Abstract
Lung cancer is the principal cause of leading cancer-related incidence and mortality in the world. Various studies have excavated the potential prognostic biomarkers for cancer patients based on gene expression profiles. However, most of these reported biomarkers lack independent validation in multiple cohorts. Herein, we collected 35 datasets with long-term follow-up clinical information from TCGA (2 cohorts), GEO (32 cohorts), and Roepman study (1 cohort), and developed a web server named OSluca (Online consensus Survival for Lung Cancer) to assess the prognostic value of genes in lung cancer. The input of OSluca is an official gene symbol, and the output web page of OSluca displays the survival analysis summary with a forest plot and a survival table from Cox proportional regression in each cohort and combined cohorts. To test the performance of OSluca, 104 previously reported prognostic biomarkers in lung carcinoma were evaluated in OSluca. In conclusion, OSluca is a highly valuable and interactive prognostic web server for lung cancer. It can be accessed at http:// bioinfo.henu.edu.cn/LUCA/LUCAList.jsp.
Keywords: survival, lung cancer, biomarker, prognosis, OSluca
Introduction
Lung cancer (LUCA) is an aggressive disease with leading mortality and incidence in the world. Based on histology, there are two types of LUCA, including non-small cell lung cancer (NSCLC), which accounts for 80% of LUCA and small cell lung cancer (SCLC), which accounts for approximately 20% of LUCA (Raponi et al., 2006; Bray et al., 2018). NSCLC can be further sub-divided into four subtypes, including adenocarcinoma, squamous cell carcinoma, large cell carcinoma, and bronchioloalveolar carcinoma (Ramalingam et al., 2011). Classical histological subtypes indeed play a dominant role in treatment and prognosis of lung cancer. Recently, reclassification of lung cancer based on tumor biomarkers improves lung cancer therapy (Beer et al., 2002; Hoadley et al., 2018).
Many studies have demonstrated that using clinical-association-prognostic biomarkers can assist the characterization of cancer subtypes and provide new insights of cancer recurrence and patients response to more precise therapies (Meyerson and Carbone, 2005; Bild et al., 2006; Raponi et al., 2006). It is worth noting that numerous single- or multi-prognostic biomarkers have been identified using high-throughput profiling methods (Raponi et al., 2006). By mining a mass of these profiling data deposited in public database, meta-analysis has exploited potential prognostic genes, such as KRT8 (Xie et al., 2019a). However, for biologists and clinicians, it is technically difficult to analyze these massive public data to screen and develop prognostic biomarkers. Previously, we have built several web servers of prognostic biomarker analysis for breast cancer, esophageal carcinoma, etc. (Wang et al., 2019a,b,c, 2020; Xie et al., 2019b,c; Yan et al., 2019; Zhang et al., 2019, 2020; Dong et al., 2020). In this current study, we have integrated bulky RNA expression profiles of lung cancer with clinical survival information, mainly from TCGA (The Cancer Genome Atlas) and GEO (the Gene Expression Omnibus) databases, and built a prognostic analysis web server named OSluca (Online consensus Survival for Lung Cancer) to analyze and evaluate prognostic potency of gene in 35 independent lung cancer cohorts.
Materials and Methods
Collection of Lung Cancer Datasets
The lung cancer cohorts for OSluca with expression profiling and clinical follow-up data were collected from PubMed, TCGA,1 and GEO2 by searching the keywords: “lung” AND “cancer” AND “survival” (Table 1). The dataset for each cohort that met these following criteria will be included in OSluca: (1) have RNA sequencing or gene microarray data; (2) have complete follow-up data, such as overall survival and status (Liu et al., 2018); (3) all the data were specific for lung cancer, not from secondary or metastatic lung tumor from other types of tumors; (4) the cohort size is no less than 30 cases. The primary clinical pathological characteristics of lung cancer patients are listed in Table 1.
TABLE 1.
Summary of clinical characteristics of lung cancer cohorts in Online Consensus Survival for Lung Cancer (OSluca).
|
NSCLC |
SCLC (N = 223) | #NA (N = 85) | |||||
| NSCLC, Total (N = 4937) | AD (N = 3345) | SCC (N = 1381) | LCC (N = 197) | NOS (N = 194) | |||
| Age, year | 64 (13–91) | 64 (13–90) | 66 (39–83) | 63 (39–81) | 62 (22–80) | 64 (40–83) | 58 (15–82) |
| Gender | |||||||
| Male, % | 52.6 | 46.9 | 68.3 | 77.2 | 12.9 | 58.1 | 50 |
| Female, % | 38.8 | 47.7 | 23.7 | 18.1 | 12.4 | 41.9 | 50 |
| #NA, % | 8.6 | 5.4 | 8.0 | 4.7 | 73.7 | 0 | 0 |
| Stage* | |||||||
| I, n | 2301 | 1,653 | 567 | 66 | 28 | 10 | 9 |
| II, n | 889 | 500 | 347 | 27 | 15 | 5 | 4 |
| III, n | 595 | 366 | 199 | 18 | 12 | 2 | 3 |
| IV, n | 101 | 73 | 13 | 2 | 13 | 0 | 0 |
| T stage 1/2/3/4/#NA | 646/1074/230/103/2884 | 468/663/102/49/2063 | 155/362/109/39/716 | 20/44/17/9/107 | 3/5/2/6/178 | 11/13/5/4/190 | 28/20/10/6/21 |
| N Stage 0/1/2/3/#NA | 1638/495/280/21 | 1038/254/198/5/1859 | 549/218/70/7/537 | 48/20/17/5/107 | 3/3/4/4/180 | 14/4/12/6/187 | 33/25/5/1/21 |
| M stage 0/1/#NA | 1685/42/3210 | 853/26/2466 | 740/8/633 | 82/2/113 | 10/6/178 | 33/4/186 | 63/2/20 |
| Smoking/non-smoking/#NA | 1839/262/2836 | 1112/256/1977 | 618/3/760 | 40/1/156 | 9/2/183 | 18/1/204 | 9/8/68 |
| OS, mo | 46 (0.03–256) | 48 (0.03–242) | 41 (0.03–256) | 46 (0.1–216) | 38 (0.5–208) | 51 (2–211) | 68 (2–244) |
| DSS, mo | 42 (0.03–256) | 43 (0.19–242) | 41 (0.03–256) | 45 (1–216) | 36 (6–76) | 24 (2–140) | 69 (2–244) |
| DFI, mo | 33 (0.16–242) | 32 (0.6–242) | 34 (0.16–159) | – | – | – | – |
| PFI, mo | 33 (0.03–242) | 36 (0.03–242) | 30 (0.03–180) | 53 (1.8–164) | 4 (0.23–54) | – | 30 (2–73) |
NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; AD, adenocarcinoma; SCC, squamous cell carcinoma; LCC, large cell cancer; NOS, NSCLC, not otherwise specified; F, female; M, male; n, number; mo, months; OS, overall survival; DSS, disease-specific survival; DFI, disease-free interval; PFI, progression-free interval or recurrence free survival. *The stage only counts stages of lung cancer patients described in the original datasets; #NA, data lost or unknown.
Construction of OSluca Web Server
Online consensus Survival for Lung Cancer is built in a tomcat server as previously described with minor modifications (Wang et al., 2019b, c; Xie et al., 2019b, c; Yan et al., 2019; Zhang et al., 2019). Briefly, front-end application was used for inputting query and displaying the results. Java and R package were used to analyze request and output the results. In addition, profiles and clinical information were stored in the SQL Server database. The prognostic significance of inputted gene is determined by analyzing the association of gene expression and survival time using the R package “survival.” In addition, a genome-wide pre-calculation of Cox proportional regression for all the human genes were performed as well, and the home page of OSluca could display the survival analysis summary with a forest plot and a table of Cox proportional regression result for inputted gene in all cohorts with P-value and HR [(95% confidence interval (CI)] with the built-in upper 25% cutoff. The R package “forestplot” was used to produce the forest plot for inputted gene in OSluca web server.
Validation of Previously Reported Prognostic Biomarkers of Lung Cancer in OSluca
Keywords including “lung cancer,” “survival,” “biomarker,” and “prognosis” were used to search biomarkers of lung cancer in NCBI PubMed. We finally obtained 104 prognostic biomarkers using the following criteria (Table 2): (1) immunohistochemistry (IHC) or qRT-PCR (qPCR) detection of biomarkers in primary cancer tissue; (2) a significant association between biomarker and survival; (3) the sample size must be above 50 cases; (4) the study was published in the English for full access.
TABLE 2.
Clinico-pathological traits of lung cancer cohorts.
NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; AD, adenocarcinoma; SCC, squamous cell carcinoma; LCC, large cell carcinoma; NOS, not otherwise specified; OS, overall survival; DSS, disease-specific survival; DFI, disease-free interval; PFI, progression-free interval.
Statistical Analysis
The association of lung cancer clinical factors and survival outcomes was analyzed by GraphPad Prism 8.0 software. The Cox proportional hazards regression and Kaplan Meier plot functions from R package “survival” were used in the OSluca to determine the association between gene expression and survival. The P ≤ 0.05 was considered statistically significant.
Results
Clinical Characteristics of Lung Cancer Patients in OSluca
To develop an online survival web server for lung cancer, we collected 35 published high-throughput profiling datasets of lung cancer with long-term follow-up information (2 TCGA datasets, 32 GEO datasets, and 1 Roepman dataset). TCGA comprises 513 lung adenocarcinoma cases and 499 squamous cell carcinoma cases (Tables 1, 2). GEO cohorts and Roepman cohort had more than 4,000 samples and 172 samples, respectively, as shown in Table 2. 4,901 patients have OS (overall survival) data; 2,176 patients have DSS (disease-specific survival) data; and 2,075 patients have PFI (progression-free interval or recurrence-free survival) data, while 608 patients have DFI (disease-free interval) data. The results showed that the patients with lung adenocarcinoma significantly survive longer than those of other histological lung cancer, and small cell lung cancer is associated with the worst prognosis compared to other types of lung cancer (Figure 1A). Moreover, other clinical characteristics can also prominently affect patients’ prognosis, such as gender (P < 0.0001), stage (P < 0.0001), p-TNM stage (P < 0.0001), and smoking status (P < 0.0001) (Figures 1B–E). Besides, these risk factors can influence other survival endpoints, such as PFI (data not shown). These results are in accordance with previous researches (Mao et al., 2016; Bray et al., 2018).
FIGURE 1.
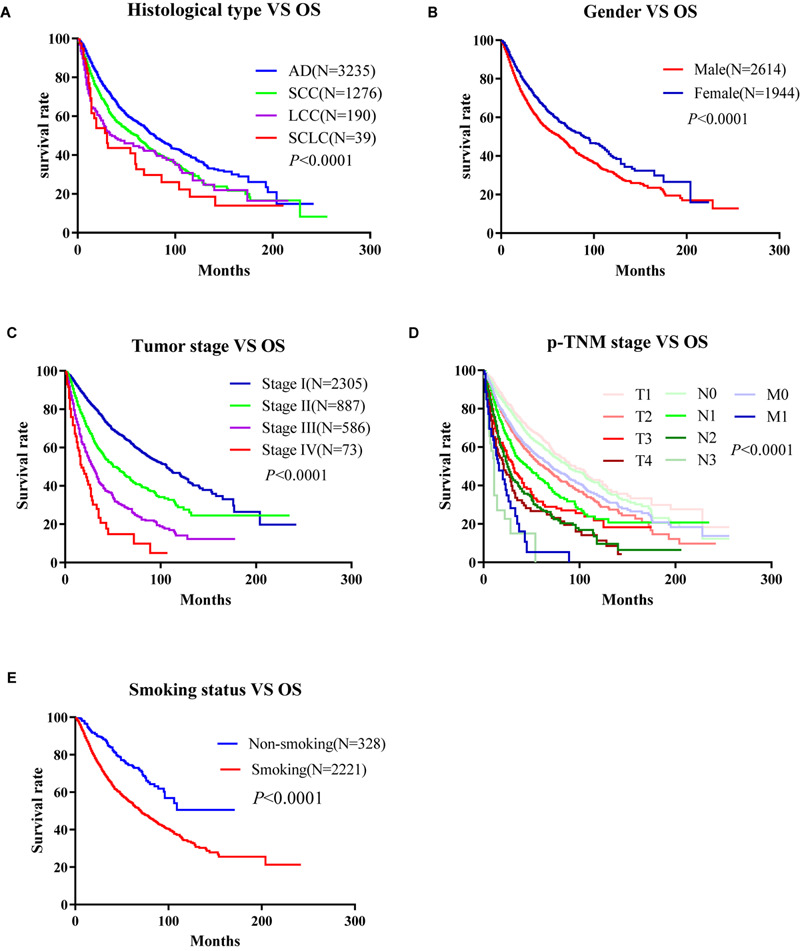
Correlation between the clinico-pathologic characteristics and overall survival of lung cancer in Online Consensus Survival for Lung Cancer (OSluca). (A) Correlation between histological types and OS. (B) Correlation between gender and OS. (C) Correlation between tumor stages and OS. (D) Correlation between p-TNM stages and OS. (E) Correlation between smoking status and OS. OS, overall survival; AD, adenocarcinoma; SCC, squamous cell carcinoma; LCC, large cell cancer.
Construction and Usage of Prognostic Web Server OSluca
Online consensus Survival for Lung Cancer includes a set of optional clinico-pathological factors, such as age, sex, histological type, grade, smoking status, and so on. Four survival endpoints can be selected basing on original patient outcomes, containing OS, DSS, DFI, and PFI (Liu et al., 2018). In order to make the user clearly see the prognostic effect of interested gene, a meta-analysis is to summarize the prognostic value for each gene on the home page of OSluca. Briefly, after the user types the official gene symbol into the input box on the home page, OSluca will display the survival analysis summary with a forest plot and a table from Cox proportional regression in each cohort and combined cohorts (combining all the datasets together). Take the tumor suppressor gene TP53 (tumor protein p53) as an example and type “TP53” into the gene symbol box and click on “Survival analysis” (Figure 2A, left). The meta-analysis results with a forest plot and a survival table for the TP53 gene, will display the P-value and HR with 95% CI of each cohort and the combined cohorts (Figure 2A, right). Then, the user can easily obtain KM plots of separate cohorts such as GSE30219 dataset by clicking on the “Go” button in the survival table (Figure 2B). In addition, it is also available to use a subgroup of certain cohort to obtain specific prognostic information with selectable risk factors, such as cutoff value, histological type, grade, etc. Briefly, OSluca can output survival rates displaying a forest plot and a survival table with KM plot and P-value to measure the association between the investigated gene and survival rate.
FIGURE 2.
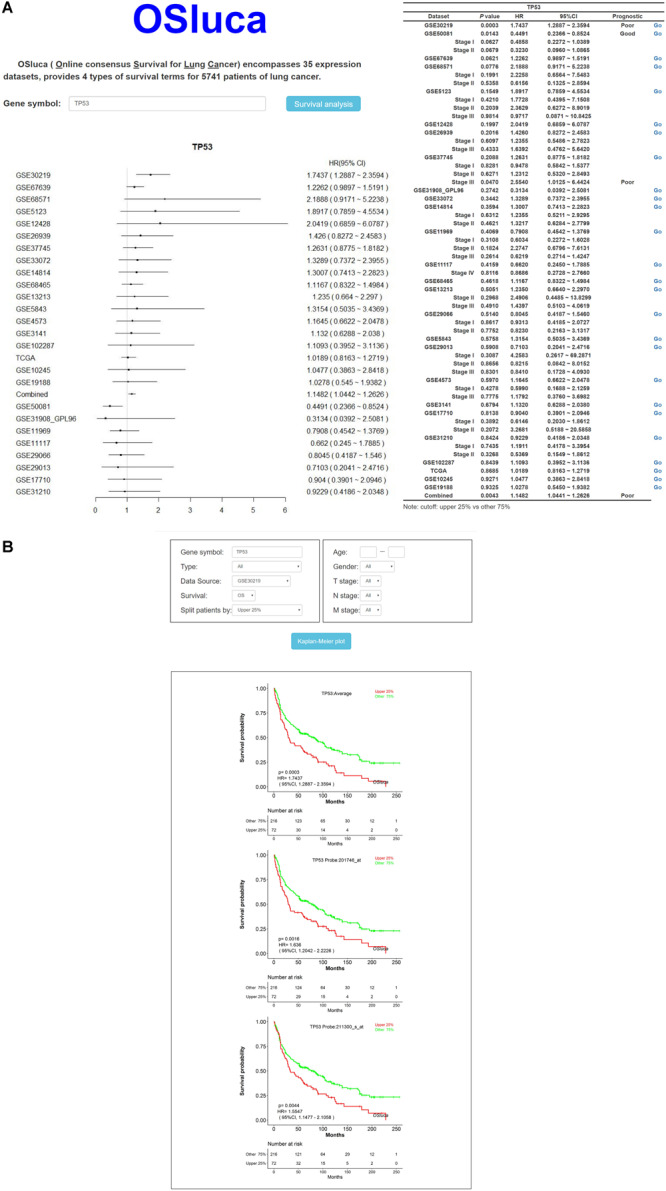
The output home page and KM output web subpage in OSluca for lung cancer. (A) Home page of OSluca with TP53 gene survival analysis, containing prognostic meta-analysis of a forest plot and a survival table. (B) KM plots of TP53 gene in the GSE30219 cohort. Note: the cutoff value is the upper 25% vs. other 75%. The “Combined” in forest plot and survival table means the overall prognostic significance of inputted gene in a pooling cohort with all the datasets. TP53, tumor protein p53.
Validation of Previously Reported Lung Cancer Prognostic Biomarkers in OSluca
A search for lung cancer biomarkers was performed using a set of keywords in NCBI PubMed, including “lung cancer,” “survival,” “biomarker,” and “prognosis.” In total, we collected 104 published lung cancer prognostic biomarkers verified by IHC or qPCR (Supplementary Table S1) to evaluate the performance of OSluca. For example, Hsu et al. reported that ERO1L (ERO1-like protein alpha, also named ERO1A) is significantly overexpressed in tumor tissue and could be as a poor prognostic biomarker for lung adenocarcinoma (Hsu et al., 2016). The prognostic analysis of ERO1L in OSluca showed that high expression of ERO1L gene is significantly associated with poor outcome in eight out of nine cohorts (Top 9 cohorts, the sample size above 150 cases) (Figures 3A–H), except the Roepman dataset (Figure 3I). Next, each published biomarker was investigated in the Top 9 cohorts in OSluca, and the results showed that approximately 66% of biomarkers (69/104) were consistent with original published findings (Supplementary Table S1). Meanwhile, OSluca can be used to perform the outcome meta-analysis of the interested gene that showed that 14% (14/104) (Supplementary Table S1) of published prognostic genes have the similar prognostic values in one or multiple OSluca cohorts as reported in the literature, but these genes also showed the opposite outcomes in some other cohorts from OSluca. These genes need further investigations, such as the DDIT3 gene (Supplementary Figure S2 and Supplementary Table S1). In contrast, there are some prognostic biomarkers, which have been shown different outcomes between OSluca and previous findings. A total of 9% of the published prognostic genes showed opposite outcome results between OSluca and literatures (9/104) (see Supplementary Table S1), suggesting that these genes need further validation. For example, the transcription factor KLF15 (Krüppel-like factor 15) had been proven to be higher in tumor tissue than that of adjacent non-tumor tissue and played an important role in promoting proliferation and carcinoma diversification in lung adenocarcinoma, associated with poor prognostic outcome (Gao et al., 2017). It was not anticipated that the patients with high expression of KLF15 have better survival than those with low expression (Supplementary Table S1 and Supplementary Figure S1). The OSluca result for the KLF15 gene was consistent with other prognostic analysis tools (Gyõrffy et al., 2013; Anaya, 2016), such as the KM plotter [P < 0.001, HR (95% CI) = 0.4 (0.28–0.58)]. In addition, the remaining 12 of 104 previously published prognostic biomarkers (11%) were not significant for prognostic analysis in the Top 9 cohorts in OSluca, but 8 of them (8/12) are significant in one or multiple datasets other than the Top 9 cohorts in OSluca (data not shown). All in all, the OSluca server is an interactive and free web server for researchers to develop potential prognostic biomarkers for lung cancer.
FIGURE 3.
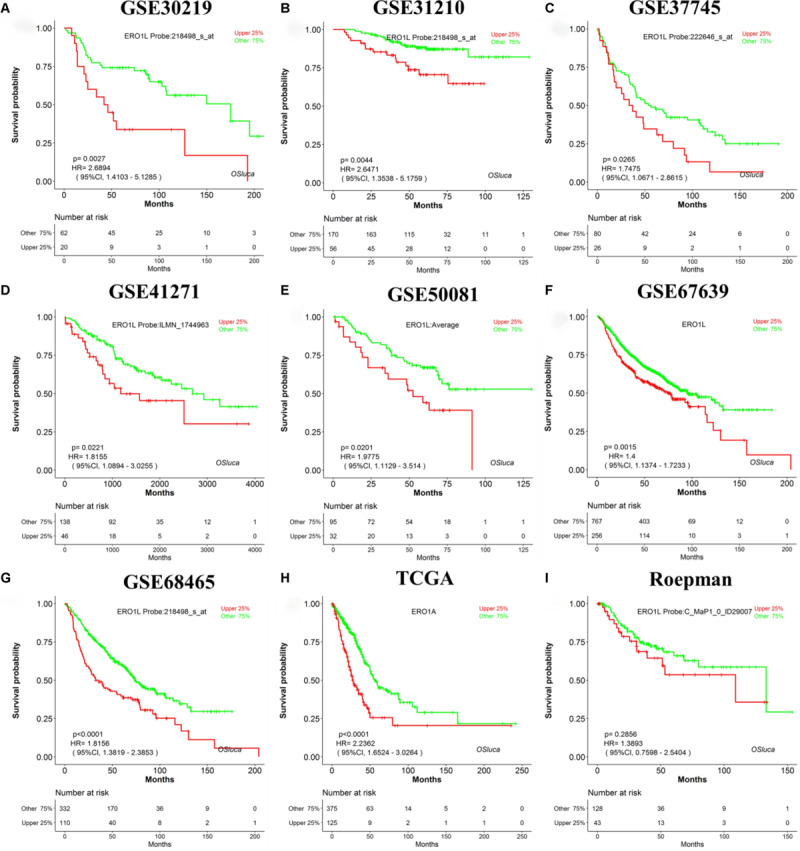
Validation of a previously reported biomarker ERO1L in OSluca. Overexpression of ERO1L in tumor tissue is suggested as a worse survival biomarker in lung adenocarcinoma. (A) Overall survival (OS) of ERO1L gene in GSE30219 cohort. (B) OS in GSE31210 cohort. (C) OS in GSE37745 cohort. (D) OS in GSE41271 cohort. (E) OS in GSE50081 cohort. (F) OS in GSE67639 cohort. (G) OS in GSE68465 cohort. (H) OS in TCGA in lung adenocarcinoma. (I) OS in Roepman cohort. The histological type of all the above cohorts is lung adenocarcinoma. ERO1L, ERO1-like protein alpha (also named ERO1A).
Discussion
Owing to tumor molecular heterogeneity, the prognosis of lung cancer patients is variable and difficult to predict. The prognosis of patients suffering from lung cancer had been demonstrated to be highly dependent on clinical factors of the patient, such as histological type, smoking status, and so on. However, it is also an imperative need to exploit novel prognostic biomarkers for determining the risk of cancerous lesions and predicting lung cancer patient outcomes by all available means, especially by high-throughput sequencing technologies. However, one major challenge to non-bioinformatics researchers is how to integrate the high-dimension profiling datasets of lung cancer and discover new biomarkers to potentially guide prognostic stratification. Previous studies had revealed that the online prognostic web servers of cancer (Elfilali et al., 2006; Mizuno et al., 2009; Goswami and Nakshatri, 2013; Gyõrffy et al., 2013; Tang et al., 2017) could substantially help researchers to discover potential biomarkers (Zheng et al., 2020). Herein, we developed a free web server OSluca to assess the prognostic value of the interesting gene in multiple cohorts of lung cancers. In OSluca, all the lung cancer cases are originated from the organ lung, not the second cancer from other cancers or organs. As a result, the prognostic specificity is only for lung cancer. Nevertheless, its prognostic significance in other types of cancers is also worth to be determined. To access the repeatability of previously reported prognostic biomarkers in OSluca, we collected 104 previously published prognostic biomarkers of lung cancer identified by qPCR or IHC, and tested their prognostic significance in OSluca. The testing results showed that most of the biomarkers were verified in OSluca and were confirmed for the published findings. Nevertheless, some genes showed different prognostic outcomes compared to previous literatures.
The advantage of OSluca over other online prognostic web servers is that the size of lung cancer samples in OSluca is large, and tens of independent cohorts are available, which is extremely valuable for the identification and validation of cancer prognostic biomarkers, since the most important part for the biomarker development is independent validation across different datasets/cohorts. The limitation of the current study is that OSluca can only test a single gene for outcome analysis. In summary, OSluca is a free web server for non-bioinformatics researchers to study potential lung cancer prognostic biomarkers, accessed at http://bioinfo.henu.edu.cn/LUCA/LUCAList.jsp.
Data Availability Statement
The datasets generated for this study can be found in the TCGA, NCBI GEO, and Roepman dataset.
Author Contributions
XG: research design. QW and XG: establish OSluca web server. ZY, ZL, and XS: deal with RNA sequencing with clinical data of lung cancer. ZY, LX, XS, LZ, YL, and XG: draft of the manuscript. YD, XS, LZ, PS, YL, TX, and JM: collect previously reported biomarkers of lung cancer. ZY, LX, LZ, WZ, YZ, and XG: critical revision of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by the following funding: The Kaifeng Science and Technology Major Project (18ZD008), the National Natural Science Foundation of China (Nos. 81602362 and 81801569), the Program for Science and Technology Development in Henan Province (Nos. 162102310391, 172102210187, and 192102310302), the Program for Young Key Teacher of Henan Province (2016GGJS-214), the supporting grants of Henan University (Nos. 2015YBZR048 and B2015151), and the Yellow River Scholar Program (No. H2016012).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00420/full#supplementary-material
References
- Anaya J. (2016). OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peerj Comput. Sci. 2:e67. [Google Scholar]
- Baty F., Facompre M., Kaiser S., Schumacher M., Pless M., Bubendorf L., et al. (2010). Gene profiling of clinical routine biopsies and prediction of survival in non-small cell lung cancer. Am. J. Respir. Crit. Care Med. 181 181–188. 10.1164/rccm.200812-1807OC [DOI] [PubMed] [Google Scholar]
- Beer D. G., Kardia S. L., Huang C. C., Giordano T. J., Levin A. M., Misek D. E., et al. (2002). Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 8 816–824. 10.1038/nm733 [DOI] [PubMed] [Google Scholar]
- Bild A. H., Yao G., Chang J. T., Wang Q., Potti A., Chasse D., et al. (2006). Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439 353–357. 10.1038/nature04296 [DOI] [PubMed] [Google Scholar]
- Boelens M. C., van den Berg A., Fehrmann R. S., Geerlings M., de Jong W. K., te Meerman G. J., et al. (2009). Current smoking-specific gene expression signature in normal bronchial epithelium is enhanced in squamous cell lung cancer. J. Pathol. 218 182–191. 10.1002/path.2520 [DOI] [PubMed] [Google Scholar]
- Botling J., Edlund K., Lohr M., Hellwig B., Holmberg L., Lambe M., et al. (2013). Biomarker discovery in Non–small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin. Cancer Res. 19 194–204. 10.1158/1078-0432.ccr-12-1139 [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 68 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Byers L. A., Diao L., Wang J., Saintigny P., Girard L., Peyton M., et al. (2013). An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 19 279–290. 10.1158/1078-0432.CCR-12-1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der S. D., Sykes J., Pintilie M., Zhu C.-Q., Strumpf D., Liu N., et al. (2014). Validation of a histology-independent prognostic gene signature for early-stage, non–small-cell lung cancer including stage IA patients. J. Thorac. Oncol. 9 59–64. 10.1097/JTO.0000000000000042 [DOI] [PubMed] [Google Scholar]
- Dong H., Wang Q., Zhang G., Li N., Yang M., An Y., et al. (2020). OSdlbcl: an online consensus survival analysis web server based on gene expression profiles of diffuse large B-cell lymphoma. Cancer Med. 9 1790–1797. 10.1002/cam4.2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfilali A., Lair S., Verbeke C., La Rosa P., Radvanyi F., Barillot E. (2006). ITTACA: a new database for integrated tumor transcriptome array and clinical data analysis. Nucleic Acids Res. 34 D613–D616. 10.1093/nar/gkj022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Qiu H., Liu J., Ma Y., Feng J., Qian L., et al. (2017). KLF15 promotes the proliferation and metastasis of lung adenocarcinoma cells and has potential as a cancer prognostic marker. Oncotarget 8 109952–109961. 10.18632/oncotarget.21972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A. J., Bratman S. V., Lee L. J., Harris J. P., Feng W., Nair R. V., et al. (2015). Integrating tumor and stromal gene expression signatures with clinical indices for survival stratification of early-stage Non-small cell lung cancer. J. Natl. Inst. 107:djv211 10.1093/jnci/djv211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami C. P., Nakshatri H. (2013). PROGgene: gene expression based survival analysis web application for multiple cancers. J. Clin. Bioinform. 3:22 10.1186/2043-9113-3-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyõrffy B., Surowiak P., Budczies J., Lánczky A. (2013). Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8:e82241 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman P. S., Lawrence M. S., Voet D., Jing R., Cibulskis K., Sivachenko A., et al. (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature 489 519–525. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley K. A., Yau C., Hinoue T., Wolf D. M., Lazar A. J., Drill E., et al. (2018). Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 Types of cancer. Cell 173 291.e6–304.e6. 10.1016/j.cell.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Aerts J., den Hamer B., van Ijcken W., den Bakker M., Riegman P., et al. (2010). Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PloS One 5:e10312 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.-H., Hsu C.-W., Hsueh C., Wang C.-L., Wu Y.-C., Wu C.-C., et al. (2016). Identification and characterization of potential biomarkers by quantitative tissue proteomics of primary lung adenocarcinoma. Mol. Cell. Proteomics 15 2396–2410. 10.1074/mcp.M115.057026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Virtanen C., Honjoh D., Miyoshi T., Satoh Y., Okumura S., et al. (2004). Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet 363 775–781. 10.1016/s0140-6736(04)15693-6 [DOI] [PubMed] [Google Scholar]
- Kuner R., Muley T., Meister M., Ruschhaupt M., Buness A., Xu E. C., et al. (2009). Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer 63 32–38. 10.1016/j.lungcan.2008.03.033 [DOI] [PubMed] [Google Scholar]
- Larsen J. E., Pavey S. J., Bowman R., Yang I. A., Clarke B. E., Colosimo M. L., et al. (2007a). Gene expression of lung squamous cell carcinoma reflects mode of lymph node involvement. Eur. Respir. J. 30 21–25. 10.1183/09031936.00161306 [DOI] [PubMed] [Google Scholar]
- Larsen J. E., Pavey S. J., Passmore L. H., Bowman R., Clarke B. E., Hayward N. K., et al. (2007b). Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis 28 760–766. 10.1093/carcin/bgl207 [DOI] [PubMed] [Google Scholar]
- Larsen J. E., Pavey S. J., Passmore L. H., Bowman R. V., Hayward N. K., Fong K. M. (2007c). Gene expression signature predicts recurrence in lung adenocarcinoma. Clin. Cancer Res. 13 2946–2954. 10.1158/1078-0432.ccr-06-2525 [DOI] [PubMed] [Google Scholar]
- Lee E.-S., Son D.-S., Kim S.-H., Lee J., Jo J., Han J., et al. (2008). Prediction of recurrence-free survival in postoperative Non–small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin. Cancer Res. 14 7397–7404. 10.1158/1078-0432.ccr-07-4937 [DOI] [PubMed] [Google Scholar]
- Liu J., Lichtenberg T., Hoadley K. A., Poisson L. M., Lazar A. J., Cherniack A. D., et al. (2018). An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173 400.e11–416.e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Lemon W., Liu P.-Y., Yi Y., Morrison C., Yang P., et al. (2006). A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 3:e030467 10.1371/journal.pmed.0030467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Yang D., He J., Krasna M. J. (2016). Epidemiology of lung cancer. Surg. Oncol. Clin. North Am. 25 439–445. 10.1016/j.soc.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Meyerson M., Carbone D. (2005). Genomic and proteomic profiling of lung cancers: lung cancer classification in the age of targeted therapy. J. Clin. Oncol. 23 3219–3226. 10.1200/JCO.2005.15.511 [DOI] [PubMed] [Google Scholar]
- Mitchell K. A., Zingone A., Toulabi L., Boeckelman J., Ryan B. M. (2017). Comparative transcriptome profiling reveals coding and noncoding RNA differences in NSCLC from african americans and european americans. Clin. Res. 23 7412–7425. 10.1158/1078-0432.ccr-17-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H., Kitada K., Nakai K., Sarai A. (2009). PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics 2:18 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R., et al. (2012). Identification of genes up-regulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72 100–111. 10.1158/0008-5472.can-11-1403 [DOI] [PubMed] [Google Scholar]
- Ramalingam S. S., Owonikoko T. K., Khuri F. R. (2011). Lung cancer: new biological insights and recent therapeutic advances. Cancer J. Clin. 61 91–112. 10.3322/caac.20102 [DOI] [PubMed] [Google Scholar]
- Raponi M., Zhang Y., Yu J., Chen G., Lee G., Taylor J. M., et al. (2006). Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 66 7466–7472. 10.1158/0008-5472.can-06-1191 [DOI] [PubMed] [Google Scholar]
- Robles A. I., Arai E., Mathé E. A., Okayama H., Schetter A. J., Brown D., et al. (2015). An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J. Thora.c Oncol. 10 1037–1048. 10.1097/JTO.0000000000000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepman P., Jassem J., Smit E. F., Muley T., Niklinski J., van de Velde T., et al. (2009). An Immune response enriched 72-gene prognostic profile for early-stage non–small-cell lung cancer. Clin. Cancer Res. 15 284–290. 10.1158/1078-0432.ccr-08-1258 [DOI] [PubMed] [Google Scholar]
- Rousseaux S., Debernardi A., Jacquiau B., Vitte A.-L., Vesin A., Nagy-Mignotte H., et al. (2013). Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 5:ra66 10.1126/scitranslmed.3005723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Larsen J. E., Lee W., Sun H., Shames D. S., Dalvi M. P., et al. (2013). Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol. Cancer Res. 11 638–650. 10.1158/1541-7786.MCR-12-0634-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedden K., Taylor J. M. G., Enkemann S. A., Tsao M.-S., Yeatman T. J., Gerald W. L., et al. (2008). Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 14 822–827. 10.1038/nm.1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staaf J., Isaksson S., Karlsson A., Jonsson M., Johansson L., Jonsson P., et al. (2013). Landscape of somatic allelic imbalances and copy number alterations in human lung carcinoma. Int. J. Cancer 132 2020–2031. 10.1002/ijc.27879 [DOI] [PubMed] [Google Scholar]
- Staaf J., Jönsson G., Jönsson M., Karlsson A., Isaksson S., Salomonsson A., et al. (2012). Relation between smoking history and gene expression profiles in lung adenocarcinomas. BMC Med. Genomics 5:22 10.1186/1755-8794-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T., Tomida S., Yatabe Y., Kosaka T., Osada H., Yanagisawa K., et al. (2006). Expression profile–defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J. Clin. Oncol. 24 1679–1688. 10.1200/JCO.2005.03.8224 [DOI] [PubMed] [Google Scholar]
- Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 W98–W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature 489 519– 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (2014). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511 543–550. 10.1038/s41586-018-0228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida S., Koshikawa K., Yatabe Y., Harano T., Ogura N., Mitsudomi T., et al. (2004). Gene expression-based, individualized outcome prediction for surgically treated lung cancer patients. Oncogene 23 5360–5370. 10.1038/sj.onc.1207697 [DOI] [PubMed] [Google Scholar]
- Tomida S., Takeuchi T., Shimada Y., Arima C., Matsuo K., Mitsudomi T., et al. (2009). Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J. Clin. Oncol. 27 2793–2799. 10.1200/JCO.2008.19.7053 [DOI] [PubMed] [Google Scholar]
- Wang F., Wang Q., Li N., Ge L., Yang M., An Y., et al. (2019a). OSuvm: an interactive online consensus survival tool for uveal melanoma prognosis analysis. Mol. Carcinog. 59 56–61. 10.1002/mc.23128 [DOI] [PubMed] [Google Scholar]
- Wang F., Wang Q., Li N., Ge L., Yang M., An Y., et al. (2020). OSuvm: an interactive online consensus survival tool for uveal melanoma prognosis analysis. Mol. Carcinog. 59 56–61. [DOI] [PubMed] [Google Scholar]
- Wang Q., Xie L., Dang Y., Sun X., Xie T., Guo J., et al. (2019b). OSlms: a web server to evaluate the prognostic value of genes in leiomyosarcoma. Front. Oncol. 9:190 10.3389/fonc.2019.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang L., Yan Z., Xie L. (2019c). OScc: an online survival analysis web server to evaluate the prognostic value of biomarkers in cervical cancer. Future Oncol. 15 3693–3699. 10.2217/fon-2019-0412 [DOI] [PubMed] [Google Scholar]
- Wilkerson M. D., Yin X., Hoadley K. A., Liu Y., Hayward M. C., Cabanski C. R., et al. (2010). Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin. Cancer Res. 16 4864–4875. 10.1158/1078-0432.CCR-10-0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson M. D., Yin X., Walter V., Zhao N., Cabanski C. R., Hayward M. C., et al. (2012). Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PloS One 7:e36530 10.1371/journal.pone.0036530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Dang Y., Guo J., Sun X., Xie T., Zhang L., et al. (2019a). High KRT8 expression independently predicts poor prognosis for lung adenocarcinoma patients. Genes 10:36. 10.3390/genes10010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wang Q., Dang Y., Ge L., Sun X., Li N., et al. (2019b). OSkirc: a web tool for identifying prognostic biomarkers in kidney renal clear cell carcinoma. Future Oncol. 15 3103–3110. 10.3892/ol.2019.10440 [DOI] [PubMed] [Google Scholar]
- Xie L., Wang Q., Nan F., Ge L., Dang Y. (2019c). OSacc: gene expression-based survival analysis web tool for adrenocortical carcinoma. Cancer Manag Res. 11 9145–9152. 10.2147/cmar.s215586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Xiao G., Coombes K. R., Behrens C., Solis L. M., Raso G., et al. (2011). Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin. Cancer Res. 17 5705–5714. 10.1158/1078-0432.CCR-11-0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Wang Q., Sun X., Ban B., Lu Z., Dang Y., et al. (2019). OSbrca: a web server for breast cancer prognostic biomarker investigation with massive data from tens of cohorts. Front. Oncol. 9:1349. 10.3389/fonc.2019.01349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Wang Q., Yang M., Yao X., Qi X., An Y., et al. (2020). OSpaad: an online tool to perform survival analysis by integrating gene expression profiling and long-term follow-up data of 1319 pancreatic carcinoma patients. Mol. Carcinog. 59 304–310. 10.1002/mc.23154 [DOI] [PubMed] [Google Scholar]
- Zhang G., Wang Q., Yang M., Yuan Q., Dang Y., Sun X., et al. (2019). OSblca: a web server for investigating prognostic biomarkers of bladder cancer patients. Front. Oncol. 9:466. 10.2217/fon-2019-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Zhang G., Zhang L., Wang Q., Li H., Han Y., et al. (2020). Comprehensive review of web servers and bioinformatics tools for cancer prognosis analysis. Front. Oncol. 10:68 10.3389/fonc.2020.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.-Q., Ding K., Strumpf D., Weir B. A., Meyerson M., Pennell N., et al. (2010). Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J.Clin. Oncol. 28 4417–4424. 10.1200/JCO.2009.26.4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the TCGA, NCBI GEO, and Roepman dataset.


