Chemical modifications play an important role in modifying and regulating the functions of coding and noncoding RNAs. Noncoding RNAs mainly include microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs). A common feature of these noncoding RNAs is that they are all transcribed from the genome and can perform biological functions. Importantly, the stable expression of noncoding RNAs in vivo makes them potential biomarkers for the diagnosis, prognosis, and clinical treatment of cancers and a variety of immunological diseases.1,2
circRNAs are a new type of regulatory RNA that are widely expressed in eukaryotic cells and participate in a multitude of biological processes. Unlike linear RNAs with a 5′ cap and 3′ tail structure, circRNAs have a unique circular covalently closed structure, which protects them from exonucleases. Although the mechanisms of circRNA generation and function are not clear, emerging evidence has shown that circRNAs are involved in epithelial–mesenchymal transition, protein translation (as templates), regulation of gene expression and can also act as miRNA sponges.3 In addition, circRNAs play an important role in the initiation and progression of human diseases, especially tumorigenesis and antitumor immunity.3,4 Surprisingly, recent breakthrough investigations have demonstrated that N6-methyladenosine (m6A) modification occurs in circRNAs and promotes protein translation through recruitment of the initiation factor eIF4G2 and the m6A reader YTHDF3.4 Altered m6A modifications in messenger RNAs (mRNAs) are widely linked with numerous human diseases.1,4 In addition, m6A modification pathways have also emerged as contributors to circRNA immunity, and exogenous circRNAs potently stimulate immune signaling.5
To date, over 170 cellular RNA modifications have been identified in both coding and a variety of noncoding RNAs. m6A is the most abundant and important modification in eukaryotic mRNAs and regulates mRNA metabolism, including splicing, stability, and translation.1 The m6A modification is catalyzed by three special classes of proteins generally referred to as “writers” (m6A methyltransferases), “erasers” (m6A demethylases), and “readers” (m6A-binding proteins).6 The m6A modification is reversibly installed and removed by writers and erasers, respectively, whereas readers, which are members of the YT521-B homology (YTH) protein family, selectively bind to m6A in RNAs and affect their fate.7
A growing body of evidence indicates that m6A modification controls immune responses.4,7 A study reported that influenza virus and Rous sarcoma virus produced viral transcripts with m6A modifications, thereby inducing an antiviral innate state. Subsequently, the m6A-modified RNAs were unable to stimulate retinoic acid-inducible gene-I (RIG-I)-mediated antiviral immune signaling and induce interferon expression. Furthermore, m6A modification is also involved in the export and translation of signaling molecules, including MAVS, TRAF3, and TRAF6, thereby regulating interferon production in the antiviral innate immune response.4
Interestingly, several studies have revealed that m6A has variable functions during innate immunity and inflammation through cooperation with different m6A writers and readers.8 Another study showed that the levels of m6A dramatically increased in primary human foreskin fibroblasts infected with human cytomegalovirus (HCMV), which is required for viral propagation. Furthermore, the inhibition of the m6A writer METTL3 and reader YTHDF2 led to an increase in the induction of interferon-stimulated genes after viral infection. It was also observed that m6A of the murine Ifnb transcript accelerated its transcript degradation.9 Another study revealed that METTL14 depletion reduced viral replication and stimulated double-stranded DNA- or HCMV-induced IFNB1 mRNA accumulation by increasing both nascent IFNB1 mRNA production and stability.4 Chen et al.5 very recently showed that YTHDF2 inhibited innate immunity by recognizing m6A.
Tumor-specific neoantigens are vital biomarkers that can be used to predict the therapeutic effects of immune checkpoint blockade therapy and as potential target for cancer immunotherapy. A recent study showed that antitumor immunity is regulated by mRNA m6A methylation through the m6A-binding protein YTHDF1 in dendritic cells (DCs). Furthermore, loss of DC-specific YTHDF1 enhances the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells in a mouse model.4 In addition, YTHDF1 depletion promotes interferon-γ (IFN-γ) production, followed by an increase in PD-L1 CD8+ T cells. Therefore, combining PD-L1 checkpoint inhibitors with YTHDF1 depletion could be a potential new therapeutic target for anticancer immunotherapy.4 In a recent study, m6A modification was recognized as a crucial regulator of T cell homeostasis and the immune response to bacterial or viral infection.4 Therefore, these studies suggest that selectively altered m6A levels along with other types of immunotherapies may be efficient in the management of a variety of immunological diseases.
Other recent studies have shown that m6A modifications regulate the generation and function of noncoding RNAs, including miRNAs, lncRNAs, and circRNAs. Furthermore, m6A modification of noncoding RNAs regulates the cleavage, transport, stability, and degradation of the noncoding RNAs themselves through interactions with RNA-binding proteins.4 A recent study revealed that m6A modifications are especially abundant in circRNAs regulated by FTO and the METTL3/14 complex. A single m6A residue is sufficient to induce circRNA translation via recruitment of YTHDF3 and the translation initiation factors eIF4G2 and eIF3A.4
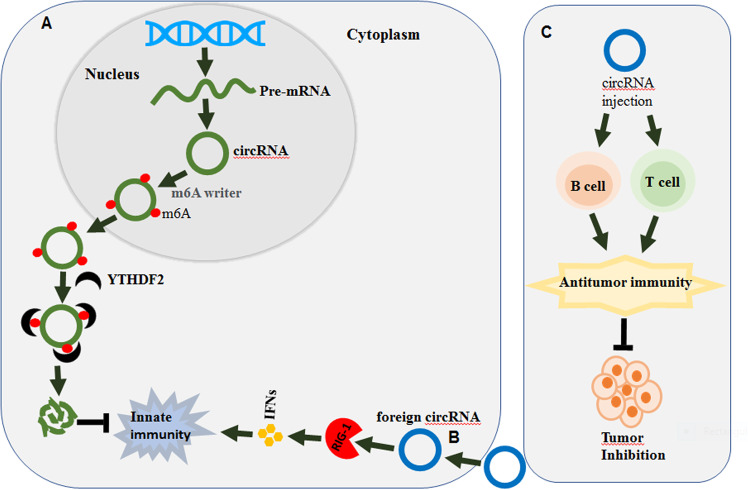
Chen et al.5 reported that m6A modifications of human endogenous circRNAs exerted an important function of suppressing innate immune responses by inhibiting RIG-I activation (Fig. 1a). The authors also showed that exogenous circRNAs activate RIG-I-mediated innate immune responses (Fig. 1b). In addition, exogenous circRNAs induced antigen-specific T and B cell activation, antibody production, and antitumor immunity in vivo (Fig. 1b). They also demonstrated that YTHDF2 was essential for inhibiting innate immunity by recognizing m6A, which might be due to the degradation of m6A-modified circRNAs (Fig. 1a) as several reports have shown that m6A-modified RNAs, including mRNAs and ncRNAs, were degraded through YTHDF2.1,2 Therefore, these results revealed that circRNAs regulate tumor progression through their m6A modifications.
Fig. 1.
The functional role of circRNAs in immune responses. a Regulation of circRNAs by m6A modifications. CircRNAs are regulated by m6A writers (METTL3, METTL14, and WTAP), which induce m6A methylation modifications of circRNAs. Furthermore, YTHDF2 (m6A reader) recognizes the m6A methylation site on the circRNA, thus leading to the degradation of circRNAs and the inhibition of the innate immune response. b Foreign circRNAs induce the innate immune response. Exogenous (foreign) circRNAs activate RIG-I-mediated innate immune responses. c Foreign circRNAs in antitumor immunity. Foreign circRNA injection induces antigen-specific T cell activation, antibody production, and antitumor immunity, which leads to in vivo tumor inhibition
NSUN2 (NOP2/Sun domain family, member 2) is a methyltransferase that plays an important role in cellular development via modification by RNA methylation. Overexpression of NSUN2 has been widely observed in human cancers and implicated in multiple biological processes, including cell proliferation, migration, and human tumorigenesis. Furthermore, experimental reports have provided substantial evidence of circNSUN2 m6A modifications, which are frequently upregulated in tumor tissues and serum samples from colorectal carcinoma (CRC) patients with liver metastasis (LM), eventually leading to poorer patient survival. The study also demonstrated that m6A modification of circNSUN2 modulates cytoplasmic export and stabilizes HMGA2 to promote CRCLM. It has also been suggested that circNSUN2 could be a critical prognostic marker and therapeutic target for such diseases.10 Collectively, these results provide strong evidence that m6A modification of circRNAs is a key regulator in the innate immune response and tumor immunity.
Competing interests
The authors declare no competing interests.
References
- 1.Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019;21:552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 2.Ma S, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019;12:121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vo JN, et al. The landscape of circular RNA in. Cancer Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C, Fu J, Zhou Y. A review in research progress concerning m6A methylation and immunoregulation. Front. Immunol. 2019;10:922. doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YG, et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paramasivam A, Vijayashree Priyadharsini J, Raghunandhakumar S. N6-adenosine methylation (m6A): a promising new molecular target in hypertension and cardiovascular diseases. Hypertens. Res. 2019;43:153–154. doi: 10.1038/s41440-019-0338-z. [DOI] [PubMed] [Google Scholar]
- 7.Paramasivam A, Priyadharsini JV, Raghunandhakumar S. Implications of m6A modification in autoimmune disorders. Cell. Mol. Immunol. 2019;16:1–2. doi: 10.1038/s41423-018-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat. Commun. 2019;10:1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler R, et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019;20:173–182. doi: 10.1038/s41590-018-0275-z. [DOI] [PubMed] [Google Scholar]
- 10.Chen RX, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]