Abstract
The discovery of long noncoding RNAs (lncRNAs) has increased our understanding of the development and progression of many cancers, but their contributions to non-small cell lung cancer (NSCLC) remain poorly understood. Here, we profiled lncRNA expression in NSCLC and investigated in detail the molecular function of one upregulated lncRNA, LINC01234. LINC01234 was overexpressed in NSCLC compared with normal lung tissue and correlated positively with poor prognosis. Downregulation of LINC01234 impaired cell proliferation in vitro and tumor growth in vivo. RNA pull-down/mass spectrometry experiments showed that LINC01234 interacted with the RNA-binding protein heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1), which, in turn, led to the recruitment of DiGeorge syndrome critical region gene 8 (DGCR8), a subunit of the microRNA (miRNA) microprocessor complex. Accordingly, depletion of either LINC01234 or HNRNPA2B1 reduced the processing of several miRNA precursors, including primary microRNA (pri-miR)-106b. miR-106b-5p enhanced NSCLC cell growth by downregulating cryptochrome 2 (CRY2), thereby increasing c-Myc expression. Finally, we found that activated c-Myc binds to the LINC01234 promoter to increase its transcription, creating a c-Myc–LINC01234–HNRNPA2B1–miR-106b-5p–CRY2–c-Myc positive-feedback loop. We identified numerous lncRNAs with dysregulated expression in NSCLC and demonstrated a novel oncogenic axis involving LINC01234, HNRNPA2B1, miR-106b-5p, CRY2, and c-Myc. Components of this axis may be potential novel targets for NSCLC.
Keywords: NSCLC, LINC01234, HNRNPA2B1, miR-106b biogenesis, c-Myc
Graphical Abstract
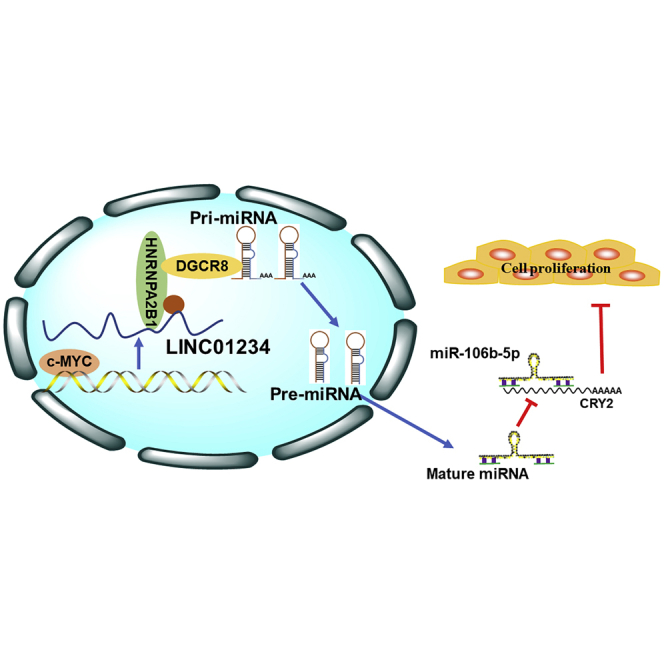
Wang and colleagues identified a novel c-Myc-activated lncRNA, LINC01234, which is dysregulated in NSCLC and associated with poor prognosis. LINC01234 promotes cell growth via interaction with HNRNPA2B1 and recruitment of DGCR8 to induce pri-miR-106b processing. These observations suggest that LINC01234 may serve as diagnostic and therapeutic targets for NSCLC.
Introduction
Lung cancer is the leading cause of cancer-related death worldwide.1 About 80% of all lung cancer diagnoses are non-small cell lung cancers (NSCLCs), the major subtypes being lung adenocarcinoma and lung squamous cell carcinoma. Despite rapid advances in diagnostic techniques, molecular-targeted drugs, and immune-checkpoint therapies, the 5-year overall survival (OS) of NSCLC patients remains less than 15%. Several genetic alterations have been reported to be “drivers” of NSCLC tumorigenesis, including mutations in epidermal growth factor receptor and KRAS genes, amplification of mesenchymal–epidermal transition-related genes, and fusion between the echinoderm microtubule protein-like 4 and anaplastic lymphoma kinase genes. Nevertheless, the mechanisms underlying NSCLC pathogenesis are not well understood.2, 3, 4, 5 There is thus a clear need to identify novel driver genes and to explore their mechanisms of action with the goal of improving the diagnosis and treatment of NSCLC patients.
Advanced whole-genome sequencing techniques, in combination with reference gene annotation databases, such as GENCODE, have revealed that the human genome encodes a large number of noncoding RNAs, including microRNAs (miRNAs), pseudogenes, and long noncoding RNAs (lncRNAs).6, 7, 8 lncRNAs are widely expressed in tissues and cells and participate in diverse cellular biological processes, such as X chromosome imprinting, immune response regulation, and cell metabolism, differentiation, and proliferation.9, 10, 11, 12, 13 Importantly, studies using high-throughput sequencing and microarray analyses have shown that thousands of lncRNAs are differentially expressed in human cancers,14 where they contribute to tumorigenesis through numerous mechanisms, including recruiting histone modification enzymes that repress or activate gene transcription,15,16 acting as competing endogenous RNAs or sponges to inhibit miRNA activity,17 regulating mRNA stability via interaction with RNA-binding proteins, such as STAU1, UPF1, and hnRNPL,10,18,19 and paradoxically encoding small active peptides.20 Our previous studies revealed that overexpression of the lncRNA HOX transcript antisense RNA (HOTAIR) promotes gastric cell proliferation by sponging microRNA (miR)-331-3p,21 and we also showed that transcription of the antisense lncRNA HOXA11-AS by transcription factor E2F1 facilitates gastric cancer cell growth and metastasis by acting as a scaffold for the chromatin-modifying factors PRC2, LSD1, and DNMT1 and reducing the transcription of PRSS8.22 In colorectal cancer, the c-Myc-regulated lncRNAs MYCLo-1 and MYCLo-2 regulate cell proliferation and progression of the cell cycle by interacting with RNA-binding proteins, such as HuR and hnRNPK and regulating expression of the c-Myc target genes CDKN1A (p21) and CDKN2B.23
Recent work has characterized the functional roles and underlying mechanisms of action of several lncRNAs in NSCLC. For instance, elevated LINC00963 expression promotes NSCLC cell migration and invasion in vitro and enhances lung metastasis in vivo by interacting with and preventing the ubiquitination of phosphoglycerate kinase 1, which leads to activation of the AKT/mammalian target of rapamycin (mTOR) signaling pathway.24 Sustained LINC00473 expression is essential for the growth and survival of LKB1-inactivated NSCLC cells through its interaction with NONO, which facilitates cyclic AMP response element-binding protein (CREB)-regulated transcription coactivator (CRTC)/CREB-mediated transcription.25 We previously showed that SNHG20 promotes NSCLC cell proliferation and migration via epigenetic silencing of the cell-cycle regulatory protein p21,11 whereas LINC00152 promotes lung adenocarcinoma cell proliferation by interacting with EZH2 and repressing expression of interleukin 24.26
In the present study, we performed comprehensive lncRNA profiling of NSCLC using publicly available datasets and identified numerous novel NSCLC-associated lncRNAs. One of these, LINC01234, was investigated in more depth to identify the molecular mechanisms underlying its dysregulation and function in NSCLC tumorigenesis.
Results
LINC01234 Is Highly Expressed in NSCLC and Correlates with Poor Prognosis
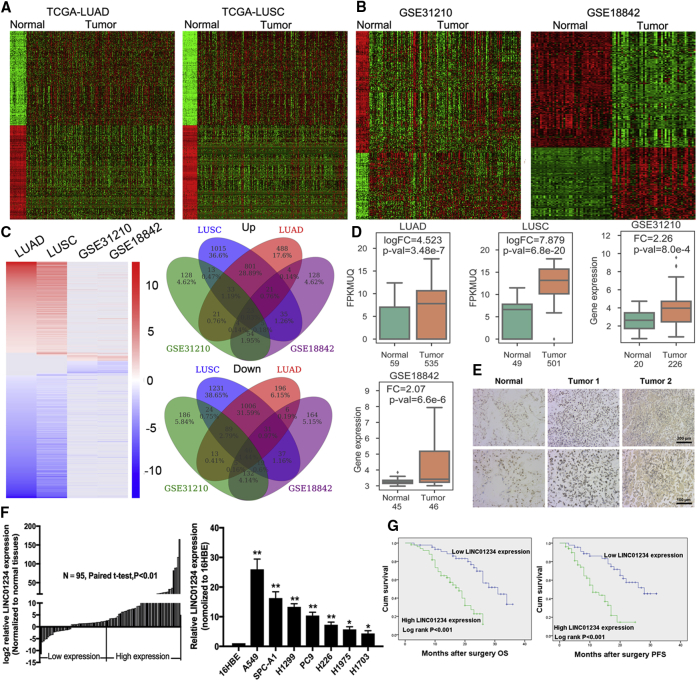
To identify differentially expressed lncRNAs in NSCLC, we downloaded and analyzed lung adenocarcinoma and lung squamous cell carcinoma RNA sequencing (RNA-seq) datasets from The Cancer Genome Atlas (TCGA) and microarray gene-profile datasets from GEO: GSE31210 and GSE18842 (Figures 1A and 1B). We identified 46 and 23 lncRNAs that were significantly downregulated and upregulated, respectively, in all four datasets (Figure 1C). The upregulated lncRNAs were analyzed further because they are likely to be of greater utility as diagnostic and prognostic markers. Among the 23 lncRNAs, LINC01234 was the most differentially upregulated, and TCGA pan-cancer analysis revealed that LINC01234 was also overexpressed in many other types of cancers (colon adenocarcinoma, esophageal carcinoma, glioblastoma multiforme, kidney chromophobe, kidney clear cell renal carcinoma, papillary renal cell carcinoma, hepatocellular carcinoma, prostate adenocarcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma) (Figures 1D and S1A). To validate these results, we performed fluorescence in situ hybridization (FISH) assays and confirmed the aberrant overexpression of LINC01234 in NSCLC tumors compared with adjacent normal tissues (Figure 1E). Similarly, quantitative reverse transcriptase PCR (qRT-PCR) analysis of 95 paired sets of NSCLC and normal tissues demonstrated that LINC01234 was significantly upregulated (fold change > 1; p < 0.01) in 78% (74/95) of cancerous tissues compared with normal tissues. Analysis of LINC01234 expression in the NSCLC cell lines gave similar results (Figure 1F).
Figure 1.
LINC01234 Is Highly Expressed in NSCLC and Correlated with Poor Prognosis
(A and B) Data mining of altered lncRNA expression in (A) TCGA dataset (LUAD and LUSC) and (B) two microarray gene profiles (GEO: GSE31210 and GSE18842). (C) Venn diagrams showing lncRNAs commonly upregulated and downregulated in all three datasets. (D) Data mining of the fold change of LINC01234 in TCGA and microarray datasets. (E) In situ hybridization shows the expression of LINC01234 in NSCLC tissues and adjacent nontumor tissues. (F) qRT-PCR analysis of LINC01234 expression in 95 pairs of NSCLC tissues and adjacent nontumor lung tissues, 16HBE cells, and NSCLC cell lines. (G) Kaplan–Meier survival analysis of the association between LINC01234 expression level and NSCLC patient overall survival and disease-free survival. ∗p < 0.05, ∗∗p < 0.01.
Next, we explored the relationship between LINC01234 expression and clinicopathological features of patients with NSCLC by assigning the 95 specimens to high (n = 50, fold change > 2.5) and low (n = 45, fold change < 2.5) LINC01234 expression groups (Figure 1F). Kaplan–Meier survival analysis showed that the 3-year overall survival and progression-free survival (PFS) rates were lower for patients with high versus low LINC01234-expressing tumors (Figure 1G). High LINC01234 expression also significantly correlated with larger tumor size (p = 0.007), TNM stage (p = 0.003), and lymph node metastasis (p = 0.005) in NSCLC patients but not with other factors, including sex and age (Table S1). Univariate Cox regression analysis identified histologic grade, tumor size, lymphatic metastasis, TNM stage, and LINC01234 expression level as significant prognostic factors. Histologic grade, lymphatic metastasis, and LINC01234 expression level remained independent prognostic factors in multivariate analysis (Table S2).
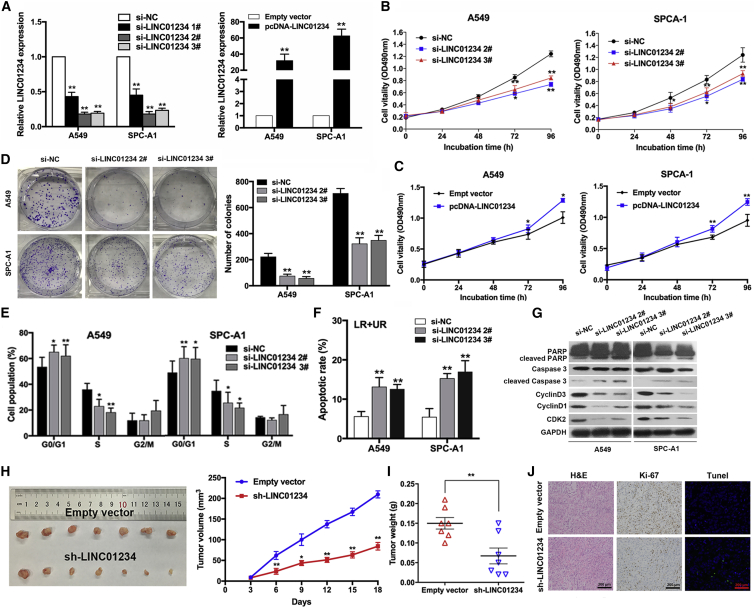
LINC01234 Promotes NSCLC Cell Proliferation and Inhibits Cell Apoptosis In Vitro and In Vivo
To explore the biological functions of LINC01234 in NSCLC, we transfected A549 and SPC-A1 cells with small interfering (si)-LINC01234, a nontargeting control siRNA (si-NC), a LINC01234 overexpression vector, or an empty vector (Figure 2A). MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) proliferation assays showed that downregulation of LINC01234 significantly inhibited NSCLC cell proliferation compared with the control cells, and conversely, LINC01234 overexpression promoted cell proliferation (Figures 2B and 2C). Similarly, colony-forming assays showed that the clonogenic survival potential was significantly decreased or increased by knockdown or overexpression of LINC01234, respectively (Figures 2D and S1B). Similar results were obtained when cell proliferation was measured with 5-ethynyl-2′-deoxyuridine (EdU) assays (Figures S1C and S1D). The block in cell proliferation induced by si-LINC01234 expression was accompanied by arrest of the cells in the G1 to G0 phase of the cell-cycle (Figures 2E and S1E) and elevated levels of apoptosis, as measured using TUNEL staining or flow cytometry assays (Figures 2F and S1F). Accordingly, A549 and SPC-A1 cells depleted of LINC01234 expressed significantly higher levels of apoptosis-related proteins, including cleaved poly (ADP-ribose) polymerase (PARP) and cleaved caspase-3, and markedly decreased levels of G1/S-phase checkpoint proteins, such as cyclin D3, cyclin D1, and cyclin-dependent kinase 2 (CDK2) (Figure 2G). These data established that LINC01234 plays a crucial role in controlling NSCLC cell proliferation, cell-cycle progression, and apoptosis. To validate these findings in vivo, we employed a mouse xenograft model. A549 cells stably expressing control or LINC01234-specific short hairpin RNAs (shRNAs) were injected subcutaneously into athymic nude mice, and tumor growth was monitored for 18 days. Indeed, tumors derived from sh-LINC01234 cells were smaller than those derived from sh-NC control cells (Figures 2H and 2I). LINC01234 expression was confirmed to be suppressed in the excised tumors by qPCR analysis (Figure S1G). Compared with the control tumors, sh-LINC01234-derived tumors expressed lower levels of the proliferation marker Ki-67 and contained more apoptotic cells, as assessed by immunohistochemical (IHC) and TUNEL staining, respectively (Figure 2J).
Figure 2.
Downregulation of LINC01234 Inhibits NSCLC Cell Growth and Induces Apoptosis In Vitro and In Vivo
(A) qRT-PCR analysis of LINC01234 expression in NSCLC cells overexpressing or depleted of LINC01234. (B and C) Growth curves show the proliferation ability of LINC01234-depleted (B) or -overexpressing (C) A549 and SPC-A1 cells. (D) Colony formation assays were used to evaluate the colony formation capacity of LINC01234-depleted A549 and SPC-A1 cells. (E and F) Fluorescence-activated cell sorting (FACS) analysis of the effect of LINC01234 downregulation on cell-cycle progression (E) and apoptosis (F). (G) Western blot analysis of apoptosis- and cell-cycle-related proteins in A549 and SPC-A1 transfected with control or LINC01234-specific siRNA. (H) Representative images of mice-bearing tumors derived from cells expressing empty vector or sh-LINC01234 and tumor volume versus time-growth curves. (I) Tumor weights from two groups are represented. (J) Representative images of tumor samples derived from cells expressing empty vector or sh-LINC01234 subjected to H&E staining, Ki-67 immunostaining, and TUNEL staining. ∗p < 0.05, ∗∗p < 0.01.
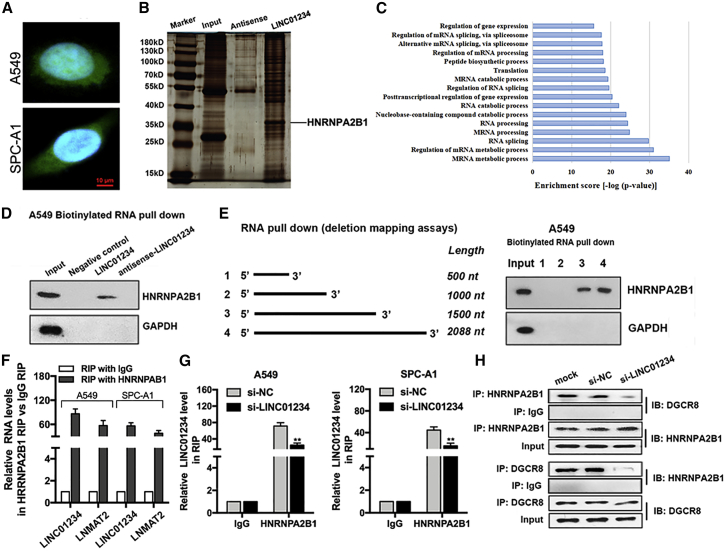
LINC01234 Interacts with HNRNPA2B1 in NSCLC Cells
To better understand the underlying mechanism of action of LINC01234, we examined its distribution and protein interactions in NSCLC cells. LINC01234 RNA was located in both the nucleus and cytoplasm, as determined by FISH analysis (Figure 3A). RNA pull-down assays combined with mass spectrometry identified HNRNPA2B1 proteins in NSCLC cell lysates that bound to biotinylated LINC01234 RNA but not to the antisense RNA (Figure 3B; Table S5). Gene Ontology (GO) and pathway analysis revealed that the LINC01234-interacting proteins were particularly enriched in mRNA metabolic, RNA splicing, and RNA processing pathways (Figure 3C). One of these, heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1), is reportedly involved in the development of diverse carcinomas via regulation of pre-mRNA processing and miRNA maturation. We confirmed the specific association between LINC01234 and HNRNPA2B1 in RNA pull-down assays, which showed that HNRNPA2B1 bound to biotinylated LINC01234 RNA and not with the biotinylated negative control or antisense RNAs (Figure 3D). Similar experiments performed with truncated LINC01234 sequences (1–500, 1–1,000, 1–1500, and 1–2,088 nucleotides) showed that the main HNRNPA2B1-binding site in NSCLC cells was located in the 1,000–1,500 sequence of LINC01234 (Figure 3E). Conversely, LINC01234 was identified in HNRNPA2B1 immunoprecipitates from A549 and SPC-A1 cells using RNA immunoprecipitation (RIP) assays. LNMAT2 was used as a positive control in RIP assays, as it was found to be interacted with HNRNPA2B127 (Figure 3F). The interaction of LINC01234 and HNRNPA2B1 was reduced by LINC01234 silencing, as expected (Figure 3G). Collectively, these results suggested that LINC01234 forms a complex with HNRNPA2B1 that may regulate downstream target expression.
Figure 3.
LINC01234 Interacts with RNA-Binding Proteins HNRNPA2B1 and Forms a Complex with DGCR8 in NSCLC Cells
(A) FISH analysis of the subcellular localization of LINC01234 in A549 and SPC-A1 cells. (B) Representative silver-staining image shows the LINC01234 RNA- and anti-sense RNA-precipitated proteins by RNA pull-down assay. (C) GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of LINC01234-interacted proteins enriched the cellular process and pathways. (D) The anti-HNRNPA2B1 western blot image shows HNRNPA2B1 binding to desthiobiotinylated LINC01234 in A549 cells. (E) Schematic representation of the full-length LINC01234 and its truncations. The anti-HNRNPA2B1 western blot image shows HNRNPA2B1 binding to desthiobiotinylated full-length and different truncations of LINC01234 in A549 cells. (F) RIP assays showing LINC01234 and LNMAT2 coimmunoprecipitation (coIP) with HNRNPA2B1 from A549 and SPC-A1 cells. (G) RIP assays showing LINC01234 coimmunoprecipitation with HNRNPA2B1 in LINC01234 or control siRNA-transfected A549 and SPC-A1 cells. (H) CoIP assays show the interaction between HNRNPA2B1 and DGCR8 in A549 and SPC-A1 cells. ∗∗p < 0.01.
A recent study demonstrated that HNRNPA2B1 can interact with DiGeorge syndrome critical region gene 8 (DGCR8), a component of the primary (pri)-miRNA microprocessor, and regulate miRNA expression.28 To examine this in NSCLC cells, we performed reciprocal immunoprecipitation of endogenous DGCR8 and HNRNPA2B1 and analyzed the coprecipitated proteins by western blotting. This analysis revealed that the two proteins did indeed interact in NSCLC cells, and importantly, binding was abolished by expression of si-LINC01234 (Figure 3H). These data point to a role for LINC01234 in pri-miRNA processing via complex formation with HNRNPA2B1 and DGCR8.
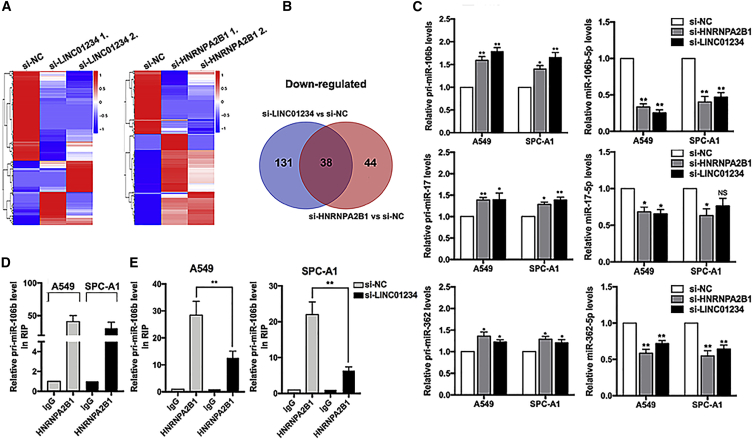
LINC01234 and HNRNPA2B1 Facilitate pri-miRNA Processing
To determine whether LINC01234 affects pri-miRNA processing in an HNRNPA2B1-dependent manner, we performed global miRNA profiling of A549 cells depleted of LINC01234 and/or HNRNPA2B1. Expression of a large number of miRNAs was reduced in cells with knockdown of either LINC01234 or HNRNPA2B1 (Figures 4A and S2A), of which 38 miRNAs were downregulated in both LINC01234- and HNRNPA2B1-depleted cells (Figure 4B).
Figure 4.
LINC01234/HNRNPA2B1 Complex Regulates miR-106b-5p Maturation
(A) Heatmaps were drawn to show the differentially expressed miRNAs in LINC01234- or HNRNPA2B1-depleted cells. (B) Venn diagram shows the overlap between LINC01234-regulated miRNAs and HNRNPA2B1-affected miRNAs. (C) qPCR analyses show the changes of pri- and mature miR-106b, miR-17, and miR-362 expression levels after transfection with LINC01234 or HNRNPA2B1 siRNA. (D) Immunoprecipitation of HNRNPA2B1 and qPCR analysis of associated pri-miR-106b. (E) Immunoprecipitation of HNRNPA2B1 and qPCR analysis of associated pri-miR-106b after transfection with LINC01234 siRNA. ∗p < 0.05, ∗∗p < 0.01.
We validated these findings by qPCR analysis of the primary and mature forms of the five most downregulated miRNAs—miR-106b-5p, miR-17-5p, miR-362-5p, miR-374-3p, and miR-425-3p—in LINC01234- and HNRNPA2B1-depleted A549 and SPC-A1 cells. The results showed that levels of the mature miRNAs were decreased by LINC01234 or HNRNPA2B1 knockdown, whereas levels of their pri-miRNAs were increased (Figures 4C, S2B, and S2C), indicative of a block in processing. The most striking changes occurred for pri-miR-106b and miR-106b-5p, and we therefore asked whether HNRNPA2B1 and DGCR8 interacted with miR-106b in NSCLC cells. We found that pri-miR-106b was enriched in anti-HNRNPA2B1 and anti-DGCR8 immunoprecipitates of NSCLC cell lysates (Figures 4D and S2D), and the associations were reduced by LINC01234 knockdown (Figures 4E and S2E). Taken together, these data suggest that HNRNPA2B1 and DGCR8 could interact with pri-miR-106b, and LINC01234 promoted its maturation to miR-106b-5p with the help of HNRNPA2B1 and DGCR8.
LINC01234, HNRNPA2B1, and miR-106b-5p Promote Tumorigenic Behaviors in NSCLC Cells
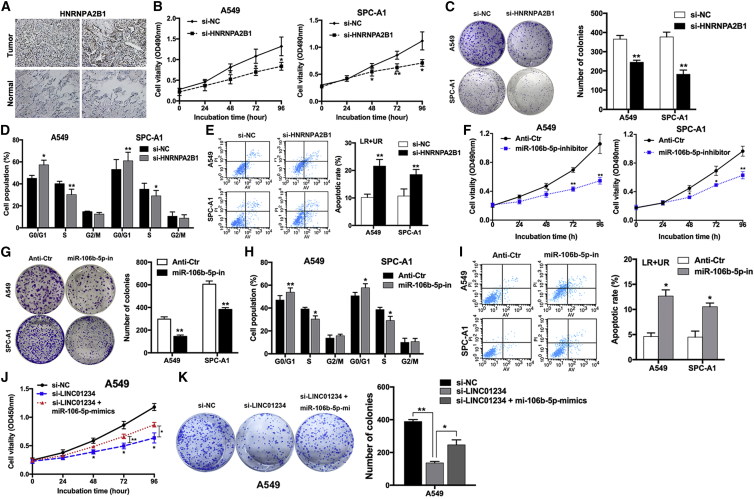
IHC analysis of HNRNPA2B1 protein expression in NSCLC and adjacent normal lung tissues indicated that the protein was aberrantly upregulated in the tumor tissues (Figure 5A), consistent with the upregulation of LINC01234. To investigate the biological roles and interdependence of LINC01234, HNRNPA2B1, and miR-106b-5p in NSCLC cells, we analyzed the effects of their modulation on cell proliferation, apoptosis, and cell-cycle progression. qRT-PCR analysis of cells expressing si-HNRNPA2B1 confirmed that HNRNPA2B1 mRNA levels were specifically reduced by ∼80% compared with si-NC-expressing cells (Figure S3A). HNRNPA2B1 knockdown significantly inhibited the proliferation of A549 and SPC-A1 cells, as assessed by MTT and colony-forming assays (Figures 5B and 5C), and additionally induced cell-cycle arrest in G0/G1 and increased cell apoptosis (Figures 5D and 5E and S3B) compared with control cells. Consistent with the LINC01234 findings, HNRNPA2B1 supports the growth of NSCLC cells.
Figure 5.
HNRNPA2B1 and miR-106b-5p Exert Oncogenic Roles in NSCLC Cells
(A) IHC staining detection of HNRNPA2B1 in NSCLC tumor and normal tissues. (B) Growth curves show the proliferation ability of HNRNPA2B1-depleted A549 and SPC-A1 cells. (C) Colony formation assays were used to evaluate the colony formation capacity of HNRNPA2B1-depleted A549 and SPC-A1 cells. (D and E) FACS analysis of the effect of HNRNPA2B1 downregulation on cell-cycle progression (D) and apoptosis (E). (F) Growth curves show the proliferation ability of miR-106b-5p inhibitor-treated A549 and SPC-A1 cells. (G) Colony formation assays were used to evaluate the colony formation capacity of miR-106b-5p inhibitor-treated A549 and SPC-A1 cells. (H and I) FACS analysis of the effect of miR-106b-5p inhibitor treatment on cell-cycle progression (H) and apoptosis (I). (J) Growth curves show the proliferation ability of the LINC01234 siRNA and miR-106b-5p mimic-cotransfected A549 cell. (K) Colony formation assays were used to evaluate the colony formation capacity of the LINC01234 siRNA and miR-106b-5p mimic-cotransfected A549 cell. ∗p < 0.05, ∗∗p < 0.01.
Because LINC01234–HNRNPA2B1 interactions increase miR-106b processing, we examined whether a miR-106b-5p inhibitor would recapitulate and a miR-106b-5p mimic would rescue the effects of LINC01234 and HNRNPA2B1 knockdown in NSCLC cells. First, we detected the efficiency of knockdown and overexpression of miR-106b-5p (Figure S3C). Indeed, transfection of A549 and SPC-A1 cells with a miR-106b-5p inhibitor reduced cell proliferation (Figures 5F and 5G), induced cell-cycle arrest in G0/G1, and increased apoptosis (Figures 5H and 5I and S3D) compared with cells transfected with a control sequence. Notably, cotransfection of a miR-106b-5p mimic into A549 cells partially rescued the effects of LINC01234 knockdown (Figures 5J and 5K), suggesting that the proproliferative effects of LINC01234 are mediated, at least in part, via upregulation of miR-106b-5p.
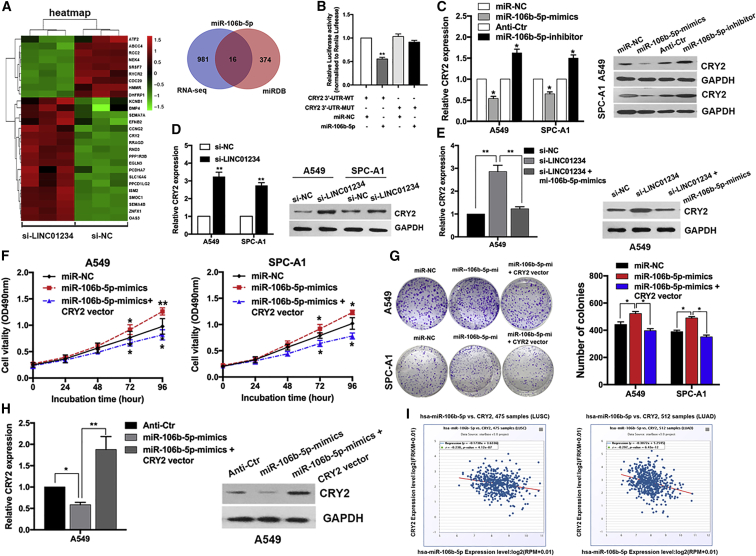
Cryptochrome 2 (CRY2) Is a Target of miR-106b-5p and LINC01234/HNRNPA2B1
Next, we sought to identify the downstream target genes regulated by LINC01234–HNRNPA2B1–miR-106b-5p that might mediate their effects in NSCLC. To this end, we performed unbiased RNA-seq of A549 cells expressing si-LINC01234 or si-NC and analyzed the differentially expressed mRNAs (Figure 6A). Gene-set enrichment analysis revealed that LINC01234 silencing affected the expression of genes enriched in pathways associated with cell-cycle arrest and apoptosis (Figure S3E). To identify miR-106b-5p- and LINC01234-coregulated targets, we performed an integrative analysis of the miRDB database and the gene-expression profiles. We found that of the 997 genes that were significantly upregulated (log2 fold change > 1.1 and p < 0.05) by LINC01234 silencing, 16 were predicted target genes of miR-106b-5p (Figure 6A; Table S6). One of these, the circadian clock protein CRY2, is of particular interest because of its remarkable expression fold change upon LINC01234 knockdown (Figure S3F) and its vital role in tumor suppression. Thus, we selected it for further analysis. To verify that CRY2 expression is indeed regulated through LINC01234 and miR-106b-5p, we performed luciferase reporter assays. HEK293T cells were transfected with a luciferase vector under the control of the wild-type (WT) 3′ untranslated region (UTR) of CRY2 (WT-CRY2) or one carrying mutations in the putative miR-106b-5p-binding sites (mut-CRY2) and then cotransfected with either a control sequence or miR-106b-5p mimic. We found that miR-106b-5p significantly decreased luciferase activity driven by WT-CRY2 but not by mut-CRY2 (Figures 6B and S3G), indicating that CRY2 expression is regulated by miR-106b-5p. Consistent with this, qRT-PCR and western blot analysis indicated that CRY2 mRNA and protein levels were reduced or elevated after transfection with a miR-106b-5p mimic or inhibitor, respectively (Figure 6C).
Figure 6.
CRY2 Is a Key Target of miR-106b-5p in NSCLC Cells
(A) Hierarchically clustered heatmap of upregulated and downregulated genes in LINC01234-depleted A549 cells. Venn diagram shows the overlap between predicted miR-106b-5p targets by the miRDB database and LINC01234 depletion upregulated genes. (B) Luciferase activities were measured in NSCLC cells cotransfected with the luciferase reporter containing CRY2 wild-type or mutant and the mimics of miR-106b-5p. (C and D) qPCR and western blot analysis of CRY2 mRNA and protein expression after transfection with miR-106b-5p mimics or inhibitors (C) and LINC01234 siRNA (D). (E) qPCR and western blot analysis of CRY2 mRNA and protein expression after cotransfection with LINC01234 siRNA and miR-106b-5p mimics. (F) Growth curves show the proliferation ability of miR-106b-5p mimics and the CRY2 overexpression vector cotransfected cell. (G) Colony formation assays were used to evaluate the colony formation capacity of miR-106b-5p mimics and the CRY2 overexpression vector cotransfected cell. (H) qPCR and western blot analysis of CRY2 mRNA and protein expression after cotransfection with miR-106b-5p mimics and the CRY2 overexpression vector. (I) The scatterplot images show the correlation between miR-106b-5p and CRY2 in TCGA dataset.∗p < 0.05, ∗∗p < 0.01.
Given that expression of LINC01234 and miR-106b-5p is positively associated in NSCLC cells, we predicted that LINC01234 knockdown may recapitulate the effects of miR-106b-5p on CRY2 expression. Indeed, si-LINC01234 expression significantly increased CRY2 mRNA and protein levels in A549 and SPC-A1 cells (Figure 6D), and these effects were reversed by coexpression of the miR-106b-5p mimic (Figure 6E). Furthermore, overexpression of CRY2 reversed the proproliferative effects of the miR-106b-5p mimic (Figures 6F and 6G), confirming that the LINC01234–HNRNPA2B1–miR-106b-5p axis negatively regulates CRY2 expression and function in NSCLC cells. The downregulation of CRY2 mRNA and protein induced by miR-106b-5p overexpression was also partly rescued by cotransfection with a CRY2 vector (Figure 6H). Finally, we analyzed the lung adenocarcinoma and lung squamous cell carcinoma TCGA datasets and identified a significant negative correlation between the expression levels of miR-106b-5p and CRY2 mRNA29 (Figure 6I). Collectively, these data indicate that CRY2 is a target gene of miR-106b-5p, and its expression in NSCLC cells is coregulated by LINC01234 and miR-106b-5p.
CRY2 Acts as a Tumor Suppressor in NSCLC Cells
To verify the potential role of CRY2 as a tumor suppressor, we examined its expression and function in NSCLC tissues and cells. IHC analysis and TCGA analysis showed that tumor tissues expressed lower CRY2 levels compared with normal lung tissues29 (Figures 7A and S4A). As expected, transfection of A549 and SPC-A1 cells with a CRY2 overexpression vector (Figure S4B) significantly inhibited cell proliferation (Figures 7B and 7C), which contrasts with the effects of LINC01234/miR-106b-5p overexpression. To determine whether manipulation of CRY2 levels could reverse the effects of LINC01234, we cotransfected NSCLC cells with CRY2 and LINC01234 overexpression or siRNA vectors. We found that CRY2 partially reversed the proproliferative effects of LINC01234 overexpression, and conversely, expression of si-CRY2 largely rescued the growth inhibition induced by LINC01234 knockdown (Figures 7D and 7E).
Figure 7.
CRY2 Exerts Tumor-Suppressor Functions via Repressing c-Myc Expression in NSCLC Cells
(A) IHC staining detection of CRY2 in NSCLC tumor and normal tissues. (B) Growth curves show the proliferation ability of the CRY2 overexpression vector or empty vector-transfected cell. (C) Colony formation assays were used to evaluate the colony formation capacity of the CRY2 overexpression vector or empty vector transfected cell. (D) Growth curves show the proliferation ability of CRY2- and LINC01234-overexpressing or -depleted cells. (E) Colony formation assays were used to evaluate the colony formation capacity of CRY2 and LINC01234-overexpressing or -depleted cells. (F) Western blot analysis of c-Myc and CRY2 levels in NSCLC cells overexpressing CRY2. (G) Western blot analysis of c-Myc and CRY2 expression in NSCLC cells with knockdown or overexpression of CRY2 and LINC01234. (H) Kaplan–Meier survival analysis of the association between CRY2 expression level and NSCLC patient overall survival survival. ∗p < 0.05, ∗∗p < 0.01.
A recent study reported that CRY2 promotes c-Myc degradation, and accordingly, CRY2 depletion stabilizes c-Myc protein levels.30 Consistent with this, we found that CRY2 overexpression and LINC01234 knockdown decreased c-Myc protein levels in NSCLC cells, whereas LINC01234 overexpression increased c-Myc levels. Moreover, this was partially reduced by coexpression of CRY2 (Figures 7F and 7G). Finally, Kaplan–Meier analysis of NSCLC patient datasets from TCGA, GEO, and the European Genome Archive using Kaplan–Meier (KM) plotter indicated that low tumor expression of CRY2 was associated with poorer prognosis31 (Figure 7H). Thus, LINC01234 and miR-106b-5p positively regulate c-Myc expression and promote NSCLC growth via negative regulation of CRY2 levels.
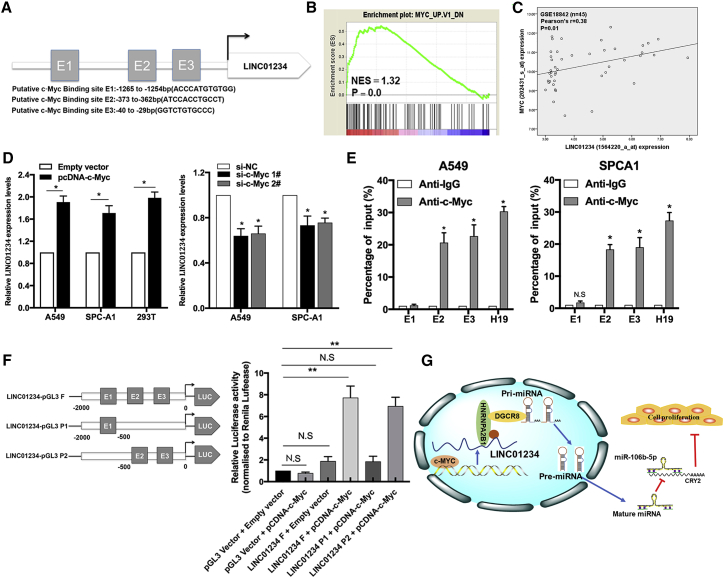
c-Myc Activates LINC01234 Transcription, Creating a c-Myc–LINC01234–CRY2 Positive-Feedback Loop in NSCLC Cells
Increasing evidence suggests that oncogenic transcription factors, such as E2F1 and c-Myc, play a role in the aberrant expression of lncRNAs in human cancers. Having demonstrated a relationship between LINC01234–miR-106b-5p–CRY2 and c-Myc in NSCLC cells, we next asked whether c-Myc or other transcription factors might contribute to the LINC01234 dysregulated expression observed in NSCLC. With the use of JASPAR, the online database of transcription factor-binding profiles, we found that the LINC01234 promoter region contained several potential binding sites for transcription factors, including c-Myc (Figure 8A). Moreover, gene-set enrichment analysis of the RNA-seq data indicated that c-Myc-regulated genes were enriched among those differentially regulated by LINC01234 expression in NSCLC (Figure 8B). Consistent with this, we observed a significantly positive correlation between LINC01234 and c-Myc expression in the GEO: GSE18842 lung cancer dataset (Figure 8C). These findings suggested that c-Myc might be an upstream regulator of LINC01234.
Figure 8.
c-Myc Promotes LINC01234 Transcription in NSCLC Cells
(A) Prediction of c-Myc-binding sites in the promoter region of LINC01234 by JASPAR. (B) Gene set enrichment analysis (GSEA) analysis shows the enrichment of LINC01234-regulated genes in the c-Myc target list. (C) Correlation between c-Myc and LINC01234 expression in the GEO: GSE18842 dataset. (D) qRT-PCR analysis of LINC01234 expression in A549, SPC-A1, and HEK293T cells transfected with a c-Myc-overexpression vector or siRNA. (E) ChIP-qPCR analysis of c-Myc occupancy of the LINC01234 promoter in A549 and SPC-A1 cells. H19 was used as a positive control and IgG as a negative control. (F) Dual-Luciferase reporter assays of c-Myc binding to the LINC01234 promoter region. (G) Proposed model illustrated the modulatory role of c-Myc-activated LINC01234 acting as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. ∗p < 0.05, ∗∗p < 0.01.
To test this hypothesis, we overexpressed or silenced c-Myc in A549 and SPC-A1 cells and analyzed LINC01234 expression (Figures S4B–S4D). We found that c-Myc overexpression or knockdown increased or decreased, respectively, LINC01234 expression (Figure 8D). To determine whether c-Myc regulates LINC01234 expression, we performed chromatin immunoprecipitation (ChIP) and luciferase reporter assays. We found that LINC01234 promoter sequences were indeed enriched in anti-c-Myc immunoprecipitates compared with control immunoglobulin G (IgG) immunoprecipitates of NSCLC cell lysates (Figure 8E), and luciferase reporter assays identified the E2 (−373 to −362) and E3 (−40 to −29) regions of the LINC01234 promoter as specific c-Myc-binding sites (Figure 8F). Finally, we examined the relationship between c-Myc expression in NSCLC tumors and patient prognosis, and we found that overall survival rates were lower for patients with c-Myc-overexpressing tumors (Figure S4E). Additionally, we also determined a negative correlation between c-Myc and CRY2 and another negative correlation between LINC01234 and CRY2 in TCGA data (Figure S4F). Collectively, these results demonstrate that the expression and activity of the LINC01234–HNRNPA2B1–miR-106b-5p–CRY2–c-Myc pathway play a role in NSCLC development and progression (Figure 8G).
Discussion
Multiple factors are involved in tumorigenesis, and numerous recent studies have implicated lncRNAs as critical regulators of this process. In the present study, we identified thousands of differentially expressed lncRNAs in human NSCLC compared with normal lung tissue. In addition to the known lncRNAs MALAT1, LINC00673, and ANRIL,32, 33, 34 we identified many new NSCLC-associated lncRNAs, including LINC01234. Importantly, high tumor expression of LINC01234 was significantly associated with poorer prognosis and shorter survival time in NSCLC patients. In addition, modulation of LINC01234 expression revealed its oncogenic activity through promotion of cell growth and inhibition of apoptosis, supporting a potential role for LINC01234 dysregulation in NSCLC pathogenesis. Finally, complementary in vivo studies using a mouse xenograft model confirmed the role played by LINC01234 in tumor growth.
Recent studies have shown that oncogenic transcription factors contribute to lncRNA dysregulation in cancer cells. For example, SP1 activates expression of the lncRNAs AGAP2-AS1 and TINCR in gastric cancer cells,35,36 whereas E2F1 promotes ANRIL and HOXA11-AS transcription in lung cancer and gastric cancer.22,34 In the present study, we verified that c-Myc binds to the promoter and facilitates LINC01234 transcription. c-Myc is a known oncogene, is frequently amplified in human cancers, and regulates transcription of genes involved in cell growth and cell-cycle progression. Several additional c-Myc-regulated lncRNAs, such as MYCLo-1 and MYCLo-2, have been shown to regulate colorectal cancer growth.23 In that study, lncRNAs bound to the RNA-binding proteins HuR and hnRNPK to influence the expression of c-Myc target genes, such as CDKN1A and CDKN2B.23 We found that LINC01234 affected the expression of not only CDKN1A but also P57, BTG2, and CRY2 in NSCLC cells. Collectively, these findings indicate that lncRNAs may be important mediators of c-Myc-induced tumorigenesis.
Current evidence suggests that lncRNAs play a variety of roles in tumorigenesis. Here, we established that LINC01234 binds to HNRNPA2B1, which has known cancer-promoting functions in a variety of tumors. For example, HNRNPA2B1 accelerates pre-mRNA processing and interacts with and regulates the activity of mutant KRASG12V and KRASG12D proteins.37 In addition, HNRNPA2B1 forms complexes with a number of lncRNAs to regulate gene expression.38,39 In this study, we demonstrated that the LINC01234–HNRNPA2B1 complex recruits DGCR8 to promote processing of pri-miRNAs, such as miR-106b-5p, miR-17-5p, and miR-362-5p, to their mature forms. Accordingly, LINC01234 depletion decreased mature miRNA levels and increased unprocessed primary miRNA levels in the nucleus. Our findings are consistent with a previous study showing that HNRNPA2B1 could regulate the alternative splicing of a set of nuclear transcripts containing the pri-miRNAs.28 Our findings demonstrate that LINC01234 complexes with HNRNPA2B1 and recruits DGCR8 to facilitate miR-106b-5p maturation in NSCLC cells.
miR-106b is highly expressed in and contributes to the development and progression of multiple cancers. Li et al.40 reported that overexpressed miR-106b-5p facilitated glioma cell proliferation via repression of retinoblastoma-like 1 (RBL1), RBL2, and caspase-8 translation. In breast cancer, high miR-106b-5p expression promotes cell invasion and proliferation through suppression of phosphatase and tensin homolog (PTEN) and activation of phosphoinositide 3-kinase/AKT signaling.40 A recent study also demonstrated that miR-106b-5p is upregulated in NSCLC,41 although that study did not investigate its function or regulation of downstream target genes. Here, we showed that miR-106b-5p promotes proliferation and inhibits apoptosis of NSCLC cells via repression of CRY2. Growing evidence supports an import role for circadian clock proteins, such as CRY1 and CRY2, in the development of malignant tumors. CRY2 is a component of the FBXL3-containing E3 ligase, which ubiquitinates and promotes the degradation of threonine 58-phosphorylated c-Myc.30 We found that reduced expression of CRY2 is associated with shorter survival of NSCLC patients. Consistent with this, the oncogenic function of LINC01234 and miR-106b-5p was at least partly dependent on downregulation of CRY2. CRY2 overexpression promoted c-Myc degradation, suggesting the existence a c-Myc–LINC01234–miR-106b-5p–CRY2 feedback loop that may play an essential role in NSCLC development.
In summary, the present study identifies a novel c-Myc-activated lncRNA, LINC01234, which is dysregulated in human NSCLC and associated with poor prognosis. LINC01234 promotes cell growth and inhibits apoptosis via interaction with HNRNPA2B1 and recruitment of DGCR8 to induce pri-miR-106b processing. These observations advance our understanding of lncRNA–miRNA–mRNA networks in NSCLC and suggest that LINC01234 may have utility as a diagnostic marker and/or therapeutic target for NSCLC and potentially other malignancies.
Materials and Methods
Microarray and TCGA Dataset Analysis
We obtained raw microarray data (CEL files) from GEO: GSE18842 and GSE31210 datasets, which are paired normal and NSCLC tissues sequenced on the Affymetrix Human Genome U133 Plus 2.0 Array platform. For lncRNA analysis, all probes were reannotated using the GENCODE Release 22 lncRNA annotation file. For analysis of TCGA data, the expression levels of all lncRNAs were calculated from aligned BAM files by using RNA-SeqQC software. GEO and TCGA analyses were summarized in Table S3.
NSCLC Specimen and Cell Lines
A cohort of 95 paired NSCLC specimens and adjacent nontumor tissues was obtained from 95 NSCLC patients at the First Affiliated Hospital of Nanjing Medical University and Second Affiliated Hospital of Nanjing Medical University between 2013 and 2015. They did not receive radiotherapy, chemotherapy, or molecular target therapy prior to surgical treatment. These specimens were immediately snap frozen and stored in liquid nitrogen until required. The clinicopathological features of all patients are summarized in Table S1. This study was approved by the Research Ethics Committee of Nanjing Medical University, and informed consent was obtained from all patients.
Five lung adenocarcinoma cell lines (A549, SPC-A1, PC9, H1299, and H1975), two lung squamous carcinoma cell lines (H226 and H1703), and a normal human bronchial epithelial cell line (16HBE) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). H1299, A549, H1703, H1975, and H226 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 basic medium with 10% fetal bovine serum, and SPC-A1, PC9, and 16HBE cells were cultured in Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum. All cells were cultured in a humidified with 5% CO2 at 37°C.
RNA Extraction and qRT-PCR Assays
The TRIzol reagent (Invitrogen) was used to extract the total RNA from NSCLC tissues and cells following the manufacturer’s protocol. Next, 1 μg of extracted RNAs was reversely transcribed to cDNA using a PrimeScript RT Reagent Kit (TaKaRa), according to the manufacturer’s manual. A real-time PCR assay was performed using the SYBR Premix Ex Taq (TaKaRa) on an Applied Biosystems (ABI) 7500 System. Housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control, and RNU 6 (RNA, U6 small nuclear 1) was chosen as the internal control for miRNA quantitation. To measure expression of LINC01234 and other genes, specific primers listed in Table S4 were used. pri-miRNAs and mature miRNAs were quantified by TaqMan assays (ABI). The qRT-PCR data were analyzed relative to threshold cycle (CT) values and then converted to fold changes.
Plasmid Construction and Cell Transfection
The full-length cDNA of human LINC01234 was synthesized; c-Myc and CRY2 sequences were synthesized, according to their coding sequences. All sequences were cloned into the expression vector pCDNA3.1 (Invitrogen). Control and LINC01234-targeting shRNAs were purchased from Invitrogen and inserted into the pLKO.1 vector. All final constructs were verified by sequencing. Plasmids were purified using DNA Midiprep Kits (QIAGEN, Valencia, CA, USA) and transfected into NSCLC cells using the X-treme GENE HP DNA transfection reagent (Roche, Basel, Switzerland). Three LINC01234-targeting siRNAs obtained from Invitrogen and other gene-targeting siRNAs, miRNA mimics, and miRNA inhibitors (GenePharma, Shanghai, China) were transfected into NSCLC cells using RNAiMAX (Invitrogen), following the manufacturer’s manual. Nucleotide sequences for the siRNAs and shRNAs are listed in Table S4. Cells were collected 48 h post-transfection and analyzed, as indicated for the individual experiments.
Cell Proliferation and Flow Cytometric Assays
Cell Proliferation Reagent Kit I (MTT assay kit; Roche Applied Science) and an EdU assay kit (Life Technologies, Carlsbad, CA, USA) were used to measure cell proliferation following the manufacturers’ instructions. Colony-forming assays were independently performed three time to examine NSCLC cell capacity for clonal growth. For cell apoptosis analysis, cells were harvested at 48 h after transfection using Accutase (Invitrogen). After that, cells were double stained with propidium iodide (PI) and fluorescein isothiocyanate (FITC)-Annexin V and then analyzed on a flow cytometer (FACScan; BD Biosciences). Cells were then classified into dead cells, viable cells, late apoptotic cells, and early apoptotic cells, and the relative ratio of apoptotic cells was calculated and compared with control cells. To analyze cell-cycle progression, NSCLC cells were stained with PI and then analyzed on a FACScan. The percentage of NSCLC cells in G0/G1, S, and G2/M phases was calculated and compared.
In Vivo Tumor Formation Experiments
All protocols were approved by the Committee on the Ethics of Animal Experiments of the Nanjing Medical University and were strictly carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. For the tumorigenicity studies, control shRNA or sh-LINC01234 stably expressed A549 cells (3 × 106) were injected subcutaneously into the ventral side of male BALB/c nude mice (4 weeks old). Tumor dimensions were examined every 3 days, and the volume was calculated as follows: V (mm3) = 0.5 × D × d2 (where V is volume; D is longest diameter, and d is diameter perpendicular to the longest diameter). Mice were euthanized 15 days after the formation of tumor, and the tumors were excised, weighed, and collected for further analysis. Sections of primary tumors were fixed and embedded for hematoxylin and eosin (H&E) staining or IHC staining, as previously described.42
FISH
A549 and SPC-A1 cells were washed with phosphate-buffered saline (PBS) and fixed with 4% formaldehyde for 15 min. Next, fixed cells washed with PBS and incubated with pepsin (1% in 10 cmM HCl) and subsequently dehydrated with 70%, 90%, and 100% ethanol. Then, the 40 -nM LINC01234 FISH probe in hybridization buffer (100 mg/mL dextran sulfate, 10% formamide in 2 × saline-sodium citrate) was added into the air-dried cells and incubated at 80°C for 2 min, hybridized at 55°C for 2 h, washed with PBS, and dehydrated. The slides were finally mounted with 4′,6-diamidino-2-phenylindole (DAPI) for detection of nuclei. The LINC01234 FISH probe was designed and synthesized by Bogu (Shanghai, China), and the sequences were listed in Table S4.
RNA Pull-Down Assays
LINC01234 or its antisense RNAs were transcribed in vitro from pcDNA3.1-LINC01234 using T7 RNA polymerase (Ambion Life) and purified using a MEGAclear Kit (Ambion Life). One aliquot of transcribed LINC01234 RNA was biotinylated with a Biotin RNA Labeling Mix (Ambion Life). Then, biotinylated RNAs were incubated with streptavidin-conjugated magnetic beads and cell lysates at room temperature. The beads were then washed, and the eluted proteins were examined by mass spectrometry and western blot analysis.
RIP
RIP assays were carried out using an EZ Magna RIP kit (Millipore), following the manufacturer’s instructions. A549 and SPC-A1 cells were lysed in complete cell lysis buffer, and the lysates were incubated with magnetic beads conjugated with the HNRNPA2B1-specific antibody or control IgGs for 3–6 h at 4°C. The beads were washed with wash buffer and incubated with proteinase K to degrade proteins, and the purified RNA was eluted and analyzed for the presence of LINC01234 by qRT-PCR. Details of the antibodies and primers are given in Table S4.
Western Blot Analysis
A549 and SPC-A1 cells were lysed with RIPA (radioimmunoprecipitation assay) Lysis and Extraction Buffer (Beyotime), supplemented with protease inhibitor cocktail (Roche). Protein in cell lysis was separated by 8% to 12% sodium dodecyl sulfate-polyacrylamide gel and transferred to a 0.2-μm Immobilon-PSQ PVDF (polyvinylidene fluoride) Membrane (Millipore). Membranes were incubated with HNRNPA2B1 and other antibodies using standard methods. Enhanced chemiluminescence (ECL) chromogenic substrate was used to image the target protein bands, which was further quantified according to densitometry (Quantity One software; Bio-Rad, Hercules, CA, USA). Antibodies against cyclin D1, cyclin D3, CDK2 caspase-3, cleaved caspase-3, PARP, and cleaved PARP (1:1,000) were purchased from Cell Signaling Technology. Antibodies against CRY2 (Abcam), c-Myc (Cell Signaling Technology), HNRNPA2B1, and GAPDH (Proteintech) were used for western blot analysis. GAPDH was probed as an internal control. Antibodies are listed in Table S4.
Luciferase Reporter Assay
The JASPAR (http://jaspar.genereg.net/) software was used to predict potential transcription factors binding the motif in the promoter region of LINC01234. The sequences of the LINC01234 promoter region containing the putative c-Myc-binding motif were synthesized, cloned into the pGL3-Basic luciferase reporter vector (Promega), and verified by Sanger sequencing. Vectors were transfected into human HEK293T cells for 48 h. Next, the luciferase activity was examined using a Dual-Luciferase kit (Promega), following the manufacturer’s manual. The data are presented as relative firefly luciferase activity, which is normalized to Renilla luciferase activity. All experiments were conducted in three independent replicates.
Statistical Analysis
SPSS 20.0 (IBM, Armonk, NY, USA) and GraphPad Prism 6.0 software (GraphPad, La Jolla, CA, USA) were used to conduct all of the statistical analyses. lncRNA expression levels in primary solid tumors and normal solid tissue samples were compared using the Mann-Whitney U test. For the remaining assays, χ2 test, Wilcoxon’s test, and paired two-tailed Student’s t test were used to assess the differences between different groups as appropriate. ANOVAs were used in those experiments with more than 3 groups. Multivariate and univariate Cox proportional hazard modeling was used to evaluate the effects of different factors on NSCLC patients’ survival. The Kaplan–Meier survival analysis was used to evaluate the OS and PFS. A p value <0.05 was defined as statistically significant.
Author Contributions
M.S. and Z.W. designed and supervised the study. Z.C. and X.C. conducted the experiment. T.L., J.G., and J.H. contributed to acquisition of results. Y.G. and X.C. performed the data analysis. Z.C., X.C., and M.S. wrote the manuscript. Z.W. and B.L. provided technical and administrative support. All of the authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Lung adenocarcinoma and lung squamous cell carcinoma RNA sequencing datasets were obtained from TCGA, and microarray gene profile datasets from GEO: GSE3121043 and GSE1884244. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. This work was supported by grants from the National Natural Science Foundation of China (nos. 81672307 and 81871871 to Z.W., and 81902333 to X.C.); Key Research and Development Plan (Social Development) of Science and Technology, Department of Jiangsu Province (no. BE2019760 to Z.W.); Medical Innovation Team Foundation of the Jiangsu Provincial Enhancement Health Project (no. CXTDA2017021 to Z.W.); “333 High Class Talented Man Project” (no. BRA2016509, 2016-II-426 to Z.W.); Scientific Research Foundation of Jiangsu Province Health Department (no. H201310 to B.L.); Key Young Medical Talents of Jiangsu Province (no. QNRC2016662 to B.L.); Science and Technology Development Fund of Nanjing Medical University(no. NMUB2018035 to X.C.); and Postgraduate Research & Practice Innovation Program of Jiangsu Province (no. KYCX19_1170 to Z.C.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.03.010.
Contributor Information
Ming Sun, Email: msun7@mdanderson.org.
Zhaoxia Wang, Email: zhaoxiawang66@126.com.
Supplemental Information
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Koivunen J.P., Mermel C., Zejnullahu K., Murphy C., Lifshits E., Holmes A.J., Choi H.G., Kim J., Chiang D., Thomas R. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janku F., Stewart D.J., Kurzrock R. Targeted therapy in non-small-cell lung cancer--is it becoming a reality? Nat. Rev. Clin. Oncol. 2010;7:401–414. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 4.Roberts P.J., Stinchcombe T.E. KRAS mutation: should we test for it, and does it matter? J. Clin. Oncol. 2013;31:1112–1121. doi: 10.1200/JCO.2012.43.0454. [DOI] [PubMed] [Google Scholar]
- 5.Sadiq A.A., Salgia R. MET as a possible target for non-small-cell lung cancer. J. Clin. Oncol. 2013;31:1089–1096. doi: 10.1200/JCO.2012.43.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borel C., Gagnebin M., Gehrig C., Kriventseva E.V., Zdobnov E.M., Antonarakis S.E. Mapping of small RNAs in the human ENCODE regions. Am. J. Hum. Genet. 2008;82:971–981. doi: 10.1016/j.ajhg.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pei B., Sisu C., Frankish A., Howald C., Habegger L., Mu X.J., Harte R., Balasubramanian S., Tanzer A., Diekhans M. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang R., Tang J., Chen Y., Deng L., Ji J., Xie Y., Wang K., Jia W., Chu W.M., Sun B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 2017;8:15129. doi: 10.1038/ncomms15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z., Chen X., Chen P., Yu S., Nie F., Lu B., Zhang T., Zhou Y., Chen Q., Wei C. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. 2017;8:e3092. doi: 10.1038/cddis.2017.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P., Xu J., Wang Y., Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358:1051–1055. doi: 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda A., Tomikawa J., Miura T., Hata K., Nakabayashi K., Eggan K., Akutsu H., Umezawa A. The role of maternal-specific H3K9me3 modification in establishing imprinted X-chromosome inactivation and embryogenesis in mice. Nat. Commun. 2014;5:5464. doi: 10.1038/ncomms6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan X., Hu Z., Feng Y., Hu X., Yuan J., Zhao S.D., Zhang Y., Yang L., Shan W., He Q. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cifuentes-Rojas C., Hernandez A.J., Sarma K., Lee J.T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen L., Wang Q., Liu R., Chen Z., Zhang X., Zhou P., Wang Z. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46:717–729. doi: 10.1093/nar/gkx1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Chen Z., Fan R., Jiang B., Chen X., Chen Q., Nie F., Lu K., Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol. Cancer. 2017;16:82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niknafs Y.S., Han S., Ma T., Speers C., Zhang C., Wilder-Romans K., Iyer M.K., Pitchiaya S., Malik R., Hosono Y. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C., Luo Y., He W., Zhao Y., Kong Y., Liu H., Zhong G., Li Y., Li J., Huang J. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Invest. 2020;130:404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X.H., Sun M., Nie F.Q., Ge Y.B., Zhang E.B., Yin D.D., Kong R., Xia R., Lu K.H., Li J.H. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun M., Nie F., Wang Y., Zhang Z., Hou J., He D., Xie M., Xu L., De W., Wang Z., Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 23.Kim T., Jeon Y.J., Cui R., Lee J.H., Peng Y., Kim S.H., Tili E., Alder H., Croce C.M. Role of MYC-regulated long noncoding RNAs in cell cycle regulation and tumorigenesis. J. Natl. Cancer Inst. 2015;107:dju505. doi: 10.1093/jnci/dju505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu T., Zhao Y., Hu Z., Li J., Chu D., Zhang J., Li Z., Chen B., Zhang X., Pan H. MetaLnc9 Facilitates Lung Cancer Metastasis via a PGK1-Activated AKT/mTOR Pathway. Cancer Res. 2017;77:5782–5794. doi: 10.1158/0008-5472.CAN-17-0671. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z., Li J.L., Lin S., Cao C., Gimbrone N.T., Yang R., Fu D.A., Carper M.B., Haura E.B., Schabath M.B. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J. Clin. Invest. 2016;126:2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q.N., Chen X., Chen Z.Y., Nie F.Q., Wei C.C., Ma H.W., Wan L., Yan S., Ren S.N., Wang Z.X. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol. Cancer. 2017;16:17. doi: 10.1186/s12943-017-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C., Luo Y., He W., Zhao Y., Kong Y., Liu H., Zhong G., Li Y., Li J., Huang J. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Invest. 2020;130:404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber A.L., Papp S.J., Chan A.B., Henriksson E., Jordan S.D., Kriebs A., Nguyen M., Wallace M., Li Z., Metallo C.M., Lamia K.A. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol. Cell. 2016;64:774–789. doi: 10.1016/j.molcel.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi X., Ma C., Zhu Q., Yuan D., Sun M., Gu X., Wu G., Lv T., Song Y. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558–25575. doi: 10.18632/oncotarget.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie F.Q., Sun M., Yang J.S., Xie M., Xu T.P., Xia R., Liu Y.W., Liu X.H., Zhang E.B., Lu K.H., Shu Y.Q. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol. Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 35.Li W., Sun M., Zang C., Ma P., He J., Zhang M., Huang Z., Ding Y., Shu Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu T.P., Wang Y.F., Xiong W.L., Ma P., Wang W.Y., Chen W.M., Huang M.D., Xia R., Wang R., Zhang E.B. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837. doi: 10.1038/cddis.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barcelo C., Etchin J., Mansour M.R., Sanda T., Ginesta M.M., Sanchez-Arevalo Lobo V.J., Real F.X., Capellà G., Estanyol J.M., Jaumot M. Ribonucleoprotein HNRNPA2B1 interacts with and regulates oncogenic KRAS in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:882–892.e8. doi: 10.1053/j.gastro.2014.06.041. [DOI] [PubMed] [Google Scholar]
- 38.Wang H., Liang L., Dong Q., Huan L., He J., Li B., Yang C., Jin H., Wei L., Yu C. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8:2814–2829. doi: 10.7150/thno.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y., Yang X., Chen Z., Tian L., Jiang G., Chen F., Li J., An P., Lu L., Luo N. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer. 2019;18:87. doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li N., Miao Y., Shan Y., Liu B., Li Y., Zhao L., Jia L. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017;8:e2796. doi: 10.1038/cddis.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Tian J., Guo Z.J., Zhang Z.B., Xiao C.Y., Wang X.C. Expression of microRNAs-106b in nonsmall cell lung cancer. J. Cancer Res. Ther. 2018;14(Supplement):S295–S298. doi: 10.4103/0973-1482.235344. [DOI] [PubMed] [Google Scholar]
- 42.Sun T.T., He J., Liang Q., Ren L.L., Yan T.T., Yu T.C., Tang J.Y., Bao Y.J., Hu Y., Lin Y. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 43.Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R., Furuta K., Tsuta K., Shibata T., Yamamoto S. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez-Palencia A., Gomez-Morales M., Gomez-Capilla J.A., Pedraza V., Boyero L., Rosell R., Fárez-Vidal M.E. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int. J. Cancer. 2011;129:355–364. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.