Abstract
Yeast surface display empowers selection of protein binding ligands, typically using recombinant soluble antigens. However, ectodomain fragments of transmembrane targets may fail to recapitulate their true, membrane-bound form. Direct selections against adhered mammalian cells empower enrichment of genuine binders yet benefit from high target expression, robustly adherent mammalian cells, and nanomolar affinity ligands. This study evaluates a modified format with mammalian cells immobilized to magnetic beads; yeast-displayed fibronectin domain and affibody ligands of known affinities and cells with expression ranges of epidermal growth factor receptor (EGFR) and CD276 elucidate important parameters to ligand enrichment and yield in cell suspension panning with comparison to adherent panning. Cell suspension panning is hindered by significant background of non-displaying yeast but exhibits yield advantages in model EGFR systems for a high affinity (KD = 2 nM) binder on cells with both high (106 per cell) target expression (9.6 ± 0.6% vs. 3.2 ± 0.4%, p < 0.0001) and mid (105) target expression (2.3 ± 0.5% vs. 0.41 ± 0.09%, p = 0.0008), as well as for a low affinity (KD > 600 nM) binder on high target expression cells (2.0 ± 0.5% vs. 0.017 ± 0.005%; p = 0.001). Significant enrichment was observed for all EGFR systems except the low-affinity, high expression system. The CD276 system failed to provide significant enrichment, indicating that this technique may not be suitable for all targets. Collectively, this study highlights new approaches that yield successful enrichment of yeast-displayed ligands via panning on immobilized mammalian cells.
Keywords: yeast display, biopanning, ligand, cell suspension panning, epidermal growth factor receptor, protein engineering
Graphical Abstract
Introduction
Protein ligands have been engineered1–3 to clinically treat diseases with altered biomarker expression through direct inhibition, targeted drug/radioisotope delivery, and immune system engagement.4,5 Similar ligands have been used for diagnostic purposes, such as targeted molecular imaging6 and ex vivo biomarker detection in blood and urine.7–9 Recent advances in clinical biomarker discovery such as next-generation sequencing,10 mass spectrometry-based proteomics,11,12 and other chemical biology techniques have drastically increased the number of well-characterized, clinically relevant targets. This has resulted in a growing demand for engineered ligands to target these new biomarkers.
Common high-throughput methods, such as yeast13–15 and phage16–18 display technologies linking genotype to phenotype, coupled with selections via magnetic bead capture19–23 and fluorescence activated cell sorting (FACS),24,25 have been used to address this demand. Experimental screening strategies require the use of the biomarker to isolate binding ligands; however, many clinically relevant biomarkers possess hydrophobic transmembrane domains, making them difficult to work with in an aqueous environment. In lieu of full-length proteins, recombinantly produced soluble ectodomains are often used as analogs during ligand selections. While this strategy has proven successful26–32, many engineered ligands fail to bind to genuine antigen on target-expressing cells.33 As negative results may be less likely to be published,34 there is a lack of research thoroughly investigating this problem, thus understating the need for more reliable methods of translatable ligand discovery.
The failure of recombinant soluble ectodomain-based selection methods indicates that these ectodomain fragments may not fully recapitulate the protein in its true, membrane-bound form. While this issue remains largely unstudied, a number of factors may cause this failure, including the improper folding and aggregation of the soluble ectodomains,35–38 non-natural post-translational modification due to the production host cell type,39,40 the addition of biological or chemical tags for purification and/or selection,41 and the absence of a transmembrane domain or cell membrane. All of these factors may create epitopes that are masked or not present in the true, membrane-bound antigen.
To avoid these issues, a number of selection techniques utilizing target-expressing cells have been developed, including fluorescence activated cell sorting (FACS) using detergent-solubilized cell lysate15,42,43 or whole cells44,45 and panning on adherent cell monolayers43,46,47; however, these techniques leave room for improvement. While FACS with cell lysate or whole cells is an effective method for enrichment, its throughput (107−108 per hour) is substantially below current display library sizes22,31,48,49 and its lack of avidity limits the enrichment of weaker affinity ligands, such as those isolated from some de novo libraries. While adherent cell panning has been employed successfully in multiple scenarios,18,51–59 a study using EGFR-binding fibronectins found that enrichment is hindered when using cells with low-to-moderate (≤105 targets/cell) expression or ligands of micromolar or weaker affinity.47
The use of cells in suspension has been investigated as an alternative method of cell panning in phage display libraries44,60–62 and has shown success in isolating binding ligands to cell biomarkers.63,64 There is also some evidence that cell suspension panning may outperform adherent cell panning in the phage display context, as two phage library campaigns against cells in suspension succeeded after equivalent campaigns against adherent cells failed.65,66 This success motivates adaptation of these methods to yeast surface display libraries; however, cell suspension panning methods for phage display use differential centrifugation to separate cell-bound phages from nonbinding phages, which is incompatible with yeast surface display. Selections using mammalian cells conjugated to magnetic beads is an attractive alternate method of sorting due to the effectiveness of conjugation67 as well as the high capacity (106 binding yeast per 106 magnetic beads), throughput (1010 cells per selection), and established reliability of isolating specific binders to the immobilized target associated with magnetic bead selections.19,68
This study aims to evaluate a new method of ligand selection using suspended mammalian cells immobilized to magnetic beads as a selection agent in comparison to traditional adherent mammalian cell panning. It is hypothesized that the use of suspended mammalian cells may aid in enrichment and enhance capacity by increasing the exposed mammalian-cell surface area for ligand binding while potentially enabling ligand campaigns against cell lines that cannot be cultured adherently for traditional cell panning.
To optimize and better understand the parameters for successful cell-based selections using yeast surface display in this new system, washing stringency, target expression on mammalian cells, ligand binding affinity, ligand protein scaffold, and target of interest were systematically varied to understand how each parameter affects enrichment ratio and yield of ligand-displaying yeast in EGFR-expressing and CD276-expressing systems. While a significantly increased background yield of non-displaying yeast limits the effectiveness of cell suspension panning, it shows a significant increase in the yield of binding yeast compared to adherent panning. In addition, cell suspension panning demonstrates substantially increased throughput, which allows the sorting of larger libraries in shorter timespans. The use of a CD276-expressing model failed to provide effective enrichment, indicating that not all targets may be suitable for this technique. Together, these results advance understanding and development of protocols for ligand selection experiments.
Results
Two different magnetic beads were used to generate bead-mammalian cell conjugates for testing in yeast-displayed ligand selections: a carboxylic acid-functionalized magnetic bead that was covalently conjugated to amines present on the mammalian cell surface and a streptavidin-functionalized magnetic bead that was noncovalently conjugated to biotinylated cells. These bead-cell conjugates were used as a pulldown agent to test enrichment of high affinity binder-displaying yeast from non-displaying yeast. In parallel, conventional adherent mammalian cell panning was used to compare the ability of cell suspension panning to enrich ligand-displaying yeast relative to established techniques.
Streptavidin and Carboxylic Acid Beads both Effectively Conjugate Mammalian Cells
A431 cells were conjugated, in a 1:1 mixture at 5×106 cells or beads per mL, to carboxylic acid-functionalized magnetic beads in moderate, albeit highly variable, yield (29 ± 7) (Figure 1A). Examination of conjugated cells visualized by phase contrast microscopy showed individual cells attached to several (3.3 ± 0.4) beads with no observable aggregation of cells and beads (Figure 1B). A431 cells conjugated in this manner showed fluorescence similar to unconjugated cells when labeled with an anti-EGFR antibody, indicating that EGFR expression was not appreciably blocked (Figure 1C).
Figure 1.
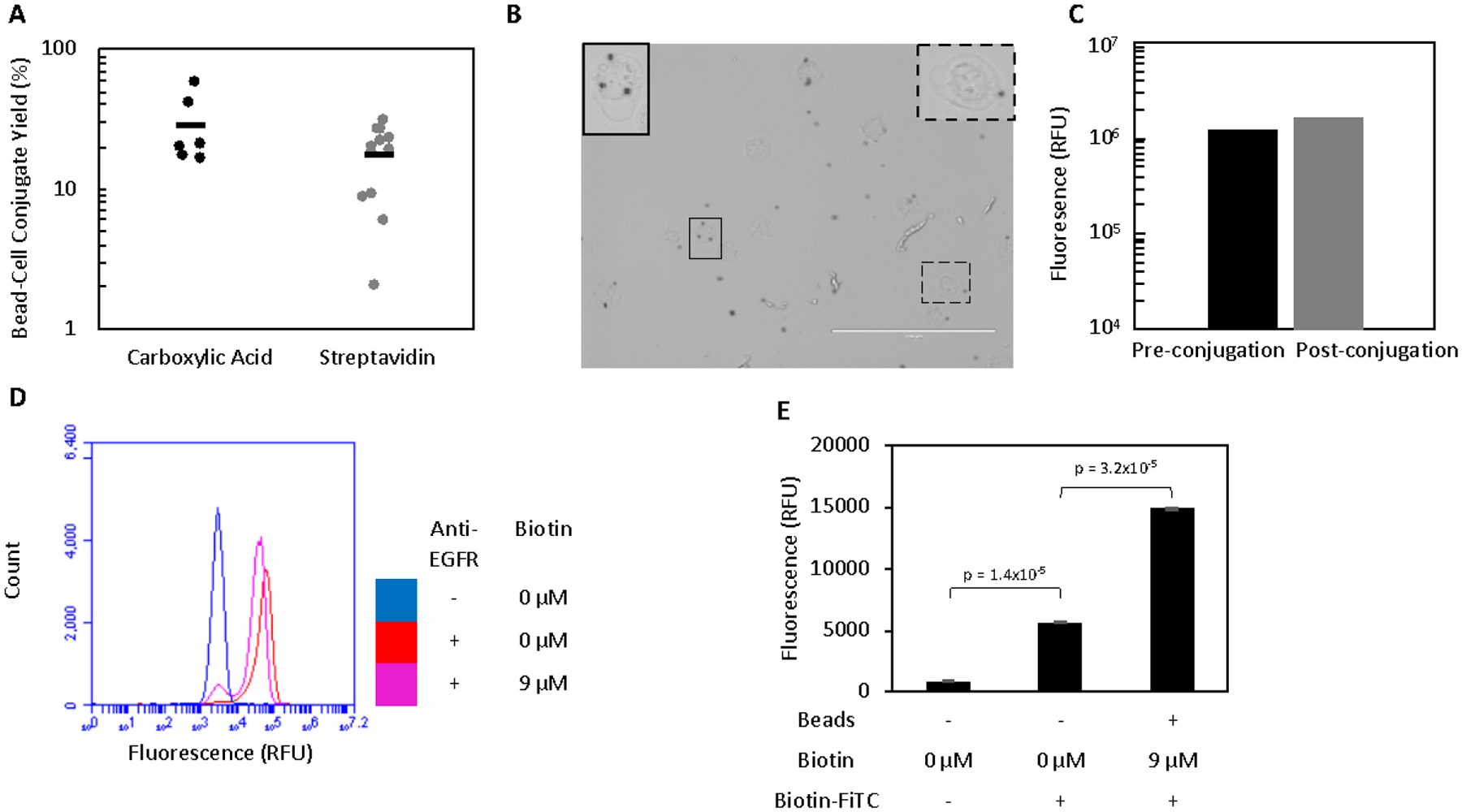
Mammalian cells can be efficiently conjugated to magnetic beads. A. Beads and A431 cells were incubated in a 1:1 ratio prior to washing on a magnet to remove all unbound cells. The yield of conjugates was quantified by flow cytometry. B. Bead-A431 conjugates (n = 39) were visualized to ensure proper conjugation. Carboxylic acid beads (black spheres) can be seen attached to the majority of A431 cells. Scale bar is 200 μm. Two cells (outlined) are enlarged and contrast-enhanced to show detail. C. A431 cells were labelled for with an anti-EGFR antibody and their fluorescence quantified by flow cytometry before and after conjugation to carboxylic acid beads to assess whether EGFR expression was extensively masked by conjugation to beads. D. A431 cells were labelled with an anti-EGFR antibody and their fluorescence quantified by flow cytometry before and after biotinylation to assess whether EGFR expression was extensively masked. A small number of cells had their expression masked by the labelling with 9 μM biotin. E. Unbiotinylated A431 cells with no beads and biotinylated A431 cells conjugated to streptavidin beads were labelled with biotin-FITC and the fluorescence of the cell population (as gated by scatter) was analyzed to detect the presence of streptavidin beads attached to A431 cells. While biotin-FITC significantly labels the cell surface in the absence of streptavidin beads, a further significant increase in signal is observed after conjugation occurs. Fluorescence is presented as mean ± standard error of three trials.
To explore an alternative method of conjugation, A431 cells were biotinylated for capture by streptavidin-functionalized magnetic beads. Sulfo-NHS-LC-biotin with a 22.3 Å linker failed to provide substantial conjugation to streptavidin beads (data not shown), despite substantial biotinylation of the cell surface. The use of an analogous sulfo-NHS biotin with a 20,000 Da PEG linker provided significantly enhanced conjugate yield, perhaps due to the increased accessibility of the biotin moiety to the extracellular space. This increase allowed streptavidin bead conjugation to become comparable to the carboxylic acid beads (Figure 1A). Varying biotin labelling concentrations from 0.9 – 90 μM were tested, with all conditions providing similar yield (SI Figure 1A). 9 μM was chosen as a conservative balance between limiting material consumption and ensuring sufficient biotinylation. No significant difference in yield was observed for labelling times varied from 30 minutes to 2 hours (SI Figure 1B). Thus, a 30-minute labelling time was selected for subsequent experiments. Biotinylated A431 cells showed a similar fluorescence to unbiotinylated cells when labelled with an anti-EGFR antibody, indicating that EGFR expression was not masked on the majority of cells (Figure 1D). Streptavidin bead-cell conjugates exhibited significantly increased signal relative to unbiotinylated cells when labelled with biotin-FITC and analyzed for fluorescence by flow cytometry (gating for mammalian cells by scatter), confirming the presence to streptavidin beads (Figure 1E).
In summary, both carboxylic acid bead capture using native cellular amines and streptavidin bead capture using biotinylated cells were relatively effective. In cases where cell supply is limited, thereby motivating higher yield, increased concentrations or stoichiometric excess of beads could be evaluated.
Cell Suspension Panning Enriches Yeast-Displayed EGFR-Binding Fibronectin Domains
Target-expressing mammalian cells conjugated to magnetic beads were assessed for their ability to effectively enrich yeast displaying binding ligands. A mixture of 106 yeast harboring display plasmids encoding for a fibronectin domain with high affinity for EGFR (E6.2.6’ KD = 2 ± 2 nM)26 and 108 plasmidless EBY100 were panned against bead-immobilized A431 cells, which express 4 ± 1 ×106 EGFR/cell. The mixture of yeast and bead-cell conjugates was incubated, placed on a magnet to collect bead-cell conjugates along with bound yeast and washed twice. This baseline condition resulted in an enrichment that was highly variable but significantly greater than unity (9 ± 1 with carboxylic acid beads, p < 0.0001; 11 ± 2, p = 0.0004 with streptavidin beads) of binding yeast relative to non-displaying yeast with high yield of binding yeast (19 ± 6% with carboxylic acid beads; 10 ± 2% with streptavidin beads). Binder yields were comparable or moderately lower for the less stringent one-wash condition as well as with three- to five-washes (Figure 2A). Surprisingly, the yield of non-displaying yeast did not decrease as expected across sequential washes in the streptavidin bead system and only slightly decreased in the carboxylic acid bead system, suggesting some form of interaction prevented the effective removal of non-displaying yeast from the system by washing (Figure 2B). As a result, the enrichment did not exhibit a substantial increase at any alternate washing conditions (Figure 2C).
Figure 2.
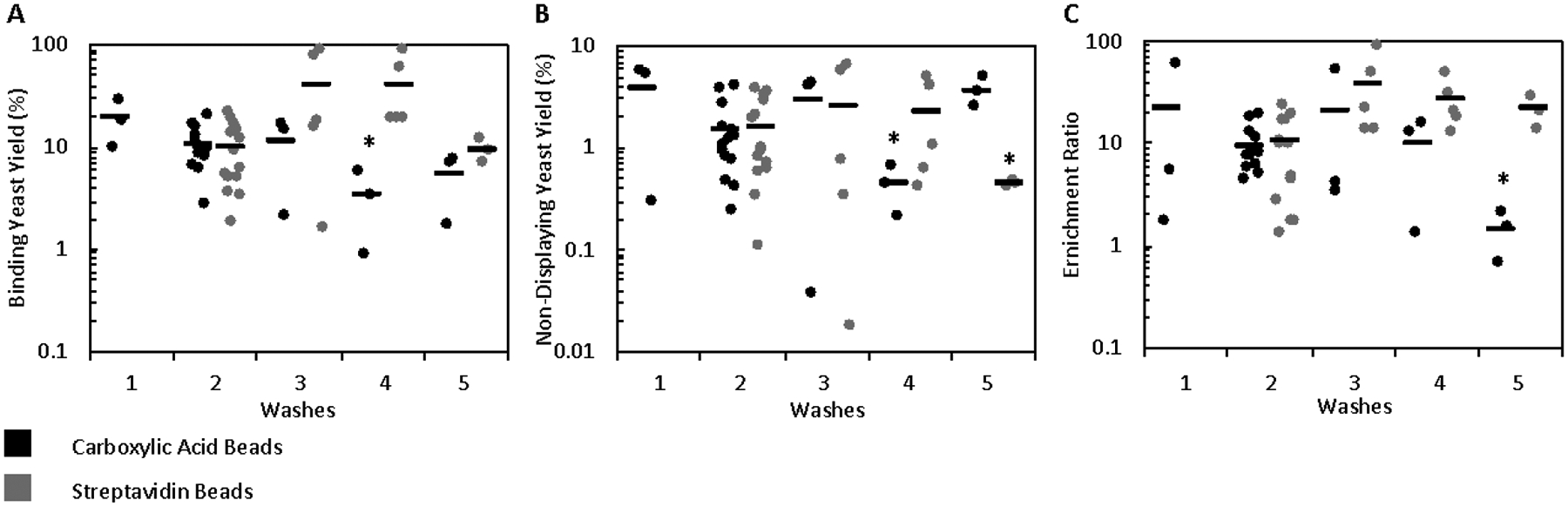
Cell suspension panning enrichment is relatively insensitive to washing conditions. Mixtures of yeast displaying E6.2.6’ and non-displaying EBY100 yeast were incubated with bead-A431 conjugates and washed 1–5 times. (A) Binding yeast yield, (B) non-displaying yeast yield, and (C) enrichment ratio were quantified. A (*) indicates p < 0.05 (with Bonferroni correction) relative to the 2-wash baseline.
To further probe the unexpected result of sustained yield of non-displaying yeast even with extensive washing, 108 non-displaying yeast were mixed with bare beads and the yield quantified after washing. In the case of the carboxylic acid bead system, bare beads resulted in a high yield of non-displaying yeast compared to the streptavidin bead system (2.2 ± 0.8% vs. 0.19 ± 0.06%), indicating that the retention of non-displaying yeast on the carboxylic acid bead system may be due to yeast-bead interactions, despite extensive quenching of beads after mammalian cell conjugation (Figure 3A). When non-displaying yeast were mixed with bead-A431 conjugates, the yield of non-displaying yeast rose significantly (2.8 ± 0.9%) in the streptavidin system while remaining similar in the carboxylic acid system (1.2 ± 0.1%) relative to bare beads (Figure 3B). This appears to indicate that an underlying yeast-A431 interaction may also be preventing proper removal of non-displaying yeast. While these results were obtained with 0.1% (w/v) bovine serum albumin to limit nonspecific interactions, more complex inhibitors, 1% (w/v) milk powder or 1% (v/v) fetal bovine serum, were tested to further screen any potential nonspecific interactions. Only the addition of serum in the carboxylic acid system resulted in a significant decrease in the yield of non-binding yeast, and this decrease was not nearly as substantial as expected if nonspecific interactions were being effectively screened. Combined, this indicates that even the addition of complex blocking mixtures was not sufficient to hinder nonspecific yeast recovery (Figure 3B).
Figure 3.
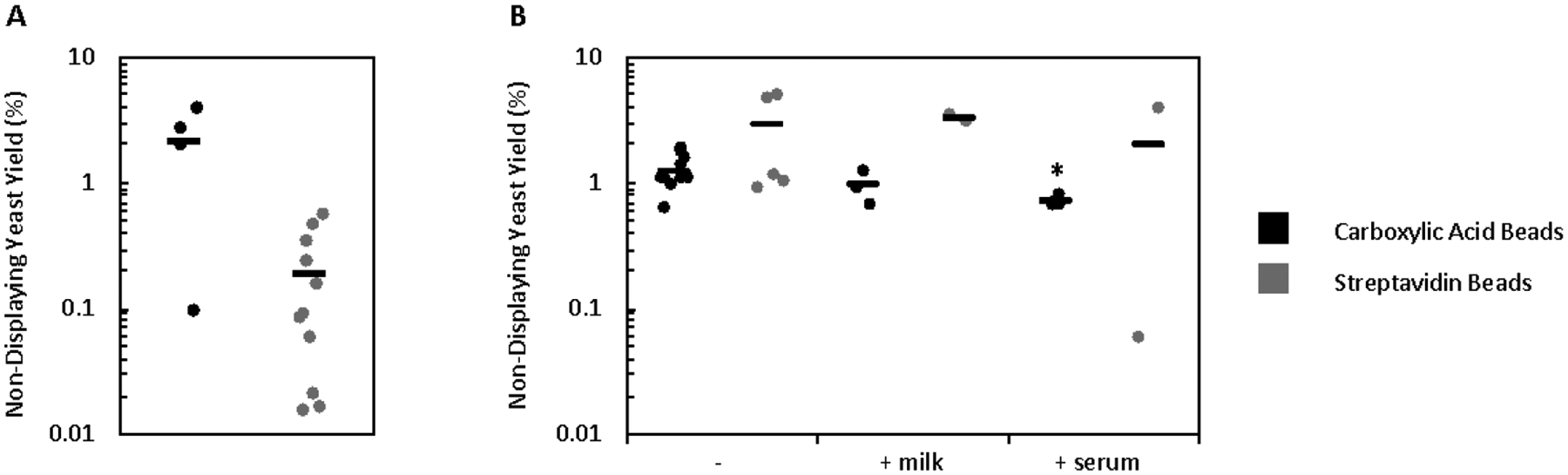
Non-displaying yeast show nonspecific interactions with both beads and bead-cell conjugates. Non-displaying yeast were incubated with (A) bare magnetic beads or (B) bead-A431 conjugates and washed twice with PBSACM or alternate buffers prior to quantifying yield. A (*) indicates p < 0.05 (with Bonferroni correction) relative to washing with PBSACM.
Cell Suspension Panning Enriches EGFR-Binding Fibronectins with High Throughput and Capacity
One of the potential benefits of the suspension cell panning approach is to efficiently process a large library of yeast cells. To assess the throughput of the bead-cell conjugate system, increasing amounts of non-displaying yeast were incubated with bead-cell conjugates while holding the number of binding yeast constant at 106, which mimics an initial sort of a naïve library with a moderate number of binders within a large pool of non-functional clones. The highest tested condition (1010 non-displaying yeast) showed no significant difference in the yield of binding yeast compared to 100-fold fewer non-displaying yeast (16 ± 3% vs. 15 ± 2%, p = 0.67; Figure 4A). Moreover, the yield of non-displaying yeast decreased slightly, thereby resulting in nominally increased enrichment given constant binder yield (Figure 4B). This result of up to 1010 yeast panned per 106 target mammalian cells compares favorably to adherent panning recommendations which limit the number of panned yeast to 5×107 yeast per cm2, or about 4×108 yeast per 106 mammalian cells, given an estimate of 1.2×106 cells per well in a 6-well plate.56 This is comparable to magnetic bead selections with soluble recombinant target.19 The capacity for recovering binding yeast was also tested by incubating increasing amounts of binding yeast with bead-cell conjugates while holding the number of non-displaying yeast constant. While a 10-fold increase in the amount of binding yeast resulted in a similar yield, the use of 108 binding yeast did have a significantly decreased yield relative to baseline (9 ± 2% vs. 2.1 ± 1.2%, p = 0.004; Figure 4C), meaning that there are limitations to panning populations containing greater than 107 binding yeast per 106 input mammalian cells. While there is a significant increase in the number of binding yeast recovered across all tested conditions, the reduction in yield at 108 input binding yeast may indicate that a recovery of roughly 106 binding yeast per 106 input mammalian cells may be at or approaching the system capacity (Figure 4D).
Figure 4.
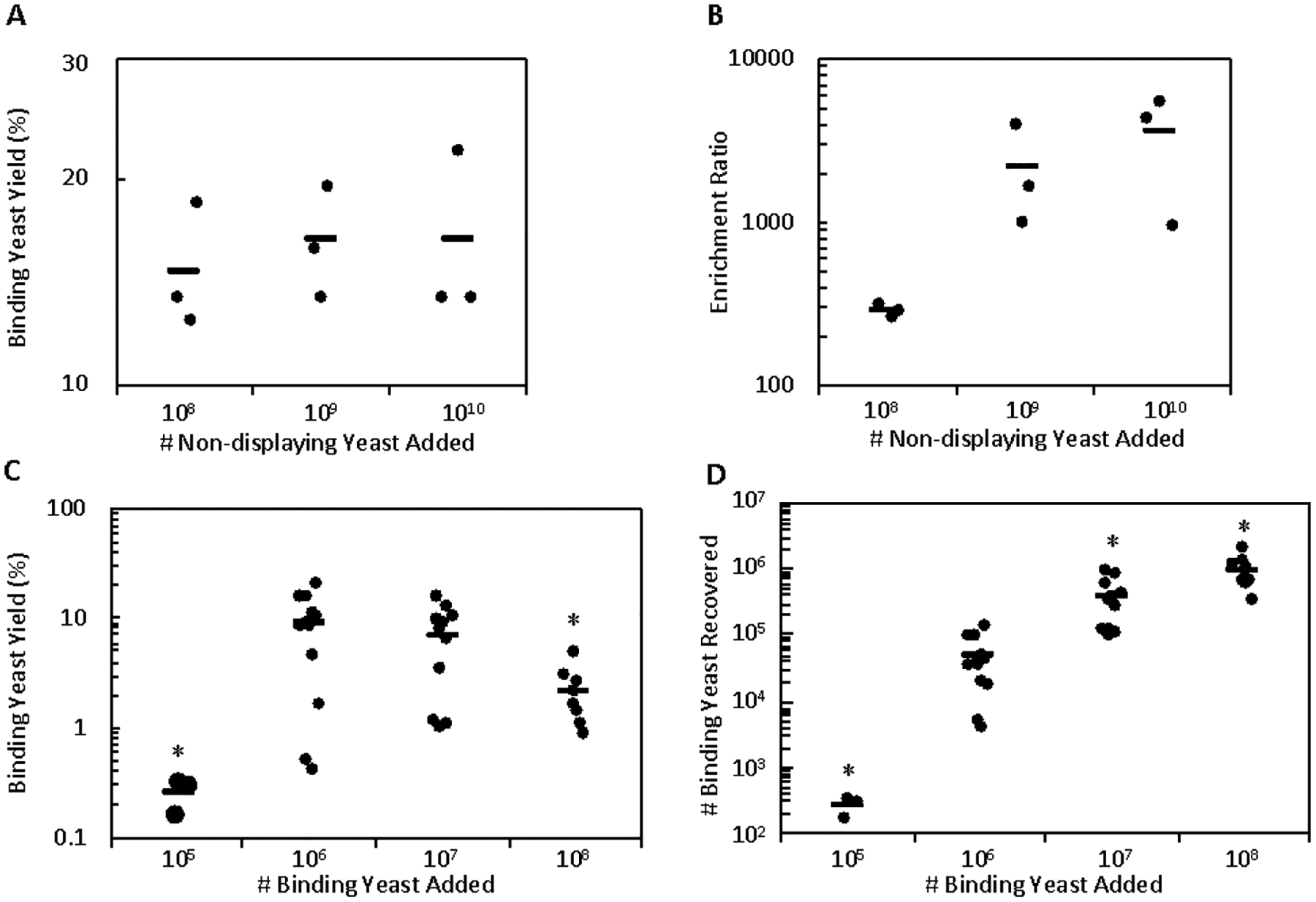
Cell suspension panning shows high binding yeast capacity and throughput. (A and B) Mixtures of 106 binding yeast and increasing numbers of non-displaying yeast were sorted against bead-A431 conjugates and the yield of binding yeast and enrichment ratio was quantified. (C and D) Mixtures of 108 non-displaying yeast and increasing numbers of binding yeast were sorted against bead-A431 conjugates and the yield of binding yeast and absolute number of binding yeast was quantified. A (*) indicates p < 0.05 (with Bonferroni correction) compared to the baseline conditions. The reduced yield of the 105 binding yeast conditions is hypothesized to be due to a small nonspecific loss of yeast due to binding to the tube surface that is negligible at all other conditions.
Cell Suspension Panning Provides Higher Binder Yield and Lower, but Effective, Enrichment Relative to Adherent Cell Panning
To compare the efficacy of this system relative to traditional adherent cellular panning, 18 replicates of both cell suspension and adherent cell panning were conducted in parallel over three days using the highly EGFR-expressing cell line A431 and high affinity E6.2.6’ ligand with carboxylic acid beads. Compared to adherent panning, cell suspension panning showed higher yield of binding yeast (12 ± 3% vs. 3.4 ± 0.4%, p = 0.005; Figure 5A) and lower yield of non-displaying yeast (0.043 ± 0.009% vs. 0.15 ± 0.03%, p = 0.001; Figure 5B), resulting in an increase in enrichment (500 ± 200 vs. 160 ± 50, p = 0.01; Figure 5C).
Figure 5.
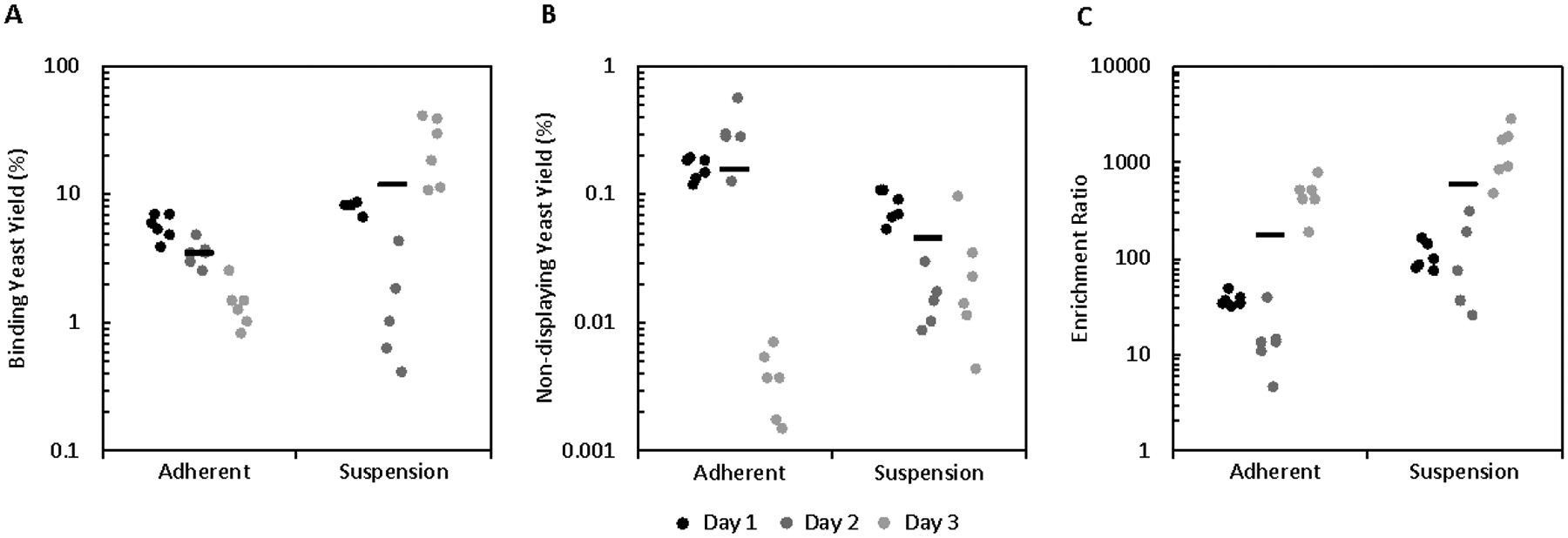
Initial cell suspension panning data demonstrates enhanced yield and enrichment compared to adherent cell panning in a high affinity, high target expression system. Mixtures of yeast displaying E6.26.’ and non-displaying yeast were sorted in suspension and adherently in parallel using bead-A431 conjugates as a pulldown agent. (A) Binding yeast yield, (B) non-displaying yeast yield, and (C) enrichment ratio were quantified.
Initially, this data suggested that, while variable, suspension cell panning could provide more effective enrichment compared to traditional adherent panning. However, the exceptionally low yield of non-displaying yeast in the cell suspension panning system contradicted our earlier results (Figures 2 and 3). To investigate this discrepancy, two additional sets of experiments were run using both carboxylic acid and streptavidin beads. These results were consistent with the earlier analysis, with cell suspension panning showing an increase in the yield of binding yeast (9.6 ± 0.6% vs. 3.2 ± 0.3% with carboxylic acid beads, p < 0.0001; 9 ± 1% vs. 3.8 ± 0.2% with streptavidin beads, p = 0.002; Figure 6A, D) along with an increase in the yield of non-displaying yeast (1.0 ± 0.1% vs. 0.08 ± 0.01% with carboxylic acid beads, p < 0.0001; 2.0 ± 0.3% vs. 0.051 ± 0.007% with streptavidin beads, p < 0.0001; Figure 6B, E), which results in a decrease in the enrichment (11 ± 2 vs. 60 ± 20 with carboxylic acid beads, p = 0.006; 10 ± 3 vs. 100 ± 20 with streptavidin beads, p < 0.001; Figure 6C, F) relative to adherent cell panning but still allows for enrichment significantly greater than unity (p < 0.001 with carboxylic acid beads, p = 0.0006 with streptavidin beads), enabling practical use of this system.
Figure 6.
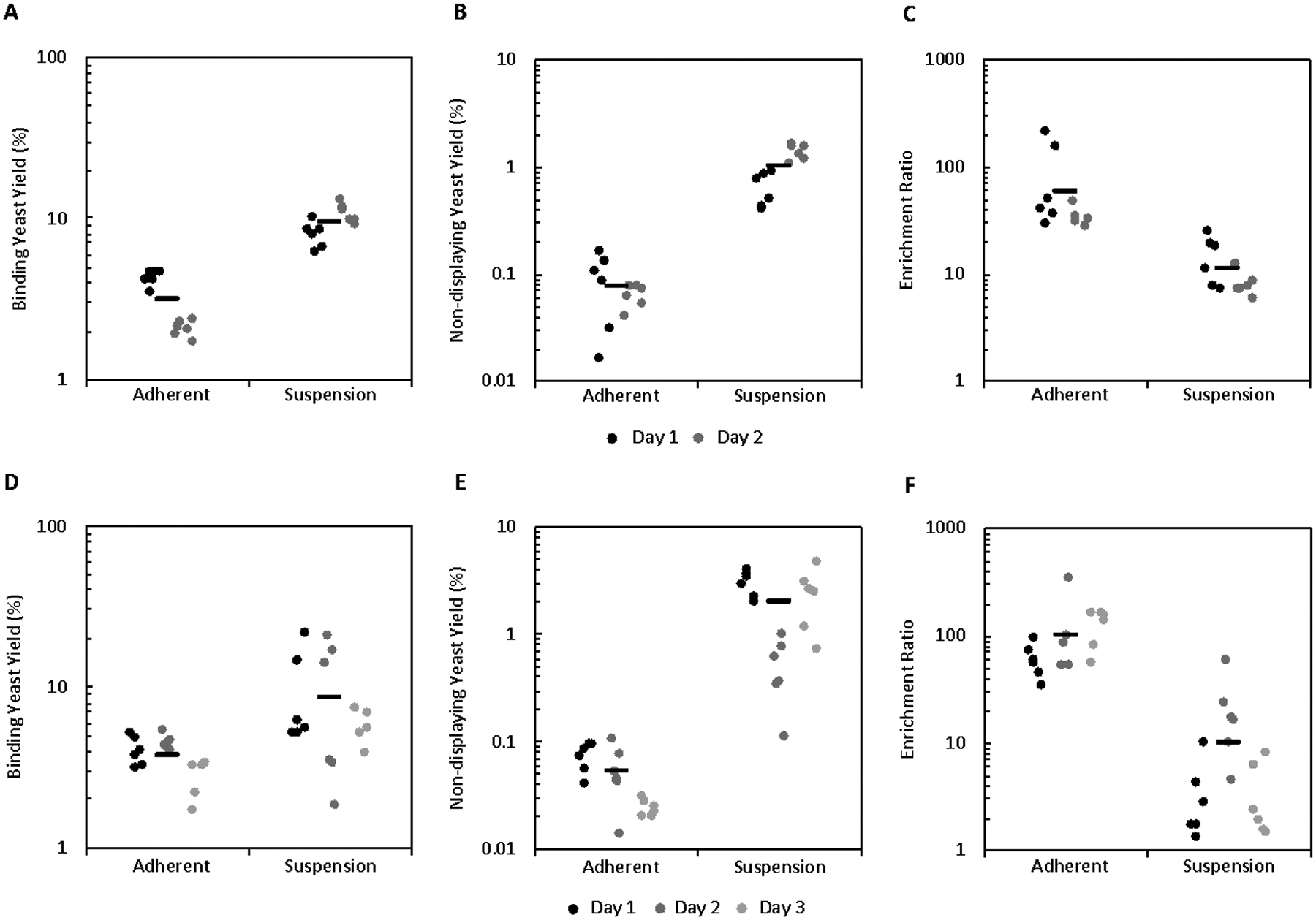
Further cell suspension panning data shows enhanced yield relative to adherent cell panning but does not recapitulate the increased enrichment. Mixtures of yeast displaying E6.26.’ and non-displaying yeast were sorted in suspension and adherently in parallel using bead-A431 conjugates using (A-C) carboxylic acid beads or (D-F) streptavidin beads as a pulldown agent. (A and D) Binding yeast yield, (B and E) non-displaying yeast yield, and (C and F) enrichment ratio were quantified.
The variability regarding the yield of non-displaying yeast across the multiple data sets was unable to be explained. All experiments were conducted by the same researcher, with cells displaying similar EGFR expression from the same master stock, and using the procedures listed herein. Nonetheless, the variability observed with the method is important to note. The results place a lower limit – which is still in the functional regime – on the enrichment while also offering optimism for even stronger performance. The relative bimodality of the non-displaying yield suggests impact of an undefined parameter without adequate experimental control. However, at this time, no speculation regarding this possible parameter is evident.
Cell Suspension Panning Enhances Binder Yield Relative to Adherent Cell Panning Robustly Across Ligand Affinities and Target Expressions, but not to all Targets
The effect of lower ligand affinity and/or cellular target expression on yield and enrichment in cell suspension panning was studied, as well as how this effect compared to adherent panning. Due to the similar results observed in the high affinity, high target expression system these experiments were conducted only in the carboxylic acid bead system. Similar to the high expression system, yeast displaying E6.2.6’ panned on mid-expressing cells MDA-MB-231 (2.9 ± 1.7 ×105 EGFR/cell) showed a higher binding yeast yield compared to adherent panning (2.3 ± 0.5% vs. 0.41 ± 0.09%, p = 0. 0008; Figure 7A) but lower enrichment (2.0 ± 0.3 vs. 8 ± 2, p = 0.006; Figure 7C). Though lower, the enrichment was still significantly greater than unity (p=0.007). Low affinity ligand AASV (KD > 600 nM for EGFR) panned on high-expressing A431 cells yielded more binding yeast than observed for adherent panning (2.0 ± 0.5% vs. 0.017±0.005%, p = 0.001; Figure 7D), but a higher yield of non-displaying yeast resulted in similar enrichment compared to adherent panning (1.5 ± 0.3 vs. 1.7 ± 0.8, p = 0.79; Figure 7F), neither of which was significantly greater than unity (p = 0.053 and p = 0.20, respectively). Thus, weaker affinity ligands remain challenging for cellular panning enrichment.
Figure 7.
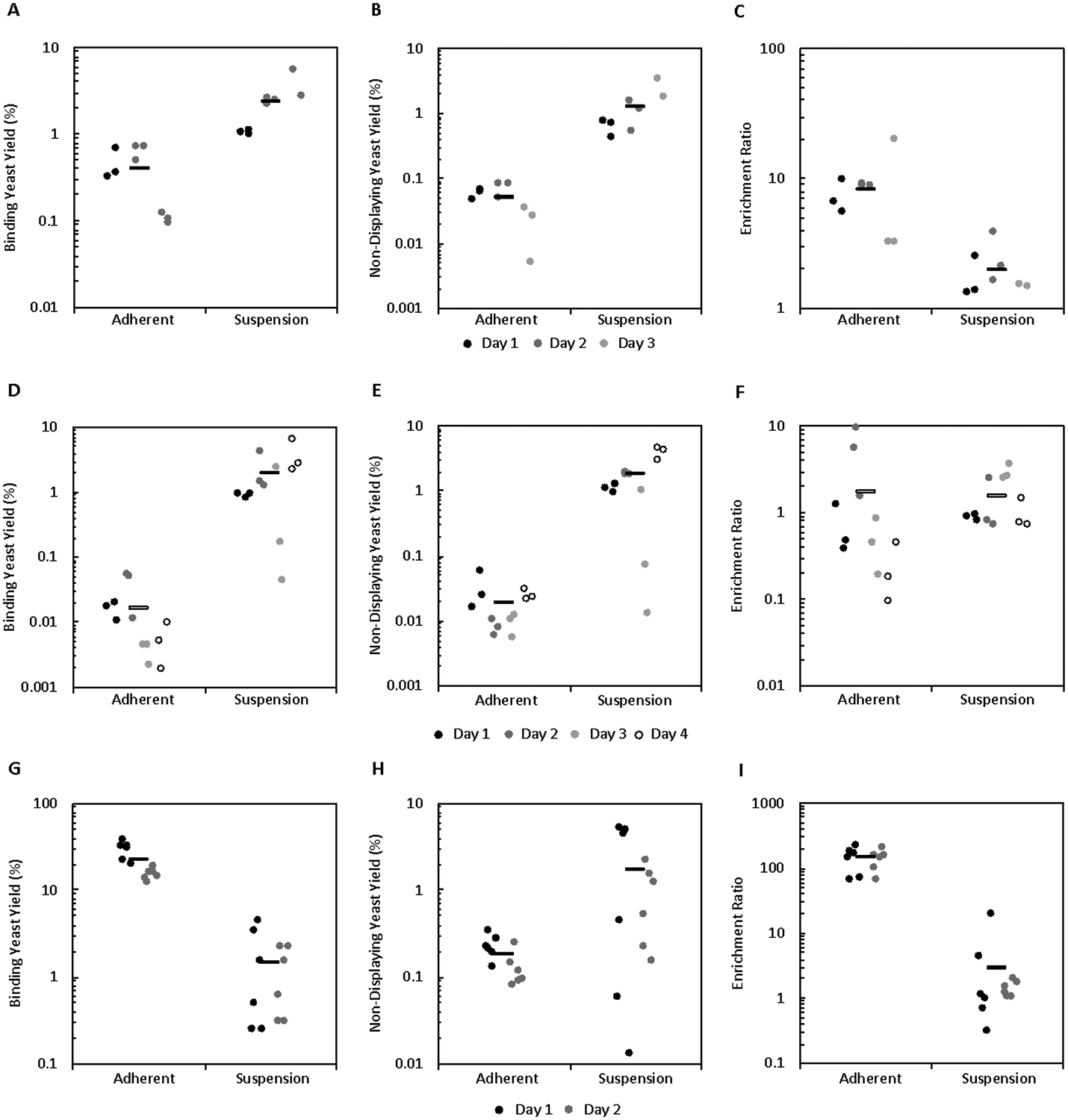
The effect of ligand affinity, target expression, and target/protein type on yield and enrichment. Mixtures of binding and non-binding yeast were sorted in parallel by cell suspension and adherent panning and binding yeast yield, non-displaying yeast yield, and enrichment ratio were quantified. (A-C) Ligand: E6.2.6’ Cell Line: MDA-MB-231 (D-F) Ligand: AASV Cell Line: A431 (G-I) Ligand: AC2 Cell Line MS1-CD276
To test how cell suspension panning compares to adherent panning in a system with an alternative protein scaffold and target, sorts in parallel with a cell line that highly expresses CD276 (MSI-CD276; 1.2 ± 0.1 ×106 CD276/cell) and a mid-affinity affibody ligand AC2 (KD = 310 ± 75 nM for CD276) were conducted. Surprisingly, cell suspension panning displayed both lower binding yeast yield (1.5 ± 0.4% vs. 22 ± 4%, p < 0.0001 Figure 7G) and enrichment (3 ± 1 vs. 140 ± 20, p < 0.0001; Figure 7I) relative to adherent panning and was unable to provide significant effective enrichment (p = 0.11). This result suggests that while cell suspension panning provides a yield advantage to fibronectin domains in an EGFR model system, this might not translate to all protein scaffold or targets.
Discussion
Existing high-throughput methods have successfully isolated binding ligands to soluble ectodomain analogs; however, difficulties translating binding to genuine antigen on target-expressing cells have hampered the clinical development and use of such molecules. The failure of such recombinant soluble ectodomain-based selection methods to generate ligands that bind to full-length, membrane-bound target indicate that these ectodomains may not fully recapitulate protein expressed on the cell surface. Surface display library selection techniques using target-expressing cells have been developed and successfully employed in many cases to overcome translatability issues but could be further improved. FACS-based selections employing target-expressing mammalian cells or cell lysate provide effective enrichment but are limited by throughput and lack avidity for weak affinity ligand enrichment. Adherent cell panning, on the other hand, provides high-throughput, avid selections with effective enrichment in a variety of cases; although prior work with EGFR-binding fibronectins indicate limited effectiveness in weak affinity and/or low expression conditions, motivating further improvement.47,56 Likewise, the effective selection of binding ligands from a phage display library using cells in suspension and anecdotal evidence of cell suspension panning campaigns succeeding in cases where adherent panning failed provide motivation to adapt these methods to yeast surface display libraries.65,66 This adaptation, utilizing target-expressing mammalian cells immobilized to magnetic beads, was compared to adherent cell panning and displayed yield advantages and functional enrichment using a high affinity binder on cells with both high and mid target expression and provided only a yield advantage using a low affinity binder on cells with high target expression.
The conjugation of A431 cells to both carboxylic acid- and streptavidin-functionalized magnetic beads was accomplished in sufficient yield to produce a functional number of conjugates with limited optimization. No extensive networking of cells and beads was observed, and the target expression was essentially maintained across both conjugation methods. In both cases, it remains possible that higher concentrations or stoichiometric excess of beads could enhance conjugate yield. Additionally, while the 20,000 Da PEG biotin employed aided in enhancing conjugate yield, the optimal linker length to enable a biotinylated cell-bead linkage was not studied. Combined, this provides several avenues for further improving conjugation yield.
Cell suspension panning provided functional enrichment in an EGFR system using a high affinity binder and a high expression cell line with increased throughput relative to adherent panning. The high yield and relative insensitivity of non-displaying yeast to washing was apparently due to a combination of bead-yeast and mammalian cell-yeast interactions that were not appreciably mitigated by buffers that have been previously used to screen non-specific interactions. This limits the possible enrichment of the system which, while still functional, could be improved by further depletion of non-displaying yeast. While not tested in this study, it is possible that the use of mild surfactants or a pre-sort against bare beads or target-negative bead-cell conjugates could help mitigate these interactions.
The first set of experiments performed using carboxylic acid beads in the high affinity, high target expression system showed a consistent, lower yield of non-displaying yeast that was not replicated across any other experiment, which all showed a consistent, moderate yield of non-binding yeast. Despite not knowing the precise parameter inducing this bimodality in the data, the observed variability provides a lower bound for performance with the potential that further optimization can promote even higher enrichment.
Follow-up experiments conducted with a high affinity ligand on high and mid target-expression cell lines and a low affinity ligand on a high target-expression cell line showed a significant increase in the yield of binding yeast across all conditions compared to adherent panning. In addition, all conditions with the exception of low affinity, high target-expression produced functional enrichment. This indicates that, while enrichment of weaker affinity ligands remains challenging in cell panning selections, cell suspension panning is a practical alternative selection across a wide range of ligand affinities and target expressions.
When testing cell suspension panning using a mid-affinity CD276-binding affibody and a high CD276-expressing cell line, we found that cell suspension panning failed to provide the yield advantage or functional enrichment observed in the EGFR system. It is unclear whether the protein scaffold, cellular target, or another uncontrolled parameter is responsible for this reversal in the observed trend of binding yeast yield, but this result indicates that cell suspension panning may not be appropriate for all protein scaffolds or targets.
While expression was measured biweekly over the course of this study and observed to remain consistent, it is possible that the use of pre-sort analysis of mammalian cell target expression could yield additional insight regarding the inter-day variability observed in cell suspension panning. Target expression on mammalian cells has been previously shown to strongly impact the yield of binding yeast in adherent mode panning.47 Additionally, the use of sequential sorts is expected to also aid in the reduction of inter-day variability through the averaging of enrichment values.
Conclusion
In conclusion, the above work presents and characterizes a new system for translatable ligand selection using suspended mammalian cells immobilized on magnetic beads. Both streptavidin and carboxylic acid beads can be used to effectively conjugate mammalian cells without forming large aggregates or masking native cell expression. Optimization of washing conditions enabled a system that can effectively enrich binding yeast relative to non-displaying yeast; however, an unexplained high background of non-displaying yeast is present that was not able to be reduced with increased washing or alternate buffers to screen nonspecific interactions. While these interactions seem to be both yeast-bead and yeast-cell driven, their exact cause is still unknown. Despite this, cell suspension panning provides practical enrichment to high affinity ligands on cell lines expressing 105-106 targets per cell, while low affinity ligands fail to be effectively enriched due to the increased background. Cell suspension panning also compared favorably to traditional adherent panning with a significant yield advantage in both high and low affinity ligand cases, as well as increased throughput allowing up to 1010 cells to be sorted in a single tube. However, cell suspension panning failed to provide a benefit in a CD276 system, indicating that this panning approach may not be suitable for all targets or protein scaffolds. Ultimately, the findings of this study provide an alternate, higher throughput method for translatable selection to increase the efficiency of translatable ligand discovery and motivating further study into the limiting factors of this mode of selection.
Materials and Methods
Cells and Cell Culture
MDA-MB-231 were provided by Professor Jayanth Panyam (Department of Pharmaceutics, University of Minnesota – Twin Cities). A431 were provided by Professor Daniel Vallera (Department of Therapeutic Radiology, University of Minnesota – Twin Cities). Miles Sven 1 cells stably transfected to express human CD276 (MS1-CD276)69 were provided by Juergen Willmann (Department of Radiology, Stanford University). All cell lines were grown with DMEM containing 4.5 g/L D-glucose, sodium pyruvate, and L-glutamine and supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) 10,000 U/mL penicillin-streptomycin. All cell lines were stored in an incubator at 37 °C in a humidified atmosphere with 5% CO2.
Yeast surface display was performed essentially as previously described.70 Expression plasmids, detailed below, were transformed into Saccharomyces cerevisiae yeast strain EBY100 by EZ-Yeast Transformation (Zymo Research, Irvine, CA). Yeast harboring expression plasmids were grown in SD-CAA medium (20.0 g/L dextrose, 16.8 g/L sodium citrate dihydrate, 6.7 g/L yeast nitrogen base, 5.0 g/L casamino acids, and 3.9 g/L citric acid in deionized H2O) at 30 °C with shaking. Protein expression was induced by transferring yeast cells in logarithmic phase (OD600nm<6) into SG-CAA medium (19.0 g/L galactose, 10.2 g/L sodium phosphate dibasic heptahydrate, 8.6 g/L sodium phosphate monobasic monohydrate, 6.7 g/L yeast nitrogen base, 5.0 g/L casamino acids, and 1.0 g/L dextrose in deionized H2O) and growing at 20 °C overnight. EBY100 without plasmid were grown in YPD medium (20.0 g/L peptone, 20.0 g/L dextrose, and 10.0 g/L yeast extract in deionized H2O) at 30 °C with shaking.
Expression Plasmids
The pCT-80 plasmid was used as the expression vector for yeast surface display on the C-terminus of Aga2p. The vector encodes for Aga2p followed by an 80-amino acid linker (comprising a Factor Xa cleavage site, an HA epitope, a proline/alanine/serine peptide based on the PAS#1 motif,71 and a glycine-rich peptide), the ligand, and a C-terminal Myc epitope. In essence, this is the classic pCT yeast surface display plasmid13 with the addition of the 40-amino acid proline/alanine/serine peptide (as such, it was previously named pCT-4047). Fibronectin domain genes were cloned into pCT-80 vector by NheI and BamHI restriction sites: EGFR-binding E6.2.6’26 and E6.2.6’ AASV47, and CD276-binding affibody clone33 AC2.
Receptor Expression Quantification
Polystyrene beads with known quantities of immobilized anti-mouse IgG (Bangs Laboratories, Inc., Fishers, IN) were used to construct a calibration curve from which the EGFR and CD276 expression of cell lines was quantified. Beads or cells were labelled with either mouse anti-EGFR clone ab30 (4 μg/mL) or mouse anti-CD276 clone 185504 (4 μg/mL) for 30 minutes at 4 °C. Beads or cells were washed once with phosphate-buffered saline with 0.1% (w/v) bovine serum albumin, 1 mM CaCl2 and 0.5 mM Mg2SO4 (PBSACM) and pelleted at 500g for 3 minutes. The beads or cells were then labelled with goat anti-mouse Alexa Fluor 647 conjugate (Life Technologies, Carlsbad, CA) for 30 minutes at 4 °C, washed once with PBSACM, and again pelleted at 500 g for 3 minutes. Fluorescence was analyzed by flow cytometry using an Accuri C6 Plus (BD Biosciences, San Jose, CA).
Mammalian Cell – Magnetic Bead Conjugation
Mammalian cells were grown to approximately 70–90% confluency in 75 cm2 tissue culture-treated flasks. The culture medium was removed, and the cells were washed once with 5 mL of PBS. The cells were then detached by trypsin-EDTA treatment for 10 minutes, followed by the addition of culture medium with serum. The detached cells were then pelleted at 500g for 3 minutes. The trypsin-containing medium was removed, and the cells were resuspended in fresh culture medium for counting using a Countess II FL (Thermo Fisher Scientific). The cells were pelleted at 500g for 3 minutes at 4 °C and washed twice with ice cold PBS before being pelleted again and the supernatant removed.
For streptavidin bead conjugation, washed cells were resuspended in 1 mL of a 9 μM solution of 20 kDa biotin-PEG-SVA (Laysan Bio Inc., Arab, AL) in PBS per 106 mammalian cells and rotated for 30 minutes at room temperature. The biotinylated cells were washed five times with ice cold PBSACM to quench and remove excess biotin prior to incubation with an equimolar amount of streptavidin-coated Dynabeads (Thermo Fisher Scientific, Cat: 11205D) in 50 μL of PBSACM at 4 °C for an hour. The bead-cell conjugates were then placed on a DynaMag-2 magnet (Thermo Fisher Scientific, Cat: 12321D) for two minutes to capture all conjugated cells and washed once with PBSACM to remove any unconjugated cells.
For carboxylic acid bead conjugation, carboxylic-acid-coated Dynabeads (Thermo Fisher Scientific, Cat: 14305D) were mixed with 200 μL of a 10 mM sodium hydroxide solution per 106 beads and rotated at room temp for 10 minutes. The beads were placed on a Dynamag-2 magnet for 2 minutes and washed twice with 200 μL of 4 °C deionized water per 106 beads, then resuspended in 200 μL of a 4 °C, 50 mg/mL 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide solution per 106 beads. This mixture was allowed to rotate at room temperature for 30 minutes before being placed again on the magnet and washed once with 200 μL of 4 °C deionized water and twice with 200 μL of 100 mM MES. An equimolar amount of activated beads was added to the pelleted cells and mixed thoroughly prior to rotating at room temperature for 30 minutes. The bead-cell conjugates were placed on the magnet and washed once with 500 μL of 0.05 M Tris and once with 500 μL of PBSACM.
For both types of bead-cell conjugation, reaction yield was quantified by flow cytometry on an Accuri C6 Plus to count the number of cells in both the pre- and post-conjugation mixtures.
Cell Suspension Panning
Yeast mixtures containing 1×108 plasmidless EBY100 and 1×106 yeast harboring ligand display plasmids were washed in PBSACM and added to a 1.7 mL tube containing 106 mammalian cells conjugated to either streptavidin or carboxylic-acid-coated beads in 1 mL of ice cold PBSACM. This mixture was rotated for 15 minutes at 4 °C. The mixture was placed on a magnet, unbound yeast were aspirated, and bead-cell conjugates were washed twice with 1 mL of ice cold PBSACM. The yield of plasmid-harboring yeast was quantified by dilution plating on SD-CAA plates, while the yield of total yeast was quantified by dilution plating on YPD. Enrichment ratio was calculated as the yield of plasmid-harboring yeast divided by the yield of plasmidless yeast.
Adherent Mammalian Cell Panning
Adherent cell panning selections were carried out essentially as described.47 Mammalian cells were grown in 12-well plates to approximately 70–90% confluency. The culture medium was aspirated, and the cells were washed once with ice cold PBSACM. Yeast mixtures containing 1×108 plasmidless EBY100 and 1×106 yeast harboring ligand display plasmids were washed in PBSACM and added dropwise to each well in 1 mL of ice cold PBSACM. Cells were incubated without shaking for 15 minutes at 4 °C and unbound yeast were aspirated. Cells were washed with 1 mL of ice cold PBSACM four times with 25 gentle tilts and 5 nutations and one time with 10 nutations. Bound yeast were removed by scraping cell monolayers and resuspending them in 1 mL of PBSACM. The yield of plasmid-harboring yeast was quantified by dilution plating on SD-CAA plates, while the yield of total yeast was quantified by dilution plating on YPD. Enrichment ratio was calculated as the yield of plasmid-harboring yeast divided by the yield of plasmidless yeast.
Optimization of Washing Conditions
The washing conditions of adherent mammalian cell panning have been optimized in prior work.33,43,47 To determine the optimal number of washes for the cell suspension panning, the baseline conditions described above were repeated while varying the number of washes from one to five. Yield and enrichment were quantified by dilution plating on YPD and SD-CAA plates.
Quantification of System Capacity and Non-binding Yeast Blocking
The capacity of cell suspension panning was determined by conducting sorts using the baseline conditions above while varying the number of initial displaying yeast from 105 to 108. The ability of non-binding yeast to block binding yeast was determined by conducting sorts using the baseline conditions above while varying the number of initial non-displaying yeast from 108 to 1010. In both cases, yield and enrichment were quantified by dilution on YPD and SD-CAA plates.
Statistical Analysis
Washing data and functional enrichment were tested for significance using Welch’s t-test, while comparisons between cell suspension and adherent panning were binned by day and method prior to significance testing by two-way ANOVA with only main effects. Samples containing greater than six replicates were subjected to Grubb’s test to remove potential outliers. All statistics are reported as the mean ± standard error.
Supplementary Material
Acknowledgements
We appreciate research support from the American Cancer Society (130418-RSG-17-110-01-TBG) and the National Institutes of Health (R01 EB023339, R01 EB028274).
Footnotes
Supporting Information
Cell-bead conjugate yield as a function of biotin concentration and incubation time
References
- (1).Stern LA; Case BA; Hackel BJ Alternative Non-Antibody Protein Scaffolds for Molecular Imaging of Cancer. Curr. Opin. Chem. Eng 2013, 2 (4), 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Banta S; Dooley K; Shur O Replacing Antibodies: Engineering New Binding Proteins. Annu. Rev. Biomed. Eng 2013, 15, 93–113. [DOI] [PubMed] [Google Scholar]
- (3).Binz HK; Amstutz P; Plückthun A Engineering Novel Binding Proteins from Nonimmunoglobulin Domains. Nat. Biotechnol 2005, 23 (10), 1257–1268. [DOI] [PubMed] [Google Scholar]
- (4).Leader B; Baca QJ; Golan DE Protein Therapeutics: A Summary and Pharmacological Classification. Nat. Rev. Drug Discov 2008, 7 (1), 21–39. [DOI] [PubMed] [Google Scholar]
- (5).Kamta J; Chaar M; Ande A; Altomare DA; Ait-Oudhia S Advancing Cancer Therapy with Present and Emerging Immuno-Oncology Approaches. Front. Oncol 2017, 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).James ML; Gambhir SS A Molecular Imaging Primer: Modalities, Imaging Agents, and Applications. Physiol. Rev 2012, 92 (2), 897–965. [DOI] [PubMed] [Google Scholar]
- (7).Dijkstra S; Mulders PFA; Schalken JA Clinical Use of Novel Urine and Blood Based Prostate Cancer Biomarkers: A Review. Clin. Biochem 2014, 47 (10–11), 889–896. [DOI] [PubMed] [Google Scholar]
- (8).Husseinzadeh N Status of Tumor Markers in Epithelial Ovarian Cancer Has There Been Any Progress? A Review. Gynecol. Oncol 2011, 120 (1), 152–157. [DOI] [PubMed] [Google Scholar]
- (9).Yotsukura S; Mamitsuka H Evaluation of Serum-Based Cancer Biomarkers: A Brief Review from a Clinical and Computational Viewpoint. Crit. Rev. Oncol. Hematol 2015, 93 (2), 103–115. [DOI] [PubMed] [Google Scholar]
- (10).Desai AN; Jere A Next-Generation Sequencing for Cancer Biomarker Discovery. Next Gener. Seq. Cancer Res 2015, 2, 103–125. [Google Scholar]
- (11).Surinova S; Schiess R; ttenhain R; Cerciello F; Wollscheid B; Aebersold R On the Development of Plasma Protein Biomarkers. J. Proteome Res 2011, 10 (1), 5–16. [DOI] [PubMed] [Google Scholar]
- (12).Crutchfield CA; Thomas SN; Sokoll LJ; Chan DW Advances in Mass Spectrometry-Based Clinical Biomarker Discovery. Clin. Proteomics 2016, 13 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Boder ET; Wittrup KD Yeast Surface Display for Screening Combinatorial Polypeptide Libraries. Nat. Biotechnol 1997, 15 (6), 553–557. [DOI] [PubMed] [Google Scholar]
- (14).Hackel BJ; Wittrup D Yeast Surface Display in Protein Engineering and Analysis. Protein Eng. Handbook, Vol. 1 Vol. 2 2011, 621–648. [Google Scholar]
- (15).Tillotson BJ; Lajoie JM; Shusta EV Yeast Display-Based Antibody Affinity Maturation Using Detergent-Solubilized Cell Lysates. Methods Mol. Biol 2015, 1319, 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Smith GP Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science (80-. ) 1985, 228 (4705), 1315–1317. [DOI] [PubMed] [Google Scholar]
- (17).Bratkovic T Progress in Phage Display: Evolution of the Technique and Its Application. Cell. Mol. Life Sci 2010, 67 (5), 749–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Even-Desrumeaux K; Chames P Phage Display and Selections in Cells. Methods Mol. Biol 2012, 907, 225–235. [DOI] [PubMed] [Google Scholar]
- (19).Ackerman M; Levary D; Tobon G; Hackel B; Orcutt KD; Wittrup KD Highly Avid Magnetic Bead Capture: An Efficient Selection Method for de Novo Protein Engineering Utilizing Yeast Surface Display. Biotechnol. Prog 2009, 25 (3), 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Orlova A; Magnusson M; Eriksson TLJ; Nilsson M; Larsson B; Höiden-Guthenberg I; Widström C; Carlsson J; Tolmachev V; Ståhl S; et al. Tumor Imaging Using a Picomolar Affinity HER2 Binding Affibody Molecule. Cancer Res 2006, 66 (8), 4339–4348. [DOI] [PubMed] [Google Scholar]
- (21).Horiya S; Bailey JK; Krauss IJ Directed Evolution of Glycopeptides Using MRNA Display. Methods Enzymol 2017, 597, 83–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Dreier B; Plückthun A Rapid Selection of High-Affinity Binders Using Ribosome Display. Methods Mol. Biol 2012, 805, 261–286. [DOI] [PubMed] [Google Scholar]
- (23).Doerner A; Rhiel L; Zielonka S; Kolmar H Therapeutic Antibody Engineering by High Efficiency Cell Screening. FEBS Lett 2014, 588 (2), 278–287. [DOI] [PubMed] [Google Scholar]
- (24).Beerli RR; Bauer M; Buser RB; Gwerder M; Muntwiler S; Maurer P; Saudan P; Bachmann MF Isolation of Human Monoclonal Antibodies by Mammalian Cell Display. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (38), 14336–14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).VanAntwerp JJ; Wittrup KD Fine Affinity Discrimination by Yeast Surface Display and Flow Cytometry. Biotechnol. Prog 2000, 16 (1), 31–37. [DOI] [PubMed] [Google Scholar]
- (26).Hackel BJ; Ackerman ME; Howland SW; Wittrup KD Stability and CDR Composition Biases Enrich Binder Functionality Landscapes. J. Mol. Biol 2010, 401 (1), 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Friedman M; Nordberg E; Höidén-Guthenberg I; Brismar H; Adams GP; Nilsson FY; Carlsson J; Ståhl S Phage Display Selection of Affibody Molecules with Specific Binding to the Extracellular Domain of the Epidermal Growth Factor R Eceptor. Protein Eng. Des. Sel 2007, 20 (4), 189–199. [DOI] [PubMed] [Google Scholar]
- (28).Horak E; Heitner T; Robinson MK; Simmons HH; Garrison J; Russeva M; Furmanova P; Lou J; Zhou Y; Yuan Q-A; et al. Isolation of ScFvs to In Vitro Produced Extracellular Domains of EGFR Family Members. Cancer Biother. Radiopharm 2005, 20 (6), 603–613. [DOI] [PubMed] [Google Scholar]
- (29).Kruziki MA; Sarma V; Hackel BJ Constrained Combinatorial Libraries of Gp2 Proteins Enhance Discovery of PD-L1 Binders. ACS Comb. Sci 2018, 20 (7), 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chan JY; Hackel BJ; Yee D Targeting Insulin Receptor in Breast Cancer Using Small Engineered Protein Scaffolds. Mol. Cancer Ther 2017, 16 (7), 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Woldring DR; Holec PV; Zhou H; Hackel BJ High-Throughput Ligand Discovery Reveals a Sitewise Gradient of Diversity in Broadly Evolved Hydrophilic Fibronectin Domains. PLoS One 2015, 10 (9), 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Woldring DR; Holec PV; Stern LA; Du Y; Hackel BJ A Gradient of Sitewise Diversity Promotes Evolutionary Fitness for Binder Discovery in a Three-Helix Bundle Protein Scaffold. Biochemistry 2017, 56 (11), 1656–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Stern LA; Lown PS; Kobe AC; Abou-Elkacem L; Willmann JK; Hackel BJ Cellular-Based Selections Aid Yeast-Display Discovery of Genuine Cell-Binding Ligands : Targeting Oncology Vascular Biomarker CD276. ACS Comb. Sci 2019, 21 (3), 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Suñé P; Suñé JM; Montoro JB Positive Outcomes Influence the Rate and Time to Publication, but Not the Impact Factor of Publications of Clinical Trial Results. PLoS One 2013, 8 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Baneyx F; Mujacic M Recombinant Protein Folding and Misfolding in Escherichia Coli. Nat. Biotechnol 2004, 22 (11), 1399–1407. [DOI] [PubMed] [Google Scholar]
- (36).Gasser B; Saloheimo M; Rinas U; Dragosits M; Rodríguez-Carmona E; Baumann K; Giuliani M; Parrilli E; Branduardi P; Lang C; et al. Protein Folding and Conformational Stress in Microbial Cells Producing Recombinant Proteins: A Host Comparative Overview. Microb. Cell Fact 2008, 7, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mazzei GJ; Edgerton MD; Losberger C; Lecoanet-Henchoz S; Graber P; Durandy A; Gauchat JF; Bernard A; Allet B; Bonnefoy JY Recombinant Soluble Trimeric CD40 Ligand Is Biologically Active. J. Biol. Chem 1995, 7025–7028. [DOI] [PubMed] [Google Scholar]
- (38).Singer E; Landgraf R; Horan T; Slamon D; Eisenberg D Identification of a Heregulin Binding Site in HER3 Extracellular Domain. J. Biol. Chem 2001, 276 (47), 44266–44274. [DOI] [PubMed] [Google Scholar]
- (39).Gomord V; Faye L Posttranslational Modification of Therapeutic Proteins in Plants. Curr. Opin. Plant Biol 2004, 7 (2), 171–181. [DOI] [PubMed] [Google Scholar]
- (40).Demain AL; Vaishnav P Production of Recombinant Proteins by Microbes and Higher Organisms. Biotechnol. Adv 2009, 27, 297–306. [DOI] [PubMed] [Google Scholar]
- (41).Kruziki MA; Bhatnagar S; Woldring DR; Duong VT; Hackel BJA 45-Amino-Acid Scaffold Mined from the PDB for High-Affinity Ligand Engineering. Chem. Biol 2015, 22 (7), 946–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Cho YK; Shusta EV Antibody Library Screens Using Detergent-Solubilized Mammalian Cell Lysates as Antigen Sources. Protein Eng. Des. Sel 2010, 23 (7), 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Tillotson BJ; Cho YK; Shusta EV Cells and Cell Lysates: A Direct Approach for Engineering Antibodies against Membrane Proteins Using Yeast Surface Display. Methods 2013, 60 (1), 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Jones ML; Alfaleh MA; Kumble S; Zhang S; Osborne GW; Yeh M; Arora N; Hou JJC; Howard CB; Chin DY; et al. Targeting Membrane Proteins for Antibody Discovery Using Phage Display. Sci. Rep 2016, 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Yang Z; Wan Y; Tao P; Qiang M; Dong X; Lin CW; Yang G; Zheng T; Lerner RA A Cell–Cell Interaction Format for Selection of High-Affinity Antibodies to Membrane Proteins. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (30), 14971–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wang XX; Shusta EV The Use of ScFv-Displaying Yeast in Mammalian Cell Surface Selections. J. Immunol. Methods 2005, 304, 30–42. [DOI] [PubMed] [Google Scholar]
- (47).Stern LA; Schrack IA; Johnson SM; Deshpande A; Bennett NR; Harasymiw LA; Gardner MK; Hackel BJ Geometry and Expression Enhance Enrichment of Functional Yeast-Displayed Ligands via Cell Panning. Biotechnol. Bioeng 2016, 113 (11), 2328–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Sachdev SS; Lowman HB; Cunningham BC; Wells JA Phage Display for Selection of Novel Binding Properties. Methods Enzymol 2000, 328, 333–363. [DOI] [PubMed] [Google Scholar]
- (49).Ho M; Pastan I Mammalian Cell Display for Antibody Engineering. Methods Mol. Biol 2009, 525, 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Jones AR; Stutz CC; Zhou Y; Marks JD; Shusta EV Identifying Blood-Brain-Barrier Selective Single-Chain Antibody Fragments. Biotechnol. J 2014, 9 (5), 664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Cai X; Garen A Anti-Melanoma Antibodies from Melanoma Patients Immunized with Genetically Modified Autologous Tumor Cells: Selection of Specific Antibodies from Single-Chain Fv Fusion Phage Libraries. Proc. Natl. Acad. Sci. U. S. A 1995, 92 (14), 6537–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Watters JM; Telleman P; Junghans RP An Optimized Method for Cell-Based Phage Display Panning. Immunotechnology 1997, 3 (1), 21–29. [DOI] [PubMed] [Google Scholar]
- (53).Dangaj D; Lanitis E; Zhao A; Joshi S; Cheng Y; Sandaltzopoulos R; Ra H-J; Danet-Desnoyers G; Powell DJ; Scholler N Novel Recombinant Human B7-H4 Antibodies Overcome Tumoral Immune Escape to Potentiate T-Cell Anti-Tumor Responses. Cancer Res 2013, 73 (15), 4820–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Williams RM; Hajiran CJ; Nayeem S; Sooter LJ Identification of an Antibody Fragment Specific for Androgen-Dependent Prostate Cancer Cells. BMC Biotechnol 2014, 14 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Zorniak M; Clark PA; Umlauf BJ; Cho Y; Shusta EV; Kuo JS Yeast Display Biopanning Identifies Human Antibodies Targeting Glioblastoma Stem-like Cells. Sci. Rep 2017, 7 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Wang XX; Cho YK; Shusta EV Mining a Yeast Library for Brain Endothelial Cell-Binding Antibodies. Nat. Methods 2007, 4 (2), 143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Umlauf BJ; Clark PA; Lajoie JM; Georgieva JV; Bremner S; Herrin BR; Kuo JS; Shusta EV Identification of Variable Lymphocyte Receptors That Can Target Therapeutics to Pathologically Exposed Brain Extracellular Matrix. Sci. Adv 2019, 5 (5), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Xu MY; Xu XH; Chen GZ; Deng XL; Li J; Yu XJ; Chen MZ Production of a Human Single-Chain Variable Fragment Antibody against Esophageal Carcinoma. World J. Gastroenterol 2004, 10 (18), 2619–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Shen Y; Yang X; Dong N; Xie X; Bai X; Shi Y Generation and Selection of Immunized Fab Phage Display Library against Human B Cell Lymphoma. Cell Res 2007, 17 (7), 650–660. [DOI] [PubMed] [Google Scholar]
- (60).Eisenhardt SU; Schwarz M; Bassler N; Peter K Subtractive Single-Chain Antibody (Scfv) Phagedisplay: Tailoring Phage-Display for High Specificity against Function-Specific Conformations of Cell Membrane Molecules. Nat. Protoc 2007, 2 (12), 3063–3073. [DOI] [PubMed] [Google Scholar]
- (61).Giordano RJ; Cardó-Vila M; Lahdenranta J; Pasqualini R; Arap W Biopanning and Rapid Analysis of Selective Interactive Ligands. Nat. Med 2001, 7 (11), 1249–1253. [DOI] [PubMed] [Google Scholar]
- (62).Alfaleh M; Jones M; Howard C; Mahler S Strategies for Selecting Membrane Protein-Specific Antibodies Using Phage Display with Cell-Based Panning. Antibodies 2017, 6 (10), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Yoon H; Song JM; Ryu CJ; Kim YG; Lee EK; Kang S; Kim SJ An Efficient Strategy for Cell-Based Antibody Library Selection Using an Integrated Vector System. BMC Biotechnol 2012, 12 (62), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Ridgway JBB; Ng E; Kern JA; Lee J; Brush J; Goddard A; Carter P Identification of a Human Anti-CD55 Single-Chain Fv by Subtractive Panning of a Phage Library Using Tumor and Nontumor Cell Lines. Cancer Res 1999, 59 (11), 2718–2723. [PubMed] [Google Scholar]
- (65).Keller T; Kalt R; Raab I; Schachner H; Mayrhofer C; Kerjaschki D; Hantusch B Selection of ScFv Antibody Fragments Binding to Human Blood versus Lymphatic Endothelial Surface Antigens by Direct Cell Phage Display. PLoS One 2015, 10 (5), 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Hoogenboom HR; Lutgerink JT; Pelsers MMAL; Rousch MJMM; Coote J; Van Neer N; De Bruïne A; Van Nieuwenhoven FA; Glatz JFC; Arends JW Selection-Dominant and Nonaccessible Epitopes on Cell-Surface Receptors Revealed by Cell-Panning with a Large Phage Antibody Library. Eur. J. Biochem 1999, 260 (3), 774–784. [DOI] [PubMed] [Google Scholar]
- (67).Neurauter AA; Bonyhadi M; Lien E; Nøkleby L; Ruud E; Camacho S; Aarvak T Cell Isolation and Expansion Using Dynabeads. Adv. Biochem. Eng. Biotechnol 2007, 106, 41–73. [DOI] [PubMed] [Google Scholar]
- (68).Yeung YA; Wittrup KD Quantitative Screening of Yeast Surface-Displayed Polypeptide Libraries by Magnetic Bead Capture. Biotechnol. Prog 2002, 18 (2), 212–220. [DOI] [PubMed] [Google Scholar]
- (69).Lutz AM; Bachawal SV; Drescher CW; Pysz MA; Willmann JK; Gambhir SS Ultrasound Molecular Imaging in a Human CD276 Expression-Modulated Murine Ovarian Cancer Model. Clin. Cancer Res 2014, 20 (5), 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Chen TF; De Picciotto S; Hackel BJ; Wittrup KD Engineering Fibronectin-Based Binding Proteins by Yeast Surface Display. Methods Enzymol 2013, 523, 303–326. [DOI] [PubMed] [Google Scholar]
- (71).Schlapschy M; Binder U; Börger C; Theobald I; Wachinger K; Kisling S; Haller D; Skerra A PASylation: A Biological Alternative to PEGylation for Extending the Plasma Half-Life of Pharmaceutically Active Proteins. Protein Eng. Des. Sel 2013, 26 (8), 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


