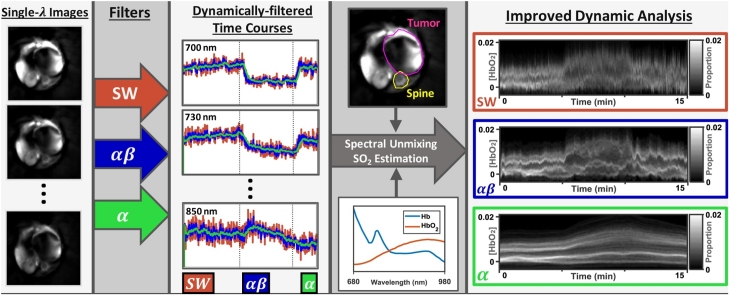
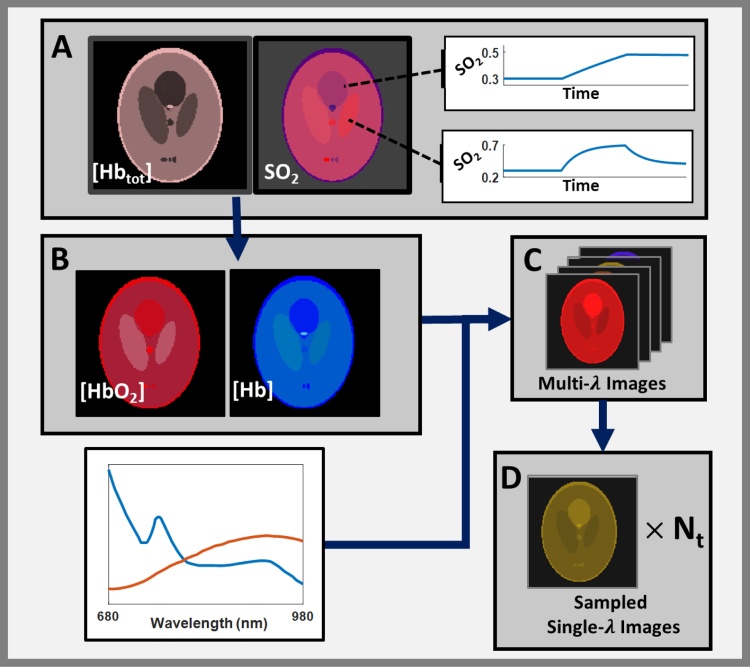
Graphical abstract
Keywords: Photoacoustic imaging, Oxygen, Hemoglobin, Dynamic response, Noise suppression, Kidney tumor
Highlights
-
•
Simple and filters effectively suppress noise in dynamic spectral images.
-
•
A multispectral extension of the Shepp-Logan phantom was developed.
-
•
Practical application is demonstrated in distinct renal cell carcinoma models.
-
•
filters enabled effective separation of gas breathing challenge response.
Abstract
Multispectral optoacoustic tomography (MSOT) is an emerging imaging modality, which is able to capture data at high spatiotemporal resolution using rapid tuning of the excitation laser wavelength. However, owing to the necessity of imaging one wavelength at a time to the exclusion of others, forming a complete multispectral image requires multiple excitations over time, which may introduce aliasing due to underlying spectral dynamics or noise in the data. In order to mitigate this limitation, we have applied kinematic and filters to multispectral time series, providing an estimate of the underlying multispectral image at every point in time throughout data acquisition. We demonstrate the efficacy of these methods in suppressing the inter-frame noise present in dynamic multispectral image time courses using a multispectral Shepp-Logan phantom and mice bearing distinct renal cell carcinoma tumors. The gains in signal to noise ratio provided by these filters enable higher-fidelity downstream analysis such as spectral unmixing and improved hypothesis testing in quantifying the onset of signal changes during an oxygen gas challenge.
1. Introduction
Imaging has become a powerful tool in diverse fields such as medical diagnostics, high-throughput screening, and remote sensing [[1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11]]. The imaging toolkit is becoming progressively more sophisticated, moving beyond spatial discrimination with additional dimensions: Spectral imaging allows the capture of multiple imaging channels, in the hope of using the spectral fingerprints of underlying components to learn more about the system under study [1,[7], [8], [9], [10],[12], [13], [14], [15], [16], [17], [18], [19]]; and dynamic imaging captures many single images over time in order to resolve changes in the observed system [12,13,[20], [21], [22], [23], [24]]. Imaging methods are subject to inherent trade-offs between spatial, spectral, and temporal resolution, as well as the discrimination of signal versus noise. The quality of data is fixed once the data have been acquired by instrument hardware, but it is possible to take advantage of the inherent coherence and structure within a dataset in order to mitigate the effects of noise and finite sampling.
Multispectral optoacoustic tomography (MSOT) is an imaging modality, characterized by the use of optical excitation and ultrasonic detection, which is able to capture spectral images at high spatial and temporal resolution [7,17,20,22]. MSOT intrinsically acquires data based on sequential wavelength excitation such that images are captured one channel at a time. There may therefore be substantial spectral distortion due to aliasing in imaging scenarios where the rate of spectral dynamics is comparable to the multispectral sampling rate. Further exacerbating aliasing is the presence of noise, which may vary spatially, temporally, and spectrally, resulting in the reduction of measurement fidelity.
One experimental motif which exemplifies these difficulties is the gas-breathing challenge, wherein the inhaled gas of a subject under observation is varied in order to induce a change in the subject’s blood oxygen saturation (SO2) [25,26]. In order to determine or estimate the oxygen saturation by MSOT, images must be acquired at multiple wavelengths and spectrally unmixed to provide oxy- and deoxy-hemoglobin images, whereupon the ratio of oxy-hemoglobin to total hemoglobin provides an estimate of the oxygen saturation of blood. The propagation of image noise through this process can result in extremely noisy SO2 estimates, limiting the amount of useful information which can be extracted from noisy data.
It is therefore desirable to devise a scheme to mitigate the corrupting effects of noise while still accurately capturing dynamic phenomena. Similarly, it is desirable to be able to perform such filtering in an online manner, as data are acquired, such that the filtering may be implemented for real-time imaging or efficient post-acquisition analysis. This last criterion imposes a strong restriction on the complexity of any algorithm, since many photoacoustic imaging systems gather data at rates exceeding 10 Hz (.
We briefly establish general notation for equations throughout this work: is the current time index, refers to the states of interest, here the pixel values for the spectral image; is the acquired data at time , is the estimate of at time k considering all data up to and including the data at time , is the so-called prior residual, and is the estimate of after integration of the data acquired at time point .
A simple, efficient approach to recursively estimating the state of a multispectral image is to perform online averaging on a per-channel basis according to the following update equations:
| (1) |
| (2) |
For stationary distributions, online averaging performs exactly as the usual average, converging to the true mean as , i.e., the variance of sampling noise is proportional to the square root of the number of samples averaged [27]. For non-stationary distributions such as those encountered in dynamic imaging, will not, in general, converge to the true mean even after acquisition of an infinite number of samples. In the context of a gas challenge, will be heavily biased towards the initially acquired data and will not effectively follow dynamics.
To accommodate nonstationary distributions, the term may be replaced with a constant . This gives the averaging process a time-invariant form which may be analyzed as a recursive filter, often referred to as the exponentially weighted moving average or filter:
| (3) |
Observation indicates that varying between 0 and 1 biases the estimate of towards the previous estimate and the new data, respectively. converges to the same structure as the online average as the sampling time tends to infinity. Conversely, ignores all previous data in favor of the new data, but without any noise rejection. This trade-off between noise rejection and dynamical sensitivity is irrelevant if there are no underlying dynamics, but for values of the estimate will always lag the true mean of dynamic systems [28]. The limiting case of results in the filter taking on the form of a sliding-window (SW) filter, where old data is entirely overwritten by new data as it is acquired:
| (4) |
Clearly, a simple filter is unable to fully accommodate nonstationary data and is therefore generally inappropriate for dynamic imaging. Many more sophisticated algorithms such as the broad class of Bayesian filters can incorporate large amounts of prior knowledge, including the presence of dynamics, and have exceptional performance in tracking the state of dynamical systems [28,29]. However, Bayesian filters can require large amounts of data to undergo computationally intensive processing and are particularly challenging to apply to spectral images due to the large size of associated datasets.
Though full Bayesian filters are computationally prohibitive, they may be simplified by assuming Gaussian noise statistics and linear dynamics. These so-called Kalman filters operate in a two-stage prediction-correction cycle, wherein the prediction phase propagates the state estimate in time according to the dynamical model and the correction phase incorporates new data observations into the state estimate. Kalman filters have been shown to be useful in integrating multi-domain measurements, including multi-channel images, due to their inherently dynamical nature [[30], [31], [32], [33], [34]]. Nevertheless, the size of the covariance matrices necessary for the operation of Kalman filters requires offline processing or substantial computing resources.
If the assumption is made that the Kalman filter is operating at steady state, it degenerates to a form which is temporally invariant. Further assumption that the state of interest has a known second-order process variance (i.e., that the process noise is in the acceleration term of a Newtonian kinematic model) yields a family of recursive filters indexed by the order of their Taylor series expansion, with associated constant coefficients [35,36]. Here, we describe the form of the 1st and 2nd order filters, the former of which is the previously-described filter and the latter correspondingly named the filter.
As before, the state of interest is denoted by , with first temporal derivative given by . The measurement noise variance, describing the ‘noisiness’ of each measurement, is denoted by with units determined by the measurement device (here arbitrary units a.u.), and the process noise variance, describing the volatility of the underlying dynamics, as with units (a.u./s2). We assume that each pixel evolves independently and thus the filter is applied to each pixel of the acquired image separately.
The filter coefficients may be optimally found using prior knowledge of , , and via a single scalar parameter, the tracking index:
| (5) |
We assume that the values of , , and are homogenous across the entire image such that each image has a single tracking index. In practice, these parameters may vary between channels and over time, which we address in Section 4.
The filtering equations for an filter are identical to the exponentially weighted moving average described in Eqs. 1 and 3. As the filter tracks only position information, the prediction step is degenerate and only the correction phase updates the state estimate:
Prediction
| (6) |
Correction
| (7) |
The filtering equations for an filter propagate the state estimate over time by assuming constant-velocity dynamics and are:
Prediction
| (8) |
| (9) |
Correction
| (10) |
| (11) |
The tracking index formalism enables analytic determination of the and coefficients (Appendix A) in an optimal manner, and requires knowledge of only three parameters , , and , which may themselves be determined from prior knowledge of the system, or updated recursively by examining the prior and posterior residuals of the filtering process [37]. In environments where the sampling rate is inhomogeneous, such as when oversampling each channel before sampling another, is variable. This requires that the tracking index and thus the and coefficients must be updated at each time point.
We compare the and filters against the sliding window (SW) filter equivalent to : A circular buffer of data is maintained and updated on a per-channel per-time-point basis as data are acquired, following a ‘sample and hold’ pattern. There is thus no incorporation of previous data into the current estimate, nor is there any accommodation for dynamic evolution of the state.
2. Methods
2.1. Numerical phantom
We developed a dynamic, multispectral numerical phantom to act as a known ground-truth for testing the ability of the various filters to track spectral changes while rejecting noise. The full process of creating the phantom is described in Appendix B and illustrated in Fig. 1. Briefly, a Shepp-Logan phantom [38] was modified to include time-varying concentrations of endmembers with known spectra. Sampling the phantom image at specific wavelengths at each point in time provided a sample train to emulate the acquisition of one channel at a time. We directly sampled the phantom, rather than simulating the photoacoustic imaging process, to limit the possibility of introducing implementation and modeling errors. After sampling, zero-mean white Gaussian noise with standard deviation was added to each image, corresponding to an SNR of approximately 20. The dynamics of the phantom were designed to mimic a gas challenge due to the variety of states and rates of change which can be present in a single time course.
Fig. 1.
Workflow for creating numerical phantom. A) A Shepp-Logan phantom was created with ellipses defined by various constant values for total hemoglobin [Hbtot] and time-varying SO2. The time-varying images of [Hb] and [HbO2] were then calculated (B) and their spectra used to create time-varying spectral images (C). The spectral image was then sampled at predetermined wavelengths to create a time course of single-wavelength images (D), which were used as input for the filtering methods described in this work.
2.2. Experimental data
Three mice from ongoing studies were used for in vivo validation, with all data acquired using an MSOT InVision 256-TF photoacoustic imaging system [39] (iThera Medical GmbH, Munich, Germany). The water bath used for imaging was maintained at a temperature of 35 C for all experiments. All animal work was conducted in accordance with the UT Southwestern Institutional Animal Care and Use Committee guidelines as well as all superseding federal guidelines. Two distinct wavelength sampling patterns were used to demonstrate the applicability of the filters to general acquisition conditions.
The first mouse was a female BALB/c mouse (UT Southwestern Animal Breeding Core) with a RENCA-luc [40] mouse kidney tumor implanted in the right kidney following published protocols [41], with data acquired 28 days after implantation. Prior to imaging, the mouse was shaved and any remaining hair was removed from the animal using depilation cream. Five wavelengths were sampled [λ = 700, 730, 756, 800, 850 nm] to correspond with major features of the hemoglobin spectrum, including local spectral peaks and the isosbestic point. Each wavelength was oversampled 5 times and averaged prior to image reconstruction, leading to an overall acquisition time of 2.5 s per multispectral sampling cycle. All images were acquired at the same axial cross-section. During imaging, the anesthetized mouse underwent a gas challenge from air-oxygen-air, with 10.5 min on air, 10 min on oxygen, followed by another 5 min on air, while breathing 2.0 % isoflurane at a rate of 2 L/min.
The remaining two mice were female Hsd:Athymic Nude-Foxn1nu mice (Envigo) with XP373 human renal cell carcinoma tumor xenografts [42] implanted in the right kidney following published protocols [41], with two imaging sessions acquired for Mouse #1 at 31 and 36 days, and one session at for Mouse #2 at 31 days after tumor implantation. Prior to imaging, hair was removed from the animal using depilation cream. Seven wavelengths were sampled sequentially [λ = 680, 734, 757, 798, 850, 922, 980 nm] to correspond with major features of the hemoglobin spectrum, including local spectral peaks and the isosbestic point. Each wavelength was sequentially oversampled 3 times and averaged prior to image reconstruction, leading to an overall acquisition time of 2.1 s per multispectral sampling cycle. All images were acquired at the same axial cross-section. During imaging, each anesthetized mouse underwent a gas breathing challenge from air-oxygen-air, with 5 min on each gas, while breathing 2.0 % isoflurane at a rate of 2 L/min.
2.3. Image reconstruction
After acquisition, each single-wavelength image was preprocessed by correcting for laser power variation and deconvolving the signal impulse response using Wiener deconvolution with a regularization constant of 0.001. Each image was then reconstructed using a curve-driven model matrix as a forward model [43] and a non-negative accelerated projected conjugate gradient method as the inverse solver [44].
2.4. Multispectral state filtering
All analysis was performed using code developed in MATLAB 2017b, with production code available at https://git.biohpc.swmed.edu/PIRL/kalata-filters-matlab. Archived version is available at https://doi.org/10.5281/zenodo.3836682.
The single-wavelength images from the numerical and experimental datasets were used as input to each of the SW, , and filters. For each method, an multispectral image with pixels per channel and channels was maintained, to store each filter’s estimate of the multispectral image. At each point in time, all channels of the entire multispectral image were updated according to the prediction step for each method, using either Eq. 6 for the SW and filters, or Eqs. 8 and 9 for the filter. Following the prediction step, a single-channel image, acquired at wavelength , was loaded into memory, corresponding to in Eq. 1. This image was used to update channel in each filter’s multispectral state estimate, using either Eq. 7 for the SW and filters or Eqs. 10 and 11 for the filter.
The numerical phantom data were filtered using , , the RENCA-luc data with , , and the XP373 data with ,
2.5. Post-processing
At each time point, the multispectral images estimated by each filtering method were spectrally unmixed using an unconstrained linear model to yield images of oxy- and deoxy-hemoglobin. The total hemoglobin and oxygen saturation in each pixel were estimated as:
| (12) |
We note that the resultant values are estimates, and are thus indicated by the superscript MSOT as used by others [26]; truly quantitative measures of endmember concentration would require additional processing using, e.g., fluence correction [18,19,45,46] or Bayesian methods [47]. In general, the filtering methods presented here cannot correct for errors in quantitation due to modelling errors.
2.6. Quantification methods
2.6.1. Image quality
The deviation of each estimated multispectral image from the known ground truth image was calculated using two metrics: the mean-squared error (MSE) and the structural similarity index (SSIM).
The MSE was used to examine the effects of each approach on noise rejection, but includes some deviation due to differences in the image mean. It is calculated as:
| (13) |
The SSIM was used to examine images on a holistic basis, accounting for structure, noise, and constant value offsets. It is calculated as:
| (14) |
where and represent the mean of each image, and represent the variance of each image, and represents the covariance between the two images. and are positive constants chosen to improve numerical stability and are related to the dynamic range of the images.
2.6.2. ROI analysis
For the phantom data, a 5 × 5 region of interest (ROI) was defined in the same location for all images within ellipse (e) (Appendix B, Table B1), and the average value within that region determined for all time points for each channel. This was performed for all filtering methods, for each channel of the multispectral image, as well as for each of the [Hb]MSOT, [HbO2]MSOT, [Hbtot]MSOT, and SO2 MSOT images, to yield time courses of each parameter.
Manually selected ROIs were defined over the tumor and spine for each mouse and used as the ROIs for both single-channel data and the unmixed parameters. Within each ROI, for each filter, for each parameter, at each time point, a histogram was calculated using 256 equally-spaced bins, with lower and upper limits defined by the 0.1st and 99.9th percentile of the values for that parameter in that ROI for all time points. Each one-dimensional histogram was then used as a column of pixels in a dynamic histogram image. Three time points for each tumor type were chosen prior to, during, and after the transition period from air to oxygen. For each condition, the histograms from 11 time points centered on those selected time points were averaged together and used as representative time slices of the dynamic histogram image to improve visualization.
Further analysis was performed on each filter to compare the statistical effects of each, and this is described in Appendix C.
3. Results
3.1. Phantom data
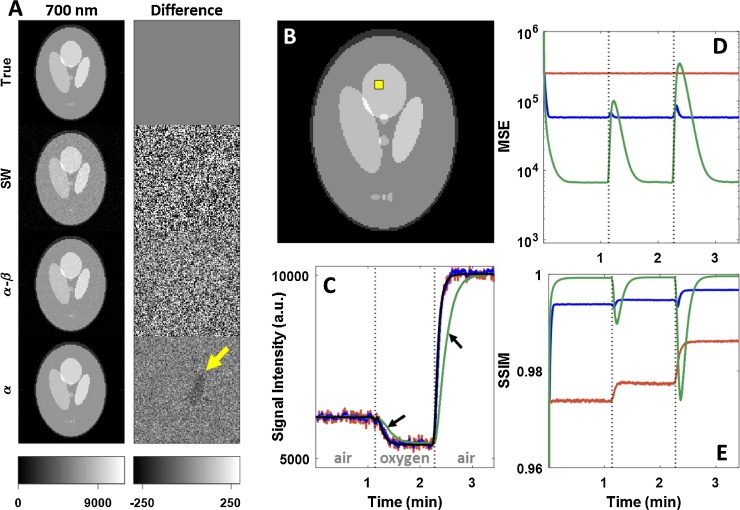
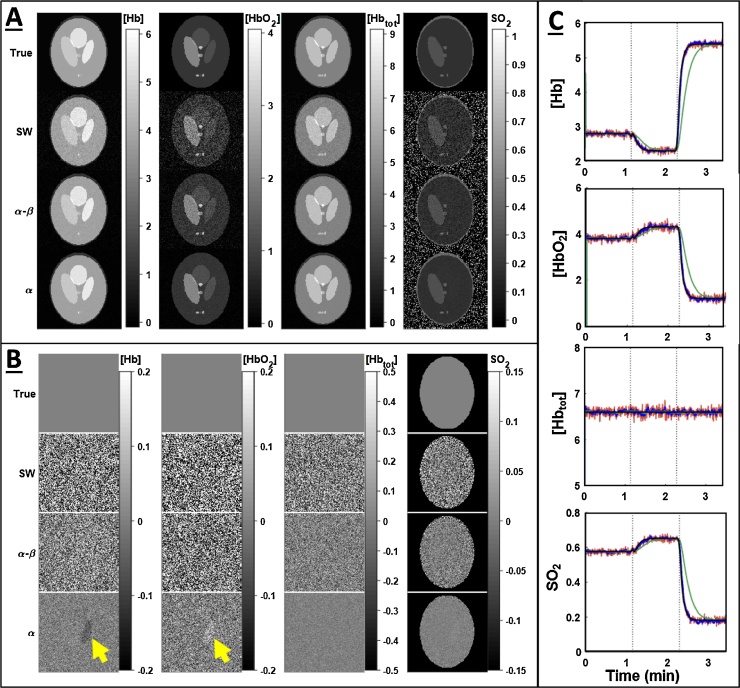
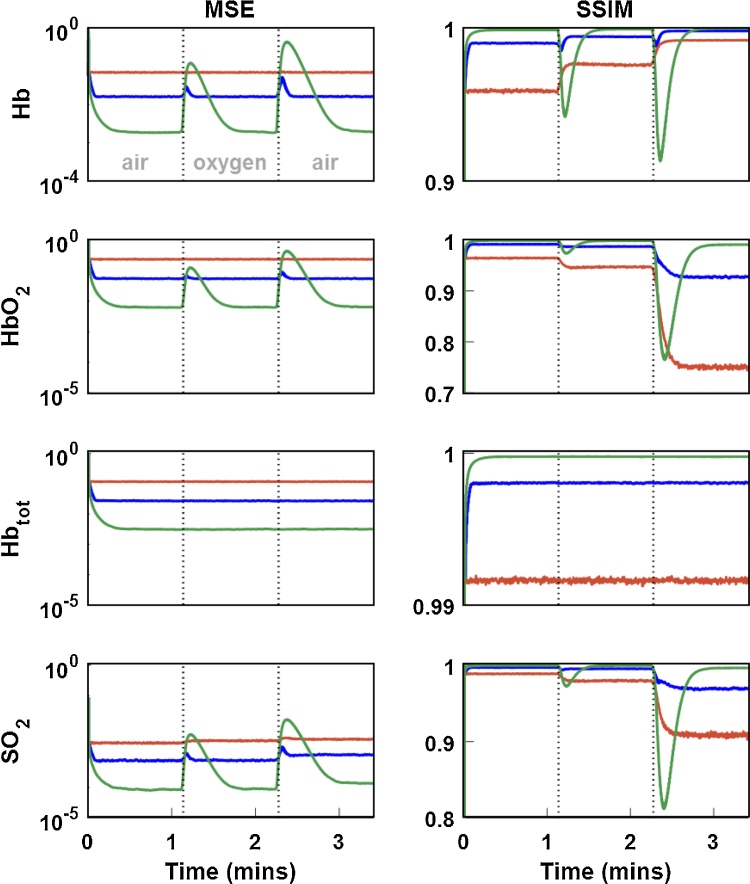
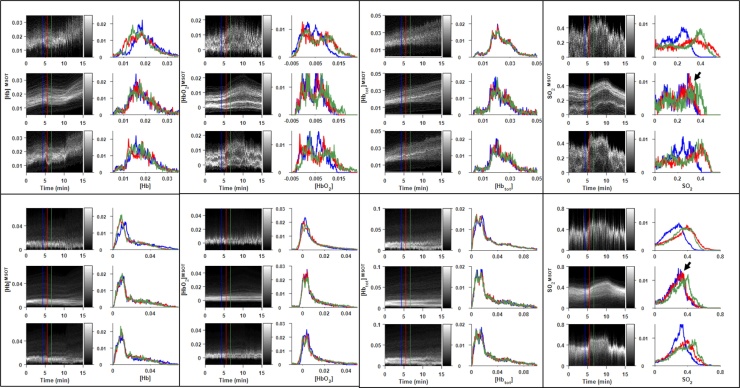
When applied to the simulated data, both and filters effectively suppressed noise in the multispectral images as compared to the SW filter. As seen in Fig. 2, the filter rejected a large amount of noise from the signal, but failed to follow the rapid dynamics of the simulated gas challenge, causing visible artifacts (Fig. 2A, bottom right). By comparison, the filter both rejected noise and accurately tracked dynamic changes throughout the image. The relative performance of each of the filtering methods can be seen in the single-channel ROI time course and quality metric time courses. Overall, the filter performed extremely well unless challenged by sudden dynamic changes. Due to the filter’s intrinsic time lag, there was a substantial resettling time before it returned to pre-challenge performance. The properties of each filter are retained upon spectral unmixing of the multispectral images, and still further when calculating the derived [Hbtot]MSOT and SO2MSOT images as seen in Fig. 3. The filter demonstrated superior performance on the unchanging [Hbtot]MSOT channel, but once again suffered from a time lag following dynamic changes. These observations are validated by the time courses of error metrics in Fig. 4. Notably, the unmixed images using the SW were extremely noisy, with the [HbO2]MSOT SSIM dropping below 0.8 towards the end of the challenge. The filter is able to achieve excellent noise rejection at baseline, yielding small MSE and SSIM very close to 1, but during transitions its performance fell below that of the SW.
Fig. 2.
Effect of each filter on dynamic images in numerical phantom. A) Single-wavelength images (left) and their differences (right) from the known ground truth image (left, top) at the final time point of the simulated gas challenge. A clear reduction in noise is seen for both the and filters when compared to the SW filter. The filter suffers from a visible artifact due to its intrinsic time lag, as indicated by the yellow arrow. B) Image showing ROI for signal intensity calculation in C. C) Mean signal intensity over the yellow ROI in B for each time point during challenge. Conspicuous lag is visible in the filtered data. Red: SW, blue: , green: black: Ground truth. D) Mean squared error of entire multispectral image at each point in time. Degradation of filter performance is clearly seen accompanying gas challenge changes. E) Structural similarity of entire multispectral image at each point in time, matching D.
Fig. 3.
Effect of each filter on time courses of derived parameters. A) Derived images from filtered data for each filter. The effects of noise in the multispectral images are magnified in the unmixed data. Filtering prevents noise from being propagated to this stage of analysis, leading to qualitatively clearer images for both the and filters. B) Difference of each derived parameter image from the corresponding ground truth (A, top row). Both and demonstrate decreased noise relative to SW, but the filter’s lag has resulted in an inaccurate unmixing result, overestimating [HbO2] and underestimating [Hb] as indicated by yellow arrows. C) Time courses of each parameter for different filters. Red: SW, blue: , green: black: Ground truth. Though dynamic changes cause the filter to deviate from ground truth, it has excellent performance in filtering the constant [Hbtot] channel.
Fig. 4.
Image quality metrics over time for unmixed parameter images in phantom. Red: sliding window filter, green: , blue: . As in the multispectral case, the filter achieves excellent noise reduction in steady-state contexts, such as the initial baseline or the entirety of the [Hbtot] time course. Overall performance of each filter in the unmixed data is similar to that seen in the multispectral case (Fig. 2).
3.2. Experimental data
The αβ filter effectively captured transient spectral dynamics while rejecting noise in both tumor models (Fig. 5). This is in contrast to the SW’s lack of noise rejection, illustrated by the highly variable baseline signal, and the α’s lack of dynamic sensitivity, seen in the rounding-off of the gas challenge transition in the 700 nm channel (Fig. 5B, D: green line).
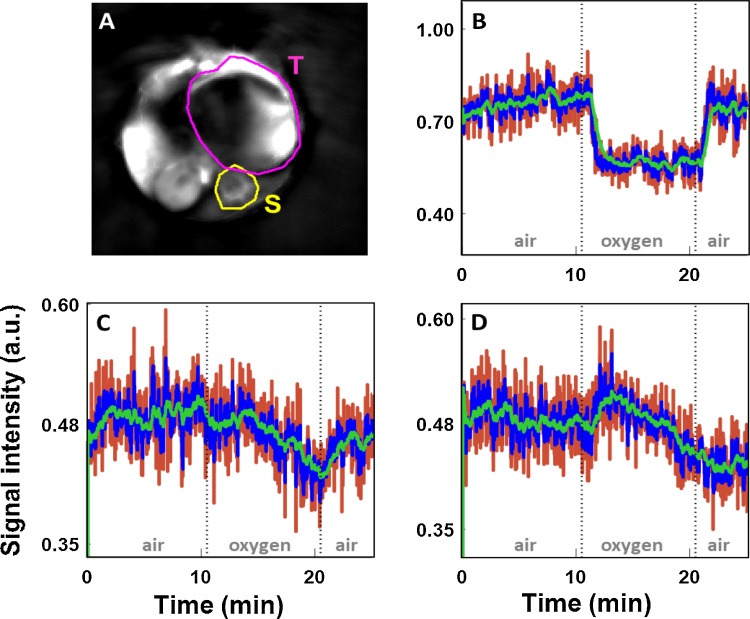
Fig. 5.
Time courses of MSOT spine signal for representative channels in RENCA-luc mouse. A) Sum image of all channels at final time point, with ROIs used for analysis indicated. T: tumor, S: spine. B, C, D) Time courses of signal in selected channels based on respective filters. B) 700 nm, C) 800 nm, D) 850 nm. Red: sliding window filter, green: , blue: . The filter removed a substantial proportion of the noise from the dynamic signal while still accurately following the gas challenge. All channels are shown in Supp. Figures S1 (image comparisons) and S2 (time courses).
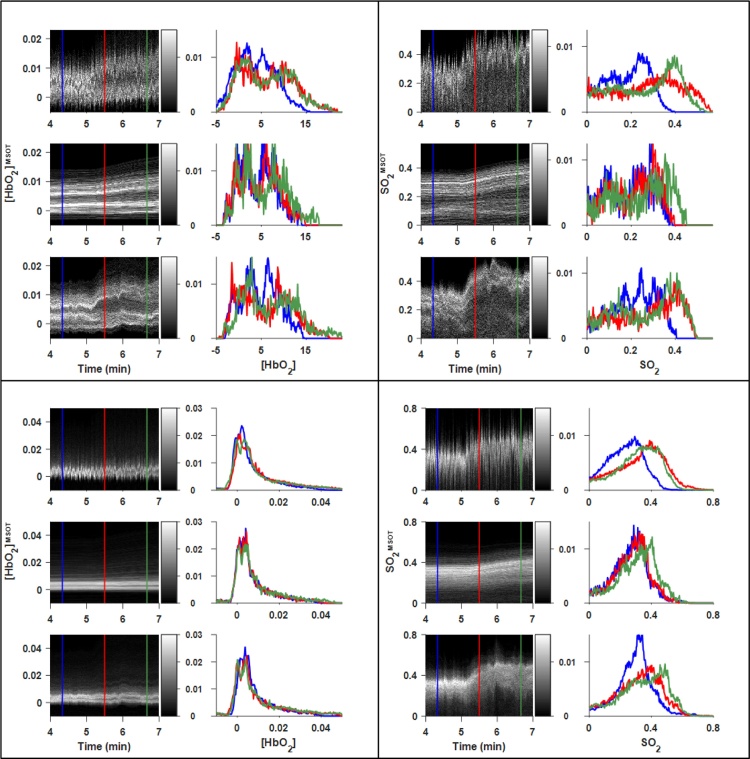
The dynamic performance of each filter for the RENCA-luc data is shown in Fig. 6, Fig. 7: Whereas the sliding window data are extremely noisy, preventing straightforward characterization of the response to gas challenge, the α filter clearly separated distinct subpopulation time courses within each ROI. However, the α filter introduced a substantial response time, taking minutes to reach steady state after each gas change, and consequently destroying quantitative information regarding the rate constant of the transition, as well as the transition time itself. In comparison, the αβ filter revealed conspicuous differences in both the rates and degree of response to gas challenge across both simulated and experimental data. These effects are more obvious in the transition region highlighted in Fig. 7: There was a dramatic decrease in the amount of noise in both the α and αβ filtered data, but only the filter preserved the rate and degree of response at each time point.
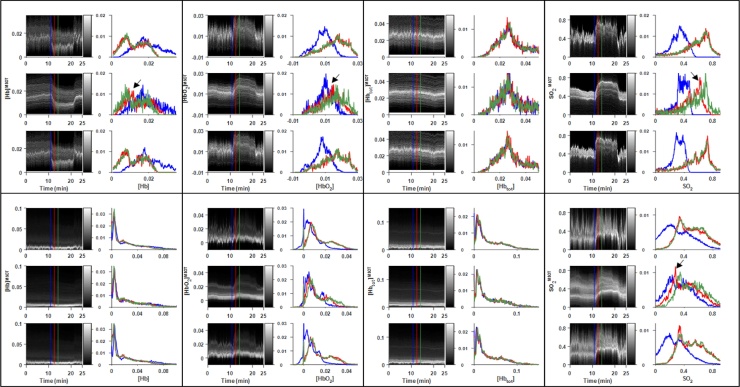
Fig. 6.
Dynamic histogram images and selected histograms for each parameter-filter combination in RENCA-luc tumor-bearing mouse. ROIs defined in Fig. 5A. Upper panels: spine ROI. Lower panels: Tumor ROI. Each panel: Dynamic histogram images (left column) and corresponding histograms at selected time points (right column) for sliding window (top), (middle), and (bottom) filters. For all parameters, the use of either the or filters reduced the noise in the dynamic histograms, but the filter lagged behind across the entire ROI, as seen by the misalignment in the red and green histograms highlighted by the black arrows.
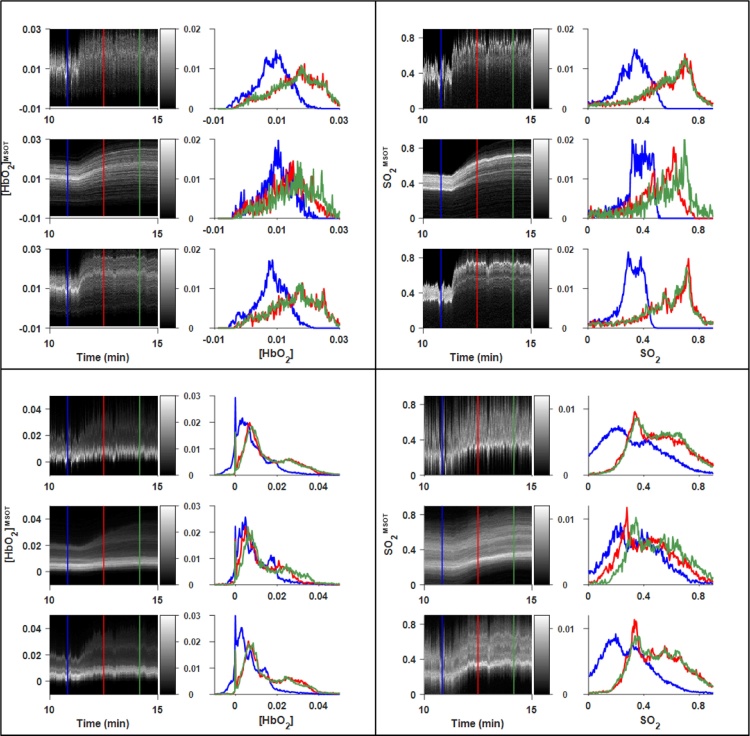
Fig. 7.
Expanded time courses of air-oxygen transition in RENCA-luc tumor-bearing mouse. ROIs defined in Fig. 5A. Upper panels: Spine ROI. Lower panels: Tumor ROI. Left panels: [HbO2]MSOT. Right panels: [SO2]MSOT. A conspicuous reduction in noise is seen between the sliding window (each panel, top) and (each panel, bottom) time courses, allowing one to resolve the distinct evolution of different subpopulations within the ROI.
When applied to the XP373 tumor (Fig. 8, Fig. 9), the filter reveals heterogenous response to the gas challenge which is not visible in the sliding window data. Indeed, it is clear in the upper-left panel of Fig. 9 that there are both responsive and nonresponsive subpopulations of pixels within the spine ROI. The αβ filter also performed well on relatively constant data such as the [HbO2]MSOT channel in the tumor ROI, delineating a bimodal distribution which was obscured by noise in the SW filter. These general between-filter patterns are sustained when applying the filters to additional XP373 gas challenge datasets, as seen in Supplementary Figs. S3–5.
Fig. 8.
Dynamic histogram images and selected histograms for each parameter-filter combination in XP373 tumor-bearing Mouse #1. ROIs defined in Supp. Fig. 3A. Upper panels: Spine ROI. Lower panels: Tumor ROI. For all derived parameters, the use of either the or filters reduced the amount of noise in the dynamic histograms, though the filter lagged behind across the entire ROI, as seen by the misalignment in the red and green histograms highlighted by the black arrows.
Fig. 9.
Expanded time courses of air-oxygen transition in XP373 tumor-bearing mouse (Fig. 8). Upper panels: Spine ROI. Lower panels: Tumor ROI. Left panels: [HbO2]MSOT. Right panels: [SO2]MSOT. A conspicuous reduction in noise can be seen between the sliding window (each panel, top) and (each panel, bottom) time courses, allowing one to resolve the distinct evolution of different subpopulations within the ROI.
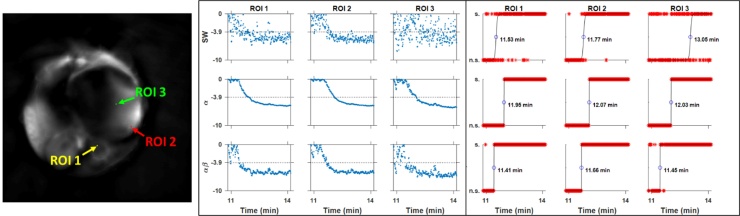
Both the and filters provided improved statistical performance in assessing whether an oxygen-induced change is significantly different from the air baseline, in both spatial and temporal dimensions (Fig. 10, Table 1). Areas where the signal to noise ratio is low are deemed as insignificant changes under analysis of the SW-filtered data, but both the and filters resolve those same areas as significant (Supplementary Fig. S6). In examining the transition over time from insignificant to significant, fitting of a logistic function to the binary classification time course yields an effective transition time. The analysis provided by the SW filter provided inconsistent fits for transition times, which were highly variable depending on the SNR of the local region. Though the filter provided a much sharper transition from non-significant to significant, it occurred at a substantial delay from the known switching time of 10.5 min. The filter yields consistent and sensible transition times between all ROIs.
Fig. 10.
Significance testing of HbO2MSOT change due to gas challenge in RENCA-luc tumor-bearing mouse. Left: 4-pixel ROIs used. ROIs 1 and 2 are significant via the pixel-wise analysis for all filters, while ROI 3 is not significant for the SW filter (Table 1, Supplementary Fig. S2). Center: log10(p) values over time for each filter in each ROI. Right: Binary classification of change as significant (s.) or not (n.s.), as well as logistic curve fit and indicated switching time. The SW filter is conspicuously noisy throughout time for all ROIs, but particularly so in ROI 3, causing an unreasonable fit of the switching time. The filter denoises effectively and has a very sharp transition from non-significant to significant, but incurs a 0.5 min delay in the switching time. The filter denoises effectively while preserving the dynamic transition, giving a consistent switching time for all ROIs (see Table 1).
Table 1.
Comparison of filter performance based on spatial and temporal analyses. The spatial analysis refers to the significance of the [HbO2]MSOT difference before and after the gas challenge transition within each ROI inFig. 10. The temporal analysis refers to the significance of the difference between a single [HbO2]MSOT image at a particular time point before the gas challenge transition and each successive [HbO2]MSOT image during the gas challenge transition.
| ROI | Filter | Spatial analysis |
Temporal analysis |
|||
|---|---|---|---|---|---|---|
| p-value | Scaled p-value | Significant | Mean p-value of | tswitch (min) | ||
| 1 | SW | Yes | 11.53 | |||
| Yes | 11.95 | |||||
| Yes | 11.41 | |||||
| 2 | SW | Yes | 11.77 | |||
| Yes | 12.07 | |||||
| Yes | 11.66 | |||||
| 3 | SW | No | 13.05 | |||
| Yes | 12.03 | |||||
| Yes | 11.45 | |||||
4. Discussion
The filter provides an effective balance between fidelity to spectral dynamics and noise rejection in a wide variety of contexts and under different spectral sampling patterns. Notably, the filter is robust to variations in the parameters and , and thus requires little tuning in order to achieve substantially improved results.
In scenarios where no appreciable change is expected, or where the dynamics are very slow relative to the sampling rate, the filter provides superior performance over the filter due to its effective noise-suppression. This may be used to accommodate piecewise-stationary statistics wherein large changes in the underlying signal occur in between periods of steady-state behavior, or when the noise power is large relative to the true signal variation.
Accommodating high-noise, low-variation environments may be accomplished by setting the measurement noise variance high relative to a lower process noise variance. This causes the filters to bias themselves towards previous estimates, providing benefits for such contexts as measuring hemodynamic cycling. Other dynamic imaging modalities such as real-time functional MRI [48,49] or calcium imaging [50] may benefit from the use of these filters as well, as experimental motifs such as neurofeedback require reduced data noise without incurring substantial analysis delays [51].
Due to the ability of the and filters to incorporate information from previous samples, it may be desirable to choose sampling patterns which acquire one image at each wavelength, rather than oversampling each wavelength sequentially. Although the latter case enables averaging to improve signal to noise ratio, it inherently degrades time resolution and can introduce substantial blurring due to motion [24]. Further, this work only explores the application of these filters to reconstructed images – it is probable that image quality itself could be improved by applying the methods to raw photoacoustic data, in order to better condition the reconstruction process itself, and we will investigate this matter in a forthcoming publication.
Motion may present a challenge for these filters due to their recursive structure. As the and filters are infinite impulse response (IIR) filters, an arbitrarily extreme observation or outlier will affect the output data at any later time. Rejection of spurious data could be accomplished by identifying frames which are misregistered and down-weighting the associated residuals. This identification process may be accomplished via a number of methods, ranging from simple differences of sequential frames for online analysis [24] to sophisticated clustering methods when post-processing is an option [[52], [53], [54]].
The work presented here assumes that , and are constant through time, as well as being homogeneous throughout the image and across channels. As measurement noise may vary between imaging channels and across a field of view, and different regions may have distinct dynamics, a single global tracking index may not be optimal. The filter performs well even with the use of global parameters, but it may nevertheless be desirable to enable these parameters to vary throughout the temporal, spatial, and spectral dimensions.
Although other methods such as Bayesian or Kalman filters might provide better quantitative and qualitative results, the filters described here are very efficient both in terms of computational complexity and required storage, and can be readily implemented in any processing or acquisition pipeline with minimal performance impact. Indeed, the SW and filters each require only a single copy of the spectral image, while the filter only requires the additional storage occupied by the image for substantially improved performance.
As these filters are extremely efficient, it is possible to simultaneously maintain several filters with different parameters, using the data likelihood for each in order to provide an improved estimate of the state [28,29]. This, in turn, may be used to modify the parameters for each filter, giving an efficient adaptive update scheme.
Table 2 shows the time complexities and storage requirements for each of the presented methods, as well as comparisons against the full Kalman filter (KF). We assume an overall multispectral image size of pixels over channels, a measured image size of N, and the use of global parameters for both and filters. For the KF, we assume non-sparse matrices and no control inputs. Timing for the various filters was performed using a single thread on an Intel Xeon E5-2680 v3 processor operating at 2.5 GHz. The algorithm-specific memory footprint was less than 5 MB for all filters tested.
Table 2.
Comparison of filter performance for various filtering methods.
| Time Complexity | Storage Requirement | Time/frame (s) | |
|---|---|---|---|
| SW | O(N) | N*M | 8.41 ( 9.54) x 10−5 |
| O(N) | N*M + 3 | 6.83 (2.26) x 10−4 | |
| O(N) | 2*N*M + 3 | 1.3 ( 0.42) x 10−−3 | |
| Kalman | O(N3M3) | 4N2M2 + N2M + 2N2 +NM+2N | Not tested |
The SW retains only the copy of the multispectral image, and overwrites the values in a given imaging channel with the measured values. It therefore has a time complexity of O(N) and a storage requirement of N*M. The filter retains a copy of the multispectral image as well as a single floating-point value to store the tracking index, or three values to represent the , , and values. The calculations are performed elementwise, and so the calculation has a time complexity of O(N), and a storage of N*M + 3. The filter retains a copy of the multispectral image as well as the dx image, giving it a storage requirement of 2*N*M + 3. The calculations still operate using basic arithmetic, and so the overall cost of an iteration is O(N).
The Kalman filter requires the storage of a N*M x N*M state-transition matrix F, a N*M x N*M covariance matrix P, a N*M x N*M process noise covariance matrix Q, measurement matrices H for each channel, yielding M * (N x N*M), a measurement noise covariance matrix R, of size N*N, the innovation covariance S, of size N*N, and must have storage for the Kalman gain matrix of size N*M x N. There must also be storage for the previous estimate, the measurement itself, and the innovation residuals, adding another M*N+2*N elements. The time complexity is dominated by matrix multiplications and the finding of a matrix inverse, and exceeds O(N3M3). Various optimizations may be performed in order to limit the complexity of the KF, but many induce their own overhead and do not lower the complexity to the linear time seen in these filters [55].
5. Conclusion
The and filters, originally developed for target tracking and now applied to spectral imaging, are able to effectively preserve the spatial, spectral, and temporal structure of MSOT data while improving the signal to noise ratio, and are able to do so in a highly efficient, online fashion. These filters are simple and easily implemented, theoretically optimal, and practically effective. The resultant increases in image quality enable new capabilities and better insight into highly dynamic biological systems, allowing improved hypothesis testing and decision making.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The research was supported in part by the National Institutes of Health (NIH) Grant No. 1 P50 CA196516, and the assistance of the Southwestern Small Animal Imaging Resource through the NIH National Cancer Institute Cancer Center Support Grant No. 1P30 CA142543. The iThera MSOT was purchased under NIH Grant 1 S10 OD018094-01A1. We are grateful to Jeni Gerberich for her expert surgical assistance. XP373 tumor tissue was kindly provided by the UT Southwestern Kidney Cancer SPORE and we appreciate the collegial support of Drs. Kapur, Brugarolas and their staff in acquiring the XP373 tumor line. The RENCA-luc tumor line was kindly provided by Dr. Hans Hammers.
Biographies

Devin O’Kelly is currently a PhD student at the University of Texas Southwestern Medical Center (UTSW) pursuing a degree in Biomedical and Molecular Imaging as well as Computational and Systems Biology. He received his B.S. in Biomedical Engineering with high honors from the University of Texas at Austin. His major research interest is the development and application of experimental techniques and numerical methods in order to probe complex biological systems using high-information imaging. He is a Biomedical High-Performance Computing Fellow at the UTSW BioHPC, developing computational frameworks to enable scalable analyses of massive imaging datasets.

Dr. Yihang Guo is currently a visiting fellow of the Small Animal Imaging Resource at the University of Texas Southwestern Medical Center (UTSW), as well as a surgical doctor of the Department of Gastrointestinal Surgery at The Third XiangYa Hospital of Central South University in China. He received his M.D and Ph.D in Clinical Medicine from Central South University. His major research interest is the development and application of molecular animal imaging for predicting optimal cancer therapy and assessing early response to treatment. He has extensive experience with building multiple animal orthotopic tumor models in kidney, lung, colon, pancreas and tibia.

Dr. Ralph Mason, a chemist by training, has over 25 years’ experience in cancer imaging, therapy, and tumor pathophysiology. He is Professor of Radiology and directs the Small Animal Imaging Resource. His primary research interest is prognostic radiology - developing and implementing methods for predicting optimal cancer therapy and assessing early response to treatment ("precision medicine"). Dr. Mason has experience with diverse imaging modalities and has published extensively regarding technology development and applications to assessing hypoxia, radiation therapy and the effects of vascular disrupting agents.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:10.1016/j.pacs.2020.100184.
Appendix A
Calculation of coefficients for and filters
The calculation of the and coefficients for the tracking filters depends on the order of the filter; the calculated for a 1st order filter ( filter) is different from that calculated for a 2nd order filter ( filter). The equations here were derived in the paper by Kalata [36].
In each case, the value of the tracking index is the same and is calculated using Eq. 5.
(A.1) Filter
| (A.1) |
(A.2) Filter
| (A.2) |
| (A.3) |
| (A.4) |
Appendix B
Dynamic Spectral Phantom
In order to provide a test bench for quantifying the efficacy of the filters, we created well-defined datasets with specific properties to present challenges for filtering in spatial, spectral, and temporal dimensions.
We used a modified form of the Shepp-Logan phantom with all-positive entries, built up from the sum of non-negative parts. This is done as imaging is usually concerned with measuring quantities that are strictly positive. Each of the ellipses which comprise the phantom was given its own time course of spectral components. This allowed us to test the effects of multiple basis elements j present in a given pixel, all of which may follow different dynamics.
| (B.1) |
To test the dynamics, was created as a pair of step functions convolved with a particular exponential kernel for each ellipse.
| (B.2) |
| (B.3) |
| (B.4) |
Where denotes convolution and H is the Heaviside step function. For this work, t1 and t2 were defined to be 1/3 and 2/3 of the total simulation time, respectively.
Definition of oxy-deoxy hemoglobin time courses was performed by first defining the time courses of total hemoglobin (Hbtot) and oxygen saturation (SO2) and then multiplying each concentration time course by the corresponding absorption spectrum as in Eq. B.1
| (B.5) |
| (B.6) |
Total hemoglobin concentration was defined to be constant within each ellipse for all time.
Based on the ground-truth multispectral images, one single-wavelength image was sampled at each point in time to provide a sampled multispectral phantom. The phantom created for this paper used the parameters in Table B1.
Table B1.
Parameters for dynamic multispectral phantom.
| Ellipse | Center (x,y) | Major Axis | Minor Axis | Theta | Hbtot | c0 | c1 | c2 | kresponse |
|---|---|---|---|---|---|---|---|---|---|
| a | (0,0) | 0.69 | 0.92 | 0 | 1 | 0.4 | 0.6 | 0.4 | 0.2 |
| b | (0, −0.0184) | 0.6624 | 0.874 | 0 | 0.8 | 1.0 | 0.5 | 0.0 | 0.3 |
| c | (0.22,0) | 0.11 | 0.31 | −18° | 0.2 | 0.2 | 0.5 | 0.1 | 0.02 |
| d | (−0.22, 0) | 0.16 | 0.41 | 18° | 0.2 | 0.5 | 0.5 | 0.6 | 0.04 |
| e | (0, 0.35) | 0.21 | 0.25 | 0 | 0.1 | 0.2 | 0.9 | 0.2 | 0.2 |
| f | (0, 0.1) | 0.046 | 0.046 | 0 | 0.1 | 0.1 | 0.1 | 0.9 | 0.1 |
| g | (0, −0.1) | 0.046 | 0.046 | 0 | 0.1 | 0.9 | 0.8 | 0.7 | 0.05 |
| h | (−0.08, −0.605) | 0.046 | 0.023 | 0 | 0.1 | 1.0 | 1.0 | 1.0 | 0.4 |
| i | (0, −0.605) | 0.023 | 0.023 | 0 | 0.1 | 0.7 | 0.6 | 0.2 | 0.2 |
| j | (0.06, −0.605) | 0.023 | 0.046 | 0 | 0.1 | 0.9 | 0.2 | 0.9 | 0.1 |
Appendix C
Statistical Comparisons of Different Filtering Approaches
In order to compare the efficacy of the different filtering approaches, we considered the onset of the air-oxygen gas challenge and the quantification of the significance of a given change on both a pixel-wise basis and on an ROI-wise basis. Both analyses were done using the [HbO2]MSOT image time courses from the RENCA-luc mouse data; the XP373 data was analyzed using the pixel-wise approach only.
Pixel-wise comparison
Two image populations from the RENCA-luc mouse image time-series were considered for each filter: , consisting of the [HbO2]MSOT images from filtered frames 1297–1303, such that ; and , consisting of the [HbO2]MSOT images from filtered frames 1697–1703. These time points were chosen to correspond with the blue and green histograms considered in Fig. 6, Fig. 7, and assuming that the dynamic changes over each population’s time range are small relative to the level of noise.
For each pixel , we performed a one-tailed T-test with a null hypothesis that the difference in the means of and is positive: . This test was chosen as we expect the level of oxy-hemoglobin to increase when transitioning from air to oxygen. We assume that issues of spectral coloring are negligible. As the test is repeated across every pixel in the RENCA tumor images, the number of hypotheses tested . The Bonferroni-corrected significance threshold is thus . Repeating this analysis for each pixel results in p-value maps which are used to assign a binary classification of significant if the p-value laid below , and non-significant otherwise. Results are depicted in Supplementary Fig. S6.
For each of , , and as described below, the median significance classification and mean p-value were calculated and used as figures of merit for each filtering method.
The same approach was repeated for each of the XP373 mice, with consisting of the [HbO2]MSOT images from filtered frames 2597–2603, such that ; and , consisting of the [HbO2]MSOT images from filtered frames 3997–4003. Results are depicted in Panel A of Supplementary Figs. S3–S5.
ROI-wise comparison
Three ROI populations within each image were considered for each filter: was chosen to lie in the spine, in a set of pixels whose change was deemed significant for all methods by the pixel-wise comparison. was chosen to lie in the tumor bulk, in a set of pixels whose change was significant for all methods by the spatial comparison. was chosen to lie in the tumor bulk, in a set of pixels deemed significant by the spatial analysis using both the and filters, but was non-significant using the SW filter. consisted of the pixels , consisted of the pixels , and consisted of the pixels .
For each ROI and each time-point between 1300 and 1700, we performed a one-tailed T-test with a null hypothesis that the difference in the means of and is positive: . As the test is repeated for every time-point between 1300 and 1700, the number of hypotheses tested . The Bonferroni-corrected significance threshold is thus .
The classification of the difference in means for each ROI was assigned a value of 1 if the null hypothesis was rejected, and 0 if it was not. This yielded a time course of binary classification for each method for each ROI. A logistic function was then fit to these classification time courses using the equation
and tswitch, the time coordinate of the half-max of the logistic function, used as the indicated switching time.
The mean p-value of the last 6 time points was used to report the degree of significance revealed by each of the filtering methods.
Appendix D. Supplementary data
The following is Supplementary data to this article:
References
- 1.Roger R., Arnold J. Reliably estimating the noise in AVIRIS hyperspectral images. Int. J. Remote Sens. 1996;17:1951–1962. [Google Scholar]
- 2.Sorg B.S., Moeller B.J., Donovan O., Cao Y., Dewhirst M.W. Hyperspectral imaging of hemoglobin saturation in tumor microvasculature and tumor hypoxia development. J. Biomed. Opt. 2005;10 doi: 10.1117/1.2003369. [DOI] [PubMed] [Google Scholar]
- 3.Lutzweiler C., Meier R., Rummeny E., Ntziachristos V., Razansky D. Real-time optoacoustic tomography of indocyanine green perfusion and oxygenation parameters in human finger vasculature. Opt. Lett. 2014;39:4061–4064. doi: 10.1364/OL.39.004061. [DOI] [PubMed] [Google Scholar]
- 4.Oraevsky A.A., Buehler A., Wang L.V., Herzog E., Razansky D., Ntziachristos V. Visualization of mouse kidney perfusion with multispectral optoacoustic tomography (MSOT) at video rate. Proc. SPIE 7899, Photons Plus Ultrasound: Imaging and Sensing. 2011;789914:2011. doi: 10.1117/12.875248. [DOI] [Google Scholar]
- 5.Dean-Ben X.L., Ozbek A., Razansky D. Volumetric real-time tracking of peripheral human vasculature with GPU-accelerated three-dimensional optoacoustic tomography. IEEE Trans. Med. Imaging. 2013;32:2050–2055. doi: 10.1109/tmi.2013.2272079. [DOI] [PubMed] [Google Scholar]
- 6.Hendargo H.C., Zhao Y., Allenby T., Palmer G.M. Snap-shot multispectral imaging of vascular dynamics in a mouse window-chamber model. Opt. Lett. 2015;40:3292–3295. doi: 10.1364/OL.40.003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallidi S., Larson T., Aaron J., Sokolov K., Emelianov S. Molecular specific optoacoustic imaging with plasmonic nanoparticles. Opt. Express. 2007;15:6583–6588. doi: 10.1364/oe.15.006583. [DOI] [PubMed] [Google Scholar]
- 8.Silva A.C., Morse B.G., Hara A.K., Paden R.G., Hongo N., Pavlicek W. Dual-energy (Spectral) CT: applications in abdominal imaging. RadioGraphics. 2011;31:1031–1046. doi: 10.1148/rg.314105159. [DOI] [PubMed] [Google Scholar]
- 9.Novo D., Gregori G., Rajwa B. Generalized unmixing model for multispectral flow cytometry utilizing nonsquare compensation matrices. Cytometry A. 2013;83:508–520. doi: 10.1002/cyto.a.22272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz D.C., Chein I.C. Fully constrained least squares linear spectral mixture analysis method for material quantification in hyperspectral imagery. IEEE Trans. Geosci. Remote. Sens. 2001;39:529–545. doi: 10.1109/36.911111. [DOI] [Google Scholar]
- 11.Heylen R., Burazerovic D., Scheunders P. Fully constrained least squares spectral unmixing by simplex projection. IEEE Trans. Geosci. Remote. Sens. 2011;49:4112–4122. doi: 10.1109/TGRS.2011.2155070. [DOI] [Google Scholar]
- 12.Luís Deán-Ben X., Razansky D. Adding fifth dimension to optoacoustic imaging: volumetric time-resolved spectrally enriched tomography. Light: Sci. Appl. 2014;3:e137. doi: 10.1038/lsa.2014.18. https://www.nature.com/articles/lsa201418#supplementary-information [DOI] [Google Scholar]
- 13.Song W., Tang Z., Zhang D., Burton N., Driessen W., Chen X. Comprehensive studies of pharmacokinetics and biodistribution of indocyanine green and liposomal indocyanine green by multispectral optoacoustic tomography. RSC Adv. 2015;5:3807–3813. doi: 10.1039/c4ra09735a. [DOI] [Google Scholar]
- 14.Ntziachristos V., Deán-Ben X.L., Buehler A., Ntziachristos V., Razansky D., Lin C.P. Vol. 8800. 2013. p. 88000H. (Optical Attenuation Correction in Multispectral Optoacoustic Tomography with Logarithm Unmixing). [DOI] [Google Scholar]
- 15.Wankhede M., Agarwal N., Fraga-Silva R., Raizada M., Oh S.P., Sorg B.S. Spectral imaging reveals microvessel physiology and function from anastomoses to thromboses. J. Biomed. Opt. 2010;15 doi: 10.1117/1.3316299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz R.R., Stevenson R.L. Stochastic modeling and estimation of multispectral image data. IEEE Trans. Image Process. 1995;4:1109–1119. doi: 10.1109/83.403416. [DOI] [PubMed] [Google Scholar]
- 17.Beard P. Biomedical photoacoustic imaging. Interface Focus. 2011;1:602–631. doi: 10.1098/rsfs.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox B.T., Arridge S.R., Beard P.C. Estimating chromophore distributions from multiwavelength photoacoustic images. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 2009;26:443–455. doi: 10.1364/josaa.26.000443. [DOI] [PubMed] [Google Scholar]
- 19.Cox B.T., Laufer J.G., Beard P.C., Arridge S.R. Quantitative spectroscopic photoacoustic imaging: a review. J. Biomed. Opt. 2012;17 doi: 10.1117/1.JBO.17.6.061202. [DOI] [PubMed] [Google Scholar]
- 20.Taruttis A., Morscher S., Burton N.C., Razansky D., Ntziachristos V. Fast multispectral optoacoustic tomography (MSOT) for dynamic imaging of pharmacokinetics and biodistribution in multiple organs. PLoS One. 2012;7:e30491. doi: 10.1371/journal.pone.0030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deán-Ben X.L., Bay E., Razansky D. Functional optoacoustic imaging of moving objects using microsecond-delay acquisition of multispectral three-dimensional tomographic data. Sci. Rep. 2014;4:5878. doi: 10.1038/srep05878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hupple C.W., Morscher S., Burton N.C., Pagel M.D., McNally L.R., Cárdenas-Rodríguez J. A light-fluence-independent method for the quantitative analysis of dynamic contrast-enhanced multispectral optoacoustic tomography (DCE MSOT) Photoacoustics. 2018;10:54–64. doi: 10.1016/j.pacs.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buehler A., Kacprowicz M., Taruttis A., Ntziachristos V. Real-time handheld multispectral optoacoustic imaging. Opt. Lett. 2013;38:1404–1406. doi: 10.1364/OL.38.001404. [DOI] [PubMed] [Google Scholar]
- 24.O’Kelly D., Zhou H., Mason R.P. Tomographic breathing detection: a method to noninvasively assess in situ respiratory dynamics. J. Biomed. Opt. 2018;23:1–6. doi: 10.1117/1.jbo.23.5.056011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallac R.R., Zhou H., Pidikiti R., Song K., Stojadinovic S., Zhao D., Solberg T., Peschke P., Mason R.P. Correlations of noninvasive BOLD and TOLD MRI with pO2 and relevance to tumor radiation response. Magn. Reson. Med. 2014;71:1863–1873. doi: 10.1002/mrm.24846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomaszewski M.R., Gonzalez I.Q., O’Connor J.P.B., Abeyakoon O., Parker G.J.M., Williams K.J., Gilbert F.J., Bohndiek S.E. Oxygen enhanced optoacoustic tomography (OE-OT) reveals vascular dynamics in murine models of prostate cancer. Theranostics. 2017;7:2900–2913. doi: 10.7150/thno.19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince J.L., Links J.M. Pearson Prentice Hall; Upper Saddle River: 2006. Medical Imaging Signals and Systems. [Google Scholar]
- 28.Labbe R. 2018. Kalman and Bayesian Filters in Python.https://github.com/rlabbe/Kalman-and-Bayesian-Filters-in-Python (Accessed 13 September 2019. [Google Scholar]
- 29.Gelman A., Carlin J.B., Stern H.S., Dunson D.B., Vehtari A., Rubin D.B. Chapman and Hall/CRC; 2013. Bayesian Data Analysis. [Google Scholar]
- 30.Beezley J.D., Mandel J. Morphing ensemble Kalman filters. Tellus A. 2008;60:131–140. [Google Scholar]
- 31.Galatsanos N.P., Chin R.T. Restoration of color images by multichannel Kalman filtering. IEEE Trans. Signal Process. 1991;39:2237–2252. [Google Scholar]
- 32.Kim K., Kim B., Kim M., Lee Y., Vauhkonen M. Image reconstruction in time-varying electrical impedance tomography based on the extended Kalman filter. Meas. Sci. Technol. 2001;12:1032. [Google Scholar]
- 33.Mandel J. University of Colorado at Denver and Health Sciences Center, Center for Computational Mathematics Reports; 2006. Efficient Implementation of the Ensemble Kalman Filter. [Google Scholar]
- 34.Ott E., Hunt B.R., Szunyogh I., Zimin A.V., Kostelich E.J., Corazza M., Kalnay E., Patil D., Yorke J.A. A local ensemble Kalman filter for atmospheric data assimilation. Tellus A. 2004;56:415–428. [Google Scholar]
- 35.Blair W. Fixed-gain two-stage estimators for tracking maneuvering targets. IEEE Trans. Aerosp. Electron. Syst. 1993;29:1004–1014. [Google Scholar]
- 36.Kalata P.R. The tracking index: a generalized parameter for α-β and α-β-γ target trackers. IEEE Trans. Aerosp. Electron. Syst. 1984:174–182. [Google Scholar]
- 37.Akhlaghi S., Zhou N., Huang Z. Adaptive adjustment of noise covariance in Kalman filter for dynamic state estimation. 2017 IEEE Power & Energy Society General Meeting. 2017:1–5. [Google Scholar]
- 38.Shepp L.A., Logan B.F. The Fourier reconstruction of a head section. IEEE Trans. Nucl. Sci. 1974;21:21–43. [Google Scholar]
- 39.Dima A., Burton N.C., Ntziachristos V. Multispectral optoacoustic tomography at 64, 128, and 256 channels. J. Biomed. Opt. 2014;19:36021. doi: 10.1117/1.jbo.19.3.036021. [DOI] [PubMed] [Google Scholar]
- 40.Ellis L., Shah P., Hammers H., Lehet K., Sotomayor P., Azabdaftari G., Seshadri M., Pili R. Vascular disruption in combination with mTOR inhibition in renal cell carcinoma. Mol. Cancer Ther. 2012;11:383–392. doi: 10.1158/1535-7163.mct-11-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavía-Jiménez A., Tcheuyap V.T., Brugarolas J. Establishing a human renal cell carcinoma tumorgraft platform for preclinical drug testing. Nat. Protoc. 2014;9:1848. doi: 10.1038/nprot.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W., Hill H., Christie A., Kim M.S., Holloman E., Pavia-Jimenez A., Homayoun F., Ma Y., Patel N., Yell P., Hao G., Yousuf Q., Joyce A., Pedrosa I., Geiger H., Zhang H., Chang J., Gardner K.H., Bruick R.K., Reeves C., Hwang T.H., Courtney K., Frenkel E., Sun X., Zojwalla N., Wong T., Rizzi J.P., Wallace E.M., Josey J.A., Xie Y., Xie X.J., Kapur P., McKay R.M., Brugarolas J. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Wang K., Peng D., Li H., Zhu Y., Zhang S., Liu M., Tian J. Curve-driven-Based acoustic inversion for photoacoustic tomography. IEEE Trans. Med. Imaging. 2016;35:2546–2557. doi: 10.1109/TMI.2016.2584120. [DOI] [PubMed] [Google Scholar]
- 44.Ding L., Luis Dean-Ben X., Lutzweiler C., Razansky D., Ntziachristos V. Efficient non-negative constrained model-based inversion in optoacoustic tomography. Phys. Med. Biol. 2015;60:6733–6750. doi: 10.1088/0031-9155/60/17/6733. [DOI] [PubMed] [Google Scholar]
- 45.Mitcham T., Taghavi H., Long J., Wood C., Fuentes D., Stefan W., Ward J., Bouchard R. Photoacoustic-based sO(2) estimation through excised bovine prostate tissue with interstitial light delivery. Photoacoustics. 2017;7:47–56. doi: 10.1016/j.pacs.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brochu F.M., Brunker J., Joseph J., Tomaszewski M.R., Morscher S., Bohndiek S.E. Towards quantitative evaluation of tissue absorption coefficients using light fluence correction in optoacoustic tomography. IEEE Trans. Med. Imaging. 2017;36:322–331. doi: 10.1109/TMI.2016.2607199. [DOI] [PubMed] [Google Scholar]
- 47.Olefir I., Tzoumas S., Yang H., Ntziachristos V. A bayesian approach to eigenspectra optoacoustic tomography. IEEE Trans. Med. Imaging. 2018;37:2070–2079. doi: 10.1109/tmi.2018.2815760. [DOI] [PubMed] [Google Scholar]
- 48.Kelly M.E., Blau C.W., Griffin K.M., Gobbo O.L., Jones J.F., Kerskens C.M. Quantitative functional magnetic resonance imaging of brain activity using bolus-tracking arterial spin labeling. J. Cereb. Blood Flow Metab. 2010;30:913–922. doi: 10.1038/jcbfm.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lurie D.J., Kessler D., Bassett D.S., Betzel R.F., Breakspear M., Kheilholz S., Kucyi A., Liégeois R., Lindquist M.A., McIntosh A.R. Questions and controversies in the study of time-varying functional connectivity in resting fMRI. Netw. Neurosci. 2020;4:30–69. doi: 10.1162/netn_a_00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy M.C., Chan K.C., Kim S.-G., Vazquez A.L. Macroscale variation in resting-state neuronal activity and connectivity assessed by simultaneous calcium imaging, hemodynamic imaging and electrophysiology. Neuroimage. 2018;169:352–362. doi: 10.1016/j.neuroimage.2017.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sulzer J., Haller S., Scharnowski F., Weiskopf N., Birbaumer N., Blefari M.L., Bruehl A.B., Cohen L.G., DeCharms R.C., Gassert R. Real-time fMRI neurofeedback: progress and challenges. Neuroimage. 2013;76:386–399. doi: 10.1016/j.neuroimage.2013.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taruttis A., Claussen J., Razansky D., Ntziachristos V. Motion clustering for deblurring multispectral optoacoustic tomography images of the mouse heart. J. Biomed. Opt. 2012;17 doi: 10.1117/1.jbo.17.1.016009. [DOI] [PubMed] [Google Scholar]
- 53.Schwarz M., Garzorz-Stark N., Eyerich K., Aguirre J., Ntziachristos V. Motion correction in optoacoustic mesoscopy. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung J., Nguyen L. Motion estimation and correction in photoacoustic tomographic reconstruction. SIAM J. Imaging Sci. 2017;10:216–242. doi: 10.1137/16m1082901. [DOI] [Google Scholar]
- 55.Raitoharju M., Piché R. On computational complexity reduction methods for Kalman filter extensions. Ieee Aerosp. Electron. Syst. Mag. 2019;34:2–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.