Abstract
Background
Coinfection with avian leukosis virus subgroup J (ALV-J) and reticuloendotheliosis virus (REV) is common in chickens, and the molecular mechanism of the synergistic pathogenic effects of the coinfection is not clear. Exosomes have been identified as new players in the pathogenesis of retroviruses. The different functions of exosomes depend on their cargo components.
Objectives
The aim of this study was to investigate the function of co-regulation differentially expressed proteins in exosomes on coinfection of ALV-J and REV.
Methods
Here, viral replication in CEF cells infected with ALV-J, REV or both was detected by immunofluorescence microscopy. Then, we analyzed the exosomes isolated from supernatants of chicken embryo fibroblast (CEF) cells single infected and coinfected with ALV-J and REV by mass spectrometry. KEGG pathway enrichment analyzed the co-regulation differentially expressed proteins in exosomes. Next, we silenced and overexpressed tripartite motif containing 62 (TRIM62) to evaluate the effects of TRIM62 on viral replication and the expression levels of NCK-association proteins 1 (NCKAP1) and actin-related 2/3 complex subunit 5 (ARPC5) determined by quantitative reverse transcription polymerase chain reaction.
Results
The results showed that coinfection of ALV-J and REV promoted the replication of each other. Thirty proteins, including TRIM62, NCK-association proteins 1 (NCKAP1, also known as Nap125), and Arp2/3-5, ARPC5, were identified. NCKAP1 and ARPC5 were involved in the actin cytoskeleton pathway. TRIM62 negatively regulated viral replication and that the inhibition of REV was more significant than that on ALV-J in CEF cells coinfected with TRIM62. In addition, TRIM62 decreased the expression of NCKAP1 and increased the expression of ARPC5 in coinfected CEF cells.
Conclusions
Collectively, our results indicated that coinfection with ALV-J and REV competitively promoted each other's replication, the actin cytoskeleton played an important role in the coinfection mechanism, and TRIM62 regulated the actin cytoskeleton.
Keywords: Avian leukosis virus subgroup J, reticuloendotheliosis virus, coinfection, TRIM62, actin cytoskeleton
INTRODUCTION
Avian leukosis virus subgroup J (ALV-J) and reticuloendotheliosis virus (REV) are 2 oncogenic avian retroviruses that mainly cause myelocytomas and reticuloendotheliosis, respectively [1,2,3]. Coinfection with ALV-J and REV is common in chickens and induces more serious pathogenic effects, such as immunosuppression growth retardation, accelerated neoplasia progression, and causes serious losses to the poultry industry [4,5,6]. The molecular mechanism of the synergistic effects of this coinfection is not clear.
Exosomes are endosome-derived nanovesicles involved in intercellular crosstalk through the transfer of proteins and genetic material. It has been reported that exosomes play an important role in the immune suppression associated with the identified gene expression and miRNA signatures in exosomes [7]. Several recent publications have identified exosomes as new players in the pathogenesis of human immunodeficiency virus (HIV) infection [8]. Our previous study also demonstrated that exosomes play an important role in ALV-J infection [9]. ALV-J and REV synergistically increased the accumulation of exosomal miRNAs, which shed light on the synergistic molecular mechanism of ALV-J and REV [10].
In addition to miRNAs, exosomes carry proteins, but the roles of exosomal proteins in ALV-J and REV coinfection are unclear. Proteins do not function independently in organisms. Instead, different proteins coordinate to perform a series of biochemical or biological functions. In the present study, to further reveal the roles of proteins in coinfection with ALV-J and REV, exosomal proteins were extracted from chicken embryo fibroblast (CEF) cells infected with ALV-J, REV or both at the optimal coinfection time for analysis by mass spectrometry (MS). The key proteins obtained from MS were validated to be present in exosomes by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Furthermore, we evaluated the protein-protein interactions related to the synergistic effects of ALV-J and REV coinfection.
MATERIALS AND METHODS
Cells culture and viral infection
CEF cells were maintained in DMEM supplemented with 10% fetal bovine serum, 0.1 mg/mL streptomycin and 100 U/mL penicillin at 37°C in a 5% CO2 atmosphere. Cells were used in subsequent assays when they were in the logarithmic growth phase. The stock SNV strains of ALV-J (Chinese strain NX0101) and REV (Chinese strain JX0927) were maintained in our laboratory. We performed a single infection of ALV-J or REV or a coinfection of ALV-J and REV at a multiplicity of infection of 0.1.
Immunofluorescence microscopy
CEF cells were collected at different times after infection (24 h, 48 h, 72 h and 96 h) and then fixed with cold acetone:ethanol solution (3:2). The monoclonal antibodies (prepared by our lab) against gp85 of ALV-J and env of REV were used as primary antibodies. A FITC-labeled goat anti-mouse IgG antibody (or Cy3-labeled goat anti-mouse IgG) was used as a secondary antibody. The infected cells were stained with DAPI staining solution and observed by immunofluorescence microscopy (Leica SP8; Leica, Germany).
Isolation and identification of exosomes
At 72 h after infection, CEF cell supernatants were collected, and debris was removed. Exosomes were separated and purified from the supernatants according to the manufacturer's protocol for total exosomes isolation (from cell culture media) kits (Invitrogen, USA). The exosomal proteins concentration was measured with BCA Protein Assay Kit (Thermo Fisher Scientific, USA). The morphology of isolated exosomes was observed under transmission electron microscopy (TEM) [9]. The isolated exosomes were examined for exosomal and contamination markers by western blot (WB).
Proteomic analysis of exosomes
In this study, a high-resolution mass spectrometer Q Exactive (Thermo Fisher Scientific) was used for quantitative proteomic analysis. MS analysis was performed according to a previously reported protocol [9] for the detection of polypeptide sequence of proteins in isolated exosomes. Proteins were identified using Protein Prospector [11] against the Gallus chicken database, ALV database, and REV database. The proteins with a fold change of expression > 1.2 (up/downregulation) and a p value < 0.05 were regarded as differentially expressed proteins. Quantitative sequence information for the proteins was extracted from the UniProtKB database (version number 201602).
Bioinformatics analysis of differentially expressed proteins
In a previous study, our laboratory identified 5 upregulated miRNAs (miRNA-184-3p, miRNA-146a-3p, miRNA-146a-5p, miRNA-3538 and miRNA-155) in exosomes from CEF cells coinfected with ALV-J and REV [10]. According to the obtained miRNA sequences and how they aligned to Gallus and virus genomes, target gene prediction was conducted using miRanda [12].
To investigate the biological function of the differentially expressed proteins and miRNA target genes, KEGG pathway enrichment analyses were performed using the DAVID tool (https://david.ncifcrf.gov/). False discovery rate < 0.05 and p value < 0.05 were considered statistically significant. The key proteins, miRNAs, and common pathways were identified. The expression levels of key exosomal proteins were verified by Real-Time qRT-PCR.
TRIM62 plasmid and short hairpin RNA (shRNA) lentiviral vector construction
TRIM62 plays an important role in antiviral processes and we have confirmed that TRIM62 inhibit ALV-J replication [13]. TRIM62 was identified from the differentially expressed proteins. To confirm the role of TRIM62 in the coinfection of ALV-J and REV and the regulation of other identified proteins or signaling pathways, overexpression and knockdown of TRIM62 were performed. The full-length chicken TRIM62 (XM_015297235.2) was cloned into the lentiviral vector GV492 (JikaiGene Technology, Inc., China). The lentiviral empty vector was used as a control. ShRNA targeting TRIM62 (shTRIM62) for knockdown (sequence: GCAGTACACC ATCTG GAAGTC) and one nonspecific scramble shRNA as a negative control were cloned into the lentiviral vector GV493 (JikaiGene Technology, Inc.). Then, the pGV492-TRIM62/pGV493-shTRIM62 plasmid was cotransfected with packaging plasmids into 293T cells to produce a lentivirus. After 48 h of transfection, viral supernatants were collected and further used to infect CEF cells for transfection. All constructs were identified through sequencing.
TRIM62 and shRNA transfection
CEF cells were seeded on 6-well plates for 12 h and then transfected with TRIM62 for 12 h or shTRIM62 for 24 h using a lentiviral vector. The pTRIM62, shTRIM62 and empty vector were purchased from JikaiGene Technology, Inc. After stably expressing pTRIM62/shTRIM62, CEF cells were infected with ALV-J, REV or both. After 72 h of infection, the RNA expression levels of virus and identified proteins were detected by qRT-PCR.
qRT-PCR
The isolated exosomes and transfected CEF cells were collected. Exosomes RNA was isolated using total exosomes RNA and protein isolation kit (Invitrogen). Total cellular RNA isolation, reverse transcription to cDNA, and quantitative PCR were performed according to a previously described protocol [14,15]. Glyceraldehyde 3-phosphate dehydrogenase was used as a control for basal RNA levels. The specific primer sequences of the virus and identified proteins are described in Table 1. Three independent experiments were conducted for statistical analysis.
Table 1. Primers used to detect miRNA expression using qRT-PCR.
| Gene target | Primer sequence | Fragment size (bp) |
|---|---|---|
| ALV-J (env) | Forward: TGCGTGCGTGGTTATTATTTC | 144 |
| Reverse: AATGGTGAGGTCGCTGACTGT | ||
| REV (env) | Forward: TTGTTGAAGGCAAGCATCAG | 330 |
| Reverse: GAGGATAGCATCTGCCCTTT | ||
| TRIM62 | Forward: TACTGGGAGGTGGTGGTGTC | 246 |
| Reverse: CGTCGGCGTTGTAGAAGATG | ||
| ITGα1 | Forward: TAAGTTCATAGCGAGCGACC | 125 |
| Reverse: TCAGCACAG CCCCAT TCC | ||
| NCKAP1 | Forward: TTGTCTTTTCGGTCGTTG | 126 |
| Reverse: TGCCACCTTCATGTCAGT | ||
| ARPC5 | Forward: TGGACGAGTACGACGAGA | 254 |
| Reverse: TGAGGACCTTCAGGA | ||
| GAPDH | Forward: GAACATCATCCCAGCGTCCA | 132 |
| Reverse: GAGGATAGCATCTGCCCTTT |
qRT-PCR, quantitative reverse transcription polymerase chain reaction; ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus; TRIM62, tripartite motif containing 62; ITGα1, integrinα1; NCKAP1, NCK-association proteins 1; ARPC5, actin-related 2/3 complex subunit 5; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
WB
Equivalents of exosomes (20 μg) were separated on 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Exosomes from infected DF-1 cells with ALV-J and REV single infection or co-infection were analysis by WB with anti-glucose-regulated protein 78 (GRP78) and anti-Hsp70 primary antibodies (Bioss, China) at both 1:200.
Statistical analysis
Data were expressed as the mean ± SD of results from 3 independent replicates. The significance of variability was determined by t-test and 2-way analysis of variance using SPSS 19.0 statistical software (IBM Corp., USA). A p value less than 0.05 was considered statistically significant.
RESULTS
Coinfection with ALV-J and REV enhanced virus replication
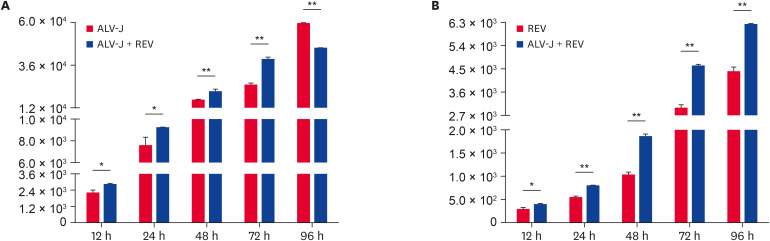
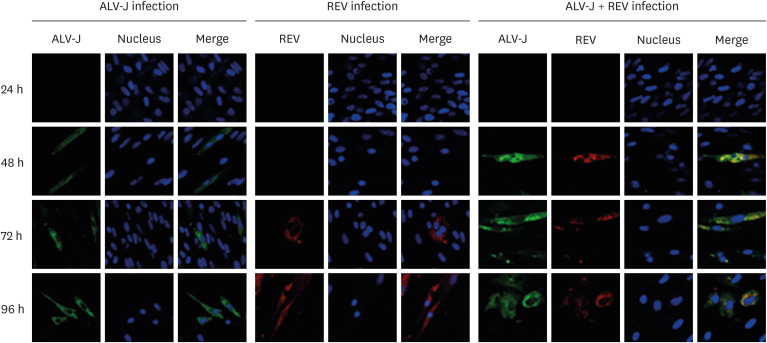
To investigate the effects of ALV-J and REV coinfection on each other's replication, we detected the transcriptional levels of virus in coinfected and single infected cells by qRT-PCR. The mRNA expression of ALV-J in co-infected cells was significantly higher than that in single infected cells before 72 h of infection (Fig. 1A, p < 0.01). However, after infected 96 h, the replication of ALV-J in co-infected cells decreased significantly. The mRNA level of REV in co-infected cells was significantly higher than that in single infected cells (Fig. 1B, p < 0.01). Furthermore, immunofluorescence microscopy was used to monitor the distribution of the fluorescence signal of the virus. As shown in Fig. 2, we found that the green fluorescence signal of ALV-J in coinfected cells was stronger than that in ALV-J only-infected cells at 48 h after infection. The red fluorescence of REV was also found at 48 h after infection in the coinfection group and earlier than that in the REV infection group (72 h). These results indicated that ALV-J and REV coinfection synergistically promoted the viral protein expression of the other virus. Our result was consistent with a previous report using qRT-PCR and WB [10].
Fig. 1. The transcriptional levels of ALV-J and REV in infected cells at different times. (A) The transcriptional level of ALV-J in co-infected and single infected cells. (B) The transcriptional level of REV in co-infected and single infected cells.
ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus.
Statistically significant changes were calculated using Kruskal-Wallis with Dunn's multiple comparison test: *p < 0.05; **p < 0.01.
Fig. 2. ALV-J and REV coinfection competitively promoted viral replication in CEF cells. ALV-J and REV staining was observed in CEF cells at 24 h, 48 h, 72 h, and 96 h post infection by immunofluorescence microscopy.
ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus; CEF, chicken embryo fibroblast.
Proteomic analysis of exosomes
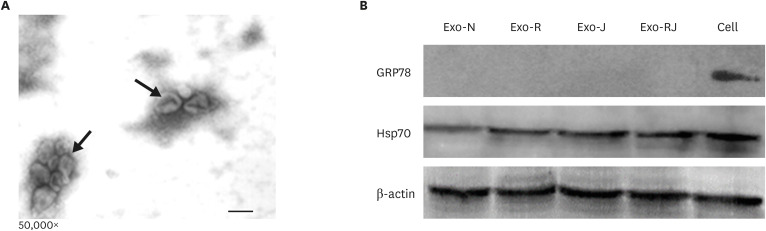
The morphology and size of purified exosomes were assessed by TEM. As shown by the TEM results in Fig. 3A, exosomes were typically cup-shaped with an average diameter ranging from 80 to 120 nm. Exosomes contain several proteins that serve as exosomes markers, such as Hsp70, CD9, CD63, and CD81 [16]. To further characterize the exosomes purified in our experimental conditions, we evaluated the expression of Hsp70. As shown in Fig. 3B, Hsp70 was detected in exosomes and cell lysates. In addition, GRP78 was only detected in cell lysates. GRP78 is a signal protein of endoplasmic reticulum stress. Our results indicated that the purified exosomes were not contaminated by organelles originating from the endoplasmic reticulum.
Fig. 3. Characterization of isolated exosomes. The electron microscopy analysis showed that exosomes were typically cup-shaped, with an average diameter ranging from 80 to 120 nm (scare bar: 100 nm, black arrow) (A). Western blot analysis of exosomes and cell lysates showed that GRP78 was only detected in cell lysates, and a band for Hsp70 was observed in exosomes and cell lysates (B). Exo-N represent exosomes from un-infected cells, Exo-R represent exosomes from REV infected cells; Exo-J represent exosomes from ALV-J infected cells; Exo-RJ represent exosomes from REV and ALV-J co-infected cells.
ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus; GRP78, glucose-regulated protein 78.
We further analyzed the protein composition of exosomes and found that exosomes from infected cells carried key components of viruses. As shown in Table 2, exosomes from coinfected cells (Exo-RJ) carried components both of ALV-J and REV. Furthermore, compared with both Exo-J and Exo-R, there were 30 differentially expressed proteins identified with 16 upregulated and 14 downregulated in Exo-RJ (Table 3) [17]. The differentially expressed proteins in exosomes may be related to the synergistic effect of REV and ALV-J coinfection.
Table 2. Exosomes from co-infection cells carrying the structural proteins both of ALV-J and REV.
| Virus | Protein | Accession | Description |
|---|---|---|---|
| ALV-J | Gag | Q64996 | Gag proteins OS = avian leukosis virus HPRS103 |
| ALV-J | Env | Q64997 | Envelope protein subgroup J OS = avian leukosis virus HPRS103 GN = env |
| REV | Gag | C7FGW7 | Gag polyprotein OS = avian reticuloendotheliosis virus GN = gag |
| REV | Pol | Q462Q9 | Polymerase (fragment) OS = avian reticuloendotheliosis virus GN = pol |
| REV | Env | Q462Q8 | Envelope glycoprotein OS = avian reticuloendotheliosis virus GN = env |
ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus; OS, organism species; GN, gene name.
Table 3. The statistic of co-regulation differentially expressed proteins.
| Number | Accession | Protein | R + J/J fold change | R + J/R fold change | Change |
|---|---|---|---|---|---|
| 1 | F1NBOB | C3H6ORF103 | 4.018 | 2.556 | Up |
| 2 | A8VHZ1 | Interleukin 6 | 2.569 | 1.410 | Up |
| 3 | F1NK52 | RHO-J | 2.257 | 1.581 | Up |
| 4 | P02701 | AVD | 2.130 | 1.389 | Up |
| 5 | F1NHI4 | SOD3 | 2.501 | 1.597 | Up |
| 6 | R4GHA7 | CRLF1 | 2.263 | 1.840 | Up |
| 7 | Q155F6 | TNFIP6 | 1.896 | 1.797 | Up |
| 8 | P08317 | Interleukin 8 | 1.733 | 1.514 | Up |
| 9 | Q5F3X4 | EFTUD2 | 1.709 | 1.470 | Up |
| 10 | R4GHA7 | TRIM62 | 1.557 | 2.083 | Up |
| 11 | F1P417 | VNN1 | 1.573 | 1.801 | Up |
| 12 | E1C6G9 | NCKAP1 (Nap125) | 1.436 | 1.415 | Up |
| 13 | F1N9E1 | MASP1 | 1.419 | 1.423 | Up |
| 14 | Q9DEQ5 | P37NB | 1.377 | 1.380 | Up |
| 15 | F1NUF8 | AMBP | 1.378 | 1.905 | Up |
| 16 | FIPIG6 | KIAA1199 | 1.593 | 1.413 | Up |
| 17 | P48440 | DDOST | 0.664 | 0.677 | Down |
| 18 | Q98TF9 | RPL14 | 0.623 | 0.504 | Down |
| 19 | F1NU61 | PCSK5 | 0.643 | 0.697 | Down |
| 20 | P41125 | RPL13 | 0.601 | 0.608 | Down |
| 21 | Q09121 | EIF5A1 | 0.553 | 0.608 | Down |
| 22 | P32760 | PTN | 0.642 | 0.572 | Down |
| 23 | E1C4W3 | PITPNM2 | 0.508 | 0.661 | Down |
| 24 | C7G540 | OCX32 | 0.415 | 0.676 | Down |
| 25 | F1NUA2 | ARPC5 (Arp2/3) | 0.414 | 0.668 | Down |
| 26 | E1BYD4 | NDRG1 | 0.488 | 0.488 | Down |
| 27 | R4GKL8 | C1QTNF3 | 0.679 | 0.679 | Down |
| 28 | E1BW00 | WISP2 | 0.672 | 0.672 | Down |
| 29 | F1NME8 | ZMYND8 | 0.656 | 0.656 | Down |
| 30 | E1C2U6 | PRKAR1B | 0.589 | 0.590 | Down |
NCKAP1 (Nap125), NCK-association proteins 1; ARPC5 (Arp2/3), actin-related 2/3 complex subunit 5. The bold words were we focus on in the next study.
KEGG annotation of identified proteins and miRNA target genes
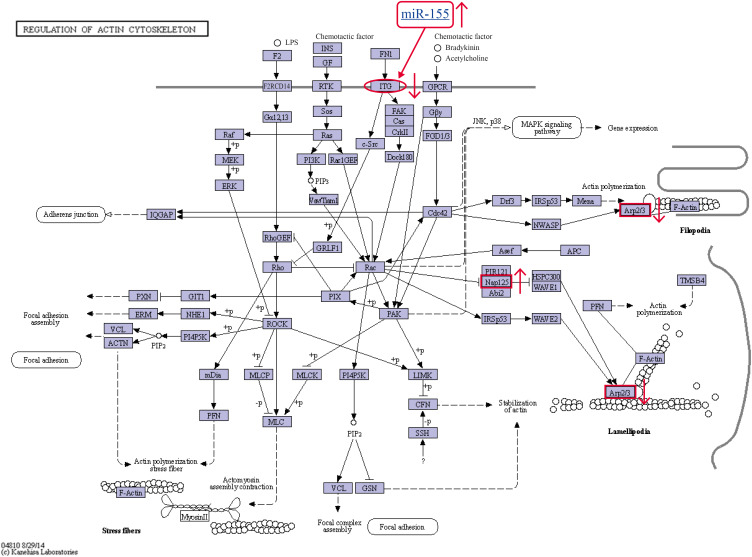
To further explore the role of differentially expressed proteins in the synergistic effect of REV and ALV-J coinfection, we annotated the 30 identified proteins and the target genes of 5 miRNAs (miRNA-184-3p, miRNA-146a-3p, miRNA-146a-5p, miRNA-3538 and miRNA-155) using the KEGG database. According to the results of the predicted miRNA target genes, integrinα1 (ITGα1) is one target gene of miRNA-155 [10]. The upregulation of miRNA-155 induced the downregulation of ITGα1. After classification, the 2 proteins NCK-association proteins 1 (NCKAP1, also known as Nap125) and actin-related 2/3 complex subunit 5 (Arp2/3-5, ARPC5) and one miRNA target gene (ITGα1) were shown to be involved in the regulation of the actin cytoskeleton signaling pathway (Fig. 4). Coinfection with ALV-J and REV may achieve a synergistic effect via regulation of the actin cytoskeleton.
Fig. 4. Three identified differentially expressed proteins were involved in the actin cytoskeleton signaling pathway. The actin cytoskeleton signaling pathway (from http://www.kegg.jp/kegg-bin/show_pathway?ko04810+K05750+K05754, map ID: ko04810). In the signaling pathway of coinfected CEF cells, the expression of ITGα1 was decreased, NCKAP1 was increased, and ARPC5 was decreased; (red arrow).
ITGα1, integrinα1; NCKAP1, NCK-association proteins 1; ARPC5, actin-related 2/3 complex subunit 5; CEF, chicken embryo fibroblast.
Validation of exosomal protein expression
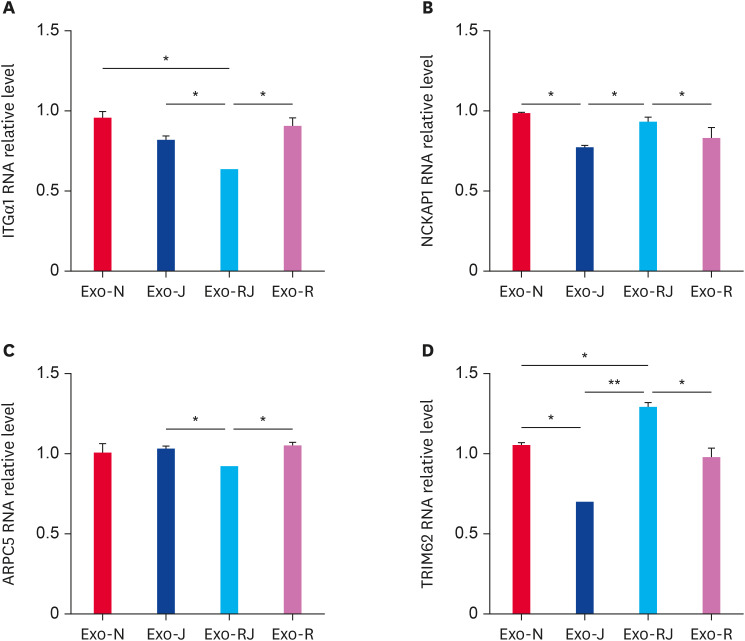
Of the 30 identified proteins in Exo-RJ, we focused on TRIM62 and the 2 proteins involved in the actin cytoskeleton. To validate the MS results, 4 proteins, ITGα1, NCKAP1, ARPC5, and TRIM62, were analyzed by qRT-PCR with the primers listed in Table 3. We found that ITGα1 and ARPC5 RNA expression in Exo-RJ was significantly lower than that in Exo-J and Exo-R (Fig. 5A and C) (p < 0.05), while NCKAP1 and TRIM62 RNA expression in Exo-RJ was significantly higher than that in Exo-J and Exo-R (Fig. 5B and D) (p < 0.05). The decreased RNA expression of ITGα1 matched the upregulation in miRNA-155 expression [10]. The results demonstrated that the RNA expression of these 4 proteins in exosomes was consistent with the proteomic analysis data and deep sequencing data [10], which indicated that the RNA expression of the 4 identified proteins reflected the protein expression.
Fig. 5. The expression of identified critical proteins in exosomes was validated by qRT-PCR. The RNA expression of ITGα1 (A) and ARPC5 (C) was significantly decreased, while that of NCKAP1 (B) and TRIM62 (D) was significantly increased in exosomes from coinfected CEF cells compared with monoinfected cells. Exo-N represent exosomes from un-infected cells, Exo-R represent exosomes from REV infected cells; Exo-J represent exosomes from ALV-J infected cells; Exo-RJ represent exosomes from REV and ALV-J co-infected cells.
qRT-PCR, quantitative reverse transcription polymerase chain reaction; ITGα1, integrinα1; ARPC5, actin-related 2/3 complex subunit 5; NCKAP1, NCK-association proteins 1; TRIM62, tripartite motif containing 62; ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus.
*p < 0.05; **p < 0.01; ***p < 0.001.
TRIM62 negatively regulated viral replication
TRIM62 is a member of the TRIM family of proteins, also known as DEAR1 (ductal epithelium-associated RING chromosome) [18]. Based on the presence of a RING domain, TRIM62 has also been defined as an E3 ubiquitin ligase [19]. RING domain E3 ubiquitin ligase has been associated with the antiviral activity of the TRIM protein [20,21]. However, the role of TRIM62 in regulating coinfection with ALV-J and REV is not clear.
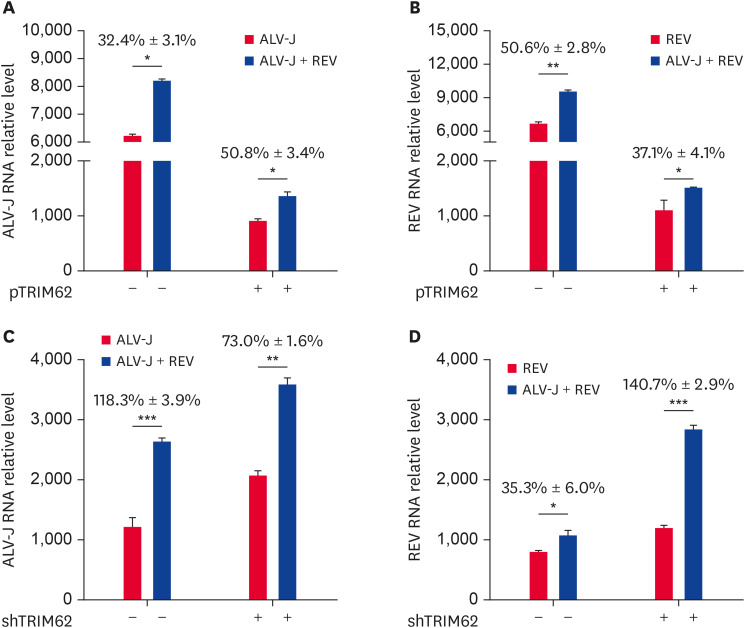
To explore the role of TRIM62 effect on the synergistic effect of ALV-J and REV coinfection, we overexpressed or silenced TRIM62 in CEF cells and further detected viral replication in coinfected cells compared with monoinfected cells at 72 h post infection. We found that TRIM62 overexpression decreased the expression of ALV-J and REV in CEF cells both with single infection and coinfection (Fig. 6A and B). In addition, compared with that in the single infection group, overexpression of TRIM62 enhanced the expressional upregulation of ALV-J in the coinfection group (Fig. 6A) and decreased that of REV (Fig. 6B). On the other hand, knockdown of TRIM62 with specific shRNA increased the expression of both ALV-J and REV in CEF cells both with single infection and coinfection (Fig. 6C and D). Furthermore, we found that silencing TRIM62 reduced the expressional upregulation of ALV-J in the coinfection group (Fig. 6C) and increased that of REV (Fig. 6D).
Fig. 6. The effects of TRIM62 on viral replication were analyzed by qRT-PCR. CEF cells were transfected with plasmid containing TRIM62 or empty vector for 12 h before single infection or coinfection with ALV-J and REV (A, B). CEF cells were transfected with shTRIM62 or empty vector for 24 h before infection (C, D). The RNA expression of ALV-J (A, C) and REV (B, D) was assessed.
TRIM62, tripartite motif containing 62; qRT-PCR, quantitative reverse transcription polymerase chain reaction; CEF, chicken embryo fibroblast; ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus; shTRIM62, short hairpin RNA targeting tripartite motif containing 62.
*p < 0.05; **p < 0.01; ***p < 0.001.
Our results demonstrated that TRIM62 negatively regulated viral replication in CEF cells, and the inhibition on REV was more significant than that on ALV-J in coinfected CEF cells. These results indicated the presence of a competitive effect between the 2 viruses in the context of coinfection.
TRIM62 regulated actin cytoskeletal dynamics
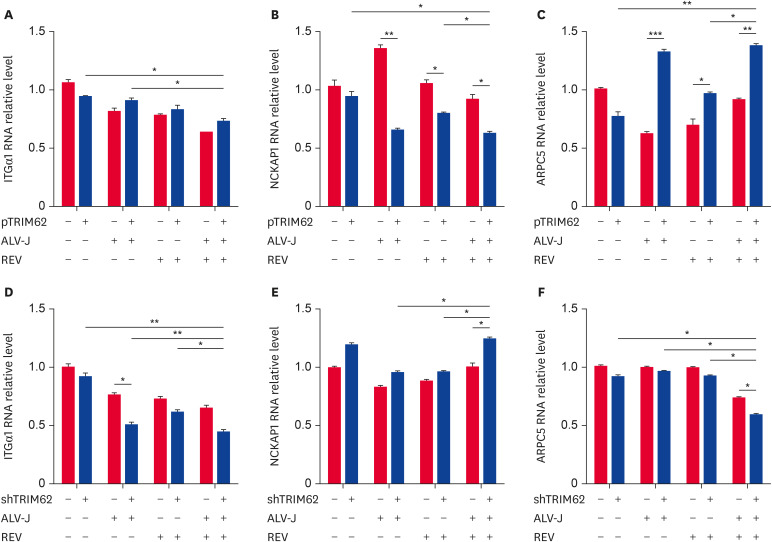
To evaluate the effect of TRIM62 on actin cytoskeletal dynamics, we used qRT-PCR to detect the regulation of the expression of the 3 identified proteins involved in the actin cytoskeletal pathway by TRIM62. Compared with the empty vector group, in CEF cells with ALV-J and REV single infection or coinfection, overexpression of TRIM62 decreased NCKAP1 expression (Fig. 7B) and increased ARPC5 expression (Fig. 7C) but did not affect ITGα1 (Fig. 7A).
Fig. 7. TRIM62 regulation of proteins involved in the actin cytoskeleton pathway. CEF cells were transfected with plasmid containing TRIM62 or empty vector for 12 h before single infection or coinfection with ALV-J and REV (A-C). CEF cells were transfected with shTRIM62 or empty vector for 24 h before infection (D-F). After 72 h of infection, the expression levels of ITGα1 (A, D), NCKAP1 (B, E), and ARPC5 (C, F) were measured. The black columns mean cells transfected with pTRIM62 (A-C) and with shTRIM62 (D-F); the grey columns mean cells transfected with empty vector (A-F).
TRIM62, tripartite motif containing 62; CEF, chicken embryo fibroblast; ALV-J, avian leukosis virus subgroup J; REV, reticuloendotheliosis virus; ITGα1, integrinα1; NCKAP1, NCK-association proteins 1.
*p < 0.05; **p < 0.01; ***p < 0.001.
However, compared with the empty vector, shTRIM62 significantly decreased ITGα1 expression both in the ALV-J single infection and coinfection groups, and compared with that in the single infection groups, the expression of ITGα1 was significantly decreased in the coinfection group (Fig. 7D). Importantly, compared with the empty vector, shTRIM62 significantly increased NCKAP1 expression (Fig. 7E) and decreased ARPC5 expression (Fig. 7F), which was observed only in the coinfection group. In addition, compared with expression in the single infection groups, the expression of NCKAP1 (Fig. 7E) was significantly increased and that of ARPC5 was decreased (Fig. 7F) in the coinfection group.
Collectively, these results indicated that TRIM62 was modified and affected actin cytoskeletal dynamics by inhibiting NCKAP1 expression and promoting ARPC5 expression to negatively regulate virus replication in the context of coinfection.
DISCUSSION
The results described here showed that ALV-J and REV coinfection synergistically promoted the replication of each virus. Compared with single infection, ALV-J replication was increased more significantly than that of REV before 72 h post infection, while REV replication was upregulated more significantly than that of ALV-J after 96 h post infection. The results indicate that 72 h post infection is an important time point in CEF cells coinfected with ALV-J and REV, which is consistent with the previous study [10]. Both ALV-J and REV can cause immunosuppression, which promotes the replication of other viruses. It is not clear why there was predominance of ALV-J replication over REV replication. In addition, viral RNA expression in TRIM62-overexpressing or silenced CEF cells was detected after 72 h of infection, and our results showed that TRIM62 affected REV more significantly than ALV-J. The effects of TRIM62 on viral replication demonstrated the presence of a competitive effect between the 2 viruses in the process of coinfection. These results indicate that the 2 viruses synergistically promote each other but also display a competitive relationship in the process of coinfection.
Exosomes are rich in endosome-associated proteins but also carry different molecules in their lumen, including proteins, RNA (i.e., miRNA), and pathogen-derived cargo. The different biological functions of exosomes depend on their cargo components [8]. Furthermore, our data show that coinfection with ALV-J and REV induced changes in the exosomal proteins and miRNAs. Thus, exosomes may provide microenvironmental conditions for ALV-J and REV infection and play an important role in their synergistic effects. In addition to viral components (Table 2), we focused on TRIM62, NCKAP1 and ARPC5 of the 30 identified proteins. TRIM62 is an innate immune regulator [22]. We previously confirmed that TRIM62 restricts ALV-J replication [13]. KEGG annotation of the identified proteins and miRNA target genes [10] demonstrated that 2 of the identified proteins (NCKAP1 and ARPC5) and 1 target gene of miRNA-155 (ITGα1) were involved in the actin cytoskeleton signaling pathway. We hypothesize that TRIM62 and the actin cytoskeleton play important roles in ALV-J and REV coinfection.
To reveal the role of TRIM62 regulation on the synergistic effect of ALV-J and REV coinfection, we then carried out TRIM62 overexpression and silencing to modulate its effect on the actin cytoskeleton. We found that TRIM62 inhibited the expression of NCKAP1 and promoted the expression of ARPC5 in coinfected CEF cells. The results further indicated that TRIM62 may affect ALV-J and REV coinfection by regulating the actin cytoskeleton. Our study provides a possible new antiviral strategy targeting this novel regulator. It has been reported that retroviruses HIV and Moloney murine leukemia virus utilize the cell cytoskeleton to expedite their assembly and budding [23]. Further research is needed on how ALV-J and REV utilize the cytoskeleton synergistically to promote the replication of each other.
Furthermore, it is usually considered that ALV-J and REV induced tumor by insertional mutagenesis of host DNA [24]. It has been reported that ALV-J carried complete v-fps oncogene [25]. NCKAP1 (Nap125) has been associated with prognosis in hepatocellular carcinoma as a target gene of the tumor promoting miRNA-34c-3p [26]. ARPC5 is one subunit of the Arp2/3 complex [27]. The Arp2/3 complex as a regulator that plays a pivotal role in actin filament formation [28]. Plk4 promotes cancer invasion and metastasis through regulation of Arp2/3 complex-mediated actin cytoskeletal rearrangement [29]. These results demonstrate that the actin cytoskeleton is closely related to cancer. Human TRIM62 acts as a tumor suppressor and is involved in lung cancer [30], breast cancer [18], acute myeloid leukemia [31], and cervical cancer [32]. Therefore, TRIM62, NCKAP1, and ARPC5 may be related to the tumor formation induced by coinfection with ALV-J and REV.
Footnotes
Funding: This work was supported by the Natural Science Foundation of Shandong Provincial (ZR2017MC011) and the National Natural Science Foundation of China (31772703).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Li L.
- Data curation: Li L, Zhuang P.
- Formal analysis: Yang J.
- Funding acquisition: Wang G.
- Investigation: Bi J.
- Methodology: Zhuang P.
- Project administration: Wang G.
- Resources: Wang G.
- Software: Bi J.
- Supervision: Yang J.
- Validation: Cheng Z.
- Visualization: Li L.
- Writing - original draft: Li L.
- Writing - review & editing: Cheng Z, Wang G.
References
- 1.Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol. 1991;72(Pt 4):801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 2.Witter RL, Glass SE. Reticuloendotheliosis in breeder turkeys. Avian Dis. 1984;28(3):742–750. [PubMed] [Google Scholar]
- 3.Zavala G, Cheng S, Barbosa T, Haefele H. Enzootic reticuloendotheliosis in the endangered Attwater's and greater prairie chickens. Avian Dis. 2006;50(4):520–525. doi: 10.1637/7655-052806R.1. [DOI] [PubMed] [Google Scholar]
- 4.Cui Z, Sun S, Zhang Z, Meng S. Simultaneous endemic infections with subgroup J avian leukosis virus and reticuloendotheliosis virus in commercial and local breeds of chickens. Avian Pathol. 2009;38(6):443–448. doi: 10.1080/03079450903349188. [DOI] [PubMed] [Google Scholar]
- 5.Dong X, Ju S, Zhao P, Li Y, Meng F, Sun P, Cui Z. Synergetic effects of subgroup J avian leukosis virus and reticuloendotheliosis virus co-infection on growth retardation and immunosuppression in SPF chickens. Vet Microbiol. 2014;172(3-4):425–431. doi: 10.1016/j.vetmic.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Dong X, Zhao P, Chang S, Ju S, Li Y, Meng F, Sun P, Cui Z. Synergistic pathogenic effects of co-infection of subgroup J avian leukosis virus and reticuloendotheliosis virus in broiler chickens. Avian Pathol. 2015;44(1):43–49. doi: 10.1080/03079457.2014.993359. [DOI] [PubMed] [Google Scholar]
- 7.Huber V, Vallacchi V, Di Guardo L, Cova A, Canevari S, Santinami M, Bollati V, Rodolfo M, Rivoltini L. Role of exosomes in immune suppression. Eur J Cancer. 2015;51(Supplement 1):S13. [Google Scholar]
- 8.Poveda E, Freeman ML. Hot news: exosomes as new players in HIV pathogenesis - New data from the IAS 2017. AIDS Rev. 2017;19(3):173–175. [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Wang Z, Zhuang P, Zhao X, Cheng Z. Exosomes carring gag/env of ALV-J possess negative effect on immunocytes. Microb Pathog. 2017;112:142–147. doi: 10.1016/j.micpath.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Zhou D, Xue J, He S, Du X, Zhou J, Li C, Huang L, Nair V, Yao Y, Cheng Z. Reticuloendotheliosis virus and avian leukosis virus subgroup J synergistically increase the accumulation of exosomal miRNAs. Retrovirology. 2018;15(1):45–56. doi: 10.1186/s12977-018-0427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheruiyot C, Pataki Z, Williams R, Ramratnam B, Li M. SILAC based proteomic characterization of exosomes from HIV-1 infected cells. J Vis Exp. 2017;3(121):e54799. doi: 10.3791/54799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Feng W, Cheng Z, Yang J, Bi J, Wang X, Wang G. TRIM62-mediated restriction of avian leukosis virus subgroup J replication is dependent on the SPRY domain. Poult Sci. 2019;98(11):6019–6025. doi: 10.3382/ps/pez408. [DOI] [PubMed] [Google Scholar]
- 14.Dai M, Feng M, Liu D, Cao W, Liao M. Development and application of SYBR Green I real-time PCR assay for the separate detection of subgroup J avian leukosis virus and multiplex detection of avian leukosis virus subgroups A and B. Virol J. 2015;12:52. doi: 10.1186/s12985-015-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan H, Wang Y, Li Y, Cui Z, Chang S, Zhao P. Development of a real-time quantitative RT-PCR to detect REV contamination in live vaccine. Poult Sci. 2016;95(9):2023–2029. doi: 10.3382/ps/pew147. [DOI] [PubMed] [Google Scholar]
- 16.Kumar D, Gupta D, Shankar S, Srivastava RK. Biomolecular characterization of exosomes released from cancer stem cells: possible implications for biomarker and treatment of cancer. Oncotarget. 2015;6(5):3280–3291. doi: 10.18632/oncotarget.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Zhuang PP, Wang XM, Zhou DF, Xue JW, Cheng ZQ, Wang GH. Effects of miRNA-155 on the cytoskeletal pathways in REV and ALV-J co-infection. Chin J Cell Biol. 2018;40(10):1706–1718. [Google Scholar]
- 18.Lott ST, Chen N, Chandler DS, Yang Q, Wang L, Rodriguez M, Xie H, Balasenthil S, Buchholz TA, Sahin AA, Chaung K, Zhang B, Olufemi SE, Chen J, Adams H, Band V, El-Naggar AK, Frazier ML, Keyomarsi K, Hunt KK, Sen S, Haffty B, Hewitt SM, Krahe R, Killary AM. DEAR1 is a dominant regulator of acinar morphogenesis and an independent predictor of local recurrence-free survival in early-onset breast cancer. PLoS Med. 2009;6(5):e1000068. doi: 10.1371/journal.pmed.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang F, Xiao H, Sun BL, Yang RG. Characterization of TRIM62 as a RING finger E3 ubiquitin ligase and its subcellular localization. Biochem Biophys Res Commun. 2013;432(2):208–213. doi: 10.1016/j.bbrc.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Fridell RA, Harding LS, Bogerd HP, Cullen BR. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209(2):347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 21.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4(2):e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008;4(2):e16. doi: 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladnikoff M, Shimoni E, Gov NS, Rousso I. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys J. 2009;97(9):2419–2428. doi: 10.1016/j.bpj.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Justice J, 4th, Malhotra S, Ruano M, Li Y, Zavala G, Lee N, Morgan R, Beemon K. The MET gene is a common integration target in avian leukosis virus subgroup J-induced chicken hemangiomas. J Virol. 2015;89(9):4712–4719. doi: 10.1128/JVI.03225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Li J, Li Y, Fang L, Sun X, Chang S, Zhao P, Cui Z. Identification of ALV-J associated acutely transforming virus Fu-J carrying complete v-fps oncogene. Virus Genes. 2016;52(3):365–371. doi: 10.1007/s11262-016-1301-6. [DOI] [PubMed] [Google Scholar]
- 26.Xiao CZ, Wei W, Guo ZX, Zhang MY, Zhang YF, Wang JH, Shi M, Wang HY, Guo RP. MicroRNA-34c-3p promotes cell proliferation and invasion in hepatocellular carcinoma by regulation of NCKAP1 expression. J Cancer Res Clin Oncol. 2017;143(2):263–273. doi: 10.1007/s00432-016-2280-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vartiainen MK, Machesky LM. The WASP-Arp2/3 pathway: genetic insights. Curr Opin Cell Biol. 2004;16(2):174–181. doi: 10.1016/j.ceb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 29.Kazazian K, Go C, Wu H, Brashavitskaya O, Xu R, Dennis JW, Gingras AC, Swallow CJ. Plk4 promotes cancer invasion and metastasis through Arp2/3 complex regulation of the actin cytoskeleton. Cancer Res. 2017;77(2):434–447. doi: 10.1158/0008-5472.CAN-16-2060. [DOI] [PubMed] [Google Scholar]
- 30.Quintás-Cardama A, Post SM, Solis LM, Xiong S, Yang P, Chen N, Wistuba II, Killary AM, Lozano G. Loss of the novel tumour suppressor and polarity gene Trim62 (Dear1) synergizes with oncogenic Ras in invasive lung cancer. J Pathol. 2014;234(1):108–119. doi: 10.1002/path.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintás-Cardama A, Zhang N, Qiu YH, Post S, Creighton CJ, Cortes J, Coombes KR, Kornblau SM. Loss of TRIM62 expression is an independent adverse prognostic factor in acute myeloid leukemia. Clin Lymphoma Myeloma Leuk. 2015;15(2):115–127.e15. doi: 10.1016/j.clml.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu TY, Chen J, Shang CL, Shen HW, Huang JM, Liang YC, Wang W, Zhao YH, Liu D, Shu M, Guo LY, Hu Z, Yao SZ. Tripartite motif containing 62 is a novel prognostic marker and suppresses tumor metastasis via c-Jun/Slug signaling-mediated epithelial-mesenchymal transition in cervical cancer. J Exp Clin Cancer Res. 2016;35(1):170. doi: 10.1186/s13046-016-0445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]