Abstract
Background:
Few epigenome-wide association studies (EWAS) on air pollutants exist, and none have been done on transportation noise exposures, which also contribute to environmental burden of disease.
Objective:
We performed mutually independent EWAS on transportation noise and air pollution exposures.
Methods:
We used data from two time points of the Swiss Cohort Study on Air Pollution and Lung and Heart Diseases in Adults (SAPALDIA) from 1,389 participants contributing 2,542 observations. We applied multiexposure linear mixed-effects regressions with participant-level random intercept to identify significant Cytosine-phosphate-Guanine (CpG) sites and differentially methylated regions (DMRs) in relation to 1-y average aircraft, railway, and road traffic day-evening-night noise (Lden); nitrogen dioxide (); and particulate matter (PM) with aerodynamic diameter (). We performed candidate (CpG-based; cross-systemic phenotypes, combined into “allostatic load”) and agnostic (DMR-based) pathway enrichment tests, and replicated previously reported air pollution EWAS signals.
Results:
We found no statistically significant CpGs at false discovery rate . However, 14, 48, 183, 8, and 71 DMRs independently associated with aircraft, railway, and road traffic Lden; ; and , respectively, with minimally overlapping signals. Transportation Lden and air pollutants tendentially associated with decreased and increased methylation, respectively. We observed significant enrichment of candidate DNA methylation related to C-reactive protein and body mass index (aircraft, road traffic Lden, and ), renal function and “allostatic load” (all exposures). Agnostic functional networks related to cellular immunity, gene expression, cell growth/proliferation, cardiovascular, auditory, embryonic, and neurological systems development were enriched. We replicated increased methylation in cg08500171 () and decreased methylation in cg17629796 ().
Conclusions:
Mutually independent DNA methylation was associated with source-specific transportation noise and air pollution exposures, with distinct and shared enrichments for pathways related to inflammation, cellular development, and immune responses. These findings contribute in clarifying the pathways linking these exposures and age-related diseases but need further confirmation in the context of mediation analyses. https://doi.org/10.1289/EHP6174
Introduction
Transportation-related noise (including road traffic, railway, and aircraft noise) and air pollution [including nitrogen dioxide () and particulate matter (PM) with aerodynamic diameter ()] both make the greatest contribution to the global environmental burden of disease (Fritschi et al. 2011; Hänninen et al. 2014; Vienneau et al. 2015). Both exposure groups have been linked to cross-systemic phenotypes, including respiratory (Adam et al. 2015; Eze et al. 2018; Hoek et al. 2013), cardiovascular (Beelen et al. 2014; Fiorito et al. 2018; Foraster et al. 2017; Heritier et al. 2019; Kempen et al. 2018), and metabolic diseases (Eze et al. 2015; Zare Sakhvidi et al. 2018); cancers (Andersen et al. 2018; Hegewald et al. 2017; Raaschou-Nielsen et al. 2013); and neurological disturbances (Clark and Paunovic 2018; Stafoggia et al. 2014; Zhang et al. 2018). The potential mechanisms linking air pollution and these phenotypes include inflammatory, immune, and oxidative stress responses following inhalation (Münzel et al. 2017a), whereas noise is thought to act through annoyance reactions, sleep disturbances, stress, and activation of the hypothalamic–pituitary–adrenal (HPA) axis and sympathetic nervous system, with subsequent release of stress hormones and inflammatory molecules (Daiber et al. 2019).
DNA methylation alterations may mediate part of the physiological and biochemical changes leading from these traffic-related exposures to their associated preclinical and clinical phenotypes. Increasing evidence has linked short- and long-term air pollution exposure to various measures of DNA methylation in both children and adults (Abraham et al. 2018; de FC Lichtenfels et al. 2018; Gondalia et al. 2019; Gruzieva et al. 2017; Lee et al. 2019; Mostafavi et al. 2018; Panni et al. 2016; Plusquin et al. 2017, 2018; Sayols-Baixeras et al. 2019). Although findings from these studies were largely inconsistent with regard to single Cytosine-phosphate-Guanine (CpG) sites, the systematic review by Alfano et al. (Alfano et al. 2018) highlighted that air pollution was consistently linked to global hypomethylation in children (Breton et al. 2016; Cai et al. 2017; Janssen et al. 2015) and in adults (De Nys et al. 2018; De Prins et al. 2013) and accelerated epigenetic aging in adults (Nwanaji-Enwerem et al. 2016, 2017; Ward-Caviness et al. 2016). Overarching epigenomic effects of air pollution identified across these studies include inflammation, mitochondrial and DNA damage responses, and accelerated biological aging (Alfano et al. 2018).
Transportation noise, like air pollution, may also influence health outcomes via DNA methylation. First, they might share mechanistic pathways, because there is growing evidence for the inflammatory and oxidative downstream effects of noise exposure (Daiber et al. 2019; Münzel et al. 2017). Second, oxidative DNA damage, a correlate of altered DNA methylation and gene expression (Russo et al. 2016) was linked to occupational noise (Bagheri Hosseinabadi et al. 2019; Nawaz and Hasnain 2013) and traffic noise (Hemmingsen et al. 2015). Third, DNA methylation changes were reported for noise-related stressors such as vehicular traffic (Commodore et al. 2018) and insufficient sleep (Gaine et al. 2018). Methylation changes were also reported in circadian rhythm genes in conditions of acute sleep loss (Cedernaes et al. 2015) and night-shift work (Zhu et al. 2011). A study investigating brain tissue DNA methylation in relation to noise exposure in animal models reported gene-specific methylation changes, which were in turn associated with metabolic health (Guo et al. 2017).
No transportation noise epigenome-wide association study (EWAS) has been performed so far in population-based studies, and previous air pollution EWAS studies did not account for concurrent noise exposure (Eze and Probst-Hensch 2019). This partial scope of research is a limitation because both exposures share common sources and might be potential mutual confounders (Tetreault et al. 2013). As demonstrated by previous studies (Franklin and Fruin 2017; Heritier et al. 2019; Tetreault et al. 2013), the effect estimates of air pollution might be overestimated if noise level is unaccounted for, and vice versa. In addition, a parallel consideration of both exposure groups might elucidate directly shared and independent pathways of individual exposures and provide an improved understanding of mechanisms linking these exposures to disease.
In this paper, we aimed within the Swiss Cohort Study on Air Pollution and Lung and Heart Diseases in Adults (SAPALDIA) to
conduct a multiexposure EWAS (single CpGs and genomic regions) involving long-term exposure to source-specific transportation noise (aircraft, railway, and road traffic) and air pollution ( and );
assess in a candidate approach, using a priori-curated CpGs, the pathway enrichment for “allostatic load”-related cardio-metabolic, immunological, and renal phenotypes (Johnson et al. 2017; McCrory et al. 2019; Seeman et al. 2010), given that both traffic-related exposures are chronic stressors of several physiological systems;
assess in an agnostic approach, the functional pathway and network enrichment of EWAS-derived differentially methylated regions (DMRs); and
replicate previously reported CpGs associated with long-term exposure to and in the LifeLines (de FC Lichtenfels et al. 2018), EPIC-ITALY and EPIC-NL (Plusquin et al. 2017), and Korean COPD (Lee et al. 2019) cohorts.
Methods
Study Population
The SAPALDIA cohort has been described elsewhere in detail (Ackermann-Liebrich et al. 2005; Martin et al. 1997). SAPALDIA is a population-based study that recruited 9,651 adults from 8 geographically diverse Swiss areas (Aarau, Basel, Davos, Geneva, Lugano, Montana, Payerne, and Wald) in 1991 (SAP1) to investigate the respiratory effects of air pollution exposure. The first (SAP2) and second (SAP3) follow-up surveys occurred in 2002 and 2010; included 8,047 and 6,088 participants, respectively; and additionally, assessed cardio-metabolic and quality of life phenotypes. At each survey, participants had physical examinations and health- and lifestyle-related interviews. Measures of transportation noise and air pollution exposures were modeled at participants’ residences. At both SAP2 and SAP3, whole blood samples were collected, preprocessed, and stored in a biobank () for biomarker assessments, including DNA methylation. In the context two SAPALDIA nested studies—Aging Lungs in European Cohorts study (ALEC; www.alecstudy.org) and the EXPOsOMICS study (Vineis et al. 2017)—SAP2 and SAP3 repeat blood samples from 987 representative participants (ALEC), and an independent 405 nonsmoking participants (for at least 1 y before SAP2; EXPOsOMICS), were applied toward DNA methylation assessment. The EXPOsOMICS sample was an asthma case–control sample of nonsmokers (204 cases and 201 controls), where the asthma cases had methylation assessed only in the SAP3 blood sample and the controls had methylation assessment in both the SAP2 and SAP3 blood samples. Following exclusions due to methylation data quality and covariate data availability, we included SAP2 data from 972 ALEC and 198 EXPOsOMICS (asthma controls) participants and SAP3 data from 970 ALEC and 402 EXPOsOMICS (204 asthma cases and 198 controls) participants.
The present study therefore includes 1,389 participants contributing 2,542 observations, with an average of 1.8 observations per participant. Figure 1 presents the details of participant selection and inclusion. The SAPALDIA study complies with the Declaration of Helsinki. All participants provided informed written consent before participating in any aspect of the SAPALDIA surveys and ethical approvals were obtained from the Swiss Academy of Medical Sciences and the Ethics committees of the participating cantons.
Figure 1.
Selection of participants included in the present study. SAPALDIA: Swiss cohort study on air pollution and lung and heart diseases in adults. ALEC: Aging lungs in European cohorts. ALEC and EXPOsOMICS are European cohort consortia in which SAPALDIA participates. DNA methylation was measured using Illumina Infinium 450K BeadChip and processed in the same manner across the ALEC and EXPOsOMICS samples to derive the residuals of the beta values (corrected for technical bias) of overlapping 430,477 CpGs, which were subsequently applied to the present epigenome-wide association study.
Assessment of DNA Methylation
DNA methylation was measured in the ALEC and EXPOsOMICS samples separately in different laboratories, but using consistent methodology, using Illumina Infinium 450K BeadChip and processed as previously described (Imboden et al. 2019; Jeong et al. 2019). In brief, paired blood samples from the same participant were randomized across the arrays and placed next to each other on the arrays. Dye-bias correction (Triche et al. 2013) and absolute methylation level (-values, defined as the ratio of methylation intensity over total intensity, with offset of 100) were computed using the minfi R-package (R Development Core Team) (Aryee et al. 2014). Quality control criteria included call rate , detection -value , sex consistency, autosomal chromosome location of probes, and restriction to probes identified in both ALEC and EXPOsOMICS samples. We applied beta mixture quantile normalization (BMIQ) of the -values to correct for the Illumina probe design bias (Teschendorff et al. 2013). We further excluded 37,882 CpGs that are cross-reactive or target polymorphic (Chen et al. 2013). For technical bias (batch effect) correction, these -values were then regressed on the first 30 principal components derived from a principal component analysis of the control probes incorporated on the methylation chip (Lehne et al. 2015). Figure S1 shows the distributions of the BMIQ-normalized -values and technical bias-corrected residuals. In line with previous studies (Imboden et al. 2019; Jeong et al. 2019), we applied these residuals (in place of the -values) in subsequent EWAS covering 430,477 CpG sites that overlapped between ALEC and EXPOsOMICS samples.
To minimize bias due to extreme values (EVs) of the residuals—the dependent variables in the present EWAS—while retaining the distribution of DNA methylation at the nonskewed CpG sites, we performed modified winsorization of the CpG sites containing EVs. We defined EVs as values lying beyond three times the interquartile range (IQR) below and above the first (Q1) and third quartiles (Q3), respectively (Tukey 1977). We replaced each EV with the corresponding threshold of detection, in a CpG-specific manner, that is:
If , then EV is replaced with the value of at the CpG site;
If , then EV is replaced with the value of at the CpG site.
Among the 430,477 included CpGs, 393,397 CpGs (91%) had at least one EV. The EVs comprised 0.04%–22% of observations across the “winsorized” CpG sites. We used these “winsorized” data as the primary outcome variables in subsequent analyses (including EWAS, enrichment, and replication), and only used the “nonwinsorized” data for sensitivity testing of the top CpG signals identified in the EWAS.
Assessment of Transportation Noise Exposure
As detailed elsewhere (Karipidis et al. 2014), annual average day-evening-night noise level [Lden, with respective 5 dB and 10 dB penalties for evening (19–23 h) and nighttime (23–07 h; Lnight)] were calculated for 2001 and 2011 (corresponding to SAP2 and SAP3, respectively), for aircraft, railway, and road traffic noise at the maximum-exposed façade of the residential floors of participants. This calculation was done in the framework of the Short and Long Term Effects of Transportation Noise Exposure (SiRENE) project (Röösli et al. 2017). Aircraft noise was modeled with the FLULA2 version 4 software (EMPA 2010) with input data covering four airports in Basel, Geneva, Zurich, and Payerne. Railway noise emission and propagation were modeled using the sonRAIL (Thron and Hecht 2010) and the SEMIBEL (FOEN 1990) models, respectively, whereas road traffic noise emission and propagation were separately modeled using sonROAD (Heutschi 2004) and StL-86 (FOEN 1987). Validation of noise calculations was done by comparing the calculated noise levels with measured levels from the field. Taking all measurements into account, a mean Lden difference of was observed (Schlatter et al. 2017). Lden and Lnight values were respectively truncated at and for railway and aircraft noise, and and for road traffic noise. Calculated Lden and Lnight values below these limits were replaced by the respective truncation values. Participants with truncated values were assigned a truncation indicator (yes/no) for subsequent modeling, in line with previous analyses in the context of the SiRENE study (Eze et al. 2017; Foraster et al. 2017). Given the high correlations of source-specific Lden and Lnight, (Spearman’s rank correlation, , , and ), and the generally lower Lnight levels (Table S1 and Figure S2); we focused on source-specific Lden as the noise parameters of interest in this EWAS.
Assessment of Air Pollution Exposure
Traffic-related air pollutants— and —were also modeled at participant’s residences at SAP2 and SAP3 as outdoor mean exposures. For SAP2, annual mean was estimated (year 2003) using a hybrid model that regressed passive sampler measurements against dispersion model estimates, seasonal and climatic variables, as well as traffic and land use characteristics. Adjusted of the hybrid model was 0.8 (Liu et al. 2012). was derived from the Swiss PolluMap dispersion model (year 2000). Modeled values were validated against measured values with an of 0.9 (Heldstab et al. 2003). For SAP3, and were modeled as average biennial exposures (2010/2011) using land use regression models. The best models were used in each case: the combined with an adjusted of 0.6, and the area-specific models with adjusted of 0.5–0.9 across SAPALDIA study areas (Eeftens et al. 2016). We included both pollutants in our analyses, examining their associations in both single- and multiexposure models.
Assessment of Covariates
At the level of the individual, we considered groups of potential confounders measured at both SAP2 and SAP3, which might be associated with transportation noise, air pollution exposures, and DNA methylome. We considered sociodemographic factors including age (y), sex (male/female), and educational attainment (primary education/secondary education or apprenticeship/tertiary education). We considered lifestyle factors such as smoking status (never/former/current), smoking pack-years (calculated from cigarettes per day and duration of smoking), passive smoking (yes/no), alcohol consumption (beers, wines, liquors and spirits; ), frequency of fruit intake (citrus or noncitrus fruits in any form; ), vegetable intake (raw or cooked; ), body mass index (BMI; ), and sufficient moderate-to-vigorous physical activity ( of engagement in activities that makes one at least moderately sweat or breathless). To minimize the between-nested study differences, including residual batch effects and asthma status, we considered the contributing nested study (ALEC/EXPOsOMICS) and asthma status, defined as ever having a diagnosis of asthma. We also considered estimates of leukocyte composition for B cells, CD4T cells, CD8T cells, eosinophils, monocytes, natural killer cells, and neutrophils (derived from the DNA methylation data using the “estimatecellcounts” function in the minfi R package (Houseman et al. 2012; Reinius et al. 2012), to control the influence of cell proportions on methylation level (Adalsteinsson et al. 2012).
On the contextual scale, we considered some commonly indicated potential confounders of environmental health such as study area, a neighborhood index of socioeconomic position (SEP; %), and greenness index within a residential buffer. SEP was derived from a principal component analysis of household characteristics (education, occupation of household head, occupancy and median rent of household) based on 2001 census data (Panczak et al. 2012), and assigned to residential geo-coordinates at SAP2 and SAP3. Greenness index was calculated for 2014 as normalized difference vegetation index based on surface reflectance and assigned to participants’ geo-coordinates at SAP2 and SAP3 (Vienneau et al. 2017).
Statistical Analyses
EWAS.
We described the characteristics of included participants by survey and by nested study, summarizing categorical variables as counts and proportions and summarizing continuous variables as medians and IQR. Using the combined sample, we performed EWAS by linear mixed-effects regressions on the “winsorized” technical-bias corrected residuals of 430,477 CpGs, applying random intercepts at the level of participants. We used the “lmer” function of the lme4 R package for the regressions (Bates et al. 2015). We performed single- as well as multiexposure EWAS for aircraft, railway, and road traffic Lden, and , adjusting for age; sex; education; smoking status; pack-years; passive smoking; fruit, vegetable, and alcohol intake; study area; SEP; greenness index; survey; nested study; asthma; Lden truncation indicator; and Houseman estimates of leukocyte composition in the main model. We identified genome-wide significant CpGs at false discovery rate (FDR) and Bonferroni-corrected -value thresholds of 0.05 and , respectively.
We tested sensitivity of the top 10 CpGs to further adjustment for the potential mediators BMI and physical activity, which have been associated with transportation noise (An et al. 2018b; Foraster et al. 2016; Roswall et al. 2017) and air pollution exposures (An et al. 2018a, 2018c). We further stratified these CpGs by nested study to assess consistency in effect direction, limited these models to participants who reported regular nighttime opening of windows, and assessed the robustness of top signals in models using the “nonwinsorized” methylation data.
We tested for DMRs, using the “dmrcate” function in the DMRcate R package (Peters et al. 2015) and the multiexposure EWAS-derived parameters for each CpG as input file. Thus, we performed the DMR analyses using the individual estimates of 430,477 CpGs from the main multiexposure model. We defined DMRs as significant if they contained at least two CpGs, within at least 1,000 base pairs, and had a minimum FDR . Except when unannotated, all CpGs and DMRs are reported with gene annotation in parenthesis.
Pathway Enrichment
In a candidate pathway approach, we identified previously published EWAS signals for different physiological systems (captured by selected phenotypes) of potential relevance to transportation-related noise and air pollution effects. These systems (phenotypes) included a) immunological (C-reactive protein, CRP); b) metabolic [glycemia, insulin secretion/sensitivity, lipoprotein cholesterol, BMI, waist circumference (WC)/visceral adipose tissue mass, and metabolic syndrome]; c) renal (estimated glomerular filtration rate, eGFR); and d) cardiovascular and autonomic nervous systems [blood pressure and cardiac autonomic responses (CAR)]. For all phenotypes, we curated CpGs from the EWAS atlas (Li et al. 2019), at a -value threshold of , except for CRP where only the genome-wide significant signals at -value of were reported (Ligthart et al. 2016). CpGs that overlapped with those of the SAPALDIA study were finally curated for enrichment analyses (Table S1).
We defined a global “allostatic load” pathway as the entirety of unique CpGs assigned to at least one of the constituent pathways (). We applied the Weighted Kolmogorov-Smirnov method (Charmpi and Ycart 2015) to test for enrichment of the candidate and “allostatic load” pathways using the absolute values of test statistics from multiexposure EWAS. These test statistics from CpGs mapped to the pathway were compared with the empirical null distribution derived by 10,000 permutation samples. If the Kolmogorov-Smirnov test statistic was larger than the 90th percentile of test statistics obtained from the permutation samples after permutation-based multiple testing correction (van der Laan et al. 2005), we declared enrichment for the pathway.
In an agnostic approach, we assessed the functional pathways of exposure-specific differential methylation, using the “Core Analysis” of ingenuity pathway analysis (IPA; Ingenuity Systems) on genes annotated to significant DMRs. For each exposure, we identified the functional pathways that were significantly enriched at -value , as well as top disease and functional networks related to these pathways.
Replication of Previously Reported Air Pollution–Associated CpGs
Using the SAPALDIA EWAS results, we looked up the single- and multiexposure EWAS signals for validation of 22 and 10 previously reported genome-wide significant CpGs for long-term exposure to and , respectively. These include signals from the LifeLines cohort [cg04908668, cg14938677, cg00344801, cg18379295, cg25769469, cg02234653, and cg08500171 (de FC Lichtenfels et al. 2018)], EPIC-ITALY [cg08120023, cg22856765, cg18164357, cg13918628, cg03870188, cg20939320, cg13420207, cg04914283, cg21156210, cg16205861, cg12790758, and cg18201392 (Plusquin et al. 2017)], and the Korean COPD cohort [cg05171937, cg06226567, and cg26583725 (Lee et al. 2019)]. -associated signals include in cg23890774 from EPIC-ITALY; and cg12575202, cg08630381, cg17629796, cg07084345, cg04319606, cg09568355, cg03513315, cg25489413, and cg00005622 from EPIC-NL (Plusquin et al. 2017). We identified CpGs as replicated in our study if they showed consistent effect direction at nominal -value threshold of 0.05.
Results
The characteristics of the study sample at SAP2 and SAP3 are presented in Table 1. In general, participants tended to gain weight and smoke less with relatively lower pollutant exposures between surveys. ALEC and EXPOsOMICS participants mainly differed by design in their smoking habits and asthma status, but their environmental exposure profiles were comparable (Table S2). Figure S2 shows the overall distributions of the aircraft, railway, and road traffic Lden and and exposures. The Spearman’s rank correlations (r) of the five exposures are shown in Table S1. Correlation of and was 0.65. had higher correlation with road traffic () than railway () and aircraft Lden (), whereas had higher correlation with aircraft () than railway () and road traffic Lden (). Correlation patterns were consistent across SAP2 and SAP3.
Table 1.
Summary of the included SAPALDIA sample.
| SAPALDIA2a | SAPALDIA3a | |
|---|---|---|
| Categorical variables [n (%)] | 1,170 (100) | 1,372 (100) |
| Women | 623 (53) | 737 (54) |
| Formal education (y) | ||
| 56 (5) | 62 (4) | |
| 10–12 | 759 (65) | 887 (65) |
| 355 (30) | 423 (31) | |
| Smoking status | ||
| Never | 539 (46) | 659 (48) |
| Former | 355 (30) | 496 (36) |
| Current | 276 (24) | 217 (16) |
| Passive smoke exposure | 289 (25) | 159 (12) |
| Alcohol intake | 456 (39) | 529 (38) |
| Fruit intake | 326 (28) | 280 (20) |
| Vegetable intake | 86 (7) | 100 (7) |
| Urban area | 689 (59) | 897 (60) |
| Prevalent asthma | 161 (14) | 398 (21) |
| 330 (28) | 354 (26) | |
| Regular nighttime opening of windows | 971 (83) | 1,131 (82) |
| Nested study | ||
| ALEC | 972 (83) | 970 (71) |
| EXPOsOMICS | 198 (17) | 402 (29) |
| Continuous variables [median (IQR)] | ||
| Age | 50 (18) | 58 (18) |
| Body mass index () | 24.9 (5) | 25.8 (6) |
| Smoking pack-years | 0.4 (15) | 0 (14) |
| Neighborhood index of socioeconomic position (%) | 64.6 (13) | 64.8 (13) |
| Greenness index within 1-km buffer | 0.61 (0.2) | 0.62 (0.2) |
| Aircraft Lnight (dB) | 20 (2) | 20.1 (5) |
| Railway Lnight (dB) | 22.9 (14) | 20 (10) |
| Road traffic Lnight (dB) | 44.9 (111) | 45.1 (11) |
| Aircraft Lden (dB) | 30 (9) | 32.7 (8) |
| Railway Lden (dB) | 30 (11) | 30 (7) |
| Road traffic Lden (dB) | 53.7 (11) | 53.9 (11) |
| () | 20.2 (14) | 16.7 (10) |
| () | 14.3 (5) | 12.9 (2) |
Note: ALEC and EXPOsOMICS are European cohort consortia in which SAPALDIA participates. ALEC, Aging Lungs in European Cohorts; MVPA, moderate to vigorous physical activity; , particulate matter with aerodynamic diameter ; , nitrogen dioxide; SAPALDIA, Swiss Cohort Study on Air Pollution and Lung and Heart Diseases in Adults.
Population included in the analysis was limited to participants with complete methylome, exposure, and covariate data.
EWAS
In the single exposure models (Table S3), we observed one genome-wide significant () -associated CpG [cg26704043 (FARS2)] and one borderline-significant () railway Lden-associated CpG [cg25201280 (ATPBD4)]. There were no genome-wide significant signals in the multiexposure model (Table 2). Here, the -associated cg26704043 (FARS2) got considerably weaker (), whereas the railway Lden-associated cg25201280 (ATPBD4) remained borderline-significant (). The top 10 CpGs of the single-exposure models were not entirely consistent with those of the multiexposure models, with varying degrees of overlap across exposures. However, overlapping CpGs were directionally consistent (for all exposures), and the top CpG was positionally consistent (except for road traffic Lden) between the single- and multiexposure models (Tables 2 and S3).
Table 2.
Top ten CpGs independently associated with source-specific transportation noise and air pollution in the SAPALDIA study, multiexposure models.
| Exposure | CpG ID | CHR | Location | Gene | Feature | Model 1 | SE | -Value | P (FDR) | Model 2 | SE | -Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | Beta | |||||||||||
| Aircraft Lden | cg02286155 | 5 | 176826262 | NA | NA | 0.001 | 0.637 | 0.001 | ||||
| cg15063530 | 2 | 17716941 | NA | NA | 0.002 | 0.659 | 0.002 | |||||
| cg16218477 | 7 | 1066167 | C7orf50 | Body | 0.001 | 0.659 | 0.001 | |||||
| cg21602842 | 18 | 46291908 | KIAA0427 | Body | 0.001 | 0.659 | 0.001 | |||||
| cg09042449 | 10 | 44064225 | ZNF239 | 5′UTR | 0.0004 | 0.659 | 0.0004 | |||||
| cg10975000a | 13 | 28371375 | NA | NA | 0.0005 | 0.659 | 0.0005 | |||||
| cg06220958 | 17 | 10452851 | MYH2 | 5′UTR | 0.011 | 0.002 | 0.659 | 0.010 | 0.002 | |||
| cg25462190a | 5 | 177547067 | N4BP3 | Body | 0.002 | 0.659 | 0.002 | |||||
| cg11944797 | 13 | 99135711 | STK24 | Body | 0.0003 | 0.659 | 0.0003 | |||||
| cg04635504 | 11 | 2829241 | KCNQ1 | Body | 0.001 | 0.664 | 0.001 | |||||
| Railway Lden | cg25201280a | 15 | 35838552 | ATPBD4 | TSS200 | 0.0002 | 0.075 | 0.0002 | ||||
| cg24653263a | 5 | 38258335 | EGFLAM | TSS200 | 0.001 | 0.193 | 0.001 | |||||
| cg16825060a | 8 | 144242342 | LY6H | TSS1500 | 0.001 | 0.256 | 0.001 | |||||
| cg23468045 | 5 | 12669584 | NA | NA | 0.001 | 0.0002 | 0.256 | 0.001 | 0.0002 | |||
| cg19270309a | 17 | 77712853 | ENPP7 | 3′UTR | 0.002 | 0.0005 | 0.266 | 0.002 | 0.0005 | |||
| cg07461273 | 7 | 99697172 | MCM7 | Body | 0.001 | 0.266 | 0.001 | |||||
| cg01301319 | 7 | 27153580 | HOXA3 | 5′UTR | 0.001 | 0.266 | 0.001 | |||||
| cg23113715a | 22 | 25800663 | NA | NA | 0.001 | 0.266 | 0.001 | |||||
| cg13402217 | 1 | 151584375 | SNX27 | TSS1500 | 0.001 | 0.266 | 0.001 | |||||
| cg24047259a | 14 | 65347275 | NA | NA | 0.0003 | 0.266 | 0.0003 | |||||
| Road traffic Lden | cg09129334 | 13 | 111837676 | ARHGEF7 | Body | 0.001 | 0.384 | 0.001 | ||||
| cg17383236a | 7 | 100167504 | NA | NA | 0.0005 | 0.384 | 0.0005 | |||||
| cg01066220 | 6 | 31696240 | DDAH2 | Body | 0.001 | 0.0001 | 0.384 | 0.001 | 0.0001 | |||
| cg23910243a | 16 | 31484618 | TGFB1I1 | Body | 0.0005 | 0.384 | 0.0005 | |||||
| cg06646021a | 1 | 229406520 | RAB4A | TSS1500 | 0.001 | 0.384 | 0.001 | |||||
| cg03066594 | 20 | 10415919 | C20orf94 | TSS200 | 0.0005 | 0.0001 | 0.386 | 0.0004 | 0.0001 | |||
| cg03966094a | 22 | 21058792 | TMEM191A | Body | 0.001 | 0.386 | 0.001 | |||||
| cg13948857 | 5 | 131763756 | C5orf56 | Body | 0.001 | 0.386 | 0.001 | |||||
| cg08351004a | 2 | 172965650 | DLX2 | Body | 0.0005 | 0.456 | 0.0005 | |||||
| cg13777730 | 1 | 234793300 | NA | NA | 0.001 | 0.458 | 0.001 | |||||
| cg04337651a | 2 | 239344738 | ASB1 | Body | 0.004 | 0.001 | 0.657 | 0.003 | 0.001 | |||
| cg18776472 | 10 | 50732819 | ERCC6 | Body | 0.0002 | 0.657 | 0.0002 | |||||
| cg18601596 | 6 | 39283313 | KCNK16 | Body | 0.006 | 0.001 | 0.657 | 0.006 | 0.001 | |||
| cg12392998 | 17 | 79550668 | NPLOC4 | Body | 0.0004 | 0.657 | 0.0004 | |||||
| cg16550606 | 13 | 50160670 | RCBTB1 | TSS1500 | 0.004 | 0.001 | 0.657 | 0.004 | 0.001 | |||
| cg25266109 | 19 | 12404608 | ZNF44 | Body | 0.0001 | 0.657 | 0.0001 | |||||
| cg01746514a | 14 | 24520922 | LRRC16B | TSS1500 | 0.0002 | 0.657 | 0.0002 | |||||
| cg15811902 | 15 | 75918385 | SNUPN | 5′UTR | 0.0005 | 0.657 | 0.0005 | |||||
| cg26898336a | 17 | 15244519 | TEKT3 | 5′UTR | 0.002 | 0.0005 | 0.657 | 0.002 | 0.0005 | |||
| cg21099332 | 5 | 39270715 | NA | NA | 0.004 | 0.001 | 0.657 | 0.004 | 0.001 | |||
| cg26704043 | 6 | 5282702 | FARS2 | 5′UTR | 0.014 | 0.003 | 0.180 | 0.014 | 0.003 | |||
| cg05157625 | 14 | 93153553 | RIN3 | Body | 0.021 | 0.004 | 0.231 | 0.021 | 0.004 | |||
| cg20099458a | 7 | 5272275 | WIPI2 | 3′UTR | 0.014 | 0.003 | 0.231 | 0.014 | 0.003 | |||
| cg06587257 | 12 | 50452135 | ACCN2 | 5′UTR | 0.022 | 0.005 | 0.292 | 0.023 | 0.005 | |||
| cg14531665 | 9 | 91058614 | SPIN1 | Body | 0.012 | 0.003 | 0.398 | 0.011 | 0.003 | |||
| cg06526020 | 6 | 34308880 | NUDT3 | Body | 0.029 | 0.006 | 0.398 | 0.028 | 0.006 | |||
| cg21058520 | 6 | 100914733 | NA | NA | 0.004 | 0.001 | 0.398 | 0.004 | 0.001 | |||
| cg16259904 | 10 | 134146220 | LRRC27 | 5′UTR | 0.027 | 0.006 | 0.398 | 0.027 | 0.006 | |||
| cg12770741 | 17 | 883776 | NXN | TSS1500 | 0.018 | 0.004 | 0.398 | 0.018 | 0.004 | |||
| cg26750893a | 2 | 38043481 | NA | NA | 0.016 | 0.004 | 0.398 | 0.016 | 0.004 |
Note: Beta coefficients represent increase or decrease in DNA methylation per increase in aircraft, railway or road traffic Lden or 10 increase in or . All estimates were from multiexposure epigenome-wide linear mixed-effects models, with random intercept at the level of participant. Multiexposure models included all five exposures (aircraft, railway, road traffic Lden and respective truncation indicators, and ) at the same time. In a preliminary step, DNA methylation -values were regressed on the Illumina control probe-derived first 30 principal components to correct for correlation structures and technical bias, and residuals of these regressions covering 430,477 CpGs were used as the technical bias-corrected methylation level at the CpG sites. Extreme values of the residuals (lying beyond three times the interquartile range below the first quartile and above the third quartile at each CpG site) were replaced with their corresponding detection threshold value (“modified winsorization”). The “winsorized” data were then used as the dependent variables in the EWAS. Model 1: adjusted for age; sex; educational level; area; neighborhood socioeconomic status; greenness index; smoking status; smoking pack-years; exposure to passive smoke; consumption of fruits, vegetables and alcohol; nested study; asthma status; noise truncation indicators; survey; and leukocyte composition (main model). Model 2: Model mass index and physical activity. CHR, chromosome; CpG, Cytosine-phosphate-Guanine; Lden, day-evening-night noise level; NA, not annotated; , particulate matter with aerodynamic diameter ; SAPALDIA, Swiss Cohort Study on Air Pollution and Lung and Heart Diseases in Adults; SE, standard error.
Location overlaps with significant differentially methylated region.
In the multiexposure model, aircraft Lden (cg02286155), railway Lden [cg25201280 (ATPBD4)] and road traffic Lden cg09129334 (ARHGEF7) were associated with reduced methylation, whereas [cg04337651 (ASB1)] and [cg26704043 (FARS2)] were associated with increased methylation, at their respective top CpG sites. Among the top 10 signals, we observed decreased methylation at 9 (90%), 8 (80%), and 8 (80%) CpGs in relation to aircraft, railway, and road traffic Lden, respectively, and increased methylation at 5 (50%) and 10 (100%) CpGs in relation to and , respectively. These CpGs were generally robust to adjustment for BMI and physical activity (Table 2) and showed consistent effect direction when stratified by nested study (Table S4). Associations were also robust in the model limited to participants reporting regular nighttime window opening (Table S5). The “nonwinsorized” model highlighted few CpGs where extreme values would potentially bias their estimates if unaddressed. These included directionally consistent but considerably weaker effects on aircraft Lden-associated cg11944797 (STK24) and cg10975000; road traffic Lden-associated cg08351004 (DLX2); -associated cg18776472 (ERCC6), cg12392998 (NPLOC2), and cg26898336 (TEKT3); and a stronger and genome-wide significant effect on -associated cg21058520 (Table S6). Results from the multiexposure main model of CpGs associated with aircraft, railway, and road traffic Lden, —at nominal —are presented in Tables S2–S6.
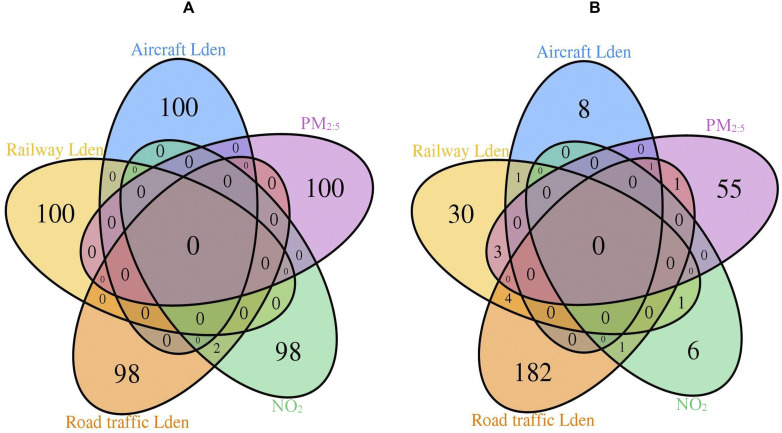
Post hoc analysis of overlap among the top 100 exposure-specific CpGs identified from the multiexposure EWAS showed minimal signal overlap between exposures (Figure 2A). We identified two overlapping signals between road traffic Lden and [cg12439232 and cg15590912 (CCSAP)]. Yet, road traffic Lden was associated with decreased methylation, whereas was associated with increased methylation at both sites (Tables S4 and S5). Complete EWAS results from the single- and multiexposure main models of 430,477 CpGs in relation to aircraft, railway, and road traffic Lden; ; and have been deposited in the Zenodo public online depository (https://zenodo.org/record/3832253#.XsLLJEKxU2x).
Figure 2.
Overlap of top 100 CpG signals (A) and genes annotated to significant differentially methylated regions (B) in relation to aircraft, railway, and road traffic Lden, , and identified from multiexposure EWAS in the SAPALDIA study. Note: EWAS, epigenome-wide association study; Lden, day-evening-night noise level; , nitrogen dioxide; , particulate matter in diameter; SAPALDIA, Swiss Cohort Study on Air Pollution and Lung and Heart Diseases in Adults. CpGs were identified by multiexposure EWAS using multivariable linear mixed-effects models with random intercepts at the level of participants, and adjusted for age, sex, educational level, area, neighborhood socioeconomic status, greenness index, smoking status, smoking pack-years, exposure to passive smoke, consumption of fruits, vegetables and alcohol, nested study, asthma status, noise truncation indicators, survey and leukocyte composition. In a preliminary step, DNA methylation -values were regressed on the Illumina control probe-derived first 30 principal components to correct for correlation structures and technical bias, and residuals of these regressions covering 430,477 CpGs were used as the technical bias-corrected methylation level at the CpG sites. Extreme values of the residuals (lying beyond three times the interquartile range below the first quartile and above the third quartile at each CpG site) were replaced with their corresponding detection threshold value (“modified winsorization”). The “winsorized” data were then used as the dependent variables in the present EWAS. CpGs (annotated gene) intersecting at the level of road traffic Lden and were cg12439232 and cg15590912 (CCSAP). Genes (DMR) intersecting at the level of road traffic Lden and was PRRT1 (chr6:32,115,964–32,117,401); and at the level of aircraft and road traffic Lden and was HOXA2 (chr7:27,141,774–27,143,806). VTRNA2-1 (chr5:135,415,129–135,416,613) intersected between aircraft and railway Lden, ZFP57 (chr6:29,648,161–29,649,084), between railway Lden and , and ZSCAN31 (chr6:28,303,923–28,304,451), between road traffic Lden and . TRIM39, TRIM39-RPP21, and HCG18 (chr6:30,296,689–30,297,941) intersected between railway Lden and , whereas SLC27A3 (chr1:153,746,588–153,747,856), B3GALT4 (chr6:33,244,976–33,246,185), EN2, and AC008060.8 (chr7:155,249,398–155,251,925) intersected between railway and road traffic Lden.
We identified independent DMRs () across all exposures in the main model (Table 3). There were 14 (10), 48 (39), 183 (189), 8 (8), and 71 (60) DMRs (genes), associated with aircraft, railway, and road traffic Lden, , and , respectively. Among the top 10 CpGs identified in the multiexposure model, two aircraft [cg10975000 and cg25462190 (N4BP3)], six railway [cg25201280 (ATPBD4), cg24653263 (EGFLAM), cg16825060 (LY6H), cg19270309 (ENPP7), cg23113715, and cg24047259], and five road traffic Lden-associated CpGs [cg17383236, cg23910243 (TGFB1I1), cg06646021 (RAB4A), cg03966094 (TMEM191A), and cg08351004 (DLX2)] were within the exposure-specific DMRs. Three [cg04337651 (ASB1), cg01746514 (LRRC16B) and cg26898336 (TEKT3)] and two -associated CpGs [cg20099458 (WIPI2) and cg26750893] were also within the exposure-specific DMRs. Top DMRs independently associated with exposures, were annotated to VTRNA2-1 (aircraft and railway Lden; Chr5:135,415,129–135,416,613), OXT (road traffic Lden; Chr20:3,051,954–3,053,196), ZSCAN31 (; Chr6:28,303,923–28,304,451), and TRIM39, HCG18, and TRIM39-RPP21 (; Chr6:30,296,689–30,297,941). Most of the CpGs within the DMRs associated with source-specific Lden showed decreased methylation (, , and ), whereas those associated with air pollution showed increased methylation ( and ). Tables S7 and S8 show the results from the multiexposure main model of DMRs associated with aircraft, railway, and road traffic Lden; ; and at .
Table 3.
Summary of EWAS-derived differentially methylated regions and enrichment in relation to transportation noise and air pollution exposure in the SAPALDIA study.
| Exposure | DMRs [Genes (n)] | Average effect on DMRs | Top DMR (Gene) | CpGs in top DMR (n) | FDR -value (top DMR) | Top enriched canonical pathway (TECP) | Genes in TECP | Top enriched disease networks |
|---|---|---|---|---|---|---|---|---|
| Aircraft Lden | 14 (10) | methylation (64%) | Chr5:135415129–135416613 (VTRNA2-1) | 19 | NA | NA | Cell cycle, embryonic development, organismal development | |
| Railway Lden | 48 (39) | methylation (69%) | Chr5:135415129–135416613 (VTRNA2-1) | 19 | Type II diabetes mellitus signaling; diphthamide biosynthesis | PRKAA1, SLC27A3, TNFRSF11B, DPH6 | cardiovascular system development and function, gene expression, organ development | |
| Road traffic Lden | 183 (189) | methylation (93%) | Chr20:3051954–3053196 (OXT) | 13 | Wnt/ signaling; cholecystokinin/gastrin-mediated signaling | CDH1, CSNK1E, SOX2, SOX8, WNT16, MEF2D, PXN, SHC1, TNF | Cell-mediated immune response, cell-to-cell signaling and interaction, cellular movement | |
| 8 (8) | methylation (63%) | Chr6:28303923–28304451 (ZSCAN31) | 11 | NA | NA | Cell cycle, cell-to-cell signaling and interaction, post-translational modification | ||
| 71 (60) | methylation (93%) | Chr6:30296689–30297941 (TRIM39, HCG18, TRIM39-RPP21) | 14 | and 4-aminobutyrate degradation I; systemic lupus erythematosus signaling | ABAT1, PRPF31, PRPF8, PTPN6 | Cell cycle, nervous system development and function, organismal injury, and abnormalities |
Note: Each DMR analysis had the corresponding multiexposure EWAS-derived parameters as input. Multiexposure EWAS derived from linear mixed-effects models, with random intercept at the level of participant, and adjusted for age; sex; educational level; area; neighborhood socioeconomic status; greenness index; smoking status; smoking pack-years; exposure to passive smoke; consumption of fruits, vegetables, and alcohol; nested study; asthma status; survey; noise truncation indicators; and leukocyte composition. In a preliminary step, DNA methylation -values were regressed on the Illumina control probe-derived first 30 principal components to correct for correlation structures and technical bias, and residuals of these regressions covering 430,477 CpGs were used as the technical bias-corrected methylation level at the CpG sites. Extreme values of the residuals (lying beyond three times the interquartile range below the first quartile and above the third quartile at each CpG site) were replaced with their corresponding detection threshold value (“modified winsorization”). The “winsorized” data were then used as the dependent variables in the EWAS. Significant () and annotated DMRs were used for canonical pathway and network enrichment in the Ingenuity Pathway Analysis software (Ingenuity Systems). ↓, decrease in methylation; ↑, increase in methylation; CpG, Cytosine-phosphate-Guanine; DMRs, limiting statistical power to detect enriched canonical pathways; EWAS, epigenome-wide association study; FDR, false discovery rate; Lden, day-evening-night noise level; NA, not applicable due to few significant and annotated; , nitrogen dioxide; , particulate matter with aerodynamic diameter ; SAPALDIA, Swiss Cohort Study on Air Pollution and Lung and Heart Diseases in Adults.
Post hoc analysis of overlap among the gene-annotated significant DMRs also showed minimal overlap between exposures (Figure 2B). SLC27A3, B3GALT4, EN2, and AC008060.8 overlapped between railway and road traffic Lden; TRIM39, TRIM39-RPP21, and HCG18 overlapped between railway Lden and ; and HOXA2 overlapped between aircraft and road traffic Lden and . Other overlapping genes include VTRNA2-1 (aircraft and railway Lden), ZFP57 (railway Lden and ), and ZSCAN31 (road traffic Lden and ), and PRRT1 (road traffic Lden and ). Overlapping DMRs showed opposing effect direction between exposures, except for consistently increased methylation at TRIM39, TRIM39-RPP21, HCG18 (railway Lden and ), and HOXA2 (aircraft Lden and ), and decreased methylation at SLC27A3, B3GALT4, EN2, and AC008060.8 (railway and road traffic Lden) (Tables S7 and S8).
Pathway Enrichment
For the candidate single phenotype and combined “allostatic load” pathways, multiple testing-corrected enrichment analyses showed varying enrichment across exposures (Table 4). Considering the results for single phenotypes, -related methylation was the most enriched, whereas -related methylation was the least enriched. Single pathways (-values) enriched for included CRP (0.0004), glycemia (0.0947), WC (0.0004), BMI (0.0004), and eGFR (0.0004). Aircraft and road traffic Lden-related methylation were enriched for CRP (0.0038; 0.0395), BMI (0.0007; 0.0008), and eGFR (0.0015; 0.0031). Railway Lden-related methylation was enriched for eGFR (0.0562) and CAR (0.0229), whereas -related methylation was enriched only for eGFR (0.0058). Considering the global “allostatic load” pathway, we found consistent enrichment across all exposures, including aircraft (0.0004), railway (0.0871), and road traffic Lden (0.0004); (0.0004); and (0.0510).
Table 4.
Pathway enrichment tests (-values) for transportation noise and air pollution exposures based on curated CpGs reported for selected cross-systemic outcomes.
| Exposure | CRP | Metabolic syndrome | Lipids | FG/HbA1c | Insulin | WC | BMI | Blood pressure | eGFR | CAR | Allostatic load |
|---|---|---|---|---|---|---|---|---|---|---|---|
| N (CpGs) | 256 | 10 | 14 | 9 | 158 | 168 | 893 | 20 | 296 | 7 | 1,626 |
| Aircraft Lden | 0.0038 | 0.5915 | 0.6326 | 0.4545 | 0.6630 | 0.2925 | 0.0007 | 0.1020 | 0.0015 | 0.9096 | 0.0004 |
| Railway Lden | 0.3163 | 0.4943 | 0.9533 | 0.5778 | 0.6284 | 0.3001 | 0.9023 | 0.6773 | 0.0562 | 0.0229 | 0.0871 |
| Road traffic Lden | 0.0395 | 0.2434 | 0.9969 | 0.3879 | 0.3461 | 0.5361 | 0.0008 | 0.2858 | 0.0031 | 0.8013 | 0.0004 |
| 0.2117 | 0.1237 | 0.7831 | 0.5733 | 0.3697 | 0.6873 | 0.8818 | 0.5355 | 0.0058 | 0.9728 | 0.0510 | |
| 0.0004 | 0.4517 | 0.1263 | 0.0947 | 0.3451 | 0.0004 | 0.0004 | 0.3759 | 0.0004 | 0.3814 | 0.0004 |
Note: Lipids includes triglycerides, high-, low- and very low-density lipoprotein cholesterol. Insulin includes measures of insulin secretion and resistance. WC also includes central obesity and adiposity. BMI also includes general obesity. Blood pressure includes systolic and diastolic blood pressure. eGFR also includes impaired renal function. CAR includes acceleration and deceleration capacity. Allostatic load combines all the phenotypes. Pathway enrichment -values derived from Weighted Kolmogorov-Smirnov method using the absolute values of test statistics from multiexposure epigenome-wide association studies (EWAS), and comparing the EWAS-derived CpGs mapped to each pathway to the empirical null distribution derived by 10,000 permutation samples. The overall procedure included permutation-based multiple testing correction. EWAS was done using linear mixed-effects models, with random intercept at the level of participant, and adjusted for age; sex; educational level; area; neighborhood socioeconomic status; greenness index; smoking status; smoking pack-years; exposure to passive smoke; consumption of fruits, vegetables, and alcohol; nested study; asthma status; noise truncation indicators; survey; and leukocyte composition. In a preliminary step, DNA methylation -values were regressed on the Illumina control probe-derived first 30 principal components to correct for correlation structures and technical bias, and residuals of these regressions covering 430,477 CpGs were used as the technical bias-corrected methylation level at the CpG sites. Extreme values of the residuals (lying beyond three times the interquartile range below the first quartile and above the third quartile at each CpG site) were replaced with their corresponding detection threshold value (“modified winsorization”). The “winsorized” data were then used as the dependent variables in the EWAS. BMI, body mass index; CAR, cardiac autonomic response; CpG, Cytosine-phosphate-Guanine; CRP, C-reactive proteins; eGFR, estimated glomerular filtration rate; FG, fasting glucose; HbA1c, glycated hemoglobin; Lden, day-evening-night noise level; , nitrogen dioxide; , particulate matter in diameter; WC, waist circumference.
Agnostic functional enrichment showed significantly enriched canonical pathways for railway, road traffic Lden and , with predominance of inflammatory and immune regulation-related pathways across exposures (Table S9). Some of the enriched canonical pathways included type 2 diabetes signaling, tight junction signaling, mTOR signaling, and lipopolysaccharide/interleukin-1 (IL-1)–mediated inhibition of retinoid X receptor (RXR) function (railway Lden); Wnt/ signaling, cholecystokinin/gastrin-mediated signaling, G-protein coupled receptor signaling, and Th1 and Th2 activation pathways (road traffic Lden); , 4-aminobutyrate degradation; and systematic lupus erythematosus signaling, and IL-4 signaling (). Network analyses showed the enrichment for disease mechanisms related to cellular signaling/interaction and embryonic/organ development (all exposures), cell-mediated immune/inflammatory responses (road traffic Lden and ) and diseases related to connective tissue development and function (railway and road traffic Lden). Pathways related to cardiovascular function, gene expression, and respiratory system disease (railway Lden); carbohydrate and small molecule transport/biochemistry, cancer, and auditory function (road traffic Lden); and hematological and nervous system function () were enriched (Table S10).
Replication of previously reported and –associated CpGs.
Using the single-exposure model, corresponding to the previous EWAS studies, we replicated the increased methylation in cg08500171 (BAT2) associated with (). This replication remained robust in the multiexposure model (). We observed a borderline significant decreased methylation in cg17629796 associated with () in the single-exposure model, which became statistically significant in the multiexposure model (). Nine (41%) -associated CpGs and 5 (50%) -associated CpGs had the same direction of effect in the SAPALDIA study (Table 5).
Table 5.
Replication of previously reported EWAS signals for long-term exposure to and , in the SAPALDIA study, single- and multiexposure models.
| Air pollutant; Cohort (Reference) | CpG ID | CHR | Location | Gene | Beta | SE | -Value | SAPALDIA (single-exposure model) | SE | -Value | SAPALDIA (multiexposure model) | SE | -Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | Beta | ||||||||||||
| ; LifeLines, de FC Lichtenfels et al. 2018 | cg04908668 | 6 | 32823941 | PSMB9 | 0.002 | 0.0003 | 0.0003 | 0.333 | 0.0004 | 0.0004 | 0.322 | ||
| cg14938677 | 7 | 127231698 | ARF5 | 0.023 | 0.004 | 0.0004 | 0.001 | 0.619 | 0.0004 | 0.001 | 0.663 | ||
| cg00344801 | 22 | 46685728 | TTC38 | 0.005 | 0.001 | 0.001 | 0.209 | 0.001 | 0.001 | 0.282 | |||
| cg18379295 | 14 | 52326155 | GNG2 | 0.020 | 0.004 | 0.001 | 0.204 | 0.001 | 0.367 | ||||
| cg25769469 | 5 | 71643841 | PTCD2 | 0.035 | 0.006 | 0.001 | 0.002 | 0.547 | 0.002 | 0.002 | 0.237 | ||
| cg02234653 | 2 | 224625080 | AP1S3 | 0.003 | 0.0005 | 0.001 | 0.553 | 0.0003 | 0.001 | 0.752 | |||
| cg08500171a | 6 | 31590674 | BAT2 | 0.024 | 0.004 | 0.002 | 0.001 | 0.042 | 0.003 | 0.001 | 0.019 | ||
| ; EPIC-ITALY, Plusquin et al. 2017 | cg08120023 | 1 | 116947203 | C1orf203 | 0.0005 | 0.001 | 0.001 | 0.586 | 0.002 | 0.001 | 0.179 | ||
| cg22856765 | 8 | 42693384 | THAP1 | 0.001 | 0.0002 | 0.960 | 0.0002 | 0.930 | |||||
| cg18164357 | 11 | 77534497 | C11orf67 | 0.001 | 0.0005 | 0.0003 | 0.042 | 0.001 | 0.0003 | 0.016 | |||
| cg13918628 | 9 | 35610380 | CD72 | 0.002 | 0.0002 | 0.184 | 0.0003 | 0.074 | |||||
| cg03870188 | 13 | 113717830 | MCF2L | 0.001 | 0.0001 | 0.0003 | 0.670 | 0.0001 | 0.0003 | 0.650 | |||
| cg20939320 | 3 | 132563279 | NCRNA00119 | 0.001 | 0.001 | 0.001 | 0.091 | 0.001 | 0.001 | 0.185 | |||
| cg13420207 | 7 | 81666278 | CACNA2D1 | 0.002 | 0.001 | 0.947 | 0.001 | 0.973 | |||||
| cg04914283 | 1 | 23181832 | EPHB2 | 0.001 | 0.001 | 0.001 | 0.298 | 0.001 | 0.001 | 0.118 | |||
| cg21156210 | 4 | 100485208 | RG9MTD2 | 0.010 | 0.002 | 0.0002 | 0.941 | 0.0001 | 0.0003 | 0.731 | |||
| cg16205861 | 12 | 54146572 | NA | 0.001 | 0.0005 | 0.788 | 0.0005 | 0.882 | |||||
| cg12790758 | 15 | 37369914 | MEIS2 | 0.001 | 0.0006 | 0.0005 | 0.287 | 0.001 | 0.898 | ||||
| cg18201392 | 1 | 185023741 | RNF2 | 0.001 | 0.0002 | 0.0001 | 0.118 | 0.0002 | 0.0001 | 0.133 | |||
| ; Korean COPD Cohort, Lee et al. 2019 | cg05171937 | 12 | 27396765 | STK38L | 0.010 | 0.002 | 0.003 | 0.001 | 0.068 | 0.002 | 0.002 | 0.125 | |
| cg06226567 | 20 | 22559676 | C20orf56 | 0.003 | 0.001 | 0.00002 | 0.0001 | 0.863 | 0.0001 | 0.0002 | 0.514 | ||
| cg26583725 | 13 | 110397643 | NA | 0.0001 | 0.0001 | 0.183 | 0.0001 | 0.0001 | 0.228 | ||||
| ; EPIC-ITALY, Plusquin et al. 2017 | cg23890774 | 19 | 36618841 | NA | 0.078 | 0.014 | 0.0001 | 0.0003 | 0.704 | 0.0003 | 0.0003 | 0.263 | |
| ; EPIC-NL never-smokers, Plusquin et al. 2017b | cg12575202 | 10 | 133331128 | NA | 0.080 | 0.003 | 0.786 | 0.003 | 0.529 | ||||
| cg08630381 | 13 | 100612277 | NA | 0.461 | 0.073 | 0.001 | 0.415 | 0.001 | 0.394 | ||||
| cg17629796a | 13 | 30707265 | NA | 0.094 | 0.001 | 0.076 | 0.001 | 0.033 | |||||
| cg07084345 | 15 | 61972967 | NA | 0.075 | 0.002 | 0.008 | 0.816 | 0.002 | 0.008 | 0.769 | |||
| cg04319606 | 2 | 26785290 | C2orf70 | 0.261 | 0.068 | 0.0002 | 0.002 | 0.893 | 0.002 | 0.989 | |||
| cg09568355 | 2 | 45228633 | NA | 0.261 | 0.049 | 0.003 | 0.002 | 0.286 | 0.004 | 0.003 | 0.179 | ||
| cg03513315 | 2 | 30988383 | PES1 | 0.307 | 0.058 | 0.001 | 0.631 | 0.001 | 0.837 | ||||
| cg25489413 | 7 | 44794343 | ZMIZ2 | 0.068 | 0.001 | 0.002 | 0.712 | 0.0002 | 0.002 | 0.933 | |||
| cg00005622 | 8 | 145180403 | NA | 0.064 | 0.001 | 0.109 | 0.003 | 0.136 |
Note: Beta coefficients represent increase or decrease in DNA methylation in relation to or exposure. In a preliminary step, DNA methylation -values were regressed on the Illumina control probe-derived first 30 principal components to correct for correlation structures and technical bias, and residuals of these regressions covering 430,477 CpGs were used as the technical bias-corrected methylation level at the CpG sites. Extreme values of the residuals (lying beyond three times the interquartile range below the first quartile and above the third quartile at each CpG site) were replaced with their corresponding detection threshold value (“modified winsorization”). The “winsorized” data were then used as the dependent variables in the present EWAS. The multiexposure model contained all five exposures at same time. All SAPALDIA estimates were derived from linear mixed-effects EWAS models, with random intercept at the level of participant, adjusted for age; sex; educational level; area; neighborhood socioeconomic status; greenness index; smoking status; smoking pack-years; exposure to passive smoke; consumption of fruits, vegetables, and alcohol; nested study; asthma status; noise truncation indicators; survey; and leukocyte composition. Multiexposure model included all five exposures (Aircraft, railway, road traffic Lden and respective truncation indicators, and ) at the same time. CHR, chromosome; COPD, chronic obstructive pulmonary disease; CpG, Cytosine-phosphate-Guanine; EPIC, European Prospective Investigation into Cancer and Nutrition; EWAS, epigenome-wide association study; N.A., not annotated; NL, Netherlands; , nitrogen dioxide; , particulate matter with aerodynamic diameter ; SAPALDIA, Swiss Cohort Study on Air Pollution and Lung and Heart Diseases in Adults; SE, standard error.
Validated in the SAPALDIA study.
SAPALDIA estimates derived from never-smoker sample comparable to the EPIC-NL never-smoker estimates.
Discussion
In this first multiexposure EWAS covering long-term aircraft, railway, and road traffic noise, as well as and exposures, we found genome-wide significant signals—at the level of genomic regions—associated with source-specific transportation noise and air pollution exposures. We demonstrated some mutual confounding of the associations of exposures with DNA methylation, where single exposure models slightly overestimated the observed methylation effect. Overall, methylation signals minimally overlapped but showed common enrichment of inflammation and immune response-related pathways across exposures. In this study, we also validated air pollution–related CpG signals identified in external EWAS.
The railway noise–related decrease in methylation at cg25201280 (transcription start site 200 of ATPBD4 gene on chromosome 15) and -related increase in methylation at cg26704043 (5′ untranslated region of FARS2 gene on chromosome 6) were the strongest single CpG signals identified in the study. ATPBD4 (or DPH6) is a protein-coding gene that regulates adenosine triphosphate (ATP) binding and diphthamide synthase activity, within the protein metabolism pathway (Chertow 1981; Young et al. 2004). Polymorphisms in this gene were associated—in various genome-wide association studies (GWAS)—with adiposity (Kichaev et al. 2019; Pulit et al. 2019), cognitive function (Lee et al. 2018; Li et al. 2015), and Crohn’s disease progression (O’Donnell et al. 2019). FARS2 is a protein-coding gene that regulates the translation of mitochondrial proteins and gene expression (Bullard et al. 1999). Variants of this gene were associated—in GWAS—with extreme obesity (Wheeler et al. 2013) and acute myeloid leukemia (Lv et al. 2017), whereas in EWAS, cg26704043 was associated with systemic lupus erythematosus (Imgenberg-Kreuz et al. 2018) and Grave’s disease (Chen et al. 2019). These findings highlight the role of DNA methylation changes in these genes as potential mechanisms by which railway noise and contribute to the resulting inflammatory and neurological phenotypes.
Our observation of numerous independent DMRs (for all five exposures), despite paucity of single CpG signals, highlights the relevance of concurrent investigation of genomic regions in EWAS. Analyses of genomic regions contextualize CpGs to identify DMRs with possible functional relevance in regulation of gene transcription (Rakyan et al. 2011). It is interesting to note that transportation noise showed mostly decreased methylation, whereas air pollution showed mostly increased methylation at the respective DMRs. This difference in direction of association was also observed at the levels overlapping CpGs and annotated DMRs. The relevance of these findings remains to be clarified because DNA hypermethylation is often associated with transcriptional gene repression, whereas hypomethylation is associated with a chromatin configuration that allows transcription (Sawalha 2008). However, concordant hypermethylation in TRIM39 (railway noise and ) and HOXA2 (aircraft noise and ) and hypomethylation in SLC27A3 and EN2 (railway and road traffic noise) protein-coding genes indicate some synergistic pathways between exposures. TRIM39 activates the apoptotic signaling pathway and plays a role in cellular signaling and response to stimuli (Zhang et al. 2012), and its hypermethylation was associated with depression (Crawford et al. 2018). HOXA2 encodes a transcriptional regulator that controls cellular differentiation during development (Akin and Nazarali 2005). Hypermethylation at this gene was associated transcriptional suppression in various cancers (Li et al. 2013). SLC27A3 is involved in lipid metabolism and brain development (Stahl 2004), and hypomethylation in this gene was associated with recurrent endometrial carcinoma (Hsu et al. 2013). EN2 functions in the development of the central nervous system and is commonly associated with autism (Lupu et al. 2018; Márquez-Valadez et al. 2018). Aberrant methylation in EN2 was recently linked to clear cell renal carcinoma (Lai et al. 2017). Although these highlight certain distinct and shared pattern of associations, they demonstrate the complex network in the mechanisms linking these exposures to disease. The integration of gene expression or transcriptomic data in future studies will improve our understanding of the mechanisms going from divergence in directions of associations to convergence on pathway levels across these exposures.
The results demonstrating enrichment for DNA methylation associated with phenotypes of “allostatic load” for both air pollution and noise exposure are novel but in line with previously hypothesized mechanisms. Chronic low-level exposure to air pollution and noise are established risk factors for cardio-metabolic disease and were previously linked to the single phenotypes making up the “allostatic load” pathway in this study (Münzel and Daiber 2018; Thomson 2019). Recent experimental evidence specifically linked PM and ozone pollution as well as transportation noise to alterations in the HPA axis (Jafari et al. 2017). The findings of “allostatic load” enrichment agree with the functional enrichment of pathways related to oxidative stress and immune responses across all exposures. Furthermore, the enriched disease networks identified for overlaps the networks that were recently reported for in another study (Lee et al. 2019). Taken together, the evidence especially supports the recent links of transportation noise to markers of inflammation and oxidative stress (Bagheri Hosseinabadi et al. 2019; Münzel et al. 2017a, 2017b; Münzel and Daiber 2018). However, our findings on the inflammatory pathway might have been influenced by the higher prevalence of asthma in this sample (22%) in comparison with that of the entire SAPALDIA study (12%), but our models contained asthma status in addition to smoking and pack-years and should minimize this potential bias. In line with recent evidence on the early-life methylome effects of air pollution (Cai et al. 2017; Gruzieva et al. 2017) and potentially noise exposures, we observed enrichment of pathways related to embryonic and organ development for all exposures. Altered DNA methylation due to these exposures might therefore explain the reported associations of PM and poor birth outcomes (Liu et al. 2019; Smith et al. 2020), as well as the early-life theory of age-related disease (Walhovd et al. 2016). This finding is even more relevant if these methylation changes are heritable and, when confirmed, improves our understanding of the relevance of this pathway in relation to these exposures.
The relevance of confounding by leukocyte composition on the association between exposures and peripheral blood DNA methylation has been of interest (Heiss and Brenner 2017; van Rooij et al. 2019). In post hoc analyses, we demonstrated the general weakening of top CpGs (with some signals not attaining nominal significance) when leukocyte composition was not considered. However, main effect sizes remained robust (Table S7). Source-specific transportation noise and air pollution exposures were also associated with various leukocyte types (Table S8). These associations indicate that changes in leukocyte composition probably drive methylation at certain sites and should be considered potential confounders in this and similar studies. Our findings in this EWAS (adjusted for leukocyte composition) are more likely to reflect actual DNA methylation changes rather than the underlying leukocyte composition changes in relation to transportation noise and air pollution exposure. This finding would otherwise be difficult to interpret, given the observed associations of exposures with leukocyte types.
The strengths of our study include being the first EWAS to assess mutually independent effects of source-specific transportation noise and air pollution exposures. Our study considered in parallel the independent effects of these exposures on DNA methylation at single CpG sites and in genomic regions. The availability of individually assigned, source-specific noise and air pollution data, derived from validated models, allowed the exploration of mutual confounding of these exposures on DNA methylation. We were able to control for potential confounders given the detailed characterization of the participants in the SAPALDIA study. The multiexposure approach also allowed the investigation of the independent pathway enrichment using both candidate and agnostic approaches to improve mechanistic understanding of transportation noise and air pollution exposures. We used a novel approach to investigate indirectly the effect of these exposures on physiological stress systems. We could also explore the influence of leukocyte composition, among other sensitivity analyses. The availability of genome-wide methylation data allowed the validation of previously reported air pollution signals, contributing to their external validity in the SAPALDIA study.
Our study is limited in its cross-sectional design, which precludes causal inferences and differentiating short-term vs. long-term exposure effects. Our noise and air pollution estimates may have been biased by errors in input data, which could be exposure-specific and potentially account for variations in observed effects. Such bias, however, would most likely be nonsystematic and nondifferential. The air pollution and noise levels in the SAPALDIA study are relatively low and may have reduced statistical power of identifying signals and limited the replication of previous findings in settings of higher exposure. Unlike road traffic noise, which was ubiquitous, railway and aircraft noise in the SAPALDIA study were less common, with only exposure to these sources. Nevertheless, we made some significant observations. We could not identify any EWAS on stress hormones that directly captured alterations in the HPA axis; thus, our definition of “allostatic load” may have been biased. However, we expect any bias to be minimal, given our inclusion of CAR and other downstream biomarkers of physiological stress. As demonstrated in a recent review (Johnson et al. 2017), there is yet no consensus on which markers best capture “allostatic load.” We find it interesting that “allostatic load” score was always associated with negative health outcomes regardless of constituent biomarkers or phenotypes (Castagné et al. 2018; Johnson et al. 2017; Ribeiro et al. 2019).
Conclusions
DNA methylation was independently associated with transportation-related noise and air pollution exposures in the SAPALDIA study, with enrichment for pathways related to inflammation and immune response. Differential methylation due to these exposures may therefore explain the link between these exposures and several age-related outcomes. More EWAS with combined exposures and gene expression or transcriptome data are needed, especially from more polluted areas (including low- and middle-income countries), to corroborate present findings and to better capture the full extent and relevance of DNA methylation changes associated with these exposures. In particular, it remains to be seen if DNA methylation related to the identified pathways mediates the association between transportation noise and air pollution exposures and incident age-related phenotypes.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation, SNF-SAPALDIA (grant numbers 33CS30-148470/1, 33CSCO-134276/1, 33CSCO-108796, 324730_135673, 3247BO-104283, 3247BO-104288, 3247BO-104284, 3247-065896, 3100-059302, 3200-052720, 3200-042532, 4026-028099, PMPDP3_129021/1, PMPDP3_141671/1); SNF-SiRENE (grant number CRSII3_147635); the European Commission Horizon 2020 research and innovation programme (ALEC study; grant number 633212); grant FP7 of the European Commission “Enhanced exposure 36 assessment and -omic profiling for high priority environmental exposures in Europe” (EXPOsOMICS study; grant number 308610).
SAPALDIA is also supported by the Swiss Federal Office for the Environment; Federal Office of Public Health; the Federal Office of Roads and Transport; the canton’s government of Aargau, Basel-Stadt, Basel-Land, Geneva, Luzern, Ticino, Valais, and Zürich; the Swiss Lung League; the canton’s Lung League of Basel Stadt/Basel Landschaft, Geneva, Ticino, Valais, Graubünden and Zurich, Stiftung ehemals Bündner Heilstätten, SUVA, Freiwillige Akademische Gesellschaft; UBS Wealth Foundation; Talecris Biotherapeutics GmbH; Abbott Diagnostics; European Commission 018996 (GABRIEL); and Wellcome Trust WT 084703MA. MF. has received support of the Beatriu de Pinós postdoctoral programme of the Government of Catalonia’s Secretariat for Universities and Research of the Ministry of Economy and Knowledge.
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization (WHO), the authors alone are responsible for the views expressed in this article, and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/WHO.
References
- Abraham E, Rousseaux S, Agier L, Giorgis-Allemand L, Tost J, Galineau J, et al. 2018. Pregnancy exposure to atmospheric pollution and meteorological conditions and placental DNA methylation. Environ Int 118:334–347, PMID: 29935799, 10.1016/j.envint.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Ackermann-Liebrich U, Kuna-Dibbert B, Probst-Hensch NM, Schindler C, Felber Dietrich D, Stutz EZ, et al. 2005. Follow-up of the Swiss cohort study on air pollution and lung diseases in adults (SAPALDIA 2) 1991–2003: methods and characterization of participants. Soz Praventivmed 50(4):245–263, PMID: 16167509, 10.1007/s00038-005-4075-5. [DOI] [PubMed] [Google Scholar]
- Adalsteinsson BT, Gudnason H, Aspelund T, Harris TB, Launer LJ, Eiriksdottir G, et al. 2012. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLoS One 7(10):e46705, PMID: 23071618, 10.1371/journal.pone.0046705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam M, Schikowski T, Carsin AE, Cai Y, Jacquemin B, Sanchez M, et al. 2015. Adult lung function and long-term air pollution exposure. ESCAPE: a multicentre cohort study and meta-analysis. Eur Respir J 45(1):38–50, PMID: 25193994, 10.1183/09031936.00130014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin ZN, Nazarali AJ. 2005. Hox genes and their candidate downstream targets in the developing central nervous system. Cell Mol Neurobiol 25(3–4):697–741, PMID: 16075387, 10.1007/s10571-005-3971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano R, Herceg Z, Nawrot TS, Chadeau-Hyam M, Ghantous A, Plusquin M. 2018. The impact of air pollution on our epigenome: how far is the evidence? (A systematic review). Curr Environ Health Rep 5(4):544–578, PMID: 30361985, 10.1007/s40572-018-0218-8. [DOI] [PubMed] [Google Scholar]
- An R, Ji M, Yan H, Guan C. 2018a. Impact of ambient air pollution on obesity: a systematic review. Int J Obes (Lond) 42(6):1112–1126, PMID: 29795462, 10.1038/s41366-018-0089-y. [DOI] [PubMed] [Google Scholar]
- An R, Wang J, Ashrafi SA, Yang Y, Guan C. 2018b. Chronic noise exposure and adiposity: a systematic review and meta-analysis. Am J Prev Med 55(3):403–411, PMID: 30122217, 10.1016/j.amepre.2018.04.040. [DOI] [PubMed] [Google Scholar]
- An R, Zhang S, Ji M, Guan C. 2018c. Impact of ambient air pollution on physical activity among adults: a systematic review and meta-analysis. Perspect Public Health 138(2):111–121, PMID: 28829249, 10.1177/1757913917726567. [DOI] [PubMed] [Google Scholar]
- Andersen ZJ, Jørgensen JT, Elsborg L, Lophaven SN, Backalarz C, Laursen JE, et al. 2018. Long-term exposure to road traffic noise and incidence of breast cancer: a cohort study. Breast Cancer Res 20(1):119, PMID: 30290832, 10.1186/s13058-018-1047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. 2014. Minfi: A flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30(10):1363–1369, PMID: 24478339, 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri Hosseinabadi M, Khanjani N, Münzel T, Daiber A, Yaghmorloo M. 2019. Chronic occupational noise exposure: effects on DNA damage, blood pressure, and serum biochemistry. Mutat Res 841:17–22, PMID: 31138406, 10.1016/j.mrgentox.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Soft 67:, 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- Beelen R, Stafoggia M, Raaschou-Nielsen O, Andersen ZJ, Xun WW, Katsouyanni K, et al. 2014. Long-term exposure to air pollution and cardiovascular mortality: an analysis of 22 European cohorts. Epidemiology 25(3):368–378, PMID: 24589872, 10.1097/EDE.0000000000000076. [DOI] [PubMed] [Google Scholar]
- Breton CV, Yao J, Millstein J, Gao L, Siegmund KD, Mack W, et al. 2016. Prenatal air pollution exposures, DNA methyl transferase genotypes, and associations with newborn LINE1 and Alu methylation and childhood blood pressure and carotid intima-media thickness in the Children’s Health Study. Environ Health Perspect 124(12):1905–1912, PMID: 27219456, 10.1289/EHP181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JM, Cai YC, Demeler B, Spremulli LL. 1999. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J Mol Biol 288(4):567–577, PMID: 10329163, 10.1006/jmbi.1999.2708. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhao Y, Liu P, Xia B, Zhu Q, Wang X, et al. 2017. Exposure to particulate air pollution during early pregnancy is associated with placental DNA methylation. Sci Total Environ 607-608:1103–1108, PMID: 28724248, 10.1016/j.scitotenv.2017.07.029. [DOI] [PubMed] [Google Scholar]
- Castagné R, Garès V, Karimi M, Chadeau-Hyam M, Vineis P, Delpierre C, et al. 2018. Allostatic load and subsequent all-cause mortality: which biological markers drive the relationship? Findings from a UK birth cohort. Eur J Epidemiol 33(5):441–458, PMID: 29476357, 10.1007/s10654-018-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedernaes J, Osler ME, Voisin S, Broman JE, Vogel H, Dickson SL, et al. 2015. Acute sleep loss induces tissue-specific epigenetic and transcriptional alterations to circadian clock genes in men. J Clin Endocrinol Metab 100(9):E1255–E1261, PMID: 26168277, 10.1210/JC.2015-2284. [DOI] [PubMed] [Google Scholar]
- Charmpi K, Ycart B. 2015. Weighted Kolmogorov Smirnov testing: an alternative for Gene Set Enrichment analysis. Stat Appl Genet Mol Biol 14(3):279–293, PMID: 26030794, 10.1515/sagmb-2014-0077. [DOI] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8(2):203–209, PMID: 23314698, 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Pu W, Guo S, Jin L, He D, Wang J. 2019. Genome-wide DNA methylation profiles reveal common epigenetic patterns of interferon-related genes in multiple autoimmune diseases. Front Genet 10:223–223, PMID: 31024609, 10.3389/fgene.2019.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow BS. 1981. The role of lysosomes and proteases in hormone secretion and degradation. Endocr Rev 2(2):137–173, PMID: 6117463, 10.1210/edrv-2-2-137. [DOI] [PubMed] [Google Scholar]
- Clark C, Paunovic K. 2018. WHO environmental noise guidelines for the European region: a systematic review on environmental noise and cognition. IJERPH 15(2):285, PMID: 29414890, 10.3390/ijerph15020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commodore A, Mukherjee N, Chung D, Svendsen E, Vena J, Pearce J, et al. 2018. Frequency of heavy vehicle traffic and association with DNA methylation at age 18 years in a subset of the Isle of Wight birth cohort. Environ Epigenet 4:dvy028, PMID: 30697444, 10.1093/eep/dvy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford B, Craig Z, Mansell G, White I, Smith A, Spaull S, et al. 2018. DNA methylation and inflammation marker profiles associated with a history of depression. Hum Mol Genet 27(16):2840–2850, PMID: 29790996, 10.1093/hmg/ddy199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiber A, Kröller-Schön S, Frenis K, Oelze M, Kalinovic S, Vujacic‐Mirski K, et al. 2019. Environmental noise induces the release of stress hormones and inflammatory signaling molecules leading to oxidative stress and vascular dysfunction—signatures of the internal exposome. Biofactors 45(4):495–506, PMID: 30937979, 10.1002/biof.1506. [DOI] [PubMed] [Google Scholar]
- de FC Lichtenfels AJ, van der Plaat DA, de Jong K, van Diemen CC, Postma DS, Nedeljkovic I, et al. 2018. Long-term air pollution exposure, genome-wide DNA methylation and lung function in the LifeLines cohort study. Environ Health Perspect 126(2):027004, PMID: 29410382, 10.1289/EHP2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nys S, Duca RC, Nawrot T, Hoet P, Van Meerbeek B, Van Landuyt KL, et al. 2018. Temporal variability of global DNA methylation and hydroxymethylation in buccal cells of healthy adults: association with air pollution. Environ Int 111:301–308, PMID: 29217223, 10.1016/j.envint.2017.11.002. [DOI] [PubMed] [Google Scholar]
- De Prins S, Koppen G, Jacobs G, Dons E, Van de Mieroop E, Nelen V, et al. 2013. Influence of ambient air pollution on global DNA methylation in healthy adults: a seasonal follow-up. Environ Int 59:418–424, PMID: 23917442, 10.1016/j.envint.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Eeftens M, Meier R, Schindler C, Aguilera I, Phuleria H, Ineichen A, et al. 2016. Development of land use regression models for nitrogen dioxide, ultrafine particles, lung deposited surface area, and four other markers of particulate matter pollution in the Swiss SAPALDIA regions. Environ Health 15:53, PMID: 27089921, 10.1186/s12940-016-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMPA (Eidgenössische Materialprüfungs und Forschunganstalt). 2010. FLULA2, Ein Verfahren zur Berechnung und Darstellung der Fluglärmbelastung. Technische Programm Dokumentation version 4. Dübendorf, Switzerland: EMPA, Akustik/Lärmminderung. [Google Scholar]
- Eze IC, Foraster M, Schaffner E, Vienneau D, Heritier H, Pieren R, et al. 2018. Transportation noise exposure, noise annoyance and respiratory health in adults: a repeated-measures study. Environ Int 121(pt 1):741–750, PMID: 30321849, 10.1016/j.envint.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Eze IC, Foraster M, Schaffner E, Vienneau D, Heritier H, Rudzik F, et al. 2017. Long-term exposure to transportation noise and air pollution in relation to incident diabetes in the SAPALDIA study. Int J Epidemiol 46(4):1115–1125, PMID: 28338949, 10.1093/ije/dyx020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze IC, Hemkens LG, Bucher HC, Hoffmann B, Schindler C, Kunzli N, et al. 2015. Association between ambient air pollution and diabetes mellitus in Europe and North America: systematic review and meta-analysis. Environ Health Perspect 123(5):381–389, PMID: 25625876, 10.1289/ehp.1307823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze IC, Probst-Hensch N. 2019. Editorial commentary: ecology of cardio-metabolic diseases: low-income countries also matter. Trends Cardiovasc Med 29(5):283–284, PMID: 30471986, 10.1016/j.tcm.2018.11.008. [DOI] [PubMed] [Google Scholar]
- Fiorito G, Vlaanderen J, Polidoro S, Gulliver J, Galassi C, Ranzi A, et al. 2018. Oxidative stress and inflammation mediate the effect of air pollution on cardio- and cerebrovascular disease: a prospective study in nonsmokers. Environ Mol Mutagen 59(3):234–246, PMID: 29114965, 10.1002/em.22153. [DOI] [PubMed] [Google Scholar]
- FOEN (Federal Office of the Environment). 1987. Computer Model for the Calculation of Street Noise, Part 1, Operating Instructions for the Computer Program STL-86, in the Environmental Protection Series. Bern, Switzerland: Swiss Federal Office for the Environment. [Google Scholar]
- FOEN. 1990. SEMIBEL: Swiss Emission and Immission Model for the Calculation of Railway Noise. Bern, Switzerland: Swiss Federal Office for the Environment. [Google Scholar]
- Foraster M, Eze IC, Schaffner E, Vienneau D, Heritier H, Endes S, et al. 2017. Exposure to road, railway, and aircraft noise and arterial stiffness in the SAPALDIA study: annual average noise levels and temporal noise characteristics. Environ Health Perspect 125(9):097004, PMID: 28934719, 10.1289/EHP1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foraster M, Eze IC, Vienneau D, Brink M, Cajochen C, Caviezel S, et al. 2016. Long-term transportation noise annoyance is associated with subsequent lower levels of physical activity. Environ Int 91:341–349, PMID: 27030897, 10.1016/j.envint.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Franklin M, Fruin S. 2017. The role of traffic noise on the association between air pollution and children’s lung function. Environ Res 157:153–159, PMID: 28558263, 10.1016/j.envres.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschi L, Brown L, Kim R, Schwela D, Kephalopoulos S. 2011. Burden of disease from environmental noise: quantification of healthy life years lost in Europe. https://apps.who.int/iris/bitstream/handle/10665/326424/9789289002295-eng.pdf. [accessed 10 May 2019].
- Gaine ME, Chatterjee S, Abel T. 2018. Sleep deprivation and the epigenome. Front Neural Circuits 12:14, PMID: 29535611, 10.3389/fncir.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondalia R, Baldassari A, Holliday KM, Justice AE, Méndez-Giráldez R, Stewart JD, et al. 2019. Methylome-wide association study provides evidence of particulate matter air pollution-associated DNA methylation. Environ Int 132:104723, PMID: 31208937, 10.1016/j.envint.2019.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Anto JM, Auffray C, et al. 2017. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ Health Perspect 125(1):104–110, PMID: 27448387, 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Li PH, Li H, Colicino E, Colicino S, Wen Y, et al. 2017. Effects of environmental noise exposure on DNA methylation in the brain and metabolic health. Environ Res 153:73–82, PMID: 27914298, 10.1016/j.envres.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Hänninen O, Knol AB, Jantunen M, Lim TA, Conrad A, Rappolder M, et al. 2014. Environmental burden of disease in Europe: assessing nine risk factors in six countries. Environ Health Perspect 122(5):439–446, PMID: 24584099, 10.1289/ehp.1206154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegewald J, Schubert M, Wagner M, Dröge P, Prote U, Swart E, et al. 2017. Breast cancer and exposure to aircraft, road, and railway-noise: a case–control study based on health insurance records. Scand J Work Environ Health 43(6):509–518, PMID: 28813586, 10.5271/sjweh.3665. [DOI] [PubMed] [Google Scholar]
- Heiss JA, Brenner H. 2017. Impact of confounding by leukocyte composition on associations of leukocyte DNA methylation with common risk factors. Epigenomics 9(5):659–668, PMID: 28470095, 10.2217/epi-2016-0154. [DOI] [PubMed] [Google Scholar]
- Heldstab J, Haan P, Künzle T, Keller M, Zbinden R. 2003. Modelling of PM10 and PM2.5 ambient concentrations in Switzerland 2000 and 2010. Environmental Documentation 169. Bern, Switzerland: Swiss Federal Office for the Environment. [Google Scholar]
- Hemmingsen JG, Møller P, Jantzen K, Jönsson BA, Albin M, Wierzbicka A, et al. 2015. Controlled exposure to diesel exhaust and traffic noise–effects on oxidative stress and activation in mononuclear blood cells. Mutat Res 775:66–71, PMID: 25898780, 10.1016/j.mrfmmm.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Heritier H, Vienneau D, Foraster M, Eze IC, Schaffner E, de Hoogh K, et al. 2019. A systematic analysis of mutual effects of transportation noise and air pollution exposure on myocardial infarction mortality: a nationwide cohort study in Switzerland. Eur Heart J 40(7):598–603, PMID: 30357335, 10.1093/eurheartj/ehy650. [DOI] [PubMed] [Google Scholar]
- Heutschi K. 2004. sonROAD: new Swiss road traffic noise model. Acta Acust united Ac 90:548–554. [Google Scholar]
- Hoek G, Krishnan RM, Beelen R, Peters A, Ostro B, Brunekreef B, et al. 2013. Long-term air pollution exposure and cardio-respiratory mortality: a review. Environ Health 12(1):43, PMID: 23714370, 10.1186/1476-069X-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86, PMID: 22568884, 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-T, Gu F, Huang Y-W, Liu J, Ruan J, Huang R-L, et al. 2013. Promoter hypomethylation of EpCAM-regulated bone morphogenetic protein gene family in recurrent endometrial cancer. Clin Cancer Res 19(22):6272–6285, PMID: 24077349, 10.1158/1078-0432.CCR-13-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imboden M, Wielscher M, Rezwan FI, Amaral AFS, Schaffner E, Jeong A, et al. 2019. Epigenome-wide association study of lung function level and its change. Eur Respir J 54(1):1900457, PMID: 31073081, 10.1183/13993003.00457-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imgenberg-Kreuz J, Carlsson Almlöf J, Leonard D, Alexsson A, Nordmark G, Eloranta M-L, et al. 2018. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann Rheum Dis 77(5):736–743, PMID: 29437559, 10.1136/annrheumdis-2017-212379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari Z, Mehla J, Kolb BE, Mohajerani MH. 2017. Prenatal noise stress impairs HPA axis and cognitive performance in mice. Sci Rep 7(1):10560, PMID: 28874680, 10.1038/s41598-017-09799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BG, Byun HM, Gyselaers W, Lefebvre W, Baccarelli AA, Nawrot TS. 2015. Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: an ENVIRONAGE birth cohort study. Epigenetics 10(6):536–544, PMID: 25996590, 10.1080/15592294.2015.1048412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong A, Imboden M, Ghantous A, Novoloaca A, Carsin A-E, Kogevinas M, et al. 2019. DNA methylation in inflammatory pathways modifies the association between BMI and adult-onset non-atopic asthma. IJERPH 16(4):600, PMID: 30791383, 10.3390/ijerph16040600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Cavallaro FL, Leon DA. 2017. A systematic review of allostatic load in relation to socioeconomic position: poor fidelity and major inconsistencies in biomarkers employed. Soc Sci Med 192:66–73, PMID: 28963986, 10.1016/j.socscimed.2017.09.025. [DOI] [PubMed] [Google Scholar]
- Karipidis I, Vienneau D, Habermacher M, Köpflii M, Brink M, Probst-Hensch N, et al. 2014. Reconstruction of historical noise exposure data for environmental epidemiology in Switzerland within the SiRENE project. Noise Mapp 1(1):3–14, 10.2478/noise-2014-0002. [DOI] [Google Scholar]
- Kempen EV, Casas M, Pershagen G, Foraster M. 2018. WHO environmental noise guidelines for the European region: a systematic review on environmental noise and cardiovascular and metabolic effects: a summary. IJERPH 15(2):379, PMID: 29470452, 10.3390/ijerph15020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. 2019. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet 104(1):65–75, PMID: 30595370, 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C-Y, Yu G-S, Xu Y, Wu X, Heng B-L, Xue Y-J, et al. 2017. Engrailed-2 promoter hyper-methylation is associated with its downregulation in clear cell renal cell carcinoma. Oncol Lett 14(6):6888–6894, PMID: 29151918, 10.3892/ol.2017.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. 2018. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 50(8):1112–1121, PMID: 30038396, 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Xu CJ, Carnes MU, Nichols CE, Ward JM, Kwon SO, et al. 2019. Genome-wide DNA methylation and long-term ambient air pollution exposure in Korean adults. Clin Epigenet 11(1):37, PMID: 30819252, 10.1186/s13148-019-0635-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, et al. 2015. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol 16:37, PMID: 25853392, 10.1186/s13059-015-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Parrado AR, Samtani MN, Narayan VA, Alzheimer’s Disease Neuroimaging Initiative. 2015. Variations in the FRA10AC1 fragile site and 15q21 are associated with cerebrospinal fluid Aβ1-42 level. PLoS One 10(8):e0134000, PMID: 26252872, 10.1371/journal.pone.0134000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HP, Peng CC, Chung IC, Huang M-Y, Huang ST, Chen CC, et al. 2013. Aberrantly hypermethylated Homeobox A2 derepresses metalloproteinase-9 through TBP and promotes invasion in Nasopharyngeal carcinoma. Oncotarget 4(11):2154–2165, PMID: 24243817, 10.18632/oncotarget.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zou D, Li Z, Gao R, Sang J, Zhang Y, et al. 2019. EWAS atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res 47(D1):D983–D988, PMID: 30364969, 10.1093/nar/gky1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, et al. 2016. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol 17(1):255, 10.1186/s13059-016-1119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Tsai M-Y, Keidel D, Gemperli A, Ineichen A, Hazenkamp-von Arx M, et al. 2012. Long-term exposure models for traffic related NO2 across geographically diverse areas over separate years. Atmos Environ 46:460–471, 10.1016/j.atmosenv.2011.09.021. [DOI] [Google Scholar]
- Liu Y, Xu J, Chen D, Sun P, Ma X. 2019. The association between air pollution and preterm birth and low birth weight in Guangdong, China. BMC Public Health 19(1):3, PMID: 30606145, 10.1186/s12889-018-6307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu D, Varshney MK, Mucs D, Inzunza J, Norinder U, Loghin F, et al. 2018. Fluoxetine affects differentiation of midbrain dopaminergic neurons in vitro. Mol Pharmacol 94(4):1220–1231, PMID: 30115672, 10.1124/mol.118.112342. [DOI] [PubMed] [Google Scholar]
- Lv H, Zhang M, Shang Z, Li J, Zhang S, Lian D, et al. 2017. Genome-wide haplotype association study identify the FGFR2 gene as a risk gene for acute myeloid leukemia. Oncotarget 8(5):7891–7899, PMID: 27903959, 10.18632/oncotarget.13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Valadez B, Valle-Bautista R, García-López G, Díaz NF, Molina-Hernández A. 2018. Maternal diabetes and fetal programming toward neurological diseases: beyond neural tube defects. Front Endocrinol (Lausanne) 9:664, PMID: 30483218, 10.3389/fendo.2018.00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BW, Ackermann-Liebrich U, Leuenberger P, Kunzli N, Stutz EZ, Keller R, et al. 1997. SAPALDIA: methods and participation in the cross-sectional part of the Swiss Study on Air Pollution and Lung Diseases in Adults. Soz Praventivmed 42(2):67–84, PMID: 9151378, 10.1007/BF01318136. [DOI] [PubMed] [Google Scholar]
- McCrory C, Fiorito G, Ni Cheallaigh C, Polidoro S, Karisola P, Alenius H, et al. 2019. How does socio-economic position (SEP) get biologically embedded? A comparison of allostatic load and the epigenetic clock(s). Psychoneuroendocrinology 104:64–73, PMID: 30818253, 10.1016/j.psyneuen.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Mostafavi N, Vermeulen R, Ghantous A, Hoek G, Probst-Hensch N, Herceg Z, et al. 2018. Acute changes in DNA methylation in relation to 24 h personal air pollution exposure measurements: a panel study in four European countries. Environ Int 120:11–21, PMID: 30055357, 10.1016/j.envint.2018.07.026. [DOI] [PubMed] [Google Scholar]
- Münzel T, Daiber A. 2018. Environmental stressors and their impact on health and disease with focus on oxidative stress. Antioxid Redox Signal 28(9):735–740, PMID: 29278923, 10.1089/ars.2017.7488. [DOI] [PubMed] [Google Scholar]
- Münzel T, Daiber A, Steven S, Tran LP, Ullmann E, Kossmann S, et al. 2017a. Effects of noise on vascular function, oxidative stress, and inflammation: mechanistic insight from studies in mice. Eur Heart J 38(37):2838–2849, PMID: 28329261, 10.1093/eurheartj/ehx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel T, Sørensen M, Gori T, Schmidt FP, Rao X, Brook FR, et al. 2017b. Environmental stressors and cardio-metabolic disease: part II-mechanistic insights. Eur Heart J 38(8):557–564, PMID: 27460891, 10.1093/eurheartj/ehw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz SK, Hasnain S. 2013. Occupational noise exposure may induce oxidative DNA damage. Pol J Environ Stud 22:1547–1551. [Google Scholar]
- Nwanaji-Enwerem JC, Colicino E, Trevisi L, Kloog I, Just AC, Shen J, et al. 2016. Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ Epigenet 2(2):dvw006, PMID: 27453791, 10.1093/eep/dvw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwanaji-Enwerem JC, Dai L, Colicino E, Oulhote Y, Di Q, Kloog I, et al. 2017. Associations between long-term exposure to PM2.5 component species and blood DNA methylation age in the elderly: the VA normative aging study. Environ Int 102:57–65, PMID: 28284819, 10.1016/j.envint.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell S, Borowski K, Espin-Garcia O, Milgrom R, Kabakchiev B, Stempak J, et al. 2019. The unsolved link of genetic markers and Crohn’s disease progression: a North American cohort experience. Inflamm Bowel Dis 25(9):1541–1549, PMID: 30801121, 10.1093/ibd/izz016. [DOI] [PubMed] [Google Scholar]
- Panczak R, Galobardes B, Voorpostel M, Spoerri A, Zwahlen M, Egger M, et al. 2012. A Swiss neighbourhood index of socioeconomic position: development and association with mortality. J Epidemiol Community Health 66(12):1129–1136, PMID: 22717282, 10.1136/jech-2011-200699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, et al. 2016. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the Normative Aging Study. Environ Health Perspect 124(7):983–990, PMID: 26731791, 10.1289/ehp.1509966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, et al. 2015. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8:6, PMID: 25972926, 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusquin M, Chadeau-Hyam M, Ghantous A, Alfano R, Bustamante M, Chatzi L, et al. 2018. DNA methylome marks of exposure to particulate matter at three time points in early life. Environ Sci Technol 52(9):5427–5437, PMID: 29597345, 10.1021/acs.est.7b06447. [DOI] [PubMed] [Google Scholar]
- Plusquin M, Guida F, Polidoro S, Vermeulen R, Raaschou-Nielsen O, Campanella G, et al. 2017. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ Int 108:127–136, PMID: 28843141, 10.1016/j.envint.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, et al. 2019. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet 28(1):166–174, PMID: 30239722, 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaschou-Nielsen O, Andersen ZJ, Beelen R, Samoli E, Stafoggia M, Weinmayr G, et al. 2013. Air pollution and lung cancer incidence in 17 European cohorts: prospective analyses from the European Study of Cohorts for Air Pollution Effects (ESCAPE). Lancet Oncology 14(9):813–822, PMID: 23849838, 10.1016/S1470-2045(13)70279-1. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. 2011. Epigenome-wide association studies for common human diseases. Nat Rev Genet 12(8):529–541, PMID: 21747404, 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D, et al. 2012. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7(7):e41361, PMID: 22848472, 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AI, Fraga S, Kelly-Irving M, Delpierre C, Stringhini S, Kivimaki M, et al. 2019. Neighbourhood socioeconomic deprivation and allostatic load: a multi-cohort study. Sci Rep 9(1):8790, PMID: 31217447, 10.1038/s41598-019-45432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röösli M, Vienneau D, Foraster M, Eze IC, Héritier H, Schaffner E, et al. 2017. Short and long term effects of transportation noise exposure (SiRENE): an interdisciplinary approach. In: Proceedings of the 12th ICBEN Congress on Noise as a Public Health Problem, Zurich.
- Roswall N, Ammitzbøll G, Christensen JS, Raaschou-Nielsen O, Jensen SS, Tjønneland A, et al. 2017. Residential exposure to traffic noise and leisure-time sports – a population-based study. Int J Hyg Environ Health 220(6):1006–1013, PMID: 28579229, 10.1016/j.ijheh.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Russo G, Landi R, Pezone A, Morano A, Zuchegna C, Romano A, et al. 2016. DNA damage and repair modify DNA methylation and chromatin domain of the targeted locus: mechanism of allele methylation polymorphism. Sci Rep 6:33222, PMID: 27629060, 10.1038/srep33222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawalha AH. 2008. Epigenetics and t-cell immunity. Autoimmunity 41(4):245–252, PMID: 18432405, 10.1080/08916930802024145. [DOI] [PubMed] [Google Scholar]
- Sayols-Baixeras S, Fernández-Sanlés A, Prats-Uribe A, Subirana I, Plusquin M, Künzli N, et al. 2019. Association between long-term air pollution exposure and DNA methylation: the REGICOR study. Environ Res 176:108550, PMID: 31260916, 10.1016/j.envres.2019.108550. [DOI] [PubMed] [Google Scholar]
- Schlatter F, Piquerez A, Habermacher M, Ragettli M, Röösli M, Brink M, et al. 2017. Validation of large scale noise exposure modelling by long-term measurements. Noise Mapp 4(1):75–86, 10.1515/noise-2017-0006. [DOI] [Google Scholar]
- Seeman T, Epel E, Gruenewald T, Karlamangla A, McEwen BS. 2010. Socio‐economic differentials in peripheral biology: cumulative allostatic load. Ann NY Acad Sci 1186:223–239, PMID: 20201875, 10.1111/j.1749-6632.2009.05341.x. [DOI] [PubMed] [Google Scholar]
- Smith RB, Beevers SD, Gulliver J, Dajnak D, Fecht D, Blangiardo M, et al. 2020. Impacts of air pollution and noise on risk of preterm birth and stillbirth in London. Environ Int 134:105290, PMID: 31783238, 10.1016/j.envint.2019.105290. [DOI] [PubMed] [Google Scholar]
- Stafoggia M, Cesaroni G, Peters A, Andersen ZJ, Badaloni C, Beelen R, et al. 2014. Long-term exposure to ambient air pollution and incidence of cerebrovascular events: results from 11 European cohorts within the ESCAPE project. Environ Health Perspect 122(9):919–925, PMID: 24835336, 10.1289/ehp.1307301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl A. 2004. A current review of fatty acid transport proteins (SLC27). Pflugers Arch 447(5):722–727, PMID: 12856180, 10.1007/s00424-003-1106-z. [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 K DNA methylation data. Bioinformatics 29(2):189–196, PMID: 23175756, 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault LF, Perron S, Smargiassi A. 2013. Cardiovascular health, traffic-related air pollution and noise: are associations mutually confounded? A systematic review. Int J Public Health 58(5):649–666, PMID: 23887610, 10.1007/s00038-013-0489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson EM. 2019. Air pollution, stress, and allostatic load: linking systemic and central nervous system impacts. J Alzheimers Dis 69(3):597–614, PMID: 31127781, 10.3233/JAD-190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thron T, Hecht M. 2010. The sonRAIL emission model for railway noise in Switzerland. Acta Acust united Ac 96(5):873–883, 10.3813/AAA.918346. [DOI] [Google Scholar]
- Triche TJ Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. 2013. Low-level processing of Illumina Infinium DNA methylation beadarrays. Nucleic Acids Res 41(7):e90, PMID: 23476028, 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. 1977. Exploratory Data Analysis. Reading, Massachusetts: Addison-Wesley. [Google Scholar]
- van der Laan MJ, Birkner MD, Hubbard AE. 2005. Empirical Bayes and resampling based multiple testing procedure controlling tail probability of the proportion of false positives. Stat Appl Genet Mol Biol 4(1):Article29, PMID: 16646847, 10.2202/1544-6115.1143. [DOI] [PubMed] [Google Scholar]
- van Rooij J, Mandaviya PR, Claringbould A, Felix JF, van Dongen J, Jansen R, et al. 2019. Evaluation of commonly used analysis strategies for epigenome- and transcriptome-wide association studies through replication of large-scale population studies. Genome Biol 20(1):235–235, PMID: 31727104, 10.1186/s13059-019-1878-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vienneau D, de Hoogh K, Faeh D, Kaufmann M, Wunderli JM, Röösli M, et al. 2017. More than clean air and tranquility: residential green is independently associated with decreasing mortality. Environ Int 108:176–184, PMID: 28863390, 10.1016/j.envint.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Vienneau D, Perez L, Schindler C, Lieb C, Sommer H, Probst-Hensch N, et al. 2015. Years of life lost and morbidity cases attributable to transportation noise and air pollution: a comparative health risk assessment for Switzerland in 2010. Int J Hyg Environ Health 218(6):514–521, PMID: 26003939, 10.1016/j.ijheh.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Vineis P, Chadeau-Hyam M, Gmuender H, Gulliver J, Herceg Z, Kleinjans J, et al. EXPOsOMICS Consortium. 2017. The exposome in practice: design of the EXPOsOMICS project. Int J Hyg Environ Health 220(2 pt A):142–151, PMID: 27576363, 10.1016/j.ijheh.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Krogsrud SK, Amlien IK, Bartsch H, Bjørnerud A, Due-Tønnessen P, et al. 2016. Neurodevelopmental origins of lifespan changes in brain and cognition. Proc Natl Acad Sci USA 113(33):9357–9362, PMID: 27432992, 10.1073/pnas.1524259113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward-Caviness CK, Nwanaji-Enwerem JC, Wolf K, Wahl S, Colicino E, Trevisi L, et al. 2016. Long-term exposure to air pollution is associated with biological aging. Oncotarget 7(46):74510–74525, PMID: 27793020, 10.18632/oncotarget.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler E, Huang N, Bochukova EG, Keogh JM, Lindsay S, Garg S, et al. 2013. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet 45(5):513–517, PMID: 23563609, 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5(10):781–791, PMID: 15459659, 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Zare Sakhvidi MJ, Zare Sakhvidi F, Mehrparvar AH, Foraster M, Dadvand P. 2018. Association between noise exposure and diabetes: a systematic review and meta-analysis. Environ Res 166:647–657, PMID: 30006240, 10.1016/j.envres.2018.05.011. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen X, Zhang X. 2018. The impact of exposure to air pollution on cognitive performance. Proc Natl Acad Sci USA 115(37):9193–9197, PMID: 30150383, 10.1073/pnas.1809474115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Mei Y, Fu NY, Guan L, Xie W, Liu HH, et al. 2012. TRIM39 regulates cell cycle progression and DNA damage responses via stabilizing p21. Proc Natl Acad Sci USA 109(51):20937–20942, PMID: 23213251, 10.1073/pnas.1214156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Stevens RG, Hoffman AE, Tjonneland A, Vogel UB, Zheng T, et al. 2011. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol Int 28(10):852–861, PMID: 22080730, 10.3109/07420528.2011.618896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Figure 1 is a flow chart with six steps. Step 1. SAPALDIA 1 1991; n equals 9,551. Step 2. SAPALDIA 2 2002; n equals 8,047. Step 3. SAPALDIA 3 2010; n equals 6,088. Step 4. SAPALDIA 2 2002; n equals 8,047 is divided into two parts ALEC and EXPOsOMICS. ALEC leads to 987 representative sample selected for D N A methylation assessment; 983 participants with methylation data overlapping C p Gs passing quality control; and 972 participants with complete data on transportation noise, air pollution, and relevant covariates. EXPOsOMICS leads to 201 nonsmoking asthma controls selected for DNA methylation assessment; 198 participants with methylation data in overlapping C p Gs passing quality control; and 198 participants with complete data on transportation noise, air pollution, and relevant covariates. ALEC and EXPOsOMICS lead to 1,170 participants included in the analysis. SAPALDIA 3 2010; n equals 6,088 is divided into two parts ALEC and EXPOsOMICS. ALEC leads to 987 participants (same SAPALDIA 2 participants) selected for DNA methylation assessment; 983 participants with methylation data in overlapping C p Gs passing quality control; and 970 participants with complete data on transportation noise, air pollution, and relevant covariates. EXPOsOMICS leads to 405 nonsmoking participants [201 non-asthma controls (same SAPALDIA 2 participants) and 204 asthma cases] selected for DNA methylation assessment; 402 participants (198 asthma controls and 204 asthma cases) with methylation data overlapping CpGs passing quality control; and 402 participants (198 asthma controls and 204 asthma cases) with complete data on transportation noise, air pollution, and relevant covariates. ALEC and EXPOsOMICS lead to 1,372 participants included in the analyses. The final outcome is 1,389 participants contributing 2,542 observations to final analyses: 985 ALEC participants contributing 1,942 observations and 404 EXPOsOMICS participants contributing 600 observations.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d2ff/7263738/5fdbe7f3ecae/ehp6174_f1.jpg)