Graphical abstract
Keywords: Transcriptomics, RNASeq, Normalization, TOMOSeq, DESeq, Median-of-ratios
Abstract
The merit of RNASeq data relies heavily on correct normalization. However, most methods assume that the majority of transcripts show no differential expression between conditions. This assumption may not always be correct, especially when one condition results in overexpression. We present a new method (NormQ) to normalize the RNASeq library size, using the relative proportion observed from RT-qPCR of selected marker genes. The method was compared against the popular median-of-ratios method, using simulated and real-datasets. NormQ produced more matches to differentially expressed genes in the simulated dataset and more distribution profile matches for both simulated and real datasets.
1. Background
RNASeq has become one of the most useful tools in molecular biology, especially as it allows for the comparison of mRNA expression levels between different conditions [1]. However, to accomplish this, it is important that the raw counts of the mapped transcripts are normalized correctly to compensate for innate biases in the technology [2], [3]. To minimize these effects, many methods have been proposed on how to normalize the data, such as by library size or adjusting the distribution of the individual read counts [4], [5]. However, each of these normalization methods have their advantages and limitations, typically being appropriate for certain experimental designs [2].
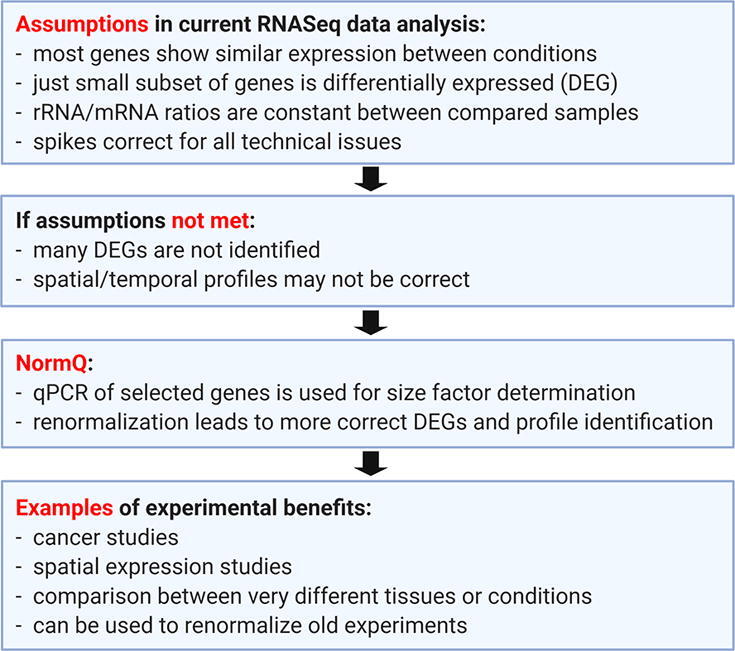
Most of these normalization methods are limited by their assumption that the majority of the transcripts is not differentially expressed. DESeq2, one of the most popular library-based tools, abides by this assumption, and utilizes the median-of-ratios method to normalize the library size by calculating a size factor that can best fit the data [6]. This size factor is calculated using either a spike-in control (control-based) or the gene counts from the biological replicates (average-bulk). However, given this innate assumption, in the case of global scale differential expression, the average-bulk method may underestimate the true number of differentially expressed genes (DEGs) [7]. Spike-in controls may help to offset this bias, as it does not create any major assumptions for the genes of interest. However, it requires that factors affecting the spike-in controls are also equally affecting all the genes and that the concentration of the spike-in is uniform between the samples, which may not necessarily be true [2], [8]. Other normalization techniques, example RUVs, can also account for technical variability by using validated stable non-DEGs [9]. However, the identification of such stable homogeneously expressed genes can be very difficult.
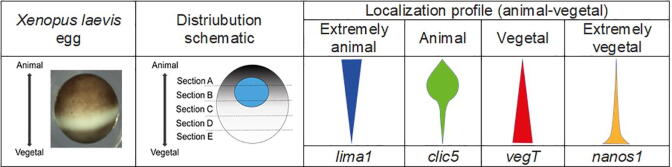
There are several scenarios where a global shift in transcript expression may be observed. This may be an issue affecting cancer studies due to overexpression of master regulator genes or the comparison between very different tissue types [7], [10], [11]. Another type of RNASeq experimentation that may also demonstrate global scale differential expression are those involving the use of TOMOSeq [12], [13]. Spatial profiling with RNASeq, based on a defined sample, such as a single cell, dissected into segments offers very unique challenges. Firstly, the amount of total RNA is not necessarily the same in all sections [14]. Secondly, if the cell has compartmentalization of selected transcripts (See Fig. 1), the assumption that the majority of genes between sections shows no differential expression is likely incorrect [12]. Therefore, given these limitations, most software for assessing DEGs may not produce the best results when analyzing cryosectioned eggs. These issues have perhaps not been blatantly apparent since many of the historic approaches to TOMOSeq have been limited primarily to separating the egg into two parts, for example the animal versus vegetal region [15]. It is possible that these types of issues have affected other RNASeq studies but have gone undetected.
Fig. 1.
Schematic of the localization profile of selected mRNAs representing the four major profiles observed in Xenopus laevis egg.
Our laboratory was one of the first to analyze sub-compartmentalization of maternal transcripts in eggs [12], [14], [16]. To do this we developed approaches to deal with some of the current limitations during normalization. Our study presents a new method for size factor estimation based on RT-qPCR data obtained from selected genes with known localization profiles (normalization by RT-qPCR – NormQ method). NormQ should also be useful for other RNASeq studies where a global shift in expression is observed or where the total transcript abundance between samples is very different.
We compared the results from NormQ with those calculated using DESeq2’s traditional average-bulk and control-based median-of-ratios methods. We analyzed both a Simulated-TOMOSeq dataset and also real datasets from TOMOSeq performed on maternal transcripts from the Xenopus laevis and Acipenser ruthenus egg models and present evidence in support for the use of RT-qPCR normalization of RNASeq data under certain conditions.
2. Results
2.1. TOMOSeq simulation
2.1.1. Effect of normalization on distribution of sections
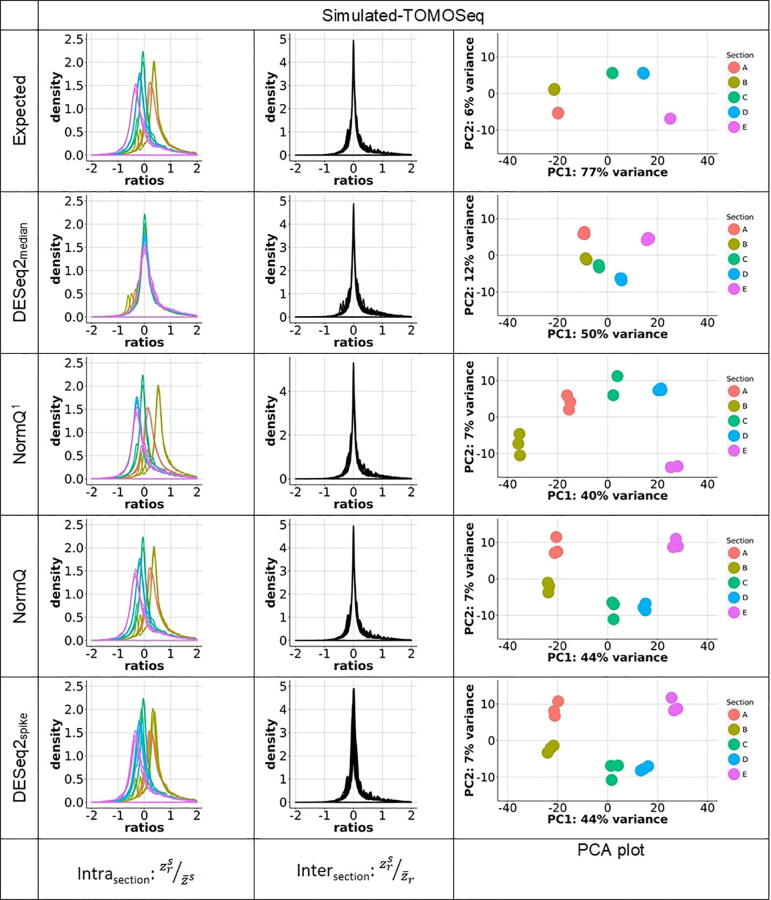
The simulation produced expected intra-sample gene counts that had a mean expression ratio that was skewed relative to each different section, reflective of the expected biased transcript population per section (See Fig. 2). Only NormQ and DESeq2spike were able to mimic the intra-section expected distribution. NormQ1 was also able to mirror most of the expected intra-section distribution, while DESeq2median was unable to provide comparable resolution (See Fig. 2). All normalization techniques were able to normalize effectively between the inter-sections when compared to the expected distribution and also showed good clustering of inter-section replicates of the top 5000 variable genes. However, only NormQ and DESeq2spike had similar intra-section separation of the 5000 genes compared to the expected, while DESeq2median had less separation and NormQ1 had more separation between sections A and B.
Fig. 2.
degCheckFactor analysis of the proportion of the normalized counts for each gene relative to its mean count across the different sections (intra-section) or all same sections (inter-section) from the Simulated-TOMOSeq data and the resulting Principle Component Analysis for the 5000 genes showing the most variance. Replicate number is represented as r, a given egg section as s and gene as z. Intra-section analysis shows how well the normalization technique maintains separation between the different sections, while the inter-section analysis shows how well the median-of-ratios method can normalize between replicates.
2.1.2. Effect of normalization on detected differentially expressed genes
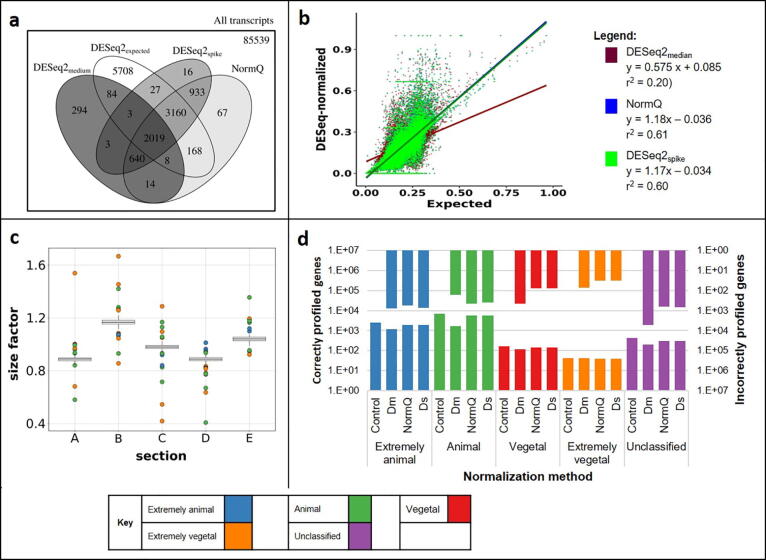
More DEGs were detected when using the NormQ (7009) versus the DESeq2median (3065) or DESeq2spike (6801) in the Simulated-TOMOSeq experiments (See Fig. 3a). However, in regard to true DEGs, DESeq2spike and NormQ were able to identify 47% and 48% of the expected DEGs (11177) respectively while DESeq2median was only able to identify 19% (See Fig. 3a). The AUC-ROC showed an improved performance when using DESeq2spike and NormQ versus the DESeq2median (Table 1). Additionally, NormQ and DESeq2spike were the only two methods that gave substantial gene localization profile matches compared to the expected profiles for the correctly identified DEGs, the DEGs that were shared by all normalization methods and for all genes (DEGs and non-DEGs) (See Table 1; Fig. 3d; Additional file 1: Table S1).
Fig. 3.
a) Differences in the number of significant (padj < 0.1) DEGs detected between sections when using different normalization techniques for the Simulated-TomoSeq. b) Correlation between each section’s gene count proportion (relative to the egg) for the normalized data, versus those from the expected proportions in the Simulated-TOMOSeq. c) Distribution of the size factors obtained from each marker gene for each replicate and section for the Simulated-TOMOSeq. d) Number of marker genes detected within each profile after use of each normalization method. The localization profile comparison for the Simulated-TOMOSeq was assessed using genes that were commonly detected in all three normalization methods. The bottom axis shows the number of genes that were correctly identified within the given profile while the top axis shows the number of genes that were incorrectly profiled. The y-axis represents the log(10) of the number of detected genes. “Dm” represents DESeq2median while Ds represents DESeq2spike.
Table 1.
Assessments of the Area under the Receiver Operating Characteristic (ROC) curve (AUC) and also the number of profile matches for correctly identified DEGs after using each normalization method on the Simulated-TOMOSeq data.
| Normalization method | AUC-ROC of DEGs | Profile matches for DEGs shared with DESeq2expecteda | Profile matches for DEGs shared amongst all normalization methodb |
|---|---|---|---|
| DESeq2spike | 0.995 | 90% (4667/5188)) | 92% (1814/1978) |
| NormQ | 0.989 | 92% (4907/5334) | 94% (1853/1978) |
| DESeq2none | 0.952 | 77% (4325/5630) | 71% (1395/1978) |
| NormQ1 | 0.76 | 77% (4460/5818) | 71% (1394/1978) |
| DESeq2mean | 0.354 | 37% (771/2060) | 37% (739/1978) |
(correct profile match ⋂ correctly identified DEGs with no missing replicate data)/correctly identified DEGs with no missing replicate data.
(correct profile match ⋂ correctly identified DEGs shared by all normalization methods with no missing replicate data)/correctly identified DEGs shared by all normalization methods with no missing replicate data.
There was a higher correlation between the relative section proportions as derived from NormQ (r2 = 0.61) marker genes and those from the expected data for the Simulated-TOMOSeq relative to those from the other normalization methods (See Fig. 3b). The DESeq2spike produced the second best fit (r2 = 0.60) with DESeq2median performing the worst (r2 = 0.20) (See Fig. 3b).
Analysis of the selection process for the marker genes when using all genes for the calculation of the NormQ size factor, found that on average there was a 0.24 probability of selecting an outlier gene from the extreme animal, 0.22 from animal, 0.24 from vegetal and 0.34 from extreme vegetal localization categories for all egg sections. AUC-ROC analysis showed that varying the number of genes used for calculating the size factor, to as low as three genes, or selecting genes from a singular localization group, did not drop the performance lower than 95% (see Additional file 2: Fig. 1S, Additional file 1: Table S2).
2.2. Real dataset
2.2.1. Effect of normalization on distribution of sections
Similar to the simulation, the NormQ normalized data showed a clearly distinct intra-section distribution while maintaining similar mean inter-section distributions (see Additional file 2: Fig. 2S, Fig. 3S). The DESeq2spike normalization which was performed only on the A. ruthenus dataset, showed an inability to adequately normalize between inter-sections (see Additional file 2: Fig. 3S). Both X. laevis and A. ruthenus DESeq2median normalization showed no intra-section separation (see Additional file 2: Fig. 2S, Fig. 3S).
Principle Component Analysis (PCA) of the 5000 most variable genes from the normalized data showed better clustering between replicates and separation between sections when using NormQ for the X. laevis data (see Additional file 2: Fig. 2S). The cluster profiles and distributions of replicates appeared consistent between all normalization methods for the A. ruthenus data (see Additional file 2: Fig. 3S). However, two replicates showed good reproducibility, while one replicate consistently remained deviated from the clusters but only within one dimension (see Additional file 2: Fig. 3S).
2.2.2. Effect of normalization on detected differentially expressed genes
More DEGs were detected when using the NormQ normalization technique versus the DESeq2median or DESeq2spike in both the X. laevis (x2.8 DESeq2median) and A. ruthenus (x2.9 DESeq2median, x1.1 DESeq2spike) experiments (see Additional file 2: Fig. 4S). Even though almost all of the DEGs for the X. laevis DESeq2median normalized data were identified using the NormQ, only 22% percent of these shared genes produced a similar gradient profile across the sections after supervised clustering analysis (Additional file 1: Table S3). A similarly relatively low proportion (62%) of genes shared related gradient profiles for the DEGs common between the NormQ and DESeq2median A. ruthenus normalized data (Additional file 1: Table S3). A larger proportion of the DEGs (91%) shared between the DESeq2spike and NormQ methods were observed to have similar profiles across the sections in A. ruthenus (Additional file 1: Table S3). Many of the shared DEGS that were of a different profile in the DESeq2median were shifted to the neighboring profile in NormQ (Additional file 1: Table S3). Additionally, many of the shared DEGS with undefined profiles when using DESeq2median were shifted to the animal profile when using the NormQ method (Additional file 1: Table S3).
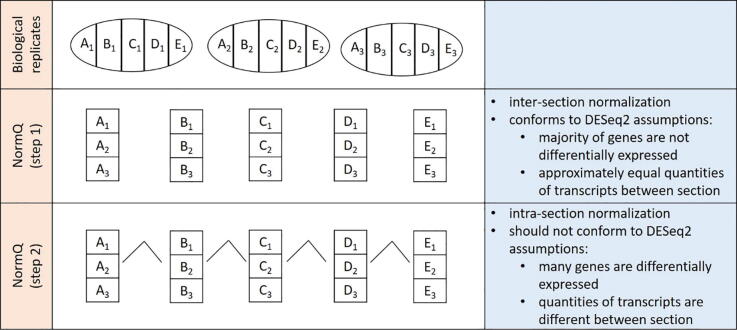
Fig. 4.
Schematic showing the normalization steps used for the NormQ method.
A larger proportion of the marker genes (X. laevis = 87.5%; A. ruthenus = 95.5%) were found to match the expected localization profile when using the NormQ method compared to the DESeq2median and DESeq2spike methods (X. laevis DESeq2median = 50%; A. ruthenus DESeq2median = 72.7%, A. ruthenus DESeq2spike = 86.4%) (t-test: p-value < 0.05) (Additional file 1: Table S1, Additional file 2: Fig. 5S, Fig. 6S). Similar to the shared DEGs, the marker genes that had a differing localization profile when using the NormQ method, only had a shift in localization profiles particularly from extremely animal to animal (see Additional file 2: Fig. 4S). Additionally, the majority of the marker gene localization profile mismatch observed for DESeq2median was into the unclassified category in X. laevis but also within the vegetal and animal categories in A. ruthenus (see Additional file 2: Fig. 4S). The assessment of the published X. laevis RT-qPCR profiles showed a higher localization profile match with the NormQ (83%) versus the DESeq2median (73%) (t-test: p-value < 0.05) (Additional file 1: Table S1, Additional file 2: Fig. 7S). The expression profiles for the extra queried genes that were assessed but not used for the NormQ normalization of the X. laevis data are shown in the Additional file 2: Fig. 8S.
There was a higher correlation between the relative section proportions as derived from the NormQ normalized data for the marker genes versus the RT-qPCR data for the X. laevis (r2 = 0.90) and A. ruthenus (r2 = 0.95) data relative to those from the other normalization methods (see Additional file 2: Fig. 9S). The DESeq2spike produced the second best fit with DESeq2median performing the worst (see Additional file 2: Fig. 9S).
There were minimal outlying size factors for the X. laevis NormQ normalization (see Additional file 2: Fig. 9S, Additional file 1: Table S4). Replicates from sections A to D showed an overall low standard deviation from the mean size factor with very few outliers (see Additional file 2: Fig. 9S, Additional file 1: Table S4). On the contrary, section E across all three replicates had a large variation with extreme outlying variables (see Additional file 2: Fig. 9S, Additional file 1: Table S4). It appears that member in the extremely vegetal profiles may not be as effective to normalize the TOMOSeq data in X. laevis (see Additional file 2: Fig. 9S, Additional file 1: Table S4). All assessed profiles for A. ruthenus contained many outliers with most of them being concentrated within sections B and E (see Additional file 2: Fig. 9S). Only 12 out of the 29 outliers were unique with four being of extreme animal profile, six vegetal, one extreme vegetal and one undefined (Additional file 1: Table S4).
3. Discussion
Normalization based on marker genes has historically been limited to genes that are expected to show no differential expression between samples. However, genes displaying this stability can be very difficult to identify between two very contrasting conditions. Analysis of the shared non-DEGs between DESeq2median and NormQ methods showed that the count data was too low for these genes and the variance too high for them to be used effectively for the calculation of size factors. Therefore, for our particular scenario a homogeneously distributed transcript of adequate quantity may not be available.
In such cases, artificial spike-ins like the ERCC sequences have been recommended for use as an exogenous control. However, the calculated size factor can be affected significantly by small-unknown variations in the quantity of the spike-in between samples. This is potentially one of the issues that may have been observed for the DESeq2spike normalization used for the A. ruthenus data, as the size factor was unable to normalize the data adequately between the same sections. Another, perhaps more relevant explanation, may be the limitation of the TATAA spike used, since it only represented fragments that were either 1000 bp or 2000 bp in length. However, other research have also encountered issues when using spike-ins [9], [17].
It was suggested by Chen and colleagues that the differences in expression levels of specific marker genes between the different cell lines may be used retrospectively to normalize old RNASeq datasets that did not contain any spike RNAs [11]. However, we have not found any research to date that have utilized the known fold changes between DEGs as controls for normalization, even though there have been several researches that have demonstrated the correlation between fold changes of RT-qPCR and RNASeq data [18], [19]. It is understandable that the variation between gene counts for highly expressed genes can be variable. However, as observed it still allowed for a better representation of the data when verified against the expected RT-qPCR profiles. The DESeq2spike was able to capture more perturbation of genes than the DESeq2median. However, the NormQ method was able to capture the majority of this data from both the DESeq2spike and also the DESeq2median. In addition, unlike DESeq2median and DESeq2spike, the NormQ was also able to adequately normalize between inter-sections while maintaining the variation between intra-sections. This intra-section variation is reduced when using DESeq2median and results in the inability to detect the minor sub-localization profiles. In addition, it results in a disproportion of the transcripts per section as can be observed from the limited fit of the gene localization profiles when compared to NormQ.
Other factors that favor NormQ over DESeq2median lies in the innate requirements for the use of normalization based on the average-bulk median-of-ratios method. One of these is the reliance that the transcript quantity and composition should be relatively the same between conditions. To ensure that this is achieved, the total RNA is usually normalized to the same volume and concentration. This is essential for most differential analysis programs as they typically assume that the total transcript number is the same between conditions. However, this is not always the case, and has been highlighted as a major issue especially when comparing between two very different cell types or between two conditions showing varying global expressions [7]. This issue also affects TOMOSeq analysis, as each section is of a different volume and therefore can contain more transcripts just through passive diffusion [14]. Additionally, the active accumulation of transcripts within a region will also create transcript asymmetry within the egg, affecting the transcript composition per section [14]. The library preparation methodology utilized in this experiment still normalizes each section to have the same total RNA. However, by using the scaling factors derived from the second part of the normalization technique, we should have effectively scaled the library back to the original section size. This unfortunately would not be achieved using DESeq2median as the size factor would be unable to scale back the data to compensate for the original section size differences as it assumes an equal total transcript quantity.
Selection of an appropriate number and type of DEG to use for normalization may significantly affect the normalization process. The more marker genes from varying localization profiles that can be included for size factor calculation will always be beneficial. The Simulated-TOMOSeq data did not show a major decrease in the performance of detecting DEGs, even when varying the number of selected marker genes to three. However, as shown from the analysis of the outlying size factors (when using all genes) for the NormQ analysis of the Simulated-TOMOSeq, the probability of selecting a random gene that may contribute an outlying size factor ranged between 0.22 and 0.34. Therefore, an adequate number of genes should be selected to prevent inadvertently choosing a gene that contributes to an outlying size factor. Everaert and colleagues found that only a small proportion of their analyzed gene-set showed discordance between RNASeq and RT-qPCR [19]. They found that on average these particular genes had lower expression, shorter lengths, had fewer exons, lower read quality, and mapped to multiple regions [19]. Therefore, these parameters may also be useful when selecting for marker genes.
It is worth noting that there may be potentially other technical and biological factors that are still hindering the normalization process. These factors may include gene length, GC content and over abundant transcripts that create a bias that cannot be effectively captured and explained by the current normalization method. The use of a completed genome with detailed annotation should also be beneficial, especially for correct mapping and quantifying the expressed genes. Both, X. laevis and A. ruthenus have limited genome annotations, with A. ruthenus suffering from both an incomplete genome sequence and no reference annotation. This limitation of the absence of an annotated transcriptome for the A. ruthenus, may reflect the marginal improvement of normalization by both the NormQ and DESeq2spike.
The proposed method appears to work well on both the TOMOSeq for X. laevis and the Simulated-TOMOSeq while showing some improvement for the A. ruthenus. This method may be most suitable for research assessing the distribution of transcripts during spatial and temporal profiling, especially under conditions where the transcripts are expected to show asymmetry or overexpression. In theory, this technique should also be valid for other RNASeq application even for experiments where the majority of the transcripts are not differentially expressed. However, it would require that the assessed differentially expressed transcripts show similar relative ratios between conditions, with low variations between biological repeats (Table 2). This may be difficult for conditions that show DEGs but a high biological variation. However, it is possible that performing RT-qPCR on the same RNA extract as used for the RNASeq may help to alleviate this issue. Another benefit of this method is that many previous RNASeq data can be re-analyzed without having to redo a completely new RNASeq experiment. Additionally, it offers a simple approach for size factor calculation when both spike-ins and average-bulk normalization methods fail.
Table 2.
Recommendations for the selection of NormQ for RNASeq normalization.
| Recommendations | |
|---|---|
| 1 | Select well established marker genes that have a known distribution. If no marker genes are known, use DESeq2median or DESeq2spike to select at least five DEGs from each derived cluster profile, so as to reduce the probability (<0.005) of selecting outlier marker genes. |
| 2 | Ensure that the marker gene count across all replicates and sample section/condition are adequate (example >100). |
| 3 | Assess the relative abundance of the marker genes within each sample section/condition using RT-qPCR. |
| 4 | Use NormQ to renormalize the data. |
| 5 | Use degCheckFactor to assess the effectiveness of the size factors used. If the distribution between different sample sections/conditions are not well separated, then DESeq2median or DESeq2spike may be more appropriate methods as there is no asymmetry of your data. |
| 6 | Compare the NormQ, DESeq2median or DESeq2spike normalized data to the RT-qPCR derived profile to determine which technique best fits the data. |
4. Conclusion
NormQ offers a simple but still effective method to normalize RNASeq data. It relies on scaling and normalizing the count data based on the proportional distributions observed for a few marker genes as assessed by the more sensitive RT-qPCR method. As a result, it does not inherently assume that the majority of the genes are not differentially expressed. Using this method, we were able to correctly identify and profile more differentially expressed genes within a simulated dataset and also correctly profile more genes in our real RNASeq dataset, when compared to the commonly used median-of-ratios method. This method should be particularly helpful in situations where there is either overexpression between conditions or the presence of asymmetrical transcript distributions. Given the way NormQ works, it should also be valid even when the majority of genes are not differentially expressed as well.
5. Methods
5.1. Sample preparation for RNASeq
The workflow and experimentation for preparation of egg samples, transcript libraries, RNASeq sequencing and data post processing for transcripts from X. laevis eggs has been described previously [12]. Twenty eggs were embedded into a block of optimal cutting temperature medium (OCT) and cut into 30-µm slices. About 35 slices were prepared and sequentially pooled into 5 sections. In total, three biological replicates (each containing pool from 20 eggs) were prepared for sequencing. Total RNA was isolated using 500 µl of TRI Reagent (Sigma-Aldrich), followed with LiCl precipitation to remove inhibiting substances (more details about quality control are available in reference [12]). The RNASeq libraries were prepared using GeneRead rRNA Depletion Kit (Qiagene) to remove rRNA and TruSeq RNA Sample Preparation v.2 kit (Illumina). Library sequencing was performed at BGI (Shenzen, China) using HiSeq 2500 (Illumina), 50 bp pair-end. Ribosomal RNA reads were filtered out from the data using Sortmerna (v. 2.1), low quality reads were filtered out using Trimmomatic and final reads were mapped to the reference genome using STAR (v. 2.4.2a) [20], [21], [22]. A final count table was then generated using htseq-count [23].
A similar procedure was used for the processing of A. ruthenus eggs. The individual mature unfertilized egg was embedded in OCT and then cryosectioned into five sections (Sections A-E) (see Fig. 1) along their animal-vegetal axis and stored at −80 °C. Total RNA was isolated using 1 ml of TRI Reagent (Sigma, T9424), and purified using LiCl precipitation. RNA samples were diluted in 20 µl of 1xTE buffer (Invitrogen, 12090–15). The concentration of RNA was measured using the Nanodrop 2000 (Thermo Scientific), and the quality assessed using a Fragment Analyzer (AATI, Standard Sensitivity RNA analysis kit, DNF-471). In total, three biological replicates (three eggs) were prepared for sequencing. Libraries for RNASeq were comprised of 200 ng of total RNA with the addition of 50x diluted RNA Spike I (TATAA, RS25SI) and 50,000× diluted RNA Spike II (TATAA, RS25SII). Ribosomal RNAs were depleted using Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) (Illumina, MRZH116) and sequencing libraries prepared using SureSelect Strand-Specific RNA Library Prep for Illumina Multiplexed Sequencing (Agilent, G9691), beginning from Step 2 (fragmentation of RNA). The libraries were sequenced on a NextSeq 500 instrument in PE75 high-output mode.
Approximately 19 million raw sequencing reads per sample were obtained. Adaptor sequences and low quality reads were filtered using TrimmomaticPE (v. 0.36) with parameters, “CROP:70 HEADCROP:12 ILLUMINACLIP: TruSeq-PE3.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36” [20]. SortMeRNA (v. 2.1b) was used to remove remaining rRNA and mtRNA reads [22]. A de novo transcriptome was created using Trinity (v. 2.3.2) with default parameters and SS_lib_type specification as RF [24]. A genome guided de novo transcriptome was also created using an in-lab sequenced draft genome of A. ruthenus. The reads were first aligned using STAR (v. 2.5.2b) and then the genome guided de novo transcriptome was generated using Trinity [21], [24]. These two transcriptomes were merged using EvidentialGene (v. 17mar10) and a count table generated using kallisto (v. 0.43.1) [25], [26]. The data was deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO), accession GSE104848 (X. laevis TOMOSeq) and GSE125819 (A. ruthenus TOMOSeq).
5.2. RT-qPCR analysis
RT-qPCR analysis was performed on the X. laevis and A. ruthenus models using the protocol described previously [12], [14]. RT-qPCR was carried out on selected marker genes with known localization profiles in X. laevis or A. ruthenus that showed either extremely animal, animal, vegetal and extremely vegetal distribution profiles after conventional DESeq2 differential analysis (Additional file 1: Table S5) [12]. The RT-qPCR utilized four biological replicates and was performed on a separate set of egg sections than those used for the RNASeq. However, they were treated and sectioned exactly as described for the RNASeq samples.
The reverse transcription was performed using SuperScriptTM III Reverse transcriptase kit (Invitrogen) according to the manufacturer’s manual. 10 ng of RNA in 5 µl of nuclease free water was mixed with 0.5 µl of oligo-dT and random hexamers (mixture 1:1, 50 µM each), 0.5 µl of dNTPs (10 mM each) and 0.5 µl of spike (TATAA Universal RNA Spike, TATAA Biocenter) and incubated for 5 min at 75 °C, 20 s at 25 °C and cooled to 4 °C. In the next step, 2 µl of First strand synthesis buffer, 0.5 µl of DTT, 0.5 µl of SuperScript III enzyme and 0.5 µl of RNaseOUT were added. The mixture was incubated at 25 °C for 5 min, after which reverse transcription was performed for 60 min at 50 °C and 15 min at 55 °C, followed with enzyme inhibition for 15 min at 75 °C and then cooling at 4 °C. Then, the mixture was diluted with 40 µl of nuclease free water and stored at −20 °C. The qPCR analysis was performed using iQ™ SYBR® Green Supermix (Bio-Rad). 2 µl of diluted cDNA was mixed with primers (final concentration 500 nM), 5 µl of iQ™ SYBR® Green Supermix and nuclease free water to final volume of 10 µl. The qPCR was performed on the CFX384 cycler (Bio-Rad) using a program of: initial denaturation: 95 °C, 2 min; 40 cycles: 95 °C for 5 s − 60 °C for 20 s − 72 °C for 20 s, followed by melting curve analysis. The complete list of all primers used for qPCR is available in Additional file 1: Table S6.
The normalized cycle for critical threshold detection was used to find the proportion of transcripts in each section relative to the total number of transcripts in the cell (Eq. (1)).
| (1) |
= quantification cycle
= section of the egg (A to E)
= proportion of transcript that is present in section relative to the egg
5.3. NormQ analysis
To normalize the raw counts obtained from the TOMOSeq, the size factor was calculated to normalize between the inter-section (same sections) replicates and then between the intra-section (different sections) replicates (see Fig. 4). In the first step (herein called NormQ1), the raw counts for the inter-section replicates were normalized using DESeq2’s (v. 1.18.1/1.22.1) average-bulk median-of-ratios method [6], [27]. Each inter-section replicate should follow true to DESeq2’s assumption that the majority of the genes does not show differential expression.
The next step normalizes between the intra-sections to compensate for read depth and library size differences, while scaling the library to reflect the biologically observed proportion between sections as measured with the RT-qPCR. The proportion of the NormQ1 normalized counts for the marker genes within a given section, relative to the whole egg, was then calculated (Eq. (2)). Next, the relative difference between this proportion and the one determined from the RT-qPCR data was calculated (Eq. (2). The median of the relative proportions for all the assessed marker genes was then determined per section for each replicate (Eq. (2). This value represents the size factor fold change required to normalize the data between sections. The median of these scaling factors for members of the same section was then calculated (Eq. (2)). The median was used instead of separate size factors for each replicate, so that the library depth amongst inter-sample section replicates would be consistent. The size factors to normalize between inter-sections and intra-sections were then merged into a single size factor and manually entered into DESeq2 for normalization of all genes. The R script used to calculate the size factors can be found in Additional file 3: NormQ_script and example associated simulated dataset Additional file 4: simulated_dataset.
| (2) |
= proportion of marker gene z that is present in section , relative to the egg as derived from the RT-qPCR data (shown in Eq. (1)).
= proportion of marker gene z that is present in section of replicate , relative to the egg as derived from the RNASeq data.
= ratio of marker gene between and in section .
= median of z for all marker genes for a given section and replicate .
= median of for all replicates of a given section . This represents the intra-section size factor, to normalize between section versus the other sections.
5.4. TomoSeq simulation
The R package, Polyester (v. 1.9.7) was used to produce simulated RNASeq data modelled from the X. laevis transcriptome (Xenbase Version: 9.2) [28], [29]. The X. laevis NormQ normalized counts for 13,877 genes was used to help model the expected fold change (Additional file 1: Table S7). Simulations were done to produce three biological replicates each consisting of five sections (Sections A–E). The simulation also considered potential bias due to transcript length (meanmodel), fragmentation (bias = rnaf) and sequencer error (error_model = illumina5) when generating the data. RNA fragment lengths were selected from a normal distribution with a mean length of 250 bp. The simulation was non-strand specific and produced paired-end reads each with a read length of 50 bp. The library size was varied for each sample, whereby the counts for a replicate were multiplied by a scaling factor of either 0.05x, 0.06x, 0.07x or 0.08x (Additional file 1: Table S8). Twenty genes were assigned as non-differentially expressed across all sections for use in control-based median-of-ratios normalization (Additional file 1: Table S7). The first ten of these genes were assigned fold changes equal to the mean distribution of the mRNA expression and the other ten were assigned two times higher levels than the expected distribution (Additional file 1: Table S7). The simulated reads were then pseudo-aligned to the X. laevis transcriptome using kallisto (v. 0.44.0) and the count data imported into DESeq2 (v. 1.18.1) using the R package tximport (v. 1.8.0) [9], [25]. The simulated counts were then normalized using NormQ, bulk-gene median-of ratios method and control-based median-of ratios method. The results of these normalization methods were then compared to those from the expected gene counts. A maximum of ten genes for use as markers for NormQ normalization were randomly selected from each localization category that were also identifiable as differentially expressed by DESeq2median and had a gene count greater than 100 in any given section (Additional file 1: Table S5). These criteria resulted in ten genes each, from the extremely animal, animal and vegetal localization categories, but only four from the extremely vegetal category.
5.5. RNASeq data analysis
All RNASeq data were normalized using the standard DESeq2 average-bulk median-of-ratios method (herein called DESeq2median), the custom normalization technique as described above (herein called NormQ), the first step of NormQ (herein called NormQ1), no normalization (herein called DESeq2none), and the control-based/spike-ins (herein called DESeq2spike) when available. The true counts and distribution from the Simulated-TOMOSeq is herein referred to as DESeq2expected. Differential analysis was performed amongst the sections using DESeq2 default parameters, followed by multiple hypothesis testing using “Benjamini & Hochberg” correction (padj < 0.1). The performance of the Simulated-TOMOSeq was assessed using the Area under the Receiver Operating Characteristic (ROC) curve (AUC) using the R package metaseqR (v. 1.24.0) [30].
Exploratory analysis was performed using both PCA plots from the DESeq2 package and also degCheckFactors from the DEGreport package (v. 1.20.0) [31]. PCA plots, for each normalization method, were used to visually assess how well the top 5000 most variable genes clustered together relative to the known biological parameters. degCheckFactors, was used to determine how well the techniques reduced the gene variance within a section relative to its mean expression across replicates or sections [31]. A library that has been normalized adequately, should show mRNA expression ratios that approximate a normal distribution and also should typically not be skewed, unless there is some other confounding biological/technical factors [31].
The localization profiles of the genes were compared against the expected profile from the Simulated-TOMOSeq or from the expected profiles from the RT-qPCR. This was achieved by first manually clustering the genes into particular localization profiles using the criteria as defined in our previous study [12]. Genes were placed into the localization categories: extremely animal, animal, vegetal, extremely vegetal and unclassified. The proportion of localization profile matches relative to the RT-qPCR or Simulated-TOMOSeq profile was assessed for each normalization method.
In the Simulated-TOMOSeq, the proportion of true DEGs, DEGs that were detected by all the normalization method, and all genes (DEGs and non-DEGs) that contained count data after normalization, were assessed for correct profiling. A two-tailed t-test using unequal variances was used to assess for significant differences between the matched localization profiles (relative to the RT-qPCR/simulated data) of the marker genes or all simulated genes between the different normalization techniques. Linear regression analysis was used to assess the correlation between the relative section proportions (; where = counts from egg section of replicate ), for each replicate of each normalization method against the equivalent data from the expected simulated data.
The outlier size factors during the second step of NormQ was assessed to determine the probability of selecting a gene that contributes to an outlying size factor. The effect of the number of genes used for the calculation of the NormQ size factor was also assessed using the Simulated-TOMOSeq data. Similarly, as described above, the DESeq2median data was used to aid in the selection of marker genes. A maximum of 30 genes were randomly selected from each localization category. The exception was the extreme vegetal group, where all four gene representatives were used. Iteratively, one gene from each group was randomly removed (without replacement) and the size factor recalculated. Additionally, the effect of selecting all genes or all members from the same localization profile was analyzed. The performance of each size factor was assessed using AUC-ROC of the obtained DEGs.
The NormQ method was also assessed using data from our previously published RT-qPCR derived gene distributions for the X. laevis model, that was obtained in a similar manner as described in our methodology (Additional file 1: Table S5) [16]. The RT-qPCR localization profile for these genes were compared to the localization profile for the same genes but as determined using NormQ on our TOMOSeq dataset. The primers designed in this publication were however based on an older X. laevis gene annotation. Therefore, before using this secondary data, the primers from this publication were assessed using Primer-BLAST, and all primers that did not align 100% to their target gene, or were complementary to both homeologous forms of the gene, were removed from the dataset [32]. Additionally, eleven extra X. laevis genes that were not used for the NormQ normalization were assessed using RT-qPCR, and their given distributions then compared to those from the NormQ and DESeq2median normalization methods (Additional file 1: Table S5).
6. Declarations
6.1. Ethics statement
All experimental procedures involving A. ruthenus and X. laevis were carried out in accordance with the Czech Law 246/1992 on animal welfare. Protocols involving A. ruthenus were reviewed by the Animal Research Committee of the Faculty of Fisheries and Protection of Waters, South Bohemian Research Center of Aquaculture and Biodiversity of Hydrocenoses, Research Institute of Fish Culture and Hydrobiology, Vodnany, Czech Republic. Protocols involving X. laevis were approved by the animal committee of the Czech Academy of Sciences.
6.2. Data availability
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO), accession GSE104848 (X. laevis TOMOSeq) and GSE125819 (A. ruthenus TOMOSeq).
6.3. Competing interest
The authors declare that they have no competing interests.
Funding
This work was supported by the Czech Science Foundation [GA17-24441S, GA19-11313S]; [RVO: 86652036]; Project Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University (BIOCEV) [CZ.1.05/1.1.00/02.0109] from the European Regional Development Fund (ERDF); and the Charles University [GAUK No. 414217].
Author contributions
RN wrote the manuscript, formulated methods and performed Bioinformatics analysis. PA formulated methods, performed library preparation for RNASeq analysis and Bioinformatics analysis. MS, ST and OS contributed to sample preparation and RT-qPCR. KP and MP contributed to sample preparation for Acipenser ruthenus. MK and RS supervised and reviewed the project.
CRediT authorship contribution statement
Ravindra Naraine: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Pavel Abaffy: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing - review & editing. Monika Sidova: Investigation, Writing - review & editing. Silvie Tomankova: Investigation, Writing - review & editing. Kseniia Pocherniaieva: Investigation, Resources, Writing - review & editing. Ondrej Smolik: Investigation, Writing - review & editing. Mikael Kubista: Resources, Writing - review & editing, Funding acquisition. Martin Psenicka: Investigation, Resources, Writing - review & editing. Radek Sindelka: Resources, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.05.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans C, Hardin J, Stoebel D. Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions 2016:1–32. https://doi.org/10.1093/bib/bbx008. [DOI] [PMC free article] [PubMed]
- 3.Bullard J.H., Purdom E., Hansen K.D., Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. UC Berkeley Div Biostat Pap Ser. 2009;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillies M.A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 5.Jiang L., Schlesinger F., Davis C.A., Zhang Y., Li R., Salit M. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovén J., Orlando D.A., Sigova A.A., Lin C.Y., Rahl P.B., Burge C.B. Revisiting global gene expression analysis Jakob. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012.Revisiting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lun A.T.L., Calero-Nieto F.J., Haim-Vilmovsky L., Göttgens B., Marioni J.C. Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data. Genome Res. 2017;27:1795–1806. doi: 10.1101/gr.222877.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotech. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Q., Zhang X. The influence of the global gene expression shift on downstream analyses. PLoS ONE. 2016;11:1–13. doi: 10.1371/journal.pone.0153903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen K, Hu Z, Xia Z, Zhao D, Li W. The overlooked fact: fundamental need of spike-in controls for. Mol Cell Biol May 2017;1:662–7. https://doi.org/10.1128/MCB.00970-14.Address. [DOI] [PMC free article] [PubMed]
- 12.Sindelka R., Abaffy P., Qu Y., Tomankova S., Sidova M., Naraine R. Asymmetric distribution of biomolecules of maternal origin in the Xenopus laevis egg and their impact on the developmental plan. Sci Rep. 2018;8:1–16. doi: 10.1038/s41598-018-26592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junker J.P., Noël E.S., Guryev V., Peterson K.A., Shah G., Huisken J. Genome-wide RNA tomography in the Zebrafish embryo. Cell. 2014;159:662–675. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Sindelka R., Sidova M., Svec D., Kubista M. Spatial expression profiles in the Xenopus laevis oocytes measured with qPCR tomography. Methods. 2010;51:87–91. doi: 10.1016/j.ymeth.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Claussen M., Lingner T., Pommerenke C., Opitz L., Salinas G., Pieler T. Global analysis of asymmetric RNA enrichment in oocytes reveals low conservation between closely related Xenopus species. Mol Biol Cell. 2015;26:3777–3787. doi: 10.1091/mbc.E15-02-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sindelka R., Jonák J., Hands R., Bustin S.A., Kubista M. Intracellular expression profiles measured by real-time PCR tomography in the Xenopus laevis oocyte. Nucleic Acids Res. 2008;36:387–392. doi: 10.1093/nar/gkm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010 doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandramohan R, Po-Yen Wu, Phan JH, Wang MD. Benchmarking RNA-Seq quantification tools. 2013 35th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc., vol. 118, IEEE; 2013, p. 647–50. https://doi.org/10.1109/EMBC.2013.6609583. [DOI] [PMC free article] [PubMed]
- 19.Everaert C., Luypaert M., Maag J.L.V., Cheng Q.X., DInger ME, Hellemans J, Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopylova E., Noé L., Touzet H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 23.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert DG. Gene-omes built from mRNA-seq not genome DNA. 7th Annu Arthropod Genomics Symp 2013:47405. https://doi.org/10.7490/f1000research.1112594.1.
- 27.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimi K., Fortriede J.D., Lotay V.S., Burns K.A., Wang D.Z., Fisher M.E. Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2018;46:D861–D868. doi: 10.1093/nar/gkx936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazee A.C., Jaffe A.E., Langmead B., Leek J.T. Polyester: Simulating RNA-seq datasets with differential transcript expression. Bioinformatics. 2015;31:2778–2784. doi: 10.1093/bioinformatics/btv272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moulos P., Hatzis P. Systematic integration of RNA-Seq statistical algorithms for accurate detection of differential gene expression patterns. Nucleic Acids Res. 2015;43:1–12. doi: 10.1093/nar/gku1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pantano L, Hutchinson J, Barrera V, Piper M, Khetani R, Daily K, et al. DEGreport: Report of DEG analysis 2017. https://doi.org/10.18129/B9.bioc.DEGreport.
- 32.Coulouris Y., Zaretskaya I., Cutcutache I., Rozen S., Madden T. Primer-BLAST: A tool to design target-specific primers for polymerse chain reaction. BMC Bioinf. 2012;18(13):134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO), accession GSE104848 (X. laevis TOMOSeq) and GSE125819 (A. ruthenus TOMOSeq).