Abstract
Compounds with dihydroquinoline-4(1H)-one nuclei have been reported in the literature for being important in the development of medicines due to their broad spectrum of activities. In this way, the structural knowledge of this class becomes relevant for obtaining new materials with desired biological properties. This study presents the structural elucidation of five halogenated dihydroquinolines, as well as the discussion about the effect on the molecular conformation of the type and position of halogen atom on aromatic rings. Compounds I and IV differ in halogen substitution on 2-phenyl ring, while compounds III and V differ in halogen substitution on the benzylidene ring. Moreover, compound II has a para-substituted 2-phenyl ring in their molecular structure. The crystal packing of all five molecules is mainly ruled by C–H⋯O and C–H···halogen interactions that form dimers and chains. The shift in position and the kind of the halogen in ring C shows a starring role in the conformation of the studied compounds, and the packaging of these compounds is more susceptible to variations when the halogen position changes.
Keywords: Dihydroquinoline, Chlorine, Fluorine, Crystal structure
Graphical abstract
Highlights
-
•
Five dihydroquinoline-4(1H)-ones have been synthesized by varying the halogenated substituents.
-
•
Different substituents lead to distinct number of molecules at asymmetric unit.
-
•
The Molecular packing is stabilized by C–H⋯O and C–H···Halogen hydrogen bonds.
-
•
The change in position and the halogen atom attached in ring C play a significant role in the conformation.
1. Introduction
Quinolines are a class of N-heterocyclic compounds, also known as benzopyridines, obtained both from natural and synthetic sources. They have attracted great attention in the scientific community being used in several industrial processes, becoming increasingly important in the development of medicines, pesticides and also due to their notable biological activities [[1], [2], [3]]. Among the compounds of this class, we are interested in those with the dihydroquinoline-4(1H)-one moiety; in this way, the recent studies conducted by our research group involve structural elucidation [[4], [5], [6]], anti-cancer properties [7,8] and their potential application as pesticides [9]. Also, other groups have explored the cytotoxic properties of this class [[10], [11], [12], [13]] and as potential Middle East respiratory syndrome coronavirus (MERS-CoV) inhibitors [14].
The biological activity of a substance is dominated by its properties, which are determined by its chemical structure. Structural elucidation is essential since it allows understanding and proposing explanations for the mechanisms of action at the molecular level, helping to design and develop new materials with desirable biological properties [15,16]. A search in the CSD version 5.41 (November 2019) database showed 132 reported structures with the dihydroquinoline-4(1H)-one nucleus. Furthermore, given the biological potential of these molecules, a need to enlarge elucidated structures of this class will contribute to the applications of dihydroquinoline-4(1H)-ones.
Thus, in this paper, we investigate and report a comprehensive single-crystal analysis of five halogenated dihydroquinoline-4(1H)-ones, namely (E)-3-(2-chlorobenzylidene)-2-(2-chlorophenyl)-1-(phenylsulfonyl)-2,3-dihydroquinolin-4(1H)-one (I), (E)-3-(2-chlorobenzylidene)-2-(4-chlorophenyl)-1-(phenylsulfonyl)-2,3-dihydroquinolin-4(1H)-one (II), (E)-2-(2-bromophenyl)-3-(2-chlorobenzylidene)-1-(phenylsulfonyl)-2,3-dihydroquinolin-4(1H)-one (III), (E)-2-(2-chlorophenyl)-3-(2-fluorobenzylidene)-1-(phenylsulfonyl)-2,3-dihydroquinolin-4(1H)-one (IV) and (E)-2-(2-bromophenyl)-3-(2-fluorobenzylidene)-1-(phenylsulfonyl)-2,3-dihydroquinolin-4(1H)-one (V). The title compounds were divided into two groups: chlorinated and fluorinated; and their molecular geometry, intermolecular interactions and crystal packing were all analyzed and discussed in terms of the effect of halogen substitution in their scaffold.
2. Experimental
2.1. Synthesis and crystallization
Compounds I to V were obtained from sulfonamide chalcones reacted with benzaldehydes in an alkaline reaction environment for 24 h (Scheme 1 ). The subsequent precipitates were purified by slow recrystallization from dichloromethane and ethanol (4:1), after drying at room temperature.
Scheme 1.
Synthesis of compounds I to V.
Nuclear magnetic resonance (NMR) spectra were acquired on a Bruker Avance III 500 spectrometer (Rheinstetten, Germany) operating at 11.75 T with a 5 mm inverse detection three-channel (1H, 2H and X-nucleus) BBI probe. The samples (ca. 10 mg) were dissolved in 600 μL of deuterated dimethylsulfoxide (DMSO‑d 6), containing tetramethylsilane (TMS) as the internal standard. The unambiguous signal assignment was achieved by correlation spectroscopy (COSY), heteronuclear multiple bond correlation (HMBC), heteronuclear single quantum correlation (HSQC), and DEPT-135 experiments, in addition to 1H and 13C analyses (Table 1 ).
Table 1.
1H NMR spectral data assignments for the compounds I, II, III, IV and V in DMSO‑d6.
| Compound |
I |
II |
III |
IV |
V |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Atom | 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C |
| C1 | 6.81 (m) | 58.4 | 6.52 (m) | 58.9 | 6.81 (s) | 60.6 | 6.79 (m) | 58.6 | 6.73 (s) | 60.7 |
| C2 | – | 132.1 | – | 131.3 | – | 132.3 | – | 132.1 | – | 130.0 |
| C3 | – | 181.2 | – | 181.1 | – | 181.2 | – | 181.2 | – | 181.1 |
| C4 | – | 129.3 | – | 127.4 | – | 129.6 | – | 129.3 | – | 129.2 |
| C5 | 7.80 (ddd, 7.6; 1.6; 0.5) | 127.7 | 7.80 (ddd, 7.6; 1.6; 0.5) | 127.8 | 7.78 (ddd, 7.6; 1.6; 0.5) | 127.6 | 7.82 (ddd, 7.7; 1.7; 0.4) | 127.6 | 7.80 (dd, 7.6; 1.4) | 127.3 |
| C6 | 7.45 (ddd, 7.6; 7.6; 1.1) | 128.5 | 7.42 (m) | 127.8 | 7.39 (ddd, 7.6; 7.6; 1.1) | 128.2 | 7.46 (m) | 128.5 | 7.45 (ddd, 7.6; 7.6; 1.0) | 128.5 |
| C7 | 7.65 (ddd, 7.6; 7.6; 1.6) | 135.3 | 7.70 (m) | 135.4 | 7.65 (ddd, 7.6; 7.6; 1.6) | 135.3 | 7.65 (m) | 135.2 | 7.65 (m) | 135.2 |
| C8 | 7.41 (m) | 128.1 | 7.65 (m) | 127.4 | 7.37 (ddd, 7.6; 1.1; 0.5) | 127.7 | 7.39 (m) | 128.4 | 7.37 (dd, 8.2; 1.0) | 128.6 |
| C9 | – | 138.1 | – | 138.1 | – | 138.1 | – | 138.1 | – | 138.0 |
| C10 | 7.88 (m) | 136.2 | 7.88 (m) | 136.1 | 7.88 (m) | 136.2 | 7.67 (m) | 132.4 | 7.68 (m) | 132.4 |
| C11 | – | 131.1 | – | 130.8 | – | 131.2 | – | 121.0 | – | 121.0 |
| C12 | – | 134.5 | – | 134.9 | – | 134.5 | – | 160.7 | – | 160.5 |
| C13 | 7.73 (dd, 8.0; 1.2) | 130.5 | 7.72 (dd, 8.0; 1.4) | 130.6 | 7.73 (dd, 8.0; 1.1) | 130.5 | 7.45 (m) | 116.5 | 7.46 (dd, 7.6; 1.1) | 116.5 |
| C14 | 7.54 (m) | 132.1 | 7.56 (ddd, 8.0; 7.70; 1.6) | 132.2 | 7.54 (ddd, 8.0; 7.6; 1.1) | 132.1 | 6.85 (td, 7.7; 1.7) | 129.1 | 6.85 (dd, 7.6; 7.6; 1.5) | 129.2 |
| C15 | 7.35 (m) | 127.6 | 7.40 (m) | 127.8 | 7.44 (ddd, 7.8; 7.6; 1.1) | 128.5 | 7.28 (td, 7.7; 1.2) | 125.0 | 7.28 (ddd, 7.6; 7.6; 1.1) | 125.1 |
| C16 | 6.72 (dd, 7.9; 1.5) | 128.9 | 7.00 (dd, 7.9; 1.6) | 128.8 | 6.72 (dd, 7.8; 1.7) | 128.9 | 7.60 (m) | 133.1 | 7.60 (m) | 133.1 |
| C17 | – | 135.0 | – | 136.3 | – | 136.3 | – | 134.7 | – | 135.8 |
| C18 | 6.79 (dd, 7.9; 1.5) | 129.8 | 7.35 (m) | 128.9 | 6.77 (dd, 7.7; 1.5) | 130.1 | 6.76 (dd, 7.7; 1.6) | 129.7 | 6.75 (dd, 7.7; 1.5) | 130.1 |
| C19 | 7.14 (m) | 127.5 | 7.42 (m) | 129.5 | 7.17 (td, 7.7; 1.5) | 128.0 | 7.12 (td, 7.7; 1.4) | 127.4 | 7.15 (td, 7.7; 1.5) | 127.9 |
| C20 | 7.35(m) | 130.8 | – | 133.4 | 7.25(ddd, 7.9; 7.7; 1.5) | 131.0 | 7.33 (td, 7.7; 1.6) | 130.8 | 7.23 (ddd, 7.9; 7.7; 1.5) | 130.9 |
| C21 | 7.60 (dd, 7.9; 1.2) | 131.0 | 7.42 (m) | 129.5 | 7.77 (dd, 7.9; 1.2) | 134.5 | 7.60 (m) | 131.0 | 7.76 (dd, 7.9; 1.2) | 134.6 |
| C22 | – | 133.3 | 7.35 (m) | 128.9 | – | 123.6 | – | 133.4 | – | 123.7 |
| C23 | – | 135.6 | – | 136.0 | – | 135.4 | – | 135.6 | – | 135.5 |
| C24 | 7.13 (m) | 127.4 | 7.16 (m) | 126.6 | 7.13 (m) | 127.6 | 7.05 (m) | 127.7 | 7.04 (m) | 127.5 |
| C25 | 7.41 (m) | 129.2 | 7.42 (m) | 129.3 | 7.40 (m) | 129.1 | 7.39 (m) | 129.0 | 7.38 (m) | 129.0 |
| C26 | 7.64 (m) | 134.0 | 7.66 (m) | 134.1 | 7.63 (m) | 134.0 | 7.66 (m) | 134.1 | 7.65 (m) | 134.1 |
| C27 | 7.41 (m) | 129.2 | 7.42 (m) | 129.3 | 7.40 (m) | 129.1 | 7.39 (m) | 129.0 | 7.38 (m) | 129.0 |
| C28 | 7.13 (m) | 127.4 | 7.16 (m) | 126.6 | 7.13 (m) | 127.6 | 7.05 (m) | 127.7 | 7.04 (m) | 127.5 |
2.2. Crystallographic characterization
Appropriate single crystals of compounds I, II, III, IV and V were carefully chosen. They were mounted in a Bruker APEX II CCD diffractometer with graphite-monochromated Mo-Kα radiation (λ = 0.71073 Å), and data were measured at 120 K. Using Olex2 [17], the structure solutions were determined by Direct Methods with SHELXS [18] and refined by full-matrix least-squares on F 2 with SHELXL [19]. All the hydrogen atoms were placed in calculated positions and refined with fixed individual displacement parameters [Uiso(H) = 1.2Ueq (C) or 1.5Ueq (C)] according to the riding model (C–H bonds equal 0.93 Å for aromatic). Ring D in IIIb was found to be disordered and modeled over two equal occupancy positions. Lastly, the validation of chemical parameters were made using PARST [20] and PLATON [21]. Data collection and structure refinement details are summarized in Table 2 . The structures I to V were deposited in the Cambridge Crystallographic Data Centre (CCDC) with the code number 1994320, 1994319, 1994318, 1994317, and 1994316, respectively.
Table 2.
Crystallographic and refinement details for I, II, III, IV and V.
| I | II | III | IV | V | |
|---|---|---|---|---|---|
| Chemical formula | C28H19Cl2NO3S | C28H19Cl2NO3S | C28H19BrClNO3S | C28H19ClFNO3S | C28H19BrFNO3S |
| Mr | 520.40 | 520.40 | 564.86 | 503.95 | 548.41 |
| Crystal system, space group | Triclinic, P-1 | Monoclinic, P21/n | Triclinic, P-1 | Triclinic, P-1 | Monoclinic, P21/c |
| Temperature (K) | 120 | 120 | 120 | 120 | 120 |
| a, b, c (Å) | 8.0771 (2), 16.4324 (5), 19.2435 (6) | 9.8048 (4), 14.2198 (6), 17.2824 (7) | 8.0141 (3), 16.6545 (7), 19.3781 (8) | 7.9709 (11), 21.980 (3), 24.471 (3) | 29.026 (5), 16.304 (3), 15.322 (2) |
| α, β, γ (°) | 109.114 (2), 94.537 (1), 97.435 (1) | 90, 95.552 (1), 90 |
108.605 (1), 95.549 (1), 97.078 (1) | 63.784 (2), 81.456 (2), 82.227 (2) | 90, 95.775 (3), 90 |
| V (Å3) | 2372.81 (12) | 2398.25 (17) | 2407.00 (17) | 3791.9 (9) | 7214 (2) |
| Z | 4 | 4 | 4 | 6 | 12 |
| μ (mm−1) | 0.39 | 0.39 | 1.94 | 0.27 | 1.84 |
| Crystal size (mm) | 0.17 × 0.17 × 0.09 | 0.37 × 0.30 × 0.27 | 0.19 × 0.18 × 0.14 | 0.19 × 0.14 × 0.08 | 0.17 × 0.17 × 0.16 |
| Tmin, Tmax | 0.675, 0.745 | 0.683, 0.712 | 0.594, 0.641 | 0.951, 0.978 | 0.940, 0.984 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 35818, 9758, 6875 | 36054, 5954, 5242 | 54875, 11976, 9107 | 68765, 16965, 12459 | 194960, 18117, 13854 |
| Rint | 0.045 | 0.019 | 0.037 | 0.046 | 0.064 |
| (sin θ/λ)max (Å−1) | 0.627 | 0.668 | 0.668 | 0.644 | 0.670 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.038, 0.089, 1.00 | 0.031, 0.085, 1.04 | 0.043, 0.101, 1.04 | 0.040, 0.096, 1.03 | 0.043, 0.100, 1.04 |
| No. of reflections | 9758 | 5954 | 11976 | 16965 | 18117 |
| No. of parameters | 631 | 316 | 661 | 946 | 946 |
| Δρmax, Δρmin (e Å−3) | 0.36, −0.42 | 0.39, −0.38 | 1.13, −2.61 | 0.33, −0.36 | 1.90, −0.84 |
3. Results and discussion
3.1. Spectroscopy characterization
Complete assignment of the signals of 1H and 13C NMR spectra are presented in Table 1 and the infrared spectra are available in the Support Information. The most deshielded carbon signal, near δ 181 and assigned to carbonyl carbon (C3), was a good starting point to confirm the proposed structure and NMR signal assignments. The strong correlations with C3 in the HMBC experiment (3 J CH) allowed the identification of hydrogens H8, H5, H10 and H1 (Fig. S1). Also, the correlations of these hydrogens with their respective carbons, by HSQC experiment (1 J CH), enabled the assignment of the carbons C10 and C1 (Fig. S2). H10 signal is a broad singlet (Fig. S3) since it presented long-range scalar coupling with the other hydrogens of the benzylidene group. Cross peaks with C1 and H1 in the HMBC spectrum provided information for C22 and C18 assignments (Fig. S1). C9 was identified by its strong correlations with H1, H5, and H7, and weak correlation with H8 (Fig. S1). H16 (and H12 when present) was assigned based on 3 J CH with C10 in HMBC.
The multiplet signal near to δ 7.1 in the 1H NMR spectrum was assigned to H24/28, since it had an integration of 2 in all the structures and presented 1 J CH and 3 J CH correlations only with the carbons C24/28 and C26, respectively. H25/27 was assigned by a cross peak with H24/28 signal in the COSY experiment (Fig. S4), and their HSQC correlation allowed C25/27 assignment as the carbon signal near to δ 129. H26 and C26 correlation were observed in HSQC and this correlation was supported by correlation with H25/27 in the COSY experiment (Fig. S4). H25/27 shared a strong HMBC cross peak with a carbon signal close to δ 135, assigned as C23. C4 and C23 were differentiated by DEPT-135, in the same way as other quaternary carbons close to tertiary carbons. Thus, all the atoms of the phenyl ring attached to the sulfonamide group were assigned.
The 3 J CH correlations of H10 and H1 were good entry points for the assignments of the rings B and C signals (C11 to C22). In structures IV and V, the presence of fluorine made the assignments of ring C easier because the neighboring fluorine carbons (1 J CF ∼ 250 Hz; 2 J CF ∼ 22 Hz; 3 J CF ∼ 9 Hz; 4 J CF ∼ 4 Hz) were observed as doublets, due to coupling between C and F. The hydrogen H16 was assigned by cross peaks observed in HMBC experiment, especially the correlation with C10. The signal for C22 was observed near to δ 123.6 for compounds III and V, which was markedly shielded when compared to I and IV (near to δ 143.3). This shielding was due to the attached bromine on the former two compounds and chlorine on the latter two compounds. This kind of strategy allowed a complete assignment of the 1H and 13C nuclei (see S.I.), and agrees with our previous report [22].
3.2. Crystallographic characterization
3.2.1. Chlorine dihydroquinolinones
Compound I belongs to the class of dihydroquinolinones having three substituents groups in its motif. A sulfonylbenzene group attached to N atom; an ortho-chlorobenzene attached to C1 atom and a chloro-2-vinylbenze attached to C2 atom (Scheme 1). Compound I crystallizes in the triclinic system (P) with two independent molecules in the asymmetric unit (ASU), labeled as Ia and Ib (Fig. 1 ). To clarify, in this paper we chose arbitrarily all the molecules within the ASU have R configuration about the stereogenic center. To understand the geometrical differences between these molecules their structures have been overlaid using the atoms C1, C3, and C5 as anchor points (Fig. S48). The primary variances noted within these structures are related to the orientations of rings B, C and D with respect to the A ring (defined on Table 3 ). These variances were measured using the following parameters: the torsion angles: C2–C10–C11–C12 (ϕ1), C2–C1–C17–C22 (ϕ2) and N1–S1–C23–C28 (ϕ3), and the dihedral angle between the planes formed by ring A and ring B (∠AB) (Table 3).
Fig. 1.
The two independent molecules of I showing the atom-labeling scheme: (a) molecule Ia, (b) molecule Ib. To clarify, in (b) the labeling scheme shows only non-carbon atoms. The labeling scheme for C atoms in (b) follows the same way as presented in (a). Displacement ellipsoids are drawn at the 50% probability level and H atoms have been omitted.
Table 3.
Torsion angles and least-square planes of aromatic rings of I, II, and III.
| Molecule | ϕ1 | ϕ2 | ϕ3 | ∠AB |
|---|---|---|---|---|
| Ia | −146.9(3) | 145.9(2) | 87.46(16) | 54.40(7) |
| Ib | −147.4(2) | 168.88(19) | 77.35(16) | 62.55(7) |
| II | 137.5(2) | 167.77(12) | −84.58(13) | 60.12(4) |
| IIIa | −148.9(3) | 164.83(2) | −104.43(2) | 61.00(9) |
| IIIb | −147.7(3) | 147.28(2) | −87.85(4) | 55.62(9) |
ϕ1 = C2–C10–C11–C12; ϕ2 = C2–C1–C17–C22; ϕ3 = N1–S1–C23–C28.
The ∠AB dihedral angle is ca 8⁰ larger in Ib than in Ia, evidencing that the rings in these two molecules have different orientation. This characteristic is also observed in other crystal structures of dihydroquinolinones derivatives [[23], [24], [25]]. Except for compound II, these characteristics are the same for all compounds studied here. The orientation of ring B (ϕ1), could be considered the same in Ia and Ib, it assumes an anti-clinal orientation. Furthermore, the values of ϕ2 show that ring C assumes an anti-clinal and an anti-periplanar orientation in Ia and Ib, respectively, with a difference of 23⁰. Finally, ϕ3 shows a syn-clinal orientation of ring D with a difference of ca 10⁰ between the two molecules.
Compound II, (Fig. 2 ), is a positional isomer of I, having a chloro-4-vinylbenzene attached to C2 atom (Scheme 1). It is the structure with ∠AB angle smaller than Ib and larger than Ia. In its molecular structure, unlike Ia, ϕ1 shows that ring B assumes an anti-clinal orientation. The change of position of the chlorine atom causes a significant geometric change concerning compounds I and III. In II the rings C and D are oppositely oriented compared with the same rings in the other compounds studied here. The values of ϕ2 indicate similarity in the molecular set and can be divided into two groups, one containing the molecules Ib, II and IIIa and the other containing the molecules Ia and IIIb. In compound II, ϕ2 and ϕ3 assume anti-periplanar and a syn-clinal orientations, respectively.
Fig. 2.
The molecular structure of II showing the atom-labeling scheme. Displacement ellipsoids are drawn at the 50% probability level and H atoms have been omitted.
Compound III differs from I only in the presence of the bromo-2-vinylbenzene bonded to the C2 atom (Scheme 1). The two independent molecules in the ASU were labeled as IIIa and IIIb (Fig. 3 ). The overlay of these structures indicates a difference of 14⁰ in ∠AB angle for IIIa and IIIb (Fig. S49). When compounds I, II and III are compared (∠AB angle), it is possible to distinguish two sets with similar values: first with IIIb and Ia, and second with Ib, II, and IIIa molecules. The decreasing of ∠AB angle value regarding chlorine dihydroquinolinones is Ia, IIIb, II, IIIa, and Ib (Table 3). There is no significant difference between ϕ1 in the molecules of compounds I and III, but this torsion in II is, ca 10⁰ less twisted. The values of ϕ2 show that ring C assumes an anti-periplanar orientation in IIIa while in IIIb it is anti-clinal with a difference of ca 17⁰. ϕ2 values are similar for Ib, II and IIIa and being ca 20⁰ smaller for Ia and IIIb. The orientation of ring D (ϕ3) in IIIa is anti-clinal while in IIIb is syn-clinal with a difference ca 17⁰.
Fig. 3.
The two independent molecules of compound III, showing the atom-labeling scheme: (a) molecule IIIa, (b) molecule IIIb. To clarify, in (b) the labeling scheme shows only non-carbon atoms. The labeling scheme for C atoms in (b) follows the same way as presented in (a). Displacement ellipsoids are drawn at the 50% probability level and H atoms have been omitted.
The crystal structure of these chlorine dihydroquinolinones is stabilized by C–H⋯O and C–H···halogen hydrogen bonds listed in Table 4 . Although these interactions are weak, we are interested in how they affect the packing and if the different substituents on ring C lead to different arrangements. In the molecular packing in the unit cell of compound I, two Ib molecules are involved in C27A—H27A···O3A interactions leading to the formation of a dimer along the b axis. Meanwhile, one Ia molecule links to these dimer, along the c axis, through three discrete contacts [26],: C8A—H8A···O3, C13A—H13A···O2, C14–H14⋯O2A (Fig. 4 ).
Table 4.
Hydrogen-bond geometry (Å, °) of I, II and III.
| D—H···A | D—H | H···A | D···A | D—H···A |
|---|---|---|---|---|
| I | ||||
| C14–H14⋯O2Ai | 0.95 | 2.46 | 3.374 (3) | 162 |
| C8A—H8A···O3ii | 0.95 | 2.36 | 3.138 (3) | 138 |
| C13A—H13A···O2i | 0.95 | 2.38 | 3.326 (3) | 175 |
| C27A—H27A···O3Aii | 0.95 | 2.50 | 3.175 (3) | 128 |
| II | ||||
| C6–H6⋯O2iii | 0.95 | 2.44 | 3.3531 (16) | 160 |
| III | ||||
| C8–H8⋯O3Ai | 0.95 | 2.40 | 3.161 (3) | 137 |
| C13–H13⋯O2Aiv | 0.95 | 2.31 | 3.245 (3) | 169 |
| C21–H21⋯O1Av | 0.95 | 2.55 | 3.458 (4) | 159 |
| C25–H25⋯O3i | 0.95 | 2.50 | 3.305 (3) | 142 |
| C14A—H14A···O2iv | 0.95 | 2.46 | 3.385 (3) | 165 |
| C19–H19⋯Br1iii | 0.95 | 2.82 | 3.626 (3) | 143 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, −y, −z+1; (iii) x+1, y, z; (iv) −x+1, −y+2, −z+1; (v) x, y, z+1.
Fig. 4.
A partial packing view of I showing the dimer formed by C27A—H27A···O3A interaction (1), and the discrete contacts C8A—H8A···O3 (2), C13A—H13A···O2 (3), and C14–H14⋯O2A (4). For clarity, H atoms not involved in the motif have been omitted.
Because this is a discrete, localized contact, there is no propagation to an extended structure through this contact. The molecular packing of compound II is not as varied as that of compound I. This arrangement can be described as chains that extend along the a axis by molecules that are associated through a C6–H6⋯O2 interaction (Fig. 5 ). The molecular packing of compound III seems remarkably similar to compound I. The differences indicate the carbonyl groups are not related to interactions in the packing of compound I. Two molecules of IIIa are arranged by C25–H25⋯O3 interaction as a dimer along the b axis, and by C19–H19⋯Br1 interaction as a chain along the a axis. The molecules of IIIb are connected to this arrangement by discrete contacts: C13–H13⋯O2A, C14–H14A ···O2, and C–H8⋯O3A along to the b axis, while the C21–H21⋯O1A interaction grows the packing along to the c axis. (Fig. 6 ).
Fig. 5.
A partial packing view of II showing the chains formed by C6–H6⋯O2 interaction. For clarity, H atoms not involved in the motif have been omitted.
Fig. 6.
A partial packing view of III, showing the dimer and the chain formed by C25–H25⋯O3 and C19–H19⋯Br1 interactions, respectively (a). The discrete contacts C13–H13⋯O2A (1), C14A—H14A···O2 (2), C8–H8⋯O3A (3), and C21–H21⋯O1A (4) join molecules of IIIa and IIIb in the packing (b); the motif in (a) is presented in (b) in light grey to better visualization. For clarity, H atoms not involved in interactions have been omitted.
3.2.2. Fluorine dihydroquinolinones
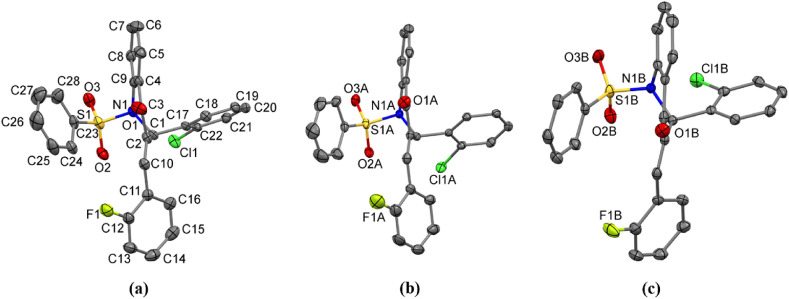
Compound IV has an ortho-chlorobenzene attached to C1 atom and a fluoro-2-vinylbenzene attached to the C2 atom (Scheme 1). The three independent molecules in ASU were labeled as IVa, IVb and IVc (Fig. 7 ). As listed in Table 5 , these three structures present a variation of 10⁰ in their ∠AB angle being the descending order equal to IVb, IVa, and IVc (Fig. S50). The torsion related to ring B (ϕ1) is syn-clinal oriented for IVb and IVc, and anti-clinal oriented for IVa, with a variation of ca 12⁰ between them. The values for ϕ2 show that ring C has more mobility than ring B, with a ca 28⁰ of variation in this torsion. The orientation of ring D (ϕ3) varies ca 19⁰, in IVa it is almost perpendicular to SO2 group while in IVb and IVc it is almost parallel to O2a and O2b atoms, respectively.
Fig. 7.
The three independent molecules of IV showing the atom-labeling scheme: (a) molecule IVa, (b) molecule IVb, and (c) molecule IVc. To clarify, in (b) and (c) the labeling scheme shows only non-carbon atoms. The labeling scheme for C atoms in (b) and (c) follow the same way as presented in (a). Displacement ellipsoids are drawn at the 50% probability level and H atoms have been omitted.
Table 5.
Torsion angles and least-square planes of aromatic rings of IV and V.
| Molecule | ϕ1 | ϕ2 | ϕ3 | ∠AB |
|---|---|---|---|---|
| IVa | −141.7(2) | 169.45(17) | 91.06(19) | 58.77 |
| IVb | −153.9(2) | 150.24(17) | 72.24(16) | 49.30 |
| IVc | −150.1(2) | −178.22(16) | 75.28(16) | 59.71 |
| Va | −157.7(3) | 175.3(2) | 78.1(2) | 44.53 |
| Vb | −144.4(3) | 173.7(2) | 72.1(2) | 62.80 |
| Vc | −149.7(3) | 161.1(2) | 82.7(2) | 52.79 |
ϕ1 = C2–C10–C11–C12; ϕ2 = C2–C1–C17–C22; ϕ3 = N1–S1–C23–C28.
Compound V is like compound IV but has an ortho-bromobenzene attached to C1 atom. The three independent molecules in the ASU were labeled as Va, Vb and Vc (Fig. 8 ). These molecules present a variation of 18⁰ in their ∠AB angle being the descending order equal to Va, Vc and Vb (Fig. S51). When ∠AB angle values for all fluorine dihydroquinolinones are compared, it is observed a decreasing order Va, IVb, Vc, IVa, IVc, and Vb (Table 5). Just like for chlorine dihydroquinolinones, it is not possible to figure out a clear correlation between the substitution of halogens and the AB ring orientation of these fluorine dihydroquinolinones. The torsion ϕ1 is anti-periplanar oriented in Va while in Vb and Vc it is anti-clinal. The values of ϕ2 show that ring C assumes an anti-periplanar orientation in all molecules of compound V with a difference of ca 14⁰. The torsion ϕ3 shows ring D in syn-clinal orientation in all molecules of compound V, with a change of ca 10⁰. These values, when compared with compound IV, shows it having more mobility in the molecule of IVa, being ca 9⁰ larger than in Vc.
Fig. 8.
The three independent molecules of compound V, showing the atom-labeling scheme: (a) molecule Va, (b) molecule Vb, and (c) molecule Vc. To clarify, in (b) and (c) the labeling scheme shows only non-carbon atoms. The labeling scheme for C atoms in (b) and (c) follow the same way as presented in (a). Displacement ellipsoids are drawn at the 50% probability level and H atoms have been omitted.
The molecular packing in the unit cell of compound IV and V is stabilized by C–H⋯O and C–H···halogen hydrogen bonds (Table 6 ). In compound IV, the contacts C5B–H5B⋯O3B, C18A—H18A···O2A, and C19A —H19A···Cl1A form and chains along the a axis. Molecules of IVa are linked through C26B–H26B⋯O3 and C15–H15⋯O3A interactions, leading to crystal packing along the b axis (Fig. 9 a). These motifs (molecules of IVb and IVc) are attached by the discrete contacts: C14A—H14A···O2B, C14B–H14B⋯O2A, C27A—H27A···O1, and C14B–H14B⋯O2A leading the packing to grow along to c axis (Fig. 9b).
Table 6.
Hydrogen-bond geometry (Å, °) for compounds IV and V.
| D—H···A | D—H | H···A | D···A | D—H···A |
|---|---|---|---|---|
| IV | ||||
| C5B–H5B⋯O3Bi | 0.95 | 2.48 | 3.104 (2) | 123 |
| C14A—H14A···O2Bii | 0.95 | 2.47 | 3.378 (3) | 161 |
| C14B–H14B⋯O2Aiii | 0.95 | 2.58 | 3.518 (3) | 168 |
| C15–H15⋯O3Aiv | 0.95 | 2.57 | 3.463 (3) | 156 |
| C18A—H18A···O2Aiv | 0.95 | 2.57 | 3.289 (2) | 133 |
| C26B–H26B⋯O3 | 0.95 | 2.60 | 3.326 (2) | 134 |
| C27A—H27A···O1v | 0.95 | 2.56 | 3.238 (2) | 129 |
| C19A—H19A···Cl1Aiv | 0.95 | 2.71 | 3.437 (3) | 155 |
| V | ||||
| C7A—H7A···O1B | 0.95 | 2.59 | 3.474 (3) | 155 |
| C8A—H8A···F1B | 0.95 | 2.54 | 3.381 (3) | 147 |
| C18A—H18A···O2Avi | 0.95 | 2.56 | 3.283 (3) | 133 |
| C18–H18⋯O2vii | 095 | 2.60 | 3256 (2) | 131 |
| C26A—H26A···O1Bviii | 0.95 | 2.39 | 3.335 (3) | 171 |
| C14B–H14B⋯O2 | 0.95 | 2.38 | 3.257 (3) | 154 |
| C18B–H18B⋯O2B8ix | 0.95 | 2.44 | 3.261 (3) | 145 |
| C24A—H24A···O1Avii | 0.95 | 2.62 | 3.346 (4) | 133 |
| C26–H26⋯O1x | 0.95 | 2.47 | 3.256 (4) | 140 |
| C27B–H27B⋯O1Aviii | 0.95 | 2.49 | 3.357 (3) | 152 |
| C27–H27⋯F1x | 0.95 | 2.52 | 3.342 (3) | 144 |
Symmetry codes: (i) x+1, y, z; (ii) x+1, y−1, z; (iii) x−1, y+1, z; (iv) x−1, y, z; (v) −x+1, −y+1, −z; (vi) x,-y+3/2,+z+1/2; (vii) x,-y+3/2,+z-1/2; (viii) -x+1,-y+1,-z+2; (ix) x,-y+1/2,+z+1/2; (x) -x,-y+2,-z+1.
Fig. 9.
A partial packing view of IV showing (a) the two chains formed by C18A—H18A ···O2A (1) and C19A —H19A···Cl1A (2), and C5–H5B⋯O3B (3) interactions. In addition, the interactions C15–H15⋯O3A (4) and C26B–H26B⋯O3 (5) responsible to connect molecules of IVa to the chains. In (b) are shown the discrete contacts C14A—H14A···O2B (6), C14B–H14B⋯O2A (7), C27A—H27A···O1 (8), C14B–H14B⋯O2A (9), responsible for grow the packing along the c axis. The motif represented in (a), is presented in light grey. For clarity, H atoms not involved in the motif have been omitted.
In the crystal packing of compound V, two molecule of Va are arranged as dimers through C26–H26⋯O1, , and C27–H27⋯F1, , interactions. C14B–H14B⋯O2 interaction attaches those dimers. Along to a axis there is a molecular pair formed by the following interactions: C8A—H8A···F1B and C7A—H7A···O1B (Fig. 10 ). The interaction C18–H18⋯O2, and C18A—H18A···O2A create two independent chains while C24A—H24A···O1A create a chain, along to c axis. In the same orientation to these chains there is one more arrangement formed by C18B–H18B⋯O2B interaction. This chain connects to others along to a axis, via C26A—H26A···O1B and C27B–H27B⋯O1A interactions (Fig. 10).
Fig. 10.
A partial packing view of V showing the dimer and molecular pair in light grey, interactions C26–H26⋯O1 (1), C27–H27⋯F1 (2), C14B–H14B⋯O2 (3), C8A—H8A···F1B (4), nC7A—H7A···O1B (5). The chains formed by C18–H18⋯O2 (6) (grey); C18A—H18A···O2A (7) and C24A—H24A···O1A (8) (dark grey); and C18B–H18B⋯O2B (11) (light blue). The interactions C27B–H27B⋯O1A (9) and C26A—H26A···O1b (10) are discrete and connect two chains. For clarity, H atoms not involved in the motif have been omitted.
4. Final remarks
In this paper we briefly presented a comparison between chlorine and fluorine dihydroquinolinones. Due to the molecular similarities, we discuss the comparison of compounds I and IV, and compounds III and V. Concerning I and IV, the difference is in the halogen at the ortho position of ring B, chlorine, and fluorine, respectively. The decreasing order of ∠AB angle is IVb, Ia, IVa, IVc and Ib. The torsion ϕ1 in I shows a smaller variation when compared to compound IV, ca. 0.3⁰ and 12⁰, respectively. Regarding torsion ϕ2, both compounds present similar values (23⁰ for I and 19⁰ for IV). The torsion angle ϕ3 in IV is larger than I (10⁰ and 19⁰, respectively). Regarding molecular packing, the chlorine compound forms and chains, while the fluorine compound forms chains. All those chains grow along the a axis. Finally, discrete contacts join these chains along the b and c axis.
In contrast, compounds III and V differ in the halogen in the ortho position of ring B, chlorine, and fluorine, respectively. In terms of ∠AB values, the decreasing order is Va, Vc, IIIb, IIIa, and Vb. The torsion ϕ1 in III is smaller than V (1⁰ and 13⁰, respectively). Regarding the torsion ϕ2, both compounds present similar values (17⁰ in III and 14⁰ in V). Moreover, the torsion ϕ3 to III is larger than V (17⁰ and 10⁰, respectively). These small alterations occur likely due to the change in radius of the halogen atoms. In terms of molecular packing both compounds have dimers but in III it is related to SO2 group while in the compound V it is related to the carbonyl group. In compound III these dimers are arranged along the c axis while in compound V they are along the b axis. Also, the molecular packing in V present two and one chain along the c axis. All those observations show that the change in position and kind of halogen atom attached in ring C play a significant role in the conformation of the studied compounds. On the other hand, the packing of these compounds is more susceptible to variations when the substituent position changes.
CRediT authorship contribution statement
Wesley F. Vaz: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Lidiane J. Michelini: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing - original draft, Writing - review & editing. Gerlon A.R. Oliveira: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing - original draft, Writing - review & editing. Luciano M. Lião: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing - original draft, Writing - review & editing. Caridad N. Perez: Data curation, Formal analysis, Investigation, Methodology, Visualization, Resources, Writing - original draft, Writing - review & editing. Allen G. Oliver: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Hamilton B. Napolitano: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil); Grant ID: 88881.190472/2018-01 - Finance Code 001, Conselho Nacional de Desenvolvimento Científico e Tecnológico and Fundação de Amparo à Pesquisa do Estado de Goiás (FAPEG); Grant ID: 20170267000634.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molstruc.2020.128559.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jain S., Chandra V., Kumar Jain P., Pathak K., Pathak D., Vaidya A. Comprehensive review on current developments of quinoline-based anticancer agents. Arab. J. Chem. 2019;12:4920–4946. doi: 10.1016/j.arabjc.2016.10.009. [DOI] [Google Scholar]
- 2.Felczak A., Bernat P., Różalska S., Lisowska K. Quinoline biodegradation by filamentous fungus Cunninghamella elegans and adaptive modifications of the fungal membrane composition. Environ. Sci. Pollut. Res. 2016;23:8872–8880. doi: 10.1007/s11356-016-6116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng Y.-M., Zeng H.-H., Liu F.-M. Synthesis and structure characterization of 1,5-benzothiazepine derivatives bearing quinoline moiety. J. Heterocycl. Chem. 2016;53:887–893. doi: 10.1002/jhet.1519. [DOI] [Google Scholar]
- 4.Moreira C.A., Custódio J.M.F., Vaz W.F., D’Oliveira G.D.C., Noda Perez C., Napolitano H.B. A comprehensive study on crystal structure of a novel sulfonamide-dihydroquinolinone through experimental and theoretical approaches. J. Mol. Model. 2019;25:205. doi: 10.1007/s00894-019-4091-7. [DOI] [PubMed] [Google Scholar]
- 5.Michelini L.J., Vaz W.F., D’Oliveira G.D.C., Pérez C.N., Napolitano H.B. Analysis of two novel 1–4 quinolinone structures with bromine and nitrobenzyl ligands. J. Mol. Model. 2019;25:55. doi: 10.1007/s00894-019-3937-3. [DOI] [PubMed] [Google Scholar]
- 6.Michelini L., Vaz W., Naves L., Pérez C., Napolitano H. Synthesis, characterization and conformational analysis of two novel 4(1H)-Quinolinones. J. Braz. Chem. Soc. 2020;31:66–78. doi: 10.21577/0103-5053.20190124. [DOI] [Google Scholar]
- 7.d’Oliveira G.D.C., Custodio J.M.F., Moura A.F., Napolitano H.B., Pérez C.N., Moraes M.O., Prókai L., Perjési P. Different reactivity to glutathione but similar tumor cell toxicity of chalcones and their quinolinone analogues. Med. Chem. Res. 2019;28:1448–1460. doi: 10.1007/s00044-019-02384-8. [DOI] [Google Scholar]
- 8.Vaz W.F., Custodio J.M.F., D’Oliveira G.D.C., Neves B.J., Junior P.S.C., Filho J.T.M., Andrade C.H., Perez C.N., Silveira-Lacerda E.P., Napolitano H.B. Dihydroquinoline derivative as a potential anticancer agent: synthesis, crystal structure, and molecular modeling studies. Mol. Divers. 2020 doi: 10.1007/s11030-019-10024-x. [DOI] [PubMed] [Google Scholar]
- 9.Vaz W.F., D’Oliveira G.D.C., Perez C.N., Neves B.J., Napolitano H.B. Machine learning prediction of the potential pesticide applicability of three dihydroquinoline derivatives: syntheses, crystal structures and physical properties. J. Mol. Struct. 2020;1206:127732. doi: 10.1016/j.molstruc.2020.127732. [DOI] [Google Scholar]
- 10.Jean J., Farrell D.S., Farrelly A.M., Toomey S., Barlow J.W. Design, synthesis and evaluation of novel 2,2-dimethyl-2,3-dihydroquinolin-4(1H)-one based chalcones as cytotoxic agents. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pejović A., Drabowicz J., Cieslak M., Kazmierczak-Baranska J., Królewska-Golińska K. Synthesis, characterization and anticancer activity of novel ferrocene containing quinolinones: 1-Allyl-2-ferrocenyl-2,3-dihydroquinolin-4(1H)-ones and 1-allyl-2-ferrocenylquinolin-4(1H)-ones. J. Organomet. Chem. 2018;873:78–85. doi: 10.1016/j.jorganchem.2018.08.004. [DOI] [Google Scholar]
- 12.Koszuk J., Bartosik T., Wojciechowski J., Wolf W.M., Janecka A., Drogosz J., Długosz A., Krajewska U., Mirowski M., Janecki T. Synthesis of 3-Methylidene-1-tosyl-2,3-dihydroquinolin-4(1 H )-ones as potent cytotoxic agents. Chem. Biodivers. 2018;15 doi: 10.1002/cbdv.201800242. [DOI] [PubMed] [Google Scholar]
- 13.Politanskaya L., Rybalova T., Zakharova O., Nevinsky G., Tretyakov E. p-Toluenesulfonic acid mediated one-pot cascade synthesis and cytotoxicity evaluation of polyfluorinated 2-aryl-2,3-dihydroquinolin-4-ones and their derivatives. J. Fluor. Chem. 2018;211:129–140. doi: 10.1016/j.jfluchem.2018.04.005. [DOI] [Google Scholar]
- 14.Yoon J.H., Lee J.Y., Lee J., Shin Y.S., Jeon S., Kim D.E., Min J.S., Song J.H., Kim S., Kwon S., Jin Y., Jang M.S., Kim H.R., Park C.M. Synthesis and biological evaluation of 3-acyl-2-phenylamino-1,4-dihydroquinolin-4(1H)-one derivatives as potential MERS-CoV inhibitors. Bioorg. Med. Chem. Lett. 2019;29:126727. doi: 10.1016/j.bmcl.2019.126727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todeschini R., Consonni V., Gramatica P. Chemometrics in QSAR. In: Brown S., Tauler R., Walczak R., editors. Compr. Chemom. Vol. 4. Elsevier; Oxford: 2009. pp. 129–172. [Google Scholar]
- 16.a Martins J.P., Ferreira M.M.C. Qsar modeling: a new open source computational package to generate and validate qsar models. Quim. Nova. 2013;36 doi: 10.1590/S0100-40422013000400013. 554-U250. [DOI] [Google Scholar]
- 17.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009;42:339–341. doi: 10.1107/S0021889808042726. [DOI] [Google Scholar]
- 18.Sheldrick G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 19.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nardelli M. Parst 95 – an update to PARST : a system of Fortran routines for calculating molecular structure parameters from the results of crystal structure analyses. J. Appl. Crystallogr. 1995;28 doi: 10.1107/S0021889895007138. 659–659. [DOI] [Google Scholar]
- 21.Spek A.L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003;36:7–13. doi: 10.1107/S0021889802022112. [DOI] [Google Scholar]
- 22.d’Oliveira G., Moura A., de Moraes M., Perez C., Lião L. Synthesis, characterization and evaluation of in vitro antitumor activities of novel chalcone-quinolinone hybrid compounds. J. Braz. Chem. Soc. 2018;29:2308–2325. doi: 10.21577/0103-5053.20180108. [DOI] [Google Scholar]
- 23.Kim J.H., Ryu H.W., Shim J.H., Park K.H., Withers S.G. Development of new and selective trypanosoma cruzi trans-sialidase inhibitors from sulfonamide chalcones and their derivatives. Chembiochem. 2009;10:2475–2479. doi: 10.1002/cbic.200900108. [DOI] [PubMed] [Google Scholar]
- 24.Ma H., Zhou X., Wei D., Cao J., Shi C., Fan Y., Huang G. KHCO 3 - and DBU-promoted cascade reaction to synthesize 3-Benzyl-2-phenylquinolin-4(1 H)-ones. Chem. Asian J. 2016;11:2829–2833. doi: 10.1002/asia.201600901. [DOI] [PubMed] [Google Scholar]
- 25.de Castro M.R.C., Naves R.F., Bernardes A., da Silva C.C., Perez C.N., Moura A.F., de Moraes M.O., Martins F.T. Tandem chalcone-sulfonamide hybridization, cyclization and further Claisen–Schmidt condensation: tuning molecular diversity through reaction time and order and catalyst. Arab. J. Chem. 2020;13:1345–1354. doi: 10.1016/j.arabjc.2017.11.005. [DOI] [Google Scholar]
- 26.Grell J., Bernstein J., Tinhofer G. Graph-set analysis of hydrogen-bond patterns: some mathematical concepts. Acta Crystallogr. Sect. B Struct. Sci. 1999;55:1030–1043. doi: 10.1107/S0108768199007120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.