Dear Editor,
Spinal muscular atrophy (SMA) is a devastating autosomal recessive motor neuron disease.1,2 Infants with more severe forms of type I SMA die before the age of 2 if no intervention is provided.1,2 Spinraza and zolgensma have been approved by the FDA as SMA therapeutics for pediatric patients.1,2 However, as an antisense oligonucleotide (ASO) based therapy, spinraza requires four loading doses, followed by three annual maintenance doses. The patients would be subjected to repeated intrathecal injections in the procedure. Zolgensma is a single-dose gene-replacement therapy for SMA, but unfortunately is unreliable in maintaining a high, stable level of gene expression. These issues have limited the therapeutic effects of the two approved drugs for SMA.
Genetically, in more than 98% of patients, SMA is caused by homozygous deletion of the survival motor neuron (SMN) 1 gene, resulting in a deficiency of functional full-length SMN (SMN-FL) protein.1,2 SMN2, a human gene paralogous to SMN1, has a translationally silent C-to-T transition at position 6 (C6T) in its 7th exon which converts an exonic splicing enhancer (ESE) to an exonic splicing silencer (ESS), thus ~95% of the SMN2 transcripts produce truncated, non-functional SMN2-Δ7 protein.1,2 Genome editing is already broadening our ability to directly correct genetic mutations in affected cells/tissues to treat diseases,3 and an optimized genome-editing strategy for SMA therapy would be highly desirable. Hua et al. reported that inclusion of SMN2 exon 7 could be enhanced by targeting the ESS in exon 7 via ASO.4 We therefore examined whether this strategy could also be achieved by genome editing (Fig. 1a).
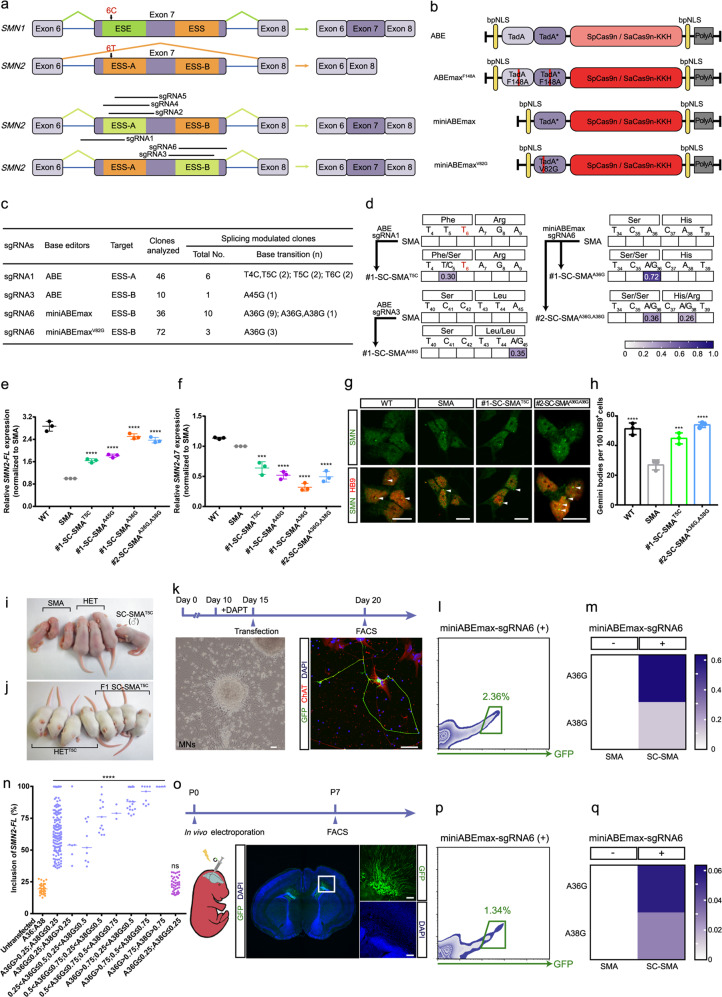
Fig. 1. Base editing-mediated conversion of exonic splicing silencers rescues SMA.
a Schematic of the base editing-mediated conversion of ESS-A and ESS-B in exon 7 of SMN2. Black arrows indicate the difference of a single nucleotide (C6T) between SMN1 and SMN2. Horizontal bars represent sgRNAs. b Maps of the plasmids used in this study. c Base editing efficiencies targeting ESS-A and ESS-B. d Heat map showing the editing efficiencies for an ABE-sgRNA1-edited SMA iPSC, an ABE-sgRNA3-edited SMA iPSC, and two miniABEmax-sgRNA6-edited SMA iPSCs. e, f TaqMan qPCR analysis of full-length SMN2 (SMN2-FL) mRNA (e) and truncated SMN2-Δ7 mRNA (f). Gene expression was normalized to the SMA iPSCs, which was arbitrarily set to 1 (n = 3). Data represent means ± SD. g Nuclear gemini body (white arrows) localization in the MNs derived from iPSCs of four genotypes: SC-SMAA36G,38G, SC-SMAT5C, SMA, and WT, co-stained with SMN (green) and HB9 (red) antibodies. Scale bar, 10 μm. h Quantification of SMN+ gemini bodies in 100 HB9+ motor neurons (n = 3). Data represent means ± SD. i The SC-SMAT5C founder mice were similar in size to the heterozygous littermates (Smn+/−, SMN22TG/0) (termed HET) at postnatal day 9. j SC-SMAT5C progeny mice exhibited no clear phenotypic differences compared to the HETT5C mice. k Timeline for base editing in SMA iPSC-derived postmitotic MNs. Left to right: The morphology of postmitotic MNs at day 15, and the treated postmitotic MNs (at day 20) were co-stained against ChAT (red), GFP (green), and DAPI (blue). Scale bar, 50 μm. l FACS of successfully transfected SMA iPSC-derived postmitotic MNs. m Heat map showing the editing efficiencies for miniABEmax-sgRNA6-edited SMA iPSC-derived postmitotic MNs. n High-throughput sequencing analysis of SMN2-FL transcript expression levels of SMA iPSC-derived single untransfected (far left) and miniABEmax-sgRNA6 transfected postmitotic MNs. Scatter dot plot; the center line indicates the median. o Timeline for base editing in vivo in SMNΔ7 SMA mouse. Immunohistochemistry of transfected neurons with staining against GFP (green) and co-staining with DAPI (blue). Insets in the images are enlarged (original magnification, ×5.0) to the right. Scale bar, 100 μm. p FACS of successfully transfected SMNΔ7 SMA mouse neurons. q Heat map showing the editing efficiencies of miniABEmax-sgRNA6 in neurons derived from SMNΔ7 SMA mouse. ns, not significant; ***P < 0.001; ****P < 0.0001; one-way ANOVA.
Given the potential generation of random sequences caused by non-homologous end joining repair,5 and the low editing efficiency mediated by the CRISPR/Cas9 homology-directed repair pathway,5 we chose to use base editors (Fig. 1b), which can install a precise nucleotide change in a specific genetic locus without inducing double-stranded breaks.6 To this end, we began by designing three single-guide RNAs (sgRNAs) (sgRNAs 1-3) for the KKH variant of Staphylococcus aureus Cas9 nickase (SaCas9n-KKH) (recognizing NNNRRT) with adenine base editor (ABE, generating A-to-G or T-to-C transitions) to target either ESS-A or ESS-B of SMN2 exon 7 locus (Supplementary information, Fig. S1a). We tested the ability of these sgRNAs to restore SMN-FL expression in an SMA patient-derived induced pluripotent stem cells (iPSCs) line harboring three SMN2 copies (Supplementary information, Fig. S1b–d). We also used healthy individual-derived iPSCs as a wild-type (WT) control (two SMN1 copies and three SMN2 copies) (Supplementary information, Fig. S1d, e).
High-throughput sequencing revealed that sgRNA1 and sgRNA3 caused editing activity (Supplementary information, Fig. S1f–h). According to the sequencing and TaqMan-based qPCR analyses, we obtained six splicing correction SMA (SC-SMA) clones (e.g., #1-SC-SMAT5C; Phe-to-Ser) targeting ESS-A and one clone (#1-SC-SMAA45G; Leu-to-Leu) targeting ESS-B (Fig. 1c–f; Supplementary information, Figs. S1i, j, S2a, b). To validate any effects from the missense T5C transition (i.e., T-to-C transition at position 5 of SMN2 exon 7), we selected the #1-SC-SMAT5C clone for further investigation, which showed characteristic expression of pluripotency markers and normal karyotypes (Supplementary information, Fig. S3a, b). A DNA off-target analysis of whole genome sequencing (WGS) data revealed no detectable off-target effects (Supplementary information, Table S1).
To test the effects of correction on splicing in SMA motor neurons (MNs), we differentiated spinal MNs from iPSCs of three genotypes: SC-SMAT5C, SMA, and WT; all of which displayed typical MN differentiation (Supplementary information, Fig. S3e–i). Moreover, immunocytochemistry analysis showed that #1-SC-SMAT5C MNs had more SMN protein gemini bodies than SMA MNs (~1.6-fold increase, P < 0.001) (Fig. 1g, h). Since SMN protein exerts slight anti-apoptotic effects;7 we found that #1-SC-SMAT5C MNs were less sensitive to the apoptosis inducers camptothecin and tunicamycin than SMA MNs (Supplementary information, Fig. S4).
We next generated an edited severe SMA mouse model (Smn−/−, SMN22TG/0) carrying the T5C transition (termed SC-SMAT5C; Smn−/−, SMN2T5C/0) through co-injection of mRNA encoding ABE and sgRNA1 into the cytoplasm of zygotes harvested from mild SMA mice (Smn−/−, SMN22TG/2TG) crossed with heterozygous Smn knock-out mice (Smn+/−) (Supplementary information, Fig. S5a). At postnatal day (P) 9, the SC-SMAT5C founder mouse appeared healthier than its unedited, emaciated, age-matched SMA littermates (Fig. 1i). The male SC-SMAT5C founder mouse was bred with female heterozygous Smn+/− mice, generating 12 live SMA pups, all carrying the T5C transition (SC-SMAT5C) (Supplementary information, Fig. S5b, c), and none of which exhibited any gross phenotypic differences from heterozygous littermates carrying the T5C transition (termed HETT5C; Smn+/−, SMN2T5C/0) (Fig. 1j; Supplementary information, Fig. S5c). However, compared to unedited SMA mice (died within 14 days), SC-SMAT5C mice had extended lifespans (>400 days), elevated body weights, and better motor function (righting-reflex test) (Supplementary information, Fig. S5d–f). Moreover, immunocytochemistry analysis revealed striking increases in the number of gemini bodies present in spinal MNs of SC-SMAT5C mice compared to unedited SMA mice (P < 0.001; Supplementary information, Fig. S5g, h).
Previous work has established that the ABE we initially used does not generate detectable DNA off-target effects;8 however, it generates tens of thousands of off-target RNA single nucleotide variations (resulting from TadA over-expression).9,10 Thus, to minimize RNA off-targets and any biological effects from the T5C transition, we used three recently developed, high-fidelity base editors—TadAF148A-TadA*F148A-ABEmax (termed ABEmaxF148A),10 TadA*-ABEmax (termed miniABEmax),11 and TadA*V82G-ABEmax (termed miniABEmaxV82G)11—along with sgRNAs 1–6 to screen for high-efficiency sgRNAs and to identify any potential target sites for splicing-correction in HEK293T cells, severe SMA mouse-derived embryonic stem cells (mESCs), and SMA iPSCs (Fig. 1b; Supplementary information, Figs. S1a and S6). This screen enabled us to successfully identify a new synonymous A36G transition (Ser-to-Ser) using the combination of miniABEmax and sgRNA6 to target ESS-B of SMN2 exon 7 (Fig. 1c–f; Supplementary information, Figs. S2c–f and S7). Notably, we obtained a #2-SC-SMAA36G,A38G clone bearing two transitions, the aforementioned A36G (Ser-to-Ser) and also A38G (His-to-Arg) (Fig. 1d; Supplementary information, Fig. S3c, d), with no detectable DNA off-target effects (Supplementary information, Table S1). Furthermore, we found that the iPSC-derived #2-SC-SMAA36G,A38G MNs were less sensitive to the apoptosis inducers camptothecin and tunicamycin than SMA MNs (Fig. 1g, h; Supplementary information, Figs. S3e–i and S4).
To assess the feasibility of base editing in postmitotic neurons, we transfected postmitotic MNs derived from SMA iPCSs with miniABEmax and sgRNA6 (Fig. 1k), resulting in successful editing of A36G and A38G transitions (Fig. 1l, m). High-throughput sequencing of single-transfected MNs showed that edited MNs had significantly increased SMN2-FL expression, with higher A36G transition editing efficiency than A38G (Fig. 1n). Furthermore, in vivo neuronal electroporation of miniABEmax-sgRNA6 of SMNΔ7 SMA mouse revealed an editing efficiency of 3~5% for the base transitions (A36G, A38G) (Fig. 1o–q).
In summary, we successfully performed splicing correction of ESS-A and ESS-B of SMN2 exon 7 using base editing, and thereby achieved an efficient and synonymous A36G transition. The biological function of the SMN protein induced by the A36G and A38G synchronized transitions was preliminarily verified by in vitro apoptotic assays; however, the in vivo effect still requires further validation. An efficient and safe system enabling delivery of base editors (e.g., adeno-associated virus (AAV) or nonviral vectors) in vivo should be deployed prior to any feasible gene therapy trials. Our proof-of-principle study illustrates a new therapeutic strategy for treating SMA patients by the base editing-mediated splicing correction.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81771230, 31522037, U1905210, and 31922048), the Joint Funds for the Innovation of Science and Technology of Fujian Province (2017Y9094 and 2018Y9082), the National Key Clinical Specialty Discipline Construction Program, the Key Clinical Specialty Discipline Construction Program of Fujian, the National Science and Technology major project (2017YFC1001302), the Shanghai City Committee of Science and Technology project (16JC1420202), and the Agricultural Science and Technology Innovation Program.
Author contributions
W.-J.C., H.Y. and E.Z. designed this study. X.L., H.C., Y.-Q.L., S.H. and X.H. wrote the initial manuscript and constructed the figures. W.-J.C., H.Y., E.Z., N.W. and L.M. contributed to the editing of the manuscript, figures, and tables. X.L., H.C., Y.-Q.L., S.H. and J.-J.L. performed genome editing in HEK293T cells, SMA mESCs, and SMA iPSCs. X.L. and L.-L.L. performed experiments on motor neurons. X.H., S.H. and Y.G. performed experiments on animals. W.Y. and E.Z. transferred embryos. Z.W. performed the data analysis regarding high-throughput sequencing. H.C. and S.H. performed the statistical analysis. All authors have read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xiang Lin, Haizhu Chen, Ying-Qian Lu, Shunyan Hong, Xinde Hu
Contributor Information
Erwei Zuo, Email: erweizuo@163.com.
Hui Yang, Email: huiyang@ion.ac.cn.
Wan-Jin Chen, Email: wanjinchen75@fjmu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0304-y.
References
- 1.Finkel RS, et al. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 2.Mendell JR, et al. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 3.Cox DB, Platt RJ, Zhang F. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hua Y, et al. PLoS. Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ran FA, et al. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudelli NM, et al. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng SY, et al. Cell Stem Cell. 2015;17:569–584. doi: 10.1016/j.stem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo E, et al. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grünewald J, et al. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C, et al. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 11.Grünewald J, et al. Nat. Biotechnol. 2019;37:1041–1048. doi: 10.1038/s41587-019-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.