Abstract
Introduction
The identification of tumour mutational burden (TMB) as a biomarker of response to programmed cell death protein 1 (PD-1) immunotherapy has necessitated the development of genomic assays to measure this. We carried out comprehensive molecular profiling of cancers using the Illumina TruSight Oncology 500 (TSO500) panel and compared these to whole-genome sequencing (WGS).
Methods
Cancer samples derived from formalin-fixed material were profiled on the TSO500 panel, sequenced on an Illumina NextSeq 500 instrument and processed through the TSO500 Docker pipeline. Either FASTQ files (PierianDx) or vcf files (OncoKDM) were processed to understand clinical actionability.
Results
In total, 108 samples (a mixture of colorectal, lung, oesophageal and control samples) were processed via the DNA panel. There was good correlation between TMB, single-nucleotide variants (SNVs), indels and copy-number variations as predicted by TSO500 and WGS (R2 > 0.9) and good reproducibility, with less than 5% variability between repeated controls. For the RNA panel, 13 samples were processed, with all known fusions observed via orthogonal techniques. For clinical actionability, 72 tier 1 variants and 297 tier 2 variants were detected, with clinical trials identified for all patients.
Conclusions
The TSO500 assay accurately measures TMB, microsatellite instability, SNVs, indels, copy-number/structural variation and gene fusions when compared to WGS and orthogonal technologies. Coupled with a clinical annotation pipeline, this provides a powerful methodology for identification of clinically actionable variants.
Electronic supplementary material
The online version of this article (10.1007/s40291-020-00462-x) contains supplementary material, which is available to authorized users.
Key Points
| Formalin-fixed, paraffin-embedded tumour samples represent a significant diagnostic challenge for diagnostic analysis in molecular pathology |
| The Illumina TruSight Oncology 500 assay provides an option to detect relevant cancer gene mutations and fusions |
| These variants can also be annotated easily with clinical interpretation software for ease of clinical interpretation |
Introduction
Recent developments in next-generation sequencing (NGS) and tumour immunology have allowed the discovery that targeting the CTLA4, programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) receptors using therapeutic monoclonal antibodies [1] can unmask cancer to the immune system, facilitating its immune-mediated destruction. Although initial trials of PD-1 inhibitors had mixed results [2, 3], as with previous targeted therapies, it was determined that a specific tumour genotype was required in order for these inhibitors to be effective, leading to the finding that dramatic regression of tumours could occur with the correct genotype.
In order for tumours to become immunogenic, a high neoepitope load must be generated via hypermutation [4–6], ideally indel/frameshifts or non-synonymous mutations that generate novel proteins that can be recognised by the immune system. These neoepitopes can then be presented via major histocompatibility complex (MHC) in order to aid immune killing [7].
The CHECKMATE [8–10] series of trials have suggested that a specific threshold of “tumour mutational burden” (TMB) must be reached in order for PD-1 blockade to become effective, although this has not been adopted formally, due to a lack of association between TMB and response, although other similar markers such as clonal TMB have shown promise. Although TMB has variable definitions, it is broadly accepted [9] as the number of missense mutations in the tumour genome, either divided by the size of the exome panel (35–45 Mb) or via the size of the human genome for whole-genome sequencing (WGS) (3.3 Gb). Based on the CHECKMATE trials, the suggested TMB threshold is greater than 10 mutations (mut)/Mb, based on the objective response rates of the tumours in these studies not improving much beyond this threshold.
Initially, TMB was measured using WGS and whole-exome sequencing [11]; however, these technologies are not cost-effective currently for routine use in the clinic. Despite the falling cost of NGS reagents, the volume of data required for sufficient coverage of either WGS (200–300 Gb for 60× read depth) or whole-exome sequencing (4–5 Gb for 100× read depth) make these techniques impractical except for dedicated sequencing cores. Secondly, even with high read depth, sufficiently deep coverage in order to identify rare subclonal [12] mutations that may contribute to the neoantigen load is required, of the order of 500×. Thus, whole-genome/exome coverage is not cost-effective [13].
Rizvi et al. [13] demonstrated that in order to accurately measure TMB using an NGS-based assay, a panel size of at least 1.5 Mb is required. This panel size offers opportunities for a pan-cancer assay, as a panel of this size could cover the majority of known driver genes across multiple cancer types. In designing an oncology assay, ideally other types of variations would be included. Recent studies [14, 15] have shown the potential utility of selecting targeted therapies using large gene panels, and therefore a panel should include mutations associated with targeting therapies.
An additional advantage of panel-based designs is the ability to enrich RNA targets. Recent studies have shown the importance of RNA fusions such as the TMPRSS-ERG fusion in prostate cancer [16], the FGFR2 fusion in cholangiocarcinoma [17] and the NTRK fusion in lung and other cancers [18]. These fusions are either targetable with molecularly targeted agents (e.g. larotrectinib [19] or pemigatinib [20]) or are prognostically relevant (i.e. TMPRSS-ERG).
An ideal oncology panel-based assay would have several characteristics [21]: enrichment chemistry rather than polymerase chain reaction (PCR) chemistry for identification of rare alleles with straightforward library preparation; a broad panel that targets the majority of DNA and RNA alterations in cancer; rapid run time; prediction of novel biomarkers such as TMB; and a standardised, reproducible analysis pipeline that can be used in a clinical setting.
In this study, we present our initial results using the Illumina TruSight Oncology 500 (TSO500) assay across a range of cancer types. We benchmarked it against WGS and whole-exome sequencing and determined its ability to detect RNA fusions and copy-number variants.
Materials and Methods
Patient Samples and Ethics
Patient samples were from three cohorts—colorectal, oesophageal and lung—in order to provide a variety of mutations for study across multiple tissue types. For the colorectal cohort, fresh frozen tumours were obtained from an internal cohort of patients that had undergone WGS as part of a pre-pilot prior to the introduction of the 100,000 Genomes project. Matched formalin-fixed, paraffin-embedded (FFPE) blocks from the tumour were obtained and used for the assay. For the oesophageal cohort, sequential oesophageal cancers underwent whole-exome sequencing from a cohort collected at the Oxford Cancer Centre. For the lung cohort, patient samples were obtained from those undergoing routine testing for EGFR mutation status at the Molecular Pathology department of the University Hospitals Birmingham NHS Foundation Trust.
Ethics approval for the study was obtained from the Oxford Ethics committee (reference 05/Q1605/66).
Nucleic Acid Extractions and Quality Assessment
DNA and RNA were extracted from 2 × 5-µm FFPE scrolls on the Covaris E220 evolution (520220, Covaris Ltd, Woodingdean, Brighton, UK) using the truXTRAC FFPE total NA Kit—Column Purification (520220, Covaris Ltd, Woodingdean, Brighton, UK) following the manufacturer’s protocol.
Sixty-five per cent isopropanol was used during RNA purification. On-column DNA digestion was performed after the first wash during RNA purification using the TURBO DNA-free kit (AM1907, Invitrogen, ThermoFisher Scientific, Paisley, UK) following the Covaris protocol. DNA and RNA concentrations were measured on the Qubit 3 Fluorometer (ThermoScientific, Paisley, UK) and the percentage of fragments > 200 nucleotides in size (DV200) was assessed using Tapestation 2200 (Agilent, Cheshire, UK). DNA quality was determined by the Infinium HD FFPE quality control (QC) Assay Protocol (15020981, Illumina, Cambridge, UK).
RNA samples with a DV200 of ≥ 30% and DNA samples with a Delta Cq value of ≤ 5 were used for downstream applications.
Library Preparation
DNA libraries were prepared using the hybrid capture-based TruSight Oncology 500 Library Preparation Kit (Illumina, San Diego, CA, USA) following Illumina’s TruSight Oncology 500 Reference Guide (document # 1000000067621 v00, Illumina Cambridge, UK) with the following modifications:
Genomic DNA (gDNA) was sheared using the Covaris E220 evolution (Covaris Ltd, Woodingdean, Brighton, UK), 8 micro TUBE—50 AFA Fiber Strip V2 (520174, Covaris Ltd, Woodingdean, Brighton, UK) and Rack E220e 8 microTUBE Strip V2 (500437, Covaris Ltd, Woodingdean, Brighton, UK). The size of double-stranded DNA (dsDNA) fragments (90–250 bp) was confirmed using Tapestation 2200 (Agilent, Cheshire, UK) after shearing. A HorizonDx HD753 control (Horizon Discovery, Cambridge, UK) was included with every set of seven test samples. When no beads were involved, reagents were mixed by pipetting up and down ten times. Before the bead-based normalisation, libraries were quantified and sized on the Qubit 3 Fluorometer (ThermoScientific, Paisley, UK) and Tapestation 2200 (Agilent, Cheshire, UK), respectively.
Ten microlitres of each normalised DNA library (maximum of eight libraries per pool) was pooled and incubated at 96 °C for 2 min. The tube containing the library pool was immediately inverted two times to mix, centrifuged briefly and placed on ice for 5 min. Ten microlitres of the library pool was mixed with 190 µL HT1 to make a 1:20 dilution (DIL1). Forty microlitres of DIL1 were mixed with 1360 µL HT1 (for a final library concentration of 1.5 pM), and 2.5 µL of denatured 20 pM PhiX was added (1%). Libraries were sequenced on an Illumina NextSeq 500 instrument.
For WGS libraries, 1 µg of DNA was prepared using the TruSeq DNA library preparation kit (Illumina, San Diego, CA, USA) and sequenced across four lanes of a HiSeq 2500 (Illumina, San Diego, CA, USA).
Bioinformatics
The raw sequencing output was transferred from the sequencing instrument to a bioinformatics server running Ubuntu 18.04LTS. A pre-supplied Docker image (the TSO500 pipeline; Illumina, San Diego, CA, USA) was used to generate TMB and microsatellite instability (MSI) calls. The pipeline consists of several steps. Initially, raw bcl files were converted to sample-specific FASTQ files as specified by the sample index. FASTQ files were then aligned against the hg19 reference genome using Isaac 4; local realignment to indels was performed, and paired-end reads were stitched together, followed by variant calling with the somatic sample caller Pisces. Germline variants were filtered using a proprietary database; then the called variants were annotated to identify synonymous and non-synonymous variants. Actual coverage of the panel compared to the reference coverage was computed, and TMB was calculated based on the number of synonymous and non-synonymous mutations detected divided by the size of the panel successfully sequenced.
Small variants were exported from the TSO500 pipeline and annotated using VEP, then converted using vcf2maf and imported into the maftools module of R/Bioconductor.
TMB calls for whole-genome sequenced control data were carried out using the Genomics England v3 pipeline for calling tumour-normal pairs and used to compare to calls from the TSO500 pipeline. In brief, this pipeline utilised Isaac v3 to align sequence data to the hg19 genome, followed by copy-number variant calling using Canvas and structural variant calling using Manta. Copy-number variation (CNV) calls for the TSO500 files were obtained using the Craft copy-number caller set in somatic tumour only mode. Overlaps were computed using bedtools. Structural variant calls for the TSO500 files were obtained using the Manta structural variant caller set in tumour only mode with a custom modification to the C++ code of the Manta structural variant caller to enable detection with less read support and on amplicon sequencing data. Structural variant overlaps were computed using bedtools.
For clinical actionability, raw FASTQ files (CGW, PierianDx, St. Louis, MO, USA) and UMI collapsed vcf files obtained from the TSO500 v1 Docker image (OncoKDM, OncoDNA, Gosselies, Belgium) were uploaded to their respective data portals and run in their standard analysis mode. The Clinical Genomic Workspace (CGW; PierianDx, St Louis, MO, USA) is a secure web-based Health Insurance Portability and Accountability Act- and General Data Protection Regulations-compliant platform for clinical decision support management. Initially developed by one of the very first medical institutes to launch a routine clinical NGS service for cancer and complex inherited diseases, the CGW encompasses a rules engine built on a curated knowledgebase that is updated weekly. Information from over 18 million publications, including Food and Drug Administration (FDA) and European Medicines Agency (EMA) approvals, National Comprehensive Cancer Network (NCCN), Association of Molecular Pathology (AMP) and European Society for Molecular Oncology (ESMO) guidelines and PubMed articles is coupled with public data sources such as population databases, dbSNP, The Cancer Genome Atlas (TCGA), ClinVar and COSMIC in order to annotate and pre-classify variants for interpretation. Uniquely, the CGW utilises the world's largest clinical interpretation-sharing network that provides variant interpretations in the context of the specific disease defined for the patient at time of accessioning. Although no patient data are transferred, network members can view the clinical interpretations supplied to the clinical team of the provider institution (giving the most up-to-date information with true clinical provenance). Actionability calls were downloaded according to standard AMP tiers. The CGW platform is configurable to accept bcl, FASTQ or vcf files and can process all variant types, including TMB and MSI biomarkers, complex variants, CNVs and fusions.
OncoKDM is a secure web-based ISO27001, IS013485 and GDPR-compliant platform for clinical decision support management and clinical report sharing. Initially developed for its proprietary OncoDEEP products that have been on the market since 2013, OncoKDM encompasses a proprietary daily/weekly curated knowledge database of 22,000 genes, 3,886,000 variants, 792 drugs (including FDA and EMA approvals, NCCN, Compermed and ESMO guidelines), 5000 associated clinical trials and 7000 associated publications. Coupled with several public data sources, OncoKDM accurately retrieves biological and clinical information for proper data interpretation and has already been used for 6 years thanks to the sharing platform OncoSHARE, used by 6500 healthcare professionals in 50 countries worldwide.
Results
DNA Quality Metrics
In total, 108 samples were profiled using the assay, with a median sample age of 2 years (range 4 months–10 years). All samples were from FFPE blocks. All samples were examined by haematoxylin staining and had tumour content > 50%. The input for all assays was 40 ng of DNA and 40 ng of RNA. The suitability of samples for sequencing was determined using the real-time PCR-based Illumina FFPE QC assay. For a sample to pass initial QC, the delta Cycle threshold (dCt) must be under 5. Study samples had a median dCt of 2.46 [interquartile range (IQR) 1.73–4.3]. The maximum dCt run was 13.57, and all samples bar one passed TMB calling. Eight samples failed MSI calling due to poor sample quality (dCt = 5–13.57). Samples were all sequenced despite the initial QC metrics to determine the validity of this measure in determining samples for sequencing.
In terms of DNA sequencing metrics, the median insert size was 92.5 bp (IQR 80–112 bp), the median exon coverage was 185× (IQR 123–247×), and 98.1% and 90.5% of all samples were covered at least 50× and 100×, respectively. The median reads per sample was 126 M (IQR 105–138 M reads); there was a median of 0.9% (IQR 0.25–2.7%) chimeric reads per sample. Median read enrichment was 82.1% (IQR 79–85%).
Mutational Coverage and Spectrum
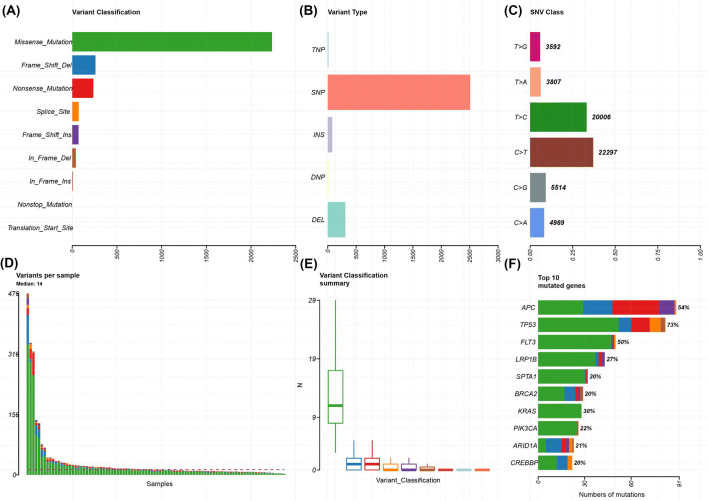
In all 108 samples, mutational coverage of the panel was successfully performed, with little probe drop out. Figures 1, 2 and 3 show the variant classification results (for cancer type variant results, see supplementary Fig. 1 in the electronic supplementary material). There was a median of 14 variants per sample (range 2–479). The predominant mutation type was a missense single-nucleotide polymorphism (SNP), followed by frameshift deletion. The top ten most commonly mutated genes were TP53 (73%), APC (54%), FLT3 (50%), LRP1B (27%), SPTA1 (20%), BRCA2 (20%), KRAS (30%), PIK3CA (22%), ARID1A (21%) and CREBBP (20%). Within the sample subgroups, colorectal cancer was significantly enriched for APC mutations (40/54 colorectal cancer samples), ZFHX3 (13/54 samples) and FBXW7 (13/54). Oesophageal cancer was significantly enriched for EPHA7 mutations (3/9 samples) and TP53 (9/9 samples). For lung, RBM10 (5/22 samples) and MGA (7/22 samples) were significantly enriched.
Fig. 1.
Plot of variant classification for all samples using TSO500. a Total number of variants detected by variant classification. b Total number of variants detected by variant type. c Total number of variants detected by SNV class. d Number of variants per sample. e Box plot of the number of variants within each classification per sample. f Top 10 mutated genes. For d–f, the colours are equivalent to a. DEL deletion, DNP dinucleotide polymorphism, INS insertion, SNP single-nucleotide polymorphism, SNV single-nucleotide variant, TNP trinucleotide polymorphism, TSO500 TruSight Oncology 500
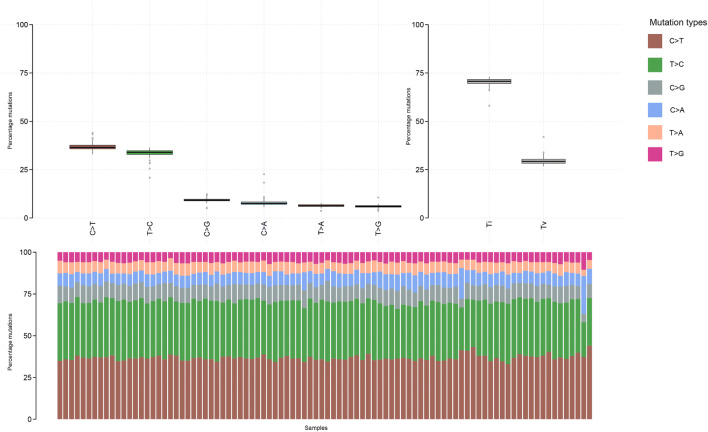
Fig. 2.
Mutational plot for all samples in TSO500 assay. Top left panel: Type of mutation (frequency in percent). Top right panel: Transitions (Ti) vs. transversion (Tv). Bottom panel: Proportion of mutations. TSO500 TruSight Oncology 500
Fig. 3.
OncoStrip plot of common variants across samples. Genes on y-axis. Samples on x-axis
In terms of mutational spectrum, a predominance of C > T transversions was seen, as the samples were all derived (with the exception of controls) from FFPE tissue blocks. The predominant mutational signatures [22] seen in the samples were signature ten (defects in polymerase POLE), signature five (due to tobacco smoke) and signature six (defective mismatch repair), which fit well with the source of the samples (lung, colorectal and oesophageal) as well as the fact that several hypermutant samples were deliberately chosen for the project.
Precision of Control Calls
In order to understand the ability of the assay to detect low variant allele frequencies (VAFs), assessment of the VAF was performed for two known low VAF mutations in the HD753 cell line (Fig. 4). This cell line has validated mutations in AKT1 (E17K, chr14: 105246551C > T), with a VAF of 0.05, and in PIK3CA (E545K, chr3: 178936091G > A), with a VAF of 0.056. The same control was run across 12 runs, with AKT1 median VAF = 0.059 (IQR 0.037–0.072) and PIK3CA median VAF = 0.036 (IQR 0.033–0.0493).
Fig. 4.

Box and whisker plot of repeated measures of VAF of known low VAF variants in a HD753 control. Known VAF is 0.054 for the AKT E17K mutation and 0.05 for the PIK3CA E545K mutation. VAF variant allele frequency
Copy-Number Calls
A subset of 24 samples underwent copy-number calling with the Craft pipeline. A variety of copy-number gains and losses were detected in the 520 genes profiled on the TSO500 panel. The HD753 control was used to determine whether the observed copy-number calls (Fig. 5) correlated with known copy-number changes: amplifications in MET (CNV = 4.5) and MYC-N (CNV = 9.5). The amplification in MET was observed in all control samples, with an average copy number of 4, and in MYC-N, with an average copy number of 9.1. No whole-gene deletions were present in the control samples, but were observed in a variety of samples in the tumour cohort.
Fig. 5.

Copy-number (CN) heat map of detected CN in HD753 samples with known amplification in MYCN and MYC. Sample IDs on y-axis
Structural Variant Calls
The HD753 control is known to have a variety of structural variants, including an SLC34A2/ROS fusion (VAF = 5.6%) and CCDC6/RET fusion (VAF = 5.0%). With use of a custom pipeline, there was evidence for detection of both fusions: 7/506 reads supported the SLC34A2/ROS fusion and 5/498 reads supported the CCDC6/RET fusion. A variety of structural variants were observed in the tumour cohort. In addition, long indels were successfully detected by the Manta pipeline, specifically a 14-bp deletion in EGFR (NM_005228.5:c.2235_2249del), known to be present in the HD753 control.
Tumour Mutational Burden and Microsatellite Instability
TMB calling was successfully performed in 107/108 samples. The one failure was a sample with very poor quality that failed hybridisation. There was good correlation between TMB determined by TSO500 and WGS (R2 = 0.9, Fig. 6). The median TMB was 8.6 mut/Mb (range 0.85–325 mut/Mb, Fig. 7). Several known hypermutant tumour samples were deliberately run first, including a somatic POLE mutant colorectal cancer (reported TMB 261.71 mut/Mb in WGS sample), a somatic MLH1 mutant colorectal cancer (reported TMB 67.43 mut/Mb) and a somatic MSH6 mutant colorectal cancer (reported TMB 104.0 mut/Mb). As the HD753 control was run in each experiment, we compared the reproducibility of the TMB measurement for this control sample. There was a median TMB of 311 mut/Mb (range 289–325) in the HD753 control, a variance in TMB score of ± 5%. Comparison to TCGA tumour cohorts was performed in mafTools and is shown in Fig. 8.
Fig. 6.

Correlation between TMB as measured by TSO500 assay vs. PCR-free WGS on colorectal samples (R2 = 0.98, p < 0.001). PCR polymerase chain reaction, TMB tumour mutational burden, TSO500 TruSight Oncology 500, WGS whole-genome sequencing
Fig. 7.

Distribution of mutations/megabase of DNA by tumour type. Y-axis: NS-TMB in mutations per megabase of DNA. NS-TMB non-synonymous tumour mutational burden
Fig. 8.

Plot of mutational burden in TSO500 trial tumour set as compared to tumours from TCGA. TSO500 trial tumour set shown in black. Y-axis Log10(variants per sample), TCGA The Cancer Genome Atlas, TSO500 TruSight Oncology 500
For MSI (Fig. 9), the threshold for classification as microsatellite instability high (MSI-H) was > 10% of microsatellite sites being unstable. Using this threshold, both known somatic mismatch repair mutant (MLH1 and MSH6) cancers were MSI-H, with 55% and 67% of sites being unstable, respectively. Reassuringly, the POLE mutant cancer had 2% of MSI sites being unstable, meaning it was microsatellite stable (MSS), as is typical in POLE mutant cancer.
Fig. 9.

Percentage of microsatellite unstable probes against tumour type. Colorectal = red, oesophageal = green and lung = blue. MSI microsatellite instability
RNA Fusions
RNA fusion analysis was carried out on 13 samples, of which six had known fusions. Fusions were detected between ETV6 and NTRK3 (three samples), RBPMS and NTRK3 (one sample), EML4 and ALK (one sample) and TG and RET (one sample). All fusions that had previously been identified by fluorescence in situ hybridisation (FISH) were detected using this methodology. A fusion was detected in one sample between ETV6 and NTRK3 that had not been identified via FISH; however, the fusion was supported by 12,627 reads in the sequencing run, which we felt was unlikely to be a false positive, and therefore, we labelled it as a true fusion.
Clinical Actionability
In order to recover as many clinically actionable variants as possible, mutational calls were fed into the OncoDNA OncoKDM (Gosselies, Belgium) and PierianDx CGW (St. Louis, MO, USA) pipelines. OncoDNA was provided with the vcf files coming from the Illumina pipeline, and PierianDx started the analysis directly from the FASTQ files. Also, in order to take an overview of pathway mutations and potential targets, the OncogenicPathways function of mafTools was used to generate a list of druggable pathways. The rat associated sarcoma pathway had 39/85 genes mutated (45.9%), the phosphoinositide 3-kinase pathway had 20/29 (69.0%), and transforming growth factor beta had 6/7 (85.7%).
Using this combined approach, more than 72 tier 1 variants and more than 297 tier 2 variants were identified between the two pipelines. Twenty-one samples were classified as TMB high, 19 as TMB medium and the remainder (64) TMB-low. Clinical trials could also be identified for all samples, with a median of 22 (range 4–105) trials suggested per sample. The most common actionable mutations observed were BRAF p.V600E (18 samples), KRAS p.G13D (14 samples) and KRAS p.G12D (12 samples). For other tiers, there were 8175 tier 3 mutations and 17,649 tier 4 mutations detected.
PierianDx and OncoKDM pipelines were not directly comparable in this study because of the differing inputs (FASTQ for PierianDx, vcf for OncoKDM), but the combination of both platforms provided a comprehensive variant overview.
Discussion
We utilised the TSO500 assay in order to understand its utility and accuracy in determining both the TMB and druggable mutation calls in cancer. One of the key challenges with patient testing [23] is the ability to take a patient biopsy sample with limited input material and produce sequencing data and mutational calls of sufficient quality in order to make decisions on target selection and drug therapy [24].
The assay was designed in its first iteration to measure TMB as a surrogate marker for response to anti–PD-1 immunotherapy, as multiple studies have shown a correlation between TMB and response to this type of therapy [8, 13]. The TSO500 assay performs well in this respect, with accurate measurement of TMB when compared to WGS. Taking a threshold of 10 mut/Mb as “TMB high” (i.e. that which would have benefit for immunotherapy), we found that the TSO500 assay was able to classify samples with 100% accuracy. The precision of the calls varies at the extremes of TMB values, undoubtedly as a factor of panel size in calling TMB at extremely high levels. We conclude that the TSO500 pipeline is usable in the clinical determination of TMB status across a range of clinical sample types and DNA inputs.
We successfully detected MSI in all samples that were known to be MSI-H using TSO500. MSI detection using NGS has been shown to be feasible [25] previously, using a variety of software solutions, usually relying on off-target reads [26], but other assays have used dedicated MSI probes (like the TSO500). We have found that the performance of this approach is variable, as the probes are vulnerable to drop out in FFPE samples. We propose that TMB instead may be a good surrogate biomarker for MSI, as a range of 30–80 mut/Mb is typically seen in MSI tumours, as opposed to MSS POLE/POLD1 tumours, which typically have greater than 150 mut/Mb.
A key requirement for clinical specimens is the ability to process low-input specimens as well as the ability to detect the low VAFs associated with these specimens [27]. Reassuringly, we found that the TSO500 assay performed well at its recommended input concentration and also below these levels. Within our control samples with known VAF (of approximately VAF = 0.05), we determined that there was good precision and reproducibility with minimal variability. Another advantage to tolerance of low sample input is the possibility of using input levels seen in circulating tumour DNA (ctDNA), which are typically 1 ng/mL plasma in most cancers. This would allow derivation of blood TMB [28], which has been shown to be a better biomarker of response in PD-1/PD-L1 inhibitor therapy. The assay is also performed at sufficiently high read depth to allow calculation of clonal TMB [29, 30], another marker associated with more accurate identification of potential response to immunotherapy.
In terms of identifying druggable mutations for targeted therapy selection, the TSO pipeline presents an attractive platform, especially when coupled with a clinical annotation engine such as the two used here (OncoKDM and PierianDx CGW [31]). We found good correlation between mutations detected in WGS experiments, and the identification of druggable mutations was made straightforward by the use of integrated clinical pipelines to produce reproducible data.
CNVs, especially amplifications, represent important therapeutic targets. The TSO500 assay detected the known amplifications in a control sample, meaning that patients can potentially undergo therapeutic targeting. A unique advantage of the TSO500 system is the ability of a partner targeted RNA-seq assay that can detect RNA fusions. We found that the assay reliably detected NTRK [32], ALK [33] and RET [34] fusions that had previously been identified by FISH, as well as a novel fusion not previously detected using other technologies. Intriguingly, we also successfully detected known fusions at the DNA level de novo in the HD753 control sample, suggesting that this methodology may also be valid for future use, although DNA-based fusion calling has a high false negative rate. Fusion genes represent good drug targets, and a number of novel agents [19, 32] have been shown to be active against fusion genes. Detection of circulating RNA for these fusion genes may also be possible [35] using this assay and could be explored further.
Direct comparison of analysis pipelines can be difficult, particularly when the data files fed into the systems have different sources. Although PierianDx hosts Illumina’s secondary analysis for the TSO500 panel and OncoDNA were provided with UMI collapsed vcf files from an Illumina pipeline, there is no indication that the pipeline versions were the same. This, along with the possibility that the same selection criteria and filter settings were not used by the two systems, could account for the fact that the PierianDx CGW platform returned more clinical associations and clinical trials, but this was not investigated here. For the purpose of this study, this difference was not explored as it was clear that both systems were suitable for determining clinical actionability in a routine clinical setting.
The UK 100,000 Genome project has recently been completed, and analysis and reporting are ongoing. The use of WGS for tumour-normal pairs using fresh frozen material still has significant challenges from a cost perspective as well as the practicalities of obtaining fresh frozen tissue over readily available paraffin-embedded material. The TSO500 assay costs approximately one third of the price of a WGS assay, requires no germline DNA control, allows RNA fusion detection and can be implemented on bench-top sequencers. Its main limitations include more laborious library preparation and enrichment chemistry that is vulnerable to drop out.
In conclusion, we believe that the TSO500 assay offers a cost-effective, accurate, pan-cancer assay that can derive SNP, CNV, and gene fusion information across the majority of cancers using a standardised pipeline and therefore is suitable for routine use in precision oncology as a comprehensive genomic profiling solution. In addition, several commercial decision support tools, two of which were successfully tested here, are available to assist in the interpretation of the increased number of variants that are derived from such a large gene set.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Professor Ian Tomlinson (University of Edinburgh) for the contribution of oesophageal cancer DNA samples for this assay.
Author Contributions
Study conception: AB and GM. Molecular analysis: VP, TS, MS, PT and JF. Data analysis: AB, GM and J-FL. Manuscript: All authors.
Data Availability Statement
All data are available in the European Genome Phenome Archive.
Compliance with Ethical Standards
Funding
AB is funded by a Cancer Research UK Advanced Clinician Scientist Fellowship (C31641/A23923). Open access was funded through the Charities Open Access Fund (COAF).
Conflict of interest
AB has received travel and conference funding from Illumina Inc. J-FL is an employee of OncoDNA. GM is an employee of PierianDx. The rest of the authors, VP, MS, TS, JMF, GM and PT, declare no competing interests. Neither Illumina, PierianDx nor OncoKDM had editorial control over the manuscript.
Ethics approval
Ethics approval for the study was obtained from the Oxford Ethics committee (reference 05/Q1605/66).
References
- 1.Couzin-Frankel J. Cancer immunotherapy. Science. 2013;342(6165):1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 2.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosaloglu-Yalcin Z, Lanka M, Frentzen A, Premlal A, Sidney J, Vaughan K, et al. Predicting T cell recognition of MHC class I restricted neoepitopes. Oncoimmunology. 2018;7(11):e1492508. doi: 10.1080/2162402X.2018.1492508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ready N, Hellmann MD, Awad MM, Otterson GA, Gutierrez M, Gainor JF, et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol. 2019;37(12):992–1000. doi: 10.1200/JCO.18.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33(5):853–861. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan J, Hegde PS, Clynes R, Foukas PG, Harari A, Kleen TO, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer. 2016;4:3. doi: 10.1186/s40425-016-0107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fittall MW, Van Loo P. Translating insights into tumor evolution to clinical practice: promises and challenges. Genome Med. 2019;11(1):20. doi: 10.1186/s13073-019-0632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36(7):633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodon J, Soria J-C, Berger R, Miller WH, Rubin E, Kugel A, et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med. 2019;25(5):751–758. doi: 10.1038/s41591-019-0424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Middleton G, Crack LR, Popat S, Swanton C, Hollingsworth SJ, Buller R, et al. The National Lung Matrix trial: translating the biology of stratification in advanced non-small-cell lung cancer. Ann Oncol. 2015;26(12):2464–2469. doi: 10.1093/annonc/mdv394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31(6):882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 17.Jain A, Borad MJ, Kelley RK, Wang Y, Abdel-Wahab R, Meric-Bernstam F, et al. Cholangiocarcinoma with FGFR genetic aberrations: a unique clinical phenotype. JCO Precis Oncol. 2018;2:1–12. doi: 10.1200/PO.17.00080. [DOI] [PubMed] [Google Scholar]
- 18.Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1(2):e000023. doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman DM, Laetsch TW, Kummar S, DuBois SG, Farago AF, Pappo AS, et al. The efficacy of larotrectinib (LOXO-101), a selective tropomyosin receptor kinase (TRK) inhibitor, in adult and pediatric TRK fusion cancers. J Clin Oncol. 2017;35(18_suppl):LBA2501-LBA. doi: 10.1200/JCO.2017.35.18_suppl.LBA2501. [DOI] [Google Scholar]
- 20.Bekaii-Saab TS, Valle JW, Borad MJ, Melisi D, Vogel A, Féliz L, et al. Trial design for a phase 3 study evaluating pemigatinib (INCB054828) versus gemcitabine plus cisplatin chemotherapy in first-line treatment of patients with cholangiocarcinoma with FGFR2 rearrangement. J Clin Oncol. 2019;37(4_suppl):TPS462-TPS. doi: 10.1200/JCO.2019.37.4_suppl.TPS462. [DOI] [Google Scholar]
- 21.Kamps R, Brandao RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18(2):308. doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Einaga N, Yoshida A, Noda H, Suemitsu M, Nakayama Y, Sakurada A, et al. Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation. PLoS ONE. 2017;12(5):e0176280. doi: 10.1371/journal.pone.0176280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lih CJ, Takebe N. Considerations of developing an NGS assay for clinical applications in precision oncology: The NCI-MATCH NGS assay experience. Curr Probl Cancer. 2017;41(3):201–211. doi: 10.1016/j.currproblcancer.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60(9):1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 26.Bonneville R, Krook MA, Chen HZ, Smith A, Samorodnitsky E, Wing MR, et al. Detection of microsatellite instability biomarkers via next-generation sequencing. Methods Mol Biol. 2020;2055:119–132. doi: 10.1007/978-1-4939-9773-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin HT, Choi YL, Yun JW, Kim NKD, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017;8(1):1377. doi: 10.1038/s41467-017-01470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 2019;5(5):696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berland L, Heeke S, Humbert O, Macocco A, Long-Mira E, Lassalle S, et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis. 2019;11(Suppl 1):S71–S80. doi: 10.21037/jtd.2018.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olfson E, Cottrell CE, Davidson NO, Gurnett CA, Heusel JW, Stitziel NO, et al. Identification of medically actionable secondary findings in the 1000 Genomes. PLoS ONE. 2015;10(9):e0135193. doi: 10.1371/journal.pone.0135193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricciuti B, Brambilla M, Metro G, Baglivo S, Matocci R, Pirro M, et al. Targeting NTRK fusion in non-small cell lung cancer: rationale and clinical evidence. Med Oncol. 2017;34(6):105. doi: 10.1007/s12032-017-0967-5. [DOI] [PubMed] [Google Scholar]
- 33.Woo CG, Seo S, Kim SW, Jang SJ, Park KS, Song JY, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol. 2017;28(4):791–797. doi: 10.1093/annonc/mdw693. [DOI] [PubMed] [Google Scholar]
- 34.Kohno T, Tsuta K, Tsuchihara K, Nakaoku T, Yoh K, Goto K. RET fusion gene: translation to personalized lung cancer therapy. Cancer Sci. 2013;104(11):1396–1400. doi: 10.1111/cas.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong Y, Zhao Z, Liu B, Bao A, Zheng H, Gu J, et al. 5'/ 3' imbalance strategy to detect ALK fusion genes in circulating tumor RNA from patients with non-small cell lung cancer. J Exp Clin Cancer Res. 2018;37(1):68. doi: 10.1186/s13046-018-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the European Genome Phenome Archive.