SUMMARY
The organization and number of microtubules (MTs) in a cell depends on the proper regulation of MT nucleation. Currently, the mechanism of nucleation is the most poorly understood aspect of MT dynamics. XMAP215/chTOG/Alp14/Stu2 proteins are MT polymerases that stimulate MT polymerization at MT plus ends by binding and releasing tubulin dimers. Although these proteins also localize to MT organizing centers and have nucleating activity in vitro, it is not yet clear whether these proteins participate in MT nucleation in vivo. Here, we demonstrate that in the fission yeast Schizosaccharomyces pombe, the XMAP215 ortholog Alp14 is critical for efficient MT nucleation in vivo. In multiple assays, loss of Alp14 function led to reduced nucleation rate and numbers of interphase MT bundles. Conversely, activation of Alp14 led to increased nucleation frequency. Alp14 associated with Mto1 and γ-tubulin complex components, and artificially targeting Alp14 to the γ-TuRCs stimulated nucleation. In imaging individual nucleation events, we found that Alp14 transiently associated with a γ-tubulin particle shortly before the appearance of a new MT. The TACC ortholog Alp7 mediated the localization of Alp14 at nucleation sites but not plus ends, and was required for efficient nucleation but not for MT polymerization. Our findings provide the strongest evidence to date that Alp14 serves as a critical MT nucleation factor in vivo. We suggest a model in which Alp14 associates with the γ-tubulin complex in an Alp7-dependent manner to facilitate the assembly or stabilization of the nascent MT.
eTOC Blurb
Microtubules are dynamic polymers that help to organize cellular contents and divide the cell. New microtubules arise by a process of nucleation, in which tubulin subunits are stitched together to begin forming a hollow tube. Flor-Parra et al. identify XMAP215/Alp14 as a nucleation factor that may facilitate the assembly of the microtubule.
INTRODUCTION
Microtubules (MTs) are hollow tubules of αβ-tubulin responsible for cellular processes such as chromosome segregation, nuclear positioning, cytokinesis, cell migration, and membrane transport. Regulation of MT nucleation underlies the spatial distribution and density of MTs in the cell. The molecular mechanism of MT nucleation remains one of the most poorly understood aspects of MT regulation. The γ-tubulin ring complex has been identified as a key MT nucleation factor [1, 2]. Current models suggest that this complex acts as a template for the formation of the αβ-tubulin MT [1, 3]. However by themselves, purified γ-tubulin complexes have low activity in vitro [1, 4], suggesting that additional factors are needed for full nucleation activity. A current priority in the MT nucleation field is to identify and characterize additional nucleation factors. Nucleation activators may induce conformational changes within the γ-tubulin complex to bring the γ-tubulins into the proper configuration to template the MT [5]. Nucleation may also be stimulated in principle by other factors that promote the addition and stabilization of αβ-tubulin dimers onto the γ-tubulin ring.
XMAP215/chTOG/Alp14/Stu2 family proteins are conserved MT regulatory proteins that have been generally implicated as MT polymerases that associate with growing MT plus ends and increase polymerization rates [6, 7]. These proteins contain multiple TOG domains that bind tubulin dimers and other regions that mediate binding to the MT lattice [7–9]. Current models suggest that these proteins bind and release tubulin dimers into the MT lattice at the growing plus end to promote MT polymerization [6, 7, 10, 11]. Recent studies defined the roles of XMAP215 and TACC orthologs at the kinetochore during mitosis [12, 13]. In addition to their localization at MT plus ends, early studies also found that XMAP215 family proteins also localize to MT organizing centers (MTOCs) and associate with Transforming Acidic Coiled Coil (TACC) proteins, which are implicated in MT nucleation[14, 15] However, the function of XMAP215 at MTOCs remains unclear. Purified XMAP215 and Alp14 exhibit MT nucleation activity in vitro even by themselves [16–18], although, the biological relevance of these observations has been questioned, as they could reflect artifacts of in vitro conditions. In vitro, XMAP215 also stimulates growth of MTs from MT seeds and centrosomes [16, 19]. In general, whether XMAP215 orthologs function in MT nucleation in vivo remains to be established.
The fission yeast Schizosaccharomyces pombe is a potent model for studying MT nucleation because of its simple MT cytoskeleton, genetic tractability, limited number of regulators, and amenability to assays of individual nucleation events. In fission yeast, MTs are nucleated from MTOCs at the spindle pole body (SPB), the equatorial division site (eMTOC), the nuclear envelope, and pre-existing MT bundles [20]. Efficient nucleation requires components of the γ-tubulin ring complex and the proteins Mto1 and Mto2, which may recruit and activate γ-tubulin ring complexes at cytoplasmic MTOCs [20, 21]. MTs in cytoplasmic bundles are often nucleated by γ-tubulin complex-containing particles termed “satellites” which reside on interphase MTs and occasionally nucleate a MT that incorporates into the pre-existing MT bundle [20, 22, 23].
Fission yeast has two orthologs of XMAP215: Alp14 and Dis1. alp14 null mutants are viable but display spindle defects and contain fewer interphase MT bundles, with MTs that grow about half as rapidly as MTs in wild-type cells [24, 25]. Alp14 tracks MT plus ends in vivo [25]. Purified Alp14 tracks MT plus ends in vitro and stimulates MT polymerization rate by 2–3-fold [18, 25]. In contrast, Dis1 does not appear to track MT plus ends and does not affect the dynamics of interphase MTs [25, 26]. As with other XMAP215 orthologs, previous studies of Alp14 focused on its function as a polymerase [18, 25].
Here, we investigated the role of Alp14 in MT nucleation. Genetic phenotypes revealed that a primary cytoplasmic function of Alp14 is MT nucleation. We found that Alp14 associates with components of the γ-tubulin complex, and that increasing the affinity of Alp14 to this complex increases the nucleation frequency. Further, Alp14 is recruited transiently at sites of MT nucleation by the TACC Alp7, promoting efficient MT nucleation. These findings establish the significance of Alp14/XMAP215 and its interacting protein Alp7/TACC in MT nucleation in the context of a living cell.
RESULTS
alp14Δ Mutants are Defective in Microtubule Nucleation
Fission yeast cells lacking Alp14 (alp14Δ) display fewer interphase MT bundles [18, 25]. To investigate the decrease in MT bundle number, we measured the effect of Alp14 on MT nucleation rates using four assays.
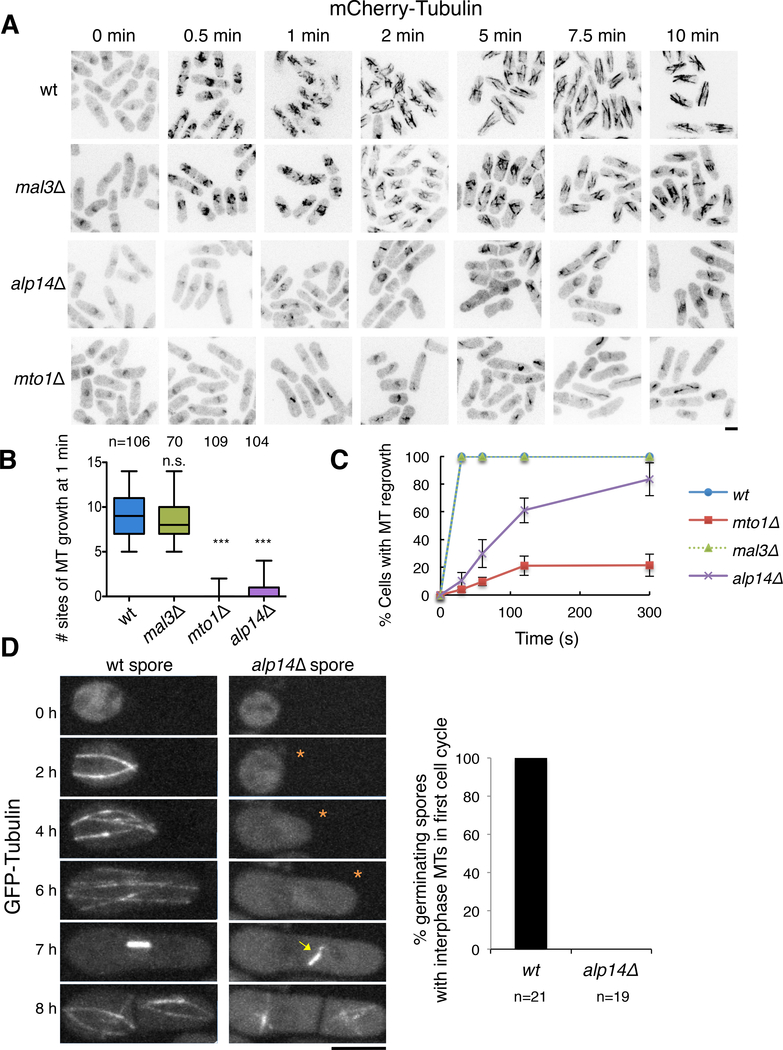
First, we performed a standard nucleation assay in which we assayed MT regrowth after depolymerization by cold treatment [27]. When wildtype cells were chilled on ice and then returned to 30˚C, MTs reappeared within 30 s, with the full MT cytoskeleton restored by 5–10 min (Figure 1A–C) [27, 28]. alp14Δ cells showed a delay in MT regrowth, similar to that displayed by the MT nucleation mutant mto1Δ (Figure 1A–C) [29]. In contrast, cells deleted for the MT plus end protein mal3 (EB protein) exhibited no delay (Figure 1A–C). These initial observations suggest a role of Alp14 in MT nucleation.
Figure 1. Alp14 Is Required For Efficient Nucleation of Cytoplasmic MTs After Cold Treatment and During Spore Germination.
A. alp14Δ cells are defective in MT regrowth after cold treatment. S. pombe cells expressing mCherry-Tubulin were incubated in ice-cold water for 30 min to depolymerize MTs, returned to 25 °C, and fixed at the indicated times. Confocal inverse images are shown.
B. Boxplots of the number of sites of MT growth after 1 min recovery from cold treatment in cells from (A).
C. Percentage of cells with detectable MTs upon recovery from cold treatment in cells from (A). Mean and standard deviation from two independent experiments are shown. Note that the wild-type (wt; blue) and mal3Δ (green) curves overlap.
D. alp14Δ germinating spores lack interphase MTs during the first cell cycle. wt and alp14Δ spores expressing GFP-Tubulin were placed on rich medium at 30 °C at time 0 to initiate germination, and were imaged at the indicated times. All wt cells formed cytoplasmic interphase MT bundles after 2 h, while none of the alp14Δ spores did so in the first cell cycle (asterisks) until after the mitotic spindle formed (yellow arrow head). Scale bars = 5 μm.
Second, we observed that alp14Δ cells have a strong defect in MT formation in germinating spores (Figure 1D and Video S1). In wild-type spores, MTs were initially not apparent in spores, and assembled 1–2 h after hydration of the spore during the first interphase after germination (21/21 cells; Figure 1D). At 4–6 h, the cells exhibited polarized growth, entered mitosis, formed a mitotic spindle, and divided. In alp14Δ germinating spores, none of the interphase cells (0/19 cells) exhibited cytoplasmic MTs in the first cell cycle (Figure 1D). However, the mutants assembled spindle MTs in the nucleus during the first mitosis and displayed interphase MTs in subsequent generations (Figure 1D). Thus, alp14Δ mutants are strongly defective in MT nucleation in the first cell cycle. It is possible that in subsequent cell cycles, cytoplasmic MTs are seeded by MTs nucleated from inside the nucleus [29] or from other MTOCs in an alp14-independent manner.
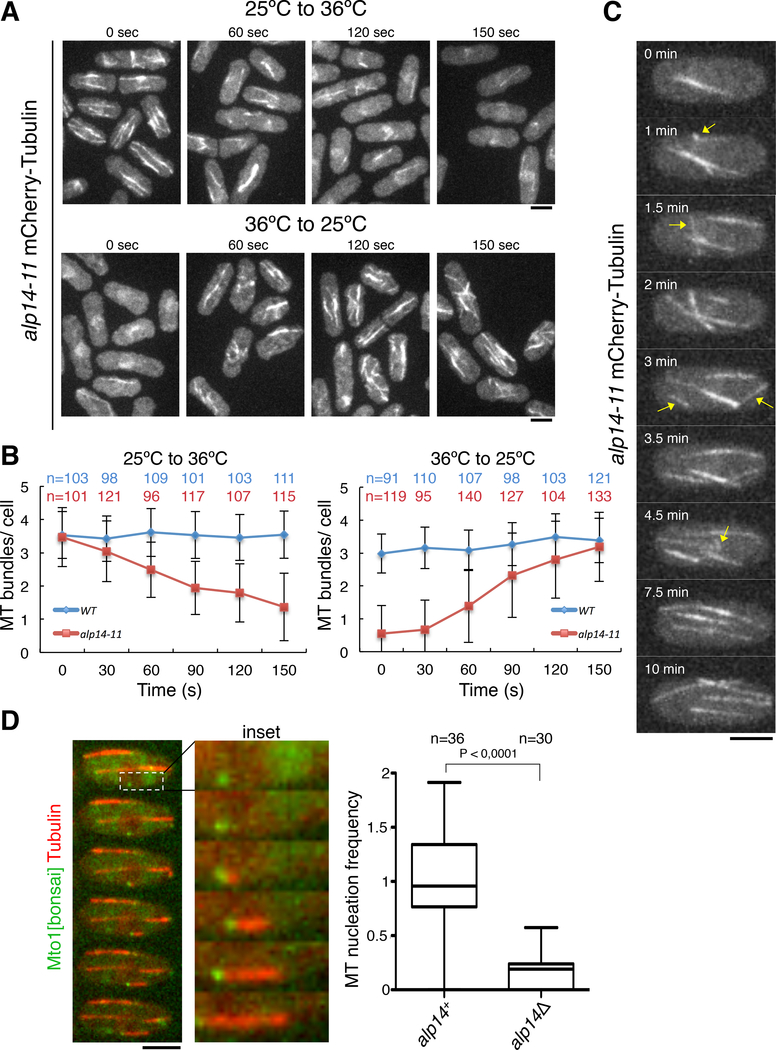
Third, to test whether Alp14 exerts a direct or indirect effect on MT nucleation, we generated a temperature-sensitive allele of alp14 to control its activity (STAR Methods). The alp14–11 allele contains two point mutations in the TOG2 domain (C350R and K525T) and one point mutation in the serine-lysine-rich (SK) region (E568G). Cells harboring this temperature-sensitive allele as the only copy of Alp14 displayed wild-type interphase MT behavior at the permissive temperature of 25 °C and recapitulated a defective MT phenotype like that of alp14Δ cells at the restrictive temperature of 36 °C (Figure S1A). Upon inactivation of Alp14, MT bundles were lost progressively in the first 2.5 min (Figure 2A, B). Conversely, when cells were temperature-shifted to activate Alp14, the wild-type distribution of interphase MT bundles was restored in ~3 min (Figure 2A, B). Temperature shifts did not affect the MT cytoskeleton in wild-type cells (Figure 2B and S1B). Time-lapse imaging revealed that many new bundles arose from apparent MT nucleation events in the cytoplasm (Figure 2C). The rapid timing of these responses suggests that the reactivation of Alp14 directly stimulates the formation of new MT bundles.
Figure 2. Alp14 Regulates the Number of MT Bundles and the Frequency of MT Nucleation in Interphase Cells.
A. Cells with an alp14–11 thermosensitive allele expressing mCherry-Tubulin were shifted from the permissive (25 °C) to the restrictive (36 °C) temperature and vice versa. Cells at each time point were fixed and imaged for MTs.
B. Quantification of interphase cytoplasmic MT bundles in wt and alp14–11 cells after temperature shift. Mean and standard deviation from two independent experiments are shown.
C. Time-lapse images of a single alp14–11 cell upon temperature shift from 36 °C to 25 °C at time 0. Arrows indicate the appearance of new MT bundles occurring within 10 min.
D. alp14Δ mutants are defective in MT nucleation in a quantitative assay. Left, time lapse imaging of cells expressing mCherry-Tubulin and Mto1-[bonsai]-GFP (1 min/frame) show cytoplasmic nucleation events in which a MT emanates from Mto1 signal. Right, normalized frequencies of these nucleation events in imaging every 30 s over 20 min. Scale bars = 5 μm.
See also Figure S1
Fourth, we quantitated the frequency of MT nucleation using a truncated Mto1 (131–549) construct, Mto1-[bonsai] [21]. This construct is not targeted to MTOCs, and thus promotes nucleation events in the cytoplasm that are less ambiguous to score than ones occurring at established MTOCs and pre-existing MTs normally seen in wild-type cells. Using this system, we visualized events in which a MT grew from an isolated dot of Mto1-[bonsai]-GFP (Figure 2D). In wild-type (alp14+) cells, MTs were nucleated at a frequency of 0.26/min ± 0.02, while in alp14Δ cells, nucleation was 5-fold lower (0.05/min ± 0.01; Figure 2D).
Together, these four assays show that the loss of Alp14 activity leads to a significant decrease in MT nucleation of cytoplasmic MTs, and that activation of Alp14 immediately increases nucleation frequency.
Alp14 Localizes to Sites of MT Nucleation and Interacts with the γ-tubulin Complex
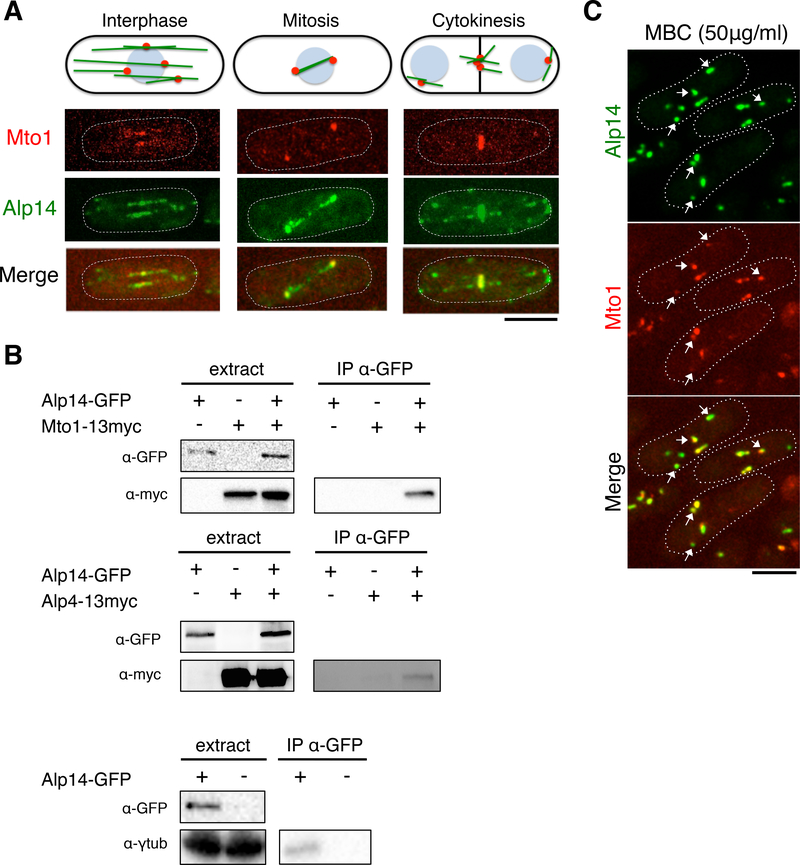
Next, we tested whether Alp14 is present at sites of MT nucleation. In addition to MT plus ends, Alp14-GFP co-localized with γ-tubulin complex proteins Alp4 and Mto1 at the MTOCs at the SPB, the nuclear envelope, and the eMTOC at the cell-division site (Figure 3A and S2A). Thus, like other XMAP215 family members [14], Alp14 localizes to MTOCs.
Figure 3. Alp14 is a Component of MTOCs and Immunoprecipates with Components of the γ-tubulin Complex.
A. Top, schematic of MTs (green) and MTOCs (red) in different stages of the cell cycle. Blue sphere, nucleus. Bottom, maximum projection confocal images of representative cells expressing Mto1-Tomato and Alp14-GFP. In addition to its localization at MT plus ends, Alp14 also localizes to multiple MTOCs.
B. Mto1–13myc, Alp4–13myc, and γ-tubulin co-immunoprecipitate (IP) with Alp14-GFP. Extracts were made from yeast cells with tagged proteins expressed at endogenous levels. IPs were pulled down with anti-GFP antibody and immunoblotted with the indicated antibodies (see Experimental Procedures). A representative experiment from three independent replicates is presented.
C. Images of cells expressing Mto1-Tomato and Alp14-GFP after 10 min treatment with 50 μg/mL MBC, a MT inhibitor. Note that Alp14 and Mto1 co-localize in dots in the cytoplasm (arrows), only some of which contain tubulin (Figure S2B). Scale bars = 5 μm.
See also Figures S2 and S3
We next examined whether Alp14 physically interacts with the γ-tubulin complex. Mto1, Alp4, and γ-tubulin co-immunoprecipitated with Alp14 from yeast extracts (Figure 3B). In cells treated with methyl benzimidazol-2-yl-carbamate (MBC) to depolymerize MTs, Mto1 and Alp14 co-localized in discrete cytoplasmic clusters (Figure 3C). A subset of these clusters contained tubulin and may represent stable MT stubs, as previously shown [28, 30]; however, many Alp14 clusters lacked detectable tubulin (Figure S2B), and therefore may represent a cluster of Alp14 and other MTOC components in the absence of MTs.
Alp14 also shows genetic interactions with components of the γ-tubulin complex. Double mutants showed that alp14Δ displayed synthetic phenotypes with mutants in γ-tubulin ring complex components and mto1 in terms of viability, sensitivity to MT inhibitors, and effects on MT bundles (Figure S3A–C). These results together suggest that Alp14 associates and functions with the γ-tubulin complex in MT nucleation.
Alp14 Transiently Associates with the γ-tubulin Complex During Nucleation
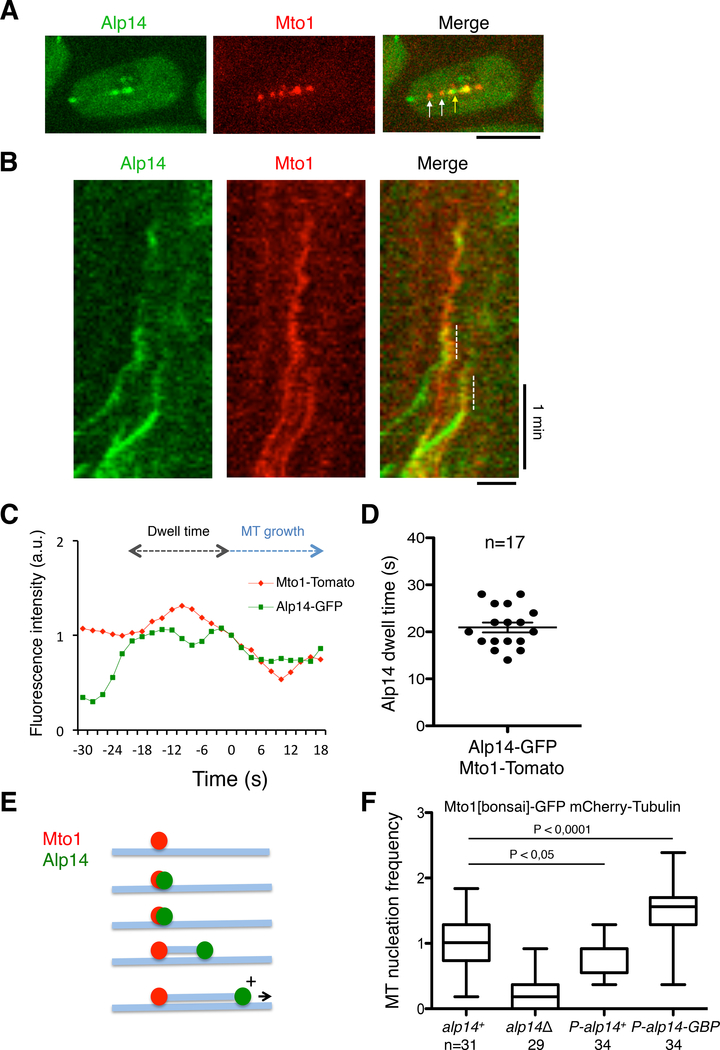
We next tested whether Alp14 associates with the γ-tubulin complex in individual MT nucleation events. Commonly, visualization of MT nucleation in MTOCs is limited by the clustering of many γ-tubulin complexes and nucleation events in one structure. In fission yeast, however, we can image individual nucleation events at discrete γ-tubulin-containing particles on interphase MTs. Many of these satellites appear inactive for nucleation, as only a subset of these particles is active for MT nucleation at any point [22]. Alp14-GFP associated with only a subset of Mto1-mTomato particles on MTs (Figure 4A). Time-lapse imaging revealed that Alp14 associates only transiently with Mto1 particles. Notably, Alp14-GFP arrived at an Mto1-mTomato particle consistently 20 ± 5 s before a MT plus end grew (Figure 4B–E). During this period, the fluorescence intensities of Alp14 and Mto1 remained relatively constant (Figure 4C). We observed some cases in which Alp14 left the nucleation site with the growing MT plus end, while in others, it stayed associated with both the MT plus and minus ends. This transient association of ~20 s is notable as it may represent a step of MT nucleation that occurs prior to MT elongation. Similar lag times have been observed during nucleation events in vitro [19].
Figure 4. Alp14 Transiently Co-localizes with Mto1 in Individual Nucleation Events and Increasing Its Affinity to Mto1 Increases MT Nucleation.
A. Images of cells expressing Mto1-Tomato and Alp14-GFP. Note that Alp14-GFP co-localizes to a subset (yellow arrow) of Mto1-Tomato satellites (white arrows).
B. Kymograph of Alp14-GFP and Mto1-Tomato localization over 3 min. Dashed lines highlight the period of co-localization of these proteins before the MT begins to visibly grow, as depicted by diagonal green lines.
C. Normalized fluorescence intensities of Alp14-GFP and Alp4-mCherry at a non-SPB MTOC during a single MT nucleation event shown in B. Dwell time is defined as the period when Alp14 co-localizes Alp4-mCherry (dashed lines in B), prior to MT growth initiation (set as time 0).
D. Period of dwell time that Alp14-GFP associates with Alp4-mCherry before initiation of MT growth.
E. Schematic of a MT nucleation event over time. A γ-tubulin complex (marked by Mto1) is situated on pre-existing MT (blue). Alp14 associates with the complex; after ~20 s, a new MT begins to grow from the site with Alp14 on the plus end.
F. Increasing affinity between Alp14 and Mto1 by co-expressing Alp14-GBP and Mto1-GFP. Quantitation of cytoplasmic MT nucleation in the indicated genotypes. Data are normalized to wildtype and are shown as boxplots. Scale bars = 5 μm.
Targeting Alp14 to the γ-tubulin Complex Increases MT Nucleation Frequency
Our observations suggest a working model in which Alp14 functions with the γ-tubulin complex to promote MT nucleation. The transient association between Alp14 and the γ-tubulin complex prior to MT elongation indicates that Alp14 binding may be a rate-limiting step in activating MT nucleation. Thus, we hypothesized that increasing the affinity of Alp14 for the complex would increase the rate of MT nucleation. To test this prediction, we used a system that promotes protein-protein interactions via GFP and the GFP-binding protein (GBP) [31]. We expressed Alp14-GBP-mCherry (at below wildtype levels; Figure S4) with Mto1-[bonsai]- GFP. Cells also expressed endogenous Alp14 so that they could continue to grow and divide. These fusion proteins localized in cytoplasmic dots that mostly co-localized (Figure S5A). Using the Mto1-[bonsai]-GFP nucleation assay, we found that Alp14-GBP increased nucleation frequency by 56% over alp14+ cells (0.42/min ± 0.02 vs. 0.27/min ± 0.02; Figure 4F). Controls indicated that this increase in MT nucleation was dependent on the presence of the GFP and GBP fusions (Figure 4F).
To test if the association of Alp14 with the γ-tubulin complex component Alp4 also promotes nucleation, we co-expressed Alp4-GFP and Alp14-GBP-mCherry at endogenous levels as sole copies of these genes (Figure S5B). In this strain, Alp4-GFP was present in multiple foci that were of similar intensity as the SPB and were localized on cytoplasmic MTs often near the nuclear envelope in interphase cells (Figure S5B–D). Although one of these foci represented the SPB, additional ones are likely to be non-SPB foci not normally seen in wildtype cells. Kymographs revealed a significant increase in the frequency of MTs growing out of the foci (Figure S5B). Co-expression of Alp14-GBP-mCherry and Mto1-GFP also produced multiple foci associated with MTs (Figure S5C). Thus, the forced association of Alp14 to Mto1 and Alp4 led to the formation of multiple highly active MTOCs and increased nucleation rates. Taken together, these findings demonstrate that the physical association of Alp14 with the γ-tubulin complex stimulates MT nucleation.
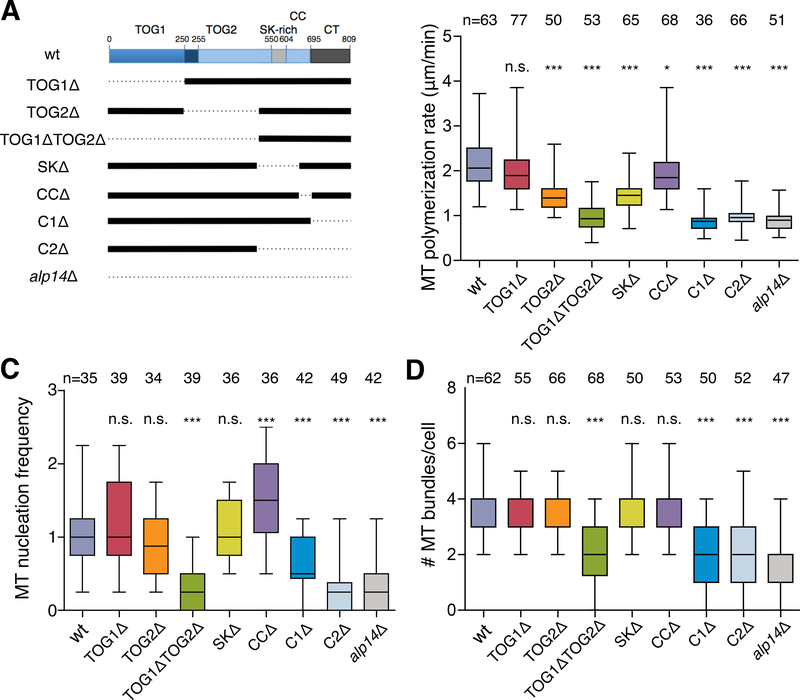
Regions of the Alp14 Protein Required for MT Polymerization and Nucleation
To begin testing the mechanisms of Alp14 in MT nucleation and polymerization, we analyzed a set of Alp14 truncation alleles (Figure 5A) [32]. These truncations disrupt specific domains or regions of Alp14: 1) TOG1 and TOG2 domains, 2) a medial SK-rich domain that contributes to MT lattice binding, 3) a coiled-coil domain (CC) that may contribute to homo-dimerization, and 4) the C-terminal region that contributes to the Alp7 interaction. These mutant alleles replace the endogenous alp14+ gene, and are expressed from the endogenous promoter as the only Alp14 in the cell [32]. The effects of such Alp14 (and Stu2) mutant proteins on in vivo MT polymerization and nucleation rates have not been reported previously.
Figure 5. MT Nucleation and Polymerase Functions of Alp14 Require TOG Domains and a C-terminal Region.
A. Schematic of Alp14 protein domains and truncation mutants. TOG1 and TOG2, Tog domains; SK, medial serine-lysine-rich domain; CC, coiled-coil domain; C1 and C2, C-terminal regions.
B. MT polymerization rates of the indicated alp14 alleles, as determined via time-lapse imaging of cytoplasmic MTs in interphase cells expressing mCherry-Tubulin. ***p < 0.001; **p<0.01; NS, not significant (Student’s t-test).
C. Quantitation of cytoplasmic MT nucleation in the cytoplasm in the indicated alp14 genotypes in interphase cells expressing Mto1-[bonsai]-GFP and mCherry-Tubulin. Frequencies were normalized to those of wild-type cells.
D. Number of cytoplasmic MT bundles in interphase cells expressing Mto1-[bonsai]-GFP and mCherry-Tubulin.
See also Figure S6
We measured the growth rates of interphase MTs in these alp14 truncation mutants (Figure 5B). alp14Δ mutants exhibited a two-fold decrease in interphase MT elongation rates relative to wild-type alp14+ cells (Figure 5B) [25]. We detected similar decreases in MT elongation in alp14 TOG2Δ, TOG1ΔTOG2Δ, and SKΔ truncations, as well as in cells carrying truncations of the C-terminal domains (Figure 5B). The TOG1Δ allele showed a smaller reduction in MT polymerization that was not statistically significant (Figure 5B). The ability of these proteins to localize to MT plus ends was maintained in TOG1Δ, TOG2Δ, and TOG1ΔTOG2Δ proteins. Localization was partially lost in cells with truncations lacking the coiled-coil domain or the SK domain, and was lost in cells with C-terminal truncations (Figure S6A).
We next assayed for MT nucleation in these truncation mutants by measuring the frequency of nucleation events from Mto1-[bonsai], MT regrowth after cold treatment, and the steady-state number of interphase MT bundles (Figure 5C,D and Figure S6B,C). These assays showed that nucleation occurred at wild-type frequencies in TOG1Δ, TOG2Δ, and SKΔ strains, but was decreased in TOG1ΔTOG2Δ and the C-terminal mutants. Thus, nucleation activity requires one of the TOG domains and the C-terminal region. In summary, the general trend suggests a rough correlation between MT nucleation and polymerization activities. However, Alp14-TOG2Δ and Alp14-SKΔ proteins were significantly more deficient in MT elongation than in MT nucleation, which suggests that nucleation is less sensitive to loss of Alp14, or that polymerization and nucleation represent separable but related functions.
TACC Alp7, an Alp14-interacting Protein, Functions with Alp14 for MT Nucleation but Not Growth
The TACC ortholog Alp7 (also known as Mia1) is a well-characterized binding partner of Alp14 [33]. In general, TACCs are localized at centrosomes, spindles, and kinetochores, and participate in spindle assembly [15]. In fission yeast, the TACC ortholog Alp7 is localized to SPBs and kinetochores, and the role of the Alp7-Alp14 complex at these sites during mitosis has been well characterized [32, 34, 35]. However, the function of Alp7 in MT regulation remains controversial. It has been reported that the alp7Δ mutant has a defect in MT bundling but not in nucleation [36, 37], while another report suggests that Alp7 stimulates MT polymerization in vivo and in vitro [18].
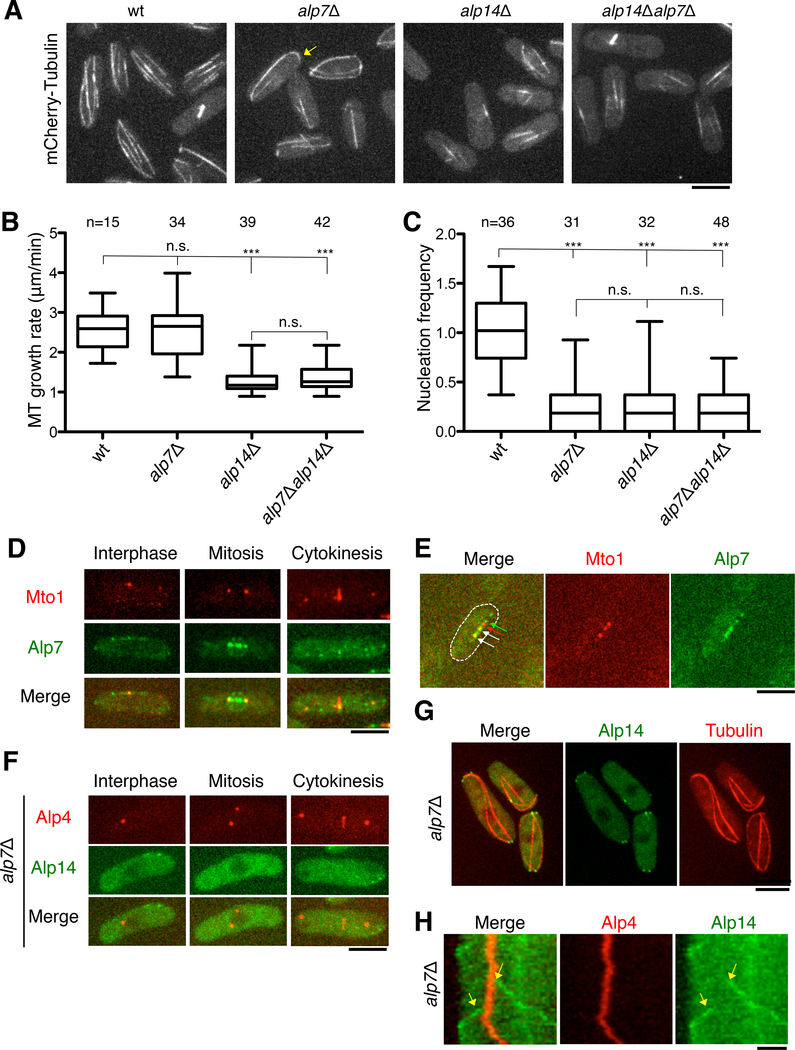
We found that alp7Δ mutants exhibited a defect in MT nucleation but not MT polymerization. Reminiscent of mto1 and γ-tubulin complex mutants [29, 38–40], interphase MT bundles in alp7Δ mutant cells were decreased in number and often extra long so that they curled around the cell tips (Figure 6A; although the extent of this MT curling was less than in mto1 mutants). This phenotype was different from that of an alp14Δ mutant, which did not exhibit such long curly MT bundles (Figure 6A). MTs in alp7Δ cells grew at wild-type rates (Figure 6B), in contrast to a recent report that alp7Δ mutants have a MT growth defect [18]; the difference between the two studies is not known, but may be due to strain or experimental differences. Recovery from cold treatment of alp7Δ cells revealed a large delay in MT recovery, similar to alp14Δ mutants (Figure S7A). In the Mto1-[bonsai] nucleation assay, nucleation frequencies in alp7Δ mutants (0.070/min ± 0.011) were similar to those of alp14Δ cells (0.051/min ± 0.010), and significantly lower than those of wild-type cells (0.269 ± 0.016) (Figure 6C). Thus, Alp7 and Alp14 are both required for efficient cytoplasmic MT nucleation, but in contrast to Alp14, Alp7 is not required for optimal MT polymerization.
Figure 6. TACC Alp7 is Required for Nucleation but Not For the Polymerase Functions of Alp14.
A. Confocal images (maximum intensity projection) of MTs in wild-type (wt), alp7Δ, and alp14Δ cells. Arrow shows an overly long MT, which is a general feature of MT nucleation mutants [29, 38–40].
B. Quantitation of cytoplasmic MT growth rates in interphase cells expressing GFP-Tubulin with the indicated genotypes.
C. Quantitation of cytoplasmic MT nucleation in the cytoplasm of interphase cells of the indicated genotypes expressing Mto1-[bonsai]-GFP and mCherry-Tubulin. Frequencies were normalized to wt.
D. Images of representative cells expressing Mto1-Tomato and Alp7-GFP reveal co-localization of the tagged proteins at MTOCs at various stages of the cell cycle. E. Mto1-Tomato and Alp7-GFP co-localize at a subset of satellites.
F. Images of representative cells expressing Alp14-GFP and Mto1-Tomato in alp7Δ cells in different cell cycle stages.
G. Images of representative cells expressing Alp14-GFP and mCherry-Tubulin in alp7Δ cells.
H. Kymograph of Alp14-GFP and Alp4-mCherry in a representative interphase alp7Δ cell over 5 min. Alp4 marks a SPB, while Alp14-GFP dots emanate from the SPB in rare MT nucleation events (arrows). These Alp14 moving dots are likely to be at growing MT plus ends (as shown in G), as they move at the speed of MT polymerization. In contrast to wt cells (Figure 4), in alp7Δ mutants, Alp14 does not associate with the SPB prior to outgrowth of the MT. Scale bars = 5 μm.
See also Figure S7
We tested if Alp7 and Alp14 function in the same genetic pathway by analyzing properties of the double mutant alp14Δ alp7Δ. This double mutant had phenotypes similar to those of the single alp14Δ mutant, including growth rate (Figure S7B) [33], distribution of interphase MTs and MT growth rates (Figure 6A, B), and MT nucleation frequency (0.057/min ± 0.008) (Figure 6C). This epistatic relationship suggests that Alp7 and Alp14 function together in MT nucleation, and do not, for instance, operate in parallel pathways.
Alp7 Brings Alp14 to Sites of MT Nucleation
We tested sought to determine whether Alp7 mediates Alp14 localization to MTOCs. It has been previously shown that Alp7 is necessary for the localization of Alp14 to the SPB, and that the localization of Alp7 at SPBs is independent of Alp14 [33]. We found that Alp7 co-localized with Mto1 at SPBs, the eMTOC, and a subset of satellites (Figure 6D, E). In alp7Δ mutant cells, Alp14-GFP was not detectable at multiple MTOCs but still tracked growing MT plus ends (Figure 6F,G and S7C). Moreover, Alp14-GFP did not form cytoplasmic clusters in alp7Δ cells treated with MBC (Figure S7D). In time lapse analyses of individual nucleation events, in contrast to wild-type cells in which Alp14 transiently associated with the nucleation site 20 s before MT outgrowth (Figure 4C and D), in alp7Δ cells, Alp14 did not exhibit this dwell period and only appeared on the growing MT plus end (18/18 nucleation events; Figure 6H). These findings suggest that in the absence of Alp7, Alp14 is not loaded at the MTOC during the initial nucleation phase, but binds subsequently to the growing plus end after the MT has been formed. This localization behavior of Alp14 in alp7Δ cells is consistent with the observed defects in nucleation but not MT polymerization. Thus, the phenotype in the alp7Δ mutant reveals the separable roles of Alp14 in MT nucleation and polymerization.
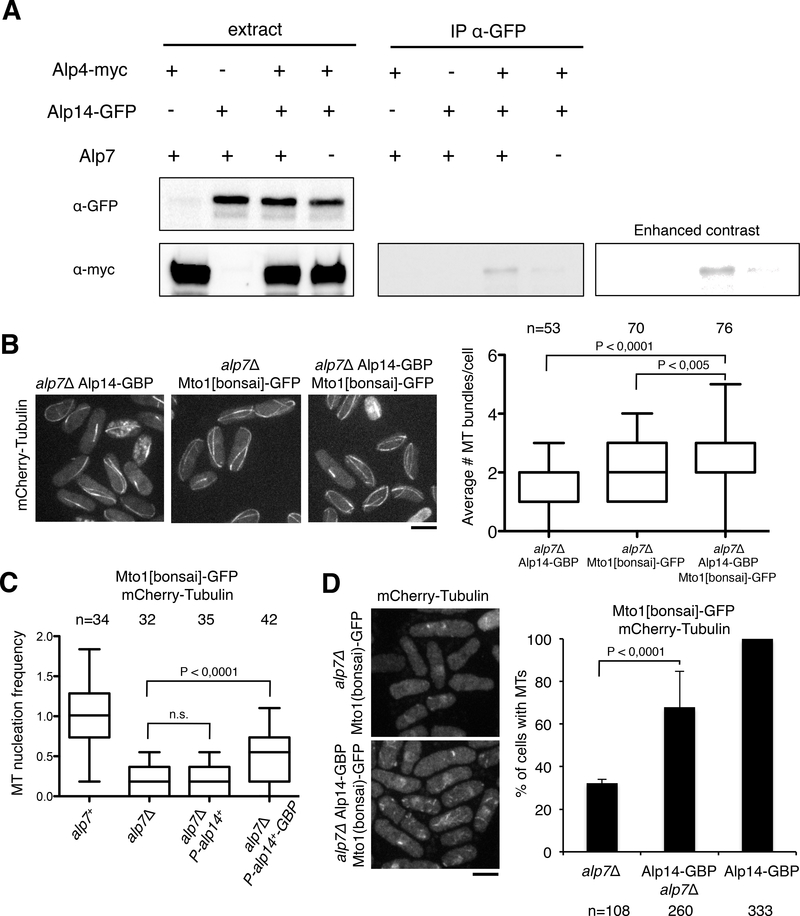
We hypothesized that the primary function of Alp7 is to recruit Alp14 to the MT nucleation site to stimulate nucleation. Immunoprecipitations showed that the biochemical association of Alp14 with Alp4 was strongly impaired in the alp7Δ strain (Figure 7A). Our hypothesis further predicts that artificially targeting Alp14-GBP to Mto1-GFP would bypass the need for Alp7 in MT nucleation. Similar to the assay described in Figure 4E, we ectopically expressed Alp14-GBP or untagged Alp14 in cells expressing Mto1-[bonsai]-GFP in alp7Δ cells (Figure 7B). Expression of Alp14-GBP produced a 2-fold increase in nucleation frequency (0.13/min ± 0.02; Figure 7C) as well as an increase in the number of MT bundles per cell versus the Alp14 control without GBP (Figure 7B). Similarly, Alp14-GBP induced MT formation in alp7Δ cells expressing Mto1-[bonsai]-GFP after cold treatment: cells were filled with many short MTs within 1 min of temperature shift (Figure 7D), although these MTs often shrank before reaching full length. Thus, the artificial association of Alp14 with Mto1 partially suppresses the requirement of Alp7 for MT nucleation.
Figure 7. Alp7 Mediates the Association Between Alp14 and the γ-tubulin Complex.
A. Co-immunoprecipitation of Alp14 and Alp4 is dependent on Alp7. Alp14-GFP pulls down Alp4-myc in wild-type cells but not in alp7Δ cells.
B. Expression of Alp14-GBP and Mto [bonsai]-GFP partially suppresses the nucleation defect of alp7Δ mutants. Histograms show the number of MT bundles in the indicated genetic backgrounds.
C. Quantitation of cytoplasmic MT nucleation in the cytoplasm from Mto1-[bonsai]-GFP in the indicated backgrounds. Data are normalized to wildtype and shown as boxplots.
D. Appearance of many short MTs after 1 min recovery after cold treatment in cells expressing Alp14-GBP and Mto1-[bonsai]-GFP. Graph shows percentage of cells with detectable MTs after 1 min. Average and standard deviation from two independent experiments are shown. Scale bars = 5 μm.
Together, these findings lead to a model in which Alp7 recruits Alp14 to the nucleation site for efficient MT nucleation. In the absence of Alp7, Alp14 is not recruited initially to the nucleation site, and thus nucleation efficiency is reduced; however, Alp14 can still associate with the growing MT plus end of the mature MT to promote MT polymerization. Thus, Alp7 may facilitate a specific role of Alp14 in promoting MT nucleation.
DISCUSSION
In this study, we present strong evidence that Alp14, an XMAP215 family member, is a MT nucleation factor that functions with the γ-tubulin complex in vivo. Loss of Alp14 function led to a decrease in MT nucleation rate in a variety of assays; most impressively, alp14 null mutants completely lacked cytoplasmic MTs in the first cell cycle after spore germination (Figure 1). Nucleation defects may explain the predominant MT defects in alp14 mutants. Conversely, activation of a conditional Alp14 mutant led to the assembly of many new MT bundles within minutes (Figure 2). Co-localization, biochemical, and genetic data demonstrated that Alp14 interacts with the γ-tubulin complex, and that this association that may be independent of MTs (Figures 3, 4). Artificially forcing this association increases MT nucleation frequency (Figure 6, 7). Together, these findings constitute the strongest evidence to date that a XMAP215 family member functions with the γ-tubulin complex in MT nucleation in vivo.
How XMAP215/Alp14 and the γ-tubulin complex work together to nucleate a MT remains unclear. Our data show that at least one of the TOG domains and the C-terminal region of Alp14 are needed for nucleation (Figure 5). Numerous models for nucleation can be imagined. First, Alp14 could initiate nucleation by increasing the local concentration of tubulin dimers and stabilizing tubulin oligomers. In this scenario, instead of a nucleator, the γ-tubulin complex could act as primarily as a stabilizer of the minus end. Second, Alp14 could help to insert and/or stabilize α/β tubulin dimers onto the γ-tubulin ring template. Third, Alp14 could insert and/or stabilize α/β tubulin dimers onto other α/β tubulin molecules on the incipient MT. Fourth, Alp14 could also contribute to conformational changes in the γ-tubulin complex that activate it [1]. More detailed understanding of the nucleation process will be needed to test these different possibilities.
Our findings here clarify the function of TACC Alp7. Our data indicate that a primary role of Alp7 is in MT nucleation, and that it may be responsible for bringing Alp14 to MTOCs (Figure 6, 7). In alp7Δ cells, Alp14 does not localize to MT nucleation sites, and the rate of nucleation in alp7Δ cells is similar to that of alp14Δ cells; however, Alp14 is still able to localize to MT plus ends and stimulate polymerization. Alp7 was partially bypassed by artificially linking Alp14 to elements of the γ-tubulin complex (Figure 7C, D), supporting a model in which the main role of Alp7 is to bring Alp14 close to the nucleation site. This nucleation-specific defect of alp7Δ cells shows that nucleation and polymerization may represent distinct functions for Alp14.
This work provides new molecular insights into the mechanism of MT nucleation. A recent study of MT nucleation in vitro reported that the process is kinetically unfavorable, requiring minutes before steady-state elongation begins, but addition of XMAP215 reduces this time lag [19]. We propose that MT nucleation follows a time-line composed of a series of steps in which MT-associated proteins modulate the timing of these events. In individual nucleation events in fission yeast, most γ-tubulin complexes contain the activators Mto1 and Mto2 but appear to be inactive [21, 22]. We found that the formation of MTs at these foci is preceded by the arrival of Alp14 at the site ~20 s before MT elongation (Figure 4). Notably, in alp7 mutants with reduced nucleation frequency, this Alp14 dwell period no longer occurs (Figure 6H). We speculate that Alp14 contributes to step(s) in nucleation during this 20-s period to promote the assembly of the new MT. For instance, this period could represent the time of loading of α/β tubulin dimers onto the γ−tubulin complex. More broadly, this finding is the first indication of a discrete timeline for MT nucleation in vivo.
The function of XMAP215 and TACC proteins in MT nucleation is likely to be conserved. XMAP215 and TACC proteins localize to centrosomes in animal cells [14, 15]. Early studies reported that XMAP215 is needed for nucleation in Xenopus extracts and, when coupled to beads, it can generate MT asters in a γ-tubulin-dependent manner [16, 41]. Titration of XMAP215 activity has been shown to set spindle length and mass in Xenopus extracts [42]. In a recent study concurrent with this one, XMAP215 was shown in extracts and with purified proteins to stimulate MT nucleation synergistically with γ-TuC and associate with the complex [43]. XMAP215 stimulates nucleation synergistically with TPX2 in vitro [44]. A recent paper indicated that condensates of SPD-5 recruit XMAP215 and TPX2 that concentrate tubulin and reconstitute MT aster formation even in the absence of γ-tubulin [45].
In conclusion, our study demonstrates that XMAP215/Alp14 proteins should not only be regarded as polymerases, but as important components of the MT nucleation machinery. In fission yeast, this nucleation function is the major in vivo function of Alp14. These proteins may be somewhat analogous to formins, which both nucleate and promote the polymerization of actin filaments by locally increasing the concentration of actin monomers [46]. Further study of XMAP215, TACC, and the γ-tubulin complex will reveal fundamental mechanisms responsible for MT nucleation and its regulation.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fred Chang (Fred.Chang@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Experiments were performed using Schizosaccharomyces pombe yeast strains listed in Table S1.
Culture conditions
Standard methods for S. pombe growth and genetics were used [47]. Cells were typically grown in exponential phase in liquid YE5S medium at 25 °C with shaking for 18–24 h.
Methyl benzimidazol-2-yl-carbamate (MBC) was dissolved in dimethyl sulfoxide to make a 10 mg/mL stock solution. Geneticin (G418) was dissolved in pure water (Sigma-Aldrich #W4502) to make a 50 mg/mL stock solution. ClonNAT was dissolved in pure water (Sigma-Aldrich #W4502) to make a 200 mg/mL stock solution.
METHOD DETAILS
Strain construction
In general, strains were constructed using a PCR-based homologous recombination method to insert markers into the yeast chromosome [48]. Constructs were checked via PCR and sequencing, and strains were outcrossed at least three times. Double mutants were generated by tetrad analyses
For the extra copy of Alp14-GBP-mCherry (Figures 4, 7, S3 and S4), we amplified GBP-mCherry-hph with primers Alp14-CTag-FW and Alp14-CTag-RV from the plasmid pJM559: pMS-GBP-mCherry-hph (a gift from Dr. James Moseley) and integrated in the endogenous locus of alp14 using a PCR-based homologous recombination method [48]. Primers Alp14_−400_F and Alp14_+100_R were used to generate an amplicon containing 400 bp of DNA upstream of the ATG of alp14 and 100 bp downstream of the end of the sequence encoding GBP-mCherry-hph. This amplicon was digested with XmaI and SalI and cloned into the integrative plasmid pJK148 [49]. The resulting plasmid was digested and integrated into an alp14+ strain (FC420) and backcrossed three times. Figure S3 depicts the localization, behavior, and relative expression levels of both constructs. Note that the expression of the exogenous Alp14-GBP is 4-fold lower than that driven by the endogenous locus. The same strategy was used to generate a strain harboring an extra copy of wild-type Alp14 using genomic DNA of the wild-type strain FC421 as template.
Generation of alp14-ts Alleles
Template DNA for mutagenizing PCR containing 400 bp of DNA upstream and 100 bp of DNA downstream of the Alp14-GFP-KanMX cassette was amplified from genomic DNA from strain FC1907 using primers Alp14_−400_F and Alp14_+100_R and cloned into the pGemT-easy vector (Promega #A1380). Random mutagenesis by PCR using BioTaq™ DNA polymerase (Bioline) in decreased dATP concentration was carried out using primers Alp14_−400_F and Alp14_+100_R in. S. pombe mal3Δ cells (FC2342 mal3::NatMX ura4 ade6 leu1–32 ura4-D18). The resulting DNA was transformed into yeast cells at 25 °C, grown for 24 h under non-selective conditions, and cells were replicated onto plates containing 100 μg/mL geneticin (G418) to select for transformants. Colonies were replicated on YE5S plates at 25 °C and 36 °C, and colonies displaying impaired growth at 36 °C but not 25 °C were isolated. Mutations were analyzed via DNA sequencing of alp14. Candidate alleles were amplified and integrated in strain FC421, and genetic crosses with strain FC2342 confirmed that mutant phenotypes were linked to these mutations. To remove the GFP-KanMX cassette, we generated a strain with the NatMX marker 123 bp downstream of alp14 (out of the 3’UTR) through homologous recombination using the primers Alp14–3UTR-Tag-FW and Alp14–3UTR-TagRV. Genomic DNA from this strain was amplified by PCR with the primers Alp14–3UTR-Check-FW and RV and used to make a marker switch in the strain containing the alp14–11-GFP-KanMX.
Imaging S. pombe Cells
For live cell imaging, cells were typically grown in exponential phase in liquid YE5S medium at 25 °C with shaking for 18–24 h. In some experiments, cells were mounted in liquid YE5S medium directly on glass. For long-term imaging, cells were placed on micro-slide wells (Ibidi #80821) coated with soybean lectin (Sigma #1395). Spores were mounted on 1% agarose YE5S pads under a glass coverslip.
Images were generally acquired using a spinning-disk confocal fluorescence NikonTI-based microscope system (Nikon Instruments, Yokagawa, Solamere Technology) with an EM CCD camera (Hamamatsu Corp.) and a 100X 1.4 N.A. objective, or with a spinning-disk confocal microscope (IX-81; Olympus; CoolSnap HQ2 camera) with a Plan Apochromat 100× 1.4 NA objective (Roper Scientific). A wide-field Nikon Eclipse 800 microscope with a 60X 1.4 N.A. objective was used for some studies. Temperature was stably controlled in the room during imaging at 25 °C unless otherwise indicated.
For MT regrowth (Figures 1A, 7D, S5B, and S6A), cells were chilled in Eppendorf tubes in an ice-water slurry for 30 min and then shifted to a shaking water bath at 25 °C. Samples were rapidly fixed in 100% methanol at −80 °C and later washed with phosphate-buffered saline for fluorescence microscopy. To quantify MT nucleation frequency in various mto1 strains (Figures 2D, 4F, 5C, 6C and 7C), movies of mCherry-Tubulin in cells were examined and MT nucleation events were scored [21].
To study germinating spores (Figure 1D), cells of opposite mating types (FC1971 and FC2322) were mated on Malt Extract plates. The resulting asci were digested with 0.6% β-glucuronidase® (Sigma#G4259)/dH2O solution and incubated for 12 h at 30 °C. Spores were germinated on 1% agarose pads with selective media.
alp14–11 thermo-sensitive cells (Figures 2 and S1) were grown at 25 °C and placed in an FCS2 chamber (Bioptechs) coated with soybean lectin (Sigma #L1395). An objective heater (Bioptechs) was used for temperature control during imaging. For counting MT bundles (Figure 2A, 2B, and S1B), cells were grown at 25 °C (or 36 °C) and placed in a shaking water bath at 36 °C (or 25 °C). Samples for each time point were quickly fixed in 100% methanol at −80 °C, and later washed with phosphate-buffered saline for fluorescence microscopy.
To quantify expression levels of Alp14-GBP-mCherry constructs (Figure S3D), cells (NF693 and NF751) were grown at 25 °C and placed in an FCS2 chamber coated with soybean lectin. To determine the relative expression levels of Alp14-GBP-mCherry (endogenous locus) and pJK148-Alp14-GBP-mCherry (ectopic integration), we measured the florescence intensities of single dots and subtracted cytoplasmic background.
Co-immunoprecipitation and Immunpblotting
For co-immunoprecipitation (Figures 3B and 7A), cells were collected via centrifugation and bead-beaten at 4°C four times during 30 seconds in the FastPrep-24 (MP Biomedicals) in CoIP buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor (Roche #04693132001)). The lysates were cleared by centrifugation at 13,000 rpm for 10 m at 4˚C. Whole cell extracts were collected as supernatants, and protein concentration was determined by Bradford assay. Cell extracts (1–10mg) were incubated with 10 μl of Anti-GFP (0.4 mg/ml) for 1 h at 4˚C, and then 25 μl of anti-Dynabeads® M-280 Sheep Anti-Mouse IgG (ThermoFisher) were added. Alternatively, GFP tagged proteins were inmunoprecipitated using GFP trapped magnetic agarose beads (Chromotek). Samples were incubated overnight with orbital rotation at 4˚C. Magnetic beads were washed 6 times with Co-IP Washing Buffer (50 mM Tris-HCl, pH 7.5; 500 mM sodium chloride; 0.1% Nonidet P-40). Bound proteins were solubilized by the addition of 20μl 2X Laemmli Sample Buffer (Biorad #1610737), incubated at 95°C for 10 min, run on gels using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted. Anti-Myc antibody (9E10, Santa Cruz), anti-GFP antibody (Roche), and anti γ-tubulin antibody (T6557, Sigma) were used for immunoblotting.
Image Analysis and Measurement of Microtubule Dynamics
Image Z-stacks were analyzed using ImageJ [50].
For Alp14-GFP residence time at Mto1-Tomato satellites (Figure 4), time-lapse movies were taken every 2 s with 3 Z sections of 0.5 μm. Maximum intensity projections were used for generating kymographs.
For analyses of MT dynamics (Figure 6B), kymographs were constructed for each MT bundle. MT assembly and disassembly rates were measured as slopes in the kymographs of individual dynamic MTs, usually at the end of the bundle-facing cell tips. The number of MT bundles was counted manually from images with 13 stacks of 0.5 μm.
QUANTITATION AND STATISTICAL ANALYSIS
At least three independent biological replicates were performed for each experimental condition. n values are described in each figure (for MT bundles, MT regrowth, and MT nucleation experiments, n = number of cells; for MT growth rates, n = number of MTs). One-tailed unpaired Student’s t-tests were performed using Prism 5 (Graphpad). P-values are described in each figure (***p<0,0001, **p<0,001, *p<0,05, n.s. stands for not significant). Box-and-whisker plots depict box plots with Tukey whiskers.
Supplementary Material
Table S1. Schizosaccharomyces pombe strain list. Related to STAR methods.
Video S1. alp14Δ Germinating Spores are Defective for MT Nucleation. Related to Figure 1.
In this time-lapse movie (5 min/frame) of wild-type and alp14Δ spores, MTs emerge in wild-type cells soon after hydration, but are not visible in alp14Δ cells until the mitotic spindle forms.
Highlights.
Alp14 (XMAP215) and Alp7 (TACC) promote microtubule nucleation in vivo.
Alp14 interaction with Mto1 and γ-tubulin complex is dependent on Alp7.
Increasing the affinity of Alp14 for Mto1 increases nucleation frequency.
Timed appearance of Alp14 at the nucleation site suggests a nucleation timeline.
ACKNOWLEDGMENTS
We thank members of the Chang and Flor-Parra labs for support and discussion. We thank Ken Sawin, Masamitsu Sato, Takashi Toda, and James Moseley for strains and reagents, and Jawdat Al-Bassam for discussion, guidance, and sharing of unpublished results. This work was supported by National Institutes of Health Grant R01GM115185 to F. C. and by an MEC/Fulbright Postdoctoral Award, the Andalucía Talent Hub Program, and funding from the research program of Universidad Pablo de Olavide to I. F-P.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kollman JM, Merdes A, Mourey L, and Agard DA (2011). Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petry S, and Vale RD (2015). Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 17, 1089–1093. [DOI] [PubMed] [Google Scholar]
- 3.Kollman JM, Polka JK, Zelter A, Davis TN, and Agard DA (2010). Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466, 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinh DB, Kern JW, Hancock WO, Howard J, and Davis TN (2002). Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol. Biol. Cell 13, 1144–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong KK, Fernandez JJ, Sali A, Kilmartin J, Davis TN, et al. (2015). Ring closure activates yeast gammaTuRC for species-specific microtubule nucleation. Nat. Struct. Mol. Biol. 22, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, and Hyman AA (2008). XMAP215 is a processive microtubule polymerase. Cell 132, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Bassam J, and Chang F (2011). Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21, 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, and Howard J (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. USA 108, 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrnes AE, and Slep KC (2017). TOG-tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. J. Cell Biol. 216, 1641–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayaz P, Munyoki S, Geyer EA, Piedra FA, Vu ES, Bromberg R, Otwinowski Z, Grishin NV, Brautigam CA, and Rice LM (2014). A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. eLife 3, e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayaz P, Ye X, Huddleston P, Brautigam CA, and Rice LM (2012). A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 337, 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang NH, Takada H, Hsu KS, and Toda T (2013). The internal loop of fission yeast Ndc80 binds Alp7/TACC-Alp14/TOG and ensures proper chromosome attachment. Mol. Biol. Cell 24, 1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller MP, Asbury CL, and Biggins S (2016). A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell 165, 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gard DL, Becker BE, and Josh Romney S (2004). MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int. Rev. Cytol. 239, 179–272. [DOI] [PubMed] [Google Scholar]
- 15.Peset I, and Vernos I (2008). The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 18, 379–388. [DOI] [PubMed] [Google Scholar]
- 16.Popov AV, Severin F, and Karsenti E (2002). XMAP215 is required for the microtubule-nucleating activity of centrosomes. Curr. Biol, 12, 1326–1330. [DOI] [PubMed] [Google Scholar]
- 17.Slep KC, and Vale RD (2007). Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell 27, 976–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussmann F, Drummond DR, Peet DR, Martin DS, and Cross RA (2016). Alp7/TACC-Alp14/TOG generates long-lived, fast-growing MTs by an unconventional mechanism. Sci. Rep. 6, 20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieczorek M, Bechstedt S, Chaaban S, and Brouhard GJ (2015). Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 17, 907–916. [DOI] [PubMed] [Google Scholar]
- 20.Sawin KE, and Tran PT (2006). Cytoplasmic microtubule organization in fission yeast. Yeast (Chichester, England) 23, 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch EM, Groocock LM, Borek WE, and Sawin KE (2014). Activation of the gamma-tubulin complex by the Mto1/2 complex. Curr. Biol. 24, 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janson ME, Setty TG, Paoletti A, and Tran PT (2005). Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J. Cell Biol. 169, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmerman S, Tran PT, Daga RR, Niwa O, and Chang F (2004). Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev. Cell 6, 497–509. [DOI] [PubMed] [Google Scholar]
- 24.Garcia MA, Koonrugsa N, and Toda T (2002). Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 21, 6015–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, and Chang F (2012). Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Mol. Biol. Cell 23, 2878–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roque H, Ward JJ, Murrells L, Brunner D, and Antony C (2010). The fission yeast XMAP215 homolog Dis1p is involved in microtubule bundle organization. PloS one 5, e14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawin KE, Lourenco PC, and Snaith HA (2004). Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763–775. [DOI] [PubMed] [Google Scholar]
- 28.Tran PT, Marsh L, Doye V, Inoue S, and Chang F (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmerman S, and Chang F (2005). Effects of {gamma}-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol, Cell 16, 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bratman SV, and Chang F (2007). Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell 13, 812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, and Leonhardt H (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282–289. [DOI] [PubMed] [Google Scholar]
- 32.Okada N, Toda T, Yamamoto M, and Sato M (2014). CDK-dependent phosphorylation of Alp7-Alp14 (TACC-TOG) promotes its nuclear accumulation and spindle microtubule assembly. Mol. Biol, Cell 25, 1969–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato M, Vardy L, Angel Garcia M, Koonrugsa N, and Toda T (2004). Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol. Biol, Cell 15, 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang NH, and Toda T (2015). Alp7/TACC recruits kinesin-8-PP1 to the Ndc80 kinetochore protein for timely mitotic progression and chromosome movement. J. Cell Sci. 128, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yukawa M, Kawakami T, Okazaki M, Kume K, Tang NH, and Toda T (2017). A microtubule polymerase cooperates with the kinesin-6 motor and a microtubule cross-linker to promote bipolar spindle assembly in the absence of kinesin-5 and kinesin-14 in fission yeast. Mol. Biol, Cell 28, 3647–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng L, Schwartz C, Wee L, and Oliferenko S (2006). The fission yeast transforming acidic coiled coil-related protein Mia1p/Alp7p is required for formation and maintenance of persistent microtubule-organizing centers at the nuclear envelope. Mol. Biol, Cell 17, 2212–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thadani R, Ling YC, and Oliferenko S (2009). The fission yeast TACC protein Mia1p stabilizes microtubule arrays by length-independent crosslinking. Curr. Biol. 19, 1861–1868. [DOI] [PubMed] [Google Scholar]
- 38.Vardy L, and Toda T (2000). The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J 19, 6098–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkatram S, Tasto JJ, Feoktistova A, Jennings JL, Link AJ, and Gould KL (2004). Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol, Cell 15, 2287–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venkatram S, Jennings JL, Link A, and Gould KL (2005). Mto2p, a novel fission yeast protein required for cytoplasmic microtubule organization and anchoring of the cytokinetic actin ring. Mol. Biol, Cell 16, 3052–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groen AC, Maresca TJ, Gatlin JC, Salmon ED, and Mitchison TJ (2009). Functional overlap of microtubule assembly factors in chromatin-promoted spindle assembly. Mol. Biol, Cell 20, 2766–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, and Julicher F (2013). XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat. Cell Biol. 15, 1116–1122. [DOI] [PubMed] [Google Scholar]
- 43.Thawani A, Kadzikand RS, and Petry S (2018). XMAP215 is a microtubule nucleation factor that functions synergistically with the gamma-tubulin ring complex Nat. Cell Biol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roostalu J, Cade NI, and Surrey T (2015). Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol. 17, 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077 e1010. [DOI] [PubMed] [Google Scholar]
- 46.Goode BL, and Eck MJ (2007). Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627. [DOI] [PubMed] [Google Scholar]
- 47.Moreno S, Klar A, and Nurse P (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Method. Enzymol. 194, 795–723. [DOI] [PubMed] [Google Scholar]
- 48.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, and Pringle JR (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast (Chichester, England) 14, 943–951. [DOI] [PubMed] [Google Scholar]
- 49.Keeney JB, and Boeke JD (1994). Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics 136, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Schizosaccharomyces pombe strain list. Related to STAR methods.
Video S1. alp14Δ Germinating Spores are Defective for MT Nucleation. Related to Figure 1.
In this time-lapse movie (5 min/frame) of wild-type and alp14Δ spores, MTs emerge in wild-type cells soon after hydration, but are not visible in alp14Δ cells until the mitotic spindle forms.