Abstract
Objectives: Viral phylogenetics revealed two patterns of HIV-1 spread among MSM in Quebec. While most HIV-1 strains (n = 2011) were associated with singleton/small clusters (cluster size 1–4), 30 viral lineages formed large networks (cluster size 20–140), contributing to 42% of diagnoses between 2011 and 2015. Herein, tissue culture selections ascertained if large cluster lineages possessed higher replicative fitness than singleton/small cluster isolates, allowing for viral escape from integrase inhibitors.
Methods: Primary HIV-1 isolates from large 20+ cluster (n = 11) or singleton/small cluster (n = 6) networks were passagedin vitro in escalating concentrations of dolutegravir, elvitegravir and lamivudine for 24–36 weeks. Sanger and deep sequencing assessed genotypic changes under selective drug pressure.
Results: Large cluster HIV-1 isolates selected for resistance to dolutegravir, elvitegravir and lamivudine faster than HIV-1 strains forming small clusters. With dolutegravir, large cluster HIV-1 variants acquired solitary R263K (n = 7), S153Y (n = 1) or H51Y (n = 1) mutations as the dominant quasi-species within 8–12 weeks as compared with small cluster lineages where R263K (n = 1/6), S153Y (1/6) or WT species (4/6) were observed after 24 weeks. Interestingly, dolutegravir-associated mutations compromised viral replicative fitness, precluding escalations in concentrations beyond 5–10 nM. With elvitegravir, large cluster variants more rapidly acquired first mutations (T66I, A92G, N155H or S147G) by week 8 followed by sequential accumulation of multiple mutations leading to viral escape (>10 μM) by week 24.
Conclusions: Further studies are needed to understand virological features of large cluster viruses that may favour their transmissibility, replicative competence and potential to escape selective antiretroviral drug pressure.
Introduction
ART has transformed HIV/AIDS from a deadly pandemic to a treatable and potentially preventable disease.1,2 Expanded access to ART has resulted in 25%–50% declines in global epidemics in heterosexual populations, raising optimism that treatment-as-prevention may arrest the pandemic by 2030 (www.unaids.org).3–7 The World Health Organization has advanced global ‘90–90–90’ targets for 2020 where 90% living with HIV-1 are informed of their status, 90% of infected individuals receive antiviral treatment and 90% of those treated achieve viral suppression (www.unaids.org). HIV-1 guidelines have been revised to meet these objectives, recommending annual HIV testing, immediate treatment initiation and pre-exposure prophylaxis for high risk populations.1,3,4,8–16
As more asymptomatic patients are started on ART, treatment goals will require more robust and durable first-line regimens to prevent the development and transmission of drug resistance.17,18 The mainstay of treatment is two NRTIs with a third core agent from one of the NNRTI, boosted PI or integrase strand transfer inhibitor (INSTI) drug classes.19 In developing countries, rates of virological failure (40%) and transmitted drug resistance (5%–16%) are rising, related to the use of thymidine analogues, particularly stavudine and zidovudine, and NNRTIs.18,20–24
Integrase inhibitors, including raltegravir, elvitegravir and dolutegravir, are the preferred choice for first-line therapy, based on their improved tolerability, dosing (once daily) and higher barrier to resistance than other antiretroviral drugs (http://aidsinfo.nih.gov/guidelines).25,26 Resistance to elvitegravir and raltegravir occurs via several mutational pathways, including: (i) N155H or G140A/G148RHQ pathways conferring raltegravir and elvitegravir cross-resistance; (ii) T66I or E92Q/G elvitegravir-specific pathways; or (iii) the Y143R/H/C raltegravir-specific resistance pathway.25 Dolutegravir retains antiviral activity against HIV-1 variants resistant to raltegravir and elvitegravir.27–30 Dolutegravir is far less prone to the development of resistance in first-line therapy with isolated reported cases of the emergence of R263K, N155H or G118R.31–34 These substitutions have been shown to arise duringin vitro cell culture selection studies.27,35 Transmitted resistance to integrase inhibitors remains anecdotal.36,37
Disturbingly, HIV-1 epidemics among MSM have not declined in resource-rich nations having access to the best antiretroviral treatments.38 Viral phylogenetic linkage has revealed that epidemics among MSM have been sustained by high rates of transmission clustering related in part to episodic risk effects during highly infectious primary HIV infection (PHI).39–46 A shift towards large cluster lineages has contributed to the growth of HIV epidemics among MSM in Quebec and the Netherlands.47,48 Whereas half of the HIV-1 epidemic in MSM in Quebec can be ascribed to 2011 viral lineages leading to singleton/small cluster transmissions (cluster size 1–4), 30 viral variants strains have led to large cluster outbreaks (cluster size 20–140), rising from 13% of new diagnoses in 2004 to 42% of new infections in 2015.45,49 The rise in large 20+ clusters has offset steady declines in other cluster groups over the last decade.45,49 Transmission clustering has been implicated in the spread of resistance to thymidine analogues and NNRTIs.21,45,50–52
HIV-1 transmission constraints lead to a single monophyletic HIV-1 variant, termed the transmitted/founder virus, establishing most new infections. There is continued debate as to whether transmission bottlenecks are stochastic, randomly restricting all viruses, or selective, favouring specific transmission/founder viruses52–55 Observational data from the Montreal PHI cohort showed that cluster size and the skewed role of 30 founder viruses (1.7% of viral lineages) in 1200 forward transmissions were not directly related to patient epidemiological factors, including numbers of reported sexual partnerships and viral load.45 We postulated that HIV-1 strains associated with large clusters have unique phenotypic features in reverse transcriptase (RT) and integrase replicative processes that may have contributed to heightened infectiousness and cluster burst size.
To test this hypothesis, tissue culture selections with dolutegravir, elvitegravir and lamivudine were used to compare the barriers to resistance for viruses derived from 11 patients belonging to 10 large clusters (cluster size 20–140) and 6 persons associated with singleton/small clusters (cluster size 1–4). Sanger (population) and next-generation (deep) sequencing was performed to monitor genotypic changes under selective drug pressure over 36 weeks. HIV-1 isolates from large cluster lineages demonstrated a lower genetic barrier to resistance to dolutegravir, elvitegravir and lamivudine as compared with viruses from singleton/small cluster networks. However, the rapid acquisition of R263K or S153Y mutations with dolutegravir compromised viral replicative competence and hindered viral breakthrough. Taken together, our findings show a selection bias for large cluster viral variants showing higher replicative fitness under selective drug pressure.
Methods
Viral phylogenetic reconstruction of the HIV-1 epidemic in MSM
Viral phylogenetics was used to reconstruct patterns of HIV-1 spread among newly diagnosed treatment-naive MSM (n = 4039, 2002–15).40,44,45 HIV-1 genotyping was performed as previously described, generating partialpol sequences that span the viral protease and RT regions.40,44,45 Phylogenetic trees were built using MEGA7 integrated software (www.megasoftware.net).44,45,50,53 Clustering of viral strains was defined by high bootstrap support (>95%) and short genetic distances (<1.5%). HIV-1 strains from large clusters were resequenced across the entire integrase region as previously described to compare clustering patterns across the protease, RT and integrase regions.54
Isolation of viruses from MSM within large and small cluster groups
The Fonds de recherche Santé (FRQS) Réseau SIDA supports a cohort of newly infected persons with clinical indication of primary infection.55 In this study, HIV-1 strains were isolated from 17 MSM subjects, 11 of whom belonged to 10 large clusters (cluster size 20–140) and 6 from singleton/small cluster transmissions (cluster size 1–4). HIV-1 isolates were amplified as previously described through co-culture of patient CD8-cell-depleted peripheral blood mononuclear cells with primary human cord blood mononuclear cells (CBMCs).56,57 HIV-1 strains, integrase natural polymorphisms, clinical features and GenBank accession numbers are summarized in Table1.
Table 1.
Baseline natural polymorphisms in the integrase of subtype B HIV-1 isolates used for in vitro selections with elvitegravir, dolutegravir and lamivudine
| Lab IDa | Cluster ID (cluster size)b | Visit 1 date | Months infected (partners)c | CD4 cells/μLc | Integrase polymorphisms | GenBank number |
|---|---|---|---|---|---|---|
| Large | ||||||
| 14947 | C185.1 (n = 45) | 01/2014 | 2 (1–4) | 60 | E11D, R20K, V31I, S39N, M50I, I72V, S119T, T124N, G193E, V201I, T218ST, D286N | KT988126 |
| 14637 | C185.09 (n = 45) | 05/2013 | 3 (1–4) | 550 | E11D, R20K, V31I, S39N, M50I, S119T, T124N, G193E, V201I, D286N | KT988125 |
| 14997 | C027.28 (n = 40) | 02/2014 | 3 (1–4) | 70 | E11D, V31I, L101I, S119G, T122I T124A, T125A, F181L, I203M, K211Q, T218I, D232E, D256E, D286N | KT988128 |
| 14969 | C053.16 (n = 24) | 01/2014 | 2 (1–4) | 220 | S17N, S24D, D25E, S39C, V201I, S255Q | KT988127 |
| 10387 | C099.10 (n = 35) | 04/2008 | 2 (5–9) | 580 | V31I, L45Q, I72V, K111E, I113V, T125A, M154I, V201I, D207N, L234I | KX714013 |
| 10249 | C099.08 103N (n = 34) | 11/2007 | 5 (>20) | 1010 | V31I, L45Q, I72V, L101I, K111E, I113V, D207N, L234I | KX714014 |
| 12083 | C050.37 G190A (n = 140) | 02/2010 | 6 (>20) | 1470 | S17N, I72V, I73V, L101I, T124A, K136Q | KX714016 |
| 14624 | C050.65 G190A (n = 140) | 07/2013 | 9 (1–4) | 187 | S17N, A23V, L28I, S39C, L101I, T124A | KX714018 |
| 12608 | C163.05 (n = 90) | 05/2011 | 4 (1–4) | 300 | E11D, A21T, S39C, L45R, I72V, L101I, G163E, K244R | KX714017 |
| 10679 | C075.19 (n = 21) | 09/2008 | 3 (1–4) | 1250 | E11D, A21T, L101I, I113V, S119P, T125A, G193E, D278N | KX714015 |
| 5331 | C089.01 (n = 39) | 02/1997 | 5 (0) | 191 | E11D, E13D, I72V, L101I, I113V, S119P, T122I, D256E | KX714020 |
| Small | ||||||
| 5326 | C196.02 (n = 4) | 11/1998 | — | — | E11D, A21T, L101I, I113V, S119P, T125A, G193E, D278N | KX714021 |
| 15366 | C506.01 (n = 2) | 12/2014 | 1 (1–4) | 23 | S17N, I72V, T124A, T206S, N254H, A265V, S283G | KT988129 |
| 14514 | S630 (n = 1) | 02/2014 | 5 (1–4) | 430 | K7R, S17N, M50I, I72V, K111Q, T112V, T124A, T125V, I220L, Y227F, D256E | KT988124 |
| 14380 | C309 (n = 1) | 11/2012 | 2 (1–4) | 730 | R20K, V31I, I84V, T112M, T124N, M154I, V201I, D278E | KT988123 |
| 5833 | S347 (n = 1) | 02/2001 | — | — | T112V, G193E, V201I, S230N | KT988122 |
| 15600 | C545.01 (n = 2) | 07/2015 | 2 (1–4) | 320 | E11D, V31I, A124T, R127K, F181L, V201I, T218I, S230N, N232D, D256E | KX714019 |
Viruses were isolated from patients participating in the PHI cohort in Montreal.
Cluster size was based on phylogenetic analysis of the entire provincial genotyping programme.
Cohort data, including estimated time post-infection (months) based on biological assays, the reported number of partners 3 months prior to infection and CD4 counts, were determined at the first visit.
In vitro selection of HIV-1 strains with reduced susceptibility to antiretroviral drugs
Dolutegravir was provided by ViiV Healthcare Ltd (Research Triangle Park, NC, USA). Elvitegravir was obtained from Gilead Sciences Inc. (Foster City, CA, USA). Lamivudine was obtained from the NIH AIDS Research and Reference Reagent Program (http://www.aidsreagent.org). HIV-1 strains from 6 and 11 patients from singleton/small and large clusters, respectively, were used to infect CBMCs at a similar moi of 0.1, i.e. 1 × 106 TCID50 per 10 × 106 CBMCs, based on p24 and RT assays.57 Viruses were serial passaged in CBMCs in the presence of escalating concentrations of dolutegravir, elvitegravir and/or lamivudine over 24–36 weeks, as previously described.58,59 RT activity in culture supernatants was quantified weekly to monitor viral replication of individual viral variants in the presence and absence of drug pressure, adjusting incremental increases in drug concentrations if RT activity assays exceeded 100 000 cpm.58,59 At each passage, aliquots of cell-free supernatant were stored at −70°C for further genotypic analysis.
HIV-1 genotyping based on Sanger and deep sequencing
Genotypic analyses at selected passages evaluated the development of amino acid substitutions under selective drug pressure, conferring reduced susceptibility to antiretroviral drugs (i.e. drug resistance mutations). Sanger (population) sequencing of viral RNA or complementary DNA extracted from culture supernatants was performed as previously described.54,59 Deep sequencing was performed using a CLIA/CAP-validated HIV-1 genotyping assay (DEEPGEN™HIV) with a >1% cut-off for the detection of minority HIV-1 variants based on the intrinsic error rate of the entire system (0.3%).60 Ultradeep sequencing confirmed the homogeneity of founder variants, as well as the earliest time to emergence of resistance to integrase inhibitors.
Ethics approval
Ethics approvals for phylogenetic surveillance of the HIV-1 epidemic were obtained from the Ministry of Health (Quebec) and L’Institut national de santé publique du Québec. Blood samples from the Montreal primary infection cohort were obtained with informed consent and ethics approval from recruited individuals from participating hospitals or private clinics.55
Results
Isolation of HIV-1 strains belonging to representative large and singleton/small clusters
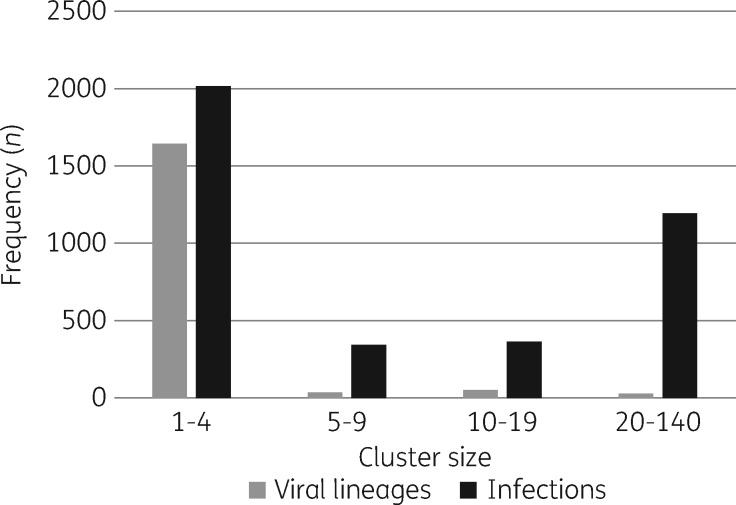
Viral phylogenetics revealed two predominant patterns of HIV-1 spread among MSM in Quebec. Half of new infections were associated with singleton/small cluster networks (cluster size 1–4), while 30 viruses (1.7% of viral lineages) resulted in micro-epidemics, averaging 40 transmissions/cluster (Figure1).45 Viruses within both cluster groups were isolated to compare viral escape from antiretroviral pressure.
Figure 1.
Patterns of spread of the HIV-1 epidemic among the MSM population in Quebec (2002–15). Phylogenetic tree analysis ascertained clustering of new infections genotyped between 2002 and 2015. The frequency of viral lineages (grey) and HIV-1 infections (black) associated with cluster groups, having 1 to 4, 5 to 9, 10 to 19, or 20 to 140 linked transmissions.
Eleven subtype B viruses belonging to 10 large 20+ clusters and 6 singleton/small clusters were derived from PHI cohort participants with estimated dates of infection of 3.7 ± 0.6 months based on Detuned/BED biological assays (Table1).55 The 11 large cluster strains were collectively associated with 600 transmissions in Quebec. The six singleton/small cluster variants included two reference clinical isolates (5326 and 5833) used in cell culture selections by our laboratory for over a decade. Table 1 shows clinical and genotypic features of the selected viruses, illustrating that cluster size was not related to the number of reported sexual partnerships by cohort participants 3 months prior to their infection, as previously reported for the entire MSM cohort.45
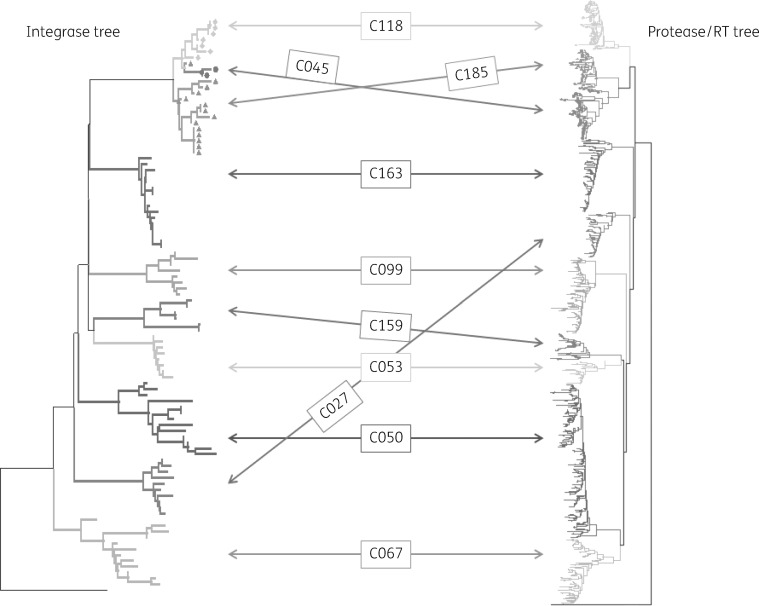
Viruses associated with the selected 10 20+ large clusters were resequenced across the integrase-coding regions. Phylogenetic trees showed similar co-clustering patterns across the integrase region as previously observed across the protease/RT domains. It was, however, noteworthy that clusters 45, 118 and 185 (cluster size = 38, 43 and 45, respectively) had a shared common branch in the integrase tree although the protease/RT regions within these three clusters were distinct (Figure2).
Figure 2.
Phylogenetic tree analysis shows 10 large clusters sequenced across the protease/RT- and integrase-coding regions. Note that viruses belonging to cluster 185 (n = 44), cluster 45 (n = 38) and cluster 118 (n = 43) share sequence homology across the integrase domain.
In vitro selection of HIV-1 variants resistant to dolutegravir, elvitegravir and lamivudine
Clinical isolates from newly infected persons typically use the CCR5 receptor, show T-cell tropism and do not grow in MT-2 cells. Tissue culture selections were performed using CBMCs to characterize possible differences in the development of drug resistance to lamivudine and integrase inhibitors.57,61 CBMCs infected with relevant HIV-1 strains (from large or singleton/small clusters) were serially passaged over the course of 24–36 weeks in the presence of increasing concentrations of dolutegravir, elvitegravir, lamivudine or dolutegravir + lamivudine dual pressure. These drugs are recommended components of choice in first-line regimens.
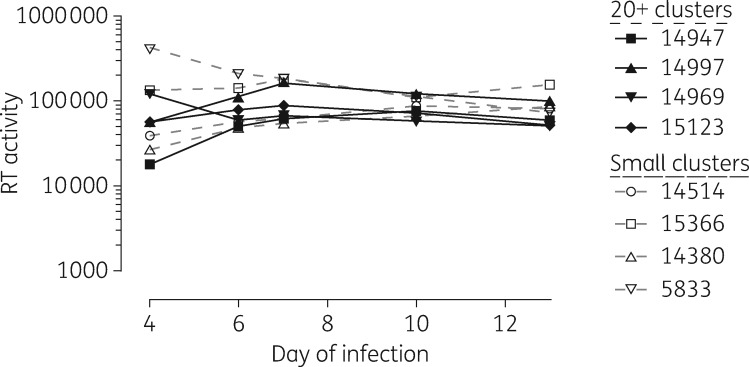
Cell culture selections using large cluster and singleton/small cluster HIV-1 strains were conducted in parallel under the same conditions. Stepwise drug dose escalations were based on weekly RT assays, performed for each isolate in the presence and absence of drug. As illustrated in Figure3, viruses in both cluster groups replicated equally well in the absence of drug pressure. The progress of viral outgrowth in the presence of stepwise increasing concentrations of inhibitors over the course of 36 weeks, comparing viruses belonging to cluster 185 (14947 and 14637), cluster 27 (14997) and cluster 53 (14969) and four singleton small cluster lineages (HIV-1 isolates 143809, 14515, 15336 and 5833), is depicted in Figure4.
Figure 3.
Levels of viral RT activity following initial infection of CBMCs with large cluster (n = 4, filled symbols) and small cluster (n = 4, open symbols) viral lineages. Levels of RT activity remained similar in subsequent serial passage in the absence of drug pressure.
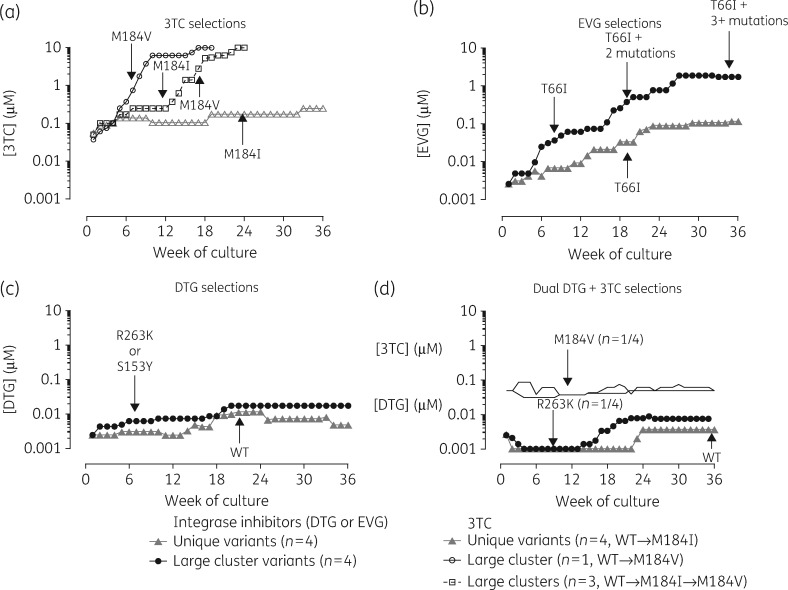
Figure 4.
Drug resistance selections with lamivudine, elvitegravir, dolutegravir and dolutegravir + lamivudine. Stepwise increases in drug concentrations under selective drug pressure were based on levels of RT enzymatic activity at each weekly passage relative to a no-drug control. The mean weekly drug concentrations were averaged for the four large cluster viral lineages (black lines) belonging to clusters 27 (14997), 53 (14969) and 185 (14947 and 14637) as compared with four singleton/small cluster variants (isolates 143809, 14515, 15336 and 5833, grey lines) grown under selective pressure with lamivudine, elvitegravir, dolutegravir or dolutegravir + lamivudine over 36 weeks. The arrows represent viral breakthrough based on the first acquisition of new resistance mutations presented in Figures4–6. Regarding lamivudine, one large cluster virus passaged with lamivudine acquired M184V leading to earlier viral escape by week 16; the remaining three large clusters developed M184I. DTG, dolutegravir; EVG, elvitegravir; 3TC, lamivudine.
As illustrated in Figure4(a), drug dose escalations and viral breakthrough, under lamivudine selective pressure, progressed at faster rates in the case of large cluster lineages as compared with singleton/small cluster strains. Drug dose escalations with lamivudine in large cluster variants were associated with the appearance at week 8 of M184V (isolate 14947) or M184I (isolates 14637, 14997 and 14637). The latter isolates shifted from M184I to M184V by weeks 15–23, leading to higher fold resistance (>100-fold) and viral escape. Small cluster isolates developed resistance more slowly, acquiring M184I infections by week 23 (Figure3a). The rapid selection of M184V by isolate 14947 parallels that observed for CXCR4-tropic pNL4.3 reference strain grown in MT-2 cells or CBMCs. The M184I substitution conferred lower fold resistance, decreased processivity and replicative fitness than the M184V mutation.62,63 Large cluster variants showed a more rapid shift from M184I to M184V when compared with small cluster lineages where selection of M184I was attenuated.
Similarly, elvitegravir drug selections performed on four large cluster variants progressed more rapidly, leading to the first acquisition of the INSTI resistance mutations T66I (n = 2), N155H (n = 1) or S147G (n = 1) at week 8, followed by the further accumulation of resistance mutations leading to viral escape (>100-fold resistance) at weeks 23 and 36 (Figure3b). In contrast, elvitegravir selections performed on singleton/small cluster viral lineages were delayed with the first appearance of T66I as a singlet mutation occurring at week 23 in only two of four small cluster strains (Figure4b).
Large cluster strains also showed a lower barrier to resistance to dolutegravir with R263K (n = 3) or S153Y (n = 1) mutations appearing by week 8 in large cluster lineages; these mutations were absent in the four small cluster viruses at week 36 (Figure 4c). It was, however, noteworthy that the acquisition of R263K and S153Y had a negative impact on RT enzymatic activity, hindering escalations in dolutegravir concentrations beyond 5–10 nM (Figure4c). Dual selections with dolutegravir and lamivudine showed the appearance of resistance in only one large cluster isolate (14947) with the first appearance of R263K at week 8 preceding the acquisition of M184I/V and M184V at weeks 15 and 23, respectively (Figure4d). Drug concentrations of both drugs could not be increased following the acquisition of R263K.
Genotypic analysis of viral outgrowth under dolutegravir and elvitegravir pressure
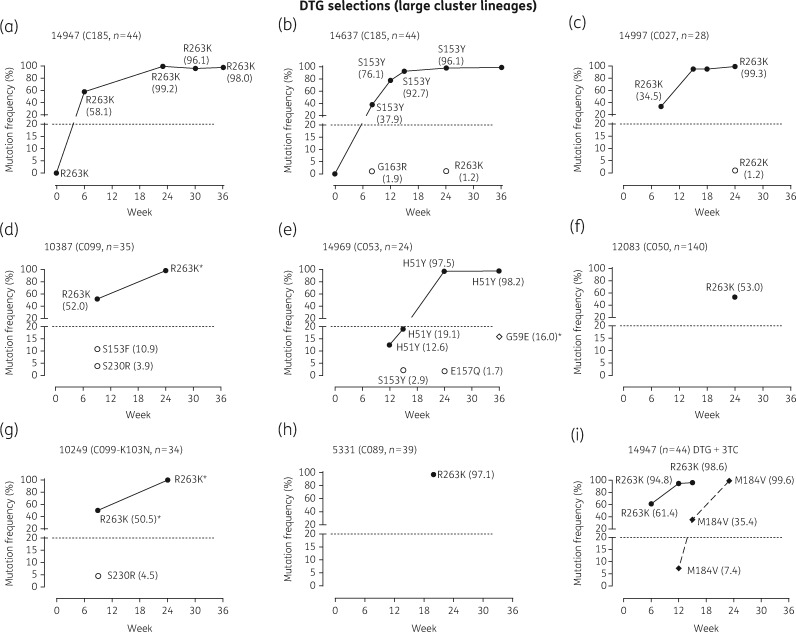
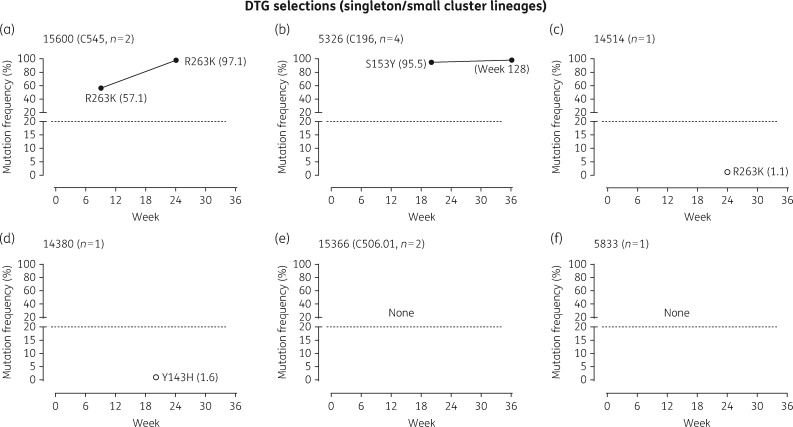
HIV-1 genotypic analysis based on deep sequencing was performed at selected passages to monitor temporal dynamics of the emergence and accumulation of mutations conferring resistance to dolutegravir in eight large cluster strains, as compared with six small cluster variants (Figures5 and6).
Figure 5.
Viral outgrowth of HIV-1 large cluster strains cultured in the presence of increasing concentrations of dolutegravir. HIV-1 isolates from eight large clusters (cluster size in parentheses) were cultured in the presence of increasing concentrations of dolutegravir. Cell-free supernatant was collected at selected time periods and viral RNA purified and genotyped across the integrase or RT domain. The frequency (%) of individual amino acid substitutions acquired in the virus population at the designated weekly passage was determined based on deep sequencing. Cluster 14947 (a and i) was selected with both dolutegravir and dolutegravir + lamivudine to ascertain the sequential appearance of R263K and M184V. DTG, dolutegravir; 3TC, lamivudine. An asterisk indicates Sanger sequencing only.
Figure 6.
Viral outgrowth of HIV-1 singleton/small cluster strains cultured in the presence of increasing concentrations of dolutegravir. HIV-1 isolates from six singleton/small clusters (cluster size in parentheses) were cultured in the presence of increasing concentrations of dolutegravir. Cell-free supernatant was collected at selected time periods and viral RNA purified and genotyped across the integrase domain. The frequency (%) of individual amino acid substitutions acquired in the virus population at the designated weekly passage was determined based on deep sequencing. DTG, dolutegravir.
As depicted in Figure5, dolutegravir selections, performed on large cluster viruses, led to rapid appearance of R263K (n = 6/8) or S153Y (n = 1/8) as dominant members of quasi-species within 6–8 weeks, while H51Y (n = 1/8) appeared by week 12 (Figure5). In general, viral outgrowth under dolutegravir pressure resulted in R263K, S153Y or H51Y as singlet mutations, representing the dominant quasi-species (>96%) at weeks 24–36 based on ultradeep sequencing. The infrequent appearance of S230R, E157Q, G163R or G59E minority species (<10%) occurred in some large cluster selections at selected weeks of passage but failed to accumulate over time. Although isolate 14637, belonging to cluster 185, selected for S153Y (Figure 5b), three subsequent dolutegravir selections using this isolate resulted in R263K as the sole mutation (B. G. Brenner and M. Oliveira, unpublished results). Isolate 10387 showed the dual appearance of R263K and S153F as 52% and 10.9% of the quasi-species at week 8, although R263K (99%) became and remained the dominant quasi-species at week 26 (Figure5d).
There was a delay in the selection of dolutegravir resistance in small cluster as compared with large cluster lineages (Figure5). Four of six small cluster lineages (cluster sizes 1, 1, 1 and 2) failed to select any resistance to dolutegravir over 36 weeks. INSTI-associated mutations were present at really low frequency as determined by deep sequencing (R263K at 1.1% and Y143H at 1.6% in 14514 and 14380, respectively) (Figure5c and d). The remaining two small clusters (cluster sizes 2 and 4) selected R263K or S153Y as singleton mutations (Figure5a and b).
Incremental increases in dolutegravir concentrations (beyond 10 nM) following acquisition of R263K, S153Y and H51Y mutations could not be attempted because the pressure on viral survival was too great based on weekly RT assays (Figures4c and5). This decline in viral replicative fitness under drug pressure is consistent with cell culture selection studies and integrase enzymatic assays previously performed by our group using site-directed mutagenesis to insert resistance substitutions into the laboratory HIV-1 pNL4.3 strain.64–66 Notably, isolate 5326 acquiring S153Y showed no further accumulation of mutations after 128 weeks of culture in the presence of dolutegravir (Figure5b).
Cluster 185 isolates 14947 and 14637 harbouring R263K and S153Y at weeks 30 and 34, respectively, were grown in the absence of dolutegravir pressure for a subsequent 19 and 17 weeks, respectively (Figure4a and b). The release of drug pressure failed to lead to a reversion of R263K or S153Y, substantiating their presence as dominant quasi-species.
Interestingly, culturing HIV-1 isolate 14947 (cluster 185) with dolutegravir led to the rapid selection of variants carrying the R263K mutation, as rapidly as M184V selection with lamivudine (Figure4a and d). Again, the acquisition of M184V/I was faster for large cluster as compared with small cluster lineages. It is also significant that R263K arose more rapidly than M184V in dolutegravir + lamivudine dual selections performed with isolate 14947 and that both mutations can coexist (Figures4d and5i).
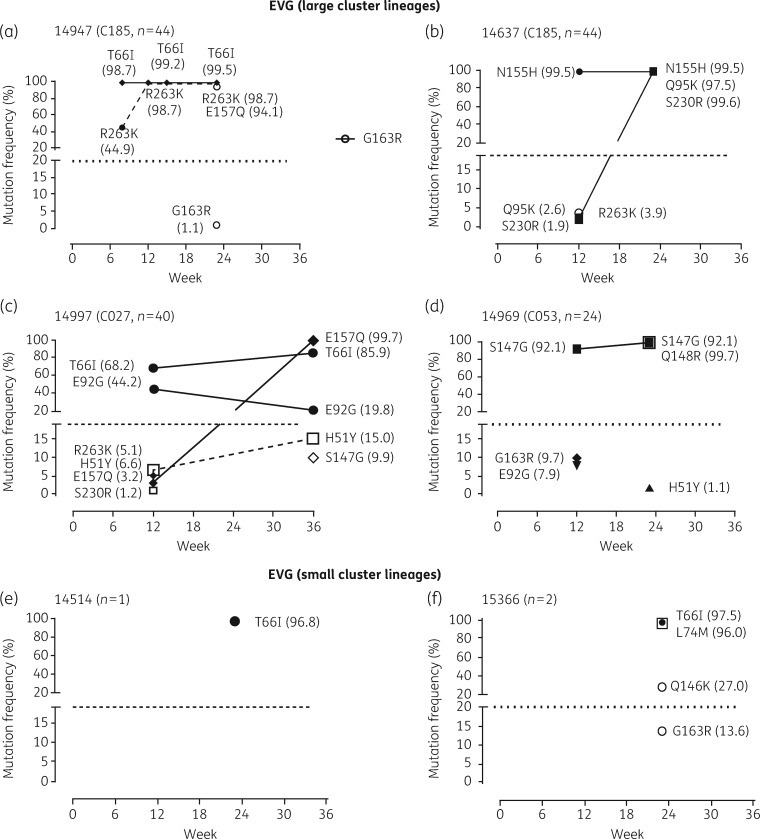
The development of resistance under elvitegravir pressure proceeded rapidly for large cluster lineages with a sequential accumulation of multiple mutations along different resistance pathways (Figure7a–d). Isolate 14947 acquired T66I and R263K within 8 weeks, representing 98.7% and 44.9% of the quasi-species, respectively (Figure6a). By week 24, the dominant virus coexpressed T66I (99.5%), R263K (98.7%) and E157Q (94.1%) (Figure6a). The second isolate, belonging to cluster 185 (14637), developed resistance with viruses coexpressing N155H (99.5%), Q95K (97.5%) and S230R (99.6%) as the dominant quasi-species (Figure6b). Isolate 14997 acquired a mixture of different viruses with T66I, E92G, E157Q, H51Y and/or S147G representing 85.9%, 19.8%, 99.7%, 15.0% and 9.9% of variants at week 36 (Figure6c). Isolate 14969 acquired S147G (92.1%) at week 8 followed by Q148R (99.7%), known to confer high levels of cross-resistance to elvitegravir and raltegravir (Figure7d).25 The accumulation of mutations along different pathways contributed to viral breakthrough and >100-fold resistance. Similarly, the acquisition of T66I followed by secondary mutations, including L74M, Q146K, and G163R, by small cluster viruses contributed to viral escape by week 24 (Figure7e and f).
Figure 7.
Genotyping analysis based on Sanger and deep sequencing of HIV-1 small cluster strains cultured in the presence of increasing concentrations of elvitegravir. HIV-1 isolates from four large clusters and two singleton/small clusters were cultured in the presence of increasing concentrations of elvitegravir. Cell-free supernatant was collected at selected time periods and viral RNA purified and the RT- and/or integrase-coding region sequenced using Sanger or deep sequencing as previously described. The frequency (%) of individual amino acid substitutions acquired in the virus population at the designated weekly passage was determined based on deep sequencing. EVG, elvitegravir.
It was rather surprising that R263K and H51Y did not adversely impact on viral replicative competence with elvitegravir as observed for dolutegravir (Figure6a). Deep sequencing-based genotyping showed T66I selection occurred prior to R263K followed by E157Q, which may have compensated for the fitness compromise of R263K (Figure6a). The presence of secondary (accessory) mutations, including T66I, E157Q, G163R and S230R, can increase levels of resistance and/or viral fitness (Figures4b and6).
Discussion
Genetic diversity is the hallmark feature of HIV-1 contributing to the divergent spread of distinct HIV-1 subtypes and recombinant forms in different regional settings. In Quebec, the subtype B HIV-1 epidemic among MSM has not declined in the era of potent and simplified ART and improved treatment-as-prevention strategies. Viral phylogenetic cluster analysis has shown that half of the subtype B HIV-1 epidemic among MSM can be ascribed to the episodic genesis and expansion of 30 viral lineages, contributing to 13%, 25% and 42% of new diagnoses over the periods 2004–07, 2008–11 and 2012–15, respectively.45,67 Here we proposed that the disproportionate contribution of these 30 strains over more than a decade may be related to distinct virological features beyond the effects of risk behaviour and the high infectiousness of acute/recent stage infection. Novel findings observed in this study usingin vitro selections revealed that HIV-1 large cluster lineages show a lower barrier to resistance when compared with viruses from singleton/small cluster networks.
The rapid acquisition of R263K, S153Y or H51Y with dolutegravir was unexpected given the isolated number of reported cases of resistance in the clinic. Indeed, the appearance of these dolutegravir-related mutations within 6–12 weeks using HIV-1 strains from large clusters was far more rapid than previous studies by our group using laboratory strains where resistance arises after 20 weeks of culture with dolutegravir.68,69 Furthermore, viruses obtained by site-directed mutagenesis of the laboratory strain pNL4.3 with R263K or H51Y are severely compromised in their ability to acquire or coexist with other mutations, such as M184V and T66I.69–71 Findings reported herein revealed the emergence of viral variants coexpressing R263K/M184V and R263K/T66I as dominant (>98%) quasi-species under selective pressure with dolutegravir + lamivudine and elvitegravir, respectively. The acquisition of E157Q (94% and 99%) by two viruses (and) coexpressing T66I/R263K (>98%, isolate 14947) or a mixed T66I (85.9%) and H51Y (15.9%) quasi-species (isolate 14997) under elvitegravir pressure suggests that E157Q may serve as an accessory mutation that restores viral replicative fitness and increases resistance.72
Given the high potency of dolutegravir in the clinic, it has proven difficult to document and characterize dolutegravir resistance profiles. Cell culture selections performed using replicatively resilient large cluster variants highlighted novel aspects regarding emergent resistance to dolutegravir that could not be otherwise ascertained.66 We observed that the rate of appearance of R263K with dolutegravir was as rapid as that observed for the appearance of M184V with lamivudine. The emergence of select mutations under dolutegravir pressure, including R263K (AGA→AAA or AGG→AAG), G163R (GGA→AGA or GGG→AGG) and E138K (GAA→AAA, GAG→AAG), may be related to APOBEC-mediated G→A hypermutation.73,74 Our previous studies also showed the development of G118R (GGA→AGA) in two patients failing dolutegravir monotherapy.35 These patients harboured a rare GGA natural polymorphism (1.5% prevalence) compared with the majority of clinical viral isolates harbouring GGC (79%) or GGT (18%) glycine motifs where G118R acquisition would require two point mutations (GGG→AGA or GGU→AGA) or single unfavourable transversions (GGG→CGG orGGU→CGT), respectively.35 The generation of M184I (ATG→ATA) can also arise from G to A hypermutation although M184V leads to higher fold drug resistance. Although APOBEC3G-mediated G to A hypermutation may contribute to the development of drug resistance, it can also inhibit viral replication by driving viruses into error catastrophe mode.75
The development of resistance to antiretroviral drug regimensin vivo is predicated on the degree and fitness costs conferred by individual mutations, alone and in combination with other resistance mutations. Dolutegravir is a drug for which thein vitro acquisition of singlet substitutions, including R263K, S153Y and H51Y, interfered with further acquisition of secondary resistance mutations.64,72 These mutations confer minimal resistance (<3-fold) but impose enormous constraints on viral replicative fitness, decreasing viral infectiousness, replication capacity, DNA integration and integrase strand-transfer activity.63,64,72 The high potency for dolutegravir resulted in EC50s of 1 nM (0.4 ng/ml) and drug concentrations could not be increased beyond 5 to 10 nM with R263K or S153Y to sustainin vitro viral replication in CBMCs.63,64,76 Thesein vitro drug concentrations are consistent with therapeutic concentrations of unbound (17 ng/mL) and bound (3400 ng/mL) dolutegravir in plasma.77,78 This contrasts with elvitegravir selections, such as the cluster 185 isolate, 14947, where acquisition of T66I preceded R263K, resulting in higher levels of resistance, the acquisition of the compensatory E157Q and viral escape.72 The combination of R263K with viruses containing M184I or M184V mutations in dolutegravir + lamivudine selections further reduced viral replication competence compared with when only single mutations were present.63 Taken together, ourin vitro findings incorporating magnitude of resistance and fitness costs of individual mutations provide an explanation for the remarkable clinical success of dolutegravir and bictegravir in recent clinical trials.78–80 The high potency of second-generation integrase inhibitors may prevent or attenuate the development of replicatively fit large cluster lineages.
Large cluster outbreaks can contribute to the onward spread of transmitted drug resistance, principally involving NNRTIs.45,50 Indeed, the frequency of these resistance mutations in treatment-naive persons has surpassed that observed in persons failing therapy since 2011.38,50 Notably, cluster 50 (cluster size 140) and cluster 99-K103N (cluster size 34) remain persistent sub-epidemics harbouring G190A and K103N, respectively. Genetic differences between viral strains may contribute to their relative transmissibility, replicative competence and propensity towards the development of resistance. Our findings show that R263K and S153Y, once acquired, may not revert. The negative viral fitness costs of these mutations may, however, limit their development and potential transmissibility. In this regard the prevalence of resistance in patients failing integrase-based regimens was 8% in Quebec (January 2014–April 2016), largely associated with raltegravir- and elvitegravir-associated resistance mutations (I. Hardy and B. G. Brenner, unpublished results). Resistance to dolutegravir has, however, been limited to the G118R case and a novel resistance pathway from a raltegravir-experienced patient harbouring the N155H mutation.30,35
Bottlenecks constrain HIV-1 transmission, such that a single monophyletic founder variant is responsible for establishing individual infections. Studies have compared the ‘signature’ genetic motifs and phenotypic properties of newly transmitted viruses to those of chronic-stage viruses.81,82 Transmitted viruses are most often CCR5-tropic, possess signature leader sequences (codon 11), show less envelope glycosylation and have shorter variable envelope loops than chronic stage viruses.81,82 Here, we also show an accelerated development of resistance to integrase and RT inhibitors on the part of viruses belonging to 10 different large 20+ clusters, perhaps suggesting a potential replicative and/or enzymatic fitness advantage in the subset of large cluster viruses. Studies are ongoing to ascertain RT and integrase activity for WT and resistant (R263K or S153Y) cluster 185 isolates, compared with pNL4.3. Direct study of the infection and replication properties of clinically relevant large cluster viruses in cell culture may help define distinct features of transmitted viruses and early events following infection.
In summary, viral phylogenetic cluster analysis of the HIV-1 epidemic among MSM in Quebec shows an evolutionary shift towards large cluster lineages. These viruses appear to be able to overcome severe transmission bottlenecks and circumvent genetic barriers to resistance to dolutegravir.64,68,79 These findings have implications for novel treatment paradigms under investigation, including simplified dolutegravir + lamivudine dual therapy and dolutegravir monotherapy regimens.35,79 Large cluster viruses may assist in assessing as yet unknown pathways implicated in emergent resistance to new second-generation INSTIs in clinical trial, including bictegravir and cabotegravir.27,83,84
Acknowledgements
The Montreal PHI cohort includes Mario Legault, coordinator and all participating physicians from Clinique médicale L’Actuel, Clinique médicale Quartier Latin, Centre hospitalier Université de Montréal, Clinique Roger Leblanc/Opus and Centre hospitalier Université McGill, including Jean-Guy Baril, Louise Charest, Marc-André Charron, Pierre Côté, Alexandra de Pokomandy, Serge Dufresne, Claude Fortin, Jason Friedman, Norbert Gilmore, Emmanuelle Huchet, Marina Klein, Louise Labreque, Richard Lalonde, Roger Leblanc, Bernard Lessard, Catherine Milne, Marie Munoz, Martin Potter, Danielle Rouleau, Jean-Pierre Routy, Jason Szabo, Réjean Thomas, Cecile Tremblay, Benoît Trottier and Sylvie Vézina. We thank all the patient participants.
Funding
This project was supported by the Canadian Institutes for Health Research (CIHR), Fonds de Recherche du Québec (FRQ) Réseau SIDA and FRQ/Research Foundation Flanders (FWO) bilateral programme (202685). M. A. W. received a Large-Scale applied research contract in genomics and personalized healthcare from Genome Canada (HIV142). M. E. Q.-M. was partially supported by the Case Western Reserve University (CWRU)/University Hospitals (UH) Center for AIDS Research (P30 AI036219) and by funding from University Hospitals Cleveland Medical Center (UHCMC) for the University Hospitals Translational Laboratory (UHTL).
All funders of research played no decision-making role in the design, execution, analysis or reporting of the research.
Transparency declarations
B. G. B. has received fees for speaking from Gilead Sciences. M. A. W. has received funds from Gilead Sciences and ViiV Healthcare for speaking and attending symposiums. All other authors: none to declare.
All authors have read and approved the text.
References
- 1. Montaner JS,Lima VD,Barrios R. et al. Association of highly active antiretroviral therapy coverage, population viral load, and yearly new HIV diagnoses in British Columbia, Canada: a population-based study.Lancet 2010;376: 532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones A,Cremin I,Abdullah F. et al. Transformation of HIV from pandemic to low-endemic levels: a public health approach to combination prevention. Lancet 2014;384: 272–9. [DOI] [PubMed] [Google Scholar]
- 3. Cohen MS,Chen YQ,McCauley M. et al. Prevention of HIV-1 infection with early antiretroviral therapy.N Engl J Med 2011; 365:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grant RM,Lama JR,Anderson PL. et al. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men.N Engl J Med 2010; 363:2587–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Insight START Study Group Lundgren JD,Babiker AG. et al. Initiation of antiretroviral therapy in early asymptomatic HIV infection.N Engl J Med 2015; 373:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grinsztejn B,Hosseinipour MC, Ribaudo HJ. et al. Effects of early versus delayed initiation of antiretroviral treatment on clinical outcomes of HIV-1 infection: results from the phase 3 HPTN 052 randomised controlled trial.Lancet Infect Dis 2014; 14:281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCormack S,Dunn DT,Desai M. et al. Pre-exposure prophylaxis to prevent the acquisition of HIV-1 infection (PROUD): effectiveness results from the pilot phase of a pragmatic open-label randomised trial.Lancet 2016;387: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Labarga P. New DHHS guidelines recommend antiretroviral therapy to all HIV-infected persons.AIDS Rev 2012; 14:154.. [PubMed] [Google Scholar]
- 9. Thompson MA,Aberg JA,Hoy JF. et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. JAMA 2012;308: 387–402. [DOI] [PubMed] [Google Scholar]
- 10. Okwundu CI,Uthman OA,Okoromah CA.. Antiretroviral pre-exposure prophylaxis (PrEP) for preventing HIV in high-risk individuals.Cochrane Database Syst Rev 2012; issue7: CD007189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thigpen MC,Kebaabetswe PM, Paxton LA. et al. Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana.N Engl J Med 2012; 367:423–34. [DOI] [PubMed] [Google Scholar]
- 12. Abdool Karim Q,Abdool Karim SS,Frohlich JA. et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science 2010;329: 1168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Donnell D,Baeten JM,Kiarie J. et al. Heterosexual HIV-1 transmission after initiation of antiretroviral therapy: a prospective cohort analysis.Lancet 2010; 375:2092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jia Z,Mao Y,Zhang F. et al. Antiretroviral therapy to prevent HIV transmission in serodiscordant couples in China (2003-11): a national observational cohort study. Lancet 2013;382: 1195–203. [DOI] [PubMed] [Google Scholar]
- 15. Granich RM,Gilks CF,Dye C. et al. Universal voluntary HIV testing with immediate antiretroviral therapy as a strategy for elimination of HIV transmission: a mathematical model. Lancet 2009;373: 48–57. [DOI] [PubMed] [Google Scholar]
- 16. Bendavid E,Grant P,Talbot A. et al. Cost-effectiveness of antiretroviral regimens in the World Health Organization’s treatment guidelines: a South African analysis. AIDS 2011;25: 211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brenner B,Wainberg MA.. We need to use the best antiretroviral drugs worldwide to prevent HIV drug resistance.AIDS 2016;30: 2725–7. [DOI] [PubMed] [Google Scholar]
- 18. Wainberg MA,Zaharatos GJ, Brenner BG.. Development of antiretroviral drug resistance.N Engl J Med 2011;365: 637–46. [DOI] [PubMed] [Google Scholar]
- 19. Llibre JM,Walmsley S,Gatell JM.. Backbones versus core agents in initial ART regimens: one game, two players.J Antimicrob Chemother 2016; 71:856–61. [DOI] [PubMed] [Google Scholar]
- 20. Rhee SY,Jordan MR,Raizes E. et al. HIV-1 drug resistance mutations: potential applications for point-of-care genotypic resistance testing.PLoS One 2015;10: e0145772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rhee SY,Blanco JL,Jordan MR. et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis.PLoS Med 2015;12: e1001810.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steegen K,Carmona S,Bronze M. et al. Moderate levels of pre-treatment HIV-1 antiretroviral drug resistance detected in the first South African national survey.PLoS One 2016;11: e0166305.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Villabona-Arenas CJ,Vidal N,Guichet E. et al. In-depth analysis of HIV-1 drug resistance mutations in HIV-infected individuals failing first-line regimens in West and Central Africa. AIDS 2016;30: 2577–89. [DOI] [PubMed] [Google Scholar]
- 24. Gregson J,Kaleebu P,Marconi VC. et al. Occult HIV-1 drug resistance to thymidine analogues following failure of first-line tenofovir combined with a cytosine analogue and nevirapine or efavirenz in sub Saharan Africa: a retrospective multi-centre cohort study. Lancet Infect Dis 2017;17: 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mesplede T,Quashie PK,Zanichelli V. et al. Integrase strand transfer inhibitors in the management of HIV-positive individuals.Ann Med 2014; 46:123–9. [DOI] [PubMed] [Google Scholar]
- 26. Osterholzer DA,Goldman M.. Dolutegravir: a next-generation integrase inhibitor for treatment of HIV infection.Clin Infect Dis 2014; 59:265–71. [DOI] [PubMed] [Google Scholar]
- 27. Tsiang M,Jones GS,Goldsmith J. et al. Antiviral activity of bictegravir (GS-9883), a novel potent HIV-1 integrase strand transfer inhibitor with an improved resistance profile. Antimicrob Agents Chemother 2016;60: 7086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Canducci F,Ceresola ER, Saita D. et al. In vitro phenotypes to elvitegravir and dolutegravir in primary macrophages and lymphocytes of clonal recombinant viral variants selected in patients failing raltegravir.J Antimicrob Chemother 2013;68: 2525–32. [DOI] [PubMed] [Google Scholar]
- 29. Canducci F,Ceresola ER, Boeri E. et al. Cross-resistance profile of the novel integrase inhibitor dolutegravir (S/GSK1349572) using clonal viral variants selected in patients failing raltegravir.J Infect Dis 2011; 204:1811–5. [DOI] [PubMed] [Google Scholar]
- 30. Hardy I,Brenner B,Quashie P. et al. Evolution of a novel pathway leading to dolutegravir resistance in a patient harbouring N155H and multiclass drug resistance.J Antimicrob Chemother 2015;70: 405–11. [DOI] [PubMed] [Google Scholar]
- 31. Walmsley S,Baumgarten A, Berenguer J. et al. Dolutegravir plus abacavir/lamivudine for the treatment of HIV-1 infection in antiretroviral therapy-naive patients: week 96 and week 144 results from the SINGLE randomized clinical trial.J Acquir Immune Defic Syndr 2015;70: 515–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raffi F,Rachlis A,Brinson C. et al. Dolutegravir efficacy at 48 weeks in key subgroups of treatment-naive HIV-infected individuals in three randomized trials. AIDS 2015;29: 167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raffi F,Jaeger H,Quiros-Roldan E. et al. Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 week results from a randomised, double-blind, non-inferiority trial.Lancet Infect Dis 2013;13: 927–35. [DOI] [PubMed] [Google Scholar]
- 34. Clotet B,Feinberg J,van Lunzen J. et al. Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 week results from the randomised open-label phase 3b study. Lancet 2014;383: 2222–31. [DOI] [PubMed] [Google Scholar]
- 35. Brenner BG,Thomas R,Blanco JL. et al. Development of a G118R mutation in HIV-1 integrase following a switch to dolutegravir monotherapy leading to cross-resistance to integrase inhibitors.J Antimicrob Chemother 2016; 71:1948–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Young B,Fransen S,Greenberg KS. et al. Transmission of integrase strand-transfer inhibitor multidrug-resistant HIV-1: case report and response to raltegravir-containing antiretroviral therapy.Antivir Ther 2011; 16:253–6. [DOI] [PubMed] [Google Scholar]
- 37. Boyd SD,Maldarelli F, Sereti I. et al. Transmitted raltegravir resistance in an HIV-1 CRF_AG-infected patient.Antivir Ther 2011; 16:257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Charest H,Doualla-Bell F, Cantin R. et al. A significant reduction in the frequency of HIV-1 drug resistance in Quebec from 2001 to 2011 is associated with a decrease in the monitored viral load.PLoS One 2014;9: e109420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leigh Brown AJ,Lycett SJ,Weinert L. et al. Transmission network parameters estimated from HIV sequences for a nationwide epidemic.J Infect Dis 2011; 204:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brenner B,Wainberg MA, Roger M.. Phylogenetic inferences on HIV-1 transmission: implications for the design of prevention and treatment interventions. AIDS 2013;27: 1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kouyos RD,von Wyl V,Yerly S. et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland.J Infect Dis 2010;201: 1488–97. [DOI] [PubMed] [Google Scholar]
- 42. Bezemer D,van Sighem A,Lukashov V. et al. Transmission networks of HIV-1 among men who have sex with men in the Netherlands.AIDS 2010; 24:271–82. [DOI] [PubMed] [Google Scholar]
- 43. Brenner BG,Wainberg MA.. Future of phylogeny in HIV prevention.J Acquir Immune Defic Syndr 2013;63 Suppl 2: S248–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brenner BG,Roger M,Routy JP. et al. High rates of forward transmission events after acute/early HIV-1 infection.J Infect Dis 2007; 195:951–9. [DOI] [PubMed] [Google Scholar]
- 45. Brenner BG,Ibanescu RI, Hardy I. et al. Large cluster outbreaks sustain the HIV epidemic among MSM in Quebec.AIDS 2017;31: 707–17. [DOI] [PubMed] [Google Scholar]
- 46. Joseph SB,Swanstrom R, Kashuba AD. et al. Bottlenecks in HIV-1 transmission: insights from the study of founder viruses.Nat Rev Microbiol 2015; 13:414–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brenner BG,Oliveira M,Ibanescu RI. et al. Metamorphosis of the Montreal MSM epidemic: large cluster viruses fuel transmission. In:Abstracts of the Twenty-third Conference on Retroviruses and Opportunistic Infections, Boston, MA, USA, 2016. Abstract 218. Foundation for Retrovirology and Human Health, Alexandria, VA, USA.
- 48. Bezemer D,Cori A,Ratmann O. et al. Dispersion of the HIV-1 epidemic in men who have sex with men in the Netherlands: a combined mathematical model and phylogenetic analysis. PLoS Med 2015;12: e1001898.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brenner BG,Oliveira M,Ibanescu RI. et al. Large cluster isolates show facilitated escape from integrase inhibitors. In: Abstracts of the Twenty-third Conference on Retroviruses and Opportunistic Infections, Boston, MA, USA, 2016. Abstract 480. Foundation for Retrovirology and Human Health, Alexandria, VA, USA.
- 50. Brenner BG,Roger M,Moisi DD. et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection.AIDS 2008; 22:2509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Audelin AM,Gerstoft J,Obel N. et al. Molecular phylogenetics of transmitted drug resistance in newly diagnosed HIV Type 1 individuals in Denmark: a nation-wide study.AIDS Res Hum Retroviruses 2011;27: 1283–90. [DOI] [PubMed] [Google Scholar]
- 52. Hattori J,Shiino T,Gatanaga H. et al. Characteristics of transmitted drug-resistant HIV-1 in recently infected treatment-naive patients in Japan.J Acquir Immune Defic Syndr 2016;71: 367–73. [DOI] [PubMed] [Google Scholar]
- 53. Brenner BG,Roger M,Stephens D. et al. Transmission clustering drives the onward spread of the HIV epidemic among men who have sex with men in Quebec.J Infect Dis 2011;204: 1115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brenner BG,Lowe M,Moisi D. et al. Subtype diversity associated with the development of HIV-1 resistance to integrase inhibitors.J Med Virol 2011; 83:751–9. [DOI] [PubMed] [Google Scholar]
- 55. Routy JP,Machouf N,Edwardes MD. et al. Factors associated with a decrease in the prevalence of drug resistance in newly HIV-1 infected individuals in Montreal. AIDS 2004;18: 2305–12. [DOI] [PubMed] [Google Scholar]
- 56. Gonzalez N,Perez-Olmeda M, Mateos E. et al. A sensitive phenotypic assay for the determination of human immunodeficiency virus type 1 tropism.J Antimicrob Chemother 2010;65: 2493–501. [DOI] [PubMed] [Google Scholar]
- 57. Oliveira M,Brenner BG,Wainberg MA.. Isolation of drug-resistant mutant HIV variants using tissue culture drug selection.Methods Mol Biol 2009; 485:427–33. [DOI] [PubMed] [Google Scholar]
- 58. Brenner BG,Oliveira M,Doualla-Bell F. et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture.AIDS 2006; 20:F9–13. [DOI] [PubMed] [Google Scholar]
- 59. Brenner B,Turner D,Oliveira M. et al. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 2003;17: F1–5. [DOI] [PubMed] [Google Scholar]
- 60. Gibson RM,Meyer AM,Winner D. et al. Sensitive deep-sequencing-based HIV-1 genotyping assay to simultaneously determine susceptibility to protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism.Antimicrob Agents Chemother 2014;58: 2167–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salomon H,Belmonte A,Nguyen K. et al. Comparison of cord blood and peripheral blood mononuclear cells as targets for viral isolation and drug sensitivity studies involving human immunodeficiency virus type 1.J Clin Microbiol 1994; 32:2000–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kulkarni R,Babaoglu K,Lansdon EB. et al. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J Acquir Immune Defic Syndr 2012;59: 47–54. [DOI] [PubMed] [Google Scholar]
- 63. Pham HT,Mesplede T,Wainberg MA.. Effect on HIV-1 viral replication capacity of DTG-resistance mutations in NRTI/NNRTI resistant viruses. Retrovirology 2016;13: 31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Quashie PK,Mesplede T,Han YS. et al. Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir.J Virol 2012; 86:2696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Quashie PK,Oliviera M,Veres T. et al. Differential effects of the G118R, H51Y, and E138K resistance substitutions in different subtypes of HIV integrase.J Virol 2015;89: 3163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mesplede T,Quashie PK,Osman N. et al. Viral fitness cost prevents HIV-1 from evading dolutegravir drug pressure.Retrovirology 2013; 10:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brenner BG,Gryllis C,Wainberg MA.. Role of antibody-dependent cellular cytotoxicity and lymphokine-activated killer cells in AIDS and related diseases.J Leukoc Biol 1991;50: 628–40. [DOI] [PubMed] [Google Scholar]
- 68. Oliveira M,Mesplede T,Moisi D. et al. The dolutegravir R263K resistance mutation in HIV-1 integrase is incompatible with the emergence of resistance against raltegravir. AIDS 2015;29: 2255–60. [DOI] [PubMed] [Google Scholar]
- 69. Oliveira M,Mesplede T,Quashie PK. et al. Resistance mutations against dolutegravir in HIV integrase impair the emergence of resistance against reverse transcriptase inhibitors. AIDS 2014;28: 813–9. [DOI] [PubMed] [Google Scholar]
- 70. Anstett K,Fusco R,Cutillas V. et al. Dolutegravir-selected HIV-1 containing the N155H and R263K resistance substitutions does not acquire additional compensatory mutations under drug pressure that lead to higher-level resistance and increased replicative capacity. J Virol 2015;89: 10482–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Anstett K,Mesplede T,Oliveira M. et al. Dolutegravir resistance mutation R263K cannot coexist in combination with many classical integrase inhibitor resistance substitutions.J Virol 2015;89: 4681–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anstett K,Cutillas V,Fusco R. et al. Polymorphic substitution E157Q in HIV-1 integrase increases R263K-mediated dolutegravir resistance and decreases DNA binding activity.J Antimicrob Chemother 2016;71: 2083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rhee SY,Sankaran K,Varghese V. et al. HIV-1 protease, reverse transcriptase, and integrase variation.J Virol 2016; 90:6058–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Russell RA,Moore MD,Hu WS. et al. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA and low in virion RNA. Retrovirology 2009;6: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Okada A,Iwatani Y.. APOBEC3G-mediated G-to-A hypermutation of the HIV-1 genome: the missing link in antiviral molecular mechanisms.Front Microbiol 2016;7: 2027.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Quashie PK,Mesplede T,Han YS. et al. Biochemical analysis of the role of G118R-linked dolutegravir drug resistance substitutions in HIV-1 integrase.Antimicrob Agents Chemother 2013;57: 6223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Letendre SL,Mills AM,Tashima KT. et al. ING116070: a study of the pharmacokinetics and antiviral activity of dolutegravir in cerebrospinal fluid in HIV-1-infected, antiretroviral therapy-naive subjects.Clin Infect Dis 2014; 59:1032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Laskey SB,Siliciano RF.. Quantitative evaluation of the antiretroviral efficacy of dolutegravir.JCI Insight 2016; 1:e90033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brenner BG,Wainberg MA.. Clinical benefit of dolutegravir in HIV-1 management related to the high genetic barrier to drug resistance.Virus Res 2016; doi:10.1016/j.virusres.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 80. Sax PE,DeJesus E,Crofoot G. et al. Bictegravir versus dolutegravir, each with emtricitabine and tenofovir alafenamide, for initial treatment of HIV-1 infection: a randomised, double-blind, phase 2 trial.Lancet HIV 2017; 4:e154–e160. [DOI] [PubMed] [Google Scholar]
- 81. Sagar M. HIV-1 transmission biology: selection and characteristics of infecting viruses.J Infect Dis 2010; 202 Suppl 2:S289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ronen K,Sharma A,Overbaugh J.. HIV transmission biology: translation for HIV prevention. AIDS 2015;29: 2219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Margolis DA,Boffito M.. Long-acting antiviral agents for HIV treatment. Curr Opin HIV AIDS 2015;10: 246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Whitfield T,Torkington A, van Halsema C.. Profile of cabotegravir and its potential in the treatment and prevention of HIV-1 infection: evidence to date.HIV AIDS (Auckl) 2016;8: 157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]