The IFITM3 gene is classified as a member of the small interferon-stimulated genes (ISGs) and plays a paramount role in defense against enveloped viruses, including influenza A viruses.1–6 Previous studies have reported that the rs12252 SNP, which is located in the open reading frame (ORF) of the IFITM3 gene, impacts the splicing procedure of protein production, which makes a truncated form of the IFITM3 protein and lowers its antiviral capacity.7–9 In addition, the rs34481144 SNP genotype affects the transcriptional regulation of the IFITM3 gene and IFITM3-neighboring genes and has a potent association with the clinical results of the H1N1 influenza 2009 pandemic in three human cohorts.10 Because several studies have reported that polymorphism of the IFITM3 gene is crucial in the host immune response, fine mapping of and extracting disease-associated SNPs from the human IFITM3 gene are important baseline studies necessary to understand how the IFITM3 protein functions in innate immunity.
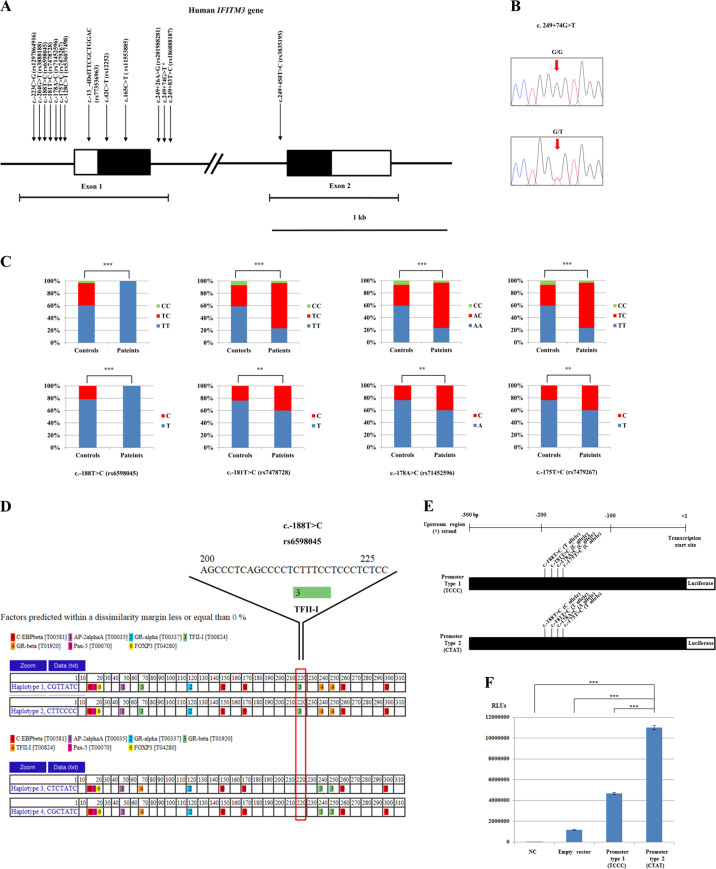
We found a total of 13 SNPs, including 1 novel SNP (Fig. 1a, b). We examined the correlation between the genetic distribution of the IFITM3 gene and the susceptibility of H1N1 pandemic influenza 2009 affected patients. Interestingly, the genotype and allele distributions were significantly different for the 4 regulatory SNPs c.−188T > C, c.−181T > C, c.−178A > C and c.−175T > C (Fig. 1c, Supplementary Table 2). We also found that 4 major haplotypes and H1N1 pandemic influenza 2009-affected patients showed a significantly different distribution (P = 0.0027, Supplementary Table 3). We interpreted the impact of regulatory SNPs, which showed significant association with susceptibility to H1N1 influenza 2009 pandemic virus infection, for the binding capacity of transcription factor using PROMO. The proximal promoter located ~300 base pairs upstream of the transcription start site was classified into 4 major haplotypes, and the binding ability of transcription factors by these haplotypes was analyzed by PROMO. Interestingly, there was a difference in the binding ability of the TFII-I transcription factor between haplotypes 1 and 2 and haplotypes 3 and 4 divided according to the alleles of SNP at the c.−188T > C (rs6598045) of the IFITM3 gene (Fig. 1d).
Fig. 1.
Polymorphisms of the interferon-induced transmembrane protein 3 gene (IFITM3). a Gene map of and polymorphisms identified in the human interferon-induced transmembrane protein 3 gene (IFITM3) on chromosome 11. The open reading frame (ORF) within the exons is indicated by shaded blocks, and the 5′ and 3′ untranslated regions (UTRs) are indicated by white blocks. Edged horizontal bars indicate the regions sequenced. Arrows indicate the polymorphisms found in this study. Asterisks denote novel single nucleotide polymorphisms (SNPs). b Electropherogram of novel SNPs of the human IFITM3 gene. A novel SNP, c.249 + 74G > T, was identified in this study. The four colors indicate individual bases of the DNA sequence using an ABI 3730 automatic sequencer (blue: cytosine, red: thymine, black: guanine, green: adenine). The upper panel indicates a guanine homozygote for c.249 + 74G > T, and the lower panel indicates a guanine/thymine heterozygote for c.249 + 74G > T. c Comparison of the genotype and allele distributions in 4 regulatory SNPs of the IFITM3 gene, which are strongly associated with the susceptibility of influenza H1N1 2009 pandemic infection. d Analysis of the binding ability of transcription factors among 4 haplotypes of proximal promoter sequences of the human IFITM3 gene. The red box indicates the difference in transcription binding between haplotypes 1 and 2 and haplotypes 3 and 4. The Y-shaped bar indicates a zoomed-in area of the c-188T > C region, which showed differences in TFII-I binding between haplotypes 1 and 2 and haplotypes 3 and 4. e Schematic map of functional promoter segments of the IFITM3 gene used in this study. Promoter type 1 consists of the T allele of c.−188T > C, the C allele of c.−181T > C, the C allele of c.−178A > C and the C allele of c.−175T > C. Promoter type 2 consists of the C allele of c.−188T > C, the T allele of c.−181T > C, the A allele of c.−178A > C and the T allele of c.−175T > C. f Promoter activity of the IFITM3 gene based on two promoter types. The symbols *, **, and *** indicate p < 0.05, p < 0.01 and p < 0.001, respectively. RLUs indicate the relative luciferase light unit. NC indicates a negative control
We investigated promoter activity according to alleles of promoter SNPs, which showed different frequencies between healthy controls and 2009 pandemic influenza-infected patients (Supplementary Table 3). Promoter type 1 consists of the T allele of c.−188T > C, the C allele of c.−181T > C, the C allele of c.−178A > C and the C allele of c.−175T > C and is more prevalent in 2009 pandemic influenza-infected patients. Promoter type 2 consists of the C allele of c.−188T > C, the T allele of c.−181T > C, the A allele of c.−178A > C and the T allele of c.−175T > C and is more prevalent in the healthy population (Fig. 1e). Transcription activities of these two promoter types were analyzed by luciferase assay. Notably, Promoter type 2 showed significantly increased mRNA expression levels compared to Promoter type 1 (Fig. 1f).
In the present study, we found a total of thirteen SNPs and one insertion/deletion polymorphism in the IFITM3 gene. However, we did not find any nonsynonymous SNPs. In addition, Korean populations have only the G allele of the rs34481144 SNP, and this polymorphism was not found (Supplementary Table 2). Notably, we found that 4 regulatory SNPs showed significantly different genotype and allele distributions between healthy controls and H1N1 pandemic influenza 2009-affected patients.
Above all, using PROMO, we found that c.−188T > C (rs6598045) induced a difference in the binding capacity of the transcription factor (Fig. 1d). Because TFII-I, which is located at c.−188T > C (rs6598045), is a general transcription factor, it is implied that the SNP at c.−188T > C (rs6598045) can cause a difference in the transcription efficiency of the IFITM3 gene. In addition, we selected two promoter types that showed significant differences between 2009 pandemic influenza-infected patients and healthy populations and analyzed promoter activity. Notably, the promoter activity of Promoter type 2, which is more prevalent in the healthy population, was increased approximately 2-fold more than that of Promoter type 1, which is more prevalent in 2009 pandemic influenza-infected patients. This implies that haplotypes can significantly affect susceptibility to H1N1 influenza 2009 pandemic virus infection based on the alleles of the SNPs of the IFITM3 gene located on the promoter region. Because the proximal promoter of the IFITM3 gene also has strong linkage with the expression levels of neighboring genes, including the IFITM1, IFITM2 and B4GALNT4 genes,3 investigation into the effect of the IFITM3 haplotype according to the expression pattern of neighboring genes is needed in the future.
Supplementary information
Acknowledgements
The biospecimens and data used in this study were provided by the Biobank of Chonbuk National University Hospital, a member of the Korea Biobank Network, which is supported by the Ministry of Health, Welfare and Family Affairs. All samples derived from the Korea Biobank Network were obtained with informed consent under institutional review board-approved protocols. This research was supported by the Basic Science Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education, Science and Technology (2018R1D1A1B07048711). This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876). Min-Ju Jeong was supported by the BK21 Plus Program in the Department of Bioactive Material Sciences. This work was supported by the NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
Author contributions
Y.C.K., M.J.J. and B.H.J. conceived and designed the experiments. Y.C.K. and M.J.J. performed the experiments. Y.C.K. and B.H.J. analyzed the data. Y.C.K., M.J.J. and B.H.J. wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version of this article (10.1038/s41423-019-0322-1) contains supplementary material.
References
- 1.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: the cell’s first line of antiviral defense. Annu Rev. Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey CC, Huang IC, Kam C, Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacey MA, et al. The antiviral restriction factor IFN-induced transmembrane protein 3 prevents cytokine-driven CMV pathogenesis. J. Clin. Invest. 2017;127:1463–1474. doi: 10.1172/JCI84889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K, et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everitt AR, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YH, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YC, Jeong BH. No correlation of the disease severity of influenza A Virus Infection with the rs12252 polymorphism of the interferon-induced transmembrane protein 3 Gene. Intervirology. 2017;60:69–74. doi: 10.1159/000479087. [DOI] [PubMed] [Google Scholar]
- 10.Allen EK, et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017;23:975–983. doi: 10.1038/nm.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.