Abstract
Identification of EGFR mutations is critical to the treatment of primary lung cancer and brain metastases (BMs). Here, we explored whether radiomic features of contrast-enhanced T1-weighted images (T1WIs) of BMs predict EGFR mutation status in primary lung cancer cases. In total, 1209 features were extracted from the contrast-enhanced T1WIs of 61 patients with 210 measurable BMs. Feature selection and classification were optimized using several machine learning algorithms. Ten-fold cross-validation was applied to the T1WI BM dataset (189 BMs for training and 21 BMs for the test set). Area under receiver operating characteristic curves (AUC), accuracy, sensitivity, and specificity were calculated. Subgroup analyses were also performed according to metastasis size. For all measurable BMs, random forest (RF) classification with RF selection demonstrated the highest diagnostic performance for identifying EGFR mutation (AUC: 86.81). Support vector machine and AdaBoost were comparable to RF classification. Subgroup analyses revealed that small BMs had the highest AUC (89.09). The diagnostic performance for large BMs was lower than that for small BMs (the highest AUC: 78.22). Contrast-enhanced T1-weighted image radiomics of brain metastases predicted the EGFR mutation status of lung cancer BMs with good diagnostic performance. However, further study is necessary to apply this algorithm more widely and to larger BMs.
Subject terms: Cancer imaging, Metastasis
Introduction
Lung cancer is one of the leading causes of cancer-related death worldwide, resulting in more than 1.18 million deaths annually1–3. Lung cancer commonly metastasizes to the brain, with 10–36% of all lung cancers developing brain metastasis (BM) during the course of the disease4. The incidence of BMs has increased in recent years, likely because of the prolonged survival of these patients. BM patients today undergo more efficient treatments and are assessed with better imaging techniques than were available previously, enabling the improved detection of BM5,6. Despite advanced therapies and improvements in survival rates, BM remains an important cause of morbidity associated with progressive neurologic deficits7.
Identification of the molecular subtypes of tumors using gene expression may allow a better understanding of their biology and patient-specific treatment: For instance, patients with gliomas with mutation of isocitrate dehydrogenase 1 gene (IDH1) or IDH2 had better outcomes that those with wild-type IDH genes8. Also, O6-methylguanine DNA methyltransferase (MGMT) methylation status might be predictive of temozolomide (TMZ) response, a standard treatment for glioblastoma9. Breast cancer can be divided into three biologic subtypes, based on biomarkers, such as the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth receptor 2 (HER2); each subtype exhibits a distinct prognostic significance10. In the past several decades, identification of epidermal growth factor receptor (EGFR) mutations has become a critical part of treatment planning in advanced lung cancer and particularly in non-small cell lung cancer (NSCLC) cases11. Many recent studies have reported that patients with lung cancer and BMs harboring EGFR mutations exhibit improved survival over patients without the mutations due to higher response rates to whole-brain radiation therapy and specific chemotherapy medications. Such medications include EGFR-associated tyrosine kinase inhibitors (TKIs)12–14. EGFR-TKIs can be used as a first-line treatment for EGFR mutation-positive advanced NSCLC15,16.
Due to its relationship with differential treatment responses, the detection of EGFR mutation status with imaging biomarkers may improve clinical treatments and decision-making. A previous study found that BM imaging using a diffusion weighted approach in NSCLC cases allowed for good prediction of EGFR mutation status17. Recently, several studies have also used radiomics to extract primary brain tumor imaging features from contrast-enhanced T1-weighted images, a commonly used imaging modality18–20. However, the application of radiomic analyses of contrast-enhanced T1-weighted images to metastasis prediction has been rarely reported.
Radiomics is a growing field of diagnostic imaging that aims to non-invasively decode habitats by extracting large amounts of information on imaging features, by feature selection, and through data mining21–23. The heart of radiomics may be the extraction of high-dimensional features to capture attributes of habitats. Radiomic features can be divided into first-, second-, or higher-order statistical outputs. First-order outputs are generally based on histogram analyses and describe the distribution of values across individual voxels without concern for spatial relationships. Second-order outputs are generally based on texture analysis and describe statistical interrelationships between voxels with similar or dissimilar contrast values21,24. For instance, gray level co-occurrence matrix and gray level run length matrix are typical texture features25,26. Higher-order methods impose filters on medical images to extract repetitive or non-repetitive patterns27–30. For example, Laplacian transformations by Gaussian bandpass filtering can extract regions with increasingly coarse texture patterns31. Minkowski filters can assess patterns across voxels with an intensity above a given threshold32. Feature selection is used to resolve the “curse of dimensionality,” which refers to the problem that highly correlated and redundant features may cause overfitting and false discovery33. The most popular and readily-available feature selection algorithms include permutation random forest34, ℓ0-norm minimization35, infinite feature selection36, feature selection via concave minimization37, minimum redundancy maximum relevance38, relief39, and Laplacian40. Data mining is also a vital part of radiomics, which refers to the process of discovering patterns in large datasets. A range of machine learning algorithms have been introduced for data mining purposes, including random forest, support vector machine, adaptive boosting trees, and regularized logistic regression, which are widely used for learning and prediction22,41.
In the present study, we hypothesized that radiomics from contrast-enhanced T1-weighted images of BMs could be applied to predict EGFR mutation status in primary lung cancers. To test this, we extracted imaging features with first-, second, and higher-order methods and subsequently used different combinations of seven feature selection methods and four classification algorithms to identify the most robust analytic models.
Materials and Methods
Participants
We retrospectively reviewed data for a total of 146 lung cancer patients with BMs who underwent gadolinium-enhanced brain MRI at Gangnam Severance Hospital between June 2012 and July 2018. We excluded 85 patients for the following reasons: (1) previous neurosurgery or brain radiation therapy (n = 21), (2) presence of other malignant disease (n = 11), (3) poor image quality (n = 7), (4) absence of EGFR mutation status (n = 20), and (5) no measurable BM (n = 26). We regarded a BM as measurable when its diameter was greater than 3 mm, as it is difficult to differentiate BMs with a diameter of less than 3 mm from adjacent vessels. A total of 61 patients with 210 measurable BMs remained after exclusion. The institutional review board of Gangnam Severance Hospital approved this retrospective study and waived any requirement for informed consent because of its retrospective nature. All data were fully anonymized, and all experiments were carried out in accordance with approved guidelines.
Pathology and EGFR mutation analysis
All patients had histopathological diagnoses of lung cancer by bronchoscopic, percutaneous needle-guided, or surgical biopsies. Genomic DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissues using the DNeasy Isolation Kit (Qiagen, Valencia, CA, USA). We used the PNA ClampTM EGFR Mutation Detection Kit (PANAGENE, Daejeon, Korea) for detection of EGFR mutations by real-time PCR42.
Image processing and extraction of radiomics features
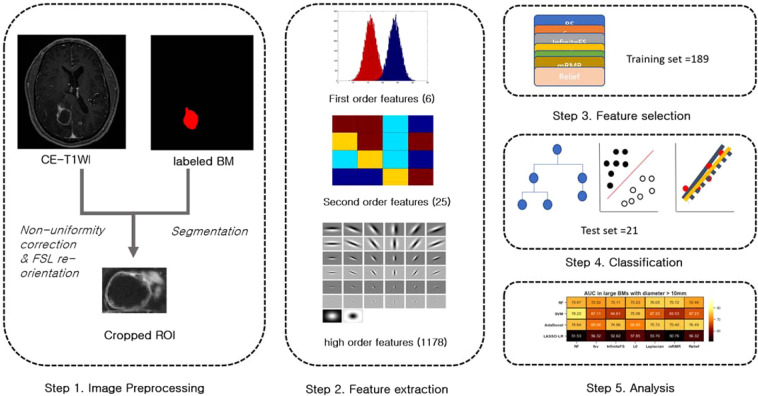
T1-enhanced images were processed with the following steps: preprocessing, feature extraction, feature selection, and classification. For preprocessing, nonuniformity was corrected using the N3 bias correction algorithm, re-orientation was applied for further analysis using FMRIB Software Library (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki), and cropped images including tumor volume were generated by a neuroradiologist (S.J.A) (Fig. 1). All imaging data were normalized to zero-mean and unit-variance to reduce bias. Radiomics features were extracted using MATLAB R2014b (MathWorks), in accordance with previous studies18. The 1209 resultant radiomics features comprised three feature groups: six first-order, 25 second-order, and 1178 higher-order features. First-order features were based on intensity profile histograms (e.g., for mean, variance, skewness, kurtosis, energy, and entropy, Supplemental Table 1). Second-order features were based on texture analysis consisting of 25 features25,43,44 (Supplemental Table 2). For higher-order features, 38 feature maps were created using the root filter set filter bank (Supplemental Table 3)45,46. Six first-order and 25 second order features were also generated for each feature map (1178 features).
Figure 1.
Flow diagram of the study design. (a) Segmentation was performed based on contrast-enhanced T1 weighted images (CE-T1WI). (b) 1209 features were extracted using first-, second- and higher-order methods. (c) Several combinations of seven selection methods and four classification algorithms were used. (d) Area under the curve, accuracy, sensitivity, and specificity were calculated.
Feature selection and classification methods
A ten-fold validation method was applied to the data set (training set = 189, test set = 21). Feature selection was performed with a training set. A two-sample t-test of positive and negative classes was used for each feature to select the most discriminative features, to prevent overfitting, and to reduce feature space dimensions. Seven different feature selection algorithms were used for further feature selection: permutation random forest34, -norm minimization35, infinite feature selection36, a feature selection via concave minimization37, minimum redundancy maximum relevance38, relief39, and Laplacian40.
Classification was performed with four different powerful algorithms to improve diagnostic performance for prediction of EGFR mutation: RF, support vector machine (SVM), adaptive boosting trees, and LASSO-regularized logistic regression47–50. These methods were chosen largely based on their common uses in previous studies and readily available implementation. Models were reestablished with features that were identified in the training set and then applied to the test set. Diagnostic performance was calculated using area under receiver operating characteristic curves (AUC), accuracy, sensitivity, and specificity. A subgroup analysis was performed depending on the size of the metastases (small vs. large). The diameter of small BMs was defined as less than 10 mm (n = 137) and that of large BMs was more than 10 mm (n = 73). For small BMs, ten-fold cross-validation was also used. However, for large BMs, the “leave one out method” was used to maintain a sufficiently large training dataset51.
Statistical analysis
To evaluate a statistical significance of the classification performances, the permutation test was performed with a similar framework performed in previous studies52,53. We randomly permuted the group labels 500 times. In each permutation, the 10-fold cross-validation process was performed based on the permutated samples to calculate the AUCs. We defined p-value as follows;
P –value = (1 + number of time achieving higher AUCs than true lables) / 501(the number of all tests including the original one)
A threshold level of 0.05 was established for significance.
Results
Patient characteristics
Patient characteristics are summarized in Table 1. No significant differences were found in clinical characteristics between EGFR-wild type and EGFR-mutation groups. The mean ages at BM diagnosis were 64.0 ± 9.8 and 62.3 ± 11.6 years (EGFR wild type and EGFR mutation, respectively, p = 0.55). 65.6% of the EGFR wild-type patients (21/33) were male, and 51.7% of the EGFR mutation patients (15/29) were male (p = 0.35). Histologically diagnosed types of primary lung cancer included adenocarcinoma (27/32, 84.3% for EGFR wild type vs. 28/29, 96.6% for EGFR mutation) and small cell (5/32, 15.7% for EGFR wild type vs. 1/29, 3.4% for EGFR mutation, p = 0.26). In patients with EGFR mutation, 14 patients (48.3%) had exon 19 mutations, 11 patients (38%) had exon 21 mutations, 3 patients (10.3%) had exon 20 mutations, and one patient (3.4%) had a combined mutation of exon 19 and 20. Majority of BMs in our cohorts were diagnosed at initial screening (48/61, 79%) and there was no significant difference between two groups (24/32, 75% vs. 24/29, 82.7%, p = 0.67). The mean numbers of measurable BMs per patient were 3.5 ± 3.3 and 3.4 ± 3.0 mm (EGFR wild type and EGFR mutation, respectively, p = 0.90). The total number of measurable BMs was 210 (116 for EGFR wild type vs. 94 for EGFR mutation). The mean diameters of measurable BMs were 10.4 ± 7.4 and 10.8 ± 9.6 mm (EGFR wild type and EGFR mutation, respectively, p = 0.72). The total number of small BMs was 137 (75 for EGFR wild type and 62 for EGFR mutation). The mean diameters of measurable BMs were 5.8 ± 1.6 and 5.5 ± 1.7 mm (EGFR wild type and EGFR mutation, respectively, p = 0.31). The total number of large BMs was 73 (41 for EGFR wild type and 32 for EGFR mutation). The mean diameters of measurable BMs were 19.6 ± 6.4 and 22.2 ± 10.5 mm (EGFR wild type vs. EGFR mutation, respectively, p = 0.24).
Table 1.
Lung cancer patient with brain metastases.
| Characteristics | EGFR wild type (N = 32) | EGFR mutation (N = 29) | P-value |
|---|---|---|---|
| Age (years) | 64.0 ± 9.8 | 62.3 ± 11.6 | 0.55 |
| Sex | 0.35 | ||
| Male | 21(65.6%) | 15(51.7%) | |
| Female | 11(34.4%) | 14(48.3%) | |
| Histology | 0.26 | ||
| Adenocarcinoma | 27(84.3%) | 28(96.6%) | |
| Small cell | 5(15.7%) | 1(3.4%) | |
| Subtype of EGFR mutation | |||
| Exon 18 | 0 | ||
| Exon 19 | 14 (48.3%) | ||
| Exon 20 | 11 (38%) | ||
| Exon 21 | 3 (10.3%) | ||
| Exon 19&Exon 20 | 1 (3.4%) | ||
| BM diagnosis at initial screening | 0.67 | ||
| Yes | 24(75%) | 24(82.7%) | |
| No | 8(25%) | 5(17.3%) | |
| Number of BMs per one patient | 3.5 ± 3.3 | 3.4 ± 3.0 | 0.90 |
| All measurable BMs | |||
| Number | 116 | 94 | |
| Diameter (mm) | 10.4 ± 7.4 | 10.8 ± 9.6 | 0.72 |
| Small BMs(Diameter ≤ 10 mm) | |||
| Number | 75 | 62 | |
| Diameter (mm) | 5.8 ± 1.6 | 5.5 ± 1.7 | 0.31 |
| Large BMs(Diameter > 10 mm) | |||
| Number | 41 | 32 | |
| Diameter (mm) | 19.6 ± 6.4 | 22.2 ± 10.5 | 0.24 |
brain metastases (BM); epidermal growth factor receptor (EGFR).
Diagnostic performance
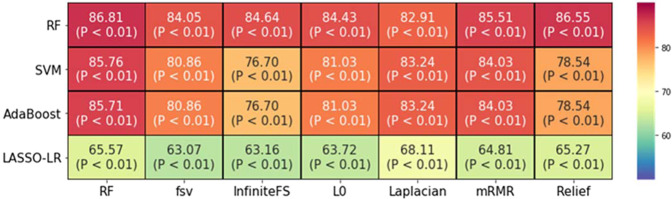
Using radiomic features, individual combinations of the seven selection features and four classification methods showed different EGFR diagnostic performances (AUC) for lung cancer BM (Fig. 2). The random forest classification using random forest selection demonstrated the highest AUC (86.81, p < 0.01). The sensitivity, specificity, and accuracy of this method were 84.41, 72.72, and 86.66, respectively. SVM and AdaBoost using the RF selection method also showed good diagnostic performances (AUC for SVM with RF: 85.76 and AUC for AdaBoost with RF: 85.71). However, LASSO-LR using Laplacian selection demonstrated a relatively poor diagnostic performance (AUC: 68.11, Table 2).
Figure 2.
Heatmap of all brain metastases (BMs) depicting areas under the curve for seven feature selection (columns) and four classification (row) methods.
Table 2.
Diagnostic performance of contrast-enhanced T1-weighted image radiomic-based prediction of EGFR mutation status in lung cancer brain metastases cases.
| Classification | Best feature selection method | Optimal feature number | AUC | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|---|
| RF | RF | 22 | 86.81 | 84.41 | 72.72 | 86.66 |
| SVM | RF | 17 | 85.76 | 82.07 | 81.81 | 86.19 |
| AdaBoost | RF | 18 | 85.71 | 83.093 | 72.72 | 85.23 |
| LASSO-LR | Laplacian | 48 | 68.11 | 55.03 | 81.81 | 69.04 |
Epidermal growth factor receptor (EGFR); area under the curve (AUC); random forest (RF); support vector machine (SVM).
Subgroup analyses
For small BMs, SVM classification using random forest selection demonstrated the highest AUC (89.09, Fig. 3a). The sensitivity, specificity, and accuracy of this method were 89.28, 100, and 89.06, respectively. AdaBoost with mRMR and RF with RF also had good diagnostic performances (AUC: 87.37 and 87.12, respectively). However, LASSO-LR using RF selection exhibited relatively poor diagnostic performance (AUC: 64.16, Table 3).
Figure 3.
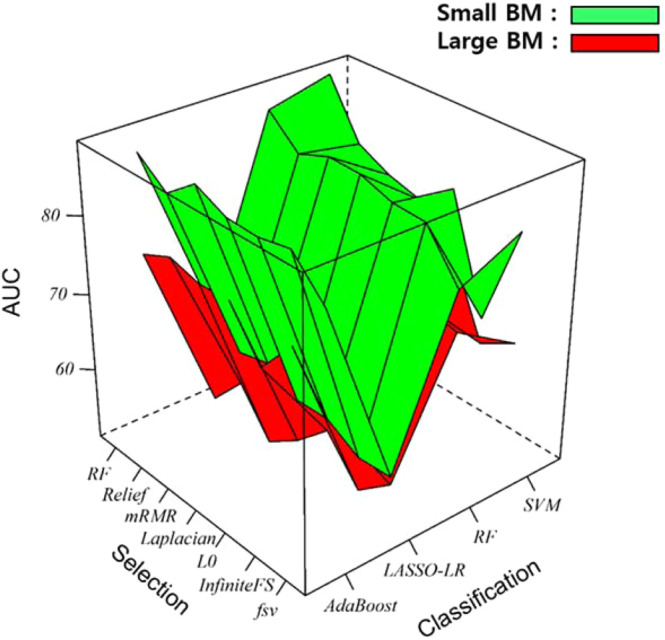
Multiple surface plots for (a) small brain metastases (BMs) (green) and (b) large BMs (red), depicting areas under the curve (AUC) for the seven feature selection (columns) and four classification (row) methods tested.
Table 3.
Subgroup analysis of diagnostic performance for EGFR status in lung cancer brain metastases cases.
| Subgroup | Classification | Best feature selection method | Optimal feature number | AUC | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|---|---|
| Small BMs | |||||||
| RF | RF | 24 | 87.12 | 86.60 | 100 | 86.92 | |
| SVM | RF | 34 | 89.08 | 89.28 | 100 | 89.06 | |
| AdaBoost | mRMR | 35 | 87.37 | 88.21 | 100 | 86.92 | |
| LASSO-LR | RF | 26 | 64.16 | 65.17 | 71.42 | 63.51 | |
| Large BMs | |||||||
| RF | Laplacian | 18 | 76.04 | 62.96 | 89.13 | 79.45 | |
| SVM | RF | 4 | 78.22 | 62.96 | 93.47 | 82.19 | |
| AdaBoost | Relief | 42 | 76.48 | 70.37 | 82.60 | 78.08 | |
| LASSO-LR | L0 | 5 | 57.85 | 22.22 | 93.47 | 67.12 | |
Brain metastases (BM), epidermal growth factor receptor (EGFR); area under the curve (AUC); random forest (RF); support vector machine (SVM).
For large BMs, SVM classification with RF selection demonstrated the highest AUC of 78.22 (Fig. 3b). The sensitivity, specificity, and accuracy of this method were 62.96, 93.47, and 82.19, respectively. AdaBoost with Relief and RF with Laplacian had similar diagnostic performances (AUC: 76.48 and 76.04, respectively). However, LASSO-LR with L0 demonstrated relatively poor diagnostic performance (AUC: 57.85, Table 3).
Discussion
Tumor radiomics utilizes advanced computational methods to convert medical tumor images into a large number of quantitative features54. In the present study, we used seven feature selection methods and four classification methods to extract 1209 features from contrast-enhanced T1 images of 210 BMs. We analyzed the potential value of these features for predicting EGFR mutation status in primary lung cancer cases. We found that radiomics could be used to predict EGFR mutation status with high diagnostic validity. However, LASSO-LR demonstrated relatively poor diagnostic performance, compared with the other classification algorithms tested. Furthermore, diagnosing EGFR mutation status in large BMs (diameter > 10 mm) was not as effective as that in small BMs.
EGFR is a transmembrane protein with cytoplasmic kinase activity that transduces important growth factor signaling from the extracellular milieu into the cell11. Patients with lung cancer and BMs harboring EGFR mutations exhibit better responses to treatment as well as different clinical features. For example, the number of BM lesions was significantly higher in patients with EGFR-mutated NSCLC than in those with wild-type NSCLC. Moreover, leptomeningeal metastases were more common in patients with EGFR-mutated NSCLC55. A recent study proposed an imaging biomarker for the non-invasive determination of EGFR mutation status. Jung et. al reported that the minimum apparent diffusion coefficient (ADC) and normalized ADC ratio of BMs could be independent predictors of EGFR mutation status17. However, diffusion weighted images, which are used to calculate ADC variables, are not a routine sequence in BM protocols and parameters may thus vary between institutions. Meanwhile, contrast-enhanced T1 imaging is a common sequence in BM protocols because it is often used to delineate tumor margins and to monitor tumor responses to therapy. The clinical relevance of our results lies in the development of a novel imaging biomarker for BM EGFR mutation status in lung cancer patients. Of particular interest, this biomarker may be extracted from a commonly used sequence.
The high performance of EGFR mutation status prediction by our model can be explained by multiple factors. First, we generated first-, second-, and higher-order features using a root filter set filter bank. Higher-order features have been reported to help with capturing characteristic features: For example, one study found effective segmentation of white matter hyperintensities using a texton filter bank56. Furthermore, high-order CT features extracted through LoG and wavelet filters were used successfully to quantify non-small cell lung cancer phenotypes21. Second, we used a combination of several feature selection and data mining methods to achieve superior diagnostic performance.
Our results indicate that RF, AdaBoost, and SVM had good diagnostic performance, while LASSO did not. RF and AdaBoost are ensemble learning paradigms, which make predictions based on a number of different decision trees. However, their methodologies differ slightly. RF trains on multiple random subsets of features in a parallel way to arrive at a final conclusion34. Meanwhile, AdaBoost is trained on a number of decision trees sequentially, and each decision tree learns from mistakes made by the previous tree57. Generally, prediction variance decreases when the number of trees in the ensemble method increases. These models are insensitive to overfitting, which might explain their good performance58. SVM classifies by finding the hyperplane59. The hyperplane is calculated from the nearest training samples, called support vectors (SVs) and is optimized by maximizing the margin between the positive and negative SVs. As predicting EGFR status is a two-class problem (wild type or mutant), SV may be best suited for the purposes of the present study. LASSO is a variable selection algorithm used in regression models50. It adds a penalty equal to the absolute value of the magnitude coefficients. LASSO is a linear method and is preferred when true decision boundaries are linear. Thus, it appeared to struggle with handling nonlinear relationships in the data here. Given that LASSO had relatively poor performance in the present study, the relationship between the radiomics of contrast-enhanced T1WI of BMs and EGFR status is likely non-linear.
We identified RF as the most powerful selection tool of those tested here, regardless of classification method. RF selected related features based on importance scores, which are derived from how pure each feature is through numerous yes-or-no questions34. This process involves numerous decision trees, each of which is built via the random extraction of multiple features. Not every tree sees all of the features, guaranteeing that trees are de-correlated and therefore less prone to overfitting, a potential strength over other selection methods.
The performance of our model for large BMs was not as good as that for small BMs, which may be explained by several reasons. First, larger BMs tend to have necrotic centers that may affect machine learning classifications17,60–62. Critically, previous radiomics studies have used different ROI exclusion methods. For instance, Kickingereder et al. excluded ROIs with necrosis, while Kotrotsou et al. insisted that necrotic portions should be included in ROIs63,64. This issue should be further investigated in future work. Second, large BMs are associated with smaller datasets, potentially resulting in overfitting. However, cross-validation techniques and the random forest method diminishes the likelihood of such overfitting34,65.
Accumulating evidence suggests that there are clinico-pathological features that are closely related with EGFR mutations. Mutations have been shown to be associated with Asian ethnicity, adenocarcinoma histology, female sex, and non-smokers11,66. On the basis of results from a large study, these clinico-pathologic features of EGFR seem to be consistent in patients with lung cancer BMs67. In our results, the EGFR mutation group comprised more females and adenocarcinomas than the EGFR wild-type group, but the differences did not reach statistical significance. Thus, a combined model of clinico-pathologic features and radiomic model may enhance diagnostic performance for predicting EGFR mutation status in lung cancer BMs from larger populations which is expected to be validated in future study.
The present study has limitations that warrant consideration. Genetic testing was performed on lung samples rather than BMs themselves. Recent studies have revealed that EGFR mutation status in metastatic lesions does not always coincide with that at primary sites55,68. Indeed, discordant rates of EGFR mutation status between primary lung cancer and BM in previous studies range from 0 to 66.7%69–75. According to meta-analysis, the EGFR discordance rate between primary tumor and central nervous system is 17.26% (95% CI = 7.64 to 29.74)76. There are several models that might explain the discordance of EGFR mutation between primary lung cancer and BM. Cancer cells with highly diverse genetic profiles might be disseminated to distant organs at an early stage, or EGFG mutation status might change though multistep metastatic progression, potentially due to influences from the microenvironment and treatment effects. Thus, further study of tissues obtained directly from brain lesions or animal model with EGFR mutation is necessary to reveal the molecular and biologic characteristics of BMs more precisely. However, we believe our result has a clinical impact because it may aid in clinical decision for first-line treatment of lung cancer. The incidence of BMs in the patients with NSCLC at initial diagnosis is approximately 10%4. On the basis of this report, routine brain MRI screening scan is performed in many institution. Majority of BMs in our cohorts were also diagnosed at initial screening scan (48/61, 79%). In this perspective, our result may provide an alternative method to non-invasively assess EGFR information of primary lung cancer and offers a great supplement to biopsy, thereby making a proper first-line treatment of lung cancer. Also, our result is novel as it provides a different approach with previous other efforts using chest CT scan77,78.
In conclusion, we demonstrated here that T1-enhanced radiomics using RF classification may predict EGFR mutation status in lung cancer BMs with a high degree of accuracy. However, further study is necessary to apply T1-enhanced radiomics to large BMs.
Supplementary information
Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2016R1A2B3016609) to J.M.L. This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2017R1C1B5014927) to S.J.A.
Author contributions
J.M.L. and S.J.A. conceived and designed the study. H.J.K. and J.J.Y. performed an image analysis. M.N.P. and S.H.S. interpreted data. Y.J.C. analyzed pathology. H.J.K. and S.J.A. performed the statistical analyses. S.J.A. and H.J.K. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sung Jun Ahn and Hyeokjin Kwon.
Supplementary information
is available for this paper at 10.1038/s41598-020-65470-7.
References
- 1.Wong MCS, Lao XQ, Ho KF, Goggins WB, Tse SLA. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep. 2017;7:14300. doi: 10.1038/s41598-017-14513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Nayak L, Lee EQ, Wen PY. Epidemiology of brain metastases. Curr Oncol Rep. 2012;14:48–54. doi: 10.1007/s11912-011-0203-y. [DOI] [PubMed] [Google Scholar]
- 4.Villano JL, et al. Incidence of brain metastasis at initial presentation of lung cancer. Neuro Oncol. 2015;17:122–128. doi: 10.1093/neuonc/nou099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Shamy G, Sawaya R. Management of brain metastases: the indispensable role of surgery. J Neurooncol. 2009;92:275–282. doi: 10.1007/s11060-009-9839-y. [DOI] [PubMed] [Google Scholar]
- 6.Bernardo G, et al. First-line chemotherapy with vinorelbine, gemcitabine, and carboplatin in the treatment of brain metastases from non-small-cell lung cancer: a phase II study. Cancer Invest. 2002;20:293–302. doi: 10.1081/CNV-120001173. [DOI] [PubMed] [Google Scholar]
- 7.Klos KJ, O’Neill BP. Brain metastases. Neurologist. 2004;10:31–46. doi: 10.1097/01.nrl.0000106922.83090.71. [DOI] [PubMed] [Google Scholar]
- 8.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegi ME, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 10.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–280. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 11.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 12.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 14.Johnson ML, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119:356–362. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masters GA, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol. 2015;33:3488–3515. doi: 10.1200/JCO.2015.62.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novello S, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27:v1–v27. doi: 10.1093/annonc/mdw326. [DOI] [PubMed] [Google Scholar]
- 17.Jung WS, Park CH, Hong CK, Suh SH, Ahn SJ. Diffusion-Weighted Imaging of Brain Metastasis from Lung Cancer: Correlation of MRI Parameters with the Histologic Type and Gene Mutation Status. AJNR Am J Neuroradiol. 2018;39:273–279. doi: 10.3174/ajnr.A5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kickingereder P, et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin Cancer Res. 2016;22:5765–5771. doi: 10.1158/1078-0432.CCR-16-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itakura H, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med. 2015;7:303ra138. doi: 10.1126/scitranslmed.aaa7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou M, et al. Radiologically defined ecological dynamics and clinical outcomes in glioblastoma multiforme: preliminary results. Transl Oncol. 2014;7:5–13. doi: 10.1593/tlo.13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coroller TP, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119:480–486. doi: 10.1016/j.radonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thawani R, et al. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer. 2018;115:34–41. doi: 10.1016/j.lungcan.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhargava R, Madabhushi A. Emerging Themes in Image Informatics and Molecular Analysis for Digital Pathology. Annu Rev Biomed Eng. 2016;18:387–412. doi: 10.1146/annurev-bioeng-112415-114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, et al. Combination of radiological and gray level co-occurrence matrix textural features used to distinguish solitary pulmonary nodules by computed tomography. J Digit Imaging. 2013;26:797–802. doi: 10.1007/s10278-012-9547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galloway, M. M. Texture analysis using grey level run lengths. NASA STI/Recon Technical Report N75 (1974).
- 27.Leung T, Malik J. Representing and recognizing the visual appearance of materials using three-dimensional textons. International journal of computer vision. 2001;43:29–44. doi: 10.1023/A:1011126920638. [DOI] [Google Scholar]
- 28.Varma, M. & Zisserman, A. Classifying images of materials: Achieving viewpoint and illumination independence in European Conference on Computer Vision 255-271 (Springer, 2002).
- 29.Varma M, Zisserman A. A statistical approach to texture classification from single images. International journal of computer vision. 2005;62:61–81. doi: 10.1007/s11263-005-4635-4. [DOI] [Google Scholar]
- 30.Liu G-H, Yang J-Y. Image retrieval based on the texton co-occurrence matrix. Pattern Recognition. 2008;41:3521–3527. doi: 10.1016/j.patcog.2008.06.010. [DOI] [Google Scholar]
- 31.Grossmann, P., Grove, O. & El-Hachem, N. Identification of molecular phenotypes in lung cancer by integrating radiomics and genomics. Sci Transl Med.
- 32.Larkin TJ, et al. Analysis of image heterogeneity using 2D Minkowski functionals detects tumor responses to treatment. Magn Reson Med. 2014;71:402–410. doi: 10.1002/mrm.24644. [DOI] [PubMed] [Google Scholar]
- 33.Trevor, H., Robert, T. & JH, F. The elements of statistical learning: data mining, inference, and prediction (New York, NY: Springer, 2009).
- 34.Breiman L. Random forests. Machine learning. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 35.Weston J, Elisseeff A, Schölkopf B, Tipping M. Use of the zero-norm with linear models and kernel methods. Journal of machine learning research. 2003;3:1439–1461. [Google Scholar]
- 36.Roffo, G., Melzi, S. & Cristani, M. Infinite feature selection in Proceedings of the IEEE International Conference on Computer Vision 4202–4210 (2015).
- 37.Bradley, P. S. & Mangasarian, O. L. Feature selection via concave minimization and support vector machines in ICML, Vol. 98 82–90 (1998).
- 38.Peng, H., Long, F. & Ding, C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis & Machine Intelligence, 1226–1238 (2005). [DOI] [PubMed]
- 39.Robnik-Šikonja M, Kononenko I. Theoretical and empirical analysis of ReliefF and RReliefF. Machine learning. 2003;53:23–69. doi: 10.1023/A:1025667309714. [DOI] [Google Scholar]
- 40.He, X., Cai, D. & Niyogi, P. Laplacian score for feature selection in Advances in neural information processing systems 507–514 (2006).
- 41.Kotsiantis SB, Zaharakis ID, Pintelas PE. Machine learning: a review of classification and combining techniques. Artificial Intelligence Review. 2006;26:159–190. doi: 10.1007/s10462-007-9052-3. [DOI] [Google Scholar]
- 42.Cho BC, et al. Phase II study of erlotinib in advanced non-small-cell lung cancer after failure of gefitinib. J Clin Oncol. 2007;25:2528–2533. doi: 10.1200/JCO.2006.10.4166. [DOI] [PubMed] [Google Scholar]
- 43.Haralick Robert M., Shanmugam K., Dinstein Its'Hak. Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics. 1973;SMC-3(6):610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 44.Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recognition Letters. 1990;11:415–419. doi: 10.1016/0167-8655(90)90112-F. [DOI] [Google Scholar]
- 45.Martin DR, Fowlkes CC, Malik J. Learning to detect natural image boundaries using local brightness, color, and texture cues. IEEE Trans Pattern Anal Mach Intell. 2004;26:530–549. doi: 10.1109/TPAMI.2004.1273918. [DOI] [PubMed] [Google Scholar]
- 46.Geusebroek J-M, Smeulders AW, Van De Weijer J. Fast anisotropic gauss filtering. IEEE Transactions on Image Processing. 2003;12:938–943. doi: 10.1109/TIP.2003.812429. [DOI] [PubMed] [Google Scholar]
- 47.Gunn SR. Support vector machines for classification and regression. ISIS technical report. 1998;14:5–16. [Google Scholar]
- 48.Kickingereder P, et al. Radiogenomics of Glioblastoma: Machine Learning-based Classification of Molecular Characteristics by Using Multiparametric and Multiregional MR Imaging Features. Radiology. 2016;281:907–918. doi: 10.1148/radiol.2016161382. [DOI] [PubMed] [Google Scholar]
- 49.Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. Journal of computer and system sciences. 1997;55:119–139. doi: 10.1006/jcss.1997.1504. [DOI] [Google Scholar]
- 50.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Methodological) 1996;58:267–288. [Google Scholar]
- 51.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21:3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 52.Ojala M, Garriga GC. Permutation tests for studying classifier performance. Journal of Machine Learning Research. 2010;11:1833–1863. [Google Scholar]
- 53.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human brain mapping. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambin P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eichler AF, et al. EGFR mutation status and survival after diagnosis of brain metastasis in nonsmall cell lung cancer. Neuro Oncol. 2010;12:1193–1199. doi: 10.1093/neuonc/noq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ithapu V, et al. Extracting and summarizing white matter hyperintensities using supervised segmentation methods in Alzheimer’s disease risk and aging studies. Human brain mapping. 2014;35:4219–4235. doi: 10.1002/hbm.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kégl, B. The return of AdaBoost. MH: multi-class Hamming trees. arXiv preprint arXiv:1312.6086 (2013).
- 58.Moradi E, et al. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. Neuroimage. 2015;104:398–412. doi: 10.1016/j.neuroimage.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20:273–297. [Google Scholar]
- 60.Pekmezci M, Perry A. Neuropathology of brain metastases. Surg Neurol Int. 2013;4:S245–255. doi: 10.4103/2152-7806.111302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi YS, et al. Incremental Prognostic Value of ADC Histogram Analysis over MGMT Promoter Methylation Status in Patients with Glioblastoma. Radiology. 2016;281:175–184. doi: 10.1148/radiol.2016151913. [DOI] [PubMed] [Google Scholar]
- 62.Yeom KW, et al. Arterial spin-labeled perfusion of pediatric brain tumors. AJNR Am J Neuroradiol. 2014;35:395–401. doi: 10.3174/ajnr.A3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kotrotsou A, Zinn PO, Colen RR. Radiomics in Brain Tumors: An Emerging Technique for Characterization of Tumor Environment. Magn Reson Imaging Clin N Am. 2016;24:719–729. doi: 10.1016/j.mric.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Kickingereder P, et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology. 2016;280:880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 65.Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Statistics surveys. 2010;4:40–79. doi: 10.1214/09-SS054. [DOI] [Google Scholar]
- 66.Sakurada A, Shepherd FA, Tsao MS. Epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer: impact of primary or secondary mutations. Clin Lung Cancer. 2006;7(Suppl 4):S138–144. doi: 10.3816/clc.2006.s.005. [DOI] [PubMed] [Google Scholar]
- 67.Shin DY, et al. EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J Thorac Oncol. 2014;9:195–199. doi: 10.1097/JTO.0000000000000069. [DOI] [PubMed] [Google Scholar]
- 68.Italiano A, et al. Comparison of the epidermal growth factor receptor gene and protein in primary non-small-cell-lung cancer and metastatic sites: implications for treatment with EGFR-inhibitors. Ann Oncol. 2006;17:981–985. doi: 10.1093/annonc/mdl038. [DOI] [PubMed] [Google Scholar]
- 69.Rau KM, et al. Discordance of Mutation Statuses of Epidermal Growth Factor Receptor and K-ras between Primary Adenocarcinoma of Lung and Brain Metastasis. Int J Mol Sci. 2016;17:524. doi: 10.3390/ijms17040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han HS, et al. EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: discordance in pleural metastases. Clin Lung Cancer. 2011;12:380–386. doi: 10.1016/j.cllc.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Gow CH, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol. 2009;20:696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- 72.Matsumoto S, et al. Frequent EGFR mutations in brain metastases of lung adenocarcinoma. Int J Cancer. 2006;119:1491–1494. doi: 10.1002/ijc.21940. [DOI] [PubMed] [Google Scholar]
- 73.Kalikaki A, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–929. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo D, et al. EGFR mutation status and its impact on survival of Chinese non-small cell lung cancer patients with brain metastases. Tumour Biol. 2014;35:2437–2444. doi: 10.1007/s13277-013-1323-9. [DOI] [PubMed] [Google Scholar]
- 75.Kim KM, et al. Discordance of Epidermal Growth Factor Receptor Mutation between Brain Metastasis and Primary Non-Small Cell Lung Cancer. Brain Tumor Res Treat. 2019;7:137–140. doi: 10.14791/btrt.2019.7.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee CC, et al. Discordance of epidermal growth factor receptor mutation between primary lung tumor and paired distant metastases in non-small cell lung cancer: A systematic review and meta-analysis. PLoS One. 2019;14:e0218414. doi: 10.1371/journal.pone.0218414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Shuo, Shi Jingyun, Ye Zhaoxiang, Dong Di, Yu Dongdong, Zhou Mu, Liu Ying, Gevaert Olivier, Wang Kun, Zhu Yongbei, Zhou Hongyu, Liu Zhenyu, Tian Jie. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. European Respiratory Journal. 2019;53(3):1800986. doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gevaert O, et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep. 2017;7:41674. doi: 10.1038/srep41674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.