal LOD threshold of 10, we determined the LOD thresholds needed to further split each LG by running SeparateChromosomes2 using all LOD thresholds between 10 and 60 and specifying the LG targeted for additional splitting using the “lg” and “map” flags (similar to Christensen et al. 2018a).
We determined that the largest 8 of the initial 30 LGs could be split using LOD thresholds ranging from 11-52, with the remaining 22 LGs remaining intact for all LOD thresholds between 10 and 60. The 8 largest LGs were split using the maximum LOD threshold that resulted in a new LG containing more than 50 SNPs, resulting in 42 LGs. Unassigned singleton SNPs were then joined to this map using JoinSingles2All run iteratively with a LOD threshold of 10 and a minimum LOD difference of 5.
The order of SNPs was initially determined by running 20 iterations of OrderMarkers2 and selecting the order with the highest likelihood for each LG. LGs were further refined by evaluating LOD matrices (output using computeLODscores = 1). For each SNP, the vector of LOD scores corresponding to possible map positions was normalized such that values ranged from 0 to 1. SNPs were removed if the maximum LOD score was less than 1 standard deviation from the mean or if more than one LOD ‘peak’ was observed for any given SNP, indicating the existence of multiple mapping positions of similar likelihood. LOD peaks were identified using the findPeaks function from the R package pracma v2.2.5, a minimum normalized peak height of 0.95 and a minimum distance between peaks greater than 25% of the length of the vector of mapping positions. RAD loci were removed from the data set if associated SNPs mapped to more than one LG. Finally, the dataset was thinned to include a single SNP for each RAD locus, with preference given to the SNP closest to the PstI restriction cut site. We opted to thin SNPs after determining which loci could be effectively mapped in order to maximize the number of unique RAD loci on the map. Maps for each LG were then reconstructed using the evaluateOrder and improveOrder = 1 options from OrderMarkers2, with SNPs that failed the above filtering criteria flagged for removal using the removeMarkers option.
Finally, LGs were inspected for possible mis-ordering using LMPlot and any LG marked with possible errors were reordered using OrderMarkers2 for an additional 60 iterations. The linkage map was further improved by trimming SNPs from the ends of LGs based on manual inspection of LOD matrix plots and alignment to rainbow trout, Arctic char, and Atlantic salmon (Salmo salar) genome sequences (see Homology section below). An additional 10 iterations of ordering were conducted after removing potential erroneously placed SNPs from the ends of LGs. Final LGs were sorted based on their number of mapped SNPs and named as Sna1-Sna42. Both male and female linkage maps were output by the program.
Centromere mapping
We identified centromeres by estimating the frequency of second division segregation (γ) across linkage groups using half-tetrad analysis conducted on gynogenetic diploid offspring from family G1 (Thorgaard et al. 1983). Cells of gynogenetic diploid offspring contain two of the four possible meiosis II products (a half-tetrad) and the frequency of heterozygous offspring can be used to estimate the frequency of recombination events between the locus in question and the centromere (Thorgaard et al. 1983). Reads for these individuals were aligned to de novo assembled RAD loci, sorted and indexed using samtools v1.3 (Li et al. 2009), and variable positions within RAD loci were genotyped using freebayes v1.1.0 (Garrison and Marth 2012). Genotypes were called without applying population or binomial observation priors, an assumed contamination probability of 1%, a minimum base quality of 20, and a minimum mapping quality of 20. Called loci were then converted to their simplest representation using vcfallelicprimatives (https://github.com/vcflib/vcflib; vcflib v1.0.0) and loci with more than 2 alleles and indels were removed, such that only SNPs remained. Genotypes were set to missing if there was less than 1 order of magnitude difference in genotype likelihoods between the called genotype and the second most likely genotype using vcftools v0.1.16 (GQ >10; Danecek et al. 2011). SNPs were removed from the dataset if more than 30% of individuals were missing genotypes or if the frequency of the minor allele was less than 0.05. SNPs were further excluded if they were not placed on the linkage map, not called heterozygous in the mother, or if both possible homozygous genotypes were not observed in offspring. The mother was removed from the dataset at this point, and observed heterozygosity for the offspring (y) was calculated using the hwe function from SeqVarTools v1.20.2 (Gogarten et al. 2017; https://github.com/smgogarten/SeqVarTools). Centromeric regions were delineated as the region between the first and last markers with y-values less than 0.1 (as in Limborg et al. 2016).
Results were cross-validated and improved upon using the RFm method (Limborg et al. 2016) applied to the phased genotypes of progeny from families S1, S2, P1, and P3. Counts of maternal recombination events were reported using OrderMarkers2 with outputPhasedData = 1 and used to calculate RFm across all maternal haplotypes and identify putative centromeric regions using a cut-off value of 0.45 as suggested in Limborg et al. (2016). The correct centromeric locations for acrocentric and telocentric chromosomes were identified by selecting the region containing, or neighboring, the lowest y-values from half-tetrad analysis.
Homology
RAD loci were aligned to the reference genomes for Arctic char (RefSeq Accession: GCF_002910315.2), Rainbow trout (Oncorhynchus mykiss; RefSeq Accession: GCF_002163495.1) and Atlantic salmon (RefSeq Accession: GCF_000233375.1) using bwa mem v7.15 (Li 2013). RAD loci were assigned to their respective linkage map positions, and male and female linkage maps were visualized relative to their order along homologous chromosomes using ggplot2 v3.2.1 (Wickham and Chang 2008). Chromosomes were considered homologous if 50 or more mapped RAD loci aligned to a chromosome with mapping qualities greater than MQ60. The map was also compared with a linkage map for brook trout (Salvelinus fontinalis; Sutherland et al. 2016) using the program MapComp (Sutherland et al. 2016; https://github.com/enormandeau/mapcomp) and the Arctic char genome as an intermediate reference in order to detect large structural variants differentiating the two species.
Putative chromosomal inversions were detected by manually inspecting plots produced by mapping the lake trout linkage map to divergent references. Inversion breakpoints were defined by the coordinates with the greatest discrepancy between the divergent physical map and the female linkage map we constructed. Inversions were classified as pericentric if putative inversion coordinates overlapped centromere mapping positions. Inversions differentiating lake trout and brook trout were detected by manually inspecting dot plots produced by MapComp.
Trait mapping
Offspring from diploid Kingscote x Killala crosses were phenotyped for fork length (FL), weight (WT), and condition factor (CF) at age 3. Additionally, photographs collected at age 3 were used to score individuals for presence-absence of spots and vermiculations (VPA) and two composite variables (PCA1 and PCA2) summarizing variation in body shape. Body shape variables were derived by performing a principal-components analysis (PCA) on the coordinates of morphometric landmarks that were normalized for slight differences in fish position and rotation using generalized Procrustes analysis. Using available photographs from families P1 and P3, we placed landmarks using tpsDIG v2 (Rohlf 2005) consistent with those described in Muir et al. (2014). Landmark coordinates were normalized and rotated using generalized Procrustes analysis conducted using the function gpagen from the R-package geomorph v3.1.1 (Adams and Otárola‐Castillo 2013). Four of 20 landmarks could not be consistently placed using available images (1,6,7,10) and were therefore excluded from the analysis. Synthetic variables PCA1 and PCA2 were calculated by performing PCA on the resulting normalized coordinates and extracting scores for the first two axes. PCA was carried out using the function prcomp from the R-package stats v3.5.3. VPA, PCA1, and PCA2 phenotypes were available for 143 of 179 individuals. Fork length, condition factor, and weight phenotypes were collected for 179 of 179 individuals.
Phased SNP genotypes for offspring were extracted from the final map files reported by OrderMarkers2 using the script map2genotypes.awk from LepMap3 v0.2. QTL mapping was then carried out for traits of interest using the R-package qtl2 v0.2 and associated functions (Broman et al. 2019). All traits were mapped to sex-averaged linkage map coordinates. Prior to QTL mapping, pseudo-markers were added to the map using insert_pseudomarkers with a step size of 1cM and genotype probabilities were calculated using calc_genoprob. A kinship matrix was calculated using the calc_kinship function using genotype probabilities. A thinned subset of markers obtained using calc_grid (step = 3) and probs_to_grid was used as input for the calc_kinship function. QTL scans were carried out using scan1 and suggestive QTL peaks were identified using find_peaks (drop = 2, peakdrop = 2, threshold = 3). Traits with approximately normal distributions (FL, CF, WT, PCA1, PCA2) were mapped using a mixed linear model with the kinship matrix included as a random effect (model = “normal” and kinship options in qtl2). Presence-absence of vermiculations and spots (VPA) was mapped as a binary trait (model = “binary” in qtl2). The kinship matrix was not included as a random effect in the binary trait mapping model because this option was not available in qtl2. For each identified LOD peak, 95% credible intervals were calculated using the function find_peaks (prob = 0.95, peakdrop = 2, threshold = 3). Finally, p-values were calculated by comparing observed LOD scores for each peak with a null distribution obtained from permuting the data 1000 times. Permutations were carried out using the function scan1perm using the same settings as the original tests and p-values were calculated using the ecdf function. The proportion of phenotypic variation explained (PVE) by each QTL peak was calculated from LOD scores and sample sizes using the equation (Broman and Sen 2009). Candidate genes for significant LOD peaks (P < = 0.05) were identified by mapping RAD loci within 95% credible intervals to the Arctic char genome and determining the three genes closest to each mapping position using the program bedtools closest v2.26 (Quinlan and Hall 2010). Genes were considered candidates if they were within 50Kb of the mapping position of a RAD locus falling within the identified QTL mapping interval.
We also mapped the sex determination region using the binary trait model using qtl2 and assessed significance using the same methodology described above. The sexually dimorphic on the Y chromosome gene (sdY) is believed to underly sex determination in lake trout and some other salmonids (Yano et al. 2013). We designed a sdY presence-absence melt curve qPCR assay (similar to Anglès d’Auriac et al. 2014) using the lake trout sdY and 18S primers described in Yano et al. (2013). 18S served as an internal amplification control. Each reaction was carried out using a 0.4 uM concentration of primers sdYE2S1 (CCCAGCACTGTTTTCTTGTCTCA) and sdYE2AS1 (TGCTCTCTGTTGAAGAGCATCAC), and a 0.04uM concentration of primers 18SS (GTYCGAAGACGATCAGATACCGT) and 18SAS (CCGCATAACTAGTTAGCATGCCG). Reaction volumes were 20uL and contained 10ul of Forget-Me-Not EvaGreen qPCR mastermix (31045, Biotium, Fremont, California), 2.5 uL of template DNA, and 7.5uL of primers eluted in water. 18S and sdY were amplified in a two-step multiplex reaction using a 2-minute heat activation step at 95° followed by 40 cycles of denaturation at 95C for 5 sec and annealing/extension at 60° for 30 sec. Melt curve analysis was carried out on PCR product for temperatures between 60° and 95° using 0.1° temperature shifts and a 3 sec pause between temperature shifts. We first tested the assay on a subset of 32 individuals of known sex (16 males and 16 females), including the parents used for crosses, in order to verify that males and females could consistently be differentiated based on the presence of a male specific sdY peak in the derivative of the melt curve. Offspring from all diploid families were subsequently genotyped using the same reaction conditions described above. At least one known male and one known female were included on each plate as a control. Sex locus mapping was carried out with sdY presence being coded as 1 and sdY absence coded as 0.
Recombination rate estimation
We estimated sex averaged recombination rates for each chromosome by performing a simple linear regression of pairwise physical distance (base pairs) against genetic distance (cMs) and requiring the intercept to pass through 0. In order to evaluate a pair of RAD loci, we required that they map to the same chromosome on the Arctic char, rainbow trout, or Atlantic salmon genome assemblies and only retained scaffolds and chromosomes with greater than 50 mapped RAD loci for which the mapping quality was 60. For each LG, 100 pairs of RAD loci mapping to the same chromosome were randomly sampled from all possible pairs and recombination rate (cM/MB) was estimated using the slope of the resulting regression. This process was repeated 100 times using alignments against the Arctic char, rainbow trout, and Atlantic salmon genomes. The mean of the distribution of estimates was reported as the chromosome specific recombination rate, and separate values were reported for alignments against the three different divergent reference genomes. Regressions were carried out using the R-package lm and recombination rate estimates were visualized using ggplot2 v3.2.1 (Wickham and Chang 2008). This process was repeated for male and female maps in order to obtain sex specific chromosomal recombination rates.
Data availability
Sequencing data for all individuals used for linkage and QTL mapping has been made publicly available in a NCBI sequence read archive (PRJNA608030). Map information, phenotypes, and genotypes used for QTL and sex locus mapping are available in supplementary material (Document S2). Supplemental material available at figshare: https://doi.org/10.25387/g3.11908326.
Results
Bioinformatics and genotyping
We obtained a mean of 2,685,178 demultiplexed paired end (PE) reads for offspring from diploid crosses (range = 660,474 – 4,317,086, SD = 598,973.3) and a mean of 4,701,286 reads for parents (range = 3,483,973 – 5,868,449, SD = 787,251.1). On average, 21.97% of reads were removed by clone_filter for these individuals. De novo assembly of RAD loci with gstacks produced 146,525 RAD loci ranging in size from 140 to 754 bp in length. Between 92.86% and 95.20% of reads were mapped to de novo assembled RAD loci (mean = 93.52%, SD = 0.33%) using bwa mem. The Lepmap3 genotyping pipeline reported genotype probabilities for 212,158 SNPs, 147,920 of which were informative for linkage mapping. Of those, 72,549 SNPs passed missingness, segregation distortion, and minor allele frequency filters.
For gynogenetic diploid offspring, we obtained an average of 3,873,649 PE reads (range = 1,517,646 – 5,789,490, SD = 1,004, 905). We generated 3,536,915 reads for the mother of this family. On average, 32.63% of reads for these samples were removed by clone_filter. Between 89.0% and 89.6% of those reads were mapped to the de novo assembly using bwa mem (mean = 89.3%, SD= 0.12%). After genotyping with freebayes and filtering data to remove non-informative markers, we identified 893 SNPs that were informative for half-tetrad analysis.
Linkage and centromere mapping
We were able to assign 15,740 RAD loci to LGs with between 878 and 113 loci mapped to each LG (Figure 2, Table 2, Table S1). The total male map length was 2043.41cM and the female map was 2842.22 cM (overall female:male map ratio = 1.391). Male LG map lengths ranged from 1.97 cM -92.87 cM, while female LG map lengths ranged from 43.07 cM – 134.64 cM (Table 2). SNPs were mapped to between 60 and 244 unique positions on linkage groups. As expected, we identified 42 LGs, 8 of which were metacentric and 34 that were acrocentric or telocentric (Table S1, Table S2). These linkage groups likely correspond to the 42 chromosomes identified by previous karyotyping studies (Phillips and Zajicek 1982; Reed and Phillips 1995). Half-tetrad analysis yielded centromere intervals for 7 of 8 metacentric chromosomes and 22 of 34 LGs identified as acrocentric or telocentric (Figure 2, Figure S8, Table S2). RFm analysis identified centromeres for 8 of 8 metacentric chromosomes and 32 of 34 acrocentric or telocentric chromosomes (Figure 2, Table S2). We were ultimately able to determine the location of centromeres for 41 out of 42 chromosomes using the two methods. We were not able to map the centromere for Sna42; however, this chromosome is likely acrocentric or telocentric based on the size of the linkage group relative to others (Table 2) and karyotyping work suggesting the existence of 34 acrocentric and telocentric chromosomes (Phillips and Zajicek 1982; Reed and Phillips 1995).
Figure 2.
Map Locations of 15,740 RAD loci along 42 lake trout linkage groups. Orange boxes highlight centromeres identified using half tetrad analysis with a y-threshold of 0.1. Blue boxes span the intervals of centromeres identified using the RFm method (Limborg et al. 2016) combined with half-tetrad analysis. Locations are in centimorgans on the female linkage map.
Table 2. Summary statistics for each of the 42 constructed linkage groups. No. Mapped Loci corresponds to the number of unique RAD contigs mapped to each linkage group. Male and Female map lengths are in centimorgans (cM). No. Unique Positions corresponds to the number of unique linkage map positions to which RAD loci were assigned. Female:Male Ratio is the ratio of Female Length and Male Length in centimorgans.
| Name | No. Mapped Loci | Male Length (cM) | Female Length (cM) | Female:Male Ratio | No. Unique Positions |
|---|---|---|---|---|---|
| Sna1 | 878 | 85.28 | 106.3 | 1.246 | 217 |
| Sna2 | 789 | 71.98 | 134.08 | 1.863 | 207 |
| Sna3 | 761 | 59.12 | 134.64 | 2.277 | 244 |
| Sna4 | 648 | 66.01 | 106.22 | 1.609 | 207 |
| Sna5 | 618 | 50.95 | 106.98 | 2.100 | 221 |
| Sna6 | 515 | 77.88 | 103.44 | 1.328 | 223 |
| Sna7 | 514 | 73.49 | 126.94 | 1.727 | 195 |
| Sna8 | 497 | 92.87 | 124.81 | 1.344 | 157 |
| Sna9 | 460 | 48.39 | 54.72 | 1.131 | 112 |
| Sna10 | 406 | 54.29 | 55.02 | 1.013 | 112 |
| Sna11 | 404 | 42.38 | 58.43 | 1.379 | 100 |
| Sna12 | 395 | 73.61 | 54.69 | 0.743 | 121 |
| Sna13 | 389 | 34.02 | 51.78 | 1.522 | 116 |
| Sna14 | 377 | 59.65 | 51.85 | 0.869 | 110 |
| Sna15 | 360 | 48.12 | 65.02 | 1.351 | 101 |
| Sna16 | 358 | 53.9 | 55.51 | 1.030 | 102 |
| Sna17 | 357 | 48.72 | 57.73 | 1.185 | 95 |
| Sna18 | 356 | 44.67 | 58.01 | 1.299 | 101 |
| Sna19 | 348 | 41.15 | 53.32 | 1.296 | 109 |
| Sna20 | 344 | 36.93 | 53.35 | 1.445 | 102 |
| Sna21 | 340 | 41.96 | 53.44 | 1.274 | 92 |
| Sna22 | 333 | 70.57 | 78.73 | 1.116 | 109 |
| Sna23 | 332 | 61.14 | 56.31 | 0.921 | 106 |
| Sna24 | 325 | 28.44 | 68.78 | 2.418 | 105 |
| Sna25 | 322 | 63.3 | 56.18 | 0.888 | 98 |
| Sna26 | 319 | 36.71 | 52.1 | 1.419 | 95 |
| Sna27 | 317 | 33.52 | 49.03 | 1.463 | 94 |
| Sna28 | 313 | 37.01 | 50.81 | 1.373 | 102 |
| Sna29 | 312 | 48.84 | 50.59 | 1.036 | 86 |
| Sna30 | 310 | 66.35 | 52.65 | 0.794 | 83 |
| Sna31 | 307 | 36.9 | 53.94 | 1.462 | 93 |
| Sna32 | 302 | 56.79 | 55.84 | 0.983 | 102 |
| Sna33 | 286 | 50.85 | 51.02 | 1.003 | 89 |
| Sna34 | 255 | 52.77 | 50.02 | 0.948 | 85 |
| Sna35 | 244 | 42.17 | 53.94 | 1.279 | 94 |
| Sna36 | 242 | 30.19 | 46.4 | 1.537 | 80 |
| Sna37 | 225 | 26.84 | 57.54 | 2.144 | 82 |
| Sna38 | 218 | 35.06 | 53.87 | 1.537 | 83 |
| Sna39 | 194 | 22.56 | 67.59 | 2.996 | 91 |
| Sna40 | 185 | 33.84 | 71.95 | 2.126 | 90 |
| Sna41 | 172 | 2.12 | 55.58 | 26.217 | 60 |
| Sna42 | 113 | 1.97 | 43.07 | 21.863 | 60 |
Homology analysis
Alignment of the linkage map to divergent salmonid reference genomes revealed that the resulting map was highly congruent with existing assembled salmonid genomes (Figures S1-S6). Large synteny blocks were detected between lake trout linkage groups and the Arctic char genome for linkage groups Sna1-Sna41 (Table 3). Alignments suggested that Sna42 is syntenic with sal34; however, fewer than 50 loci with MQ60 mapped to this chromosome from Sna42. Syntenies were detected between lake trout and all rainbow trout and Atlantic salmon chromosomes. MapComp identified homologies with all brook trout linkage groups identified by Sutherland et al. (2016) (Figure S7).
Table 3. Synteny between lake trout linkage groups and Arctic char, rainbow trout, Atlantic salmon, and brook trout genomes. Arctic char, rainbow trout, and Atlantic salmon chromosomes were recorded if more than 50 RAD contigs from the lake trout linkage group were aligned to a chromosome with a mapping quality of 60. Brook trout linkage groups were recorded if more than 10 aligned markers were detected by MapComp. Graphical depictions of alignment location vs. linkage map position are available in Supplementary Figures S1-S7.
| Lake Trout | Arctic Char | Rainbow Trout | Atlantic Salmon | Brook Trout |
|---|---|---|---|---|
| Sna1 | Sal15 | Omy6 | Ssa24, Ssa26 | BC6 |
| Sna2 | Sal1 | Omy17 | Ssa12 | BC3 |
| Sna3 | Sal20 | Omy12 | Ssa03, Ssa13 | BC8, BC14 |
| Sna4 | Sal18 | Omy16, Omy23 | Ssa01, Ssa19 | BC1 |
| Sna5 | Sal6.1, Sal6.2 | Omy2, Omy14 | Ssa05 | BC7 |
| Sna6 | Sal3 | Omy21 | Ssa07 | BC2 |
| Sna7 | Sal27 | Omy15, Omy18 | Ssa16, Ssa29 | BC5 |
| Sna8 | Sal13 | Omy4, Omy10 | Ssa04, Ssa23 | BC4 |
| Sna9 | Sal26 | Omy1 | Ssa16 | BC20 |
| Sna10 | Sal16 | Omy5 | Ssa10 | BC17 |
| Sna11 | Sal32 | Omy8 | Ssa14 | BC22 |
| Sna12 | Sal23 | Omy10 | Ssa04 | BC9 |
| Sna13 | Sal2 | Omy3 | Ssa25 | BC24 |
| Sna14 | Sal7 | Omy9 | Ssa15 | BC30 |
| Sna15 | Sal9 | Omy19 | Ssa01 | BC12 |
| Sna16 | Sal17 | Omy16, Omy20 | Ssa13, Ssa19 | BC18 |
| Sna17 | Sal8 | Omy25 | Ssa09 | BC33 |
| Sna18 | Sal33 | Omy11 | Ssa20 | BC40 |
| Sna19 | Sal36 | Omy22 | Ssa21 | BC26 |
| Sna20 | Sal11 | Omy7 | Ssa22 | BC21 |
| Sna21 | Sal4q.1:29 | Omy2 | Ssa10 | BC15 |
| Sna22 | Sal25 | Omy1 | Ssa18 | BC36 |
| Sna23 | Sal22 | Omy27 | Ssa20 | BC25 |
| Sna24 | Sal14 | Omy4 | Ssa06 | BC31 |
| Sna25 | Sal19 | Omy28 | Ssa03 | BC11 |
| Sna26 | Sal5 | Omy29 | Ssa11 | BC10 |
| Sna27 | Sal31 | Omy18 | Ssa27 | BC23 |
| Sna28 | Sal4q.2 | Omy25 | Ssa09 | BC35 |
| Sna29 | Sal28 | Omy8 | Ssa15 | BC19 |
| Sna30 | Sal10 | Omy26 | Ssa11 | BC28 |
| Sna31 | Sal4q.1:29 | Omy5 | Ssa01 | BC13 |
| Sna32 | Sal30 | Omy14 | Ssa14 | BC34 |
| Sna33 | Sal14 | Omy11 | Ssa19 | BC16 |
| Sna34 | Sal4p | Omy24 | Ssa09 | BC38 |
| Sna35 | Sal8 | Omy20 | Ssa28 | BC27 |
| Sna36 | Sal37 | Omy9 | Ssa18 | BC32 |
| Sna37 | Sal35 | Omy3 | Ssa02 | BC29 |
| Sna38 | Sal24 | Omy15 | Ssa17 | BC37 |
| Sna39 | Sal21 | Omy13 | Ssa02 | BC42 |
| Sna40 | Sal12 | Omy7 | Ssa17 | BC39 |
| Sna41 | Sal20 | Omy13 | Ssa06 | BC14 |
| Sna42 | Sal34a | Omy19 | Ssa08 | BC41 |
Sal34 appears to be homologous with Sna42, however fewer than 50 RAD contigs mapped to this chromosome.
The lake trout karyotype is differentiated from Arctic char by multiple Robertsonian translocations including one possible chromosomal fusion (Sal6.1 and Sal6.2) and four chromosomal fissions (Sal8, Sal14, Sal20, Sal4q.1.29). Sal6.1 and Sal6.2 are fused and Sal4q.1.29 is split into two LGs in lake trout, similar to the Arctic char linkage map presented by Nugent et al. (2017). The two Salvelinus species are also differentiated by at least 6 putative chromosomal inversions (Table 4), primarily on acrocentric or telocentric chromosomes. Arctic char chromosome Sal14 in particular appears to be the result of a fusion between Sna24 and Sna33. Sna24 also contains multiple chromosomal inversions that differentiate the two karyotypes (Figure 3). With the exception of inversions detected on Sna24, all putative inversions differentiating the two species were found to be near, or overlapping, the centromere (n = 5, Table 4). MapComp results suggest inversions on Sna10, Sna11, Sna24, and Sna34 are shared with brook trout; however, large inversions differentiating brook trout and lake trout were identified on Sna28 (brook trout BC35), Sna12 (brook trout BC9), and Sna23 (brook trout BC25).
Table 4. The first column is the lake trout linkage group in question and columns 2-4 list the approximate location of any detected inversions that differentiate species. The type of inversion is stated in parenthesis. Locations are listed in centimorgans on the female map. Whenever multiple inversions were detected on a chromosome, at least one was pericentric. Centromeres were not localized for Sna42, so centricity of inversions could not be determined.
| Linkage Group | Arctic Char | Rainbow Trout | Atlantic Salmon |
|---|---|---|---|
| Sna1 | — | — | — |
| Sna2 | — | — | — |
| Sna3 | — | — | — |
| Sna4 | — | — | — |
| Sna5 | — | — | — |
| Sna6 | — | — | 30-43 (Paracentric) |
| Sna7 | — | — | — |
| Sna8 | — | — | — |
| Sna9 | — | — | — |
| Sna10 | 45-55 (Pericentric) | ** | * |
| Sna11 | 0-30 (Pericentric) | 0-30 (Pericentric) | 0-30 (Pericentric) |
| Sna12 | 48-54 (Pericentric) | ** | 48-54 (Pericentric) |
| Sna13 | — | — | — |
| Sna14 | — | — | — |
| Sna15 | — | — | — |
| Sna16 | — | 12-40 (Multiple Inversions)** | 25-40 (Multiple Inversions) |
| Sna17 | — | — | |
| Sna18 | — | — | |
| Sna19 | — | 5-30 (Paracentric), 25 - 58 (Pericentric) | 30-58 (Pericentric) |
| Sna20 | — | 0-10 (Pericentric) | 0-10 (Pericentric) |
| Sna21 | — | — | — |
| Sna22 | — | — | — |
| Sna23 | — | — | — |
| Sna24 | 0-57 (Multiple inversions) | 0-12 (Pericentric) | 0-12 (Pericentric) |
| Sna25 | — | — | 0-30 (Pericentric) |
| Sna26 | — | — | — |
| Sna27 | — | — | — |
| Sna28 | — | — | 43-52 (Pericentric) |
| Sna29 | — | — | — |
| Sna30 | * | 0-10 (Pericentric) | — |
| Sna31 | — | — | 0-7 (Pericentric) |
| Sna32 | — | — | — |
| Sna33 | — | — | — |
| Sna34 | 0-30 (Pericentric) | * | * |
| Sna35 | — | 0-47 (Pericentric) | — |
| Sna36 | — | — | — |
| Sna37 | — | — | — |
| Sna38 | — | — | — |
| Sna39 | — | — | — |
| Sna40 | — | — | — |
| Sna41 | * | — | — |
| Sna42 | — | 35-43 (Unknown) | 20-43 (Unknown) |
= suggestive evidence of structure variation but unable to determine if an inversion occurred.
= Inverted region appears to be translocated to a separate chromosome.
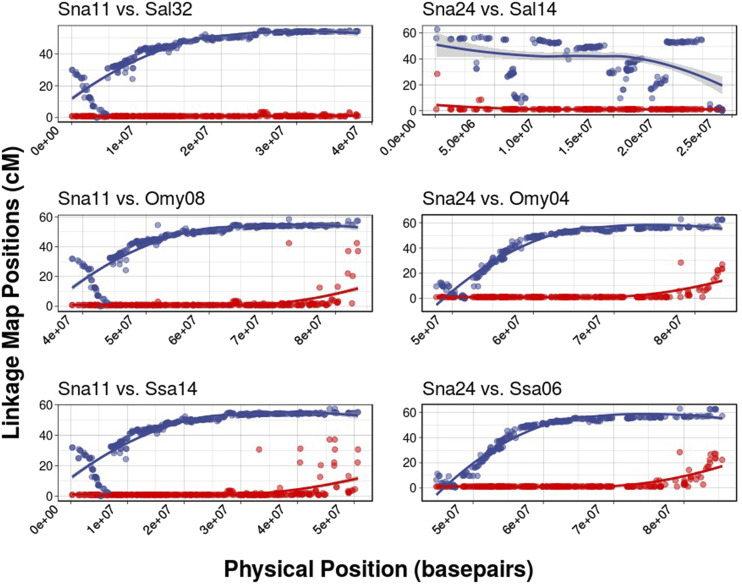
Figure 3.
Examples of two linkage groups (Sna11 and Sna24) with evidence of inversions differentiating lake trout from other salmonids. Female lake trout linkage groups are colored blue (top curves). Male lake trout linkage groups are colored red (bottom curves). Sna11(first column) is differentiated from all homologs by a single large pericentric inversion spanning 0-30cM on the female linkage map (left side of each panel). Sna24 is differentiated from Omy04 and Ssa06 by an inversion spanning 0-10cM on the female map. It is unclear if the same inversion exists in Arctic char due to extensive structural differentiation relative to lake trout and other salmonids (Sna24 vs. Sal14).
Trait mapping
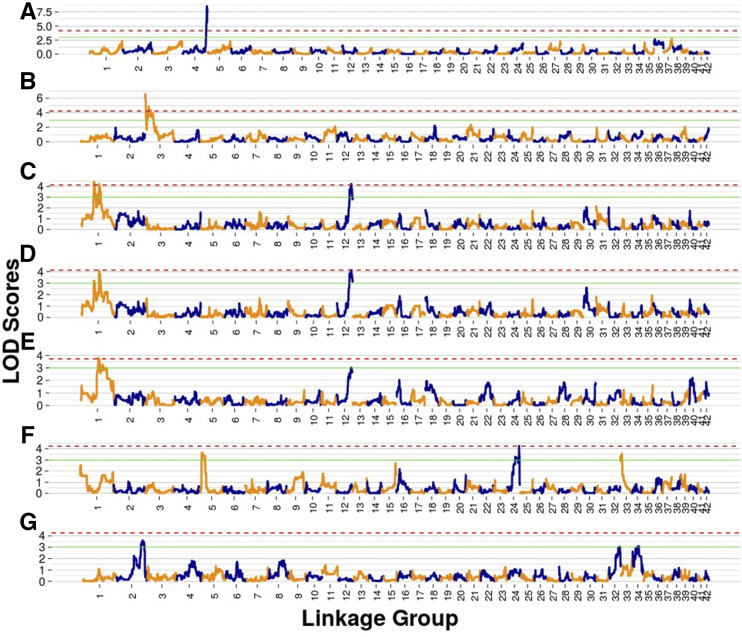
Multiple quantitative trait loci were detected for the evaluated traits (Table 5, Figure 4). A highly significant QTL for presence of spots and vermiculations mapped to a sex-averaged position of 3 cM on Sna3 (VPA1, 95% CI = 0-4.485 cM, LOD = 6.563, P = 0.001). We identified 16 candidate genes associated with this peak, including melanoregulin-like (MREG-L, Arctic char scaffold NW_019942894.1: 64734-79619; Sna3, 1.575 cM). A second QTL for this trait mapped 21.095 cM on the same linkage group (VPA2, 95% CI = 19.685 – 30.175, LOD = 4.850, P = 0.014). A total of 176 candidate genes were identified within this QTL credible interval. The four genes closest the highest LOD value were transcription factor 20-like (TCF20-L), retinoic acid induced 1-like (RAI1-L), sterol regulatory element binding factor 1-like (SREBF1-L), and calcium channel voltage-dependent T type alpha 1I subunit-like (CACNA1I-L; Table S3). These QTL explained 11.5 and 10.8% of phenotypic variance, respectively (Table 5).
Table 5. Linkage map positions (cM) of QTL peaks detected for the sex determination locus, presence-absence of vermiculations and spots, fork length, shape variable PCA1, shape variable PCA2, weight, and condition factor (Trait column). CI_Low and CI_High are the upper and lower bounds of the 95% credible interval for map positions for each QTL peak. LG is the linkage group on which the QTL was detected. Model lists the model used for QTL mapping in r/qtl2. Positions are sex averaged map positions. LOD scores are the differences in log10 likelihoods for models assuming presence or absence of a QTL at the locus in question (reported by r/qtl2). The estimates proportion of phenotypic variance explained by each QTL peak is listed in the PVE column. Estimated additive and dominant effects for the peak in question are also listed. P-values are those obtained via the permutation test described.
| Trait | LG | Position (cM) | CI_Low | CI_High | Model | LOD | Additive Effect | Dominance Effect | PVE | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Sna4 | 78.54 | 75.66 | 82.14 | Binary | 8.538 | 1.049 | −0.045 | 0.115 | <0.001*** |
| Sex | Sna4 | 84.43 | 82.14 | 86.12 | Binary | 8.04 | 1.055 | −0.044 | 0.108 | <0.001*** |
| Vermiculation | Sna3 | 3.00 | 0.00 | 4.49 | Binary | 6.563 | 1.595 | 0.278 | 0.191 | 0.001*** |
| Vermiculation | Sna3 | 21.10 | 19.69 | 30.18 | Binary | 4.855 | 0.103 | 1.048 | 0.145 | 0.014* |
| Fork Length | Sna1 | 39.00 | 36.94 | 44.60 | Normal | 4.401 | 18.905 | 8.058 | 0.107 | 0.030* |
| Fork Length | Sna1 | 60.27 | 51.48 | 66.07 | Normal | 4.224 | 15.910 | 9.172 | 0.103 | 0.043* |
| Fork Length | Sna12 | 57.63 | 51.84 | 62.03 | Normal | 4.226 | −11.693 | 10.910 | 0.103 | 0.043* |
| PCA1 | Sna5 | 11.83 | 10.80 | 16.15 | Normal | 3.651 | −0.005 | 0.011 | 0.111 | 0.156 |
| PCA1 | Sna24 | 35.99 | 27.30 | 44.50 | Normal | 4.259 | −0.003 | −0.011 | 0.128 | 0.049* |
| PCA1 | Sna33 | 4.55 | 0.00 | 6.39 | Normal | 3.554 | 0.008 | 0.007 | 0.108 | 0.184 |
| PCA2 | Sna2 | 64.47 | 45.94 | 80.32 | Normal | 3.594 | 0.006 | 0.000 | 0.109 | 0.188 |
| PCA2 | Sna32 | 45.75 | 27.93 | 50.59 | Normal | 3.041 | 0.004 | −0.001 | 0.093 | 0.451 |
| PCA2 | Sna34 | 22.82 | 0.00 | 39.41 | Normal | 3.087 | 0.005 | 0.000 | 0.095 | 0.423 |
| Weight | Sna1 | 60.27 | 37.37 | 72.40 | Normal | 4.052 | 48.657 | 29.021 | 0.099 | 0.062 |
| Weight | Sna12 | 57.67 | 50.55 | 64.15 | Normal | 4.13 | −40.950 | 29.692 | 0.101 | 0.049* |
| Condition | Sna1 | 60.27 | 52.60 | 73.11 | Normal | 3.796 | 0.050 | 0.033 | 0.093 | 0.045* |
| Condition | Sna12 | 60.10 | 47.72 | 64.15 | Normal | 3.009 | −0.053 | 0.001 | 0.074 | 0.278 |
Figure 4.
Panels display LOD values on the Y-axis vs. sex averaged map position (cM) for QTL scans for (A) the sex determination locus, (B) presence of spots and vermiculations, (C) fork length, (D) weight, (E) condition factor, (F) PCA1, and (G) PCA2. The dashed red line corresponds to the P < 0.05 significance threshold for LOD scores. The solid green line corresponds to the LOD threshold of 3 used to identify peaks putatively associated with each trait.
Significant QTL for fork length mapped to two locations on Sna1 (FL1, 39.00 cM, 95% CI = 36.94 – 44.6cM, LOD = 4.401, P = 0.03, and FL2, 60.265 cM, 95% CI = 51.475 – 66.07 cM, LOD = 4.224, P = 0.043) and one location on Sna12 (FL3, 57.630 cM, 95% CI = 51.835-63.03 cM, LOD = 4.226, P = 0.043). A significant QTL for condition also mapped to 60.265 cM on Sna1 (CF1, 95% CI = 52.6 – 73.105, LOD = 3.796, P = 0.045) and a QTL for weight mapped to 57.665 cM on Sna12 (W1, 95% CI = 50.55 - 64.15 cM, LOD = 4.13, P = 0.045). Suggestive QTL (LOD > 3, P > 0.05) were detected on Sna1 (60.265 cM, 95% CI = 37.365 – 72.4, LOD = 4.052, P = 0.062) and Sna12 (60.095 cM, 95% CI = 47.72 – 64.15 cM, LOD = 0.009, P = 0.278) for weight and condition factor, respectively. We identified 39 candidate genes associated with peak FL1, 137 genes associated with FL2, and 77 genes associated with FL3. We did not search for candidate genes for other growth and body condition related QTL (weight and condition factor) because the locations and credible intervals for these QTL overlapped almost perfectly with those detected for fork length (Table 5).
Suggestive QTL were detected for the PCA1 body shape variable on Sna5 (11.830 cM, 95% CI = 10.8 – 16.145, LOD = 3.651, P = 0.156), Sna24 (PCA1_1, 35.990 cM, 95% CI = 27.3 – 44.5 cM, LOD = 4.259, P = 0.049), and Sna33 (4.550 cM, 95% CI = 0 – 6.39 cM, LOD = 3.554, P = 0.184); however only the peak on Sna24 was statistically significant. Suggestive QTL were detected for PCA2 on Sna2 (64.464 cM, 95% CI = 45.94 – 80.32, LOD = 3.594, P = 0.188), Sna32 (45.745 cM, 95% CI = 27.925 – 50.59 cM, LOD = 3.041, P = 0.451), and Sna34 (22.820 cM, 95% CI = 0 – 39.405 cM, LOD = 3.087, P = 0.423); however, none of these QTL were found to be statistically significant. 111 candidate genes were identified for the significant QTL interval for PCA1 on Sna24.
Our sdY presence-absence assay produced a male specific peak in melt curve derivative plots at approximately 84°. A melt curve derivative peak existed for 18S at approximately 85°. Of the 16 known males used to validate the assay, 15 produced sdY peaks and 16 produced 18S peaks. The 16 known females tested all yielded 18S peaks; however, sdY peaks were absent in all females as expected. We were ultimately able to determine genotypic sex for 323 offspring produced from diploid crosses. Mapping sdY presence-absence as a binary trait using qtl2 identified strong peaks of association at 78.54 cM (95% CI = 75.66 – 82.14 cM, LOD = 8.538, P < 0.001) and 84.43 cM (95% CI = 82.14 – 86.12 cM, LOD = 8.04, P < 0.001) on Sna4.
Recombination rates
Sex averaged recombination rates estimated by alignment to the Arctic char genome ranged from 0.138 to 2.935 cM/MB with a mean of 0.817 cM/MB (Table S4; SD = 0.537) across LGs. In general, recombination rate estimates generated by mapping to the Arctic char genome were lower than those obtained from mapping to rainbow trout or Atlantic salmon genomes (Table S4, Figure S9). Consistent with previous studies on salmonids, male recombination rates were considerably lower than those observed for females (Figure S9). For example, the mean male recombination rate base on alignment to the Rainbow Trout genome was 0.203 cM/MB (SD = 0.133), while the mean female recombination rate was 1.31 cM/MB (SD = 0.602). Alignment of male and female linkage maps to divergent reference genomes demonstrated that male recombination is highly suppressed except for near telomeres (Figures S1-S3).
Discussion
Map evaluation
Multiple lines of evidence suggest this linkage map provides an accurate representation of the lake trout genome. First, we identified a single centromere for each chromosome (except Sna42), suggesting that linkage groups were appropriately split. In all cases, centromere mapping locations derived from half-tetrad and RFm analysis either overlapped or were in close proximity (Figure 2, Table S2). For acrocentric and telocentric chromosomes, centromeres always mapped to the end of the chromosome with the highest female recombination rate and lowest male recombination rate, which matches results from previous centromere mapping efforts in salmonids suggesting that male recombination occurs almost exclusively near telomeres (Moen et al. 2008; Miller et al. 2012; McKinney et al. 2016). Second, our homology analysis demonstrated a high degree of contiguity between existing genome assemblies and the map presented here (Figures S1-S7). Finally, our sex determination locus mapping results are concordant with cytogenetic studies. Previous cytogenetic analysis of male and female lake trout identified sex-specific quinacrine and C-banding patterns on a large submetacentric chromosome (Phillips and Ihssen 1985). Mapping sdY presence-absence using the linkage map demonstrated with high certainty that the sex locus exists near one of the telomeres of Sna4 (Figure 4), which is metacentric or submetacentric based on RFm and half tetrad analysis (Figure 2). Although two significant peaks were detected on this LG, they were in close proximity and credible intervals were adjacent, suggesting that they likely represent a single peak of association. This study and others suggest that Salvelinus species, and salmonids in general, have highly variable sex chromosome configurations. Specifically, the brook trout sex determination locus maps to a region that is homologous to the Arctic char sex chromosome; however, it localizes to a different arm (Sutherland et al. 2017), while the lake trout sex chromosome identified here lacks homology with those of all species examined (Sal4p.1 – Nugent et al. 2017; BC35 – Sutherland et al. 2017; Ssa02, Ssa03, and Ssa06 – Kijas et al. 2018; Omy29 – Pearse et al. 2019). Many previous studies have identified variation in sex locus mapping position both within and between salmonid species (Woram et al. 2003; Lubieniecki et al. 2015; Sutherland et al. 2017; Kijas et al. 2018), even though the same gene ultimately underlies sex determination in most cases (Yano et al. 2013). Our results add to a growing body of literature suggesting that sdY is a conserved, yet highly mobile, sex determination gene in salmonids.
Furthermore, the lake trout linkage map presented here is of similar density to those used to scaffold genome assemblies for other salmonids (Christensen et al. 2018a, 2018b) and provides valuable information on the order of loci along chromosomes and recombination rates between loci. In general, male:female map length ratios and estimated sex averaged map lengths were highly similar to those observed for other salmonids. For instance, Leitwein et al. (2018) found that chromosome specific recombination rates varied from 0.21 – 4.1 cm/MB (mean =0.88) for brown trout, compared with 0.138 to 2.935 cM/MB (mean = 0.817) for lake trout based on mapping linkage mapped RAD loci to the Arctic char reference genome. Similar to other salmonids, we observed pronounced heterochiasmy, with male recombination being almost entirely suppressed except for near telomeres (Moen et al. 2004; Moen et al. 2008; McKinney et al. 2016; Leitwein et al. 2018)
Structural variation
Combining centromere mapping locations with synteny analysis revealed that lake trout are differentiated from Arctic char, rainbow trout, and Atlantic salmon by multiple pericentric inversions, suggesting that centromeric instability (specifically on acrocentric and telocentic chromosomes) is potentially an important component of the salmonid evolutionary legacy (Table 4). Future work should evaluate whether these detected inversions are truly species diagnostic or if they are polymorphic within species. Large structural variants have previously been found to be associated with adaptive differentiation and life history variation within rainbow trout (Miller et al. 2012; Pearse et al. 2019) and inversions can contribute to pre or post-zygotic isolation between species or ecotypes (Kirkpatrick 2010). Future studies should evaluate the extent to which the structural variation detected here contribute to reproductive isolation and adaptive divergence within and between salmonid species. Sna24 presents one of the most striking examples of extensive structural variation in the genus Salvelinus, with multiple paracentric and pericentric inversions differentiating the lineages containing Arctic char and lake trout (Figure 3). With the exception of a putative inversion located between 0 and 12 cM, all other inversions on this LG were not observed in other salmonid species examined, suggesting that the other inversions on this chromosome (Sna24, Ssa14; Figure 3, column 2) are fixed or segregating within the Arctic char lineage or within the Salvelinus clade containing Arctic char, bull trout (S. confluentus), dolly varden trout (S. malma), and white char (S. albus). This hypothesis is supported by results from MapComp which suggested that brook trout, the most closely related extant species to lake trout (Crête-Lafrenière et al. 2012), and lake trout are not differentiated by any inversions on this linkage group. A large inversion spanning the centromere of Sna11 shows clear evidence of being differentially fixed between lake trout and all other salmonids except brook trout (Figure 3, Figures S1-S3, Figure S7). Inversions located on Sna10, Sna24, and Sna34 also appear to be differentially fixed between the lake trout – brook trout lineage and all other salmonids; however, interpretation is complicated by subsequent translocations and inversions that occurred in other taxa (Table 4). A large pericentric inversion on Sna12 appears to differentiate lake trout from all other salmonids, including brook trout (Figure S7). MapComp results also suggest that two large inversions on Sna28 (homologous to the brook trout sex chromosome - BC35; Sutherland et al. 2017) and Sna23 (BC25) differentiate lake trout from closely related brook trout (Figure S7). It is unclear if these structural variants are truly fixed between species, or if they might be polymorphic within lake trout or brook trout. The inversion polymorphisms identified above could be associated with chromosomal speciation within the genus Salvelinus or adaptive divergence within salmonid species and warrant further examination.
The majority of detected inversions differentiating lake trout from other salmonids are pericentric, which is not entirely unexpected. Repeat-rich eukaryotic centromeres often demonstrate exceptionally high rates of evolution (Henikoff et al. 2001) and are prone to chromosomal breakage and the accumulation of structural variation (Kalitsis and Choo 2012; Barra and Fachinetti 2018). Sutherland et al. (2016) also identified evidence for multiple inversions differentiating salmonid species, including one pericentric inversion differentiating pink, chum, and sockeye salmon from other salmonids.
Evidence for F2 inviability and reduced reproductive success between hybrids are widespread (Stebbins 1958), including for pairs of closely related species within the salmonid lineage (Renaut and Bernatchez 2011). Bull trout and brook trout, for instance, readily produce F1 offspring but F2 offspring are rarely observed (Leary et al. 1993). Hybrids between westslope cutthroat (Oncorhynchus clarkii lewisi) and rainbow trout are viable but have dramatic reductions in reproductive success (Muhlfeld et al. 2009). Future work should evaluate if instances of reduced fitness in inter or intraspecific salmonid hybrids might be linked to combined deleterious effects of recombination at multiple pericentrically inverted loci. It would also be interesting to ascertain whether centromeric regions tend to harbor signals of adaptive divergence between salmonid species and morphotypes. For example, Ellegren et al. (2012) found elevated levels of divergence between Fidecula flycatcher species near centromeres. Given the prevalence of pericentric inversions on acrocentric and telocentric chromosomes, we also might expect these loci to be associated with adaptive ecophenotypic radiations that have occurred within Salvelinus (Eshenroder 2008) and Coregonus (Lu and Bernatchez 1999).
Genomic basis for adaptive traits
Suggestive QTL for traits that differentiate lake trout morphotypes were detected on multiple linkage groups. This supports the hypothesis proposed by Perreault-Payette et al. (2017) that ecophenotypic divergence in lake trout has a polygenic basis. Our results suggest that the presence or absence of spots and vermiculations is controlled by either one or two loci on the same arm of linkage group Sna3. A search for candidate genes within the QTL mapping intervals identified melanoregulin-like (MREG-L) as a potential causal locus. The homolog of this gene is involved in the transfer of melanosomes from melanocytes to keratinocytes (Wu et al. 2012), and appears to control the distribution of pigments within mice hair (O’Sullivan et al. 2004). Pigmentation polymorphisms are common in lake trout (Wilson and Mandrak 2004; Zimmerman et al. 2007) and other trout and char (Gomez-Uchida et al. 2008), although it is unclear if the genes identified here explain skin pigmentation variation in other species and populations. Skin pigmentation variation has been shown to be associated with depth of capture in multiple lake trout populations and is hypothesized to be adaptive in some environments (Protas and Patel 2008); however, it is also possible that the trait is simply linked with some other adaptive traits. Pigmentation patterns are often linked to variation in behavior, immune response, and energy homeostasis in vertebrates, likely owing to pleiotropic effects of melanocortins (Ducrest et al. 2008). Pigmentation traits have also been linked to stress response in rainbow trout, Atlantic salmon, and Arctic char (Hoglund et al. 2000; Kittilsen et al. 2009).
Suggestive QTL for the composite body shape variable with the highest explanatory power (PCA1) were detected on Sna5, Sna24, and Sna33. Interestingly, each of these chromosomes appear to have undergone structural reorganization in relatively recent evolutionary history, based on alignment to the Arctic char genome (Figure S1). Specifically, Sna24 and Sna33 are fused in Arctic char and Sna5 is split into two chromosomes. Sna24 in particular appears to have accumulated multiple large inversions that differentiate this linkage group from the homologous region of the syntenic Arctic char chromosome. A QTL for condition factor, which is closely related to body shape (Froese 2006), has been previously detected on the brook trout linkage group homologous to Sna33 (linkage group BC16, Sutherland et al. 2017). Additional mapping crosses, ideally generated using ancestral populations with highly differentiated body shapes (leans vs. siscowet or divergent hatchery strains for example) would be valuable for further validating the existence of QTL detected here and improving our understanding of the genetic basis for adaptive divergence within lake trout.
Growth and body condition related traits all have suggestive QTL on linkage groups Sna1 and Sna12, indicating that genes on these chromosomes likely harbor variation that underlies differences in growth between populations. Linkage group Sna12 also appears to harbor an inversion that differentiates lake trout from other salmonid species examined, including brook trout. A previous study identified a putative growth rate QTL on the brook trout linkage group homologous to Sna12 (Figure S7; Sutherland et al. 2017; BC9). The same study identified a stress response QTL, measured as change in blood cortisol levels following handling stress, in brook trout on the chromosome homologous to Sna1 (Sutherland et al. 2017, BC6). Increased cortisol levels have been found to be negatively corelated with growth and condition factor in other salmonids (Barton et al. 1987; Reinecke 2010), suggesting that variation observed in our families could actually be due to variation in stress response. There is evidence for variation in fitness among lake trout hatchery strains used to supplement and restore lake trout populations in the Great Lakes, with the strain from Seneca Lake, New York appearing to have a fitness advantage (Scribner et al. 2018). Great Lakes lake trout populations are heavily impacted by predation by invasive sea lamprey and previous work has shown that larger individuals have a greater probability of surviving lamprey attack (Swink 1990). Similarly, size-selective fisheries have also been shown to impose strong natural selection on growth in multiple species (Enberg et al. 2012). Future work could examine whether the chromosomal regions identified here are associated with size-at-age or are under selection in populations experiencing lamprey predation or size-selective fisheries.
Associations between environmental conditions and phenotype have been observed across the lake trout range and across salmonid species for the afore mentioned traits, suggesting adaptive significance in some contexts. For example, patterns and intensity of skin pigmentation, along with divergence in other traits, is commonly associated with depth-of-capture in both lake trout (Zimmerman et al. 2007; Marin et al. 2016) and Arctic char populations (Gomez-Uchida et al. 2008). Variation in skin pigmentation is potentially involved with predator avoidance and camouflage, feeding behavior, mate choice (Protas and Patel 2008), or protection from ultraviolet radiation (Yan et al. 2013). Differences in age specific growth rates are also frequently observed between humper, lean, and siscowet-like lake trout morphotypes (Burnham-Curtis and Bronte 1996; Hansen et al. 2012) — as well as between Arctic char morphotypes (Jonsson et al. 1988; Snorrason et al. 1994; Adams et al. 1998). These differences in growth rate likely reflect variation in allocation of resources toward growth and reproduction, adaptation to nutrient stress (Arendt 1997), or plastic responses to environmental variation (Hindar and Jonsson 1993).
Morphotypes can also often be differentiated based on body shape differences, which are hypothesized to be optimized for different feeding behaviors and modes of locomotion (Bond 1979; Muir et al. 2014; Perreault-Payette et al. 2017). For example, the streamlined body shape of leans has been hypothesized to be adaptive for swimming and predation in shallower nearshore environments (Bond 1979; Muir et al. 2014), while the more deep-bodied shape of siscowet lake trout is believed to reflect adaptation for vertical migration and foraging in deep-water habitats (Webb 1984; Muir et al. 2014). Morphotypes with traits reflecting those observed in the species native range have the potential to emerge rapidly in some introduced invasive populations (Stafford et al. 2013), suggesting a high degree of phenotypic plasticity or exceptionally strong selection favoring divergence.
Unfortunately, many lake trout metapopulations of conservation concern have experienced reductions in abundance and decreases in ecophenotypic diversity as a result of overexploitation and introduction of invasive species (Krueger and Ihssen 1995; Hansen 1999). For example, in the Great Lakes the siscowet morphotype has been extirpated from all lakes except Lake Superior (Krueger and Ihssen 1995). The results presented here enhance understanding of the genetic architecture of traits that underlie trophic specialization in lake trout and could aid in restoring genetic and phenotypic diversity in lakes where it has been lost.
Conclusions
We identified multiple structural variants potentially involved in speciation and adaptation within the genus Salvelinus, mapped the lake trout sex determination locus, and identified QTL for traits believed to be adaptively significant in lake trout populations. Future work should use additional QTL mapping crosses and association studies in wild populations to evaluate if the QTL identified here are consistently associated with the phenotypic variation examined, as well as other phenotypes that differentiate lake trout morphotypes. Trophically specialized lake trout morphotypes and adaptively diverged populations are differentiated by multiple other traits (i.e., tissue lipid content, fin size, diet; Thurston 1962; Eschmeyer and Phillips 1965; Zimmerman et al. 2006; Zimmerman et al. 2007). QTL mapping studies using later generation crosses or genome-wide association studies in wild populations would be particularly useful for fine-scale localization of genotype-phenotype associations within QTL credible intervals identified here. Additionally, QTL mapping efforts can yield different results for different families and the genetic basis for some traits often varies across populations (Santure et al. 2015). The lake trout linkage map will allow further examination of the genetic basis for ecophenotypic variation in lake trout and will enable additional exploration of chromosomal evolution within the genus Salvelinus. Perhaps most important, this resource will allow for the assembly of a chromosome-anchored reference genome for lake trout, which will greatly facilitate future genomic research on this important species.
Acknowledgments
We would like to thank the staff of Pendills Creek National Fish Hatchery and the Codrington Fisheries Research Facility for their assistance with producing mapping crosses. We would also like to thank Simon Bernatchez for his assistance with collecting photographs and two anonymous reviewers for their thoughtful comments and suggestions. SS, SA, KS, and GL were supported by award 2017_SCR_44067 from the Great Lakes Fisheries Commission. GL was also supported in part by the National Science Foundation grant NSF-DoB-1639014.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11908326.
Communicating editor: D. Macqueen
Literature Cited
- Adams C. E., Fraser D., Huntingford F. A., Greer R. B., Askew C. M. et al. , 1998. Trophic polymorphism amongst Arctic charr from Loch Rannoch, Scotland. J. Fish Biol. 52: 1259–1271. 10.1111/j.1095-8649.1998.tb00970.x [DOI] [Google Scholar]
- Adams D. C., and Otárola‐Castillo E., 2013. geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4: 393–399. 10.1111/2041-210X.12035 [DOI] [Google Scholar]
- Alfonso N. R., 2004. Evidence for two morphotypes of lake charr, Salvelinus namaycush, from Great Bear Lake, Northwest Territories, Canada. Environ. Biol. Fishes 71: 21–32. 10.1023/B:EBFI.0000043176.61258.3d [DOI] [Google Scholar]
- Ali O. A., O’Rourke S. M., Amish S. J., Meek M. H., Luikart G. et al. , 2016. RAD capture (Rapture): flexible and efficient sequence-based genotyping. Genetics 202: 389–400. 10.1534/genetics.115.183665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S., 2014 FastQC: a quality control tool for high throughput sequence data. Version 0.11. 5. Babraham Institute, Cambridge, UK http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Anglès d’Auriac M. B., Urke H. A., and Kristensen T., 2014. A rapid qPCR method for genetic sex identification of Salmo salar and Salmo trutta including simultaneous elucidation of interspecies hybrid paternity by high‐resolution melt analysis. J. Fish Biol. 84: 1971–1977. 10.1111/jfb.12401 [DOI] [PubMed] [Google Scholar]
- Arendt J. D., 1997. Adaptive intrinsic growth rates: an integration across taxa. Q. Rev. Biol. 72: 149–177. 10.1086/419764 [DOI] [Google Scholar]
- Barra V., and Fachinetti D., 2018. The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat. Commun. 9: 4340 10.1038/s41467-018-06545-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton B. A., Schreck C. B., and Barton L. D., 1987. Effects of chronic cortisol administration and daily acute stress on growth, physiological conditions, and stress responses in juvenile rainbow trout. Dis. Aquat. Organ. 2: 173–185. 10.3354/dao002173 [DOI] [Google Scholar]
- Bay R. A., Rose N., Barrett R., Bernatchez L., Ghalambor C. K. et al. , 2017. Predicting responses to contemporary environmental change using evolutionary response architectures. Am. Nat. 189: 463–473. 10.1086/691233 [DOI] [PubMed] [Google Scholar]
- Berg P. R., Star B., Pampoulie C., Bradbury I. R., Bentzen P. et al. , 2017. Trans-oceanic genomic divergence of Atlantic cod ecotypes is associated with large inversions. Heredity 119: 418–428. 10.1038/hdy.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez S., Laporte M., Perrier C., Sirois P., and Bernatchez L., 2016. Investigating genomic and phenotypic parallelism between piscivorous and planktivorous lake trout (Salvelinus namaycush) ecotypes by means of RAD seq and morphometrics analyses. Mol. Ecol. 25: 4773–4792. 10.1111/mec.13795 [DOI] [PubMed] [Google Scholar]
- Blackie C. T., Weese D. J., and Noakes D. L. J., 2003. Evidence for resource polymorphism in the lake charr (Salvelinus namaycush) population of Great Bear Lake, Northwest Territories, Canada. Ecoscience 10: 509–514. 10.1080/11956860.2003.11682799 [DOI] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C. E., 1979. Biology of Fishes, WB Saunders Company, Philadelphia, PA. [Google Scholar]
- Bradbury I. R., Hubert S., Higgins B., Bowman S., Borza T. et al. , 2013. Genomic islands of divergence and their consequences for the resolution of spatial structure in an exploited marine fish. Evol. Appl. 6: 450–461. 10.1111/eva.12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenden T. O., Brown R. W., Ebener M. P., Reid K., and Newcomb T. J., 2013. Great Lakes commercial fisheries: historical overview and prognoses for the future, pp. 339–397 in Great Lakes fisheries policy and management, Ed. 2nd, edited by Taylor W. W., Lynch A. J., and Leonard N. J.. Michigan State University Press, East Lansing. [Google Scholar]
- Brenna-Hansen S., Li J., Kent M. P., Boulding E. G., Dominik S. et al. , 2012. Chromosomal differences between European and North American Atlantic salmon discovered by linkage mapping and supported by fluorescence in situ hybridization analysis. BMC Genomics 13: 432 10.1186/1471-2164-13-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieuc M., Waters C. D., Seeb J. E., and Naish K. A., 2014. A dense linkage map for Chinook salmon (Oncorhynchus tshawytscha) reveals variable chromosomal divergence after an ancestral whole genome duplication event. G3 (Bethesda) 4: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., and Sen S., 2009. A Guide to QTL Mapping with R/qtl, Vol. 46 Springer, New York: 10.1007/978-0-387-92125-9 [DOI] [Google Scholar]
- Broman K. W., Gatti D. M., Simecek P., Furlotte N. A., Prins P. et al. , 2019. R/qtl2: Software for Mapping Quantitative Trait Loci with High-Dimensional Data and Multiparent Populations. Genetics 211: 495–502. 10.1534/genetics.118.301595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte C. R., and Moore S. A., 2007. Morphological variation of siscowet lake trout in Lake Superior. Trans. Am. Fish. Soc. 136: 509–517. 10.1577/T06-098.1 [DOI] [Google Scholar]
- Burnham-Curtis M. K., 1994. Intralacustrine speciation of Salvelinus namaycush in Lake Superior: an investigation of genetic and morphological variation and evolution of lake trout in the Laurentian Great Lakes. PhD diss. University of Michigan.
- Burnham-Curtis M. K., and Bronte C. R., 1996. Otoliths reveal a diverse age structure for humper lake trout in Lake Superior. Trans. Am. Fish. Soc. 125: 844–851. [DOI] [Google Scholar]
- Catchen J., Hohenlohe P. A., Bassham S., Amores A., and Cresko W. A., 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22: 3124–3140. 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarie L., Howland K., Harris L., and Tonn W., 2015. Polymorphism in lake trout in Great Bear Lake: intra-lake morphological diversification at two spatial scales. Biol. J. Linn. Soc. Lond. 114: 109–125. 10.1111/bij.12398 [DOI] [Google Scholar]
- Christensen K. A., Rondeau E. B., Minkley D., Leong J. S., Nugent C. M. et al. , 2018a The Arctic charr (Salvelinus alpinus) genome and transcriptome assembly. PLoS One 13: e0204076 10.1371/journal.pone.0204076 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Christensen K. A., Leong J. S., Sakhrani D., Biagi C. A., Minkley D. et al. , 2018b Chinook salmon (Oncorhynchus tshawytscha) genome and transcriptome. PLoS One 13: e0195461 10.1371/journal.pone.0195461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crête-Lafrenière A., Weir L. K., and Bernatchez L., 2012. Framing the Salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS One 7: e46662 10.1371/journal.pone.0046662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E. et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. W., Cezard T., Fuentes‐Utrilla P., Eland C., Gharbi K. et al. , 2013. Special features of RAD Sequencing data: implications for genotyping. Mol. Ecol. 22: 3151–3164. 10.1111/mec.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Kayne R., and Feulner P., 2018. A European whitefish linkage map and its implications for understanding genome-wide synteny between salmonids following whole genome duplication. G3 (Bethesda) 8: 3745–3755. 10.1534/g3.118.200552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrest A., Keller L., and Roulin A., 2008. Pleiotropy in the melanocortin system, coloration and behavioural syndromes. Trends Ecol. Evol. 23: 502–510. 10.1016/j.tree.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Everett M. V., Miller M. R., and Seeb J. E., 2012. Meiotic maps of sockeye salmon derived from massively parallel DNA sequencing. BMC Genomics 13: 521 10.1186/1471-2164-13-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Smeds L., Burri R., Olason P. I., Backström N. et al. , 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491: 756–760. 10.1038/nature11584 [DOI] [PubMed] [Google Scholar]
- Enberg K., Jørgensen C., Dunlop E. S., Varpe O., Boukal D. S. et al. , 2012. Fishing‐induced evolution of growth: concepts, mechanisms and the empirical evidence. Mar. Ecol. (Berl.) 33: 1–25. 10.1111/j.1439-0485.2011.00460.x [DOI] [Google Scholar]
- Eshenroder R. L., 2008. Differentiation of deep-water lake charr Salvelinus namaycush in North American lakes. Environ. Biol. Fishes 83: 77–90. 10.1007/s10641-007-9265-y [DOI] [Google Scholar]
- Eschmeyer P. H., and Phillips A. M. Jr, 1965. Fat content of the flesh of siscowets and lake trout from Lake Superior. Trans. Am. Fish. Soc. 94: 62–74. 10.1577/1548-8659(1965)94[62:FCOTFO]2.0.CO;2 [DOI] [Google Scholar]
- Froese R., 2006. Cube law, condition factor and weight–length relationships: history, meta‐analysis and recommendations. J. Appl. Ichthyology 22: 241–253. 10.1111/j.1439-0426.2006.00805.x [DOI] [Google Scholar]
- Fujiwara A., Abe S., Yamaha E., Yamazaki F., and Yoshida M. C., 1997. Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma 106: 44–52. 10.1007/s004120050223 [DOI] [PubMed] [Google Scholar]
- Gagnaire P. A., Normandeau E., Pavey S. A., and Bernatchez L., 2013a. Mapping phenotypic, expression and transmission ratio distortion QTL using RAD markers in the Lake Whitefish (Coregonus clupeaformis). Mol. Ecol. 22: 3036–3048. 10.1111/mec.12127 [DOI] [PubMed] [Google Scholar]
- Gagnaire P. A., Pavey S. A., Normandeau E., and Bernatchez L., 2013b. The genetic architecture of reproductive isolation during speciation‐with‐gene‐flow in lake whitefish species pairs assessed by RAD sequencing. Evolution 67: 2483–2497. 10.1111/evo.12075 [DOI] [PubMed] [Google Scholar]
- Garrison E., and Marth G., 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907.
- Goetz F., Rosauer D., Sitar S., Goetz G., Simchick C. et al. , 2010. A genetic basis for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush). Mol. Ecol. 19: 176–196. 10.1111/j.1365-294X.2009.04481.x [DOI] [PubMed] [Google Scholar]
- Goetz F., Jasonowicz A., Johnson R., Biga P., Fischer G. et al. , 2013. Physiological differences between lean and siscowet lake trout morphotypes: Are these metabolotypes? Can. J. Fish. Aquat. Sci. 71: 427–435. 10.1139/cjfas-2013-0463 [DOI] [Google Scholar]
- Gogarten, S. M., X. Zheng, and A. Stilp, 2017 SeqVarTools: tools for variant data. R package v1 10. https://github.com/smgogarten/SeqVarTools
- Gomez‐Uchida D., Dunphy K. P., O’Connell M. F., and Ruzzante D. E., 2008. Genetic divergence between sympatric Arctic charr Salvelinus alpinus morphs in Gander Lake, Newfoundland: roles of migration, mutation and unequal effective population sizes. J. Fish Biol. 73: 2040–2057. 10.1111/j.1095-8649.2008.02048.x [DOI] [Google Scholar]
- Gonen S., Lowe N. R., Cezard T., Gharbi K., Bishop S. C. et al. , 2014. Linkage maps of the Atlantic salmon (Salmo salar) genome derived from RAD sequencing. BMC Genomics 15: 166 10.1186/1471-2164-15-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pena D., Gao G., Baranski M., Moen T., Cleveland B. M. et al. , 2016. Genome-wide association study for identifying loci that affect fillet yield, carcass, and body weight traits in rainbow trout (Oncorhynchus mykiss). Front. Genet. 7: 203 10.3389/fgene.2016.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale M. C., McKinney G. J., Bell C. L., and Nichols K. M., 2017. Using linkage maps as a tool to determine patterns of chromosome synteny in the genus Salvelinus. G3 (Bethesda) 7: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. J., 1999. Lake trout in the Great Lakes: basin-wide stock collapse and binational restoration. 417–453 in Great Lakes Fishery Policy and Management: A Binational Perspective, edited by Taylor W. W., Ferreri C. P.. Michigan State University Press, Michigam. [Google Scholar]
- Hansen M. J., Nate N. A., Krueger C. C., Zimmerman M. S., Kruckman H. G. et al. , 2012. Age, growth, survival, and maturity of Lake Trout morphotypes in Lake Mistassini, Quebec. Trans. Am. Fish. Soc. 141: 1492–1503. 10.1080/00028487.2012.711263 [DOI] [Google Scholar]
- Hansen M. J., Nate N. A., Muir A. M., Bronte C. R., Zimmerman M. S. et al. , 2016. Life history variation among four lake trout morphs at Isle Royale, Lake Superior. J. Great Lakes Res. 42: 421–432. 10.1016/j.jglr.2015.12.011 [DOI] [Google Scholar]
- Harvey C. J., Schram S. T., and Kitchell J. F., 2003. Trophic relationships among lean and siscowet lake trout in Lake Superior. Trans. Am. Fish. Soc. 132: 219–228. [DOI] [Google Scholar]
- Henikoff S., Ahmad K., and Malik H. S., 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- Hindar K., and Jonsson B., 1993. Ecological polymorphism in Arctic charr. Biol. J. Linn. Soc. Lond. 48: 63–74. 10.1111/j.1095-8312.1993.tb00877.x [DOI] [Google Scholar]
- Hoglund E., Balm P. H., and Winberg S., 2000. Skin darkening, a potential social signal in subordinate Arctic charr (Salvelinus alpinus): the regulatory role of brain monoamines and pro-opiomelanocortin-derived peptides. J. Exp. Biol. 203: 1711–1721. [DOI] [PubMed] [Google Scholar]
- Hollenbeck C. M., Portnoy D. S., and Gold J. R., 2016. A method for detecting recent changes in contemporary effective population size from linkage disequilibrium at linked and unlinked loci. Heredity 117: 207–216. 10.1038/hdy.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston R. D., Haley C. S., Hamilton A., Guy D. R., Tinch A. E. et al. , 2008. Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics 178: 1109–1115. 10.1534/genetics.107.082974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Wright J. E., and May B., 1987. Linkage relationships reflecting ancestral tetraploidy in salmonid fish. Genetics 116: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson B., Skúlason S., Snorrason S. S., Sandlund O. T., Malmquist H. J. et al. , 1988. Life history variation of polymorphic Arctic charr (Salvelinus alpinus) in Thingvallavatn, Iceland. Can. J. Fish. Aquat. Sci. 45: 1537–1547. 10.1139/f88-182 [DOI] [Google Scholar]
- Kalitsis P., and Choo K. H. A., 2012. The evolutionary life cycle of the resilient centromere. Chromosoma 121: 327–340. 10.1007/s00412-012-0369-6 [DOI] [PubMed] [Google Scholar]
- Kijas J., McWilliam S., Sanchez M. N., Kube P., King H. et al. , 2018. Evolution of sex determination loci in Atlantic salmon. Sci. Rep. 8: 5664 10.1038/s41598-018-23984-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., 2010. How and why chromosome inversions evolve. PLoS Biol. 8: e1000501 10.1371/journal.pbio.1000501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittilsen S., Schjolden J., Beitnes-Johansen I., Shaw J. C., Pottinger T. G. et al. , 2009. Melanin-based skin spots reflect stress responsiveness in salmonid fish. Horm. Behav. 56: 292–298. 10.1016/j.yhbeh.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Kodama M., Brieuc M., Devlin R. H., Hard J. J., and Naish K. A., 2014. Comparative mapping between Coho Salmon (Oncorhynchus kisutch) and three other salmonids suggests a role for chromosomal rearrangements in the retention of duplicated regions following a whole genome duplication event. G3 (Bethesda) 4: 1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger C. C., and Ihssen P. E., 1995. Review of genetics of lake trout in the Great Lakes: history, molecular genetics, physiology, strain comparisons, and restoration management. J. Great Lakes Res. 21: 348–363. 10.1016/S0380-1330(95)71109-1 [DOI] [Google Scholar]
- Küpper C., Stocks M., Risse J. E., dos Remedios N., Farrell L. L. et al. , 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48: 79–83. 10.1038/ng.3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson W. A., McKinney G. J., Limborg M. T., Everett M. V., Seeb L. W. et al. , 2015. Identification of multiple QTL hotspots in Sockeye Salmon (Oncorhynchus nerka) using genotyping-by-sequencing and a dense linkage map. J. Hered. 107: 122–133. 10.1093/jhered/esv099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary R. F., Allendorf F. W., and Forbes S. H., 1993. Conservation genetics of bull trout in the Columbia and Klamath River drainages. Conserv. Biol. 7: 856–865. 10.1046/j.1523-1739.1993.740856.x [DOI] [Google Scholar]
- Leitwein M., Garza J. C., and Pearse D. E., 2016. Ancestry and adaptive evolution of anadromous, resident, and adfluvial rainbow trout (Oncorhynchus mykiss) in the San Francisco bay area: application of adaptive genomic variation to conservation in a highly impacted landscape. Evol. Appl. 10: 56–67. 10.1111/eva.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitwein M., Guinand B., Pouzadoux J., Desmarais E., Berrebi P.. et al, 2017. A dense brown trout (Salmo trutta) linkage map reveals recent chromosomal rearrangements in the Salmo genus and the impact of selection on linked neutral diversity. G3 (Bethesda) 7: 1365–1376. 10.1534/g3.116.038497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitwein M., Gagnaire P. A., Desmarais E., Berrebi P., and Guinand B., 2018. Genomic consequences of a recent three‐way admixture in supplemented wild brown trout populations revealed by local ancestry tracts. Mol. Ecol. 27: 3466–3483. 10.1111/mec.14816 [DOI] [PubMed] [Google Scholar]
- Li H., 2013. Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. arXiv. 00: 1–3. [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Gidskehaug L., Moen T., Hayes B. J., Berg P. R. et al. , 2011. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics 12: 615 10.1186/1471-2164-12-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Koop B. F., Sandve S. R., Miller J. R., Kent M. P. et al. , 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533: 200–205. 10.1038/nature17164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limborg M. T., Waples R. K., Allendorf F. W., and Seeb J. E., 2015. Linkage mapping reveals strong chiasma interference in sockeye salmon: implications for interpreting genomic data. G3 (Bethesda) 5: 2463–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limborg M. T., McKinney G. J., Seeb L. W., and Seeb J. E., 2016. Recombination patterns reveal information about centromere location on linkage maps. Mol. Ecol. Resour. 16: 655–661. 10.1111/1755-0998.12484 [DOI] [PubMed] [Google Scholar]
- Lindner K. R., Seeb J. E., Habicht C., Knudsen K. L., Kretschmer E. et al. , 2000. Gene-centromere mapping of 312 loci in pink salmon by half-tetrad analysis. Genome 43: 538–549. 10.1139/g00-016 [DOI] [PubMed] [Google Scholar]
- Lowry D. B., and Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500 10.1371/journal.pbio.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., and Bernatchez L., 1999. Correlated trophic specialization and genetic divergence in sympatric lake whitefish ecotypes (Coregonus clupeaformis): support for the ecological speciation hypothesis. Evolution 53: 1491–1505. [DOI] [PubMed] [Google Scholar]
- Lubieniecki K. P., Lin S., Cabana E. I., Li J., Lai Y. et al. , 2015. Genomic instability of the sex-determining locus in Atlantic salmon (Salmo salar). G3 (Bethesda) 5: 2513–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin K., Coon A., Carson R., Debes P. V., and Fraser D. J., 2016. Striking phenotypic variation yet low genetic differentiation in sympatric lake trout (Salvelinus namaycush). PLoS One 11: e0162325 10.1371/journal.pone.0162325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin K., Coon A., and Fraser D., 2017. Traditional ecological knowledge reveals the extent of sympatric lake trout diversity and habitat preferences. Ecol. Soc. 22: 20 10.5751/ES-09345-220220 [DOI] [Google Scholar]
- Marsden J. E., Krueger C. C., Grewe P. M., Kincaid H. L., and May B., 1993. Genetic comparison of naturally spawned and artificially propagated Lake Ontario lake trout fry: evaluation of a stocking strategy for species rehabilitation. N. Am. J. Fish. Manage. 13: 304–317. [DOI] [Google Scholar]
- May B., Wright J. E., and Stoneking M., 1979. Joint segregation of biochemical loci in Salmonidae: results from experiments with Salvelinus and review of the literature on other species. Journal of the Fisheries Board of Canada 36: 1114–1128. 10.1139/f79-156 [DOI] [Google Scholar]
- McKinney G. J., Seeb L. W., Larson W. A., Gomez‐Uchida D., Limborg M. T. et al. , 2016. An integrated linkage map reveals candidate genes underlying adaptive variation in Chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. Resour. 16: 769–783. 10.1111/1755-0998.12479 [DOI] [PubMed] [Google Scholar]
- McKinney G. J., Pascal C. E., Templin W. D., Gilk-Baumer S. E., Dann T. H. et al. , 2019. Dense SNP panels resolve closely related Chinook salmon populations. Can. J. Fish. Aquat. Sci. 999: 1–11. [Google Scholar]
- Miller M. R., Brunelli J. P., Wheeler P. A., Liu S., Rexroad C. E. et al. , 2012. A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol. Ecol. 21: 237–249. 10.1111/j.1365-294X.2011.05305.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T., Hoyheim B., Munck H., and Gomez‐Raya L., 2004. A linkage map of Atlantic salmon (Salmo salar) reveals an uncommonly large difference in recombination rate between the sexes. Anim. Genet. 35: 81–92. 10.1111/j.1365-2052.2004.01097.x [DOI] [PubMed] [Google Scholar]
- Moen T., Hayes B., Baranski M., Berg P. R., Kjøglum S. et al. , 2008. A linkage map of the Atlantic salmon (Salmo salar) based on EST-derived SNP markers. BMC Genomics 9: 223 10.1186/1471-2164-9-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T., Baranski M., Sonesson A. K., and Kjøglum S., 2009. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics 10: 368 10.1186/1471-2164-10-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. A., and Bronte C. R., 2001. Delineation of sympatric morphotypes of lake trout in Lake Superior. Trans. Am. Fish. Soc. 130: 1233–1240. [DOI] [Google Scholar]
- Muhlfeld C. C., Kalinowski S. T., McMahon T. E., Taper M. L., Painter S. et al. , 2009. Hybridization rapidly reduces fitness of a native trout in the wild. Biol. Lett. 5: 328–331. 10.1098/rsbl.2009.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A. M., Bronte C. R., Zimmerman M. S., Quinlan H. R., Glase J. D. et al. , 2014. Ecomorphological diversity of lake trout at Isle Royale, Lake Superior. Trans. Am. Fish. Soc. 143: 972–987. 10.1080/00028487.2014.900823 [DOI] [Google Scholar]
- Muir A. M., Hansen M. J., Bronte C. R., and Krueger C. C., 2016. If Arctic charr Salvelinus alpinus is ‘the most diverse vertebrate’, what is the lake charr Salvelinus namaycush? Fish Fish. 17: 1194–1207. 10.1111/faf.12114 [DOI] [Google Scholar]
- Nugent C. M., Easton A. A., Norman J. D., Ferguson M. M., and Danzmann R. G., 2017. A SNP based linkage map of the Arctic Charr (Salvelinus alpinus) genome provides insights into the diploidization process after whole genome duplication. G3 (Bethesda) 7: 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan T. N., Wu X. S., Rachel R. A., Huang J. D., Swing D. A. et al. , 2004. dsu functions in a MYO5A-independent pathway to suppress the coat color of dilute mice. PNAS 101: 16831–16836. 10.1073/pnas.0407339101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palti Y., Genet C., Gao G., Hu Y., You F. M. et al. , 2012. A second generation integrated map of the rainbow trout (Oncorhynchus mykiss) genome: analysis of conserved synteny with model fish genomes. Mar. Biotechnol. (NY) 14: 343–357. 10.1007/s10126-011-9418-z [DOI] [PubMed] [Google Scholar]
- Pearse D. E., Barson N. J., Nome T., Gao G., Campbell M. A. et al. , 2019. Sex-dependent dominance maintains migration supergene in rainbow trout. Nat. Ecol. Evol. 3: 1731–1742. 10.1038/s41559-019-1044-6 [DOI] [PubMed] [Google Scholar]
- Peichel C. L., Nereng K. S., Ohgi K. A., Cole B., Colosimo P. F. et al. , 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414: 901–905. 10.1038/414901a [DOI] [PubMed] [Google Scholar]
- Perreault‐Payette A., Muir A. M., Goetz F., Perrier C., Normandeau E. et al. , 2017. Investigating the extent of parallelism in morphological and genomic divergence among lake trout ecotypes in Lake Superior. Mol. Ecol. 26: 1477–1497. 10.1111/mec.14018 [DOI] [PubMed] [Google Scholar]
- Pfennig D. W., and Pfennig K. S., 2012. Evolution’s wedge: competition and the origins of diversity. No. 12, Univ of California Press, Berkeley, CA: 10.1525/california/9780520274181.001.0001 [DOI] [Google Scholar]
- Phillips R. B., and Zajicek K. D., 1982. Q band chromosomal polymorphisms in lake trout (Salvelinus namaycush). Genetics 101: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R. B., and Ihssen P. E., 1985. Identification of sex chromosomes in lake trout (Salvelinus namaycush). Cytogenet. Genome Res. 39: 14–18. 10.1159/000132097 [DOI] [PubMed] [Google Scholar]
- Protas M. E., and Patel N. H., 2008. Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 24: 425–446. 10.1146/annurev.cellbio.24.110707.175302 [DOI] [PubMed] [Google Scholar]
- Qiu C., Han Z., Li W., Ye K., Xie Y. et al. , 2018. A high-density genetic linkage map and QTL mapping for growth and sex of yellow drum (Nibea albiflora). Sci. Rep. 8: 17271 10.1038/s41598-018-35583-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I. M.. . 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., Calboli F., Guo B., Shikano T., and Merilä J., 2015. Construction of ultradense linkage maps with Lep-MAP2: stickleback F 2 recombinant crosses as an example. Genome Biol. Evol. 8: 78–93. 10.1093/gbe/evv250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 33: 3726–3732. 10.1093/bioinformatics/btx494 [DOI] [PubMed] [Google Scholar]
- Reed K. M., and Phillips R. B., 1995. Molecular characterization and cytogenetic analysis of highly repeated DNAs of lake trout, Salvelinus namaycush. Chromosoma 104: 242–251. 10.1007/BF00352255 [DOI] [PubMed] [Google Scholar]
- Reinecke M., 2010. Influences of the environment on the endocrine and paracrine fish growth hormone–insulin‐like growth factor‐I system. J. Fish Biol. 76: 1233–1254. 10.1111/j.1095-8649.2010.02605.x [DOI] [PubMed] [Google Scholar]
- Renaut S., and Bernatchez L., 2011. Transcriptome-wide signature of hybrid breakdown associated with intrinsic reproductive isolation in lake whitefish species pairs (Coregonus spp. Salmonidae). Heredity 106: 1003–1011. 10.1038/hdy.2010.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B. W., and Schluter D., 2000. Natural selection and the evolution of adaptive genetic variation in northern freshwater fishes, pp. 65–94 in Adaptive Genetic Variation in the Wild, edited by Taylor W. W., Sinervo B., and Endler J.. Oxford University Press, Oxford. [Google Scholar]
- Rochette N. C., Rivera‐Colón A. G., and Catchen J. M., 2019. Stacks 2: Analytical methods for paired‐end sequencing improve RADseq‐based population genomics. Mol. Ecol. 28: 4737–4754. 10.1111/mec.15253 [DOI] [PubMed] [Google Scholar]
- Rogers S. M., and Bernatchez L., 2007. The genetic architecture of ecological speciation and the association with signatures of selection in natural lake whitefish (Coregonus sp. Salmonidae) species pairs. Mol. Biol. Evol. 24: 1423–1438. 10.1093/molbev/msm066 [DOI] [PubMed] [Google Scholar]
- Rohland N., and Reich D., 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22: 939–946. 10.1101/gr.128124.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf, F. J., 2005 tpsDig, digitize landmarks and outlines, version 2.05. Department of Ecology and Evolution, State University of New York at Stony Brook. http://life.bio.sunysb.edu/morph/. [Google Scholar]
- Rüber L., Verheyen E., and Meyer A., 1999. Replicated evolution of trophic specializations in an endemic cichlid fish lineage from Lake Tanganyika. Proc. Natl. Acad. Sci. USA 96: 10230–10235. 10.1073/pnas.96.18.10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder R. A., Kerr S. R., Taylor W. W., and Larkin P. A., 1981. Community consequences of fish stock diversity. Can. J. Fish. Aquat. Sci. 38: 1856–1866. 10.1139/f81-231 [DOI] [Google Scholar]
- Santure A. W., Poissant J., Cauwer I. D., Oers K. V., Robinson M. R. et al. , 2015. Replicated analysis of the genetic architecture of quantitative traits in two wild great tit populations. Mol. Ecol. 24: 6148–6162. 10.1111/mec.13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage C., Vagner M., Derôme N., Audet C., and Bernatchez L., 2012. Coding gene single nucleotide polymorphism mapping and quantitative trait loci detection for physiological reproductive traits in brook charr, Salvelinus fontinalis. G3 (Bethesda) 2: 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sävilammi T., Primmer C. R., Varadharajan S., Guyomard R., Guiguen Y. et al. , 2019. The chromosome-level genome assembly of European grayling reveals aspects of a unique genome evolution process within salmonids. G3 (Bethesda) 9: 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler D. E., Hilborn R., Chasco B., Boatright C. P., Quinn T. P. et al. , 2010. Population diversity and the portfolio effect in an exploited species. Nature 465: 609–612. 10.1038/nature09060 [DOI] [PubMed] [Google Scholar]
- Scribner K., Tsehaye I., Brenden T., Stott W., Kanefsky J. et al. , 2018. Hatchery strain contributions to emerging wild lake trout populations in Lake Huron. J. Hered. 109: 675–688. 10.1093/jhered/esy029 [DOI] [PubMed] [Google Scholar]
- Skulason S., and Smith T. B., 1995. Resource polymorphisms in vertebrates. Trends Ecol. Evol. 10: 366–370. 10.1016/S0169-5347(00)89135-1 [DOI] [PubMed] [Google Scholar]
- Snorrason S. S., Skúlason S., Jonsson B., Malmquist H. J., Jónasson P. M. et al. , 1994. Trophic specialization in Arctic charr Salvelinus alpinus (Pisces; Salmonidae): morphological divergence and ontogenetic niche shifts. Biol. J. Linn. Soc. Lond. 52: 1–18. 10.1111/j.1095-8312.1994.tb00975.x [DOI] [Google Scholar]
- Spruell P., Pilgrim K. L., Greene B. A., Habicht C., Knudsen K. L. et al. , 1999. Inheritance of nuclear DNA markers in gynogenetic haploid pink salmon. J. Hered. 90: 289–296. 10.1093/jhered/90.2.289 [DOI] [PubMed] [Google Scholar]
- Stafford C. P., McPhee M. V., Eby L. A., and Allendorf F. W., 2013. Introduced lake trout exhibit life history and morphological divergence with depth. Can. J. Fish. Aquat. Sci. 71: 10–20. 10.1139/cjfas-2013-0115 [DOI] [Google Scholar]
- Stebbins G. L., 1958. The inviability, weakness, and sterility of interspecific hybrids. Adv. Genet. 9: 147–215 Academic Press. 10.1016/S0065-2660(08)60162-5 [DOI] [PubMed] [Google Scholar]
- Sutherland B., Gosselin T., Normandeau E., Lamothe M., Isabel N. et al. , 2016. Salmonid chromosome evolution as revealed by a novel method for comparing RADseq linkage maps. Genome Biol. Evol. 8: 3600–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland B., Rico C., Audet C., and Bernatchez L., 2017. Sex chromosome evolution, heterochiasmy, and physiological QTL in the salmonid brook charr Salvelinus fontinalis. G3 (Bethesda) 7: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swink W. D.1990. Effect of lake trout size on survival after a single sea lamprey attack. Transactions of the American Fisheries Society 119: 996–1002. [DOI] [Google Scholar]
- Thomas S. M., Kainz M. J., Amundsen P., Hayden B., Taipale S. J. et al. , 2019. Resource polymorphism in European whitefish: Analysis of fatty acid profiles provides more detailed evidence than traditional methods alone. PLoS One 14: e0221338 10.1371/journal.pone.0221338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgaard G. H., Allendorf F. W., and Knudsen K. L., 1983. Gene-centromere mapping in rainbow trout: high interference over long map distances. Genetics 103: 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston C. E., 1962. Physical characteristics and chemical composition of two subspecies of lake trout. Journal of the Fisheries Board of Canada 19: 39–44. 10.1139/f62-002 [DOI] [Google Scholar]
- Waters T. F., 1987. The Superior North Shore: A Natural History of Lake Superior’s Northern Lands and Waters, U of Minnesota Press, Minneapolis, MN. [Google Scholar]
- Waples R. K., Seeb L. W., and Seeb J. E., 2016. Linkage mapping with paralogs exposes regions of residual tetrasomic inheritance in chum salmon (Oncorhynchus keta). Mol. Ecol. Resour. 16: 17–28. 10.1111/1755-0998.12394 [DOI] [PubMed] [Google Scholar]
- Webb P. W., 1984. Body form, locomotion and foraging in aquatic vertebrates. Am. Zool. 24: 107–120. 10.1093/icb/24.1.107 [DOI] [Google Scholar]
- Wellenreuther M., and Bernatchez L., 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 33: 427–440. 10.1016/j.tree.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Wickham, H., and W. Chang, 2008 ggplot2: An implementation of the Grammar of Graphics. R package version 0.7, URL: http://CRAN.R-project.org/package=ggplot2.
- Whiteley A. R., 2007. Trophic polymorphism in a riverine fish: morphological, dietary, and genetic analysis of mountain whitefish. Biol. J. Linn. Soc. Lond. 92: 253–267. 10.1111/j.1095-8312.2007.00845.x [DOI] [Google Scholar]
- Wilson C. C., and Mandrak N. E., 2004. History and evolution of lake trout in Shield lakes: past and future challenges. Boreal Shield watersheds: lake trout ecosystems in a changing environment, pp. 21–35 in Boreal Shield Watersheds: Lake Trout Ecosystems in a Changing Environment, edited by Gunn J., Steedman R., and Ryder R.. Lewis/CRC Press, Boca Raton, FL. [Google Scholar]
- Wilson C., and Evans D., 2010. Algonquin’s silver lake trout: highlighting the history, habitat, and concerns for a unique biodiversity element, Ontario Ministry of Natural Resources, Peterborough, ON. [Google Scholar]
- Woram R. A., Gharbi K., Sakamoto T., Hoyheim B., Holm L. E. et al. , 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13: 272–280. 10.1101/gr.578503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. S., Masedunskas A., Weigert R., Copeland N. G., Jenkins N. A. et al. , 2012. Melanoregulin regulates a shedding mechanism that drives melanosome transfer from melanocytes to keratinocytes. Proc. Natl. Acad. Sci.USA 109: E2101–E2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Liu B., Zhu C., Li K., Yue L. et al. , 2013. microRNA regulation of skin pigmentation in fish. J. Cell Sci. 126: 3401–3408. 10.1242/jcs.125831 [DOI] [PubMed] [Google Scholar]
- Yano A., Nicol B., Jouanno E., Quillet E., Fostier A. et al. , 2013. The sexually dimorphic on the Y‐chromosome gene (sdY) is a conserved male‐specific Y‐chromosome sequence in many salmonids. Evol. Appl. 6: 486–496. 10.1111/eva.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman M. S., Krueger C. C., and Eshenroder R. L., 2006. Phenotypic diversity of lake trout in Great Slave Lake: differences in morphology, buoyancy, and habitat depth. Trans. Am. Fish. Soc. 135: 1056–1067. 10.1577/T05-237.1 [DOI] [Google Scholar]
- Zimmerman M. S., Krueger C. C., and Eshenroder R. L., 2007. Morphological and ecological differences between shallow-and deep-water lake trout in Lake Mistassini, Quebec. J. Great Lakes Res. 33: 156–169. 10.3394/0380-1330(2007)33[156:MAEDBS]2.0.CO;2 [DOI] [Google Scholar]
- Zimmerman M. S., Schmidt S. N., Krueger C. C., Vander Zanden M. J., and Eshenroder R. L., 2009. Ontogenetic niche shifts and resource partitioning of lake trout morphotypes. Can. J. Fish. Aquat. Sci. 66: 1007–1018. 10.1139/F09-060 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data for all individuals used for linkage and QTL mapping has been made publicly available in a NCBI sequence read archive (PRJNA608030). Map information, phenotypes, and genotypes used for QTL and sex locus mapping are available in supplementary material (Document S2). Supplemental material available at figshare: https://doi.org/10.25387/g3.11908326.