Abstract
Coronavirus disease 19 (COVID-19) is a severe acute respiratory syndrome caused by SARS-CoV-2 (2019-nCoV). While no drugs have yet been approved to treat this disease, small molecules effective against other viral infections are under clinical evaluation for therapeutic abatement of SARS-CoV-2 infections. Ongoing clinical trials include Kaletra (a combination of two protease inhibitors approved for HIV treatment), remdesivir (an investigational drug targeting RNA-dependent RNA polymerase [RdRP] of SARS-CoV-2), and hydroxychloroquine (an approved anti-malarial and immuno-modulatory drug). Since SARS-CoV-2 replication depends on three virally encoded proteins (RdRP, papain-like proteinase, and helicase), we screened 54 FDA-approved antiviral drugs and ~3300 investigational drugs for binding to these proteins using targeted and unbiased docking simulations and computational modeling. Elbasvir, a drug approved for treating hepatitis C, is predicted to bind stably and preferentially to all three proteins. At the therapeutic dosage, elbasvir has low toxicity (liver enzymes transiently elevated in 1% of subjects) and well-characterized drug-drug interactions. We predict that treatment with elbasvir, alone or in combination with other drugs such as grazoprevir, could efficiently block SARS-CoV-2 replication. The concerted action of elbasvir on at least three targets essential for viral replication renders viral mutation to drug resistance extremely unlikely.
Author Summary
We performed in silico screens of FDA-approved and investigational drugs, to seek rapidly deployable agents that could be combined to disrupt multiple viral targets. One drug stood out with exceptionally stable binding to the three initial protein targets essential for SARS-CoV-2 replication: elbasvir, currently approved as one component of Zepatier™ (Merck) for treatment of chronic hepatitis C infections. Elbasvir was in the top decile of drugs for stable binding to each of 6 SARS-CoV-2 proteins, and thus is predicted to confer a multi-pronged defence against COVID-19, either alone or in combination with other anti-viral drugs.
Introduction
The COVID-19 pandemic has created an urgent need for effective and rapidly deployable therapeutics [1, 2]. Several studies employed in silico or in vitro drug screens to identify FDA-approved drugs that could be repurposed to treat patients infected with the SARS-CoV-2 virus. Among them, lopinavir/ritonavir (Kaletra™), chloroquine or hydroxychloroquine, and favilavir (Favipiravir™) were top candidates and are in clinical trials for the treatment of SARS-CoV-2 [3, 4]. Other studies have identified novel candidate drugs by computational screening of small-molecule libraries, but these would require extensive clinical trials prior to FDA approval.
SARS-CoV-2 belongs to the same family as SARS-CoV, coronaviridae, with the largest genome among known RNA viruses. Previous studies have indicated several key protein that can be targeted for intervention in RNA-virus diseases [5, 6]. The viral RNA-dependent RNA polymerase (RdRP) and helicase are necessary for SARS-CoV-2 replication [7, 8]. Papain-like proteinase, another critical protein required for viral replication, is also involved in blocking the host’s innate immune response [8]. Although RNA viruses are particularly known for their high mutation rates, the active-site structures of essential proteins remain highly conserved.
After the initial COVID-19 outbreak in China, the Zhang laboratory generated theoretical models of SARS-CoV-2 proteins based on the viral-genome sequence [9]. We screened libraries of 54 FDA-approved antiviral drugs and 3300 investigational drugs, against three key proteins (RdRP, helicase, and papain-like proteinase) to seek novel candidates that could be repurposed for COVID-19 therapy. Unlike other drug discovery efforts, we performed parallel screens of drugs against these three enzyme targets, in the hope of devising a highly effective cocktail to combat the disease.
Results
Targeting proteins essential for viral replication.
We selected three viral proteins that play essential roles in replication of the RNA genome of SARS-CoV-2: (1) RNA-dependent RNA polymerase (RdRP), (2) papain-like proteinase, and (3) helicase. Previous studies had shown that antiviral drugs targeting any one of these proteins will inhibit replication of RNA viruses generally, including coronaviruses [5, 7, 10]. We therefore hypothesized that disrupting these three proteins in SARS-CoV-2 should quite effectively block its replication, with little possibility of mutationally generated resistance.
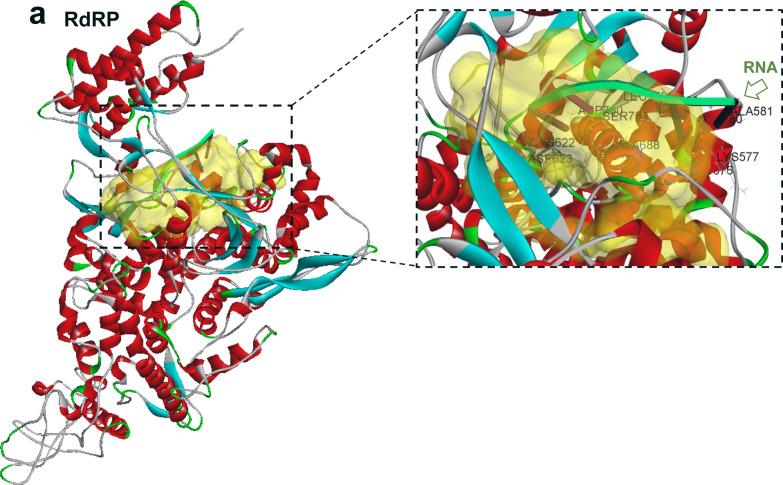
Beginning with the full-length structural models of SARS-CoV-2 proteins developed by the Zhang group [9], we used Discovery Studio Suite™ to predict the druggable pocket for each target protein (Figure 1, a–f). Molecular-dynamic (MD) simulations of SARS-CoV-2 proteins (RdRP, papain-like proteinase, and helicase) confirmed that the predicted drug-binding sites were stably maintained over time. We therefore used these predicted druggable pockets as targets to screen against libraries of approved and investigational antiviral drugs. RdRP displayed a single large pocket (Figure 1, a–b); based on an experimental model of another viral RdRP (PDB-ID 6K32), we postulated that this was likely to be the region of interaction with template RNA. Simulations of RNA docking to SARS-CoV-2 RdRP support stable RNA binding to the predicted pocket (see Figure 2a), making this a suitable target for drug-mediated disruption of viral replication. Similarly, we predicted the druggable pockets for papain-like proteinase (Figure 1, c–d), and helicase (Figure 1, e–f). The predicted helicase pocket displayed nucleic-acid binding capability (Figure 2b), supporting previous reports of nucleic-acid interactions with other viral helicases [11]. All three predicted binding cavities in candidate proteins were used as druggable targets in our subsequent screens.
Figure 1.
Structures and predicted binding pockets (arrows on left, yellow areas on right) of (a, b) viral RdRP; (c, d) viral papain-like proteinase; and (e, f) viral RNA helicase.
Figure 2.
Protein-RNA docking shows binding modes of RNA (green ribbon with nucleotide “pegs”) to (a) RdRP, and (b) helicase of SARS-CoV-2.
Elbasvir interacts with RdRP, proteinase, and helicase of SARS-CoV-2.
Because repurposing an approved drug is the most efficient strategy to quickly deploy new therapeutics against a novel pathogen, we screened FDA-approved antiviral drugs for docking potential with SARS-CoV-2 proteins. Target-based computational docking using Glide, followed by MMGBSA to assess interaction stability (see Methods), was performed sequentially against the three candidate-protein binding pockets. Lopinavir was the top-ranked drug for binding to SARS-CoV-2 RdRP, with elbasvir a very close second (Figure 3a). In preliminary clinical trials for patients with severe COVID-19, the lopinavir-ritonavir combination therapy had shown no significant benefit over the standard of care without an antiviral drug [3]. Other drugs predicted to have stable binding to RdRP include daclatasvir, delavirdine, ribavirin and sofosbuvir, but all showed lower affinity than elbasvir. We therefore focused on elbasvir as the most promising candidate for the RdRP target. Molecular modeling of the docking pose indicated that elbasvir fits well in the predicted RNA-binding pocket of RdRP (Figure 3b).
Figure 3:
Solvent-based docking energy prediction for FDA-approved antiviral drugs against target proteins. (a) Binding free energies of top-ranked drugs, docking with RdRP; (b) docking pose for elbasvir with RdRP; (c) binding scores against papain-like proteinase; (d) docking pose for elbasvir with proteinase; (e) top-ranked FDA approved drugs against helicase, (f) docking pose for elbasvir bound to helicase.
We then repeated the same docking studies for the second candidate protein, papain-like proteinase. Elbasvir showed the most stable binding to protease in solvent-based MMGBSA estimation of ΔG, the free energy of binding (Figure 3c). Docking-pose models indicate an excellent fit of elbasvir to the predicted protease cavity (Figure 3d). Other drugs, including voxilaprevir, daclatasvir and sofosbuvir (Figure 3c), bound with lower stability (higher ΔG) than elbasvir.
The third target protein is SARS-CoV-2 helicase. Drug targeting of helicase was shown to efficiently inhibit replication of other coronaviruses [8, 10]. Since helicase structure and function are conserved across the SARS family of viruses, including SARS-CoV and MERS [12], we postulated that targeting the helicase protein should inhibit viral replication, independent of other inhibitory activities. Glide docking, followed by solvent-based calculation of ΔG for drug : helicase binding, predicts that elbasvir should be a singularly effective candidate drug to target the helicase nucleic-acid-binding pocket; ΔG for elbasvir exceeds the next best drug, daclatasvir, by at least 15 kCal/mol, and zanamivir by 20 kCal/mol (Figure 3e). Docking-pose modeling supports elbasvir binding to the predicted nucleic-acid binding pocket of the helicase protein (Figure 3f). An experiment-based model of the SARS-CoV Nsp13 (helicase) protein indicates that the site of elbasvir binding coincides with the single-strand DNA (ssDNA) binding pocket established by X-ray crystallography [11, 12]. To assess the validity of these predictions, we performed ssDNA-protein docking studies with SARS-CoV-2 helicase. Results corroborate that the nucleic-acid binding site of SARS-CoV Nsp13 is likely to be structurally conserved in SARS-CoV-2. Indeed, the same DNA-interacting residues (e.g., Ser539) are predicted to interact with RNA or ssDNA, indicating conservation of helicase structure at the protein-sequence level (Figure 2b).
Molecular-dynamic simulations predict stable binding of elbasvir to target proteins
To test the stability of predicted drug-protein interactions, we performed fully solvated atomistic molecular-dynamic simulations of predicted elbasvir-bound protein complexes. Binding of elbasvir to both RdRP and helicase varied dynamically. Structural fluctuations of RdRP and its ligand are indicated by variation in the root-mean-square deviation (RMSD). The RMSD of elbasvir, shown as red tracings, initially increased and then stabilized or compacted after 20 ns (Figure 4a). However, the structure of RdRP itself remained unstable in the presence of elbasvir over 200 ns (blue RMSD tracings). The docking pose and the interacting-residue analyses at the 100-ns frame show metastable binding of elbasvir to RdRP in the predicted nucleic-acid-binding pocket (Figure 4b). To assess whether elbasvir binding would hinder RdRP interaction with RNA, we attempted to dock RNA with the elbasvir-RdRP binding complex observed at 100 ns. We found that elbasvir blocked RNA binding, although RdRP readily bound RNA in the absence of a drug (Figure 5, a & b).
Figure 4.
Molecular-dynamic simulations of protein-drug interactions. Left panels: blue tracings show RMSD of (a) RdRP, (c) helicase, or (e) protease, each bound to elbasvir; red tracings show RMSD of elbasvir bound to each protein. Right panels: Docking poses of elbasvir (green) bound to: (b) RdRP (200 ns); (d) helicase (100 ns), or (f) protease (100 ns).
Figure 5.
Protein-RNA docking shows elbasvir blocking RNA (green ribbons) from binding to RdRP (a, b), and preventing ssDNA from binding to viral helicase (c, d).
We next modeled the elbasvir-helicase complex, which appeared to stabilize after ~40 ns (Figure 4c), and retained crucial interactions such as Ser539 (Figure 4d). We retrieved the structure of helicase bound to elbasvir at 100 ns, and attempted to dock it with single-stranded DNA. Elbasvir prevented helicase from binding to ssDNA, in contrast to stable binding in the absence of drug (Figure 5, c & d). SsDNA was used in place of RNA, for consistency with structures previously reported for helicase : ssDNA [11, 12].
Papain-like proteinase showed somewhat more chaotic behavior in the presence of elbasvir, throughout the majority of the 100-ns simulation (Figure 4e); both protein and ligand appeared to stabilize only after 70–80 ns. That an elbasvir:proteinase complex was formed during modeling is supported by analysis of the docking pose at 100 ns, which shows elbasvir seated within the proteinase-docking cavity (Figure 4f).
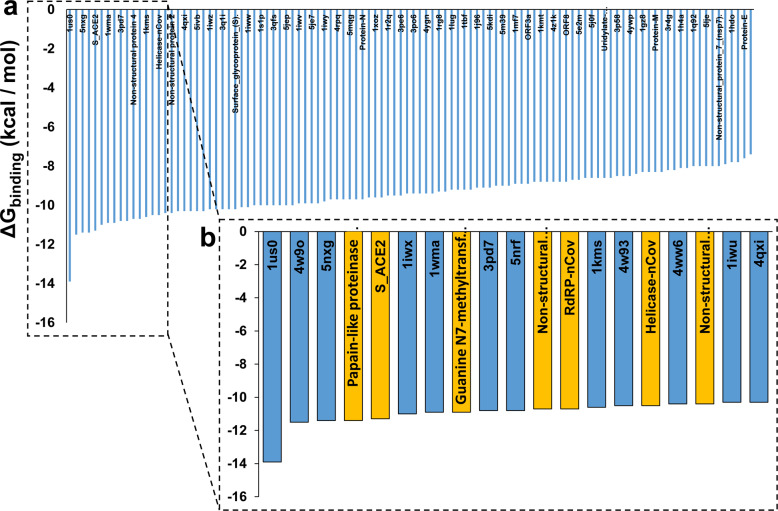
To assess the specificity of interactions between elbasvir and the targets we had selected as essential to virion replication, we compared elbasvir docking to those targets, vs. 100 proteins retrieved at random from the PDB database. Elbasvir showed surprising specificity for affinity to a variety of SARS-CoV-2 proteins: of the 100 random proteins screened, only 3 bound to elbasvir as well or better than either the papain-like proteinase or the viral S protein complex with the ACE2 host protein (S_ACE2); only 5 exceeded the Guanine-N7 methyl-transferase; 7 exceeded RdRP; and 9 surpassed the helicase (Figure 6).
Figure 6.
Elbasvir docking to SARS-CoV-2 proteins (gold bars) or 100 proteins taken at random from PDB, with structures generated by I-TASSER (blue bars).
We then asked how elbasvir compares to other antiviral drugs with respect to interaction with the full complement of SARS-CoV-2 proteins. Although several other drugs (e.g., lopinavir, raltagravir) are capable of weak interaction with several viral proteins, elbasvir is unique in its predicted high affinity for a wide variety of SARS-CoV-2 proteins, including target proteins necessary for viral replication (Figure 7). Elbasvir is predicted to bind stably (ΔG ≤ ‒10.5) to RdRP, helicase, guanine-N7 methyltransferase, papain-like proteinase, proteinase SCL-PRO, and surface glycoprotein S, alone or in a complex with ACE2 (angiotensin converting enzyme 2), a host-cell protein exploited by SARS-CoV-2 for cell entry.
Figure 7.
Elbasvir is compared to 9 other drugs, for free energy of binding to 25 SARS-CoV-2 proteins. Lopinavir shows strong binding (comparable to elbasvir) only to viral S protein in a complex with host ACE2 protein (S_ACE2), and to 2 nonstructural proteins. Raltegravir, ritonavir, and sofosbuvir are predicted to bind with only moderate avidity to viral proteins. Key (right) shows ΔGbinding range from ‒4.5 to ‒11 kcal/mol.
Discussion
Through an unbiased in silico screen, supported by molecular modeling, we have unveiled an FDA-approved drug with remarkably broad affinity for SARS-CoV-2 proteins. This drug, elbasvir, is marketed in combination with grazoprevir as Zepatier™, which was developed and approved for treatment of chronic infections with the hepatitis C virus.
We began with structure-based drug screening against viral proteins (RNA-dependent RNA polymerase, helicase, and papain-like proteinase) that are required for viral replication. Elbasvir was predicted to interact with all three proteins, with exceptionally high affinity (see Figures 3, 6 and 7). Its stability of binding to a wide variety of proteins from the same virus is rather surprising. Because elbasvir interacts with 5–10% of proteins taken at random, with binding energies comparable to those for SARS-CoV-2 proteins (Figure 6), we infer that elbasvir is a highly reactive molecule — in common with many of the most effective therapeutic drugs — and is thus likely to have many off-target interactions [13]. However, it is encouraging that it has been reported to have a relatively low incidence of deleterious side effects at the recommended antiviral dosage [14].
We note that the other component of Zepatier™, grazoprevir, showed only modest affinity for SARS-CoV-2 proteins (Figure 7), and is thus unlikely to contribute directly to therapeutic targeting of viral replication in COVID-19 despite the demonstrated benefit of combining the two drugs to treat hepatitis C [14]. On the other hand, it is possible for effective drug combinations to derive their synergy from indirect effects. In treatment of HIV, for example, the combination of protease inhibitors lopinavir and ritonavir (Kaletra™) derives most of its potency from lopinavir, with ritonavir contributing indirectly by delaying clearance of lopinavir [15]. Therefore, we propose clinical tests of elbasvir alone as well as in combination with grazoprevir or zanamivir.
The propensity of elbasvir to bind avidly to 6 distinct SARS-CoV-2 proteins, all within the top 10% of predicted affinities toward random proteins assessed for ΔGbinding (Figure 6), remains unexplained - and would occur by chance as 6 independent events at a frequency less than 1 per million (0.16). This may reflect the drug’s dimensions (similar to a short stretch of RNA or ssDNA) fitting into nucleic-acid binding pockets, along with polar atoms appropriately oriented to form electrostatic interactions with target proteins. Binding-residue analysis indicates that elbasvir interacts chiefly with positively-charged residues including arginine and lysine, and the polar residue serine. A coronavirus must assemble its constituent proteins around the RNA core prior to exit from a host cell; this essentially obliges all viral proteins to have affinity for either RNA (typically via electrostatic binding of positively-charged amino acids to the strongly negative phosphates of the RNA backbone), or for another RNA-bound viral protein. We note in this regard that the 3 viral proteins having the strongest interactions with elbasvir are guanine-N7 methyltransferase, RdRP, and helicase - all of which have nucleic-acid binding affinity. A second plausible explanation would depend upon the evolution of this RNA virus by gene duplication [16], resulting in viral protein structures that are all related and well conserved.
The strategy of targeting multiple vulnerabilities of a virus (or indeed, a cancer cell) is well established, and confers several important benefits: more effective lethality to the targeted entity, due to a multipronged attack; reduction in the requisite dose of each component, thereby reducing side effects; and elimination the potential of the pathogen to evolve resistance via mutation - an especially likely outcome for RNA viruses in view of their high mutation rates.
Conclusions
Based on target-based computational drug screening, we predict elbasvir to have very high affinity for key SARS-CoV-2 proteins, including RNA-dependent RNA polymerase, helicase, papain-like proteinase, and the viral S protein used for entry to cells decorated with ACE-2. Molecular-dynamic simulations and protein docking predict stable interactions of elbasvir with a wide range of SARS-CoV-2 targets. We therefore propose clinical tests of elbasvir as a therapeutic drug for those with demonstrated COVID-2, comparing the efficacy of elbasvir alone to its combination with grazoprevir as Zepatier™.
Methods
Structure-based targeted docking
The structures of SARS-CoV-2 proteins used in this study were retrieved from I-TASSER (neural network-based structure predictions) [9]. Proteins were preprocessed using the Protein Preparation Wizard module of Schrödinger Suite [17] before docking. Druggable pockets in target proteins, including RNA-dependent RNA polymerase, helicase, and papain-like proteinase, were predicted using BIOVIA Discovery Studio BIOVIA (Pipeline Pilot, Release 2017, San Diego: Dassault Systèmes). A library of 54 FDA-approved antiviral drugs was retrieved from DrugBank (https://www.drugbank.ca/) and preprocessed using the Schrödinger Lig-Prep module. For each target protein, a docking grid box was created based on the predicted druggable pocket, using Schrödinger’s Gridbox Generator. Docking studies were performed in two stages, as follows: (1) Glide docking was conducted in extra-precision mode, against each target protein. The top 15 drug candidates were then selected for next-level screening. (2) The Gibbs Free Energy of binding (ΔGbinding) was calculated within Schrödinger’s Prime module by the MMGBSA method, using molecular mechanics, the generalized Born solvent model, and solvent accessibility [18–20].
Molecular-dynamic simulation of protein-drug complexes
Models from MMGBSA were used for atomistic molecular-dynamic (MD) simulations using the Desmond package (Schrödinger). Each protein-drug complex was immersed in a solvent box containing Single Point-Charge (SPC) water models, neutralized with NaCl, and brought to physiological conditions by further addition of NaCl to 0.15M. The system was next equilibrated using the NPT protocol (constant number of particles, pressure and temperature) with default temperature (300°K) and pressure (1 bar). MD simulations were then conducted for 100–200 ns using Desmond’s GPU-based simulation method [18–22]. Results were analyzed using the Simulation Interactions Diagram module from Schrödinger Maestro. Drug-protein complex stabilities presented in this paper were also confirmed using a web-based GROMACS simulation tool for protein-ligand complexes (https://simlab.uams.edu/ProteinWithLigand/, data not shown).
Unbiased protein-ligand docking
Unbiased or whole-protein docking was conducted using AutoDock-Vina molecular docking package [18, 23, 24]. Briefly, proteins and drugs were prepared in Vina format prior to docking. Unlike targeted docking protocols, the docking box was constructed to include the entire protein structure [18]. Drugs were then docked against each protein of interest using the Raccoon2 graphical user interface for Vina [25]. The resulting large datasets were analyzed using an in-house developed pipeline for analysis and interpretation of docking results.
Nucleic-acid docking to proteins
Nucleic-acid structures used in this study were retrieved from the PDB databank (RNA, PDB-ID 6k32; ssDNA, PDB-ID 4N0O). The Hex docking program was used to predict the structures of complexes comprising RNA or single-strand DNA bound to a protein [26]. Docking parameters were set to “shape+electrostatic” with all other parameters left at default values. The top-ranked docking poses were retrieved and visualized using the BIOVIA Discovery Studio.
Acknowledgments
We are grateful for grants from the U.S. Department of Veteran Affairs (Merit 2 I01 BX001655 and Senior Research Career Scientist Award to RJSR); from the U.S. National Institutes of Health / National Institute on Aging (grants 2P01 AG012411 [Principal Investigator WST Griffin] and 1R01 AG062254-01 [P.I.’s RJSR and S. Ayyadevara]); and from the Inglewood Scholars Program for career-development support to MB.
References Cited
- 1.Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, et al. Drug targets for corona virus: A systematic review. Indian J Pharmacol. 2020;52(1):56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. 2020. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–71. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Clercq E, Li G. Approved Antiviral Drugs over the Past 50 Years. Clin Microbiol Rev. 2016;29(3):695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stubbs TM, Te Velthuis AJ. The RNA-dependent RNA polymerase of the influenza A virus. Future Virol. 2014;9(9):863–76. doi: 10.2217/fvl.14.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio. 2018;9(2). doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78(11):5619–32. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J Proteome Res. 2020. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frick DN. Helicases as antiviral drug targets. Drug News Perspect. 2003;16(6):355–62. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng Z, Lehmann KC, Li X, Feng C, Wang G, Zhang Q, et al. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res. 2014;42(5):3464–77. doi: 10.1093/nar/gkt1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao W, Wojdyla JA, Zhao R, Han R, Das R, Zlatev I, et al. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017;13(6):e1006474. doi: 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhari R, Tan Z, Huang B, Zhang S. Computational polypharmacology: a new paradigm for drug discovery. Expert Opin Drug Discov. 2017;12(3):279–91. doi: 10.1080/17460441.2017.1280024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson IM, Lawitz E, Kwo PY, Hezode C, Peng CY, Howe AYM, et al. Safety and Efficacy of Elbasvir/Grazoprevir in Patients With Hepatitis C Virus Infection and Compensated Cirrhosis: An Integrated Analysis. Gastroenterology. 2017;152(6):1372–82 e2. doi: 10.1053/j.gastro.2017.01.050. [DOI] [PubMed] [Google Scholar]
- 15.Zeldin RK, Petruschke RA. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J Antimicrob Chemother. 2004;53(1):4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- 16.Simon-Loriere E, Holmes EC. Gene duplication is infrequent in the recent evolutionary history of RNA viruses. Mol Biol Evol. 2013;30(6):1263–9. doi: 10.1093/molbev/mst044. [DOI] [PubMed] [Google Scholar]
- 17.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–34. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 18.Janganati V, Ponder J, Balasubramaniam M, Bhat-Nakshatri P, Bar EE, Nakshatri H, et al. MMB triazole analogs are potent NF-kappaB inhibitors and anti-cancer agents against both hematological and solid tumor cells. Eur J Med Chem. 2018;157:562–81. doi: 10.1016/j.ejmech.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakraba S, Ayyadevara S, Penthala NR, Balasubramaniam M, Ganne A, Liu L, et al. A novel microtubule-binding drug attenuates and reverses protein aggregation in animal models of Alzheimer’s Disease. Front Mol Neurosci. 2019;12:310. doi: 10.3389/fnmol.2019.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balasubramaniam M, Lakkaniga NR, Dera AA, Fayi MA, Abohashrh M, Ahmad I, et al. FCX-146, a potent allosteric inhibitor of Akt kinase in cancer cells: Lead optimization of the second-generation arylidene indanone scaffold. Biotechnol Appl Biochem. 2020. doi: 10.1002/bab.1896. [DOI] [PubMed] [Google Scholar]
- 21.Balasubramaniam M, Ayyadevara S, Shmookler Reis RJ. Structural insights into pro-aggregation effects of C. elegans CRAM-1 and its human ortholog SERF2. Sci Rep. 2018;8(1):14891. doi: 10.1038/s41598-018-33143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parcon PA, Balasubramaniam M, Ayyadevara S, Jones RA, Liu L, Shmookler Reis RJ, et al. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimers Dement. 2018;14(2):230–42. doi: 10.1016/j.jalz.2017.07.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bommagani S, Penthala NR, Balasubramaniam M, Kuravi S, Caldas-Lopes E, Guzman ML, et al. A novel tetrazole analogue of resveratrol is a potent anticancer agent. Bioorg Med Chem Lett. 2019;29(2):172–8. doi: 10.1016/j.bmcl.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61. doi: 10.1002/jcc.21334 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905–19. doi: nprot.2016.051 [pii]; 10.1038/nprot.2016.051 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie DW, Kozakov D, Vajda S. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics. 2008;24(17):1865–73. doi: 10.1093/bioinformatics/btn334 [doi];btn334 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]