Corresponding Author

Key Words: coronary artery disease, genetic risk, primary prevention, statin
Readers may have noticed a proliferation of articles on polygenic scores and risks for various medical conditions. More than likely, the deadly COVID-19 pandemic—for which a diagnosis and DNA can be had with 1 swab—will soon yield polygenic scores for susceptibility to the infection and its sequelae. Polygenic scores for coronary artery disease (CAD) have also attracted wide interest.
CAD can occur as a single-gene disorder or as a polygenic condition. In this issue of the Journal, Aragam et al. (1) show that polygenic scores based on >6 million common single nucleotide polymorphisms (SNPs) throughout the genome “robustly prognosticate coronary artery disease risk in the general population” and may be useful for prevention of CAD.
The study had >47,000 participants in 3 health care systems. Clinical data and lipid levels were from electronic health records. Disease definitions were based on billing codes and/or medication prescriptions. Pooled cohort equations were used to assign patients to clinical risk categories for management of cholesterol.
There were 16,002 people in the study who did not have atherosclerotic cardiovascular disease (CAD, peripheral artery disease, or cerebral atherosclerosis), diabetes mellitus, or severe hypercholesterolemia. And 5,890 of them had borderline or intermediate CAD risks (by pooled cohort equations). Of these borderline or intermediate risk individuals, 987 had CAD polygenic scores in the top 20% of the scores, and 652 did not have prescriptions for statins (or 652 of 16,002 = 4.1% of the primary prevention group). Thus, an additional 4.1% of the primary prevention population could be recommended for statin use, if polygenic scores were part of treatment guidelines. This is not an insignificant addition to the target population for prevention of CAD.
Polygenic Inheritance
The science of polygenic inheritance is more than 100 years old (2). Wigan (3) gave the following explanation of polygenic inheritance (4,5):
Briefly stated, the ‘quantitative’ characters, on which evolution and adaptation chiefly depend, are assumed to be under the control of numerous polygenes, with individual effects small in comparison with those caused by the environment. In the simplest case, each polygene is represented by two allelomorphs [variants], one of which has an effect in the ‘plus’ direction, tending to increase the expression of the character involved, and the other in the ‘minus,’ or opposite, direction. Plus and minus effects of different polygenes are typically balanced in an organism. That is, the polygenes are present in such combinations that the phenotype conforms relatively closely to the optimum for the environment. But this does not mean that the genotype is necessarily homozygous, since balance is expressed in the zygote by the action of the whole genotype. Therefore, in an individual heterozygous for the polygenes, recombination may give rise to new combinations of polygenes, which, on coming together in a new zygote, will give a phenotype departing more widely from the optimum and hence not as well balanced to the prevailing conditions.
In early statistical models, the number of polygenes for a trait could be as few as 3, or there could be an “infinite series” (6). One article chose 100 polygenes, for the sake of discussion, when conjecturing about genes that influence “fecundity in a sexually reproducing organism” (7).
Polygenes were “invisible” entities (8) for nearly a century. They were grasped only through complex statistical analyses—“too small in their individual effects to be separated and counted” (9). Then the genome-wide association study (GWAS) era made polygenes real—countable and identifiable—with measurable effects (called beta values). The first CAD GWAS was in 2007, involving 23,000 participants (10). By 2017, large international consortia and biobanks led to discovery of 95 CAD genes (or loci) (11). The current number may be more than 150 (12). Some of the genes play a role in lipid regulation, insulin resistance, clotting, inflammation, or vascular tone, but the mechanisms of action for most CAD polygenes are unknown (13).
Polygenic Scores
Polygenic scores (also called polygenic risk scores or genetic risk scores) are the summed effects of all the risk variants for a trait in an individual. The current method for combining risk variants into weighted polygenic scores was proposed by Horne et al. (14). Other approaches also appeared (15). Two well-known examples of CAD polygenic scores are the 27-SNP score of Mega et al. (16) and the 50-SNP score of Khera et al. (17). Mega et al. showed that individuals with the highest scores had the greatest benefit from statins. Khera et al. found that favorable lifestyles could lower coronary event rates by 50% (compared with rates for unfavorable lifestyles) among people at high polygenic risk.
Reviews on the utility of polygenic scores for CAD have been enthusiastic. For example, in a JACC editorial, Natarajan (18) called polygenic scores for CAD, “The First Risk Factor,” and said the following:
Our current [clinical] framework insufficiently identifies those likely to sustain premature CHD. Inouye et al. show that incorporation of CHD polygenic risk with clinical risk factors can improve risk prediction and may help identify individuals who are candidates for earlier preventive therapies. Additionally, this single genetic test (currently <$100) needs to be performed only once, and this framework can be applied to calculate polygenic risk for virtually any trait.
Perhaps realizing that answers to yes or no questions in titles are almost always no, Levin and Rader (19) wrote a somewhat tempered editorial for Circulation entitled, “Polygenic Risk Scores and Coronary Artery Disease: Ready for Prime Time?” The answer follows:
The current studies provide clear evidence for the value of CAD PRS in predicting recurrent cardiovascular events in patients with pre-existing CAD, even after accounting for traditional risk factors, and suggest that high CAD PRS patients may be most likely to benefit from PCSK9 inhibition. As such, they add to a growing body of evidence suggesting clinically impactful roles for genetic risk stratification in CAD. Ongoing study of the clinical utility and implementation of CAD PRS is warranted.
Around the same time, back-to-back reports in the Journal of the American Medical Association examined the ability of polygenic scores to predict future CAD events. Khan et al. (20) wrote an editorial entitled, “Do Polygenic Risk Scores Improve Patient Selection for Prevention of Coronary Artery Disease?” They concluded the following:
The available data do not support the clinical utility of CAD polygenic risk scores (in their current form) in middle-aged adults of European descent. In the meanwhile, the best approach for prevention of CAD continues to be a combination of population-wide risk factor approaches for the entire population and addition of drug therapies and lifestyle interventions according to guidelines developed by the American Heart Association and American College of Cardiology.
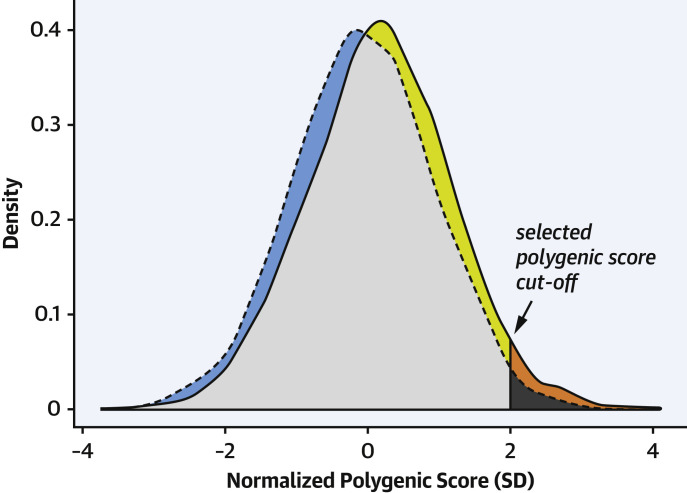
A limitation of polygenic scores as individual disease predictors was shown by Dr. Nicholas Wald (Figure 1 ). Dr. Wald argues that the odds ratios for CAD in relation to polygenic scores are robust but not high enough for the scores to be useful for general predictive screening. For example, the odds ratio is 1.9-fold for the top 20% of the polygenic scores (compared with the remaining 80%) (1). According to Wald and Old (21), “Estimates of the relative risk [odds ratios] between a disease marker and a disease have to be extremely high for the risk factor to merit consideration as a worthwhile screening test.” But, conceivably, this limitation for the entire population should not preclude use of polygenic scores from all patient care situations involving CAD.
Figure 1.
Illustration of False Positive and CAD Detection Rates for CAD Polygenic Scores
Shown are distributions of polygenic scores for people with coronary artery disease (CAD) (solid line) and controls (dashed line). The vertical line indicates a possible polygenic score cutoff for predictive testing (at scores giving a false positive rate of 5%, in this drawing). The false positive rate is the proportion of control individuals with positive scores = dark gray/[dark gray + light gray + blue]. The detection rate is the proportion of patients with CAD with positive scores = [orange + dark gray]/[orange + dark gray + yellow + light gray]. The CAD detection rate is low at the cutoff used. The false positive rate will be higher for scores with higher detection rates. The figure was adapted from Supplemental Figure 1 (all ancestries; normalized score; Partners Healthcare Biobank) in Aragam et al. (1).
Conclusions
Although we acknowledge all of the comments stated here, we believe that the cumulative data favor the view that polygenic scoring is a low-cost test that can be done once in a lifetime at any age and for a growing list of important conditions. This study extends the evidence that polygenic scores do appear to provide an added axis of risk for prevention of CAD, beyond standard clinical risk factors. The current guidelines may well not capture all individuals who could benefit from statins. There may be other specific CAD situations in which the scores are helpful—possibly among younger people. Further study will tell how best to use these new risk factors in management decisions for individual patients and will show the extent to which the scores help prevent illness and save lives.
Footnotes
Dr. Rotter has received support from National Institutes of Health grants HL151855, UL1TR001881, and DK063491. Dr. Lin has reported that he has no relationships relevant to the contents of this paper to disclose.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC author instructions page.
References
- 1.Aragam K.G., Dobbyn A., Judy R. Limitations of contemporary guidelines for managing patients at high genetic risk of coronary artery disease. J Am Coll Cardiol. 2020;75:2769–2780. doi: 10.1016/j.jacc.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher R.A. The correlation between relatives on the supposition of Mendelian inheritance. Trans Roy Soc Edin. 1918;52:399–433. [Google Scholar]
- 3.Wigan L.G. Balance and potence in natural populations. J Genet. 1944;46:150–160. [Google Scholar]
- 4.Mather K. Polygenic inheritance and natural selection. Biol Rev. 1943;18:32–64. [Google Scholar]
- 5.Mather K. Methuen and Company Ltd; London, UK: 1949. Biometrical Genetics: The Study of Continuous Variation. [Google Scholar]
- 6.Panse V.G. A statistical study of quantitative inheritance. Ann Eugen. 1940;10:76–105. [Google Scholar]
- 7.Buzzati-Traverso A.A. Quantitative traits, and polygenic systems in evolution. Cold Spring Harb Symp Quant Biol. 1959;24:41–46. doi: 10.1101/sqb.1959.024.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Mather K., Wigan L.G. The selection of invisible mutations. Proc R Soc Lond B-Biol Sci. 1942;131:50–64. [Google Scholar]
- 9.Darlington C.D. The polygene concept. Nature. 1942;150:154. [Google Scholar]
- 10.McPherson R., Pertsemlidis A., Kavaslar N. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klarin D., Zhu Q.M., Emdin C.A. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. doi: 10.1038/ng.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musunuru K., Kathiresan S. Genetics of common, complex coronary artery disease. Cell. 2019;177:132–145. doi: 10.1016/j.cell.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Khera A.V., Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horne B.D., Anderson J.L., Carlquist J.F. Generating genetic risk scores from intermediate phenotypes for use in association studies of clinically significant endpoints. Ann Hum Genet. 2005;69:176–186. doi: 10.1046/j.1529-8817.2005.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison A.C., Bare L.A., Chambless L.E. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 16.Mega J.L., Stitziel N.O., Smith J.G. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khera A.V., Emdin C.A., Drake I. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natarajan P. Polygenic risk scoring for coronary heart disease: the first risk factor. J Am Coll Cardiol. 2018;72:1894–1897. doi: 10.1016/j.jacc.2018.08.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin M.G., Rader D.J. Polygenic risk scores and coronary artery disease: ready for prime time? Circulation. 2020;141:637–640. doi: 10.1161/CIRCULATIONAHA.119.044770. [DOI] [PubMed] [Google Scholar]
- 20.Khan S.S., Cooper R., Greenland P. Do polygenic risk scores improve patient selection for prevention of coronary artery disease? JAMA. 2020;323:614–615. doi: 10.1001/jama.2019.21667. [DOI] [PubMed] [Google Scholar]
- 21.Wald N.J., Old R. The illusion of polygenic disease risk prediction. Genet Med. 2019;21:1705–1707. doi: 10.1038/s41436-018-0418-5. [DOI] [PubMed] [Google Scholar]