Abstract
Purpose
miR-877-5p has been reported as a tumor suppressor in multiple cancers. Its role in gastric cancer, however, remains unclear. Hence, the purpose of this study was to elucidate the function, and underlying molecular mechanism, of miR-877-5p in the development of gastric cancer.
Materials and Methods
We first analyzed miR-877-5p expression using the Gene Expression Omnibus (GEO) database and detected its expression in gastric cancer and gastric epithelial cells via real-time quantitative PCR (qRT-PCR). We then assessed the role of miR-877-5p in gastric cancer proliferation, apoptosis, and cell cycling. The gene targeted by miR-877-5p was predicted by bioinformatic analysis and confirmed by dual luciferase assay. Subsequently, rescue assays were carried out to validate whether the miR-877-5p effects on gastric cancer growth are dependent on the proposed target gene.
Results
miR-877-5p levels were lower in gastric cancer than in controls, based on the GEO and qRT-PCR analyses. Overexpression of miR-877-5p significantly inhibited cell growth and cell cycle progression, whereas it promoted apoptosis. Furthermore, forkhead box M1 (FOXM1) was predicted as a target of miR-877-5p, the overexpression of which diminished the suppressive effect that upregulation of miR-877-5p had on gastric cancer cells.
Conclusion
Our study results indicate that the miR-877-5p/FOXM1 axis plays an important role in gastric cancer progression, while suggesting miR-877-5p as a novel potential therapeutic target for gastric cancer.
Keywords: gastric carcinoma, microRNA-877-5p, forkhead box M1, tumor growth, cell cycle arrest, tumor suppressor function
Introduction
For many decades, digestive cancers have challenged global public health. Gastric cancer (GC), in particular, has consistently remained the most prevalent of the digestive cancers. In fact, GC incidence and mortality rates rank fourth and third, respectively, among all cancers worldwide.1 However, GC patient outcomes have gradually improved due to improved early diagnosis, surgical techniques, and chemotherapeutic drugs. In contrast, advanced GC patients often unfortunately attain only short-term benefits from these strategies. Therefore, clarity about the underlying mechanisms in GC tumorigenesis and the identity of novel potential targets for GC treatment is urgently needed.
MicroRNAs (miRNAs) are generated from introns being modified as conversed, single stranded, noncoding RNAs, and are an average length of 18–22 nucleotides. These molecules modulate target gene expression through binding to the 3′-untranslated region (UTR) of mRNA, resulting in mRNA degradation, or translational suppression.2,3 Furthermore, miRNAs have been shown to modulate the proliferation, apoptosis, and metastasis of various cancers.4 Over the past few years, increasing numbers of studies have confirmed that numerous miRNAs are dysregulated in GC, and these aberrantly expressed miRNAs exert either oncogenic promoter or tumor suppressor functions. For instance, microRNA-1284 promotes the mRNA degradation of one of its target genes, EIF4A1, to suppress the growth and metastasis of gastric carcinoma. Further, microRNA-1284 expression in GC was correlated with tumor volume and distance metastasis of patients.5 In contrast, overexpression of miR-181a was reported to activate the proliferation and migration of GC cells via caprin-1 targeting, which consequently resulted in poor prognosis for GC patients.6 Further, another study has shown that miR-877-5p inhibited the cell cycle by downregulating CDK14, thereby preventing metastasis of hepatocellular carcinoma cells.7 However, it is unclear whether miR-877-5p has the same effect and employs the same mechanism in GC as has been reported in other cancer types.
Therefore, the aim of the present study was to explore the specific function of miR-877-5p in GC following directional regulation of its expression. To this end, we analyzed data on miR-877-5p expression in GC, obtained from the Gene Expression Omnibus (GEO) public database. Additionally, cellular function was investigated, and experiments were conducted on mice, to validate the role of miR-877-5p and to explore its underlying molecular mechanism in GC. Moreover, rescue assays were performed to clarify whether this molecular mechanism directly participates in the effect of miR-877-5p in GC. These findings suggested that miR-877-5p might be a tumor-suppressive character in GC. Collectively, our results reveal a novel regulatory mechanism whereby miR-877-5p affects cell proliferation in GC, and serve to inform the development of novel GC therapies.
Materials and Methods
Human GC Dataset Extraction
Two datasets, GSE54397 and GSE61741, were downloaded from the GEO public database (http://www.ncbi.nlm.nih.gov/geo/) to analyze the levels of miR-877-5p in GC tissues and serum, respectively, from patients with GC. The dataset GSE54397 contains GC tissues and normal tissues. Another dataset, GSE61741, contains serum samples from GC patients and healthy controls.
GC Cell Lines and Culture Medium
GC cell lines (MKN-28, MKN-74, MGC-803, SGC-7901, and HGC-27), and the human gastric epithelial cell line GES-1, were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultivated in Roswell Park Memorial Institute-1640 medium (Procell, Wuhan, China) supplemented with 10% fetal bovine serum (Thermo Fisher, Waltham, MA, USA) and 1% antibiotics (100 U/mL penicillin G and 100 mg/mL streptomycin). The cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
TRIzol reagent (Thermo Fisher) was utilized to extract total RNA from cells and tissues according to the manufacturer’s protocols. Subsequently, cDNA was generated using a Reverse Transcription reagent kit (GeneCopoeia, Guangzhou, China) following the manufacturer’s instruction. qRT-PCR was carried out using a BeyoFast™ SYBR Green qPCR Mix (Beyotime, Shanghai, China), and the levels of miR-877-5p and FOXM1 were then measured on an ABI 7500 Real-Time PCR System. The primers employed for qRT-PCR were as follows: miR-877-5p sense 5′-GCCGTAGAGGAGATGGC-3′ and antisense 5′-CAGTGCGTGTCGTGGA-3′; U6 sense 5′-CTCGCTTCGGCAGCACA-3′ and antisense 5′-AACGCTTCACGAATTTGCGT-3′; FOXM1 sense 5′-CGTCGGCCACTGATTCTCAAA-3′ and antisense 5′-GGCAGGGGATCTCTTAGGTTC-3′; and GAPDH sense 5′-ACAGCCTCAAGATCATCAGC-3′ and antisense 5′-GGTCATGAGTCCTTCCACGAT-3′. The relative expressions of miR-877-5p and FOXM1 were calculated using the 2–ΔΔCt method, referencing GADPH and U6 as loading controls.
Immunoblotting Analysis
Total protein of cells and tissues was isolated using RIPA lysis buffer (Beyotime), and quantified using a BCA Protein Quantification kit (Beyotime). SDS-PAGE (Beyotime) was used to separate the denatured proteins, after which the isolated proteins were transferred onto PVDF membranes (Millipore, USA). At room temperature (approximately 20 °C), the membranes were blocked with 5% skimmed milk for 1 h, and were then incubated overnight at 4 °C with primary antibodies against FOXM1 (catalog number 13,147-1-AP; Proteintech, Wuhan, China) diluted at 1:2000, and GAPDH (60004-1-Ig; Proteintech) diluted 1:6000. All membrane bands were then labeled by HRP-labeled secondary antibody (Cell Signaling Technology, USA, diluted 1:10,000) for 1 h at approximately 20 °C, after washing with TBST. The protein bands were imaged by adding ECL reagent (Beyotime). GAPDH was employed as the internal reference.
Cell Transfection
The GC cell lines HGC-27 and MKN-28, which showed the lowest expression levels of miR-877-5p in this study, were selected for transfection to regulate miR-877-5p expression. A series of miR-877-5p mimic (miR-mimic), miR-877-5p negative control (miR-NC), pLVX-puro-FOXM1 (FOXM1), empty vector-FOXM1 (Vector), pLVX-puro-miR-877-5p (LV-miR-877-5p), and pLVX-puro-miR-NC (LV-miR-NC) was synthesized by GenePharma (Shanghai, China). Their sequences are provided in Table 1. Briefly, logarithmically grown cells were planted in 6-well plates at a density of 5×105 cells per well. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was performed to conduct the transfection according to the producer’s instruction. The cells were harvested following 48 h of transfection, for further analysis.
Table 1.
Oligonucleotide Sequences Used for Cell Transfections
| Gene Primer | Sequence (5ʹ-3ʹ) |
|---|---|
| FOXM1 Forward Primer | ctaccggactcagatctcgagatgaaaactagcccccgtcg |
| FOXM1 Reverse Primer | gtaccgtcgactgcagaattcCTACTGTAGCTCAGGAATAAACTGGG |
| miR-877-5p Forward Primer | ctaccggactcagatctcgagATGAGACTCTTGGCTCTTGAGGC |
| miR-877-5p Reverse Primer | gtaccgtcgactgcagaattcAGAGGAAAAGGGCCCCGG |
Cell Counting Kit-8 (CCK-8) Assay
Cell proliferation was detected by CCK-8 (Dojindo, Japan) assay according to manufacturer’s instructions. The transfected cells were planted in triplicate at 3×103 cells per well in 96-well plates. Next, 10 μL of CCK-8 was added to each well, followed by an additional 2 h incubation at 37 °C. Detection points were set at 0, 24, 48, and 72 h, and detection was performed using a microplate reader at 450 nm.
Flow Cytometric Detection of the Cell Cycle
To assess effects on the cell cycle, more than 3×105 transfected GC cells were collected in each tube. Next, 70% ethanol was used to fix cells at 4 °C overnight, after which 5 μL of propidium iodide (PI, Beyotime) was used to dye the fixed cells at room temperature for 30 min, in the dark. Flow cytometry (BD Biosciences, USA) was performed to determine the DNA content in each cell cycle stage. Data were analyzed using FlowJo v. 10 software.
Flow Cytometric Detection of Apoptosis
The Apoptosis Detection Kit (Keygen, Nanjing, China) was used to detect apoptotic cells, using Annexin V-FITC and PI double staining. The cells, including suspension cells, were collected and rinsed with cold PBS. The cell supernatant was carefully discarded, and cells were stained with 200 μL Annexin V-FITC mixture (195 μL binding and 5 μL Annexin V-FITC). Each tube then had 10 μL PI added, and the cells were stained at 4 °C in the dark for 10 min. The apoptotic cells, which were defined as those positive for Annexin V-FITC staining, were examined using flow cytometry (BD Biosciences).
Dual Luciferase Reporter Assay
FOXM1 was predicted as a potential target of miR-877-5p, using TargetScan (https://www.targetscan.org), an online miRNA target prediction site program. The miR-877-5p binding site on this gene was identified. The prediction was confirmed by a dual luciferase assay. The 3′-UTR of FOXM1, with or without the miR-877-5p predictive binding site, was amplified by PCR and cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA). These fragments were defined as wild-type (WT)-FOXM1 and mutant type (MUT)-FOXM1, respectively. The miR-mimic or miR-NC, along with WT-FOXM1 or MUT-FOXM1 luciferase reporter vectors, were co-transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen). After 48 h co-transfection, firefly luciferase activity was determined with the dual luciferase assay kit (Beyotime). Ranilla luciferase activity was used as a control to calculate the relative luciferase activity.
In vivo Proliferation Assay
A total of 12 female BALB/c nude mice (5-week-old) were purchased from the Vital River company (Beijing, China) and housed at the Laboratory Animal Center of Guangxi Medical University (Nanning, China). These in vivo experiments were approved by the Animal Ethics Committee of Guangxi Medical University, and followed the Laboratory Animal-Guideline for Ethical Review of Animal Welfare of the People’s Republic of China. The mice were randomly assigned into two groups and implanted subcutaneously in the left anterior flank with MKN-28 cells (2×106/mouse) transfected with LV-miR-NC or LV-miR-877-5p, to establish a xenograft model. The length and width of visible xenografts were measured using a Vernier caliper every 4 d. Tumor volume was presented as length × width2 × 0.5 cm3. The mice were killed by cervical detachment after 23 d of GC cell implantation. Tumors were then isolated, weighed, and collected to determine miR-877-5p and FOXM1 expression levels by Western blot and qRT-PCR.
Statistical Analysis
The experiments were independently repeated thrice. Results are expressed as means ± standard deviation (SD). Statistical analyses were conducted using SPSS v. 23 and GraphPad Prism v. 7. Student’s t-tests were used to compare differences between the experimental and control groups. Multiple experimental groups were analyzed by one-way ANOVA to obtain statistical differences. P values ˂ 0.05 were considered statistically significant.
Results
miR-877-5p Is Downregulated in GC
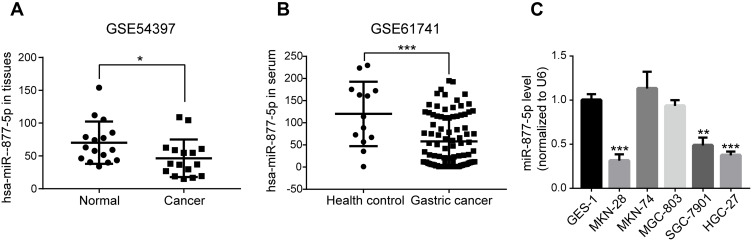
Based on the two human GC data from GEO database, the results indicated that miR-877-5p level was much lower both in GC tissues (GSE54397, Figure 1A) and serum (GSE61741, Figure 1B) than in normal tissues and healthy controls. Subsequently, qRT-PCR was carried out to screen miR-877-5p expression levels among the GC cells (SGC7-901, MKN-74, MKN-28, HGC-27, and MGC-803) and GES-1 cells. Consistent with the bioinformatics analysis results, the miR-877-5p level was lower in SGC-7901, MKN-28, and HGC-27 cells than in GES-1; however, miR-877-5p expression in MKN-74 and MGC-803 was not significantly different from that in GES-1 (Figure 1C). Among these cell lines, the lowest expression levels of miR-877-5p were found in HGC-27 and MKN-28.
Figure 1.
Down-regulation of miR-877-5p in gastric cancer (GC). Two GEO datasets were selected for validating the tendency of miR-877-5p in clinical GC samples. (A) The declined expression levels of miR-877-5p in GC and normal tissue samples were compared using the GSE54397 dataset from the GEO database. (B) Drop of miR-877-5p expression levels in the serum from patients with GC or healthy controls were compared using GSE61741. (C) The expression of miR-877-5p in GC cells (MKN-28, MKN-74, MGC-803, SGC-7901, and HGC-27) and GES-1 cells was examined by qRT-PCR, wherein expression level in MKN-28 and HGC-27 remained the most down-regulated. U6: internal control. *P < 0.05, **P < 0.01 and ***P < 0.005.
Abbreviations: GC, Gastric cancer; GEO, Gene Expression Omnibus; UTR, untranslated region; NC, Negative control.
miR-877-5p Inhibits Proliferation, Induces Apoptosis, and Arrests Cell Cycling in GC
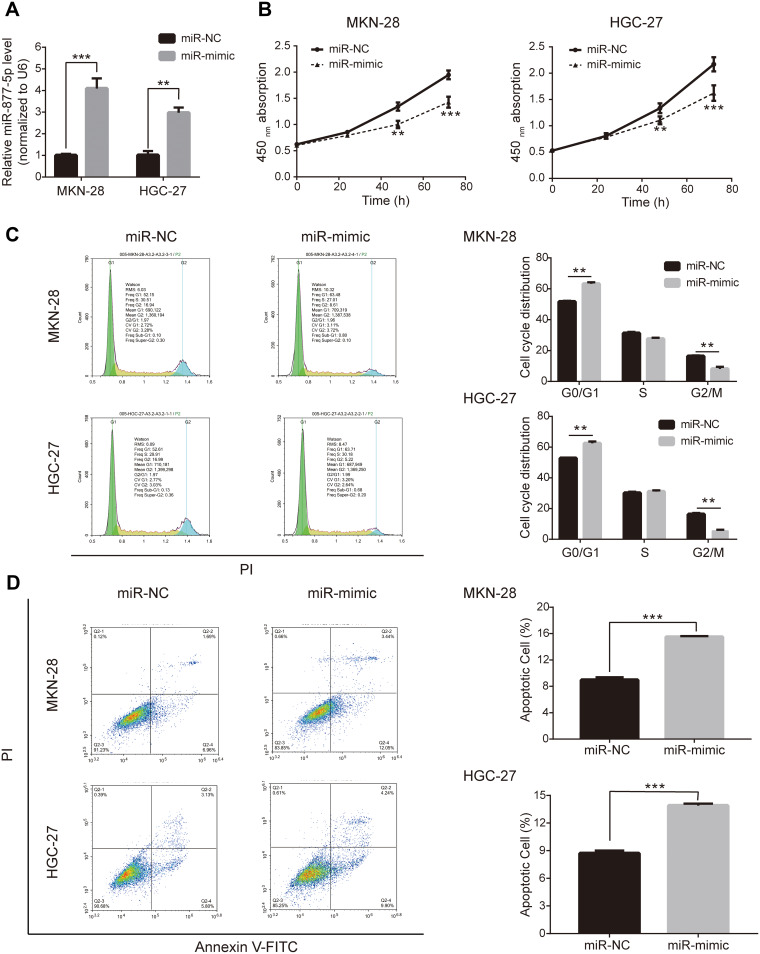
miR-877-5p mimic and miR-NC were synthesized and transfected into HGC-27 and MKN-28 cells to further investigate the effects of miR-877-5p in GC. qRT-PCR results confirmed that the miR-877-5p mimic successfully elevated miR-877-5p expression in two of the cell lines (Figure 2A). The results of the CCK-8 assay indicate that proliferative ability was reduced following upregulation of miR-877-5p (Figure 2B). Consistent with this, flow cytometry analysis revealed that upregulation of miR-877-5p arrested cells in the G0/G1 phase and inhibited their transition to the G2/M phase, whereas this did not occur in the miR-NC-expressing cells (Figure 2C). Further, flow cytometry confirmed that upregulation of miR-877-5p suppressed proliferation and induced apoptosis of GC cells (Figure 2D).
Figure 2.
Overexpression of miR-877-5p suppresses proliferation of gastric cancer (GC) cells. (A) Up-regulating mRNA expression of miR-877-5p after transfecting miR-877-5p mimic in GC cells MKN-28 and HGC-27. (B) The growth curve based on results of CCK-8 assay indicated the cell viability was inhibited when miR-877-5p was over-expressed. (C) Flow cytometry illustrated G0/G1 of cell cycle were arrested in MKN-28 and HGC-27 when over-expressing miR-877-5p. (D) The promoting apoptosis of GC cells with over-expression of miR-877-5p, being assessed by flow cytometry. **P < 0.01 and ***P < 0.005.
FOXM1 Is a Target of miR-877-5p in GC
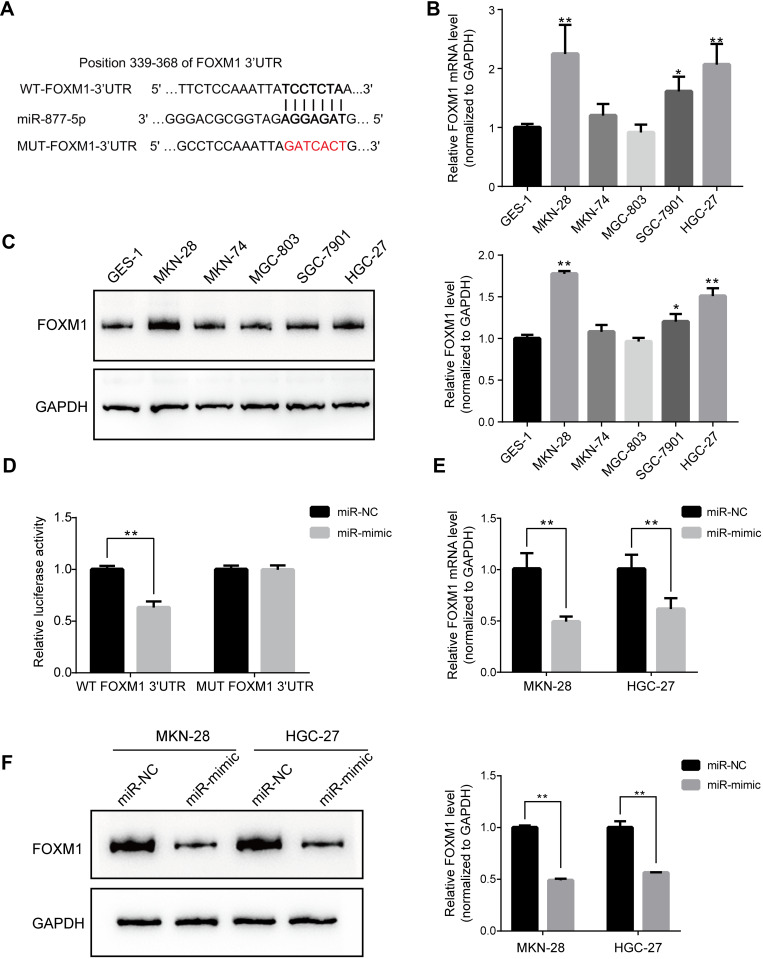
TargetScan analysis revealed that FOXM1 was predicted to be one of the direct targets of miR-877-5p (Figure 3A). Therefore, we sought to elucidate the mRNA and protein expression levels of FOXM1 in GC cells. There was greater upregulation of FOXM1 expression in GC cells than in GES-1 cells (Figure 3B and C). Further, a dual luciferase reporter assay was conducted to observe the interaction between miR-877-5p and FOXM1 mRNA. Relative luciferase activity was substantially lower in HEK293T cells that were co-transfected with miR-mimic and WT- FOXM1, relative to the control. However, miR-mimic did not significantly affect the relative luciferase activity of MUT- FOXM1, relative to miR-NC (Figure 3D). Further, qRT-PCR and immunoblotting were employed to determine whether miR-877-5p regulates FOXM1 expression in GC cell lines. Compared with the miR-NC group, FOXM1 mRNA expression was lower in GC cells transfected with miR-mimic (Figure 3E). A similar result was observed via immunoblotting (Figure 3F).
Figure 3.
FOXM1 is negatively modulated by miR-877-5p in gastric cancer (GC) cells. (A) TargetScan analysis showed the predicting binding site of FOXM1 in the sequence located at 3′-UTR for miR-877-5p. (B) The mRNA expression level of FOXM1 in various GC and GES-1 cells wherein that expression in MKN-28 and HGC-27 ranked top two highest level. GAPDH as an internal reference. (C) Protein bands showed the FOXM1 expression among GC and GES-1 cells and fold change was normalized using GAPDH as loading reference, indicating by bar chart. (D) Decrease activity of dual luciferase assay clarified that miR-877-5p bind to the 3′-UTR of FOXM1 in HEK239T cells while the mutant vector showed no changes. (E) Downtrend of FOXM1 mRNA expression in GC cells after being treated with miR-mimic towards that with miR-NC. (F) Western blotting results showed FOXM1 protein expression in GC cells with over-expressed miR-877-5p and the comparison of that with NC control. *P < 0.05 and **P < 0.01.
Overexpression of miR-877-5p Suppresses GC Growth in vivo
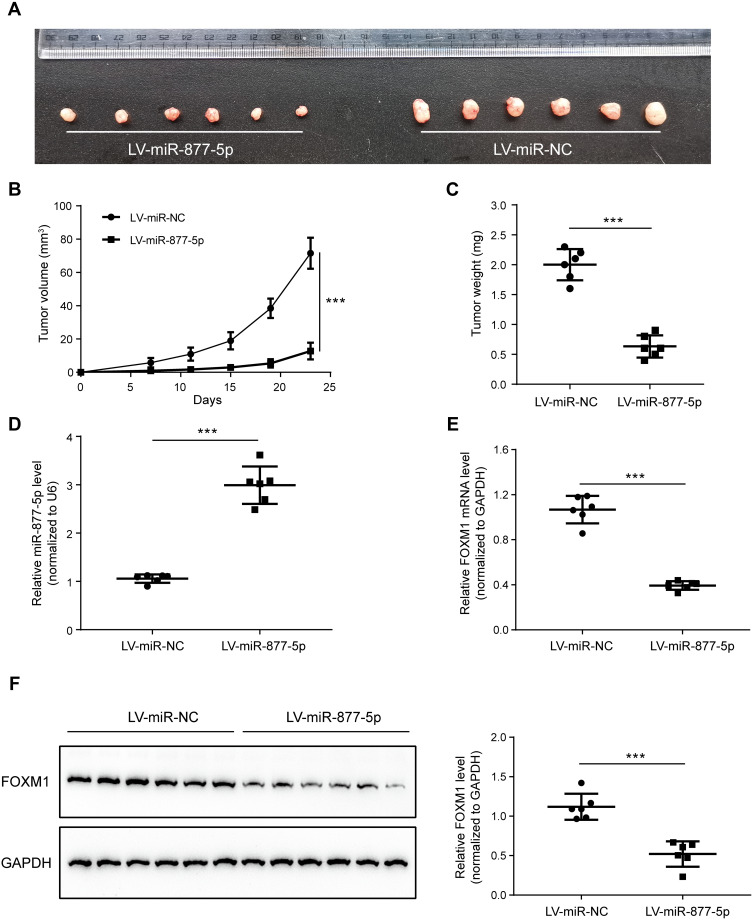
To construct a xenograft tumor model, MKN-28 cells steadily expressing miR-877-5p or miR-NC were injected subcutaneously into nude mice, and the xenograft volume was calculated every 4 d, to determine whether miR-877-5p inhibits GC proliferation in vivo. After 23 d of subcutaneous injection, all mice in both groups developed xenograft tumors at the injection site. The integrated tumor growth curve demonstrates simultaneous overexpression of miR-877-5p and reduced tumor growth, relative to the LV-miR-NC group (Figure 4A–C). In addition, qRT-PCR analysis of xenograft tumors confirmed that miR-877-5p levels were significantly elevated in the LV-miR-877-5p group relative to those in the LV-miR-NC group (Figure 4D). Moreover, FOXM1 expression was lower in the LV-miR-877-5p group than in the LV-miR-NC group (Figure 4E and F).
Figure 4.
Overexpression of miR-877-5p suppresses proliferation of gastric cancer (GC) cells in vivo, in a mouse model. (A) Individual xenograft tumors were sampled from miR-877-5p over-expressing group and the counterparts. (B) A tumor growth curve was drawn based on the tumor volume measurement every 4 d. (C) Comparison of the tumor weights which was presenting as average±SD in two groups. (D) Expression levels of miR-877-5p in the xenograft tumors from the two groups averaging from each individual. (E, F) Expression levels of tumor mRNA and protein of FOXM1 from the two groups, respectively. Both mRNA and protein level of FOXM1 was dropped when miR-877-5p was over-expressed. ***P < 0.005.
miR-877-5p Suppresses Proliferation and the Cell Cycle by Targeting FOXM1 in GC
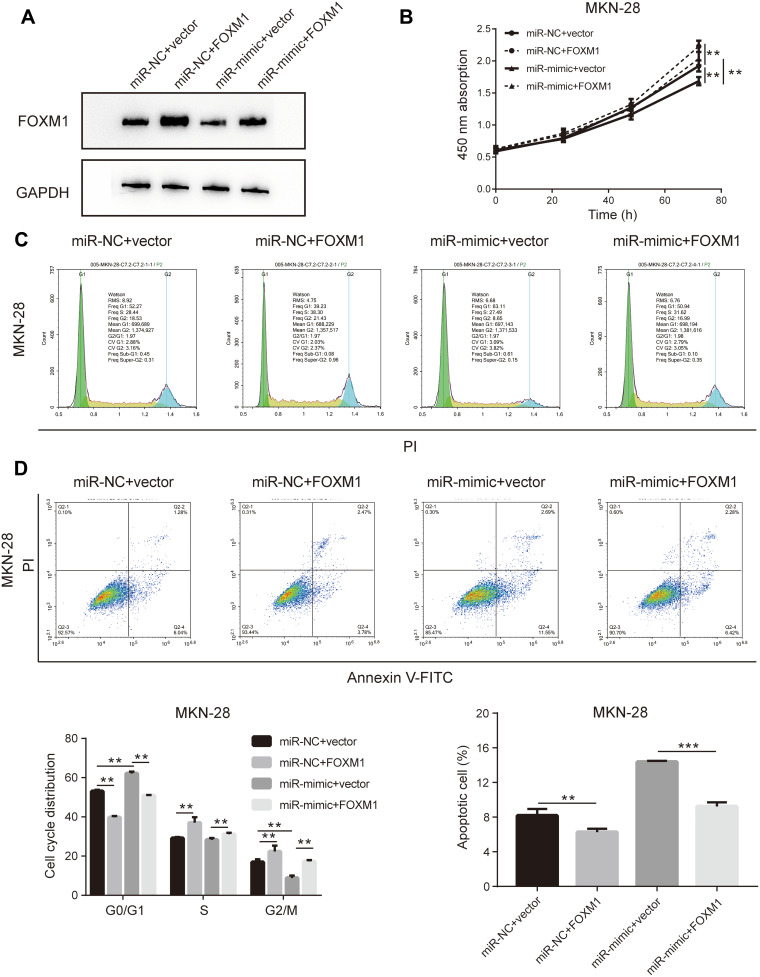
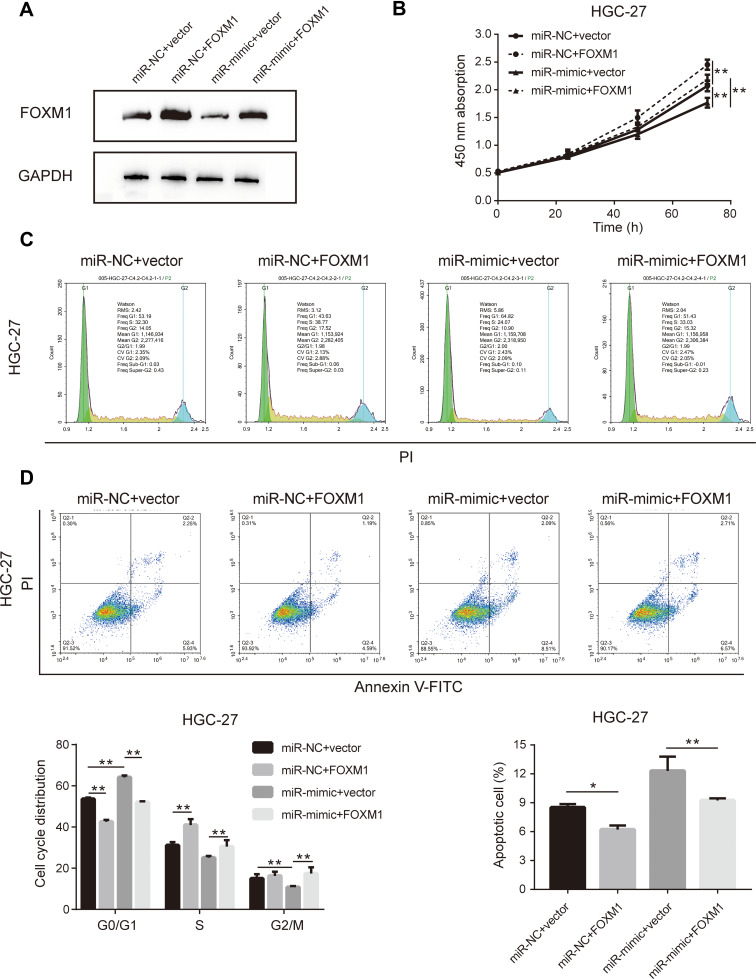
The initial findings of this study confirmed that overexpression of miR-877-5p exerted an anti-tumor effect in GC, and that FOXM1 expression was suppressed by miR-877-5p. A FOXM1-expressing plasmid, which contained only the coding sequence without the miR-877-5p binding sequence, was constructed, and rescue assays were performed to explore whether FOXM1 mediates the tumor suppressor role of miR-877-5p in GC further. The FOXM1 or Vector plasmid and miR-mimic or miR-NC were then co-transfected into HGC-27 and MKN-28 cells. FOXM1 expression levels was decreased in miR-mimic while restored by FOXM1 plasmid transfection (Figures 5A and 6A). The CCK-8 assay showed that FOXM1 restoration abolished the anti-tumor role of miR-877-5p in MKN-28 (Figure 5B) and HGC-27 (Figure 6B) cell proliferation. Moreover, FOXM1 restoration abrogated G0/G1 phase arrest and apoptosis induced by miR-877-5p overexpression in MKN-28 (Figure 5C and D) and HGC-27 cells (Figure 6C and D).
Figure 5.
Upregulation of FOXM1 abrogates the anti-tumor effect of miR-877-5p in MKN-28 cells. (A) FOXM1 expression level was presented as protein bands when administrating co-transfection FOXM1 plasmid either with combination of miR-877-5p or vector counterparts. (B) Cell viability of each group was determined using CCK-8 assay wherein the most up-trend curve indicating in over-expressing FOXM1 groups. (C) The cell cycle presenting as DNA content was assessed by flow cytometry in different co-transfection group of MKN-28. (D) Apoptosis rate of MKN-28 after transduction miR-877-5p or vector with FOXM1 plasmid, based on flow cytometry detection. Bar charts on the bottom displayed the percentage of cells in each phase (left) and percentage of cells underwent apoptosis (right), respectively. **P < 0.01 and ***P < 0.005.
Figure 6.
Upregulation of FOXM1 eliminated the anti-tumor effect of miR-877-5p in HGC-27 cells. (A) Immunoblotting was performed to detect FOXM1 level in HGC-27 when treating with FOXM1 plasmid either with combination of miR-877-5p or vector counterparts. (B) Cell proliferation curve was drawn according to the results of CCK-8 assay in each time points among different groups. (C) The distribution of experimental groups in each cell cycle was measured using flow cytometry. (D) Apoptotic cell proportion when transfected miR-877-5p or vector with FOXM1 plasmid in HGC-27 cells, applying flow cytometry detection. Bar charts located on the bottom presented the details percentage of cells in each cell cycle phase (left) and apoptotic cells rate (right) when different combination of co-transfected items were conducted. *P < 0.05 and **P < 0.01.
Discussion
Over the past few decades, dysregulation of a single miRNA molecule or a cluster of miRNAs has been found to have significant consequences for the initiation and development of various cancers, including gastric cancer.8,9 Therefore, exploring the aberrant expression of miRNAs and their functional role in GC may lead to new GC diagnostics and therapeutics. Previous studies have shown that miR-877-5p may serve as a tumor suppressor factor in cervical cancer10 and glioblastoma.11 The miR-877 family has also been shown to participate in cancer progression. Tumor suppressor miR-877-3p inhibited GC cell invasion and metastasis by targeting VEGFA.12 Similarly, miR-877 repressed colorectal cancer growth and metastasis by binding the 3′-UTR of MTDH.13 In contrast, overexpression of miR-877-3p can eliminate the inhibitory effect of long noncoding RNA UBE2R2-AS1 in glioma.14 This evidence suggests that miR-877-3p may be an oncogene in glioma. These studies suggest that miR-877 family members may serve as diverse molecular regulators in different cancers, and that they may be useful indicators and therapeutic targets for tumors. Our objective was therefore to investigate the previously uncharacterized molecular mechanisms whereby miR-877-5p acts in GC development.
Our findings reveal that miR-877-5p expression was significantly reduced in the tissues and serum of GC patients, based on data from the GEO database. Further, the similar decline trend was found in GC cells based on analysis of qRT-PCR. The results indicate that miR-877-5p may serve as a tumor suppressor in GC. In subsequent in vitro experiments, we observed that miR-877-5p upregulation led to cell cycle arrest of GC and triggered apoptotic processes, which ultimately inhibited proliferation. Furthermore, overexpression of miR-877-5p repressed xenograft tumor growth in vivo. Together, the results of the functional experiments show that miR-877-5p participates in the development of GC. However, they do not clarify how miR-877-5p elicits its anti-cancer effect in GC. Therefore, next investigated the mechanisms whereby miR-877-5p acts, and focused on its underlying mechanism.
It is well known that miRNAs modulate the expression of target genes via complementary binding of the 3′-UTR of the target mRNA, and that they further regulate biological processes.15,16 Bioinformatic analysis predicted that FOXM1 is a target of miR-877-5p, thereby providing insight into the potential molecular mechanism underlying miR-877-5p inhibition of GC cell proliferation.
FOXM1, a well-known mammalian transcription factor, is a forkhead box (FOX) protein superfamily member that contains a classical conserved winged-helix DNA-binding domain.17 As a proliferation-related transcription factor, FOXM1 is detected primarily in highly proliferative cells, such as progenitor and tumor cells.18 Accumulating evidence indicates that there is elevated expression of FOXM1 in many types of solid tumors, such as hepatocellular carcinoma,19 prostate cancer,20 non-small cell lung cancer,21 colorectal cancer,22 breast cancer,23 nasopharyngeal carcinoma,24 and GC,25 and these studies indicate that it acts as an oncogene in terms of proliferation, chemo-resistance, apoptosis, autophagy, migration, and epithelial-mesenchymal transition (EMT). These studies suggest that FOXM1 can act as an indicator for cancer prognosis and further play a vital role in tumorigenesis and development of cancer. FOXM1, as a transcription factor, not only regulates its downstream target genes but also acts as the target for epigenetic regulation. For instance, miR-214 suppressed the progression of hepatocellular carcinoma through direct targeting of FOXM1.26 Further, miR-6868-5p repressed angiogenesis by the same mechanism in colorectal cancer.27 In our study, FOXM1 expression was confirmed to be elevated in SGC-7901, MKN-28, and HGC-27 cells relative to that in GES-1. When miR-877-5p expression was upregulated by miR-mimic, the expression of FOXM1 was significantly suppressed. Moreover, the luciferase reporter assay results further confirmed a connection between miR-877-5p and the 3′-UTR of FOXM1. These results indicate that miR-877-5p was able to directly degrade FOXM1 mRNA to inhibit its expression. Even so, it remains unclear whether FOXM1 mediates the tumor suppressive effect of miR-877-5p in GC proliferation. Thus, we performed rescue experiments to further explore this question. As expected, FOXM1 restoration markedly eliminated the inhibitory effects of miR-877-5p on GC growth, partly confirming that FOXM1 may be the potential site at which miR-877-5p exerts its tumor suppressor effect. Notably, in the cell cycle assay, overexpression of miR-877-5p did not affect cell distribution in the S phase but did reduce cell distribution in the G2/M phase. It is possible that FOXM1 stimulates cell distribution not only during DNA replication in the S phase but also during the G2/M transition.28 This would provide a partial explanation for the observed cell cycle arrest induced by miR-877-5p.
Conclusion
We studied the effects of miR-877-5p, a known tumor suppressor, on GC growth. miR-877-5p expression was downregulated in GC. Further, considering that upregulating miR-877-5p effectively arrested the cell cycle in the G0/G1 phase and enhanced GC apoptosis, we conclude that miR-877-5p inhibited tumor proliferation. Further, we verified that one of the downstream target genes, FOXM1, which may be directly bound by miR-877-5p, mediates the inhibitory effects of miR-877-5p in GC. This study reveals that miR-877-5p is a potential tumor inhibitor in GC, due to its suppression of GC cell proliferation. The inhibitory effect of miR-877-5p may also prove beneficial as a therapeutic target. Based on the results of this study, our future research will focus on the clinical significance of miR-877-5p expression in the tumor tissues and serum of GC patients, providing both an experimental and theoretical basis for whether miR-877-5p can be used as a non-invasive tumor diagnostic and prognostic indicator. Further, we aim to elucidate the mechanism of the observed downregulation of miR-877-5p in GC. To this end, we will further explore the upstream regulatory mechanism of miR-877-5p, with an ultimate goal of fully elucidating the role of miR-877-5p in GC.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (NO. 81660511). The funder had no role in any of the stages from study design to submission of the paper for publication.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 4.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707 [DOI] [PubMed] [Google Scholar]
- 5.Wei W, Cao W, Zhan Z, Yan L, Xie Y, Xiao Q. MiR-1284 suppresses gastric cancer progression by targeting EIF4A1. Onco Targets Ther. 2019;12:3965–3976. doi: 10.2147/OTT.S191015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Q, Chen Y, Sun D, et al. MicroRNA-181a functions as an oncogene in gastric cancer by targeting caprin-1. Front Pharmacol. 2018;9:1565. doi: 10.3389/fphar.2018.01565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan TH, Qiu C, Sun J, Li WH. MiR-877-5p suppresses cell growth, migration and invasion by targeting cyclin dependent kinase 14 and predicts prognosis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:3038–3046. doi: 10.26355/eurrev_201805_15061 [DOI] [PubMed] [Google Scholar]
- 8.Jiang M, Shi L, Yang C, et al. miR-1254 inhibits cell proliferation, migration, and invasion by downregulating Smurf1 in gastric cancer. Cell Death Dis. 2019;10:32. doi: 10.1038/s41419-018-1262-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Z, Zhong M, Wang Y, et al. miR-381 and miR-489 suppress cell proliferation and invasion by targeting CUL4B via the Wnt/β -catenin pathway in gastric cancer. Int J Oncol. 2019;54:733–743. doi: 10.3892/ijo.2018.4646 [DOI] [PubMed] [Google Scholar]
- 10.Liang J, Zhang S, Wang W, et al. Long non-coding RNA DSCAM-AS1 contributes to the tumorigenesis of cervical cancer by targeting miR-877-5p/ATXN7L3 axis. Biosci Rep. 2020;40:BSR20192061. doi: 10.1042/BSR20193470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie H, Shi S, Chen Q, Chen Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathol Res Pract. 2019;215:152476. doi: 10.1016/j.prp.2019.152476 [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Wang YH, Yoon C, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038 [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Li C, Cao L, et al. microRNA-877 inhibits malignant progression of colorectal cancer by directly targeting MTDH and regulating the PTEN/Akt pathway. Cancer Manag Res. 2019;11:2769–2781. doi: 10.2147/CMAR.S194073 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Xu W, Hu G-Q, Da Costa C, et al. Long noncoding RNA UBE2R2-AS1 promotes glioma cell apoptosis via targeting the miR-877-3p/TLR4 axis. Onco Targets Ther. 2019;12:3467–3480. doi: 10.2147/OTT.S201732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 16.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 17.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/forkhead DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0 [DOI] [PubMed] [Google Scholar]
- 18.Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: repurposing an oncogene as a biomarker. Semin Cancer Biol. 2017;52:74–84. doi: 10.1016/j.semcancer.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 19.Hu G, Yan Z, Zhang C, et al. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res. 2019;38(1):188. doi: 10.1186/s13046-019-1202-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin JZ, Wang WW, Hu TT, et al. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett. 2020;469:481–489. doi: 10.1016/j.canlet.2019.11.014 [DOI] [PubMed] [Google Scholar]
- 21.Hsieh NT, Huang CY, Li CC, Wang IC, Lee MF. MED28 and forkhead box M1 (FOXM1) mediate matrix metalloproteinase2 (MMP2)-dependent cellular migration in human nonsmall cell lung cancer (NSCLC) cells. J Cell Physiol. 2019;234:11265–11275. doi: 10.1002/jcp.27784 [DOI] [PubMed] [Google Scholar]
- 22.Varghese V, Magnani L, Harada-Shoji N, et al. FOXM1 modulates 5-FU resistance in colorectal cancer through regulating TYMS expression. Sci Rep. 2019;9:1505. doi: 10.1038/s41598-018-38017-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park YY, Jung SY, Jennings NB, et al. Corrigendum: FOXM1 mediates dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2019;40:936. doi: 10.1093/carcin/bgz062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo W, Gao F, Li S, Liu L. FoxM1 promotes cell proliferation, invasion, and stem cell properties in nasopharyngeal carcinoma. Front Oncol. 2018;8:483. doi: 10.3389/fonc.2018.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu W, Cui X, Wan Z, Yu Y, Liu X, Jin L. Silencing forkhead box M1 promotes apoptosis and autophagy through SIRT7/mTOR/IGF2 pathway in gastric cancer cells. J Cell Biochem. 2018;119:9090–9098. doi: 10.1002/jcb.27168 [DOI] [PubMed] [Google Scholar]
- 26.Tian C, Wu H, Li C, et al. Downregulation of FoxM1 by miR-214 inhibits proliferation and migration in hepatocellular carcinoma. Gene Ther. 2018;25:312–319. doi: 10.1038/s41434-018-0029-4 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Wu M, Lei Z, et al. Dysregulation of miR-6868-5p/FOXM1 circuit contributes to colorectal cancer angiogenesis. J Exp Clin Cancer Res. 2018;37:292. doi: 10.1186/s13046-018-0970-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laoukili J, Kooistra MR, Brás A, et al. FOXM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217 [DOI] [PubMed] [Google Scholar]